Abstract
A Moloney murine leukemia virus-based single-replication-cycle assay was developed to study the effects of limiting the extent of template and primer strand complementarity on recombinogenic template switching. This system mimicked forced copy choice recombination in which nascent DNA transfers from the end of a donor template to an acceptor position on the other copackaged RNA. When acceptor target regions with different extents of complementarity to the transferring DNA were tested, efficient recombination occurred with as few as 14 complementary nucleotides. The frequencies of correct targeting, transfer-associated errors, mismatch extension, and transfer before reaching the end of the donor template were determined. All four molecular events occurred, with their proportions varying depending on the nature of acceptor/transferring DNA complementarity. When complementarity was severely limited, recombination was inefficient and most products resulted from aberrant second-strand transfer rather than from forced template switching between RNAs. Other classes of reverse transcription products, including some that resulted from template switching between virus and host sequences, were also observed when homology between the acceptor and donor was limited.
Retroviruses copackage two complete RNA genomes, and template switching between these can generate recombinant viral DNAs. During the synthesis of retroviral DNA, reverse transcriptase (RT) must perform two replicative template switches: the first and second strong stop template switches (14). It has been postulated that the requirement for these two obligatory template switches selected for RT's ability to perform non-required recombinogenic template switches (6, 48).
During recombinogenic template switching. RT begins DNA synthesis on one viral RNA and then switches to the copackaged RNA (intermolecular template switching). RT can also perform template switches between two positions on a single RNA (intramolecular template switching). Template switching between regions with high sequence similarity is far more frequent than between nonhomologous sequences (17, 57). Nonhomologous recombination does occur at low frequencies, such as in the transduction of cellular oncogenes (46).
Retroviral recombination has been measured in several ways (22, 24, 27, 28). In some assays, homologous recombination is monitored using pairs of retroviral vectors engineered so that only recombinants confer dual drug resistance (17). Other assays use a single marker that is reconstituted only when recombination occurs (58). Direct-repeat deletion has also been used to study template switching (9, 10, 19, 33, 38, 42). However, because template switching can occur over long stretches of sequence in each of these assays, none can be used to study the effects of such factors as a mismatched primer terminus or short regions of primer-terminal complementarity at the switch site.
Although homologous recombination can occur during either plus- or minus-strand synthesis, minus-strand recombination is probably more frequent (45, 59). Whether minus-strand recombination is directed more by complementarity at the primer terminus (6) or by complementarity at primer-internal regions (5, 10, 35) is unclear. One model for minus-strand recombination proposes that recombination occurs during minus-strand DNA synthesis when RT encounters a break in the template (6). Although this model has been modified to include minus-strand recombination that occurs whether or not the RNA is broken (3, 12, 15, 52), the original “forced copy choice” model provides a conceptual framework for studying recombination that occurs from a single template location.
Here we report a system to determine which regions of homology contribute to acceptor site selection during Moloney murine leukemia virus (MLV) minus-strand recombinogenic template switching. Recent studies have used minus-strand transfer to study homology requirements during replicative template switching (4, 8). We chose a system not involving strong-stop switching because there may be separate signals (in addition to donor-acceptor homology) that direct template switching to retroviral R repeats (23, 49). In the system reported here, the recombinogenic template switch was forced to occur at or near a defined position during minus-strand synthesis. The acceptor region was modified to test if the reverse transcription complex was attracted to an acceptor site more via complementarity to the terminus or to the internal positions of the nascent DNA, and how much complementarity was needed.
MATERIALS AND METHODS
Plasmid construction.
Ψ+ PURO (also called pAM 86-5, a pBabePuro derivative [21]) and pMLV Ψ− (38) plasmids have been described previously. The donor and acceptor plasmids were each constructed using several subcloning steps. The donor plasmid contained the Rous sarcoma virus (RSV) promoter from pREP8 (Invitrogen), the puromycin resistance gene to the long terminal repeat 3′ (LTR) region from pBabePuro (26) [except that the poly(A) signal in the R region was inactivated (49)], Ψ from the infectious provirus plasmid pNCA (7), and the simian virus 40 polyadenylation signal from pREP8. The initial acceptor plasmid (520 Terminal Match) contained the upstream LTR and Ψ region from pNCA, the RSV promoter from pREP8, the 5′ two-thirds of the puromycin resistance gene from pBabePuro, and the simian virus 40 polyadenylation signal from pREP8. An acceptor derivative (59 Terminal Match) was made by completely deleting the puromycin resistance gene of 520 Terminal Match such that only 59 nucleotides nt of homology remained between the donor and acceptor in the initial transcribed region (ITR) of the RSV promoter. Mutant acceptor constructs (1 Internal Mismatch; Complete Mismatch; 5 and 14 Terminal Match; 1, 3, 5, and 10 Terminal Mismatch) were made by introducing mutations into the ITR using PCR and were confirmed by sequencing entire PCR-generated inserts. The RNA probe template plasmid contained portions of the RSV promoter from pREP8, U3 from pNCA, and a short linker fragment from LITMUS29 (New England Biolabs) cloned into pBlueScript-SK(+) (Stratagene). The recovery marker plasmid contained the same LITMUS29 linker fragment cloned into pBlueScript-SK(+). Construction details for all plasmids are available on request.
Cells and viruses.
NIH 3T3 cells were grown in Dulbecco modified Eagle medium supplemented with 10% calf serum (Gibco). 293T and derivative cell lines were grown in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum (HyClone). Puromycin-resistant NIH 3T3 cells were selected in 6 μg of puromycin (Sigma) per ml. All transfections were performed using the calcium phosphate method unless otherwise indicated (38), and all infections were performed in the presence of 0.8 μg of hexadimethrine bromide (Polybrene) (Sigma) per ml (38).
To make cells that stably expressed donor RNA, 293T cells were transfected with the donor plasmid using Lipofectamine (Gibco). Unpassaged transfected cells were selected in 1 μg of puromycin per ml, and well-separated colonies were cloned. Clonal lines were cotransfected with pMLV Ψ− and 520 Terminal Match, and virus was used to infect the NIH 3T3 cells. The donor-expressing cells that gave the highest puromycin-resistant CFU titer in NIH 3T3 cells were designated the 293T:DONOR cell line.
To generate integrated viral DNA for molecular analysis, the 293T:DONOR cell line was transiently transfected with pMLV Ψ− and acceptor plasmid and virus was harvested 48 h posttransfection. NIH 3T3 cells were infected and puromycin selected. At least 50 colonies were pooled for each acceptor, and genomic DNA was isolated from these pooled cells (see below). For Complete Mismatch, 5 Terminal Match, and 10 Terminal Mismatch acceptors, puromycin-resistant clones were generated by expanding well-isolated colonies from separate plates.
Because separate introduction of donor and acceptor reduced the possibility of transfection-associated plasmid recombination, the sequential transfection approach was taken to generate proviruses subjected to product DNA analysis. However, because stably transfected plasmid expression was low, the alternate approach of transiently cotransfecting donor, acceptor, and MLV helper function (pMLV Ψ−) plasmids and subsequently analyzing virion RNA by RNase protection assay was used to determine puromycin-resistant titers per unit RNA. RNA for RNase protection assays was purified from some virus aliquots, and other aliquots from the same transfections were used in NIH 3T3 infections.
Virus quantification and titer determination.
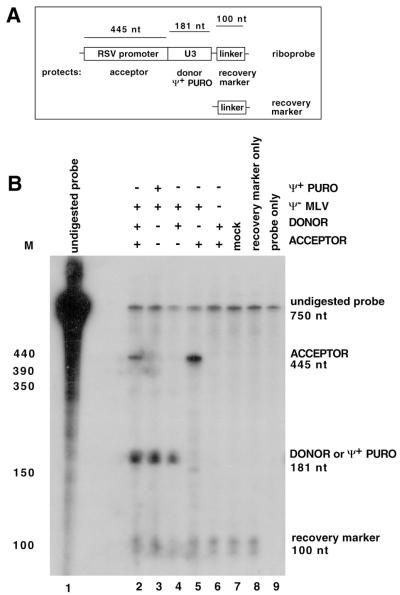
The donor and acceptor RNAs were differentiated in RNase protection assays and quantified by PhosphorImager (Molecular Dynamics) analysis. To quantify vector RNAs, background values were subtracted and molarities were normalized by dividing protected product values by the number of radiolabeled nucleoside monophosphate (CMP) residues in each. An additional calculation (called the copackaging factor) used the acceptor/donor ratio in virions to predict which portion of virions would contain both donor and acceptor RNA. For this calculation, the Hardy-Weinberg equation (A2 + 2AD + D2 = 1, where A is the amount of acceptor and D is the amount of donor) was used. The 2AD term served as the copackaging factor. Because donor/acceptor ratios were very similar for all vector pairs, in practice the copackaging factor altered values minimally. To compare the absolute amounts of RNA among samples, values were normalized to the amount of recovery marker product in each lane. The recovery marker was a short probe-complementary in vitro transcription product added to virion preparations early in processing to account for sample loss (44).
