SUMMARY
The oral pathogen, Aggregatibacter actinomycetemcomitans, uses pga gene locus for the production of an exopolysaccharide made up of a linear homopolymer of β−1,6-N-acetyl-D-glucosamine (PGA). An enzyme encoded by the pgaB of the pga operon in A. actinomycetemcomitans is a de-N-acetylase which is used to alter the PGA. The full length enzyme (AaPgaB) and the N-terminal catalytic domain (residues 25–290, AaPgaBN) from A. actinomycetemcomitans were cloned, expressed and purified. The enzymatic activities of the AaPgaB enzymes were determined using 7-acetoxycoumarin-3-carboxylic acid as the substrate. The AaPgaB enzymes displayed significantly lower de-N-acetylase activity compared with the activity of the deacetylase PdaA from Bacillus subtilis, a member of the CE4 family of enzymes. To delineate the differences in the activity and the active site architecture, the structure of AaPgaBN was determined. The AaPgaBN structure has two metal ions in the active site instead of one found in other CE4 enzymes. Based on the crystal structure comparisons among the various CE4 enzymes, two residues, Q51 and R271 were identified in AaPgaB, which could potentially affect the enzyme activity. Of the two mutants generated, Q51E and R271K, the variant Q51E showed enhanced activity compared to AaPgaB validating the requirement that an activating aspartate residue in the active site is essential for higher activity. In summary, our study provides the first structural evidence for a di-nuclear metal site at the active site of a member of CE4 family of enzymes, evidence that AaPgaBN is catalytically active and that mutant Q51E exhibits higher de-N-acetylase activity.
Keywords: Oral pathogen, biofilm, exopolysaccharide, deacetylation, kinetics
INTRODUCTION
Bacteria in a biofilm are enmeshed together in a self-synthesized matrix that enables the bacterial cells to firmly attach to the underlying surface. Biofilms significantly contribute to immune evasion and to increased resistance to antibiotics (Mah & O’Toole 2001). Exopolysaccharides, consisting of various sugars, are often a major component of the bacterial biofilm matrix (Watnick & Kolter 2000). Exopolysaccharides ensure bacterial survival by enabling abiotic surface attachment and intercellular adhesion (Agladze et al., 2005; Heilmann et al., 1996; McKenney et al., 1998; Wang et al., 2004), providing protection from host innate defenses including phagocytosis and antimicrobial peptides (Vuong et al., 2004a; Vuong et al., 2004b), and increasing virulence (Kropec et al., 2005; Rohde et al., 2010).
Among the oral bacteria, Aggregatibacter actinomycetemcomitans has been linked to localized aggressive periodontitis LAP (Zambon 1985). Recently, accumulating evidence has suggested that A. actinomycetemcomitans is required to initiate LAP, which occurs with a higher frequency not only in children of African American descent (2%) but also Hispanic (1%), Caucasian (0.1%) and Asian descent (Haubek et al., 1997; Shaddox et al., 2010). A major component of the biofilm matrix of A. actinomycetemcomitans is the exopolysaccharide, a linear homopolymer of N-acetyl-D-glucosamine (GlcNAc) residues in β-(1,6) linkage (PGA) (Kaplan et al., 2004b). This exopolysaccharide is functionally and genetically related to extracellular polysaccharide produced by Escherichia coli, Staphylococcus aureus, Staphylococcus epidermidis and Actinobacillus pleuropneumoniae (Izano et al., 2007). The exopolysaccharides from these bacteria, usually referred to as PGA, poly N-acetylglucosamine (PNAG) or polysaccharide intercellular adhesin (PIA), appear to differ in their molecular weight, in the degree of deacetylation of the GlcNAc residues, and in the presence of O-succinate substituents (Joyce et al., 2003; Mack et al., 1996; Maira-Litran et al., 2002; Sadovskaya et al., 2006).
The production of these exopolysaccharides is controlled by proteins encoded by a cluster of four tightly linked genes, icaADBC in S. epidermidis (Gerke et al., 1998) and pgaABCD in E. coli (Wang et al., 2004), A. actinomycetemcomitans and A. pleuropneumoniae (Kaplan et al., 2004a). Among these four proteins, E. coli PgaB (EcPgaB) has been previously suggested to possess a de-N-acetylase domain (Vuong et al., 2004b), which has been demonstrated experimentally to be a de-N-acetylase in vivo (Itoh et al., 2008) and in vitro (Little et al., 2012). A recent report confirmed that the S. epidermidis IcaB (SeIcaB) is also a de-N-acetylase by demonstrating the metal dependent enzyme activity on N-acetylglucosamine oligomers (Pokrovskaya et al., 2013). However, the EcPgaB enzyme is a two-domain protein possessing an additional C-terminal domain compared to SeIcaB. Unlike SeIcaB though, the N-terminal domain of EcPgaB does not possess enzymatic activity (Little et al., 2014). In this regard, the C-terminal domain of EcPgaB has been suggested to assist in the binding of the exopolysaccharide and either may be responsible in guiding the polymer to the N-terminal domain or facilitate export through interaction with PgaA (Little et al., 2014).
While most exopolysaccharides are known to be either neutral or polyanionic (Sutherland 2001), it is known that bacterial enzymes alter exopolysaccharides either during or after secretion. Such changes significantly alter the physicochemical and structural properties of the matrix (Mah & O’Toole 2001). One common mechanism is deacetylation of PGA. In this regard, PGA from A. actinomycetemcomitans is deacetylated up to 10% (Izano et al., 2007), whereas the degree of deacetylation varies in other bacterial PGA. For example, PGA in S. epidermidis occurs in a deacetylated state up to 15 to 20% (Vuong et al., 2004b), whereas E. coli PGA consists of 3–5% (Allignet et al., 2001; Darby et al., 2002; Parise et al., 2007; Wang et al., 2004). It is unclear why deacetylation of exopolysaccharide is limited in these bacteria; a possible scenario is a slowed metabolic activity in biofilms. In this regard, the low level of deacetylation by AaPgaB might be necessary for A. actinimycetemcomitans to cope with the conditions that it encounters during its life cycle in the oral biofilms. Firstly, the biofilm-degrading enzyme from A. actinomycetemcomitans, Dispersin B, is an essential enzyme for the survival of A. actinomycetemcomitans in the oral cavity to assist in the distal site colonization (Stacy et al., 2014). Because Dispersin B uses a substrate assisted hydrolytic mechanism (Ramasubbu et al., 2005), absence of the acetate group in PGA will render the enzyme ineffective. Thus, fully deacetylated or highly deacetylated PGA (highly efficient AaPgaB) may not be beneficial for bacterial survival under conditions with limited nutrients, a common environment in the oral cavity. At a low level of deacetylation, the chains are sufficiently charged to bind to extracellular DNA that is present in the A. actinomycetemcomitans biofilm matrix and assisting the planktonic cells to bind to the matrix. In addition, the charged PGA molecules could also bind to other bacterial cells in the matrix and enhance the aggregation of cells. This intercellular interaction is commonly observed in biofilms, especially in A. actinomycetemcomitans. Our earlier studies in biofilm growth characteristics of mutant strains of A. actinomycetemcomitans lacking PGA synthesis have shown that aggregation is abolished (Izano et al., 2007). Higher electrostatic interactions in highly charged PGA due to efficient deacetylation might affect membrane permeability and could become toxic to the bacterial cells. While it has not been shown that fully deacetylated PGA is either bacteriostatic or bactericidal, chitosan, the β(1,4)-linked polymer of GlcNAc has been shown to be antibacterial for several bacteria (Liu et al., 2004). Also, a mutant strain of S. aureus producing the fully acetylated polymer of PIA is shed into the medium and not retained on the cellular surface resulting in deficiencies in biofilm formation and surface attachment (Vuong et al., 2004a).
