Abstract
The Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/CRISPR-associated sequence (CRISPR/Cas) system is a cutting-edge genome-editing tool employed to explore the functions of normal and disease-related genes. The CRISPR/Cas system has a remarkable diversity in the composition and architecture of genomic loci and Cas protein sequences. Owing to its excellent efficiency and specificity, this system adds an outstanding dimension to biomedical research on genetic manipulation of eukaryotic cells. However, safe, efficient, and specific delivery of this system to target cells and tissues and their off-target effects are considered critical bottlenecks for the therapeutic applications. Recently discovered anti-CRISPR proteins (Acr) play a significant role in limiting the effects of this system. Acrs are relatively small proteins that are highly specific to CRISPR variants and exhibit remarkable structural diversity. The in silico approaches, crystallography, and cryo-electron microscopy play significant roles in elucidating the mechanisms of action of Acrs. Acrs block the CRISPR/Cas system mainly by employing four mechanisms: CRISPR/Cas complex assembly interruption, target-binding interference, target cleavage prevention, and degradation of cyclic oligonucleotide signaling molecules. Engineered CRISPR/Cas systems are frequently used in gene therapy, diagnostics, and functional genomics. Understanding the molecular mechanisms underlying Acr action may help in the safe and effective use of CRISPR/Cas tools for genetic modification, particularly in the context of medicine. Thus, attempts to regulate prokaryotic CRISPR/Cas surveillance complexes will advance the development of antimicrobial drugs and treatment of human diseases. In this review, recent updates on CRISPR/Cas systems, especially CRISPR/Cas9 and Acrs, and their novel mechanistic insights are elaborated. In addition, the role of Acrs in the novel applications of CRISPP/Cas biotechnology for precise genome editing and other applications is discussed.
Keywords: CRISPR/Cas, anti-CRISPR proteins, gene editing, enzyme inhibition, protein inhibitors, biotechnology
Introduction
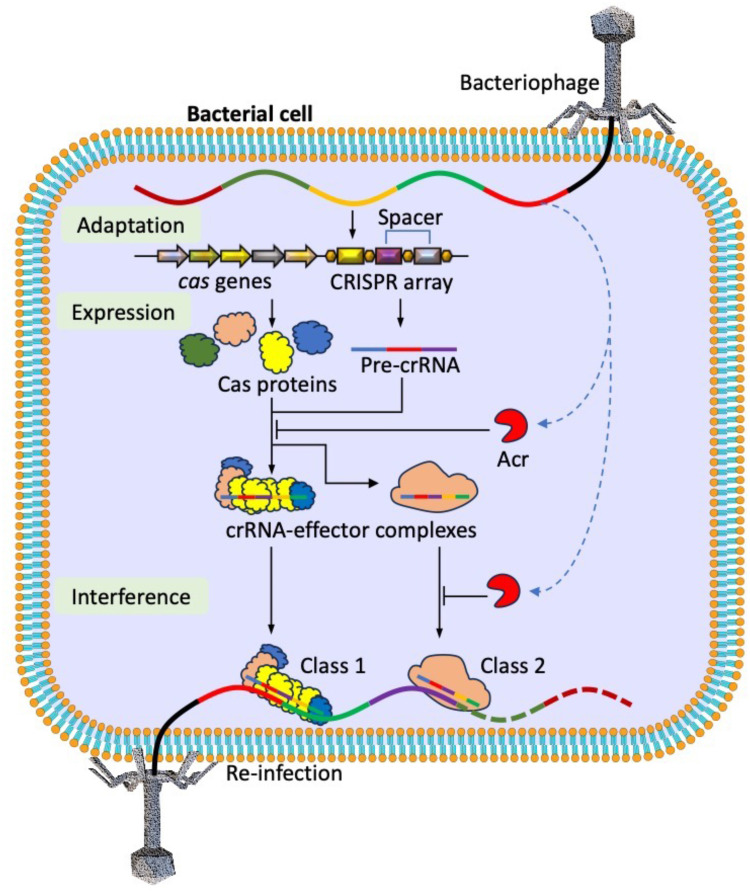
Most prokaryotes possess a Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/CRISPR-associated (Cas) sequence (CRISPR/Cas) system as a defense mechanism against phages, plasmids, and other mobile genetic elements (MGEs). It is a genome-editing system found in approximately 50% of bacteria and 90% of archaea.1 The CRISPR array is composed of several highly conserved, non-continuous DNA sequences called direct repeats that are spaced apart. These spacers are homologous DNA sequences of varying lengths that have been previously introduced into prokaryotic host cells by MGEs. Multiple effector proteins are encoded by Cas genes, which complete the cleavage of foreign nucleic acids during invasion (Figure 1).
Figure 1.
Role of CRISPR/Cas system as an adaptive immunity approach in prokaryotes and the counter role of Acrs synthesized from viral genome (blue dashed arrows) to block this genome editing approach.
The CRISPR/Cas system is divided into two classes (Class 1 and Class 2) and six types (Types I–VI) based on effector Cas proteins. Each type consists of subtypes with different architectures on the DNA or mRNA cleavage sites. Despite the differences in CRISPR/Cas systems, these genome-editing tools rely on similar basic norms of action, involving three main stages: adaptation, expression, and interference.2,3
Detailed characterization of CRISPR/Cas and anti-CRISPR proteins (Acr) has revolutionized the field of genome editing to next-generation levels. This molecular tool has been proposed as an innovative strategy for genome editing, transcriptional perturbation, epigenetic modulation, and genome imaging.4 In addition to CRISPR/Cas system applications and their limitations, the recent discovery of Acrs, which act as potent inhibitors of Cas effectors, provides a switch-off mechanism that can keep this powerful technology in check and enhance the precision with which genome perturbations can be made.
As the bacteria avoid phage and other MGEs invasion through the CRISPR/Cas system, in this arms strength race, the phages adopted new fighting strategies to escape from being targeted by this genome editing system by using anti-CRISPR proteins (Acrs). These are small proteins compared to Cas proteins (approximately 50–300 amino acid residues), providing a powerful and efficient means of combating CRISPR/Cas-based genome editing.5 To date, 122 different Acr proteins have been discovered that act against all CRISPR/Cas subtypes except type IV (Table 1). Biochemical and structural studies have revealed four specific mechanisms of action of Acrs in the CRISPR/Cas system. These include the inhibition of CRISPR/Cas complex assembly, target binding blockade, target cleavage prevention, and cyclic oligonucleotide signaling molecule degradation.6
Table 1.
Important Features of Two Classes of CRISPR/Cas Systems and Associated Anti-CRISPR Proteins
| Class | Type | Occurance | Effector Complex | Sub-type | Cas Proteins Involved | Pre-crRNA Processing | Signature Protein | Target | PAM Requirement | Nuclease | Target Molecule | Acr Proteins |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | I | Bacteria and archaea | Multi-subunit complex | I-B | Cas1, Cas2, Cas3, Cas5, Cas6, Cas7, Cas8 | Cas6 | Cas3 | dsDNA | Yes | Cas3 | dsDNA | AcrIB |
| I-C | AcrIC1, AcrIF2 (also known as AcrIC2), AcrIC3, AcrIC4, AcrIC5, AcrIC6, AcrIC7, AcrIC8, AcrIC9, AcrIC10, AcrVA3 | |||||||||||
| I-D | Cas3 and Cas 10d | AcrID1 | ||||||||||
| I-E | Cas3 | AcrIE1, AcrIE2, AcrIE3, AcrIE4, AcrIE5, AcrIE6, AcrIE7, AcrIE8, AcrIE9, AcrIE4- IF7 | ||||||||||
| I-F | AcrIF1, AcrIF2, AcrIF3, AcrIF4, AcrIF5, AcrIF6, AcrIF7, AcrIF8, AcrIF9, AcrIF10, AcrIF11, AcrIF12, AcrIF13, AcrIF14, AcrIF15, AcrIF16, AcrIF17, AcrIF18, AcrIF19, AcrIF20, AcrIF21, AcrIF22, AcrIF23, AcrIF24, AcrIF25, AcrIE4- IF7 | |||||||||||
| III | Archaea and in some bacteria | III-A | Csm in subtypes IIIA and IIID Cmr in subtype IIIB and IIIC Csm1, Csm2, Csm3, Csm4, Csm5, Csm6, |
Csm6 | Csm1 (also known as Cas10) |
ssRNA, ssDNA |
No | Csm3, Csm1 (also known as Cas10) |
ssDNA and RNA | AcrIII-1 | ||
| III-B | Cmr4, Cmr2 (also known as Cas10) |
AcrIIIB1, AcrIII-1 | ||||||||||
| IV | Bacterial plasmids and other MGEs | Csf1, Csf2, Csf3, Csf4, Csf5 | Csf5 | Unknown | Unknown | Unknown | Unknown | Unknown | Unknown | |||
| 2 | II | Bacteria | Single protein | II-A | Cas1, Cas2, Cas9 | RNaseIII and Cas9 | Cas9 | dsDNA | Yes | Cas9 RuvC and HNH domains |
dsDNA and ssRNA | AcrIIA1, AcrIIA2, AcrIIA3, AcrIIA4, AcrIIA5, AcrIIA6, AcrIIA7, AcrIIA8, AcrIIA9, AcrIIA10, AcrIIA11, AcrIIA12, AcrIIA13, AcrIIA14, AcrIIA15, AcrIIA16, AcrIIA17, AcrIIA18, AcrIIA19, AcrIIA20, AcrIIA21, AcrIIA22, AcrIIA23, BRD0539 |
| II-C | AcrIIC1, AcrIIC2, AcrIIC3, AcrIIC4, AcrIIC5 | |||||||||||
| V | Many bacteria and few archaea | V-A | Cas1, Cas2, Cas12 | RNaseIII | Cas12 | dsDNA, ssRNA |
Yes | Cas12 RuvC domain |
ssRNA | AcrVA1, AcrVA2, AcrVA3, AcrVA4, AcrVA5 | ||
| VI | Bacteria and archaea | VI-A | Cas1, Cas2, Cas13 | RNase, Cas13 | Cas13 | ssRNA | No | Cas13 hEPN domain |
ssRNA | AcrVIA1 (Lwa), AcrVIA2, AcrVIA3, AcrVIA4, AcrVIA5, AcrVIA6, AcrVIA7, AcrVIA1 (Lse) |
Owing to the limitations of the CRISPR/Cas system for full potential gene manipulation, Acrs have tremendously improved the use of this genome editing system by reducing its off-target and off-cell activities, minimizing CRISPR/Cas cell cytotoxicity, controlling gene drives, guiding dead Cas9 (dCas9) based tools, CRISPR/Cas complex detection, and minimizing CRISPR/Cas9 resistance during phage therapy.
In this review, we discuss the current updates on the CRISPR/Cas system and its classification, with a brief focus on the CRISPR/Cas9 structure and its mechanism of action. In addition, classification and novel mechanistic insights into Acrs are discussed. Furthermore, the role of different Acrs that act as versatile tools in CRISPR/Cas biotechnology to improve the future of genetic engineering approaches is discussed.
CRISPR/Cas System
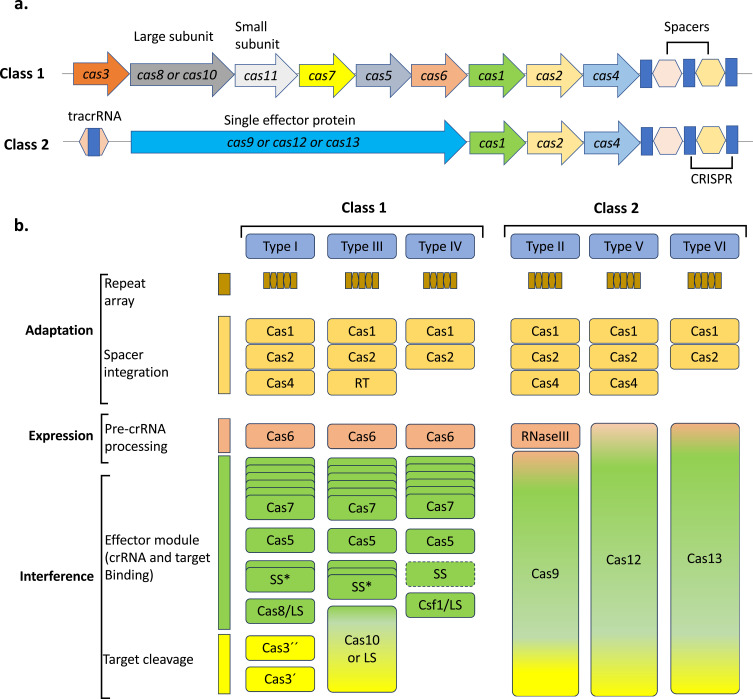
The CRISPR/Cas system functions as an adaptive immune mechanism found in bacteria and most archaea against MGEs, plasmids, and phages, which can alter the genetic makeup of these organisms.7,8 The CRISPR/Cas system exhibits remarkable diversity in the composition and architecture of genomic loci and Cas protein sequences.9–11 A set of cas genes and one or more CRISPR arrays constitutes the typical CRISPR/Cas locus. The CRISPR/Cas system consists of two classes (Class 1 and Class 2) that differ in the architecture of their effector modules involved in the processing of CRISPR RNA (crRNA) and interference (Figure 2a). Multiple Cas proteins functioning as effector modules were found in the class 1 CRISPR/Cas system, whereas a single multidomain crRNA-binding protein was found in class 2 systems (eg, Cas9 in type II systems) (Figure 2b). Furthermore, this genome editing tool is classified into six types, 33 subtypes, and 44 variants, based on the quantity and type of Cas proteins involved in genome editing.1 Despite the vast diversity of CRISPR/Cas systems, all these genome editing tools perform their actions in three stages: acquisition, expression, and interference12 (Figure 1 and 2).
Figure 2.
Effector modules organization of the two classes of CRISPR/Cas systems. (a) generic organization of Class1 and Class2 CRISPR/Cas loci. (b) Genetic, structural and functional organization of six types of CRISPR/Cas systems.
Initially during the adaptation stage, a spacer, or “memory sequence”, is created in the bacterial genome by integrating a protospacer genetic fragment into a CRISPR array. The CRISPR array is processed and transcribed during expression to produce CRISPR RNAs (crRNAs) that bind to Cas proteins to create a CRISPR/cas surveillance (Csy) complex. Each crRNA sequence contains a single spacer. Owing to their complementarity with the crRNA, the surveillance complexes can then identify the MGE that carries matching protospacers. The protospacer must be flanked by a protospacer-adjacent motif (PAM) to bind to the majority of CRISPR/Cas variants. Depending on the CRISPR/Cas system subtype, the targeted genetic material may be RNA, single-stranded DNA (ssDNA) or double-stranded DNA (dsDNA).1 Class 1 of the CRISPR/Cas system includes three type (I, III, and IV) and 16 subtypes, whereas class 2 includes three types (II, V, and VI) and 17 subtypes1,13,14 (Figure 2).