Titers were determined by end point dilution. Figure 4B shows averages from two independent infection experiments and colony counts from at least six plates per acceptor. The average titer per milliliter was divided by the sample loss factor and the copackaging factor to yield the puromycin resistant titer per unit RNA. The values in Fig 4B are percentages of the 59 Terminal Match titer per unit RNA value. The 59 Terminal Match titer was ca. 103 puromycin-resistant colonies per ml of virus-containing medium.
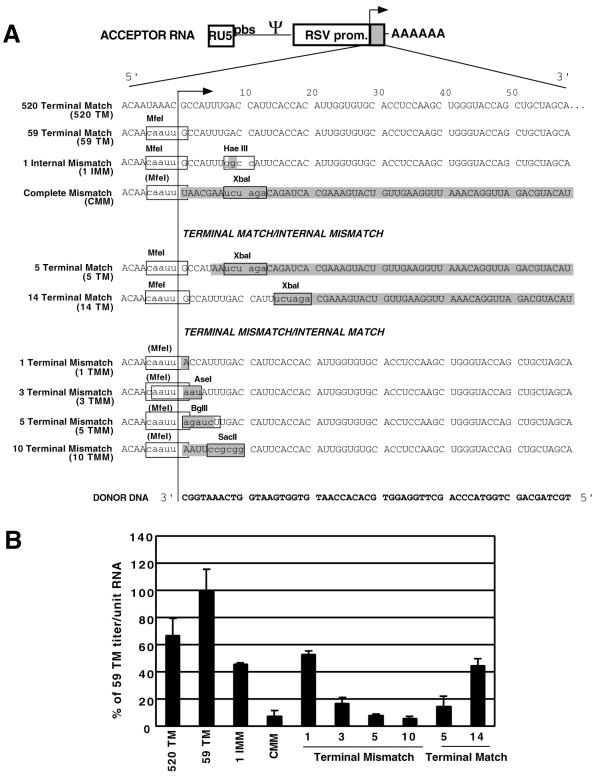
FIG. 4.
Mutant acceptor templates and puromycin-resistant titers per unit RNA. (A) Mutant acceptor templates. The diagram at the top represents the acceptor RNA (ITR is shown as a gray box); the sequences below are from the ITR. The leftmost column gives the name of each acceptor template, followed by the sequence of the ITR of that acceptor Nucleotides boxed in gray are mismatched to the donor, and relevant restriction sites are shown. The line with the arrowhead represents the position of the wild-type acceptor/donor sequence junction. Acceptor derivatives were constructed with regions of complete homology to the donor (520 and 59 Terminal Match), to have one internally mismatched nucleotide (1 Internal Mismatch), or to lack any homology to the donor (Complete Mismatch). Other derivatives contained terminal matches of 5 or 14 nucleotides (5 and 14 Terminal Match) or terminal mismatches of 1, 3, 5, or 10 nucleotides (1, 3, 5, and 10 Terminal Mismatch). The 3′-end sequence of the transferring donor DNA is shown at the bottom (in bold type). (B) Puromycin-resistant titers per unit virion RNA, determined from RNA quantification data like those in Fig. 3B and puromycin-resistant colony counts. These values are normalized to 59 Terminal Match values (see Materials and Methods).
Analysis of proviral DNA.
Genomic DNA for use in PCR and/or Southern blot analyses was isolated using the Wizard genomic DNA purification kit (Promega). For the PCR analyses in Fig. 6Δ, C, and D, DNA was amplified using an RSV promoter sense primer and an antisense primer within the puromycin resistance gene (2). PCR was performed in 10 mM Tris (pH 8.3)–50 mM KCl–1.5 mM MgCl2–250 μM each deoxynucleoside triphosphate–5% dimethyl sulfoxide–30 pmol of each primer for 30 cycles with an annealing temperature of 58°C. The products were digested with appropriate restriction enzymes, separated on 1% agarose gels, and quantified using Kodak Digital Science 1D image analysis software. For the experiment in Fig. 6B, 32P-end-labeled RSV primer (43) was used in PCR of 1 Internal Mismatch DNA. Products were digested and separated on 5% polyacrylamide gels. The dried gel was exposed to film, and products were quantified by PhosphorImager analysis.
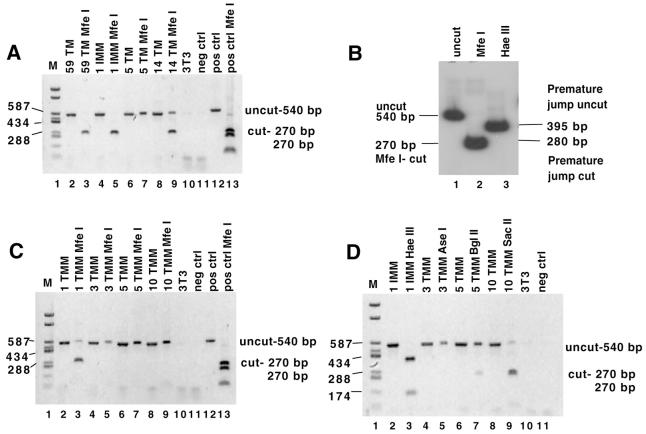
FIG. 6.
Product DNAs analyzed for switch-associated errors, premature jump, and mismatch extension. For panels A, C, and D, genomic DNA from pools of puromycin-resistant colonies was amplified using PCR primers that flanked the ITR. PCR products were digested and separated on 1% agarose gels next to undigested controls. The acceptor construct whose progeny proviruses generated the PCR products analyzed in each lane is indicated at the top. 3T3 indicates products that resulted from amplification of uninfected cell genomic DNA; neg ctrl indicates the no-template control; pos control indicates the PCR-amplified plasmid DNA which contains the restriction site of interest. (A) Errors made at the transfer site. PCR products were digested with MfeI. Error-free synthesis should generate an MfeI-digested band of 270 bp. (B) Quantitative error and premature jump rates for the 1 Internal Mismatch acceptor. Genomic DNA from 1 IMM pooled puroR colonies was PCR amplified using a 32P-labeled RSV promoter primer; the products were digested with MfeI or HaeIII and separated on a polyacrylamide gel. The products were quantified using a PhosphorImager (see Results). The uncut PCR product is 540 bp (lane 1). Digestion at the MfeI site (diagnostic of error-free transfer) gives a 270-bp band (lane 2). For the premature jump (lane 3), Premature jump uncut indicates the expected size of the band (395 bp) if no digestion was observed at the diagnostic HaeIII site; Premature jump cut indicates the expected size of the band (280 bp) if digestion occurred at the diagnostic HaeIII site. Note that in addition to the HaeIII site diagnostic of a premature jump, there is an additional HaeIII site present in the product which serves as a digestion control. (C) Mismatch extension. PCR products were digested with MfeI. Mismatch extension should generate an MfeI-digested band of 270 bp. (D) Premature jump. PCR products were digested with HaeIII, AseI, BglII, or SacII as indicated. Completely uncut products yield a 540-bp band; for most acceptors, a premature jump would yield a shorter (∼270- to 280-bp) band. Note that for 1 Internal Mismatch, an extra HaeIII site is present in the product such that digestion at the site diagnostic of premature jump gives 280-, 145-, and 115-bp bands while no digestion at the diagnostic site yields 395- and 145-bp bands.
To generate the data shown in Fig. 7, PCR products were digested with MfeI, uncut bands were purified and subcloned into LITMUS 29, and inserts were sequenced. For the PCR analysis in Fig. 10, sense primers in U3 or at the U5–primer-binding site (PBS) junction were used with the puromycin gene antisense primer to amplify pooled genomic DNA. In the PCR which yielded the results in Fig. 8 and 9, a U3 sense primer and antisense puromycin gene primer were used. Of 35 tested genomic DNA preparations, 27 yielded PCR products using these primers. Four samples that did not yield a PCR product even with primers that were both within the puromycin gene were not analyzed further. Note that DNAs for Fig. 8 and 9 were from individual cell clones, while all other PCR experiments analyzed pooled colony DNA. Southern blots were performed as described previously (37, 49), using 5% polyacrylamide gels and a probe from the puromycin resistance gene (see Fig. 5).