The importance of PGA in bacterial biofilms is well established; PGA could provide a mechanism to improve fitness in many pathogenic organisms (Cywes-Bentley et al., 2013; Shanmugam et al., 2015; Whitfield et al., 2015). Recent studies support this suggestion by demonstrating that pga mutant strain was outcompeted by the wild type strain of E. coli in a urinary tract infection model (Subashchandrabose et al., 2013), and that in A. actinomycetemcomitans, lack of PGA production affected its colonization efficiency in a rat model (Shanmugam et al., 2015). While there have been studies highlighting the genes involved in the production of PGA in A. actinomycetemcomitans (Kaplan et al., 2004a), structural or functional information about the de-N-acetylase activity of PgaB from A. actinomycetemcomitans (AaPgaB) is lacking. In the present study we investigated AaPgaB, which is a single chain polypeptide consisting of 638 amino acids and has a predicted molecular mass of 73 kDa. The AaPgaB enzyme is classified as a member of CE4 enzymes as defined by the CAZY database [http://www.cazy.org/(Coutinho & Henrissat 1999)], which includes several members that share the primary structure assigned as the “NodB homology domain” (Caufrier et al., 2003). The focus of the present report is several fold: 1) to determine whether or not the N-terminal domain of AaPgaB (AaPgaBN) exhibits catalytic activity; 2) to determine whether or not the presence of the C-terminal domain has any effect on the activity displayed by the full-length enzyme AaPgaB; and 3) to determine the three dimensional structure of AaPgaBN and to test whether or not structure-guided mutagenesis could be utilized to enhance the catalytic efficiency of the AaPgaB. Such an enhancement would partially validate the mechanism of deacetylation proposed earlier for family 4 carbohydrate esterases (CE4) enzymes (Blair et al., 2005), but also improve our understanding of the poor catalytic efficiency exhibited by the de-N-acetylases, in general.
MATERIALS AND METHODS
Cloning, expression and purification of AaPgaBN, AaPgaB and BsPdaA.
The AapgaB gene (73–1914 bp corresponding to residues 25–638 of the mature PgaB enzyme) and AapgaBN gene (73–870 bp corresponding to residues 25–290 of the mature PgaB enzyme) were amplified from the genomic DNA of A. actinomycetemcomitans (strain JP2 (Haubek et al., 1996)) using a forward primer, which inserts a NdeI restriction site at position 73 of pgaB, and a reverse primer with a XhoI restriction site (Table 1). The amplified PCR products were digested with NdeI and XhoI and ligated into the corresponding sites of the pET29b vector to yield plasmid pSM48 (AaPgaBN) and subjected to DNA sequence analysis. The plasmid was transformed into Rossetta™ cells (Novagen). We also cloned and expressed the de-N-acetylase enzyme BsPdaA from Bacillus subtilis (Strain 168) which has the critical aspartate for comparison to the enzyme activities of Aa enzymes. The BspdaA gene (70–789 bp corresponding to residues 24–263 of the mature PdaA enzyme) was cloned into pET29b using In-Fusion™ PCR Cloning method (Clontech) to generate plasmid pCP1 and transformed into Stellar competent cells (Clontech). The restriction sites (NdeI and XhoI) were introduced in the primers to facilitate cloning into pET29b vector (Novagen) and to utilize the polyhistidine tag in the vector, although two non-native amino acids will be present at the C-terminal end of the expressed proteins. Transformants were inoculated into 100 mL of Luria-Bertani broth supplemented with 50 μg of kanamycin per mL for 16 h. A 2-liter Erlenmeyer flask containing 500 mL of Luria-Bertani broth supplemented with 30 μg of kanamycin per mL was inoculated with 10 mL of an overnight culture. The flask was incubated at 37 °C with agitation (200 rpm) until the optical density at 600 nm reached 0.6 (1.5 h). IPTG was added to a final concentration of 0.1 mM and the flask was further incubated for an additional 4 h with agitation. The cells were harvested by centrifugation for 15 min at (6000 x g, 4°C) and the cell pellet was stored at –80°C until further use.
Table 1:
Primers used in the study to generate various deactylase constructs. The underlined segment corresponds to the restriction sites.
| S. No | Primer | Primer sequence (5’ to 3’) | Type |
|---|---|---|---|
| 1 | AaPgaBN25−290 | GGA ATT CCA TAT GGA TAG ATA CGG CGT GCCG CTC GAG CAC ACC ATC AAG GAA ATT TTT AAT G | Forward Reverse |
| 2 | AaPgaBN25−638 | GGA ATT CCA TAT GGA TAG ATA CGG CGT GCCG CTC GAG TGG TTT ACC TGC TTG AGT ATT C | Forward Reverse |
| 3 | Q51D (Mutation) | GAA AAA CAG TAT TTC CCG GAT ACC ATT TCC GCC AAT TTG CAA ATT GGC GGA AAT GGT ATC CGG GAA ATA CTG TTT TTC |
Forward Reverse |
| 4 | Q51E (Mutation) | GAA AAA CAG TAT TTC CCG GAA ACC ATT TCC GCC AAT TTG CAA ATT GGC GGA AAT GGT TTC CGG GAA ATA CTG TTT TTC |
Forward Reverse |
| 5 | R271K (Mutation) | GGT GAT AGA TAC GTC GGT AAA TTA TTA ATT GAC ACG GAA ACC GGT TTC CGT GTC AAT TAA TAA TTT ACC GAC GTA TCT ATC ACC |
Forward Reverse |
| 6 | BsPdaA | AAG GAG ATA TAC ATA TGG TGC CGA ATG AGC CGA TTA ATT G GGT GGT GGT GCT CGA GCA AAG ACG GCA GCC TCA TTT CTT TC |
Forward Reverse |
The cell pellet was thawed at room temperature and resuspended in 20 mL of 20 mM Tris-HCl, pH 8.5, 500 mM NaCl, 5 mM imidazole (Buffer A). The cell suspension was then sonicated on ice for 20 min (30 s pulse and 30% duty cycle) by using a Branson model 450 sonicator equipped with a microprobe to lyse the cells. The cell debris was pelleted by centrifugation at 15,000 x g for 20 min at 4°C. The supernatant containing the recombinant protein was filtered through a 0.45 μm membrane filter (Corning) and purified by immobilized metal affinity chromatography using an ÄKTA FPLC system controlled by UNICORN™ software version 5.1 (GE Healthcare). Briefly, a 5 mL HisTrap chelating HP column (GE Healthcare) prepacked with Ni2+ Sepharose was equilibrated with Buffer A prior to sample injection. Clarified supernatant (2 mL) was loaded on the column via a sample loop at a flow rate of 1.0 mL/min. Unbound proteins were eluted with a 5 column volume of Buffer A. The bound proteins were eluted with a gradient of imidazole (5 – 200 mM in Buffer A, achieved in 40 column volumes). Fractions (1 mL) of the eluate were collected and assayed for the presence of proteins by SDS-PAGE. Fractions containing protein of the appropriate molecular mass were pooled and dialyzed against 20 mM Tris-HCl, pH 8.5 by using a 10 kDa cutoff dialysis membrane at 4°C (3x buffer exchange with 1000 mL each). The dialyzed solution was concentrated using Amicon CentriPlus YM-10 (Millipore) membranes, aliquoted and stored at −80°C. To incorporate selenium to generate Se-Met derivative for structural analysis, bacteria were cultured in M9 medium supplemented with 50 mg/L SeMet, lysine, threonine, phenylalanine, leucine, and isoleucine. The Se-Met derivative (AaPgaBN25−290) was also expressed and purified as above. For kinetic experiments, full-length enzyme AaPgaBN25−638 and two mutants (Q51E and R271K) were also generated. Primers used for the mutations are given in Table 1. Mutant proteins and the BsPdaA enzyme were expressed and purified as per the procedure used for the wild type enzyme AaPgaB with some modifications: the R271K mutant and BsPdaA enzymes were expressed at 18°C and 25°C, respectively. For R271K, the kanamycin concentration was reduced to 20 μg per mL and induced with 0.05 mM of IPTG to obtain soluble protein. Molecular weight and purity of the proteins were characterized by mass spectrometry analysis and gel electrophoresis (data not shown).