Structure and Mechanistic Action of CRISPR/Cas9
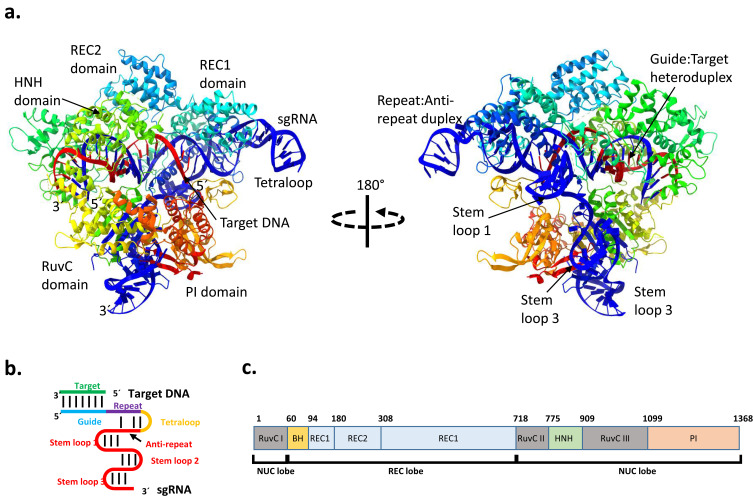
The CRISPR/Cas9 genome-editing system from Streptococcus pyogenes (SpCas9) is the most widely studies CRISPR/Cas system. It is a type IIA system, in which Cas9 is the only protein involved in the interference step (Figure 3a). This genome editing system is a complex composed of a single guide RNA (sgRNA; formed from the fusion of crRNA and tracrRNA) (Figure 3b) and Cas9, a 160 kD protein with 1368 amino acid residues (Figure 3c). Cas9 is a DNA endonuclease capable of cleaving each strand of dsDNA at precise location.14 Furthermore, for the DNA acquisition step, the CRISPR/Cas9 system involves Cas1, Cas2, and Csn2,15,16 whereas RNase III is involved in processing the pre-crRNA to mature sgRNA.17,18
Figure 3.
Three dimensional structure of Streptococcus pyogenes Cas9‐sgRNA‐DNA ternary complex. (a) Ribbon representation of Cas9-sgRNA-DNA complex at two different angles, The protein structure was obtained from protein data bank (https://www.rcsb.org, PDB with code 4OO8 and edited by using ChimeraX software. (b) Target DNA-sgRNA simplified domain organization, and (c) Protein domain organization in relation with amino acid residues numbers.
Abbreviations: NUC, nuclease; REC, recognition; PI, PAM interacting; PAM, protospacer adjacent motif; sgRNA, single‐guide RNA.
The domain architecture of SpCas9 consists of a recognition (REC) lobe (residues 56–718) and nuclease (NUC) lobe (residues 1–55, and 719–1368).14,19 The recognition lobe (the REC-I, REC-II, and REC-III domains) is responsible for nucleotide recognition.18,20 Cas9 consists of two endonuclease domains: the HNH domain (residue 766–909) and the RuvC domain (residues 1–55, 719–765, and 910–1099) (Figure 3c). Furthermore, a bridge helix (arginine rich) serves as a linker between the REC domain and RuvC-I, and is significant for cleavage initiation upon target DNA binding.14 The target DNA strand is cleaved by the HNH domain, whereas the non-target DNA strand is cleaved by the RuvC domain. The PAM-interacting domain (PI, residues 1100–1368) is responsible for conferring PAM specificity and initiating DNA binding.14,19 The positively charged residues present at the interface between the REC and NUC lobes stabilize the negatively charged sgRNA: DNA hybrid upon binding to Cas9.14 In addition, the displaced non-target DNA (ntDNA) is stabilized by positively charged residues present in the L1 and L2 linker regions between the HNH and RuvC domains.18
Conformational changes occurring in CRISPR/Cas9 have been demonstrated using X-ray crystallography during the DNA editing pathway. The different SpCas9 structures include free Cas9 (PDB 4CMQ),21 Cas9 bound to sgRNA (PDB 4ZT0),22 Cas9 bound to target DNA (tDNA), incomplete non-target (ntDNA) containing PAM (PDB 4UN3),23 and Cas9 bound to both tDNA and complete ntDNA (PDB 5F9R).22 Upon sgRNA binding, a major rearrangement occurs in the REC domain, with a significant ~65 Å shift of REC-III domain to accommodate this RNA complex.24 Melting of the target DNA occurs via PAM recognition by the Cas9: sgRNA complex.23,25
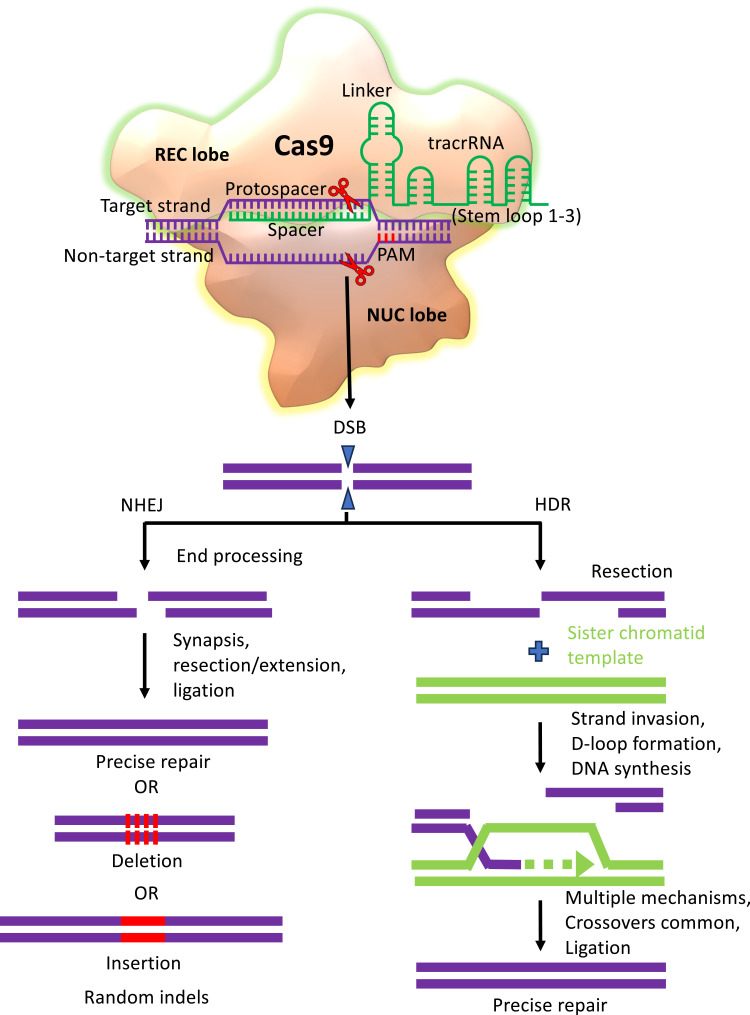
Unspecified by the protein, a 20-nt guide RNA (gRNA) sequence dictates DNA recognition and editing via the CRISPR/Cas9 system.26 By altering the gRNA sequence, the CRISPR/Cas9 system can be engineered to target the DNA sequences within the genome. The two Cas9 domains (RuvC and HNH) split the double strands of DNA upon the recognition of the target DNA. Cas9 creates a blunt-ended double-stranded break (DSB), which can be repaired by homology-directed repair (HDR) or non-homologous end-joining (NHEJ)27 (Figure 4). This causes very few insertions and/or deletions (indels) at the cleavage site. Additionally, a homologous repair template causes a predetermined genome amendment to occur at the DSB site with high fidelity. Furthermore, NHEJ-induced frameshift mutations and premature stop codon introduction may result in the early knockout and suppression of gene expression, respectively (Figure 4).
Figure 4.
The mechanism of genome editing mediated by CRISPR/Cas9. The synthetic crisprRNA-tracrRNA complex (sgRNA) directs Cas9 endonuclease to introduce a double-stranded break (DSB) in targeted DNA. The DSB generated is repaired by either non-homologous end-joining (NHEJ) or homology-directed repair (HDR) pathway.
In contrast, some proteins fully specify conventional DNA editing methods such as zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs).28 However, site-specific DNA modification using the CRISPR/Cas9 system does not require the use of recognition protein engineering.29 The impracticality of the time-consuming procedures for ZFNs and TALENs has significantly increased the usefulness of CRISPR/Cas9 for diverse genomic screening and modification. The CRISPR/Cas9 system has been widely used as a potent tool for genome editing in a variety of organisms because of its high efficiency, ease of design, and convenience of usage.30
However, some limitations, such as off-target effects, off-cell activity and immunotoxicity complications are associated with CRISPR/Cas9. These limitations have necessitated an in-depth demonstration of the role of Acrs in exploring the full potential of CRISPR/Cas9 in gene manipulation studies.
Anti-CRISPR Proteins
As an arm-race strategy, phages have developed protein inhibitors against CRISPR/Cas systems, referred to as anti-CRISPR proteins (Acrs), to counteract this genome-editing surveillance complex. These novel proteins were first discovered in 2013, and 122 Acrs belonging to 92 different subtypes have been identified to date. Currently, 98 different Acr protein families that block different CRISPR/Cas subtypes,31 and type II-A Cas9 inhibitors alone comprise 11 of these families.32
Because phage particles do not contain Acrs as a structural entity, the expression of acr genes is required at the onset of infection. Hence, acr genes are among the first to be expressed immediately after phage infection and are able to effectively block CRISPR/Cas immunity prior to phage elimination.33,34 The overexpression of acr genes is strictly controlled by an anti-CRISPR-associated (aca) gene, which, if unregulated, disturbs the expression of other phage genes.33,35
The naming of Acrs is based on a system that inhibits the discovery of these proteins were discovered.36 For example, AcrIIA4, a routinely used protein, is the fourth type of Acr that was discovered. Different Acrs have been successfully recognized to regulate gene editing in different cell types, most notably two SpyCas9 inhibitors (AcrIIA2 and AcrIIA4)37 and two Neisseria meningitidis (NmeCas9) inhibitors (AcrIIC1 and AcrIIC3).38 The classification patterns of different Acrs associated with different classes of the CRISPR/Cas system are presented in Table 1.
The origins and evolution of Acrs are not fully understood, as neither sequence nor structural comparisons yield significant similarities to Acrs.39 However, some recent approaches for protein structure prediction, such as AlphaFold2 (AF2),40 have helped to predict the structures of Acrs and their possible evolutionary ancestors. Acrs share folds with diverse unrelated proteins, suggesting that they originate from multiple sources.
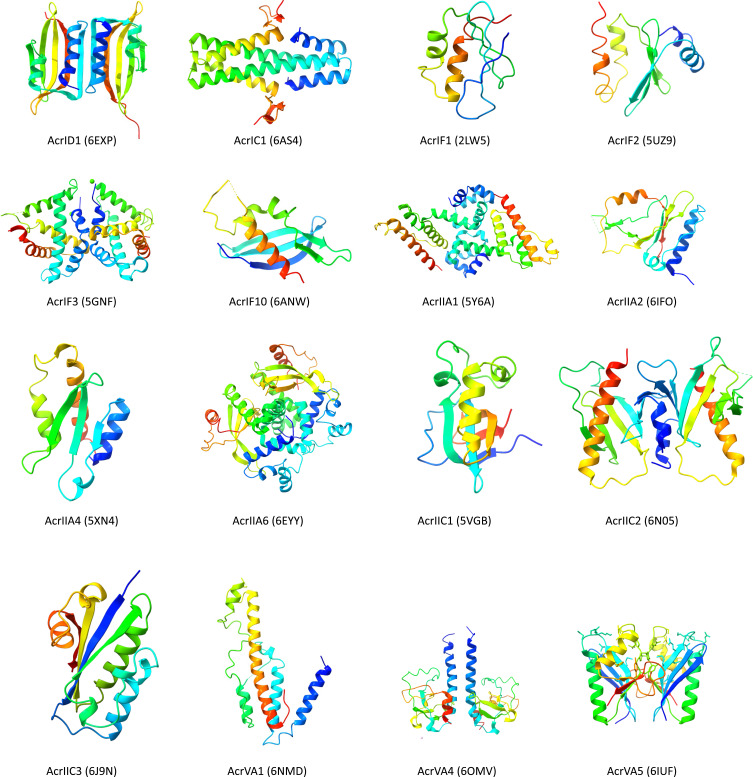
Most Acrs are small peptides with diverse structural variations, including all α-, all β-, mixed α/β, and separate α+β folds39,41 (Figure 5). The inhibition mechanisms of Acrs for CRISPR/Cas also show remarkable diversity and are not limited to the inhibition of crRNA-Cas complex formation, DNA binding by CRISPR effectors, or DNA cleavage.39,42
Figure 5.
Three-dimensional rainbow shaded ribbon structure of some type I and type II Acrs with given PDB identification codes in parentheses.
The identification of different Acrs using different methods includes the screening of phages that escape CRISPR targeting,43–45 screening and identification of genomes using self-targeting CRISPR arrays,46,47 guilt-by-association studies,48,49 and metagenome DNA screening for inhibition activity.50,51 Among these approaches, the “guilt-by-association” search approach is one of the most effective and direct. Bioinformatics tools such as AcRanker, AcrFinder, and PaCRISPR, as well as online databases such as Anti-CRISPRdb, AcrDB, AcrHub, and AcrCatalog, have facilitated the search for new Acrs.52
To counteract CRISPR/Cas systems, four main strategies have been revealed through biochemical and structural studies of these Acrs. These strategies include inhibiting the assembly of the CRISPR/Cas complex, preventing target cleavage, blocking target binding, and degrading cyclic oligonucleotide signaling molecules. Acrs also employ other recently developed tactics, such as post-translational modification of Cas effectors and Cas inactivation via dimer formation.6 Studying the mechanism of action of Acrs holds significant value for several reasons, including refining CRISPR/Cas technology, combating antibiotic resistance, unraveling the arms race in nature, and potential therapeutic applications.
Nomenclature and Database for Acrs
The names of the Acr genes and proteins were formalized in 2015.38 As a result, the type and subtype of CRISPR inhibited by Acr are indicated by the Roman numeral and alphabet that start their names. This was followed by a numerical digit representing the protein family, which was assigned according to the order of discovery. The source of this protein is indicated by a subscript at the end of its name. For instance, AcrIA5 is the name given to an Acr that exhibits inhibitory action on the type IA CRISPR system and is found at position five. Similarly, the fourth member of the AcrIIF4 family is Acr, which suppresses type IIF CRISPR system. Bondy-Denomy et al established a database for the tracking and registration of recently discovered acrs.36 This database is available at https://tinyurl.com/anti-CRISPR. Dong et al created another database of the functional arrangement of Acrs.53 This database can be accessed at http://cefg.uestc.edu.cn/anti-CRISPRdb/. It contains DNA sequences, coding sections, protein sequences, taxonomy, pathogenicity, protein interactors, and their associated three-dimensional structures. AcrFinder,54 AcRanker,55 Self-targeting Spacer Searcher,46 CRISPRminer,56 AcrCatalog,57 PaCRISPR,58 and AcrHub59 are a few bioinformatics tools that support and facilitate the research on Acrs. Table 2 provides further details of these bioinformatics tools, including their applications and websites.
Table 2.