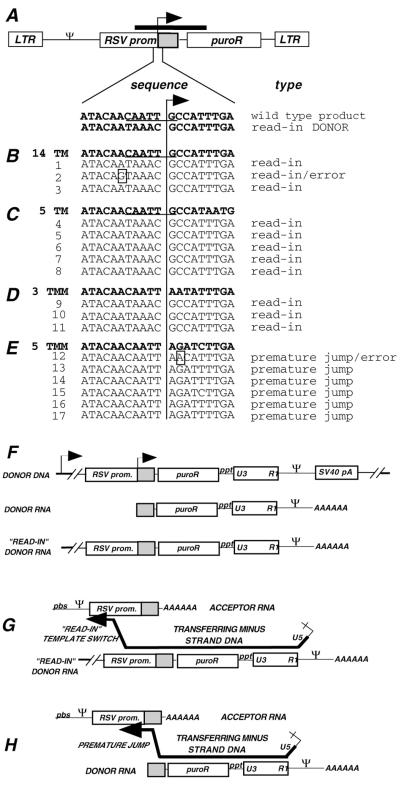
FIG. 7.
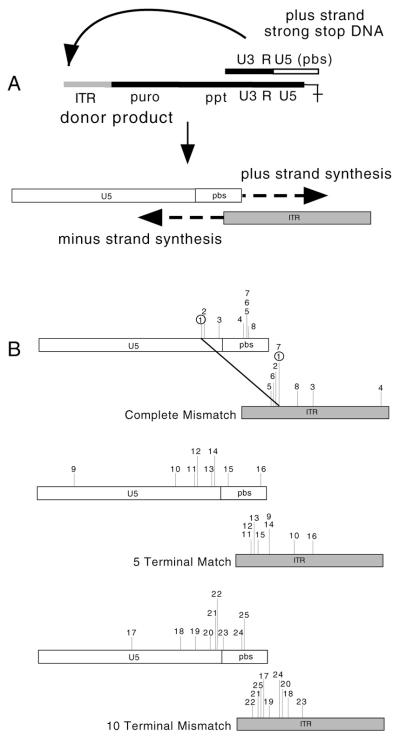
Sequences of MfeI-uncut products from acceptor provirus pools. (A) Schematic drawing of proviral DNA. The ITR is shown as a gray box. The ITR sequence that encompasses the acceptor/donor junction is enlarged below the drawing: the wild-type junction and putative read-in donor product sequences are shown. The line with the arrowhead indicates the position of the wild-type acceptor/donor junction between the RSV promoter and ITR. Primers in the RSV promoter and in the puromycin resistance gene were used to PCR amplify genomic DNA from pools. The thick bar above the schematic drawing represents the PCR-amplified region. MfeI-undigested PCR products were subcloned and sequenced. (B through E) Bold type shows the sequence of the acceptor used (14 TM, 5 TM, 3 TMM, and 5 TMM for panels B, C, D, and E, respectively), while light type below each acceptor shows individual cloned PCR product sequences, each of which is designated by a sequence number on the left. The probable mechanism of origin of each sequence is indicated on the right (see the text). Errors are shown as boxed nucleotides. Note that the sequences of products 1 and 3 for 14 TM (B) as well as 4 through 8 for 5 TM (C) were identical to one another, suggesting that in each case these products could have arisen from a single puromycin-resistant colony from the pool. Sequence 2 in panel B differed from sequences 1 and 3 only by a single-base change (A→G). Because recombination junctions for read-in products were not determined, it is not known whether the A→G change was transfer-associated is unknown. (F) Diagram of the read-in donor transcript. The integrated DNA that templates donor RNA is shown at the top. The bold lines on either end represent the flanking cellular DNA, and a putative host promoter 5′ of the integrated DNA is shown by an arrow. The intended donor transcript is called donor RNA, and the read-in donor transcript is shown at the bottom. (G) Diagram of the read in donor transcript recombination. The transferring minus-strand DNA is shown as in Fig. 1B, but in this case the extended region of homology between the read-in donor and acceptor serves as the recombination target. (H) Diagram of recombination via a premature jump. Again, the transferring minus-strand DNA is shown as in Fig. 1B, but here the jump takes place before reaching the end of the donor RNA template. Abbreviations are as in Fig. 1.
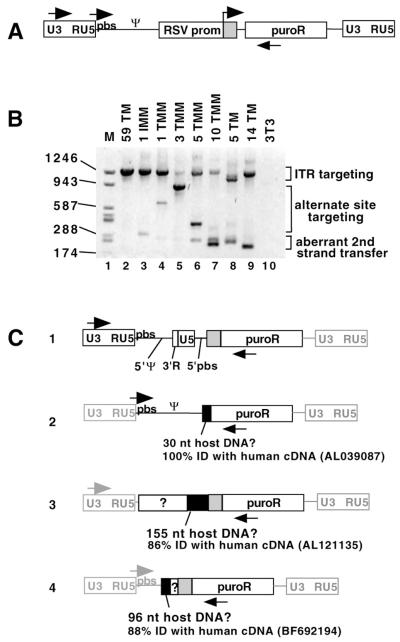
FIG. 10.
Alternate template switch sites and proviral structures. (A) Diagram of ITR-targeted proviral DNA showing locations of primers used here for PCR analysis. These primers could amplify products targeted to the ITR or alternate sites. ITR is shown as a gray box. (B) Agarose gel showing PCR products from pooled puroR colony genomic DNA. The acceptor used to generate the DNA analyzed in each lane is indicated at the top of the panel. Proviruses with ITR targeting yielded a ∼1,290-bp band, aberrant second-strand transfer yielded ∼180- to 250-bp bands, and alternate targeting yielded ∼300- to ∼900-bp bands. The U5-PBS sense primer and the puro antisense primer were used to generate the products. (C) Schematic drawings of alternate proviral products. PCR products were excised from the gel in panel B or other gels and sequenced. The primers used to amplify each structure are shown. The ITR is shown as a gray box with black outline. Regions that were not sequenced are shown as gray lines to indicate that they are presumed to have the indicated structures. A BLAST search was performed on regions without MLV homology, and regions of homology to human cDNAs are shown as black boxes. GenBank accession numbers are indicated. Boxes with “?” indicate unknown sequence without detectable homology to MLV or human cDNA. The four proviral structures shown were the only alternate structures identified in this analysis. nt, nucleotide; ID, identity.
FIG. 8.
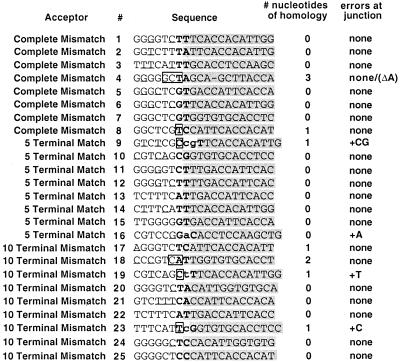
Aberrant second-strand transfer products. (A) Diagram of the aberrant second-strand transfer process. The first step shows the intermediates prior to aberrant transfer. The bottom line represents DNA that was templated by the donor RNA. The gray line represents the ITR. The top line represents the transferring plus-strand strong-stop DNA, and the open line shows U5 and the PBS. The second step shows an enlarged portion of this process after the aberrant transfer. Dotted lines with arrows indicate DNA synthesis after transfer. Little or no complementarity directed these transfers (see Fig. 9). (B) Junctions observed for aberrant second-strand transfer products. Each top box corresponds to the U5-PBS portion of plus-strand strong-stop DNA, while the bottom box corresponds to the ITR at the 3′ end of minus-strand DNA prior to the aberrant transfer for the indicated acceptors (Complete Mismatch, 5 or 10 Terminal Match). The vertical lines above the U5-PBS boxes represent junction points where homology ended, while the vertical lines for the ITR boxes show where homology to the ITR began, for each of these products. For each analyzed product, the same number is used to indicate junctions in both U5-PBS and in the ITR and the numbers correspond to sequence numbers in Fig. 9. For example, Complete Mismatch product sequence 1 (circled) departed from the plus strand DNA in U5 and joined the ITR as indicated by the diagonal line. Abbreviations are as in Fig 1.
FIG. 9.
Aberrant second-strand transfer junction sequences. PCR products were amplified from genomic DNA of individual puromycin-resistant colonies generated by viruses in which donor-acceptor homology was severely limited and were sequenced directly. Sequences in gray boxes are from the donor ITR; unshaded sequences are from the acceptor. Acceptor product numbering is the same as in Fig. 8B. For example, Complete Mismatch 1 is the product whose junctions in U5 and in the ITR are indicated by ‘1’ above the vertical lines in Fig. 8B. The precise junctions are indicated in bold type. Errors made at the junction are shown as lowercase bold letters. Regions of homology between the transferring DNA and acceptor are boxed. For Complete Mismatch 4, an internal deletion of an A is represented by a dash.
FIG. 5.