Crystallization and data collection of AaPgaBN.
Initial crystallization trials were carried out with the Mosquito robot using 100 nL of the protein (AaPgaBN25−290 or Se-MetAaPgaBN25−290) at 10 mg/mL and 100 nL of the reservoir solution from crystallization screens from Hampton Research, Molecular Dimensions and Qiagen. Several attempts to crystallize these variants or other shorter variants were otherwise unsuccessful. However, one diffraction quality crystal was obtained using the Se-Met derivative with the following condition: 20% PEG 6000, 0.01 M ZnCl2 and 0.1 M HEPES at pH 7.0. Although the Se-Met variant was successfully crystallized once, further attempts were unsuccessful. The lone Se-Met crystal was mounted on a loop (Hampton Research) and flash frozen to 100°K in liquid nitrogen. Diffraction data were collected using in house Rigaku R-axis IV++ at liquid nitrogen temperatures with an imaging plate. A total of 540 frames were collected with an exposure time of 3 minutes and an oscillation interval of 0.5° to give a 99% complete data set to 2.0 Å. Intensity data were integrated, scaled and reduced to structure factor amplitudes using HKL2000 suite of programs (Otwinowski & Minor 1997). Data collection and processing statistics are given in Table 2. The crystal is orthorhombic and belongs to the Space Group P212121 with two molecules in the asymmetric unit and the following cell parameters: a=65.2, b=67.1, c=101.0 Å.
Table 2.
Data collection and refinement statistics
| Data collection statistics | |
| Space group | P212121 |
| Unit-cell parameters (Å) | a=65.2, b=67.1, c=101.0 |
| Resolution range (Å) | 50.0–2.05(2.12–2.05) |
| Reflections (measured/unique) | 268119/28980 |
| Crystal to detector distance | 150 mm |
| Completeness (%) | 99.9(100.0) |
| Rmerge* (%) | 10.1(58.1) |
| Multiplicity | 9.4(7.8) |
| I/σ(I) | 34.1 (4.7) |
| Refinement statistics | |
| Resolution range (Å) | 42.5–2.05 |
| Rwork/Rfree (%) | 16.2/22.1 |
| Estimated coordinate error (R/Rfree) | 0.21/0.18 |
| No. of atoms | |
| Protein | 4248 |
| Zinc | 7 |
| Chloride | 2 |
| Water | 258 |
| Average B-factor (Å 2 ) | |
| Protein | 23.2 |
| Zinc | 16.4 |
| Chloride | 16.9 |
| Water | 26.4 |
| R.m.s.d. bond lengths (Å) | 0.017 |
| R.m.s.d. bond angles (°) | 1.75 |
| Ramachandran plot, residues in (%) | |
| Most favorable regions | 87.4 |
| Additional allowed regions | 10.7 |
| Generously allowed regions | 0.4 |
| Disallowed regions | 1.5 |
Values in parentheses are for the outer shell.
Rmerge = Σhkl Σi |Ii(hkl) - <I(hkl)>|/Σhkl Σi |Ii(hkl), where <Ii(hkl)> is the intensity of an observation, <I(hkl)> in the mean value for that reflection and the summations are over all equivalents.
Structure solution.
Structure was solved by molecular replacement method using E.coli N-terminal domain (PDB entry 3VUS) as the template. Before performing molecular replacement, side-chains of the template model were removed and supplied as poly-ala model to minimize any model bias. Molecular replacement was carried out using AutoMR and subsequent automated model building by AutoBuild in PHENIX software suite (Adams et al., 2010). After the initial rounds of rigid body refinement, the model consisting of two copies of AaPgaBN (Chain A, residues Met25 to Asp288 and Chain B, residues Asp26 to Leu287) could be clearly traced in the electron density maps. The model from AutoBuild produced 465 residues out of which 457 residues had electron density corresponding to the A. actinomycetemcomitans sequence with a starting Rwork and Rfree of 25% and 31%, respectively. Unlike the E. coli N-terminal domain structure (EcPgaBN; PDB Code: 3VUS) or the full length structure (EcPgaB; PDB Code: 4F9J), there were no breaks in the electron density map. To further minimize any model bias, a composite omit map was computed and used to trace the chains A and B corresponding to the two molecules of the asymmetric unit. Residue Met25 of the B chain, the C-terminal residues from 288 to 290 (Chain A) and 287 to 290 (Chain B) and the six His residues of the His-tag from both chains were not modeled in the refinement since no clear density was present in the 2Fo-Fc omit maps for these residues. These regions were assumed to be disordered and are not reported in the final structure. COOT (Emsley & Cowtan 2004) was used for manual model building and refinement was carried out by REFMAC (Murshudov et al., 2011) in CCP4 suite (Winn et al., 2011). Water molecules were added and the densities of these water molecules were carefully evaluated before being included in the refinement. XAS experiments were not attempted due to our inability to reproduce the crystallization condition that generated the crystal used in the data collection. Identity of the metal ion was deduced from difference electron density maps and anomalous difference maps. When necessary, the occupancy of the metal ions was refined and the coordination geometry was evaluated during the refinement process. Final refined model had Rwork and Rfree of 16.2% and 22.1%, respectively. PyMOL (DeLano 2002) was used to generate figures. The stereochemical quality of the structure was verified using ProCheck (Laskowski et al., 1993).
De-N-acetylation activity using a continuous assay.
The chromogenic acetyl ester, 3-carboxyumbelliferyl acetate, was used as a substrate to characterize kinetic parameters of enzymatic activity (Chibba et al., 2012). Typically, the enzyme concentration was set at 1 μM and the final substrate concentration varied from 0.16 to 1.25 mM. The substrate was solubilized using DMSO with the final concentration at 20%. The reaction mixtures were incubated with 10 μM CoCl2 for 1h before adding the substrate. Typically, the reaction mixture consisted of the substrate and enzyme in a total volume of 100 μL in PBS buffer, pH 7.4 containing 10 μM of CoCl2. We also tested the AaPgaB with ZnCl2 as counter ion for comparison purposes. For the BsPdaA enzyme, the reaction mixture consisted of 10 μM CdCl2. The reaction was monitored for 10 minutes with an excitation wavelength of 386 nm and the emission wavelength of 446 nm. A standard curve was generated using the product of the reaction, 7-hydroxycoumarin-3-carboxylic acid. Full kinetic characterization of the enzymes was carried out in triplicate (technical) and repeated twice (biological) with similar results between the biological replicates. Kinetic parameters were calculated using GraphPad Prism software (Version 6) using a non-linear regression analysis. The enzymes, AaPgaBN and AaPgaB, purified from the affinity column, were incubated separately with 10 mM EDTA (10,000 fold excess) for 1 h at room temperature as well as at 50°C. Enzymes with no addition of EDTA at room temperature and at 50°C were also prepared as controls. After incubation with EDTA, different metal ions (Co2+, Zn2+, Ni2+ as chlorides at 10 μM, final concentration) were added to the reaction mixture and incubated for 1 h. When necessary, the enzyme solution was dialyzed before metal ions were added using freshly prepared metal-free 20 mM Tris.Hcl buffer, pH 8.5. Using the EDTA treated enzymes, the de-N-acetylation activity was determined.
RESULTS
Expression of soluble de-N-acetylase domain of AaPgaB.