Listing of Current Database and Bioinformatics Tools Developed for Acr Research
| Database Name /Bioinformatic Tool | Access Address | Applications | References |
|---|---|---|---|
| AcrBank | cefg.uestc.edu.cn/anti-CRISPRdb | It is an all-inclusive web resource with integrated data on all recognized anti-CRISPR proteins. DNA sequences, coding sections, protein sequences, source organisms, taxonomy, pathogenicity, protein interactors, and their three-dimensional structures are among the data that are provided. | [53] |
| Acr assembly | https://tinyurl.com/anti-CRISPR | It is a database used to register recently found Acr. The excel file contains comprehensive details about every anti-CRISPR protein that has been found, such as the protein’s name, target CRISPR that it exhibits inhibitory activity against, origin, and sequence. | [36] |
| PaCRISPR | http://pacrispr.erc.monash.edu/ | It is an online tool that uses evolutionary relationships and sequence similarity to predict and visualize anti-CRISPR proteins. | [58] |
| AcRanker | http://acranker.pythonanywhere.com/ | It is a method that uses machine learning to directly identify novel, potentially useful anti-CRISPRs based on protein sequence. | [55] |
| AcrFinder | http://bcb.unl.edu/AcrFinder | Using three well-established methods—guilt-by-association, CRISPR-Cas self-targeting spacers, and homology search—it is an internet server for genome mining of Acr-Aca operons. | [54] |
| Self-targeting spacer searcher | – | This application was created to identify whether any spacer in a genome’s CRISPR array addresses a protospacer located inside the same genome. Self-targeting is the term for this behavior, which is used to find new Acrs. | [46] |
| AcrHub | http://pacrispr.erc.monash.edu/AcrHub/ | It is a central location for research on anti-CRISPR protein mapping, prediction, and exploration. | [59] |
| AcrCatalog | http://acrcatalog.pythonanywhere.com/ | It is a machine learning-based online resource for structural and functional research on the relationships between Acrs and CRISPR. | [57] |
| CRISPRminer | http://www.microbiome-bigdata.com/CRISPRminer | It is a web server designed to gather and explore all available information about CRISPR-Cas systems, including the locus of the CRISPR-Cas system, its classification, the identification of self-targeting events, the inference of microbe–phage interactions, and anti-CRISPR annotation. | [56] |
Inhibition of Class 1 CRISPR/Cas System
Types I, III, and IV of the class 1 CRISPR/Cas systems are characterized by effector complexes called “Cascade” that are made up of several Cas proteins. Currently, at least 46 Acrs have been detected in the type I system, two Acrs in the type III system, and no Acr in the relatively uncommon type IV system (Table 1). Acrs target different Cas proteins present in the cascade or lead to degradation of the second messenger, thus preventing CRISPR/Cas-based immunity.6
Mechanisms of Type I Acrs
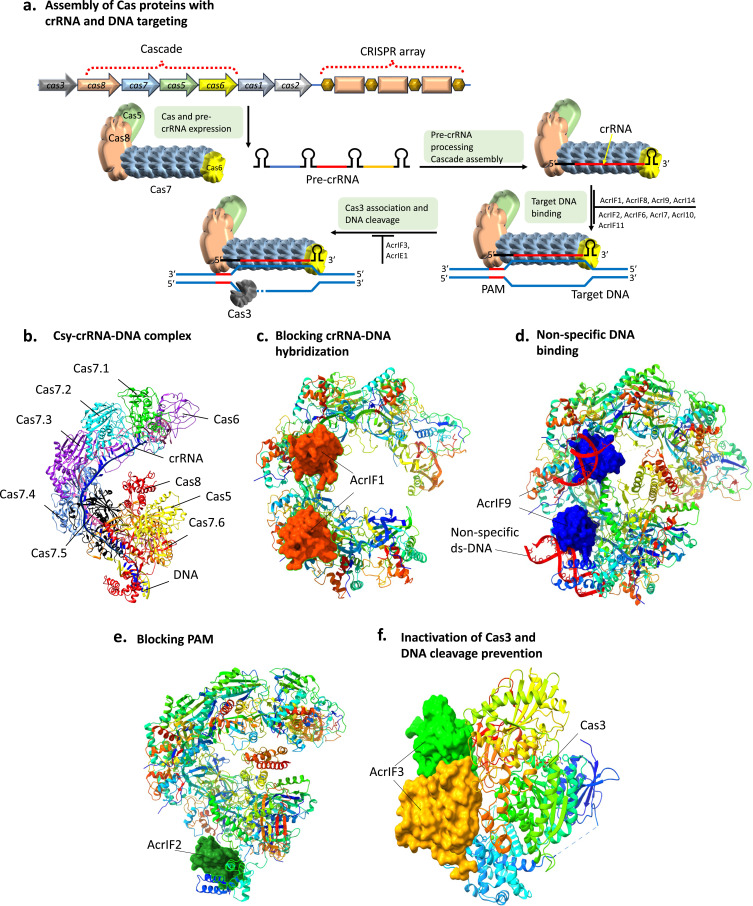
The first Acr to describe CRISPR/Cas inhibitory mechanisms was AcrIF1, AcrIF2-AcrIF5, and AcrIF1-3. Target DNA binding is sterically blocked by AcrIF1 and AcrIF2 (also called AcrIC2),60 whereas AcrIF3 inhibits Cas3.61 The crRNA-guided cascade complex and Cas3 with helicase-nuclease activity3,16 are responsible for achieving type I CRISPR/Cas immunity (Figure 6a).
Figure 6.
Functional mechanisms and domain organization of type I Acrs. (a) Assembly of Cas proteins with crRNA transcribed from CRISPR array to assemble as a multisubunit Cascade complex. The Cascade complex is guided by crRNA for the recognition of PAM and binding to target DNA followed by Cas3 recruitment that degraded both target DNA strands. (b) Structure of Csy (P. aeruginosa Cascade)-crRNA-DNA complex with PDB code 6B44. (c). Csy-crRNA complex bound with two AcrIF1 molecules (PDB code 6B46) that prevent the hybridization of crRNA guide to target DNA. (d) Structure of non-specific dsDNA binding with Csy-crRNA-AcrIF9 complex (PDB code 6WHI). (e) Structure of Csy-crRNA complex bound by AcrIF2 (PDB code 6B47). AcrIF2 blocks the PAM recognition site and inhibits target DNA binding. (f). Structure of Cas3 bound with AcrIF3 (PDB code 5B71) and prevention of the recruitment of Cas3, thus preventing target DNA cleavage.
The CRISPR array produces a precursor crRNA that is processed to mature into crRNAs by Cas6.16,62 The crRNA and Cas proteins combine to form a multisubunit cascade complex, and the spacer of the complex hybridizes to a protospacer,16,63,64 which is a complementary sequence in the target DNA. Cas8, a component of the Cascade complex, recognizes protospacer-adjacent motif (PAM) that is situated directly upstream of the DNA target (Figure 6b). Target DNA recognition by PAM is highly important before it can be unwound to hybridize with crRNA.65,66 As a continuation of this process, non-target DNA strand displacement results in R-loop formation, which initiates the recruitment of Cas3. This endonuclease breaks down both target and non-target DNA strands.67,68 The type I-A system is unique in using an “allosteric regulation mechanism“ to activate Cas3, while other type I CRISPR-Cas systems utilize a “trans-recruitment mechanism” for Cas3 activation upon R-loop formation.69
Type I Acrs most frequently function by preventing target DNA binding. Different type I Acrs inhibit the CRISPR/Cas system by directly inhibiting DNA binding. These Acrs (AcrIF1, AcrIF8, AcrIF9, and AcrIF14) perform this task by preventing the target DNA from hybridizing with the crRNA guide, whereas the other Acrs (AcrIF2, AcrIF6, AcrIF7, AcrIF10, and AcrIF11) prevent dsDNA from binding to the PAM-binding site (Figure 6a). Furthermore, to prevent Cas3 from cleaving the DNA strands, two more Acrs (AcrIE1 and AcrIF3) attach to the enzyme directly. The different mechanisms of type I Acrs inhibition by CRISPR/Cas includes disassembling of effector complex, blocking hybridization between crRNA and DNA, promotion of non-specific DNA binding, targeting of PAM recognition sites inhibiting DNA binding, and inactivation of Cas3 to prevent DNA cleavage.
Disassembling of the Effector Complex
As it is now well known that types I of the class 1 CRISPR/Cas systems are characterized by effector complexes called “Cascade” that are made up of six Cas7 proteins. The type I-F CRISPR/Cas system is inhibited by AcrIF25 by pulling apart the whole assembled effector complex. The biochemical and structural studies confirmed that starting at one end of complex, one subunit of Cas7 is removed at a time by AcrIF25 with no apparent enzymatic activity. This mechanism of action of AcrIF25 is a very recent study in which a stable and large macromolecular complex is disassembled without any external energy.70
Blockade of crRNA–DNA Hybridization to Inhibit DNA Binding
AcrIF1 binds directly to the Cas7 subunits of cascade, blocking target DNA recognition. The estimated stoichiometry of AcrIF1 molecules per crRNA-guided surveillance system (Csy; Pseudomonas aeruginosa cascade) is 2.6 ± 0.3. The cryo-EM structures revealed that the Cas7 backbone directly interferes with AcrIF1 binding at two to three different locations in the AcrIF1–Csy–crRNA complex, sterically obstructing the target DNA and blocking its hybridization to the crRNA guide71,72 (Figure 6c).
The interaction between the three AcrIF1 essential residues (Tyr6, Tyr20, and Glu31) and a conserved lysine (Lys85) in Cas7 was discovered using the cryo-EM structure of Csy–crRNA–AcrIF1.71,73 Disruption of both AcrIF1’s Cas7 binding and target DNA binding occurs when Lys85 is changed to Ala, suggesting that some Cas7 conserved residues are targeted by AcrIF1, which is necessary for crRNA guide–target DNA binding.71 Additionally, it appears that the binding of AcrIF1 may induce some Cas7 conformational changes that further limit access to target DNA, based on the configurations of the web domain of Cas7 between the AcrIF1-free and bound states that have been observed.71 Similar to AcrIF1, two molecules of AcrIF1474 and one molecule of AcrIF875 bind to the target DNA-binding site on different Cas7 subunits of the complex in a competitive manner to sterically prevent the hybridization of crRNA with DNA.
Triggering of Non-Specific DNA Binding
Two AcrIF9 molecules (Figure 6d) couple at some location in the Csy–crRNA complex, identical to that observed in AcrIF1 (Figure 6c), as reported in the cryo-EM structure of Csy–crRNA–AcrIF9. This site is likely to sterically prevent crRNA–dsDNA hybridization.75,76 Furthermore, complex sequence-non-specific dsDNA binding is triggered by AcrIF9 binding to Csy-crRNA, this non-target DNA binding sequesters the cascade complex away from the target DNA and increases Acr activity.76
Targeting the PAM Recognition Sites Inhibits DNA Binding
A proper PAM recognition is required for target DNA binding in type I CRISPR/Cas systems as a necessary initial step. Thus, another method by which Acrs evade CRISPR/Cas immunity is to bind PAM recognition sites (Figure 6e). For proper PAM recognition, Cas8 contains a specific sequence (residues 246–250), known as the “K-wedge”. Severe defects in the binding of DNA targets were observed after introducing a mutation in Lys247 (K247E).71 The Cas8-K247E mutant also demonstrated diminished binding capacity for AcrIF2 and AcrIF7.71,77 A mutation in AcrIF6 at the Cas8 Lys247-binding residues interrupts the binding interface facilitated by Lys247 and weakens the inhibition of target DNA cleavage.75 As demonstrated by in vitro biochemical experiments, a direct interaction occurred between AcrIF2 and the Cas8 and Cas5 subunits of the cascade complex.61 Furthermore, it has been demonstrated that AcrIF2 engages in the conserved PAM recognition site, as reported in the cryo-EM of Csy–crRNA–AcrIF271,73 (Figure 6e), and in the same fashion, AcrIF6,75 AcrIF7,74 and AcrIF1073 behave.
The structures of D. vulgaris type I-C cascade at different phases of dsDNA target capture have provided insights into the mechanisms underlying the recognition of PAM and the allosteric activation of cascade.78 Recently, it has been reported from the studies on Neisseria lactamica, two Acr proteins (AcrIC8 and AcrIC9) strongly inhibit the CRISPR/Cas system. The AcrIC8 inhibits the recognition of PAM through allosteric inhibition, whereas, AcrIC9 makes this inhibition through direct competition.79
As a different strategy from the previous approaches of blocking PAM, AcrIF11, a mono-ADP-ribosyl transferase specifically ADP-ribosylates Cas8 Asn250 residue, required for PAM acknowledgement. This resulted in a thorough loss of affinity for DNA binding by Csy–crRNA.80 As this mechanism is dependent on post-translational protein modifications, AcrIF11 may be a more potent inhibitor than AcrIF2, AcrIF6, or AcrIF10.
Inactivation of Cas3 to Prevent DNA Cleavage
Acrs also employs the tactic of preventing target DNA cleavage to impede CRISPR/Cas immunity. Target DNA cleavage is inhibited by AcrIF3 by blocking the recruitment of the helicase-nuclease Cas3 to the Csy–DNA complex.61 A homodimer of AcrIF3 directly binds to Cas3 (Figure 6f), locks the complex in an ADP-bound state, and prevents the DNA entrance tunnel of Cas3 from accessing the non-complementary target DNA, as per the structures obtained by X-ray crystallography and cryo-EM.81,82 Furthermore, AcrIF3 and Cas8 C-terminal domains are structurally similar; thus, it is likely that AcrIF3 mimics the Cas8 CTD, which is involved in Cas3 recruitment, thus preventing Cas3 from being recruited to the Csy–crRNA–DNA complex.71 In addition, primed spacer acquisition was inhibited when AcrIF3 blocked the Cas3-mediated DNA degradation.81,83 Similar to AcrIF3, AcrIE1 binds to Cas3 directly as a dimer to inactivate it in subtype I–E CRISPR/Cas systems; the CTD of AcrIE1 is essential for Cas3 inhibition.45
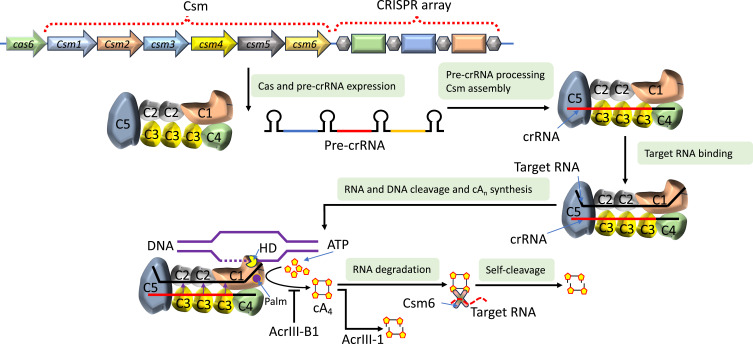
Mechanisms of Type III Anti-CRISPRs
The multiprotein effector complexes loaded with crRNA, Csm in subtype III-A and Cmr in subtype III-B,84 are responsible for achieving type III CRISPR/Cas immunity (Figure 7). Cas6 cleaves crRNA during maturation, but this enzyme is not part of the effector complex.85 Complementary RNA targets are recognized by effector complexes using crRNA.86 It triggers non-specific ssDNA cleavage by utilizing the His-Asp (HD) domain of Csm1 or Cmr2 (also known as Cas10).87,88 The palm domains of Csm1 also lead to the synthesis of cyclic oligoadenylate (cOA) from ATP.89,90 The Csm1 or Cmr2 hD palm domains are then deactivated because the Csm3 or Cmr4 subunits preferentially cut the RNA target at six-nucleotide intervals.91,92
Figure 7.
Functional mechanisms of Type III Acrs. Transcription of pre-crRNA from CRISPR array and its maturation to crRNA that assembles with Csm proteins to form effector complexes. The crRNA binding with Csm complex guides the effector complex to target RNA, cleaved by C3 at six nucleotide intervals. The target RNA binding triggers the cleavage of non-specific ssDNA by HD nuclease domain and formation of cyclic oligonucleotides, such as cyclic tetra-adenylate (cA4). The cA4 binds with Csm6 and activates its RNase activity. Subsequently, cA4 cleaves to linear diadenylates that blocks its own RNase activity and AcrIII-1 acts as a ring nuclease that degrades cA4.