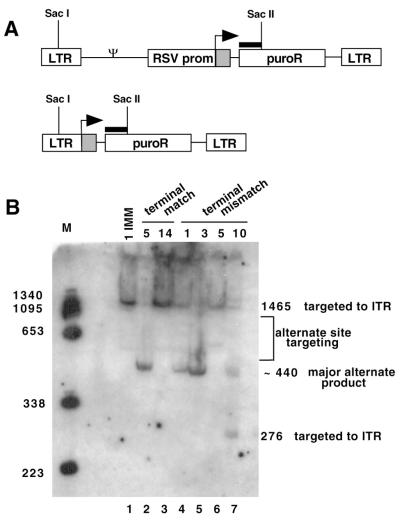
Alternate targeting assessed by Southern blotting. (A) Schematic drawings of proviral DNA structures. Genomic DNA from pooled puromycin-resistant colonies was digested with SacI and SacII and analyzed by Southern blotting. The probe (thick black bar) was from the 5′ end of puroR. The top line shows the structure of proviral DNA that would result if the ITR were used as the template switch site, while the bottom line shows the structure of a commonly observed alternate structure (labeled the major alternate product in panel B [also see Fig. 9]). (B) Southern blot. The acceptor that yielded the DNA analyzed in each lane is indicated at the top of the panel. Use of the ITR is indicated by a 1,465-bp band, while the major alternate product (bottom line of panel A) yielded ∼440-bp bands. For the 10 TMM acceptor, an extra SacII site was present if a premature jump occurred, yielding 1,465- and 276-bp bands for correct targeting.
Calculating proportions of alternate products.
The various mutant acceptor vectors generated very different spectra of products. Some reverse transcription outcomes could be distinguished based on product sizes, and others could not. In some cases, the analysis of the same alternate product required different approaches when it was generated by different vectors due to differences in prevalence and diagnostic features such as restriction sites. Thus, it was not possible to determine the proportions of all products of all vectors in a single controlled experiment. Instead, the proportion of each reverse transcription product class within the pool of all puromycin resistance-conferring products of each vector were estimated roughly as presented in Fig. 11, using the combination of approaches outlined here.
FIG. 11.
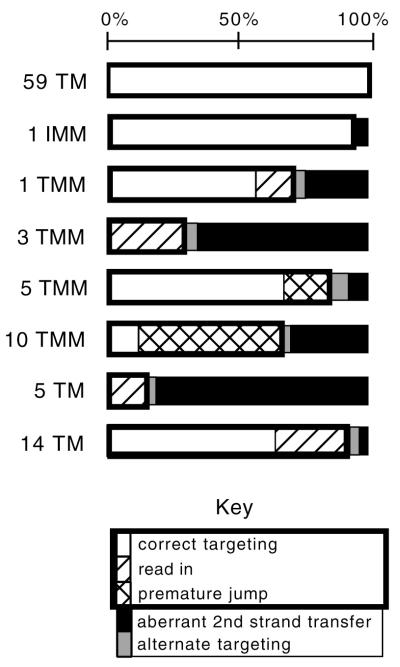
Estimated prevalence of alternate reverse transcription products. Proportions of each outcome within the puromycin resistance-conferring provirus population templated by each acceptor were calculated based on assays presented in this paper, as described in Materials and Methods. Although each acceptor yielded a different titer per unit virion RNA, here the sum of products for each vector was set at 100% (shown by the scale at the top). Open bars represent the proportion of products produced by correct targeting to the ITR; diagonal slashes represent read-in products; hatched bars represent premature jumps; black bars represent aberrant second-strand transfer; and gray bars represent minor alternate products. Combined, the products within the thick rectangle were those whose sizes corresponded to ITR targeting in the Southern blot from Fig. 5B. Values were not calculated for 520 Terminal Match or Complete Mismatch.
Product intensities from Southern blots such as that in Fig. 5 were quantified by PhosphorImager analysis and used to estimate relative amounts of ITR targeting-size products versus aberrant second-strand transfer and other alternate products. The proportions of correct transfer, read-in products, and premature transfer products among ITR targeting-size products were further analyzed by observing the digestion patterns of PCR products such as in Fig. 6 or by sequencing individually cloned products. PCR digestion products were quantified by densitometry of digital UV photographs using Kodak Digital Science 1D image analysis software. The PCR data in Fig. 10 were not used for quantification, but only to indicate the presence or absence of detectable alternate products within a provirus pool.
RESULTS
System to examine acceptor requirements for recombinogenic template switching during viral replication.
We developed a two-vector recombination assay intended to “force” a template switch to occur at or near a defined position (Fig. 1). Although the end of the donor RNA had a 5′ cap and thus differs from broken RNAs, this method was designed to mimic forced copy choice recombination at an RNA break (6).
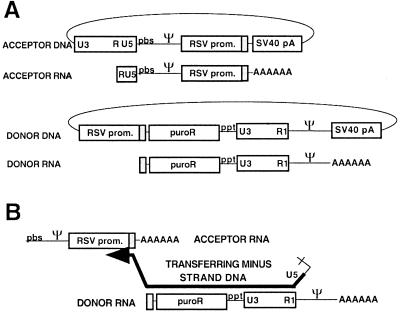
FIG. 1.
Forced-copy-choice recombination system. (A) Viral vectors. The DNA forms of the vectors are shown as circles. The acceptor RNA contains the 5′ untranslated region of MLV including R, U5, PBS, and Ψ; the RSV promoter and ITR (gray box); and poly(A) tail. The donor RNA contains (5′→3′) the ITR of the RSV promoter; the puromycin resistance gene (puroR); the 3′ untranslated region of MLV including the polypurine tract (ppt), U3, and a mutant R region (R1) that lacks polyadenylation signals; MLV Ψ; and heterologous poly (A) tail. Note that the region of sequence identity between the donor and acceptor is the ITR. (B) Recombinogenic template switch from donor DNA to acceptor RNA. DNA (thick black line) synthesis on the donor stops when it reaches the 5′ end of the RNA. The DNA then switches to the acceptor RNA at the ITR (gray box), and DNA synthesis continues. Note that reverse transcription steps that precede and follow those shown occur essentially as they do during normal replication (47). For example, prior to steps shown in this illustration, minus-strand strong-stop DNA initiates normally from the PBS on the acceptor RNA (A) and then transfers to the donor RNA R1 (in this panel, part of the minus-strand strong-stop DNA is evident as an unpaired ‘tail’ on the 3′ end of the transferring DNA). After the steps shown here, plus-strand DNA initiates from the polypurine tract (ppt) and ultimately transfers to the minus-strand DNA that results from synthesis through the PBS on the acceptor RNA.
This system generated a puromycin-resistant provirus if recombination placed the puromycin resistance gene downstream of the RSV promoter. The template switch donor was defined as the template for the minus-strand intermediate, which was subsequently forced to switch to a homologous region on the other RNA (the acceptor template; [Fig. 1A]). The region of homology was the ITR of the RSV promoter (Fig. 1B). Control experiments demonstrated that virions from cells cotransfected with helper plasmid and only the acceptor or donor did not generate any puromycin-resistant colonies (data not shown). This is probably because in addition to the recombinogenic template switch, successful DNA synthesis in this system required intermolecular first strong-stop transfer prior to the studied recombinogenic transfer. Such intermolecular strong-stop transfer can occur during reverse transcription (17, 18, 31, 50, 55).
The experimental scheme is described in Fig. 2. A 293T-based cell line stably expressing the donor RNA was transiently cotransfected with acceptor-encoding plasmid and a helper construct, pMLV Ψ−, that supplied viral proteins in trans. NIH 3T3 cells were infected with the resulting virions, and puromycin-resistant colonies were clonally expanded or pooled for integrated viral DNA analysis.
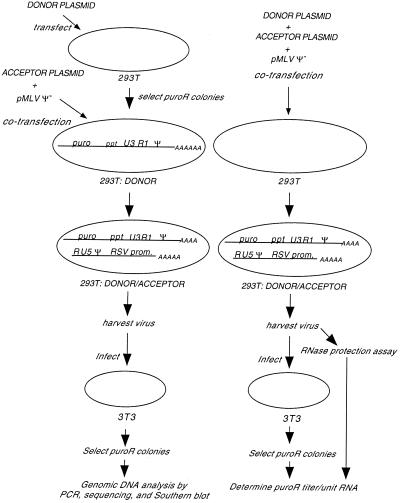
FIG. 2.
Experimental overview. The chart on the left shows the generation of proviral DNA for molecular analysis, while that on the right shows how titers per unit virion RNA were determined. (Left) To generate a stable cell line expressing donor RNA, 293T cells were transfected with the donor plasmid and a puromycin-resistant colony was chosen as the 293T:DONOR line. This line was transiently cotransfected with acceptor plasmid and pMLV Ψ−, and virus was harvested. NIH 3T3 cells were infected, and puromycin-resistant colonies were either pooled or clonally expanded. Genomic DNA from these cells was subjected to molecular analysis. (Right) Donor and acceptor plasmids and pMLV Ψ− were transiently cotransfected into 293T cells, and virus was harvested. This virus was used for RNase protection assays and for infection of NIH 3T3 cells. Puromycin-resistant colonies were counted, and the puromycin-resistant titers per unit RNA were determined. Abbreviations are as in Fig. 1.