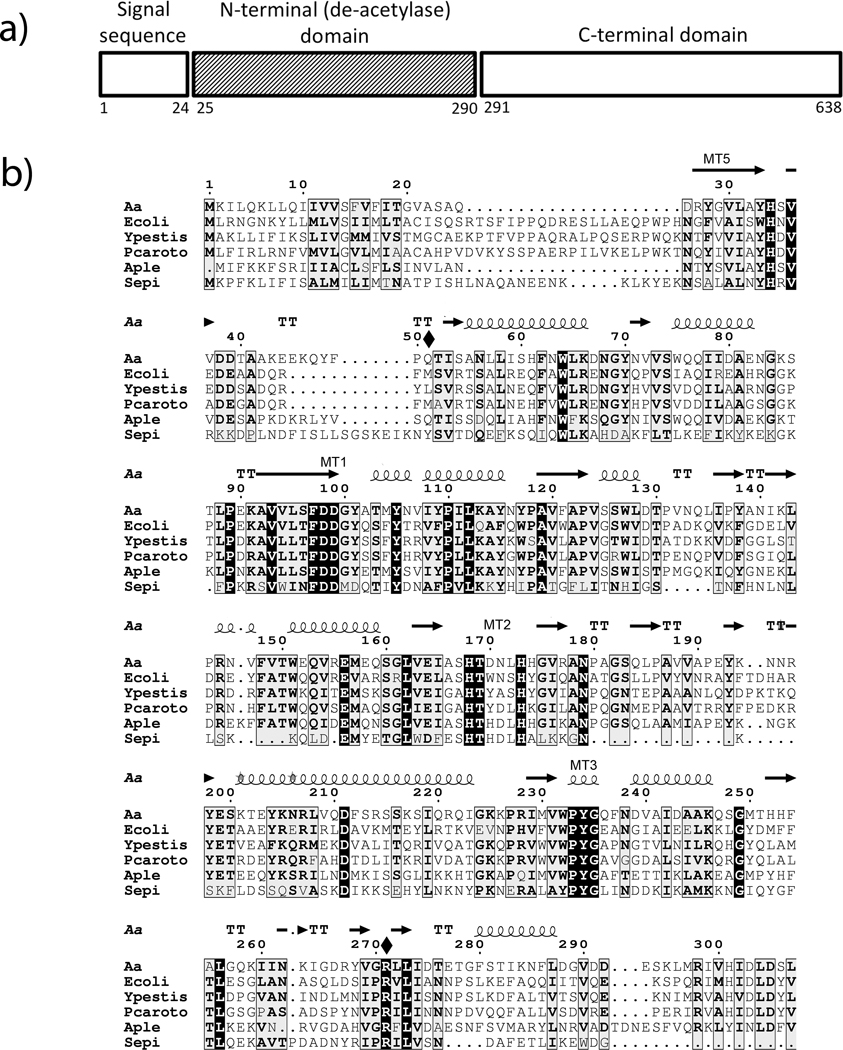
The sequence homology among PgaB proteins and the crystal structure of the EcPgaB (Nishiyama et al., 2013) was used to assign the sequence to the N-terminal domain of AaPgaB (Fig. 1). The N-terminal domain of AaPgaBN exhibits 25% identity to SeIcaB and the full length enzyme, AaPgaB shows 39% identity to EcPgaB (40% with EcPgaB N-terminal domain) suggesting that AaPgaB might also possess a de-N-acetylase domain. We chose this region to test whether or not the N-terminal domain alone exhibits catalytic activity. In this study, the pgaB gene corresponding to AaPgaB and the variant AaPgaBN were cloned and expressed in E. coli. The gene sequences of AapgaBN constructs were confirmed by nucleotide sequencing and suggested that the deduced amino acid residue at position 128 of AaPgaBN is an Asp residue instead of a Glu compared to the PgaB sequence from strain HK1651 of A. actinomycetemcomitans. After purification using Ni2+ affinity chromatography, the yield of the expressed AaPgaBN protein and that of the corresponding Se-Met derivative was estimated to be 30 mg per liter of culture. N-terminal sequence analysis was confirmed with the mature sequence with a Met at the N-terminus from the engineered NdeI restriction site. The mutants of the full-length enzyme were also expressed in a similar manner and purified to yield up to 10 mg per liter of culture. Further, to compare the kinetic parameters, the de-N-acetylase enzyme PdaA from B. subtilis (a well-studied member of the CE4 enzyme family) was also purified with yields up to 20 mg/L.
Figure 1.
Sequence homology among PgaB proteins. a) A schematic of the AaPgaB protein showing the various domains in the sequence. b) Sequence alignment of the N-terminal domain of PgaB proteins in A. actinomycetemcomitans (Aa),. E. coli (Ecoli), Y. pestis (Ypestis), P. coratovorum (Pcorato), A. pleuropneumoniae (Aple), and S. epidermidis (Sepi). The secondary structures as deduced from the AaPgaBN structure, the two mutated residues (shown with a solid diamond) and the conserved motifs are also shown above the sequence. Numbering is based on the A. actinomycetemcomitans sequence.
Overall structure of AaPgaBN.
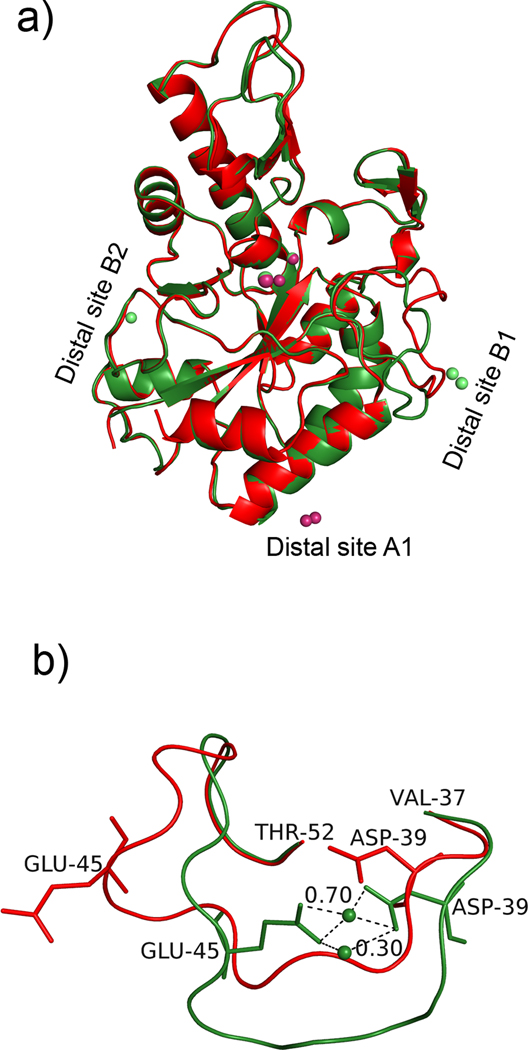
The Se-Met derivative of AaPgaBN, encompassing residues 25–290 was expressed, purified and crystallized. Initial molecular refinement methods using crystal structures of several CE4 enzymes (Example: PDB Code 2C1G; Blair et al., 2005) were unsuccessful. Later, the crystal structure of AaPgaBN was solved using the EcPgaBN structure (Nishiyama et al., 2013) using molecular replacement. The primary structure of AaPgaBN shares 40% identity with EcPgaBN (Nishiyama et al., 2013) and this level of homology is reflected in the three dimensional structures of both enzymes being similar (rmsd 1.1 Å). Model bias was minimized using the molecular replacement procedure as described in the Methods section. Like EcPgaBN (Nishiyama et al., 2013), and the corresponding full-length enzyme (Little et al., 2012), the structure of AaPgaBN adopts (β/α) barrel fold (Fig. 2a) as observed in other family 4 esterases (CE4 enzymes) and contains two molecules (chains A and B, respectively) in the asymmetric unit. The chains A and B of AaPgaBN fit well with each other (rmsd 0.27 Å) except for the loop region Val37 to Thr52 (Fig. 2b). Ordered density was missing for residue 25 (Se-Met of chain B) and few residues at the C-terminal end including the His-Tag residues.
Figure 2.
Structure of AaPgaBN. a) Cartoon representation of the structure of AaPgaBN. Chains A (green) and B (red) are superposed to show the overall fit and the location of various metal ions. Three distal metal ion sites are shown; b) Superposition of residues Val37-Thr52 in chains A and B.