The second messenger produced, known as cOA, is released from the Csm or Cmr complex.93 It then attaches to the CRISPR-associated Rossmann fold (CARF) domain of dimeric RNases, which is represented by Csm6 in subtype III-A. As a result, it allosterically stimulates RNase activity of its own higher eukaryotes and prokaryotes nucleotide-binding (HEPN) domains, which is critical for type III immunity in vivo.94,95
Interestingly, Thermococcus onnurineus Csm695 and Sulfolobus islandicus Csx196 are ring nucleases and RNases. They break down tetraadenylate rings (cA4) through their CRISPR-associated Rossmann Fold (CARF) domain, converting them into linear biadenylates with terminal 2′,3′-cyclic phosphate, thereby stopping RNase activity (Figure 7). Similarly, Enterococcus italicus97 and Streptococcus thermophilus97,98 Csm6 degrades the second messenger cA6. This mechanism limits self-toxicity by regulating RNase activity of Csm6 in a time-restricted manner.
Inhibition of the Production of Second-Messenger
Viruses also use a potential strategy to target the second messenger cOA against prokaryotic CRISPR/Cas immunity. During RNA targeting, AcrIIIB1 suppresses RNase-related mechanisms that require Csx1 RNase activity.99 The subtype III-B Cmr complex, which produces cOA, is the target of AcrIIIB1 interactions rather than Csx1, indicating that AcrIIIB1 suppresses RNase activity mediated by Csx1, most likely by mediating the Cmr complex synthesis of cOA.99
Second Messenger Degradation
A protein of viral origin called AcrIII-1 binds to cOA directly without directly interacting with the Cmr complex and degrades the secondary messenger cA4. This allows viruses to evade the type III CRISPR/Cas immunity.100 By cleaving cA4 into two linear diadenylates, Csm6-based degradation of target RNA is inhibited. The crystal structure of AcrIII-1 with cA4 showed that this cyclic tetraadenylate is covered by a movable loop and binds to the dimeric form of AcrIII-1 (Figure 7). The catalytic activity of AcrIII-1 depends on the conserved residue His47, which is 2500 times less in the AcrIII-1-H47A mutant.
Inhibition of Class 2 CRISPR/Cas Systems
Class 2 CRISPR/Cas systems include types II, V, and VI, which are distinguished from class 1 systems by the presence of a single Cas effector molecule. All three types in this class had Anti-CRISPRs with twenty-eight Acrs in the type II system, five Acrs in the type V system, and eight Acrs in the type VI system (Table 1). Target DNA (or RNA) binding, target cleavage, and interference with CRISPR/Cas complex assembly are the key mechanisms by which class 2 Acrs suppress CRISPR/Cas immunity.
Mechanisms of Type II Acrs
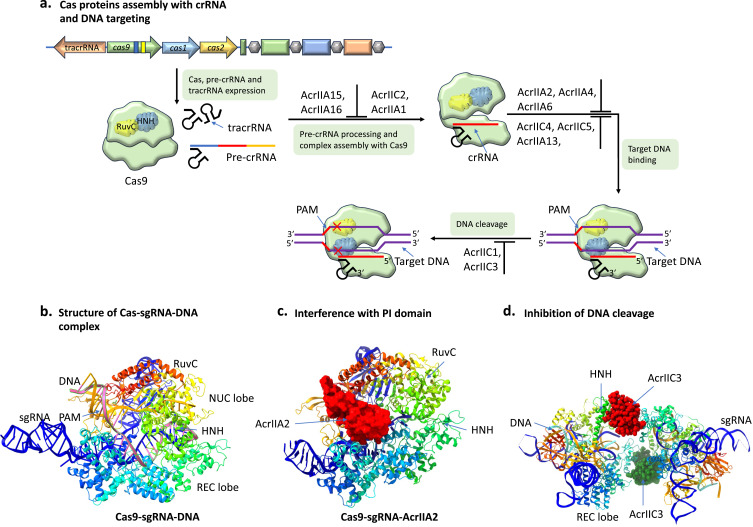
A single Cas9 protein and sgRNA (a crRNA and tracrRNA complex17,101 are responsible for the execution of type II CRISPR/Cas immunity (Figure 8a). Type II CRISPR/Cas immunity101 also requires PAM recognition, carried out by the PI domain of the NUC lobe,14 similar to type I CRISPR/Cas systems. Prior to initiating target dsDNA melting, R-loop creation, and target cleavage, the Cas9–sgRNA composite first recognize the PAM sequence of the target DNA (Figure 8b). Type II Acrs act via different approaches, such as interference with the Cas9-sgRNA complex assembly, inhibition of target DNA binding, and inhibition of DNA cleavage by direct interaction.
Figure 8.
Mechanism of action of type II Acrs. (a) Assembly of Cas9 with crRNA and tracrRNA, both transcribed from CRISPR locus. A sgRNA can be artificially obtained by covalently linking crRNA and tracrRNA. The Cas9 is guided by crRNA to recognized PAM and subsequently binding and degradation of target DNA. (b) Ribbon structure of Cas9-sgRNA-DNA complex (PDB code 5F9R). (c) Structure of Cas9-sgRNA complex bound with AcrIIA2 (PDB code 6IFO). The Cas9 PI site is occupied by AcrIIA2, thereby blocking recognition of DNA and binding by Cas9-sgRNA complex. (d) Structure of a pair of Cas9-sgRNA-DNA complex bound with dimeric AcrIIC3 (PDB code 6JE4) and prevention of DNA cleavage by locking the HNH domain.
Interference with Cas9–sgRNA Complex Assembly
The N. meningitides Cas9 (NmeCas9) bridge helix motif (positively charged portion) is bound by dimeric AcrIIC2 in an acidic groove, which inhibits the formation of an active Cas9–sgRNA complex.102,103 Furthermore, AcrIIC2 binding increases AcrIIC2’s anti-CRISPR action by keeping Cas9 in ligand-free (apo state) form, which is susceptible to cellular proteases. Subsequent structural and biochemical investigations demonstrated that AcrIIC2 might similarly target a wide range of Cas9 orthologs, which could be advantageous for biotechnological applications of the most commonly used Cas9.102
Cas9 can be degraded during the lysogenic stage,104 when AcrIIA1 binds directly to the apo-Cas9 HNH domain or sgRNA-attached Cas9. Mutagenesis revealed that Phe115 in AcrIIA1 is essential for both its association with Cas9 and its anti-CRISPR activity, and the HNH domain His840 is critically responsible for the interaction between AcrIIA1 and Cas9. Interestingly, AcrIIA1 can only cause Cas9 degradation in vivo, indicating that Cas9 degradation requires an extra stimulus from the L. monocytogenes cellular environment.
Similar to what was previously found for AcrIIC2, AcrIIA1 activates Cas9 degradation, indicating that AcrIIA1 may be destabilizing apo-Cas9 or inhibit sgRNA attached to apo-Cas9.102,103 Moreover, AcrIIA1 is a dimer that performs the dual role of a Cas9 inhibitor, and its C-terminal portion binds to Cas9 and inhibits it, whereas its N-terminal domain, with the HTH motif, represses the transcription of its own gene and functions as a Cas9 sensor, adjusting its expression to correspond with Cas9 levels.104 HTH-containing proteins have been reported to suppress Acr transcription,33,105 suggesting that HTH-containing proteins share a conserved transcriptional auto-regulation mechanism. Furthermore, it is likely that AcrIIA1 recognizes the catalytic residue His840 in the Cas9 hNH domain104 that which extensively inhibits various Cas9 orthologs.
Similar to AcrIIA1, AcrIIA13, AcrIIA14, and AcrIIA15 act as bifunctional inhibitors that bind to their own promoters to negatively regulate transcription, in addition to blocking Cas9 immunity. While AcrIIA13 prevents Cas9-mediated DNA cleavage by limiting target DNA binding, AcrIIA15 inhibits Cas9 by preventing the Cas9–sgRNA complex assembly. This results in strong suppression of Cas9-mediated genome editing in human cells.47 Although AcrIIA14 did not stop Cas9 from attaching to its target, research has shown that like AcrIIC3, AcrIIA14 causes Cas9 to dimerize47 and leads to the adoption of an inactive conformation.102,106
Furthermore, it has been demonstrated that the recently discovered AcrIIA16–AcrIIA19 inhibit CRISPRi, suggesting that they block target DNA binding.107 Different degrees of sgRNA degradation were produced by the co-expression of AcrIIA16–AcrIIA19, SpCas9, and sgRNA, indicating that they may function by modifying sgRNA levels. In particular, when incubated with sgRNA and apo-SpCas9, AcrIIA16 reduced SpCas9-mediated DNA cleavage. However, this effect was not observed when treated with the sgRNA-loaded Cas9 complex, suggesting that AcrIIA16 may obstruct sgRNA–Cas9 complex assembly.107
Inhibition of Target DNA Binding
As shown in Figure 8c, AcrIIA2108,109 occupies the PI site and locks the HNH domain, thus sterically preventing the Cas9–sgRNA complex from recognizing and binding to dsDNA. Interestingly, AcrIIA4 was attached to Cas9 at a position comparable to that of AcrIIA2, which inhabits the PI site. However, it also interacts with the RuvC domain and prevents the nontarget DNA strand from entering its active site.110,111 The binding affinities of AcrIIA2 and AcrIIA4 for Cas9-sgRNA were approximately 15 times higher than that of the Cas9-sgRNA-DNA complex.112 The affinity of AcrIIA4 for apo-Cas9 was almost 8000 times lower than that of sgRNA-loaded Cas9.111 However, structural studies have revealed that AcrIIA4 does not directly interact with sgRNA, suggesting that AcrIIA4 binding is dependent on conformational changes in Cas9 caused by sgRNA.113 AcrIIA2 and AcrIIA4 are effective tools for Cas9 regulation in gene editing applications because they can limit SpCas9 activity in both bacterial and human cells.37 Unlike AcrIIA2 and AcrIIA4, AcrIIA6 modifies the conformational dynamics required for PAM binding rather than blocking direct PAM recognition, resulting in a reduction in DNA-binding affinity.114
The cryo-EM results for the AcrIIA6–Cas9–sgRNA complex demonstrated a dimeric form of AcrIIA6 that attaches to an allosteric location on the rear face of Cas9 in relation to the target-binding surface. This causes Cas9 to dimerize, limiting the dynamics required for PAM binding.114 In mammalian cells and bacteria, AcrIIA6 can inhibit Cas9 activity, suggesting that it may be able to regulate gene editing.44 AcrIIC4 and AcrIIC5 prevent NmeCas9-mediated genome editing in mammalian and bacterial cells as well as DNA cleavage by NmeCas9-sgRNA in vitro.115 Subsequent biochemical investigations have demonstrated that AcrIIC4 and AcrIIC5 decrease the DNA affinity for Cas9-sgRNA by roughly six fold and nine fold, respectively, and inhibit stable DNA binding.115
Inhibition of DNA Cleavage by Direct Interaction
AcrIIC1 selectively binds to the active sites in the HNH domain,106 probably eliminating the divalent cation required for target DNA cleavage116 according to the structure of AcrIIC1 coupled to the Cas9 hNH domain. Interestingly, AcrIIC1 converts Cas9 into catalytically dead Cas9 (dCas9) by inhibiting conformational modifications necessary for DNA cleavage, leaving Cas9 in a DNA-bound but catalytically inactive state. AcrIIC1 is a broad-spectrum inhibitor because of its interaction with the HNH domain, the most conserved portion of Cas9.
AcrIIC3 decreased target DNA-binding affinity106 and caused Cas9 dimerization (Figure 8d). Recently, it was observed that two AcrIIC3 molecules can bind two Cas9 molecules together. Each AcrIIC3 molecule showed affinity for the REC lobe of one Cas9 molecule and the HNH domain of another Cas9 molecule, confirming the HNH domain from the target strand cleavage site102 (Figure 8d). Moreover, non-target strand cleavage was prevented by the inability of the inactive HNH state to activate the RuvC domain. It has been proposed that AcrIIC3 can suppress Cas9-mediated immunity by directly blocking DNA cleavage by the Cas9 hNH domain even in the presence of target DNA binding.
Mechanisms of Type V Acrs
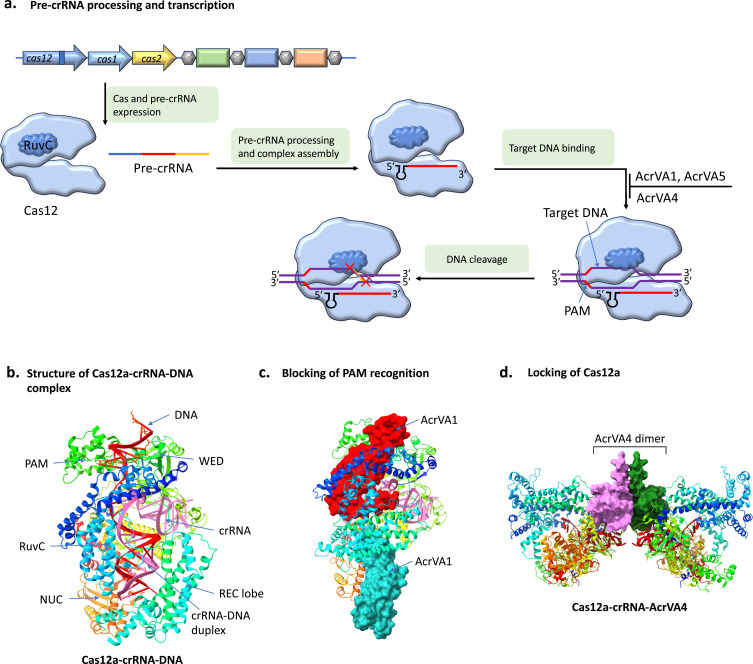
The crRNA guide-bound Cas12 complex mediates type V CRISPR/Cas immunity, resulting in dsDNA cleavage (Figure 9a) or ssRNA cleavage by the RuvC domain.117 The most well-studied Cas12 subtype is Cas12a, which consists of wedge domains118,119 connecting the REC and NUC lobes. The RuvC nuclease domain is located in the NUC lobe (Figure 9b). Target DNA is recognized by Cas12a in a PAM-dependent way.120,121 In contrast to Cas9, Cas12a exclusively uses the conserved RuvC domain to cleave target and non-target strands, resulting in a staggered DNA double-strand break (Figure 9a). The non-target strand can be cleaved in trans because the cleaved target strand is still attached to the Cas12a–crRNA complex.119 The different mechanisms of type V Acrs include the blockade of the Cas12–PAM interaction, which prevents target DNA binding, and the locking of Cas12a, leading to the inhibition of target DNA binding.
Figure 9.
Functional mechanisms of type V Acrs. (a) Transcription of pre-crRNA from CRISPR array and processing to mature crRNA, and its assembly with Cas12 protein to form the effector complex. The Cas12 is guided by crRNA to PAM of the target DNA, which subsequently degrades it by RuvC nuclease domain. (b) Structure of Cas12a-crRNA-DNA complex (PDB code 5XUS). (c) Structure of two AcrVA1 molecules bound to Cas12a-crRNA complex (PDB code 6NMC). The PAM recognition is hindered by one AcrVA1 protein that triggers the cleavage of Cas12a bound crRNA. The second AcrVA1 clashes with crRNA-single strand DNA duplex formation. (d) Structure of dimeric AcrVA4 bound with Cas12a-crRNA complex (PDB code 6NM9). The Cas12a-crRNA binding with AcrVA4 prevents conformational changes for the formation of crRNA-DNA duplex.