Puromycin-resistant provirus synthesis required copackaging of donor and acceptor RNAs. Assuming that two RNAs are copackaged, each virion produced by cells coexpressing the donor and acceptor could contain two donor RNAs, two acceptor RNAs, or one of each. The frequency of donor-acceptor copackaging was estimated by quantifying RNA ratios within the virion population by the RNase protection assays and assuming random copackaging (16, 17, 55). To determine the puromycin-resistant titer per unit encapsidated RNA, NIH 3T3 cells were infected with aliquots of the same virus preparations used in the RNase protection assays (Fig. 3). The numbers of puromycin-resistant colonies per milliliter were multiplied by normalization factors for copackaging and sample loss, as described in Materials and Methods.
FIG. 3.
Quantification of viral RNA. (A) RNA probe and recovery marker design. A single-stranded RNA probe engineered to contain portions of the RSV promoter, U3, and a short linker, all in a single probe, was used. Acceptor RNA protected a 445-base probe fragment from the RSV promoter, donor or Ψ+ PURO RNA protected 181 bases, and the recovery marker protected a 100-base fragment. (B) Representative RNase protection assay performed to determine the amounts of virion RNA, using 520 Terminal Match as the acceptor. The key at the top right shows which vectors were cotransfected to generate virus. nt, nucleotides.
Homology length effects on recombinogenic template switch frequencies, as indicated by titer.
A series of mutant acceptors was constructed with alterations in the region of homology between the acceptor and donor. Mutations that limited primer-terminal complementarity (1, 3, 5, and 10 Terminal Mismatch) or limited primer-internal complementarity (5 and 14 Terminal Match) (Fig. 4) were introduced into the acceptor. An acceptor that had no complementarity to the donor (Complete Mismatch), acceptors completely matched to the donor (for 59 to 520 bases in the 59 and 520 Terminal Match acceptors, respectively), and an acceptor that had one internal marker-introducing point mutation (1 Internal Mismatch) were also constructed. The same donor was used for all acceptor-donor pairs.
Puromycin-resistant CFU titers per unit virion RNA for the various acceptors were compared (Fig. 4B). Acceptor titers per unit RNA were reported as percentages of the 59 Terminal Match acceptor titer because, somewhat surprisingly, the acceptor with 59 bases of complementarity to the transferring DNA displayed the highest titer: slightly higher than that of the acceptor which had 520 bases of complementarity. The titer per unit RNA for 59 Terminal Match was 0.8% of the titer per unit RNA of the Ψ+ PURO vector (a positive control where recombination is not required for puromycin-resistant colony formation [data not shown]). The 1 Internal Mismatch, 1 Terminal Mismatch, and 14 Terminal Match acceptors each yielded slight titer reductions (1.5- to 2.2-fold), while titers for the Complete Mismatch, 5 Terminal Match, and 3, 5, and 10 Terminal Mismatch acceptors were all severely reduced (6- to 18-fold).
Effects of limiting the lengths of acceptor homology on the spectra of reverse transcription products.
The substantial residual titer observed for the Complete Mismatch acceptor suggested that some puromycin-resistant colonies resulted from outcomes other than template switch to the RSV ITR. To address whether or not alternate sites served as recombination acceptors, the integrated proviruses in pooled products of mutant acceptors were analyzed by Southern blotting (Fig. 5). Genomic DNA from pooled transduced cells was digested using enzymes with sites in U3 and in the puromycin resistance gene and probed with a 5′ fragment of the puromycin resistance gene (Fig. 5A). Figure 5B shows that whereas a 1,465-bp band diagnostic of ITR targeting was detectable among products of most acceptors, products of other lengths were also detected, suggestive of alternate acceptor site use. An approximately 440-bp product was frequently observed for some acceptor mutants, suggesting that much of the alternate template switching occurred to a single, fairly discrete alternate region. Additional faint bands representative of other alternate products were also visible (labeled as alternate site targeting in Fig. 5B). The structures of some of the frequent class and other alternate products are described below.
High fidelity of homology-guided recombinogenic template switch.
Several of the vectors were engineered such that outcomes of recombination could be assessed by restriction digestion of product DNAs. Thus, products that resulted from transfer to the ITR were analyzed by restriction digestion to determine whether errors occurred at the template switch site. Purified RT can add nontemplated nucleotides to the ends of transferring DNAs (35, 36, 51), and it has been proposed that incorporation of these nontemplated nucleotides may contribute to retroviral genetic variation (32, 34, 51). Nontemplated addition errors are observed among products of replicative strong-stop transfer (13, 21) but have not been observed in replication products where recombination was genetically confined to a limited region (58).
To determine if nontemplated nucleotides were added or other errors occurred when recombination was forced to occur at a single position, recombination junction structures for proviral products of acceptors with complete or partial acceptor terminal match were analyzed. The 1 Internal Mismatch and the 5, 14, and 59 Terminal Match acceptors had been engineered so that an MfeI site would be present in products if the switch targeted to the puromycin ITR were extended without error. However, errors such as extension of a nontemplated base, unless the added base were fortuitously complementary to the acceptor template, would destroy the MfeI site.
The ITR region was PCR amplified from genomic DNA isolated from pools of at least 50 proviral products of each vector. Digestion of these products with MfeI is shown in Figure 6A. 1 Internal Mismatch, 59 Terminal Match, and control products all digested to apparent completion with MfeI, indicating that few if any errors were made during recombinogenic template switching. To more accurately quantify MfeI digestion of 1 Internal Mismatch acceptor products, PCR was performed with a 32P-labeled primer and digestion products were quantified by PhosphorImager analysis (Fig. 6B, lane 2). More than 99% was digested, and the residual <1% uncut product was similar in amount to that in a positive control reaction (data not shown). This suggests that each provirus in the 65-colony pool was a product of error-free recombination.
In contrast, only 66% of the 14 Terminal Match products whose sizes were indicative of ITR targeting were MfeI- digestible, and no digestion was detectable among this size products of the 5 Terminal Match acceptor (Fig. 6A, lanes 7 and 9). Undigested bands were excised from the gel, reamplified, and redigested with MfeI. No additional digestion was observed. This suggests that most ITR-targeted products did not result from faithful transfer when primer and template complementarity was severely limited. Most of these products were subsequently determined to be recombination products of read-in transcripts, as described below.
Limited mismatch extension during recombinogenic template switching.
RT can extend mismatches both during replicative strong-stop switching (40, 49) and during template switching in purified reaction mixtures (35, 36). To determine if mismatch extension could occur during forced recombinogenic template switching, we analyzed the proviral products of 1, 3, 5, and 10 Terminal Mismatch acceptors. These acceptors had also been engineered such that an MfeI site would be present in the proviral DNA only if RT accurately extended the primer-terminal mismatch. For each acceptor, genomic DNA was isolated from pools of at least 50 puromycin-resistant colonies and analyzed. As shown in Fig. 6C, 74% cutting with MfeI was observed with 1 Terminal Mismatch, but no products of the other acceptors (with 3, 5, or 10 mismatches) showed detectable digestion. This demonstrated that one nucleotide of primer terminal mismatch was extended fairly efficiently, but that 3, 5, or 10 mismatches were not.
Alternate ITR use-sized products are generated by read-in transcription or premature template switch.
MfeI-uncut PCR products from pooled colonies of 3 and 5 Terminal Mismatch acceptors were subcloned and sequenced (Fig. 7D and E). All sequenced products of 3 Terminal Mismatch were identical (Figure 7D, 9 to 11). All were found to contain sequences specific to the donor RSV promoter just upstream of the ITR. Therefore, they were probably not products of forced transfer from the intended donor RNA 5′ ends but instead were likely to be products of recombination with read-in transcripts (7). Read-in products arise when transcription of the donor begins upstream of the intended start site such that the RSV promoter sequence is included 5′ of the intended start site in the packaged RNA (Fig. 7F). Because the RSV promoter is included in read-in donor transcripts, this extended homology between the donor and acceptor allows template switching to occur in regions of donor-acceptor homology upstream of the ITR, such as in the RSV promoter (Fig. 7G). Similar read-in products have been reported recently among aberrant first-strand transfer products (8). Note that the MfeI-uncut products for the 14 and 5 Terminal Match vectors also appear to be the result of recombination with read-in transcripts (Fig. 7B and C).