The final refined model consists of 526 amino acid residues (from chains A and B), 258 water molecules, three metal ions and a chloride ion for each chain and a fourth metal ion associated with only chain B. There are also 5 Se-Met residues in chain A (one at the N-terminus is highly disordered; <B2> = 100 Å2) and 4 in chain B. While most of the residues behaved well stereochemically, as evident from the Ramachandran plot (Laskowski et al., 1993), six residues were observed in the disallowed regions. These were, His34, Asp99, Ala139, Thr169 and Asn195 from Chains A and B. Repeated fitting and refinement of these residues did not improve their status. At least two of them are at the active site (His34 and Asp99) and one (Thr169) follows an active site residue (His168). Unlike the disordered loop region between the residues 196 and 200 in Chain A of EcPgaBN (PDB Code: 3VUS; data not shown), the corresponding loop in AaPgaBN is clearly visible in the difference electron density maps and has been fitted and refined in the AaPgaBN. The overall structure was refined to a crystallographic Rwork and Rfree of 16.2% and 22.1%, respectively, at 2.0 Å resolution.
Active site region.
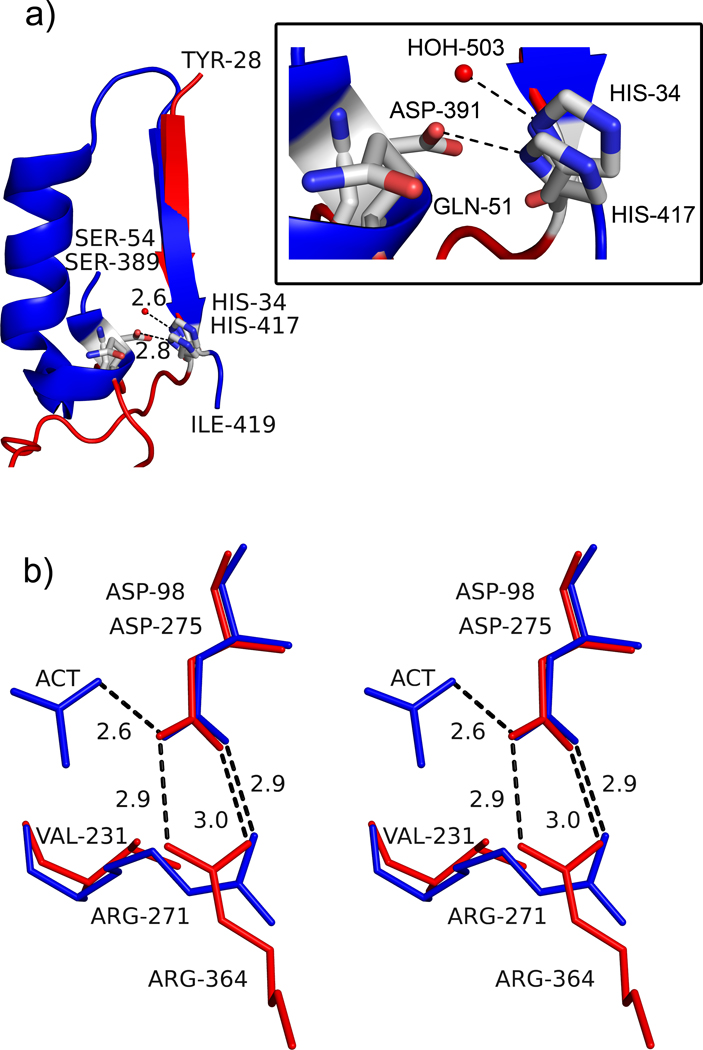
All CE4 enzymes studied thus far, through sequence alignment analysis, have been suggested to have five conserved motifs, MT1 through MT5 (Blair et al., 2005). These conserved motifs present critical amino acids in the active site pocket. Although the active site architecture of the AaPgaBN resembles that of other CE4 enzymes (Fig. 3a), AaPgaBN and EcPgaBN (Nishiyama et al., 2013) exhibit significant differences. Most important difference being the order in which residues comprising the conserved motifs MT1 through MT5 occurs (Blair et al., 2005). For example, MT5, which normally is located on the last β-strand of the TIM-barrel, is present on the first β-strand. This leads to motifs in AaPgaBN to be circularly permuted (also observed in EcPgaBN) (MT1 through MT5 (for CE4 enzymes) vs. MT5-MT1 through MT4 (for AaPgaBN)); nevertheless, such a permutation does not alter the spatial location of critical active site residues. As noted elsewhere in other studies describing the EcPgaB enzymes (Little et al., 2012; Nishiyama et al., 2013), a critical aspartate that is necessary to activate the general acid His34/56/417 (Andres et al., 2014; Blair et al., 2005; Little et al., 2012) is also absent in AaPgaBN. While this aspartate is conserved in many CE4 enzymes, its function is not clear. In a recent study, using crystal structures of apo, as well as saccharide complexes of the de-N-acetylase from Vibrio cholera (VcCDA), the mechanistic aspect of CE4 enzymes has been further clarified (Andres et al., 2014). Unlike the SpPgdA (Blair et al., 2005), the conserved aspartate in VcCDA does not interact with the His34 equivalent. In any case, in AaPgaB, like EcPgaB, the role of protonating His34, instead is taken up by a water molecule (Fig. 4a). Two other motifs, MT1 and MT2 are well conserved in sequence as well as in structure with other known CE4 enzymes. In MT3, a critical arginine that should coordinate with the conserved active site residue Asp98 is absent and is replaced with a valine. However, Arg271, which is located on a different β-strand of the barrel, provides a salt bridge to Asp98 and projects into the active site from the opposite side of the active site pocket (Fig. 4b). Such subtle changes in the active site architecture render the active site of AaPgaBN to be deeper than found in SpPgdA but retain the metal binding site intact (Fig. 3a). Altogether, the permutation in the position of MT5 motif, missing or misplaced active site residues in other motifs appears to play a role in lowering the enzyme activity compared to classical CE4 enzymes (see below).
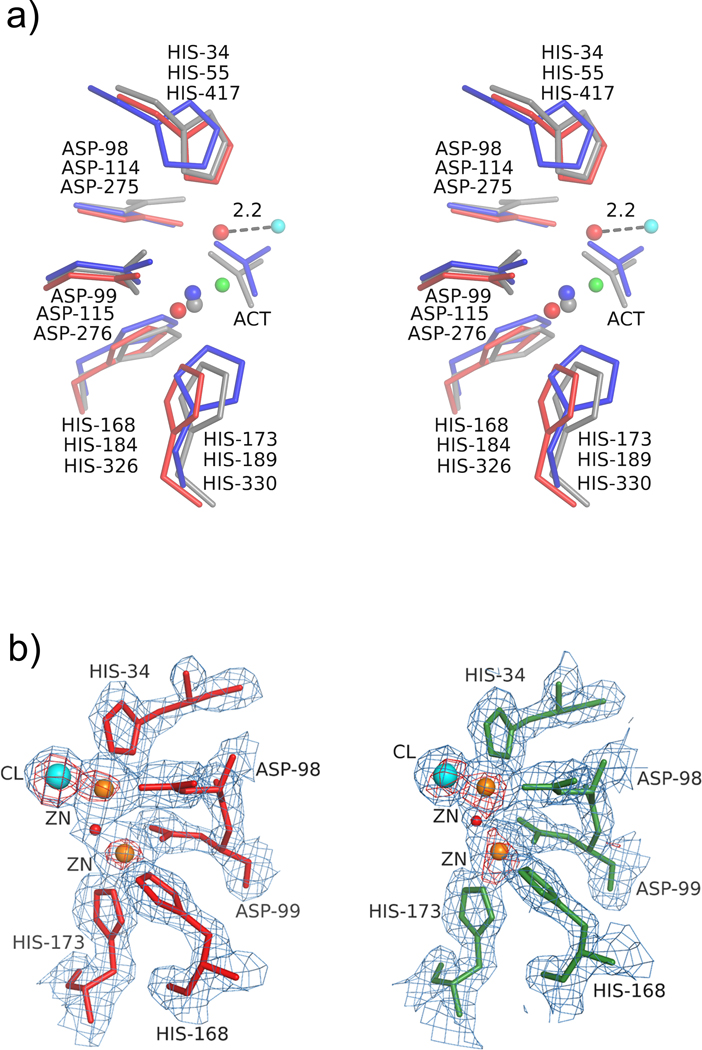
Figure 3.