Blocking of Cas12–PAM Interaction Prevents the Target DNA Binding
AcrVA1 cleaves the spacer sequence in Cas12a-bound crRNAs to irreversibly disable the Cas12a complex, therefore extensively inhibiting Cas12a orthologs in vitro and in human cells.46,48 Two AcrVA1 molecules form a complex122 linked to Cas12a-crRNA (Figure 9c), as studied by cryo-EM structure. One AcrVA1 molecule (shown in red in Figure 9c) resides in the PI region, where it obstructs Cas12a recognition of the PAM sequence and causes Cas12a-bound crRNA to cleave and deactivating the Cas12a–crRNA complex.
Since neither AcrVA1 nor Cas12a can degrade crRNA by themselves, the AcrVA1 interaction with the Cas12–crRNA complex is necessary for AcrVA1-mediated crRNA cleavage. Three positions in AcrVA1 (Arg41, His42, and His45) proximal to the crRNA were mutated, nearly eliminating crRNA cleavage, indicating that these positions may play a catalytic role in crRNA cleavage. Sterically impeding the formation of the crRNA-DNA duplex, the second AcrVA1 protein (shown in blue in Figure 9c) attached to the area of Cas12a that ordinarily binds the downstream section of the crRNA-ssDNA duplex. Thus, AcrVA1 can block Cas12a-based immunity through two approaches: inhibition of the cleavage of the crRNA guide and PAM recognition, which leads to the establishment of the crRNA–DNA duplex. On the other hand, by acetylation of Lys635 of Cas12a, AcrVA5 inhibits the binding of target DNA, which is a crucial step for the recognition of PAM.
Locking of Cas12a Leads to the Inhibition of Target DNA Binding
The dimers of AcrVA4 also promote Cas12a-crRNA dimerization (Figure 9d). This leads to blocking of Cas12a-cRNA binding to dsDNA.123 It has also been observed that, the C-terminal domain of AcrVA4, is sufficient to inhibit Cas12a,123 hence dimerization of AcrVA4 is not required for Cas12a inhibition. The Cas12a–crRNA–AcrVA4 complex cryo-EM structure showed that AcrVA4 binds to one or two Cas12a–crRNA molecules, anchors the crRNA pseudoknot, and blocks the conformational alterations necessary for crRNA–DNA duplex122,124 (Figure 9d). AcrVA4 binds to Cas12a in a crRNA-dependent manner, as evidenced by its interaction with Cas12a–crRNA but not with apo-Cas12a.122
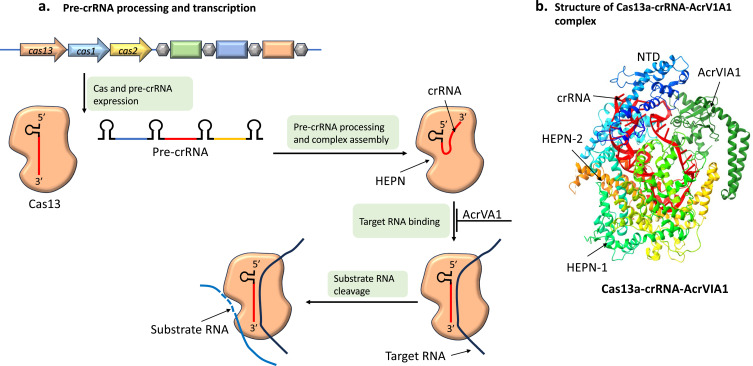
Mechanism of Type VI Acrs
Type VI CRISPR/Cas immunity is facilitated by crRNA-guided Cas13, which recognizes a crRNA- phage RNA complementary target125 and initiates non-specific destruction of host and viral “substrate” RNA, hence blocking RNA binding in type VI systems (Figure 10a). When target RNA binds to Cas13a, it undergoes a significant conformational shift that brings its two HEPN closer together and into perfect alignment, activating its composite catalytic site.126 Host transcript deterioration causes dormancy and stops viruses through multiplication.127 The ϕLS46 listeriophage genome encodes AcrVIA1, a protein that can totally evade CRISPR/Cas13 immunity.128
Figure 10.
Functional mechanism of type VI Acrs. (a) Transcription of pre-crRNA from CRISPR array and its processing into mature crRNA, and its assembly with Cas13 to form the effector complex. The Cas13 is guided by crRNA to target RNA and triggers the degradation of non-specific RNA by HEPN domain. (b) Structure of Cas13a-crRNA-AcrVIA1 complex (PDB code 6VRB).
AcrVIA1 can bind directly to Cas13a and interact directly with crRNA, as well as the HEPN-2 and Cas13a linker domains128 (Figure 10b). The target RNA obtains access to the crRNA, and conformational changes are necessary for the activation of HEPN-1 and HEPN-2 nucleases. This system has been reported to be blocked by the engagement of AcrVIA1 with both crRNA and Cas13a.
Furthermore, the entire function of AcrVIA1 has been reported to be lost due to truncation of two AcrVIA1C-terminal helices (ΔN173–N232) and mutation of critical AcrVIA1 residues Tyr39A, Ser40, Asn43, Ser93, and Gln96. In type VI systems, anti-CRISPRs found in Leptotrichia wadei F0279 prophages can regulate the activity of Cas13a in human cells.129 Therefore, CRISPR/Cas13a-based biotechnology tools may be controlled by anti-CRISPR in type VI systems.
RNA Based Anti-CRISPRs
As very different from protein inhibitors against CRISPR/Cas systems, small non-coding RNA anti-CRISPRs (Racrs), a distinct type of CRISPR/Cas inhibition strategy has been unveiled very recently.130 The CRISPR array contain different repeats which are mimicked by Racrs, encoded in viral genomes as solitary repeat units.131 It has been observed that type I-F CRISPR/Cas system is strongly inhibited by a Racr encoded from a prophage. This inhibition is based on specific interaction with Cas6f and Cas7f, that results in the establishment of an aberrant Cas sub-complex. Diverse range of plasmids and viruses encode Racrs for almost all CRISPR/Cas types.132 It is now believed that the anti-CRISPR approach performed by molecular mimicry of CRISPR repeats is a widespread strategy and this approach can benefit a lot for different biotechnological applications.133
The Applications Acrs in Biotechnology
CRISPR/Cas systems are frequently employed for gene manipulation in eukaryotic cells via gene editing,134,135 gene control,136 and imaging in eukaryotic cells.137,138 Acrs are innovative tools for modifying CRISPR/Cas function in both prokaryotes and eukaryotes.133 Understanding the molecular mechanisms underlying Acr action may help in the safe and effective use of CRISPR/Cas tools for genetic modifications, particularly in the context of medicine. Thus, attempts to regulate prokaryotic CRISPR/Cas surveillance complexes will advance the development of antimicrobial drugs and treatment of human diseases.139
Acrs Help to Reduce off-Cell Activity
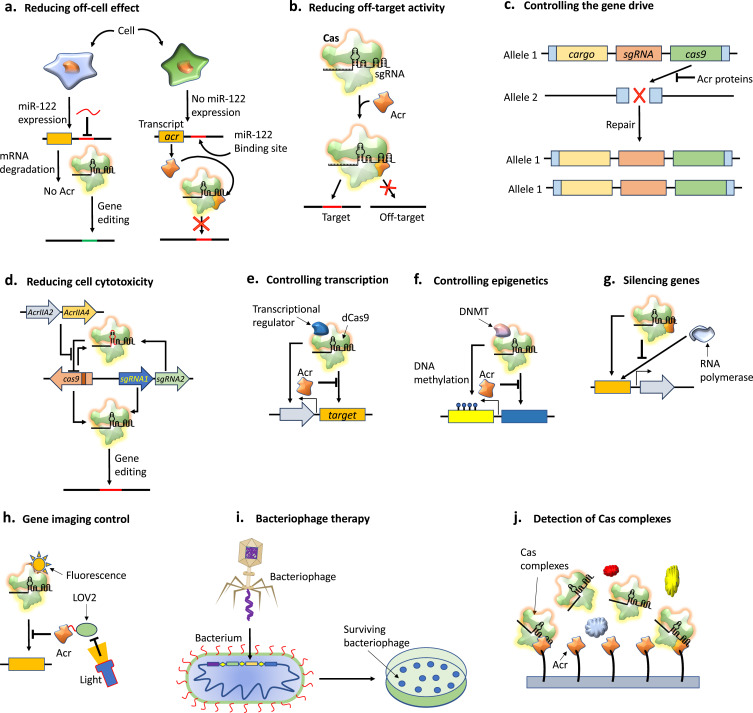
An Acr-based strategy was created to limit Cas9 activity in all auxiliary tissues to limit CRISPR/Cas function in particular tissues. This strategy is predicated on the repression of Acr expression in particular cell types, using endogenous miRNAs specific to these cell types. The expression of AcrIIA4, AcrIIC1, and AcrIIC3 is repressed by a microRNA inserted into the untranslated regions of Acr genes.140,141 To enable gene editing in the liver, the only organ in which miR-122 is expressed, and to inhibit editing in unwanted organs, such as the heart,141 miR-122-repressed AcrIIC3 was injected into adult mice using Cas9–sgRNA (Figure 11a). This approach serves as a foundation for the use of Acrs to regulate CRISPR/Cas9 gene editing in a cell-specific manner.
Figure 11.
Innovative biotechnology applications of Acrs. (a) Reducing off-cell effects. The accomplishment of cell type-specific gene editing by suppressing Acr activity with microRNA (miR). When miRNA is bound to Acr transcripts modified to contain miR-122-binding sites, it causes the transcripts to degrade in cells that express miR-122. The Acrs expressed by cells lacking miR-122 can stop gene editing. (b) Reducing off-target activity. The Acr expression reduces the cleavage of off-target DNA or RNA by CRISPR/Cas complex. (c) Controlling the gene drive. Gene drive copies the drive gene to the broken chromosome and disperses it throughout the population by using Cas9 to cut a chromosome and cause the cell to mend the break. Acrs limit such gene drives by preventing Cas9 activation. (d) Reducing cell cytotoxicity. The restriction of Cas9 expression is used to reduce the cell cytotoxicity. Cas9 self-cleavage system includes Cas9, sgRNA2 that target Cas9-encoding DNA and sgRNA1 that targets a specific genome site. AcrIIA2 and AcrIIA4, which are expressed from the producer cell and helper virus, restrict Cas9 activity during the formation of the viral vector by preventing self-cleavage. (e) Controlling transcription. Transcription control by combining a transcription regulator with catalytically dead Cas9 (dCas9). Acrs obstruct the target DNA binding and can stop the fusion of proteins intended to function at the desired gene. (f) Controlling epigenetics. Acrs that prevent target DNA binding can stop dCas9 fusions with epigenetic regulators like DNA methyltransferases (DNMTs). (g) Gene silencing. The catalytic activity of Cas9 can be rendered inactive by Acr binding without compromising its capacity to bind DNA. Transcription is hampered by the Cas9–Acr complex because it prevents RNA polymerase from binding. (h) Gene imaging control. Fusion of photosensor protein like LOV2 with Acr can be used to control the dCas9-based imaging. The presence of light degrades LOV2 and restricts dCas9 function. (i) Bacteriophage therapy. Acrs inhibit CRISPR/Cas system including spacers targeting bacteriophages. Acrs work by deactivating CRISPR/Cas systems in bacteria, which include spacers that target bacteriophages and boost the effectiveness of phage therapy against bacteria. (j). Acr-based platform can be used as an efficient alternative for the detection of CRISPR/Cas complexes.
Acr Help in Reducing off-Target Activity
In human cells, AcrIIA4 may be delivered on a specific time to minimize off-target editing while maintaining on-target editing110 (Figure 11b). In addition, AcrVIA2 and AcrVIA5 significantly decreased L. wadeii Cas13a129 off-target effects. The fusion of Acr variants with Cas9 also decreases off-targeting effects, which is practically significant.142 Although it has been demonstrated that base editing via the fusion of dCas9 with a cytidine deaminase can effectively correct point mutations related to genetic diseases without causing significant harmful double-strand breaks in DNA.143 This can also result in single-nucleotide variants in mouse embryos and rice.144,145 In human cells, AcrIIA2 and AcrIIA5 may effectively block dCas9-based base editing, and in mammalian cells, AcrIIA5 prominently decreases the off-target results of base-editing systems.146 Moreover, AcrVIA5 inhibited RNA A-to-I editing mediated by dCas13 in mammalian cells.129 Therefore, Acrs are a useful approach for reducing the off-target effects of CRISPR/Cas tools.
Acr Help in Controlling the Gene Drives
Cas9 is used to cut a chromosome at a specific location devoid of a specific set of drive genes, and is an innovative genome editing technology that causes the cell to heal the break through recombination by driving genes copying onto the impaired chromosome (Figure 11c). This novel approach can be used to treat human illnesses, including malaria and other diseases spread by insects.147,148 Safeguards are necessary for the successful use of gene drives,149 because gene drive technology can genetically modify all species. By deactivating Cas9,150 AcrIIA2 and AcrIIA4 were demonstrated to be strong inhibitors of gene expression in S. cerevisiae. One useful way to manage this exciting new gene-editing tool is to apply Acrs to gene-drive systems.
Acr Help in Reducing Cell Cytotoxicity
Limiting the activity of Cas nucleases is crucial for the effective use of Cas enzymes because their continuous activation might be harmful to cells. After the deployment of CRISPR/Cas9 in human hematopoietic stem cells, cytotoxicity and low engraftment possibilities have been reported, most likely due to the prolonged Cas9 expression period.151 Adenoviruses that express self-cleaving Cas9 have been created to limit the potential of the Cas enzyme in target cells.152 Helper-dependent adenoviruses express CRISPR/Cas9, which facilitates the transitory expression of Cas9 by directing the cleavage of the virus genome after transduction within the target cells (Figure 11d).
In the producer cells and helper virus, AcrIIA2 and AcrIIA4 were employed to block Cas9 activity to prevent self-cleavage during vector formation. Additionally, the timed delivery of AcrIIA2 and AcrIIA4 enhanced engraftment rates, decreased cytotoxicity to human hematopoietic stem cells, and modulated the duration of CRISPR/Cas9 activity151 without altering on-target genome editing rates.
Acr Help to Control dCas9-Based Tools
The presentation of CRISPR/Cas tools for transcription regulation and live-cell chromatin imaging has been expanded with the introduction of catalytically inactive Cas9 (dCas9). Numerous Acrs hinder the ability of CRISPR/Cas to bind target sequences by preventing the Cas enzyme from binding. It has been possible to disrupt gene transcription in bacterial and mammalian cells using dCas9 and to regulate CRISPR/Cas-based gene regulatory circuits153,154 using Acrs (Figure 11e).
To repress the expression of a fluorescent reporter, dSpCas9 attached to the transcriptional repressor domain of MXI1 attaches close to the promoter of the reporter. AcrIIA2 and AcrIIA4, which bind to dSpCas9–MXI1, loosen fluorescent reporter repression and boost fluorescence.153 Using such Acr-based gene circuits, tiny compounds can be identified based on the fluorescence readout. Acrs have been used to regulate epigenetic changes in addition to transcriptional regulation (Figure 11f).