For the 5 Terminal Mismatch acceptor, all ITR use-sized products displayed evidence of premature jump, i.e., a template switch that occurred before the end of the template was reached (Fig. 7E and H). This phenomenon has been studied for required template switching during replication (1, 20, 21, 39, 41, 53) and in reconstituted in vitro reactions that mimic recombination (11, 51). We do not understand why the 3 and 5 Terminal Mismatch acceptors yielded different classes of products, but this finding was reproducible.
Three different premature jump sequences were observed (Fig. 7E). Products 13, 14, 16, and 17 were identical and apparently resulted from premature transfer at the ITR −4 position; sequence 15 transferred at −5; and sequence 12 displayed evidence of a premature jump at −2 followed by nontemplated addition of an A at the transfer site.
To examine if the premature jump generated products from other acceptors, the proviral products of 1 Internal Mismatch and 3, 5, and 10 Terminal Mismatch acceptors were screened for restriction sites engineered to diagnose a premature jump. As shown in Fig. 6D, no digestion at the site diagnostic of a premature jump was seen in PCR products of pooled 1 Internal Mismatch or 3 Terminal Mismatch proviruses (lanes 3 and 5). 5 Terminal Mismatch products displayed 21% digestion (lane 7), while 10 Terminal Mismatch was digested 80% (lane 9), suggesting that premature template switching had occurred for these two acceptors. Note that this approach detected premature transfer only if it occurred 3′ of the diagnostic restriction site.
To more precisely quantify premature transfer for 1 Internal Mismatch, the ITR-containing region was PCR amplified using a 32P labeled primer, the product was digested with HaeIII, and the products were quantified by PhosphorImager analysis. A HaeIII site in the puromycin resistance gene served as a digestion control. As shown in lane 3 of Fig. 6B, no product diagnostic of a premature jump was detectable but the internal-control HaeIII site was >99% digested. This implies that none of the 65 proviruses in the 1 Internal Mismatch pool had experienced a premature jump before the ITR −8 position HaeIII site and suggests that the premature jump occurred during the synthesis of fewer than 1% of all proviruses generated by vectors with identical donor and acceptor regions.
Most alternate sized products resulted from aberrant second-strand transfer.
To estimate the frequency of non ITR targeted products, genomic DNA was isolated from 26 randomly selected puromycin-resistant colonies with proviruses templated by low-titer acceptors and amplified using primers in U3 and in the puromycin gene. One provirus was determined to be a product of a premature jump that was correctly targeted to the ITR (data not shown). All the other randomly selected products corresponded to the ∼440-bp band in the Fig. 5, which appeared to be the major product when homology was severely limited (major alternate product, Fig. 5).
Sequencing revealed that these proviruses included common features, which provided clues about their possible origins. All 25 included sequences from the 5′ untranslated region (U5 and PBS) fused directly to a truncated ITR. In the resulting proviral DNAs, puromycin resistance gene expression was presumably driven by the U3 promoter. None of the proviruses contained the full length U3-R-U5-PBS. These features suggest that these were products of aberrant second-strand transfer (Fig. 8A).
Although the aberrant second-strand transfer products were similar to one another in size, their sequences varied widely. The results of direct sequencing of the PCR products of the individual cell clone DNAs are shown in Fig. 8 and 9. No consistent junctions or apparent recombination hot spots between donor ITR and U5-PBS sequences were observed. Instead, a surprisingly heterogeneous set of both apparent donor departure sites and acceptor reassociation sites was found (Fig. 8B). In 4 of 25 sequences (sequences 9, 16, 19, and 23 as indicated in the second column of Fig. 9), nontemplated nucleotides were observed at the junction of sequences from the 5′ untranslated region and sequences from the ITR.
Minor alternate products.
In a different approach to studying alternate products, pooled colony genomic DNA was PCR amplified and analyzed on an agarose gel (Fig. 10). PCR primers were designed such that proviruses formed from both targeting to the ITR and targeting to alternate sites could be amplified (Fig. 10A). As shown in Fig. 10B, some targeting to the ITR was seen for all acceptors (∼1,290-bp bands) but several other products also were observed for most of the acceptors. Products (∼180- to 250-bp bands) with sizes that correlated with those predicted for the ∼440-bp products visible in Fig. 5B were prevalent. In addition, some PCR products ranging in size from ∼300 to ∼900 bp were seen for the 1, 3, 5, and 10 Terminal Mismatch and 5 Terminal Match acceptors.
To determine the structures of some of the proviruses that generated these minor alternate-size PCR products, individual product bands were excised from this and other gels and sequenced. Figure 10C displays the proviral structures of some of these products. In each case, multiple template switches were probably responsible for the observed structure. Three of the proviruses contained regions that are not homologous to MLV (Fig. 10C, sequences 2, 3, and 4). Portions of these sequences may have been derived from recombination with host sequences, since the viruses were generated in human 293T-based producer cells and the proviruses include segments that show partial homology to human cDNAs. For sequences 2 and 3 in Fig. 10C, 1 nucleotide of homology between the viral and host sequences is present at the recombination junction. The extent of homology at the junctions of sequences 1 and 4 from Fig. 10C could not be determined.
The proportion of puromycin-resistant proviruses for each acceptor that resulted from each of the reverse transcription outcomes described above was estimated using a combination of analytic methods described above and in Materials and Methods (Fig. 11). The total for each acceptor was set at 100%. In general, the greater the difference between the acceptor and donor templates, the greater the proportion of non-ITR-targeted products. However, correlations between the relative proportion of each alternate product and the type of acceptor alteration were not discernible in all cases.
DISCUSSION
An assay was developed to study homology requirements when a recombinogenic template switch was forced to occur at or near a defined position. Controls demonstrated that transfer before reaching the end of the donor template was ordinarily rare (Fig. 6B). Thus, the significance of specific acceptor template regions to recombinogenic acceptor template selection could be addressed by mutagenizing acceptors and then determining the effects on reverse transcription product yields and structures.
Efficient acceptor template recognition and use appeared to require more than 5 nucleotides of terminal match or a terminal mismatch of fewer than 3 nucleotides. The recombinogenic switching efficiency was quantified by measuring puromycin-resistant titers per unit virion RNA. Relatively modest decreases were observed with acceptors containing 1 internal mismatch, 1 mismatch with the primer strand terminus, or 14 terminally matched nucleotides, suggesting that these mutations were minimally detrimental for recombination. However, severe titer reductions were observed for acceptors with terminal mismatches of 3, 5, or 10 nucleotides, for an acceptor that retained only 5 terminal matches, and for a completely mismatched acceptor. The finding that more than 5 nucleotides of complementarity are required for homologous recombination is in agreement with homology requirements reported by one group for the minus-strand strong-stop replicative switch (8).
Under conditions where homology was not limited, transfers were error free and nontemplated additions were not observed. This is consistent with previous findings regarding forced (J. Jones, personal communication) and nonforced (25, 58) recombination junctions. Because mismatch extension did occur on acceptors with one terminally mismatched nucleotide, it seemed possible that if a nontemplated nucleotide were added during transfer, it would be incorporated into product DNA. However, the present studies confirmed that homologous recombinogenic switching was not error prone (note the complete MfeI digestion in Fig. 6A and B). This suggests that recombination between homologous sequences is generally genetically silent and cannot be detected based on product sequence. In contrast to the situation where the acceptor and donor were identical, alterations that reduced homology, even fairly minor acceptor alterations such as a single-primer terminal mismatch, resulted in detectable levels of aberrant or error-containing products.
The design of the vectors was intended to produce drug-resistant proviruses only when reverse transcription used the initial transcribed region of the RSV promoter (the ITR) as the recombination target site. However, some puromycin-resistant colonies resulted from alternate sites usage, especially for vectors with low titers. The major alternate product contained sequences from the 5′ untranslated region (U5 and PBS) fused directly to a truncated ITR. As proposed in Fig. 9A, recombinogenic template switching probably did not occur during synthesis of these products. Instead, it seems likely that after minus-strand DNA synthesis on the donor, newly synthesized plus-strand strong-stop DNA transferred to the 5′ end of the nascent minus-strand DNA directly using little or no homology, thus resulting in aberrant plus-strand transfer. These products lacked the ITR 5′ end, thus suggesting either that donor-directed DNA synthesis was not complete at the time of the transfer or that the transferring plus strand jumped to an internal position in the ITR. If such an internal jump occurred, a large mismatched region (5 to 50 nucleotides) would have to be extended by RT and/or excised by the host cell DNA repair machinery. There is evidence in the literature for a premature jump during second-strand transfer (25, 29, 30), and emerging evidence that host factors may complete provirus structures during viral replication (54).