Stereoview of the metal binding site in AaPgaBN. a) The critical residues in three de-N-acetylases, AaPgaBN (red, chain A), EcPgaB (grey) and SpPgdA (blue) are superposed; b) The anomalous difference density map (red contours) superposed with the 2Fo-Fc map (light blue) around the active site (Motifs MT1 and MT2) in chains A (left) and B (right) showing the metal ions (brown) and chloride (aqua). The nucleophilic water molecule is shown as a red sphere.
Figure 4.
Active site architecture in AaPgaBN. a) Part of the structure of AaPgaBN and SpPgdA showing MT5 motif and how the residues His34/His417 are presented into the active site. The red sphere represents the activating water molecule in the structure of AaPgaBN. The SpPgdA structure has a longer, more elaborate ordered region to project H417 and D391 whereas the AaPgaBN has a shorter less ordered segment to project His34 and Gln51 into the active site. The inset shows details of the interaction with residues and water molecule; b) Stereoview of the orientation and interaction of Arg271 to the catalytic base, Asp98 in AaPgaBN. Note the two electrostatic bonds between Arg271 and Asp98 (catalytic base shown in red) whereas the equivalent residues of SpPgdA, Arg364 and Asp275 (shown in blue), only have one. The direction of the interaction in AaPgaBN is altered and a Val231 is located at the position where Arg364 (SpPgdA) is present.
Metal binding sites.
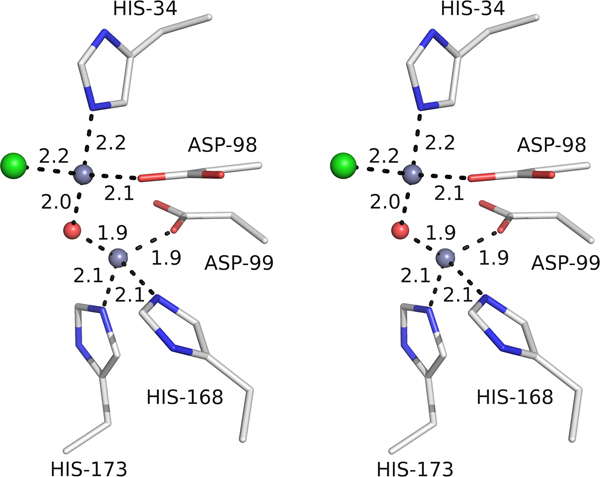
Three large peaks were observed in the difference density maps (two of which were at 24 σ and the third at 12 σ) around the active site and were assumed to be metal ions (Fig. 3a). After the initial successful attempt for crystallization of the Se-Met AaPgaBN, further attempts failed. Because of the paucity/non-availability of crystals in the condition from which Se-Met crystals were obtained, EXAFS analysis of these crystals could not be carried out to identify the bound metal ion. Nevertheless, the metal binding site in AaPgaBN, involving residues His34, Asp98, Asp99, His168 and His173, is highly conserved among the various de-N-acetylase enzymes (Fig.3b). Unique to the structure of AaPgaBN, is the presence of an additional metal ion located in the space occupied by the leaving acetyl group (as deduced in the structure of EcPgaB) during de-N-acetylation (Little et al., 2012). Thus, two of the large difference density peaks at the active site were modeled as metal ions and the third with lower difference density peak (12 σ) was modeled as a chloride ion (anion used in the crystallization buffer). The chains A and B of AaPgaBN also contain a third metal ion at a distal site (Site A1 and B1; Fig. 2a), which were included in the refinement with partial occupancy. Difference density peaks were further analyzed in the maps and one additional peak was assigned as a metal ion (site A2, chain A; not shown) and was included in the refinement. Since the identity of the metal ion is unclear, the refined structure was analyzed using the metal binding site validation server (www.csgid.org) (Zheng et al., 2014) and the metal was assigned as zinc (see discussion below).
Structure-guided mutagenesis to probe the mechanism of action of CE4 enzymes.
Structural superposition of SpPgdA, AaPgaBN and EcPgaB were analyzed to identify an aspartate that has been suggested to activate the catalytic acid (Blair et al., 2005). In both SpPgdA and AaPgaBN, the His417/His34 residue occurs in a loop connecting a β-strand and an α-helix (Fig. 4a). The helix harboring the activating residue though is conspicuously absent in AaPgaBN. We noted that residue Gln51 in AaPgaB is located in close proximity to His34. Manual modeling using COOT revealed that replacing Gln51 by a Glu would place the side chain at an optimal distance to activate His34. To test this hypothesis, a full-length mutant protein (Q51E) was expressed in E. coli and purified.
In the proposed mechanism of deacetylation (Blair et al., 2005), an aspartate is the acid base (Asp98 in AaPgaB), which has one of its oxygen atoms involved in the electrostatic interaction with an Arg residue. This renders the second oxygen atom to be involved in the catalytic process. However, in the structure of AaPgaBN and EcPgaB, the equivalent Asp (Asp98 in AaPgaB) is involved in two electrostatic interactions, which prevents either of the oxygen atoms to participate in the catalysis (Fig. 4b). In AaPgaB, because Arg271 residue is part of MT5 motif rather than MT3 motif, the Asp98 is forced to enter into two stabilizing salt bridge interactions with Arg271. We reasoned that mutating Arg271 into a Lys residue would only allow for one electrostatic interaction while approximating the length of the side chain of Arg. The mutant R271K was expressed and purified. The mutants generated were subjected to full kinetic characterization using the continuous assay and compared against the wild type enzyme (see below).
Enzymatic assay using chromogenic acetyl ester.