Because the capacity of dCas9 to bind DNA is disrupted, AcrIIA4 can be used to assess the longevity of epigenetic changes caused by dCas9 coupled with a gene modulator. By deleting cas3 nuclease gene,155 the CRISPR/Cas system type I in Escherichia coli has been modified into governable transcription repressors AcrIIC1,106 AcrIIC3,156 AcrIF3,61,81 and AcrIE145 can repurpose these CRISPR/Cas systems into CRISPRi systems to silence genes by converting the CRISPR/Cas effector complex to an inactive state akin to dCas9, compromising its capacity to bind DNA (Figure 11g).
A light-dependent Acr technique called CRISPR/Cas9 activity switching via a novel optogenetic variant of AcrIIA4 (CASANOVA) is based on the combination of an optogenetic reformed type of AcrIIA4 with the LOV2 photosensor from A. sativa. This technique has been used to track the speed at which dCas9 targets DNA in living cells.157 The cells expressed CASANOVA, a guide RNA targeting telomeres, and the dCas9–RFP fusion protein. In the absence of light, AcrIIA4–LOV2 bind and inhibits dCas9. Nevertheless, photoexcitation caused LOV2 to unfold, which in turn caused AcrIIA4 to unfold and reduces AcrIIA4 binding affinity for dCas9 (Figure 11h). Furthermore, direct and dynamic validation of dCas9-based imaging can be achieved by comparing telomere labelling in light and dark environments.
Acr Help in CRISPR/Cas Resistance in Phage Therapy
A promising non-antibiotic approach for treating bacterial resistance to antibiotics is phage treatment.158 The potential spectrum of phage therapies may be restricted by the fact that numerous bacteria possess active CRISPR/Cas systems. A possible method for generating CRISPR/Cas confrontation in phage therapy is the use of Acrs, which can naturally deactivate the CRISPR/Cas systems in bacteria (Figure 11i).
Acr Help in Detecting CRISPR/Cas Complexes
Evaluating the effectiveness, dispersion, and retention of CRISPR/Cas-based gene editing in target cells requires the detection of the CRISPR/Cas effector complex within biological materials. Acr-based biosensing technologies offer alternatives to antibodies for effector complex detection, identification, and quantification owing to the high binding affinity of these probes for effector complexes (Figure 11j). To detect the effector complex using electrochemical, fluorescence, or colorimetric techniques, AcrIIA4 can be immobilized and used as an affinity reagent. The AcrIIA4-based biosensing device can detect 8 nM CRISPR/Cas complexes under biologically relevant circumstances and 280 pM CRISPR/Cas complexes in reaction buffer.159 AcrIIC1 can be utilized as an affinity reagent to extensively detect CRISPR/Cas9 in various species because it binds to the conserved HNH domains of numerous Cas9 proteins.160 These investigations show that the use of Acr may enable the precise and efficient identification of CRISPR/Cas complexes.
Challenges and Future Prospects of Acr Applications
The field of Acrs is a recent discovery, and research on its diversity and prevalence has only recently begun. As a result, innovative applications of Acrs are still in their infancy and face numerous difficulties. In addition to Acrs, nucleic acid-base inhibitors and small-molecule inhibitors are other methods being explored to block Cas9 function.32,161 However, these two types of inhibitors have the potential to interact with other targets in mammalian cells, making it challenging to design them effectively for novel applications.32,162 In contrast, Acrs can be engineered to function as both an on-switch for Cas to facilitate the CRISPR/Cas system and an off-target switch, owing to their better affinity and specificity. Therefore, Acrs are far more effective than classical inhibitors.
The discovery of new Acrs and the elucidation of their mechanisms of action continue to pose major challenges. One of the crucial problems worth considering is the accurate and efficient distribution of Acrs to cells or tissues to limit the ability of the CRISPR/Cas system. To minimize off-target editing, the majority of earlier studies transfected Acr-encoding plasmids directly before CRISPR/Cas9 delivery. Alternatively, delivering Acr expression cassettes into CRISPR/Cas9-expressing cells via an adenovirus vector is a successful strategy. Many researchers in this field will continue to focus on developing new delivery mechanisms for Acrs, specifically their precise delivery into mammalian cells, even in living animals.
With the recent discovery of Acrs that counteract the functions of different types of CRISPR/Cas systems, it is anticipated that more effective Acrs will be found in the near future and employed to enhance gene editing-based treatment plans, synthetic biology, and CRISPR-based disease diagnosis.46 Furthermore, there are numerous instances of Acrs inhibitory functions in vitro, such as in bacteria. However, few studies have explained how this inhibition works in human cells or in vivo in animals. Further research is required to clarify the role of Acrs in vivo processes. Clarifying the unique and intricate processes would encourage the evolution of Acrs as regulators of genome editing and other uses.
Acrs function through a wide range of pathways, some of which offer new perspectives on their particular uses. Furthermore, continued discovery of various Acr mechanisms will contribute to our understanding of the competitive dynamics that exist in the ongoing evolutionary arms race between bacteriophages and their host bacteria. Even with the widespread use of CRISPR/Cas9 technology in research, off-target issues still exist and need to be considered because they carry significant risks. Acrs may function as switches to regulate the CRISPR/Cas immune system and reduce the likelihood of off-target consequences. This could result in widespread use of Acrs in genetic engineering.
Conclusion
The CRISPR/Cas system is a game changer in modern biotechnology as it has become a revolutionary tool owing to its exceptional ability to edit genes with high precision, versatility, and relative ease of use compared to other classical gene editing approaches. However, this gene-editing approach faces several challenges, such as off-target effects, sgRNA optimization, precise delivery methods, specific CRISPR/Cas variant selection, immune response, and ethical considerations. Overcoming these challenges is crucial for safe and effective translation of CRISPR/Cas technology in clinical applications. Continuous research and development efforts are underway to address these limitations and to refine the technology for broader use in biotechnology. Since their discovery in 2013, Acrs have represented a crucial aspect of the complex interplay between bacteria, viruses, and CRISPR/Cas system. The current classification system is constantly evolving, as new Acrs with diverse mechanisms of action have been discovered. Ongoing investigations of Acr systems should deepen our understanding of the roles of CRISPR/Cas and aid in the advancement of genome editing techniques. Machine learning approaches are being explored to analyze large datasets of Acr sequences and predict their functions and target specificities. As research continues to unveil their mechanisms, these proteins have immense potential for refining CRISPR/Cas technology, paving the way for advancements in gene editing and other biotechnological applications. The discovery and characterization of Acrs have also shed light on the race between phages and prokaryotes. Notably, research on the interactions between viruses and eukaryotes aids in the development of treatments for infections and human illnesses such as cancer and inflammatory diseases. Furthermore, advanced research is required to improve the creation of powerful and efficient CRISPR/Cas biotechnology tools as well as new techniques with potential therapeutic and medicinal uses.
Acknowledgments
The Researchers would like to thank the Deanship of Graduate Studies and Scientific Research at Qassim University for financial support (QU-APC-2024-9/1).
Disclosure
The authors declare no competing interests in this work.
References
- 1.Makarova KS, Wolf YI, Iranzo J, et al. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol. 2020;18(1):67–83. doi: 10.1038/s41579-019-0299-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitić D, Bolt EL, Ivančić-Baće I. CRISPR-Cas adaptation in Escherichia coli. Biosci Rep 2023;43(3):BSR20221198. doi: 10.1042/BSR20221198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mosterd C, Rousseau GM, Moineau S. A short overview of the CRISPR-Cas adaptation stage. Can J Microbiol. 2021;67(1):1–2. doi: 10.1139/cjm-2020-0212 [DOI] [PubMed] [Google Scholar]
- 4.Allemailem KS, Alsahli MA, Almatroudi A, et al. Innovative strategies of reprogramming immune system cells by targeting CRISPR/Cas9-based genome-editing tools: a new era of cancer management. Int J Nanomed. 2023;18(1):5531–5559. doi: 10.2147/IJN.S424872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song G, Tian C, Li J, et al. Rapid characterization of anti-CRISPR proteins and optogenetically engineered variants using a versatile plasmid interference system. Nucleic Acids Res. 2023;51(22):12381–12396. doi: 10.1093/nar/gkad995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jia N, Patel DJ. Structure-based functional mechanisms and biotechnology applications of anti-CRISPR proteins. Nat Rev Mol Cell Biol. 2021;22(8):563–579. doi: 10.1038/s41580-021-00371-9 [DOI] [PubMed] [Google Scholar]
- 7.H El-Ashry A. The CRISPR/Cas system: gene editing by bacterial defense. Novel Res Microbiol J. 2023;7(5):2101–2115. doi: 10.21608/nrmj.2023.317036 [DOI] [Google Scholar]
- 8.Allemailem KS, Alsahli MA, Almatroudi A, et al. Current updates of CRISPR/Cas9-mediated genome editing and targeting within tumor cells: an innovative strategy of cancer management. Cancer Commun. 2022;42(12):1257–1287. doi: 10.1002/cac2.12366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klompe SE, Sternberg SH. Harnessing “A billion years of experimentation”: the ongoing exploration and exploitation of CRISPR–cas immune systems. CRISPR J. 2018;1(2):141–158. doi: 10.1089/crispr.2018.0012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishino Y, Krupovic M, Forterre P. History of CRISPR–Cas from encounter with a mysterious repeated sequence to genome editing technology. J Bacteriol. 2018;200(20):e00580–17. doi: 10.1128/JB.00580-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allemailem KS, Almatroodi SA, Almatroudi A, et al. Recent advances in genome-editing technology with CRISPR/Cas9 variants and stimuli-responsive targeting approaches within tumor cells: a future perspective of cancer management. Int J Mol Sci. 2023;24(8):7052. doi: 10.3390/ijms24087052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allemailem KS. Recent advances in understanding the molecular mechanisms of multidrug resistance and novel approaches of CRISPR/Cas9-based genome-editing to combat this health emergency. Int J Nanomed. 2024;Volume 19:1125–1143. doi: 10.2147/IJN.S453566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Y, Li Z. CRISPR-Cas systems: overview, innovations and applications in human disease research and gene therapy. Computat Struct Biotechnol J. 2020;18:2401–2415. doi: 10.1016/j.csbj.2020.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu LF, Li YX, Wang JZ, Zhao YT, Wang Y. Controlling CRISPR‐Cas9 by guide RNA engineering. Wiley Interdiscip Rev RNA. 2023;14(1):e1731. doi: 10.1002/wrna.1731 [DOI] [PubMed] [Google Scholar]
- 15.Sasnauskas G, Siksnys V. CRISPR adaptation from a structural perspective. Curr Opin Struct Biol. 2020;65:17–25. doi: 10.1016/j.sbi.2020.05.015 [DOI] [PubMed] [Google Scholar]
- 16.Liu Y, Pinto F, Wan X, et al. Reprogrammed tracrRNAs enable repurposing of RNAs as crRNAs and sequence-specific RNA biosensors. Nat Commun. 2022;13(1):1937. doi: 10.1038/s41467-022-29604-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Behler J, Hess WR. Approaches to study CRISPR RNA biogenesis and the key players involved. Methods. 2020;172:12–26. doi: 10.1016/j.ymeth.2019.07.015 [DOI] [PubMed] [Google Scholar]
- 18.Jiang F, Doudna JA. CRISPR-Cas9 structures and mechanisms. Annu Rev Biochem. 2017;46:505–529. [DOI] [PubMed] [Google Scholar]
- 19.Ray A, Felice RD, Felice RD, Felice RD. Molecular simulations have boosted of CRISPR/Cas9: a review. J Self-Assembly Mol Electron. 2019;7(1):45–72. [Google Scholar]
- 20.Palermo G, Miao Y, Walker RC, Jinek M, McCammon JA. CRISPR-Cas9 conformational activation as elucidated from enhanced molecular simulations. Proc Natl Acad Sci U S A. 2017;114(28):7260–7265. doi: 10.1073/pnas.1707645114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhattacharya S, Satpati P. Insights into the mechanism of CRISPR/Cas9-based genome editing from molecular dynamics simulations. ACS omega. 2022;8(2):1817–1837. doi: 10.1021/acsomega.2c05583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang G, Li J. Review, analysis, and optimization of the CRISPR Streptococcus pyogenes Cas9 system. Med Drug Disc. 2021;9:100080. doi: 10.1016/j.medidd.2021.100080 [DOI] [Google Scholar]
- 23.Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513(7519):569–573. doi: 10.1038/nature13579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang F, Zhou K, Ma L, Gressel S, Doudna JA. A Cas9-Guide RNA complex preorganized for target DNA recognition. Science. 2015;348(6242):1477–1481. doi: 10.1126/science.aab1452 [DOI] [PubMed] [Google Scholar]
- 25.Yili FE, Sicheng LI, Ruodan CH, Anyong XI. Target binding and residence: a new determinant of DNA double-strand break repair pathway choice in CRISPR/Cas9 genome editing. J Zhejiang Univ Sci B. 2021;22(1):73. doi: 10.1631/jzus.B2000282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(621). doi: 10.1126/science.1258096 [DOI] [PubMed] [Google Scholar]
- 27.Yang H, Ren S, Yu S, et al. Methods favoring homology-directed repair choice in response to CRISPR/Cas9 induced-double strand breaks. Int J Mol Sci. 2020;21(18):6461. doi: 10.3390/ijms21186461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhagtaney L, Sundarrajan P. An overview of tools for genome editing: zFNs, mega nucleases, and TALENs. In: CRISPR/Cas-Mediated Genome Editing in Plants. 2023:37–64. [Google Scholar]
- 29.Hu JH, Davis KM, Liu DR. Chemical biology approaches to genome editing: understanding, controlling, and delivering programmable nucleases. Cell Chem Biol. 2016;23(1):57–73. doi: 10.1016/j.chembiol.2015.12.009 [DOI] [PubMed] [Google Scholar]
- 30.Zhang B. CRISPR/Cas gene therapy. J Cell Physiol. 2021;236(4):2459–2481. doi: 10.1002/jcp.30064 [DOI] [PubMed] [Google Scholar]