Of 25 aberrant second-strand transfer products sequenced, 4 (16%) contained errors at the transfer point (Fig. 8). Whether these errors resulted from base addition before transfer or another cause was not determined. The four junctional errors were all different (A, T, C, and CG). These errors were probably not introduced during PCR since an adjacent 170-bp region of the puromycin resistance gene from 33 different provirus-templated PCR products (a total of more than 5,600 sequenced bases) showed no errors (data not shown). Why errors accumulated at a high frequency at aberrant plus-strand transfer sites was not addressed experimentally but may indicate that template switching can be error prone when no homology is directing transfer or that errors at strong-stop template switch sites may be better tolerated than at recombinogenic junctions. Consistent with this latter notion, we previously reported that nontemplated addition followed by mismatch extension is fairly common during minus-strand strong-stop switching (21), although another report suggested that errors are not made at this replication stage (8).
Other minor products of mutant acceptors were also detected. These included premature transfer products, products generated from read-in transcripts, and grossly rearranged proviruses. Some of these latter products were examined by sequencing analysis (Fig. 10C), and several different proviral structures were observed. Three contained regions that lacked MLV homology, which probably arose by nonhomologous recombination with host RNAs. Evidence of a premature jump was commonly observed among products of acceptors with extensive terminal mismatch.
Overall, DNA synthesis using this system was inefficient: the number of integrated DNAs made per virion calculated to have one donor and one acceptor RNA was only about 0.8% of that made per virion containing RNA that would not require recombinogenic switching. This low titer could have resulted from the engineered requirements for interstrand strong-stop and recombinogenic switching, factors such as nonrandom RNA copackaging, or a combination of factors. However, it is also possible that some inefficiency resulted because donor and acceptor homology regions were much shorter (520 bases or less) than those likely to arise during replication: in this regard, the recombination system used here differs from the recombination between two similar genomes which can occur during natural infection.
This study was designed to determine whether the recombining reverse transcription complex sought homology in the acceptor template via the primer terminus or at primer-internal positions. From the data presented here, it is clear that extensive primer-terminal complementarity can target recombinogenic template switching, since the 14 Terminal Match acceptor was able to serve as a fairly efficient acceptor. However, shorter regions of primer-terminal complementarity such as the 5 Terminal Match acceptor, with complementarity lengths reminiscent of the limited regions of complementarity previously noted at nonhomologous recombination junctions (56), were not. Primer-internal complementarity also appeared important. Even the 14 Terminal Match acceptor was not as efficient an acceptor as those with more extensive primer complementarity, and the studied acceptors with less than 14 nucleotides of terminal match were unable to direct recombinogenic template switch. Additionally, premature template switch products were more common per unit RNA when terminal homology was reduced than when it was present, possibly reflecting ongoing detection of donor-acceptor homology by the elongating replication machinery.
Although previous calculations have suggested that homologous retroviral recombination is 2 to 3 orders of magnitude more frequent than nonhomologous recombination, in this study all mutant acceptors, including those that lacked any donor homology, displayed less than 20-fold-reduced titers. The requirement to generate a puromycin-resistant provirus provided strong selection for any way of generating an integration-competent DNA, and a wide variety of reverse transcription products representing several alternate DNA synthesis mechanisms were observed. These alternate products provide further evidence of the remarkable ability of retroviruses to circumvent homology limitations and normal replication processes to generate product DNAs.
ACKNOWLEDGMENTS
We thank Joshua Filter for advice on PCR; Jeff Jones and Pamela O'Neal, who also developed a forced template switch system, for useful discussion; and John Moran, David Friedman, and Mary Jane Wieland for critical reviews of the manuscript.
This work was supported by American Cancer Society grant RPG-95-058-04-MBC to A.T. and the Nancy Newton Loeb Fund and by a Rackham Predoctoral Fellowship to J.K.P.
REFERENCES
- 1.Anderson J A, Teufel R I, Yin P D, Hu W-S. Correlated template-switching events during minus-strand DNA synthesis: a mechanism for high negative interference during retroviral recombination. J Virol. 1998;72:1186–1194. doi: 10.1128/jvi.72.2.1186-1194.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons, Inc.; 1994. [Google Scholar]
- 3.Buiser R, Bambara R, Fay P. Pausing by retroviral DNA polymerase promotes strand transfer from internal regions of RNA donor templates to homopolymeric acceptor templates. Biochim Biophys Acta. 1993;1216:20–30. doi: 10.1016/0167-4781(93)90033-a. [DOI] [PubMed] [Google Scholar]
- 4.Cheslock S R, Anderson J A, Hwang V K, Pathak V K, Hu W-S. Utilization of nonviral sequences for minus-strand DNA transfer and gene reconstitution during retroviral replication. J Virol. 2000;74:9571–9579. doi: 10.1128/jvi.74.20.9571-9579.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coffin J. Retroviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fundamental virology. 3rd ed. Philadelphia, Pa: Lippincott; 1996. pp. 763–843. [Google Scholar]
- 6.Coffin J M. Structure, replication, and recombination of retrovirus genomes: some unifying hypotheses. J Gen Virol. 1979;42:1–26. doi: 10.1099/0022-1317-42-1-1. [DOI] [PubMed] [Google Scholar]
- 7.Colicelli J, Goff S P. Sequence and spacing requirements of a retrovirus integration site. J Mol Biol. 1988;199:47–59. doi: 10.1016/0022-2836(88)90378-6. [DOI] [PubMed] [Google Scholar]
- 8.Dang Q, Hu W-S. Effects of homology length in the repeat region on minus-strand DNA transfer and retroviral replication. J Virol. 2001;75:809–820. doi: 10.1128/JVI.75.2.809-820.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delviks K A, Hu W, Pathak V K. Ψ− vectors: murine leukemia virus-based self-inactivating and self-activating retroviral vectors. J Virol. 1997;71:6218–6224. doi: 10.1128/jvi.71.8.6218-6224.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delviks K A, Pathak V K. Effect of distance between homologous sequences and 3′ homology on the frequency of retroviral reverse transcriptase template switching. J Virol. 1999;73:7923–7932. doi: 10.1128/jvi.73.10.7923-7932.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeStefano J, Raja A, Cristofaro J. In vitro strand transfer from broken RNAs results in mismatch but not frameshift mutations. Virology. 2000;276:7–15. doi: 10.1006/viro.2000.0533. [DOI] [PubMed] [Google Scholar]
- 12.DeStefano J J, Mallaber L M, Rodriguez-Rodriguez L, Fay P J, Bambara R A. Requirements for strand transfer between internal regions of heteropolymer templates by human immunodeficiency virus reverse transcriptase. J Virol. 1992;66:6370–6378. doi: 10.1128/jvi.66.11.6370-6378.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gabriel A, Willems M, Mules E H, Boeke J D. Replication infidelity during a single cycle of Ty1 retrotransposition. Proc Natl Acad Sci USA. 1996;93:7767–7771. doi: 10.1073/pnas.93.15.7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilboa E, Mitra S W, Goff S P, Baltimore D. A detailed model of reverse transcription and tests of crucial aspects. Cell. 1979;18:93–100. doi: 10.1016/0092-8674(79)90357-x. [DOI] [PubMed] [Google Scholar]
- 15.Hu W-S, Temin H M. Effect of gamma radiation on retroviral recombination. J Virol. 1992;66:4457–4463. doi: 10.1128/jvi.66.7.4457-4463.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu W-S, Temin H M. Genetic consequences of packaging two RNA genomes in one retroviral particle: pseudodiploidy and high rate of genetic recombination. Proc Natl Acad Sci USA. 1990;87:1556–1560. doi: 10.1073/pnas.87.4.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu W-S, Temin H M. Retroviral recombination and reverse transcription. Science. 1990;250:1227–1233. doi: 10.1126/science.1700865. [DOI] [PubMed] [Google Scholar]