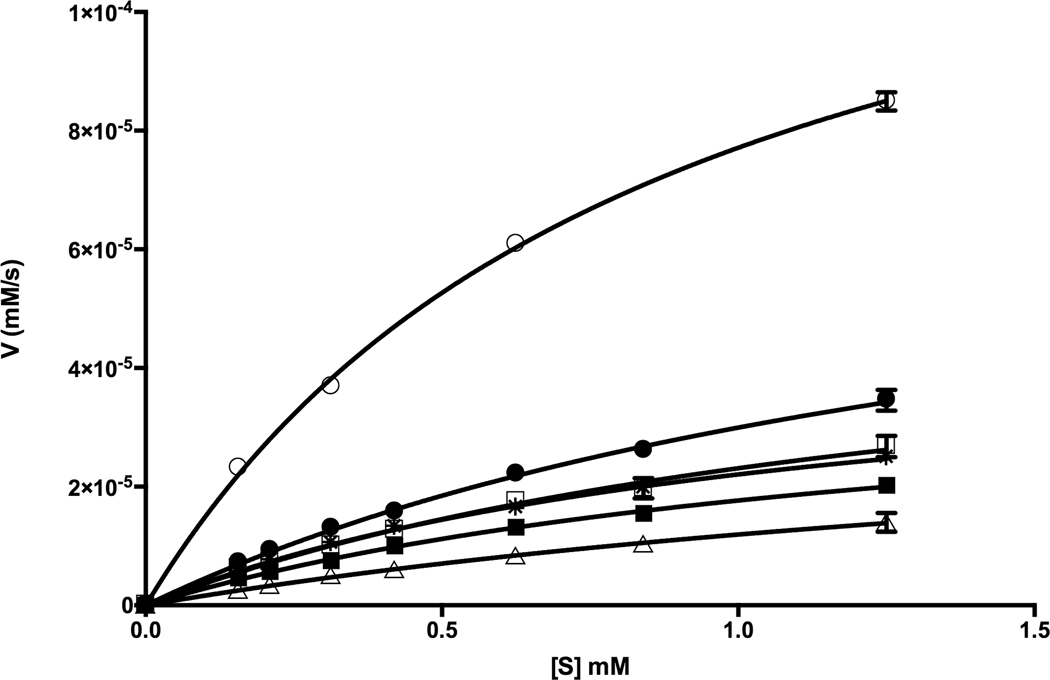
For kinetic characterization of the enzymes, a continuous assay using a chromogenic substrate 3-carboxyumbelliferyl acetate (Chibba et al., 2012) was used (Fig. 5). For AaPgaB, we used both Co2+ and Zn2+ as co-factors. The kinetic parameters for the various AaPgaBN enzymes and the full-length variants are given in Table 3. Interestingly, although the N-terminal variant, AaPgaBN has a lower efficiency compared to the full-length enzyme, this N-terminal variant is active unlike the N-terminal domain of EcPgaB (Nishiyama et al., 2013). Compared to AaPgaBN, the full-length enzyme has a two-fold higher catalytic efficiency. The kinetic efficiency of mutant Q51E of AaPgaB is 22% higher than that of AaPgaB. Thus, the presence of an acid group (corresponding to Asp391 of SpPgdA) in the vicinity of His34 could aid to carry out the acid/base catalysis in the mechanism proposed for de-N-acetylases (Blair et al., 2005). In addition, our results show that when the buried arginine, R271 is replaced with Lys (R271K), the catalytic efficiency of the mutant changed only by 5% compared to AaPgaB. Interestingly, for comparison, the efficiency of another enzyme that has the key aspartate at the active site (BsPdaA) is >300% higher than the full-length AaPgaB enzyme (Table 3; Fig. 5). In addition, the kinetic parameters are virtually unchanged when zinc is used as the cofactor (Table 3). Experiments using EDTA incubated enzymes, at room temperature or at 50°C, did not alter the activity of either the AaPgaBN or AaPgaB enzymes. Incubation with various metal ions after incubation with EDTA also did not change the enzyme activity (data not shown).
Figure 5.
AaPgaBN enzymology. Steady-state kinetics using ACC as the substrate at different substrate concentrations (0.16 −1.25 mM). Data presented were obtained from three technical replicates. AaPgaBN (Δ), AaPgaB (☐), AaPgaB (■; Zn2+), Q51E AaPgaB (●), R271K AaPgaB (*), BsPdaA (O).
Table 3.
CE4 de-N-acetylase enzyme kinetics values with ACC as substrate. The values in parentheses are standard deviations.
| Enzyme | kcat (s−1) | Km (mM) | kcat/Km (s−1 mM−1) | Reference |
|---|---|---|---|---|
| AaPgaBN WT | 0.039 (± 0.007) | 2.289 (± 0.532) | 0.017 | This study |
| AaPgaB WT | 0.056 (± 0.005) | 1.442 (± 0.185) | 0.039 | This study |
| AaPgaB Q51E | 0.079 (± 0.005) | 1.646 (± 0.152) | 0.048 | This study |
| AaPgaB R271K | 0.047 (± 0.003) | 1.124 (± 0.117) | 0.042 | This study |
| AaPgaB WT (with Zn2+) | 0.042 (± 0.003) | 1.368 (± 0.131) | 0.031 | This study |
| BsPdaA | 0.144 (± 0.003) | 0.867 (± 0.038) | 0.164 | This study |
| SlAxeA | 118 | 1 | 118 | (Taylor et al., 2006) |
| EcPgaB | 0.013 | 1.2 | 0.011 | (Little et al., 2012) |
| SeIcaB | 0.0013 | 0.5 | 0.0025 | (Pokrovskaya et al., 2013) |
DISCUSSION
De-N-acetylase domain of AaPgaB.
Biosynthesis of PGA in A. actinomycetemcomitans requires a set of four gene products, PgaA, PgaB, PgaC and PgaD. Based on multiple sequence alignment (Fig. 1) and recent structural evidence (Little et al., 2012), AaPgaB sequence can be considered to possess two domains: an N-terminal domain consisting of about 290 amino acid residues (of which the first 24 correspond to a signal sequence) and the C-terminal domain of about 350 residues. Since PGA from E. coli, S. epidermidis, A. pleuropneumoniae and A. actinomycetemcomitans have been shown to contain deacetylated GlcNAc residues (Izano et al., 2007; Mack et al., 1996; Wang et al., 2004), we hypothesized that AaPgaB might also function as a de-N-acetylase. It is evident from the results reported here that the protein AaPgaB is enzymatically active and could be considered to be a de-N-acetylase. What is interesting is that the N-terminal domain of AaPgaB (AaPgaBN) is also active. This observation is in sharp contrast to the results obtained for EcPgaB (Little et al., 2014), which required both the N- and C-terminal domains to be present. In a recent study, the N-terminal domain of BpsB (BbBpsB), the PgaB equivalent from Bordetella bronchiseptica, was shown to be catalytically active (Little, et al., 2015). Significant and unique structural features were ascribed to the activity of the N-terminal domain of BbBpsB; no such difference exists between AaPgaB and EcPgaB suggesting that subtle changes in the sequence between the two enzymes could come into play in the exhibition of activity.
Another interesting feature of the de-N-acetylase enzymes is the use of a metal cofactor for catalysis. The nature of the divalent metal appears to be enzyme-dependent. Previously, the structures of EcPgaBN and EcPgaB were crystallized with Ni2+, Co2+, Fe3+ (Little et al., 2012) or Zn2+ (Nishiyama et al., 2013). The activity of the EcPgaB is enhanced in the presence of Co2+, Ni2+ or Fe2+ ions at the active site but is attenuated in the presence of Zn2+ (Little et al., 2012). The enzyme VcCDA on the other hand has affinity to Zn2+ and is somewhat less active when bound to other divalent metals (Andres et al., 2014). In a recent study it was shown that biofilm formation in A. pleuropneumoniae, E. coli, S. aureus and S. suis was slightly inhibited by zinc ion (Wu et al., 2013). In contrast, the SeIcaB has been shown to be promiscuous in its metal dependence since addition of divalent metals to the apo-IcaB enzyme lead to similar increase in activity across a range of divalent metals (Pokrovskaya et al., 2013).
The identity of the metal ion in the Se-Met crystal of AaPgaBN could not be deduced from an analysis of the atomic force microscopy due to difficulty in getting of AaPgaBN crystals. Since AaPgaB is more akin to the EcPgaB, we considered the following ions to be likely present in AaPgaBN: Fe2+, Co2+, Ni2+ or Zn2+ (due to the crystallization buffer). To deduce the identity, we first considered the strength of the anomalous peaks; with f” values of 3.2 e− and 3.61 e−, respectively for Fe2+ and Co2+, the strength of the peaks would be expected to be greater. Secondly, the coordination geometry of the metal ions was considered; tetrahedral with M … O/N distance of 2.1 Å for Zn2+ vs. octahedral with M … O/N distance of 2.3–2.4 Å for Ni2+; Fig. 6). With no additional support from the atomic force microscopy, we have tentatively assigned the metal ions to be zinc.
Figure 6.
Identity of the metal ion. Coordination geometry around the active site metal ions showing the tetrahedral geometry with the ligand distances between 1.9 and 2.2 Å, which are normally observed for zinc ions.