- 31.Choudhary N, Tandi D, Verma RK, et al. A comprehensive appraisal of mechanism of anti-CRISPR proteins: an advanced genome editor to amend the CRISPR gene editing. Front Plant Sci. 2023;14:1164461. doi: 10.3389/fpls.2023.1164461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maji B, Gangopadhyay SA, Lee M, et al. A high-throughput platform to identify small-molecule inhibitors of CRISPR-Cas9. Cell. 2019;177:1067–79.e19. doi: 10.1016/j.cell.2019.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stanley SY, Borges AL, Chen K-H, et al. Anti-CRISPR-associated proteins are crucial repressors of anti-CRISPR transcription. Cell. 2019;178(6):1452–64.e13. doi: 10.1016/j.cell.2019.07.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meaden S, Capria L, Alseth E, et al. Phage gene expression and host responses lead to infection-dependent costs of CRISPR immunity. ISME J. 2021;15(2):534–544. doi: 10.1038/s41396-020-00794-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Birkholz N, Fagerlund RD, Smith LM, Jackson SA, Fineran PC. The autoregulator Aca2 mediates anti-CRISPR repression. Nucleic Acids Res. 2019;47(18):9658–9665. doi: 10.1093/nar/gkz721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bondy-Denomy J, Davidson AR, Doudna JA, et al. A unified resource for tracking anti-CRISPR names. CRISPR J. 2018;1(5):304–305. doi: 10.1089/crispr.2018.0043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wen S, Zhao Y, Qi X, et al. Conformational plasticity of SpyCas9 induced by AcrIIA4 and AcrIIA2: insights from molecular dynamics simulation. Comput Struct Biotechnol J. 2024;23:537–548. doi: 10.1016/j.csbj.2023.12.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hwang S, Maxwell KL. Diverse mechanisms of CRISPR-Cas9 inhibition by type II anti-CRISPR proteins. J Mol Biol. 2023;435(7):168041. doi: 10.1016/j.jmb.2023.168041 [DOI] [PubMed] [Google Scholar]
- 39.Wiegand T, Karambelkar S, Bondy-Denomy J, et al. Structures and strategies of anti-CRISPR-mediated immune suppression. Annu Rev Microbiol. 2020;74(1):21–37. doi: 10.1146/annurev-micro-020518-120107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jumper J, Evans R, Pritzel A, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583–589. doi: 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sahakyan H, Makarova KS, Koonin EV. Search for origins of anti-CRISPR proteins by structure comparison. CRISPR J. 2023;6(3):222–231. doi: 10.1089/crispr.2023.0011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y, Bondy-Denomy J. Anti-CRISPRs go viral: the infection biology of CRISPR-Cas inhibitors. Cell Host Microbe. 2021;29(5):704–714. doi: 10.1016/j.chom.2020.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bondy-Denomy J, Pawluk A, Maxwell KL, Davidson AR. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature. 2013;493(7432):429–432. doi: 10.1038/nature11723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hynes AP, Rousseau GM, Agudelo D, et al. Widespread anti-CRISPR proteins in virulent bacteriophages inhibit a range of Cas9 proteins. Nat Commun. 2018;9(1):2919. doi: 10.1038/s41467-018-05092-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pawluk A, Shah M, Mejdani M, et al. Disabling a type I-E CRISPR-Cas nuclease with a bacteriophage-encoded anti-CRISPR protein. MBio. 2017;8(6):e01751–17. doi: 10.1128/mBio.01751-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watters KE, Fellmann C, Bai HB, Ren SM, Doudna JA. Systematic discovery of natural CRISPR-Cas12a inhibitors. Science. 2018;362(6411):236–239. doi: 10.1126/science.aau5138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Watters KE, Shivram H, Fellmann C, et al. Potent CRISPR-Cas9 inhibitors from staphylococcus genomes. Proc Natl Acad Sci U S A. 2020;117(12):6531–6539. doi: 10.1073/pnas.1917668117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marino ND, Zhang JY, Borges AL, et al. Discovery of widespread type I and type V CRISPR-Cas inhibitors. Science. 2018;362(6411):240–242. doi: 10.1126/science.aau5174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Osuna BA, Karambelkar S, Mahendra C, et al. Listeria phages induce Cas9 degradation to protect lysogenic genomes. Cell Host Microbe. 2020;28(1):31–40.e40. doi: 10.1016/j.chom.2020.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Uribe RV, van der Helm E, Misiakou M-A, et al. Discovery and Characterization of Cas9 Inhibitors Disseminated across Seven Bacterial Phyla. Cell Host Microbe. 2019;25(2):233–41.e241. doi: 10.1016/j.chom.2019.01.003 [DOI] [PubMed] [Google Scholar]
- 51.Forsberg KJ, Bhatt DV, Schmidtke DT, et al. Functional metagenomics-guided discovery of potent Cas9 inhibitors in the human microbiome. Elife. 2019;8:e46540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bowen Y, Zhang J, Yin Y. AcaFinder: genome mining for anti- CRISPR-Associated genes. mSystems. 2022;7(6):e00817–22. doi: 10.1128/msystems.00817-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dong C, Hao G-F, Hua H-L, et al. Anti-CRISPRdb: a comprehensive online resource for anti-CRISPR proteins. Nucleic Acids Res. 2018;46(D1):D393–9. doi: 10.1093/nar/gkx835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yi H, Huang L, Yang B, et al. AcrFinder: genome mining anti-CRISPR operons in prokaryotes and their viruses. Nucleic Acids Res. 2020;48(W1):W358–63. doi: 10.1093/nar/gkaa351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eitzinger S, Asif A, Watters KE, et al. Machine learning predicts new anti-CRISPR proteins. Nucleic Acids Res. 2020;48(9):W4698–703. doi: 10.1093/nar/gkaa219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang J, Zhao J, Xu S, et al. Species-specific deamidation of cGAS by herpes simplex virus UL37 protein facilitates viral replication. Cell Host Microbe. 2018;24(2):234–48.e235. doi: 10.1016/j.chom.2018.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gussow AB, Park AE, Borges AL, et al. Machine-learning approach expands the repertoire of anti-CRISPR protein families. Nat Commun. 2020;11(1):3784. doi: 10.1038/s41467-020-17652-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang J, Dai W, Li J, et al. PaCRISPR: a server for predicting and visualizing anti-CRISPR proteins. Nucleic Acids Res. 2020;48(W1):W348–W356. doi: 10.1093/nar/gkaa432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang J, Dai W, Li J, et al. AcrHub: an integrative hub for investigating, predicting and mapping anti-CRISPR proteins. Nucleic Acids Res. 2021;49(D1):D630–D638. doi: 10.1093/nar/gkaa951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leon LM, Park AE, Borges AL, Zhang JY, Bondy-Denomy J. Mobile element warfare via CRISPR and anti-CRISPR in pseudomonas aeruginosa. Nucleic Acids Res. 2021;49(4):2114–2125. doi: 10.1093/nar/gkab006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang L, Zhang Y, Yin P, Feng Y. Structural insights into the inactivation of the type I-F CRISPR-Cas system by anti-CRISPR proteins. RNA Biology. 2021;18(sup2):562–573. doi: 10.1080/15476286.2021.1985347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Prakash A, Kumar M. Transcriptional analysis of CRISPR I-B arrays of Leptospira interrogans serovar lai and its processing by Cas6. Front Microbiol. 2022;13:960559. doi: 10.3389/fmicb.2022.960559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.O’Brien RE, Santos IC, Wrapp D, et al. Structural basis for assembly of non-canonical small subunits into type I-C Cascade. Nat Commun. 2020;11(1):5931. doi: 10.1038/s41467-020-19785-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schwartz EA, Bravo JP, Macias LA, et al. Assembly of multi-subunit fusion proteins into the RNA-targeting type III-D CRISPR-Cas effector complex. bioRxiv. 2022;14:2022–2026. [Google Scholar]
- 65.Wang J, Teng Y, Gong X, et al. Exploring and engineering PAM-diverse Streptococci Cas9 for PAM-directed bifunctional and titratable gene control in bacteria. Metabolic Engineering. 2023;75:68–77. doi: 10.1016/j.ymben.2022.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Teng Y, Wang J, Jiang T, Zou Y, Yan Y. Engineering a streptococcus Cas9 ortholog with an RxQ PAM-binding motif for PAM-free gene control in bacteria. ACS Synth Biol. 2023;12(9):2764–2772. doi: 10.1021/acssynbio.3c00366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tuminauskaite D, Norkunaite D, Fiodorovaite M, et al. DNA interference is controlled by R-loop length in a type I-F1 CRISPR-Cas system. BMC Biol. 2020;18(1):1–6. doi: 10.1186/s12915-020-00799-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shibata T, Iwasaki W, Hirota K. The intrinsic ability of double-stranded DNA to carry out D-loop and R-loop formation. Comput Struct Biotechnol J. 2020;18:3350–3360. doi: 10.1016/j.csbj.2020.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hu C, Ni D, Nam KH, et al. Allosteric control of type I-A CRISPR-Cas3 complexes and establishment as effective nucleic acid detection and human genome editing tools. Molecular Cell. 2022;82(15):2754–2768. doi: 10.1016/j.molcel.2022.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Trost CN, Yang J, Garcia B, et al. An anti-CRISPR that pulls apart a CRISPR–Cas complex. Nature. 2024;3:1–8. [DOI] [PubMed] [Google Scholar]
- 71.Chowdhury S, Carter J, Rollins MF, Golden SM, Jackson RN. Structure reveals mechanisms of viral suppressors that intercept a CRISPR RNA-guided surveillance complex. Cell. 2017;169(1):47–57.e11. doi: 10.1016/j.cell.2017.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peng R, Xu Y, Zhu T, Li N, Qi J. Alternate binding modes of anti-CRISPR viral suppressors AcrF1/2 to Csy surveillance complex revealed by cryo-EM structures. Cell Res. 2017;27(7):853–864. doi: 10.1038/cr.2017.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guo TW, Bartesaghi A, Yang H, Falconieri V, Rao P. Cryo-EM structures reveal mechanism and inhibition of DNA targeting by a CRISPR-Cas surveillance complex. Cell. 2017;171(2):414–426.e412. doi: 10.1016/j.cell.2017.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gabel C, Li Z, Zhang H, Chang L. Structural basis for inhibition of the type I-F CRISPR–Cas surveillance complex by AcrIF4, AcrIF7 and AcrIF14. Nucleic Acids Res. 2021;49(1):584–594. doi: 10.1093/nar/gkaa1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang K, Wang S, Li S, et al. Inhibition mechanisms of AcrF9, AcrF8, and AcrF6 against type I-F CRISPR–Cas complex revealed by cryo-EM. Proc Natl Acad Sci U S A. 2020;117(13):7176–7182. doi: 10.1073/pnas.1922638117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hirschi M, Lu W-T, Santiago-Frangos A, et al. AcrIF9 tethers non-sequence specific dsDNA to the CRISPR RNA-guided surveillance complex. Nat Commun. 2020;11(1):2730. doi: 10.1038/s41467-020-16512-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim I, Koo J, An SY, et al. Structural and mechanistic insights into the CRISPR inhibition of AcrIF7. Nucleic Acids Res. 2020;48(17):9959–9968. doi: 10.1093/nar/gkaa690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.O’Brien RE, Bravo JPK, Ramos D, Hibshman GN, Wright JT, Taylor DW. Structural snapshots of R-loop formation by a type I-C CRISPR Cascade. Molecular Cell. 2023;83(5):746–758. doi: 10.1016/j.molcel.2023.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hu C, Myers MT, Zhou X, et al. Exploiting activation and inactivation mechanisms in type I-C CRISPR-Cas3 for genome-editing applications. Molecular Cell. 2024;84(3):463–475. doi: 10.1016/j.molcel.2023.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Niu Y, Yang L, Gao T, et al. A type I-F Anti-CRISPR protein inhibits the CRISPR-Cas surveillance complex by ADP-ribosylation. Mol Cell. 2020;80(3):512–524.e515. doi: 10.1016/j.molcel.2020.09.015 [DOI] [PubMed] [Google Scholar]
- 81.Kim GE, Lee SY, Park HH. A high-resolution (1.2 Å) crystal structure of the anti-CRISPR protein AcrIF9. FEBS Open Bio. 2020;10(12):2532–2540. doi: 10.1002/2211-5463.12986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xie Y, Zhang L, Gao Z, et al. AcrIF5 specifically targets DNA-bound CRISPR-Cas surveillance complex for inhibition. Nat Chem Biol. 2022;18(6):670–677. doi: 10.1038/s41589-022-00995-8 [DOI] [PubMed] [Google Scholar]
- 83.McGinn J, Marraffini LA. Molecular mechanisms of CRISPR–Cas spacer acquisition. Nat Rev Microbiol. 2019;17(1):7–12. doi: 10.1038/s41579-018-0071-7 [DOI] [PubMed] [Google Scholar]
- 84.Sofos N, Feng M, Stella S, et al. Structures of the Cmr-β complex reveal the regulation of the immunity mechanism of type III-B CRISPR-Cas. Mol Cell. 2020;79(5):741–757.e747. doi: 10.1016/j.molcel.2020.07.008 [DOI] [PubMed] [Google Scholar]
- 85.Bhatia S, Yadav P, Yadav SK. CRISPR-Cas for genome editing: classification, mechanism, designing and applications. Int J Biol Macromol. 2023;238:124054. doi: 10.1016/j.ijbiomac.2023.124054 [DOI] [PubMed] [Google Scholar]
- 86.Burmistrz M, Krakowski K, Krawczyk-Balska A. RNA-targeting CRISPR–Cas systems and their applications. Int J Mol Sci. 2020;21(3):1122. doi: 10.3390/ijms21031122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Molina R, Sofos N, Montoya G. Structural basis of CRISPR-Cas Type III prokaryotic defence systems. Curr Opin Struct Biol 2020;65:119–129. doi: 10.1016/j.sbi.2020.06.010 [DOI] [PubMed] [Google Scholar]
- 88.Kazlauskiene M, Tamulaitis G, Kostiuk G, Venclovas Č, Siksnys V. Spatiotemporal control of type III-A CRISPR-Cas immunity: coupling DNA degradation with the target RNA recognition. Mol Cell. 2016;62(2):295–306. doi: 10.1016/j.molcel.2016.03.024 [DOI] [PubMed] [Google Scholar]
- 89.Kazlauskiene M, Kostiuk G, Venclovas Č, Tamulaitis G, Siksnys V. A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science. 2017;357(6351):605–609. doi: 10.1126/science.aao0100 [DOI] [PubMed] [Google Scholar]
- 90.Niewoehner O, Garcia-Doval C, Rostøl JT, et al. Type III CRISPR–Cas systems produce cyclic oligoadenylate second messengers. Nature. 2017;548(7669):543–548. doi: 10.1038/nature23467 [DOI] [PubMed] [Google Scholar]
- 91.Li X, Han J, Yang J, Zhang H. The structural biology of type III CRISPR-Cas systems. J Struct Biol. 2024;216(1):108070. doi: 10.1016/j.jsb.2024.108070 [DOI] [PubMed] [Google Scholar]