- 18.Jones J S, Allan R W, Temin H M. One retroviral RNA is sufficient for synthesis of viral DNA. J Virol. 1994;68:207–216. doi: 10.1128/jvi.68.1.207-216.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Julias J G, Hash D, Pathak V K. E- vectors: development of novel self-inactivation and self-activting retroviral vectors for safer gene therapy. J Virol. 1995;69:6839–6846. doi: 10.1128/jvi.69.11.6839-6846.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klaver B, Berkhout B. Premature strand transfer by the HIV-1 reverse transcriptase during strong-stop DNA synthesis. Nucleic Acids Res. 1994;22:137–144. doi: 10.1093/nar/22.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kulpa D, Topping R, Telesnitsky A. Determination of the site of first strand transfer during Moloney murine leukemia virus reverse transcription and identification of strand transfer-associated reverse transcriptase errors. EMBO J. 1997;16:856–865. doi: 10.1093/emboj/16.4.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lapadat-Tapolsky M, Gabus C, Rau M, Darlix J-L. Possible roles of HIV-1 nucleocapsid protein in the specificity of of proviral DNA synthesis and in its variability. J Mol Biol. 1997;268:250–260. doi: 10.1006/jmbi.1997.0978. [DOI] [PubMed] [Google Scholar]
- 23.Lobel L I, Goff S P. Reverse transcription of retroviral genomes: mutations in the terminal repeats. J Virol. 1985;53:447–455. doi: 10.1128/jvi.53.2.447-455.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo G X, Taylor J. Template switching by reverse transcriptase during DNA synthesis. J Virol. 1990;64:4321–4328. doi: 10.1128/jvi.64.9.4321-4328.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mikkelesen J, Lund A, Duch M, Pedersen F. Recombination of the 5′ leader of murine leukemia virus is accurate and influenced by sequence identity with a strong bias toward the kissing-loop dimerization domain. J Virol. 1998;72:6967–6978. doi: 10.1128/jvi.72.9.6967-6978.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morgenstern J P, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Negroni M, Buc H. Copy-choice recombination by reverse transcriptases: reshuffling of genetic markers mediated by RNA chaperones. Proc Natl Acad Sci USA. 2000;97:6385–6390. doi: 10.1073/pnas.120520497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Negroni M, Buc H. Recombination during reverse transcription: an evaluation of the role of the nucleocapsid protein. J Mol Biol. 1999;286:15–31. doi: 10.1006/jmbi.1998.2460. [DOI] [PubMed] [Google Scholar]
- 29.Olsen J C, Bova-Hill C, Grandgenett D P, Quinn T P, Manfredi J P, Swanstrom R. Rearrangements in unintegrated retroviral DNA are complex and are the result of multiple genetic determinants. J Virol. 1990;64:5475–5484. doi: 10.1128/jvi.64.11.5475-5484.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Rear J J, Temin H M. Spontaneous changes in nucleotide sequence in proviruses of spleen necrosis virus, an avian retrovirus. Proc Natl Acad Sci USA. 1982;79:1230–1234. doi: 10.1073/pnas.79.4.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Panganiban A T, Fiore D. Ordered interstrand and intrastrand DNA transfer during reverse transcription. Science. 1988;241:1064–1069. doi: 10.1126/science.2457948. [DOI] [PubMed] [Google Scholar]
- 32.Patel P H, Preston B D. Marked infidelity of human immunodeficiency virus type 1 reverse transcriptase at RNA and DNA template ends. Proc Natl Acad Sci USA. 1994;91:549–553. doi: 10.1073/pnas.91.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pathak V K, Temin H M. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: deletions and deletions with insertions. Proc Natl Acad Sci USA. 1990;87:6024–6028. doi: 10.1073/pnas.87.16.6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peliska J A, Benkovic S J. Fidelity of in vitro DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Biochemistry. 1994;33:3890–3895. doi: 10.1021/bi00179a014. [DOI] [PubMed] [Google Scholar]
- 35.Peliska J A, Benkovic S J. Mechanism of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Science. 1992;258:1112–1118. doi: 10.1126/science.1279806. [DOI] [PubMed] [Google Scholar]
- 36.Perrino F W, Preston B D, Sandell L L, Loeb L A. Extension of mismatched 3′ termini of DNA is a major determinant of the infidelity of human immunodeficiency virus type I reverse transcriptase. Proc Natl Acad Sci USA. 1989;86:8343–8347. doi: 10.1073/pnas.86.21.8343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pfeiffer J K, Georgiadis M M, Telesnitsky A. Structure-based Moloney murine leukemia virus reverse transcriptase mutants with altered intracellular direct repeat deletion frequencies. J Virol. 2000;74:9629–9636. doi: 10.1128/jvi.74.20.9629-9636.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pfeiffer J K, Topping R, Shin N-H, Telesnitsky A. Altering the intracellular environment increases the frequency of tandem repeat deletion during Moloney murine leukemia virus reverse transcription. J Virol. 1999;73:8441–8447. doi: 10.1128/jvi.73.10.8441-8447.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pulsinelli G A, Temin H M. Characterization of large deletions occurring during a single round of retrovirus replication: novel deletion mechanism involving errors in strand transfer. J Virol. 1991;65:4786–4797. doi: 10.1128/jvi.65.9.4786-4797.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pulsinelli G A, Temin H M. High rate of mismatch extension during reverse transcription in a single round of retrovirus replication. Proc Natl Acad Sci USA. 1994;91:9490–9494. doi: 10.1073/pnas.91.20.9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramsey C A, Panganiban A T. Replication of the retroviral terminal repeat sequence during in vivo reverse transcription. J Virol. 1993;67:4114–4121. doi: 10.1128/jvi.67.7.4114-4121.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rhode B W, Emerman M, Temin H M. Instability of large direct repeats in retrovirus vectors. J Virol. 1987;61:925–927. doi: 10.1128/jvi.61.3.925-927.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 44.Shin N H, Hartigan-O'Connor D, Pfeiffer J K, Telesnitsky A. Replication of lengthened Moloney murine leukemia virus genomes is impaired at multiple stages. J Virol. 2000;74:2694–2702. doi: 10.1128/jvi.74.6.2694-2702.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skalka A M, Boone L, Junghans R, Luk D. Genetic recombination in avian retroviruses. J Cell Biochem. 1982;19:293–304. doi: 10.1002/jcb.240190311. [DOI] [PubMed] [Google Scholar]
- 46.Sugden B. How some retroviruses got their oncogenes. Trends Biochem Sci. 1993;18:233–235. doi: 10.1016/0968-0004(93)90168-m. [DOI] [PubMed] [Google Scholar]
- 47.Telesnitsky A, Goff S P. Strong-stop strand transfer during reverse transcription. In: Skalka A M, Goff S P, editors. Reverse transcriptase. Cold Spring Harbor. N.Y: Cold Spring Harbor Laboratory; 1993. pp. 49–83. [Google Scholar]
- 48.Temin H M. Retrovirus variation and reverse transcription: abnormal strand transfers result in retrovirus genetic variation. Proc Natl Acad Sci USA. 1993;90:6900–6903. doi: 10.1073/pnas.90.15.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Topping R, Demoitie M-A, Shin N H, Telesnitsky A. Cis-acting elements required for strong stop acceptor template selection during Moloney murine leukemia virus reverse transcription. J Mol Biol. 1998;281:1–15. doi: 10.1006/jmbi.1998.1929. [DOI] [PubMed] [Google Scholar]
- 50.van Wamel J L B, Berkhout B. The first strand transfer during HIV-1 reverse transcription can occur either intramolecularly or intermolecularly. Virology. 1998;244:245–251. doi: 10.1006/viro.1998.9096. [DOI] [PubMed] [Google Scholar]
- 51.Wu W, Blumberg B M, Fay P J, Bambara R A. Strand transfer mediated by human immunodeficiency virus reverse transcriptase in vitro is promoted by pausing and results in misincorporation. J Biol Chem. 1995;270:325–332. doi: 10.1074/jbc.270.1.325. [DOI] [PubMed] [Google Scholar]
- 52.Xu H, Boeke J D. High-frequency deletion between homologous sequences during retrotransposition of Ty elements in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1987;84:8553–8557. doi: 10.1073/pnas.84.23.8553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yin P D, Pathak V K, Rowan A E, Teufel R J, Hu W S. Utilization of nonhomologous minus-strand DNA transfer to generate recombinant retroviruses. J Virol. 1997;71:2487–2494. doi: 10.1128/jvi.71.3.2487-2494.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoder K E, Bushman F D. Repair of gaps in retroviral DNA integration intermediates. J Virol. 2000;74:11191–11200. doi: 10.1128/jvi.74.23.11191-11200.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu H, Jetzt A E, Ron Y, Preston B D, Dougherty J P. The nature of human immunodeficiency virus type 1 strand transfers. J Biol Chem. 1998;273:28384–28391. doi: 10.1074/jbc.273.43.28384. [DOI] [PubMed] [Google Scholar]
- 56.Zhang J, Temin H M. 3′ junctions of oncogene-virus sequences and the mechanisms for formation of highly oncogenic retroviruses. J Virol. 1993;67:1747–1751. doi: 10.1128/jvi.67.4.1747-1751.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang J, Temin H M. Rate and mechanism of nonhomologous recombination during a single cycle of retroviral replication. Science. 1993;259:234–238. doi: 10.1126/science.8421784. [DOI] [PubMed] [Google Scholar]
- 58.Zhang J, Temin H M. Retrovirus recombination depends on the length of sequence identity and is not error prone. J Virol. 1994;68:2409–2414. doi: 10.1128/jvi.68.4.2409-2414.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang J Y, Tang L Y, Li T, Ma Y, Sapp C. Most retroviral recombinations occur during minus-strand DNA synthesis. J Virol. 2000;74:2313–2322. doi: 10.1128/jvi.74.5.2313-2322.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]