- 92.Rouillon C, Athukoralage JS, Graham S, Gruschow S, White MF. Control of cyclic oligoadenylate synthesis in a type III CRISPR system. eLife. 2018;7:e36734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jia N, Jones R, Sukenick G, Patel DJ. Second messenger cA4 formation within the composite Csm1 palm pocket of type III-A CRISPR-Cas Csm complex and its release path. Mol Cell. 2019;75(5):933–43.e936. doi: 10.1016/j.molcel.2019.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Foster K, Kalter J, Woodside W, Terns RM, Terns MP. The ribonuclease activity of Csm6 is required for anti-plasmid immunity by Type III-A CRISPR-Cas systems. RNA Biol. 2019;16(4):449–460. doi: 10.1080/15476286.2018.1493334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jia N, Jones R, Yang G, Ouerfelli O, Patel DJ. CRISPR-Cas III-A Csm6 CARF domain is a ring nuclease triggering stepwise cA4 cleavage with ApA>p formation terminating RNase activity. Mol Cell. 2019;75(5):944–56.e946. doi: 10.1016/j.molcel.2019.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Molina R, Stella S, Feng M, et al. Structure of Csx1-cOA4 complex reveals the basis of RNA decay in type III-B CRISPR-Cas. Nat Commun. 2019;10(1):4302. doi: 10.1038/s41467-019-12244-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Garcia-Doval C, Schwede F, Berk C, et al. Activation and self-inactivation mechanisms of the cyclic oligoadenylate-dependent CRISPR ribonuclease Csm6. Nat Commun. 2020;11(1):1596. doi: 10.1038/s41467-020-15334-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Smalakyte D, Kazlauskiene M, F. Havelund J, et al. Type III-A CRISPR-associated protein Csm6 degrades cyclic hexa-adenylate activator using both CARF and HEPN domains. Nucleic Acids Res. 2020;48(16):9204–9217. doi: 10.1093/nar/gkaa634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bhoobalan-Chitty Y, Johansen TB, Di Cianni N, Peng X. Inhibition of type III CRISPR-Cas immunity by an archaeal virus-encoded anti-CRISPR protein. Cell. 2019;179:448–58.e411. doi: 10.1016/j.cell.2019.09.003 [DOI] [PubMed] [Google Scholar]
- 100.Athukoralage JS, McMahon SA, Zhang C, et al. An anti-CRISPR viral ring nuclease subverts type III CRISPR immunity. Nature. 2020;577:572–575. doi: 10.1038/s41586-019-1909-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zaynab M, Sharif Y, Fatima M, et al. CRISPR/Cas9 to generate plant immunity against pathogen. Microb Pathogenesis. 2020;141:103996. doi: 10.1016/j.micpath.2020.103996 [DOI] [PubMed] [Google Scholar]
- 102.Zhu Y, Gao A, Zhan Q, et al. Diverse mechanisms of CRISPR-Cas9 inhibition by type IIC anti-CRISPR proteins. Mol Cell. 2019;74:296–309.e297. doi: 10.1016/j.molcel.2019.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Thavalingam A, Cheng Z, Garcia B, et al. Inhibition of CRISPR-Cas9 ribonucleoprotein complex assembly by anti-CRISPR AcrIIC2. Nat Commun. 2019;10:2806. doi: 10.1038/s41467-019-10577-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Osuna BA, Karambelkar S, Mahendra C, et al. Critical anti-CRISPR locus repression by a bi-functional Cas9 inhibitor. Cell Host Microbe. 2020;28:23–30. doi: 10.1016/j.chom.2020.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shehreen S, Birkholz N, Fineran PC, Brown CM. Widespread repression of anti-CRISPR production by anti-CRISPR-associated proteins. Nucleic Acids Res. 2022;50(15):8615–8625. doi: 10.1093/nar/gkac674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Harrington LB, Doxzen KW, Ma E, et al. A broad-spectrum inhibitor of CRISPR-Cas9. Cell. 2017;170(5):1224–1233.e1215. doi: 10.1016/j.cell.2017.07.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mahendra C, Christie KA, Osuna BA, et al. Broad-spectrum anti-CRISPR proteins facilitate horizontal gene transfer. Nat Microbiol. 2020;5(5):620–629. doi: 10.1038/s41564-020-0692-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jiang F, Liu -J-J, Osuna BA, et al. Temperature-responsive competitive inhibition of CRISPR-Cas9. Mol Cell. 2019;73(3):601–610.e605. doi: 10.1016/j.molcel.2018.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liu L, Yin M, Wang M, Wang Y. Phage AcrIIA2 DNA mimicry: structural basis of the CRISPR and anti-CRISPR arms race. Mol Cell. 2019;73(3):611–620.e613. doi: 10.1016/j.molcel.2018.11.011 [DOI] [PubMed] [Google Scholar]
- 110.Shin J, Jiang F, Liu -J-J, et al. Disabling Cas9 by an anti-CRISPR DNA mimic. Sci Adv. 2017;3(1):e1701620. doi: 10.1126/sciadv.1701620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kim I, Jeong M, Ka D, et al. Solution structure and dynamics of anti-CRISPR AcrIIA4, the Cas9 inhibitor. Sci Rep. 2018;8(1):3883. doi: 10.1038/s41598-018-22177-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dong Y, Guo M, Wang S, et al. Structural basis of CRISPR-SpyCas9 inhibition by an anti-CRISPR protein. Nature. 2017;546(7657):436–439. doi: 10.1038/nature22377 [DOI] [PubMed] [Google Scholar]
- 113.Jiang YH, Liu YF, Wang K, et al. Fine-tuning Cas9 activity with a cognate inhibitor AcrIIA4 to improve genome editing in Streptomyces. ACS Synth Biol. 2021;10(11):2833–2841. doi: 10.1021/acssynbio.1c00141 [DOI] [PubMed] [Google Scholar]
- 114.Fuchsbauer O, Swuec P, Zimberger C, et al. Cas9 allosteric inhibition by the anti-CRISPR protein AcrIIA6. Mol Cell. 2019;76(4):922–937.e927. doi: 10.1016/j.molcel.2019.09.012 [DOI] [PubMed] [Google Scholar]
- 115.Lee J, Mir A, Edraki A, et al. Potent Cas9 inhibition in bacterial and human cells by AcrIIC4 and AcrIIC5 anti-CRISPR proteins. mBio. 2018;9(4):e02321–18. doi: 10.1128/mBio.02321-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhu Y, Yin S, Li Z. Mechanism of inhibition of CRISPR-Cas9 by anti-CRISPR protein AcrIIC1. Biochem Biophys Res Commun. 2023;654:34–39. doi: 10.1016/j.bbrc.2023.02.065 [DOI] [PubMed] [Google Scholar]
- 117.Yan WX, Hunnewell P, Alfonse LE, et al. Functionally diverse type V CRISPR-Cas systems. Science. 2019;363(6422):88–91. doi: 10.1126/science.aav7271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wörle E, Newman A, D’Silva J, Burgio G, Grohmann D. Allosteric activation of CRISPR-Cas12a requires the concerted movement of the bridge helix and helix 1 of the RuvC II domain. Nucleic Acids Res. 2022;50(17):10153–10168. doi: 10.1093/nar/gkac767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Swarts DC, Jinek M. Mechanistic Insights into the cis- and trans-acting DNase activities of Cas12a. Mol Cell. 2019;73(3):589–600.e584. doi: 10.1016/j.molcel.2018.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Garcia-Doval C, Jinek M. Molecular architectures and mechanisms of Class 2 CRISPR-associated nucleases. Curr Opin Struct Biol. 2017;47:157–166. doi: 10.1016/j.sbi.2017.10.015 [DOI] [PubMed] [Google Scholar]
- 121.Smith CW, Kachwala MJ, Nandu N, Yigit MV. Recognition of DNA target formulations by CRISPR-Cas12a using a dsDNA reporter. ACS Synth Biol. 2021;10(7):1785–1791. doi: 10.1021/acssynbio.1c00204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang H, Li Z, Daczkowski CM, et al. Structural basis for the inhibition of CRISPR-Cas12a by anti-CRISPR proteins. Cell Host Microbe. 2019;25:815–826.e814. doi: 10.1016/j.chom.2019.05.004 [DOI] [PubMed] [Google Scholar]
- 123.Knott GJ, Thornton BW, Lobba MJ, et al. Broad-spectrum enzymatic inhibition of CRISPR-Cas12a. Nat Struct Mol Biol. 2019;26:315–321. doi: 10.1038/s41594-019-0208-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Peng R, Li Z, Xu Y, et al. Structural insight into multistage inhibition of CRISPR-Cas12a by AcrVA4. Proc Natl Acad Sci USA. 2019;116:18928–18936. doi: 10.1073/pnas.1909400116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Perčulija V, Lin J, Zhang B, Ouyang S. Functional features and current applications of the RNA‐targeting type VI CRISPR‐Cas systems. Adv Sci. 2021;8(13):2004685. doi: 10.1002/advs.202004685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Liu L, Li X, Ma J, et al. The molecular architecture for RNA-guided RNA cleavage by Cas13a. Cell. 2017;170:714–726.e710. doi: 10.1016/j.cell.2017.06.050 [DOI] [PubMed] [Google Scholar]
- 127.Meeske AJ, Nakandakari-Higa S, Marraffini LA. Cas13-induced cellular dormancy prevents the rise of CRISPR-resistant bacteriophage. Nature. 2019;570:241–245. doi: 10.1038/s41586-019-1257-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Meeske AJ, Jia N, Cassel AK, et al. A phage-encoded anti-CRISPR enables complete evasion of type VI-A CRISPR-Cas immunity. Science. 2020;369:54–59. doi: 10.1126/science.abb6151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lin P, Qin S, Pu Q, et al. CRISPR-Cas13 inhibitors block RNA editing in bacteria and mammalian cells. Molecular Cell. 2020;78:850–861.e857. doi: 10.1016/j.molcel.2020.03.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Camara-Wilpert S, Mayo-Muñoz D, Russel J, et al. Bacteriophages suppress CRISPR–Cas immunity using RNA-based anti-CRISPRs. Nature. 2023;623(7987):601–607. doi: 10.1038/s41586-023-06612-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Faure G, Shmakov SA, Yan WX, et al. CRISPR–Cas in mobile genetic elements: counter-defence and beyond. Nat Rev Microbiol. 2019;17(8):513–525. doi: 10.1038/s41579-019-0204-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Pinilla-Redondo R, Shehreen S, Marino ND, et al. Discovery of multiple anti-CRISPRs highlights anti-defense gene clustering in mobile genetic elements. Nat Commun. 2020;11(1):5652. doi: 10.1038/s41467-020-19415-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Marino ND, Pinilla-Redondo R, Csörgő B, Bondy-Denomy J. Anti-CRISPR protein applications: natural brakes for CRISPR-Cas technologies. Nat Methods. 2020;17(5):471–479. doi: 10.1038/s41592-020-0771-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Nojadeh JN, Eryilmaz NS, Ergüder Bİ. CRISPR/Cas9 genome editing for neurodegenerative diseases. EXCLI J. 2023;22:567. doi: 10.17179/excli2023-6155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Qi LS, Larson M, Gilbert L, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Adli M. The CRISPR tool kit for genome editing and beyond. Nat Commun. 2018;9:1911. doi: 10.1038/s41467-018-04252-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Qiao H, Wu J, Zhang X, Luo J, Wang H, Ming D. The advance of CRISPR-Cas9-based and NIR/CRISPR-Cas9-based imaging system. Front Chem. 2021;9:786354. doi: 10.3389/fchem.2021.786354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhang X, Bai XC, Chen ZJ. Structures and mechanisms in the cGAS-STING innate immunity pathway. Immunity. 2020;53(1):43–53. doi: 10.1016/j.immuni.2020.05.013 [DOI] [PubMed] [Google Scholar]
- 140.Hoffmann MD, Aschenbrenner S, Grosse S, et al. Cell-specific CRISPR-Cas9 activation by microRNA-dependent expression of anti-CRISPR proteins. Nucleic Acids Res. 2019;47(14):e75. doi: 10.1093/nar/gkz271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lee J, Mou H, Ibraheim R, et al. Tissue-restricted genome editing in vivo specified by microRNA-repressible anti-CRISPR proteins. RNA. 2019;25(11):1421–1431. doi: 10.1261/rna.071704.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Aschenbrenner S, Kallenberger SM, Hoffmann MD, et al. Coupling Cas9 to artificial inhibitory domains enhances CRISPR-Cas9 target specificity. Sci Adv. 2020;6(24):eaay0187. doi: 10.1126/sciadv.aay0187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhao Y, Tian J, Zheng G, et al. Multiplex genome editing using a dCas9-cytidine deaminase fusion in Streptomyces. Sci China Life Sci. 2020;63:1053–1062. doi: 10.1007/s11427-019-1559-y [DOI] [PubMed] [Google Scholar]
- 144.Zuo E, Sun Y, Wei W, et al. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science. 2019;364(6437):289–292. doi: 10.1126/science.aav9973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jin S, Zong Y, Gao Q, et al. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science. 2019;364(6437):292–295. doi: 10.1126/science.aaw7166 [DOI] [PubMed] [Google Scholar]
- 146.Liang M, Sui T, Liu Z, et al. AcrIIA5 suppresses base editors and reduces their off-target effects. Cells. 2020;9(7):1786. doi: 10.3390/cells9081786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ishizaki T, Hernandez S, Paoletta MS, Sanderson T, Bushell ES. CRISPR/Cas9 and genetic screens in malaria parasites: small genomes, big impact. Biochem Soc Trans. 2022;50(3):1069–1079. doi: 10.1042/BST20210281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kyrou K, Hammond AM, Galizi R, et al. A CRISPR-Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes. Nat Biotechnol. 2018;36(10):1062–1066. doi: 10.1038/nbt.4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Champer J, Chung J, YL Lee, et al. Molecular safeguarding of CRISPR gene drive experiments. eLife. 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Basgall EM, Goetting SC, Goeckel ME, et al. Gene drive inhibition by the anti-CRISPR proteins AcrIIA2 and AcrIIA4 in Saccharomyces cerevisiae. Microbiology. 2018;164(3):464–474. doi: 10.1099/mic.0.000635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Li C, Psatha N, Gil S, et al. HDAd5/35(++) adenovirus vector expressing anti-CRISPR peptides decreases CRISPR/Cas9 toxicity in human hematopoietic stem cells. Mol Ther Methods Clin Dev. 2018;9:390–401. doi: 10.1016/j.omtm.2018.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Palmer DJ, Turner DL, Ng P. Production of CRISPR/Cas9-mediated self-cleaving helper-dependent adenoviruses. Mol Ther Methods Clin Dev. 2019;13:432–439. doi: 10.1016/j.omtm.2019.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Li J, Xu Z, Chupalov A, Marchisio MA. Anti-CRISPR-based biosensors in the yeast S. cerevisiae. J Biol Eng. 2018;12:11. doi: 10.1186/s13036-018-0101-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Nakamura M, Srinivasan P, Chavez M, et al. Anti-CRISPR-mediated control of gene editing and synthetic circuits in eukaryotic cells. Nat Commun. 2019;10:194. doi: 10.1038/s41467-018-08158-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Luo ML, Mullis AS, Leenay RT, Beisel CL. Repurposing endogenous type I CRISPR-Cas systems for programmable gene repression. Nucleic Acids Res. 2015;43(2):674–681. doi: 10.1093/nar/gku971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kim Y, Lee SJ, Yoon H-J, et al. Anti-CRISPR AcrIIC3 discriminates between Cas9 orthologs via targeting the variable surface of the HNH nuclease domain. FEBS J. 2019;286(18):4661–4674. doi: 10.1111/febs.15037 [DOI] [PubMed] [Google Scholar]
- 157.Bubeck F, Hoffmann MD, Harteveld Z, et al. Engineered anti-CRISPR proteins for optogenetic control of CRISPR-Cas9. Nat Methods. 2018;15(11):924–927. doi: 10.1038/s41592-018-0178-9 [DOI] [PubMed] [Google Scholar]
- 158.Ssekatawa K, Byarugaba DK, Kato CD, et al. A review of phage mediated antibacterial applications. Alexandria J Med. 2021;57(1):1–20. doi: 10.1080/20905068.2020.1851441 [DOI] [Google Scholar]
- 159.Johnston RK, Seamon KJ, Saada EA, et al. Use of anti-CRISPR protein AcrIIA4 as a capture ligand for CRISPR/Cas9 detection. Biosens Bioelectron. 2019;141:111361. doi: 10.1016/j.bios.2019.111361 [DOI] [PubMed] [Google Scholar]
- 160.Phaneuf CR, Seamon KJ, Eckles TP, et al. Ultrasensitive multi-species detection of CRISPR-Cas9 by a portable centrifugal microfluidic platform. Analyst. 2019;11(3):559–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Barkau CL, O’Reilly D, Rohilla KJ, Damha MJ, Gagnon KT. Rationally designed anti-CRISPR nucleic acid inhibitors of CRISPR-Cas9. Nucleic Acid Therapeutics. 2019;29(2):136–147. doi: 10.1089/nat.2018.0758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Schneider G. Automating drug discovery. Nat Rev Drug Discov. 2018;17(2):97–113. doi: 10.1038/nrd.2017.232 [DOI] [PubMed] [Google Scholar]