Thus, our analysis of the crystal structure of AaPgaBN shows that although there is high structural conservation between AaPgaBN and EcPgaBN, there are significant differences between the structures: 1) in the number of metal ions bound to the molecule AaPgaBN; 2) in the absence of an acetate moiety at the active site in AaPgaBN. In AaPgaBN, a zinc ion is observed at the site occupied by the acetate in N-terminal domain of EcPgaB (PDB Code: 3VUS and PDB Code: 4F9D). The absence of acetate in AaPgaBN appears to be due to the counter ion (Cl−) used during crystallization trials. Both the full length and the N-terminal domain of EcPgaB used acetate in the buffer (0.2 M calcium acetate and 0.18 M sodium acetate, respectively) whereas we used chloride (0.01 M); and 3) the chains are complete with no breaks in the middle of the chain in AaPgaB.
Mechanistic aspects of de-N-acetylation.
A mechanism for the de-N-acetylation was proposed for the enzyme SpPgdA, a representative of the CE4 enzymes, based on structural, mutagenesis and molecular modeling data (Blair et al., 2005). The bound metal ion plays a central role in this mechanism that holds the water molecule from which a proton is abstracted by the catalytic base. A nearby arginine residue in turn stabilizes the catalytic base. The incipient nucleophile would attack the carbonyl carbon of the acetate moiety of the substrate to produce a tetrahedral oxyanion intermediate. A histidine, which acts as the catalytic acid, would protonate the intermediate on the amino nitrogen generating a free amine and acetate. The product acetate would be stabilized by the metal ion. In many CE4 enzymes hitherto solved by crystal structure analysis, this acetate group is bound to the metal ion at the active site.
In SpPgdA, residue Asp391 has been suggested to activate His417 (catalytic acid), but an equivalent residue is absent in AaPgaBN. In order to identify the residue that is occupying the space that should have been occupied by Asp391, we superposed the two structures. We noted that the residue Gln51 in AaPgaB, in place of Trp392 in SpPgdA, is located in close proximity to His34 in AaPgaB. The corresponding residues in EcPgaB and in PgaB of Ammonifex degensii (Little et al., 2014), at position equivalent to Q51 in AaPgaB, are Met and Tyr, respectively. In both SpPgdA and AaPgaBN, the His417/His34 residue occurs in a loop connecting a β-strand and an α-helix (Fig. 4a). This loop has been named Loop 6 when comparing the loop geometries of CE4 enzymes active on N-acetylglucosaminyl-oligosaccharides (Andres et al., 2014). The helix harboring the activating residue is conspicuously absent in AaPgaBN. Manual modeling using COOT revealed that replacing Gln51 by a Glu places the side chain at an optimal distance to activate His34. Clearly, the kinetic analysis (Table 3) showed that there is an increase in the activity only in the Q51E mutant. The second mutant, R271K, did not show an increase as expected. The higher efficiency of the full-length enzyme could be rationalized using the structure of EcPgaB (Little et al., 2012). In this structure the N- and C-terminal domains of the enzyme are juxtaposed in close proximity, which could affect the kinetic parameters. It has been suggested that the C-terminal domain might play a part in the deacetylating activity when oligomers of N-acetylglucosamine are used as substrates. While the substrate used in this study is a smaller molecule unlike oligomers of N-acetylglucosamine, it is unclear how the binding would be affected to exhibit a higher activity for the full-length enzyme.
Biological significance of de-N-acetylation.
Biologically and functionally, the low deacetylase activity of these enzymes is also not completely understood. Structurally, it is possible that circular permutation observed in the sequence of AaPgaB could also contribute to the low level of activity. Interestingly, many artificial circular permutations have been previously studied to enhance the function of an enzyme (Yu & Lutz 2011). In the case of AaPgaB, and possibly in SeIcaB and EcPgaB, this circular permutation has a lowering effect in the enzyme activity. The de-N-acetylase enzymes in AaPgaB and EcPgaB appear to be an offshoot of the other CE4 enzymes with markedly different properties due to the circular permutation: the position of the motif MT5 at the N-terminal end and the absence of the activating Asp residue in MT4. Recent studies have provided some structural evidence for the limited activity. Comparison of the crystal structure of EcPgaB with other de-N-acetylase enzymes showed the absence of a critical Asp residue in the EcPgaB enzyme (Little et al., 2014) which also could account for the reduced activity. Further, it has been suggested that poor catalytic activity might be a common feature of enzymes that act on exopolysaccharide. In comparison, the presence of an Asp residue at the active site in BsPdaA leads to higher catalytic efficiency (Table 3).
For S. pneumoniae, however, high degree of deacetylation appears to be essential since this bacterium uses the enzyme SpPgdA to modify the peptidoglycan as a defense against mammalian enzymes (Vollmer & Tomasz 2000). Interestingly, fully acetylated PNAG is a protective mechanism for bacteria including S. pyogenes, S. pneumoniae, L. monocytogenes, and N. meningitidis serogroup B since naturally occurring antibodies to the acetylated PNAG could not induce deposition of complement opsonins (Cywes-Bentley et al., 2013). In the case of BsPdaA, absence of deacetylation leads to muramic δ-lactam deficient cell wall and the spore cannot germinate (Fukushima et al., 2005; Fukushima et al., 2002).
Our demonstration of an increase in the in vitro enzyme activity in AaPgaB does raise a question about its effect in biofilm formation in a strain that harbors such mutation. The strains possessing mutations in the genomic DNA leading to either Q51E or R271K PgaB are expected to form biofilm as efficiently as the wild type strain since we have shown earlier that a mutant lacking the pga operon itself does form biofilm (Izano et al., 2008; Shanmugam et al., 2015). It should be noted that unlike E. coli, A. actinomycetemcomitans forms a complex biofilm matrix that contains capsular polysaccharide, eDNA, proteins and lipids. Therefore, drastic changes in biofilm formation is unlikely when these mutations are introduced in the genome. While enhancement of enzymatic activity has been noted through circular permutation, such an enhancement has been suggested to play a role in the evolution of calcium-dependent carbohydrate binding modules involved in xylan recognition (Montanier et al., 2010). To our knowledge, this is the first instance in which natural circular permutation has been utilized by organisms to attenuate the enzyme activity, in the case of de-N-acetylase enzymes.
ACKNOWLEDGEMENT
The project was supported by USPHS Grant DE22544 (NR) and Canadian Institutes of Health Research (CIHR) Grant 89708 (MN).
ABBREVIATIONS
- AaPgaBN
N-terminal domain of PgaB from A. actinomycetemcomitans
- ACC
7-acetoxycoumarin-3-carboxylic acid
- CE4
family 4 carbohydrate esterase
- EcPgaB
PgaB enzyme from E. coli
- GlcNAc
, N-acetyl-D-glucosamine
- IPTG
Isopropyl β-D-1-thiogalactopyranoside
- PGA
homopolymer of β-(1,6)-N-acetyl-D-glucosamine
- SpPgdA
de-N-acetylase from S. pneumoniae
- BsPdaA
de-N-acetylase from B. subtilis
Footnotes
ACCESSION NUMBERS: The coordinates and the structure factors have been deposited in the Protein Data Bank (PDB Code: 4U10).
REFERENCES
- Adams PD, Afonine PV, Bunkoczi G. et al. , (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr Sect D Biol Crystallogr 66: 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agladze K, Wang X, and Romeo T. (2005) Spatial periodicity of Escherichia coli K-12 biofilm microstructure initiates during a reversible, polar attachment phase of development and requires the polysaccharide adhesin PGA. J Bacteriol 187: 8237–8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allignet J, Aubert S, Dyke KG, and El Solh N. (2001) Staphylococcus caprae strains carry determinants known to be involved in pathogenicity: a gene encoding an autolysin-binding fibronectin and the ica operon involved in biofilm formation. Infect Immun 69: 712–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres E, Albesa-Jove D, Biarnes X, Moerschbacher BM, Guerin ME, and Planas A. (2014) Structural basis of chitin oligosaccharide deacetylation. Angew Chem Int Ed Engl 53: 6882–6887. [DOI] [PubMed] [Google Scholar]
- Blair DE, Schuttelkopf AW, MacRae JI, and van Aalten DM (2005) Structure and metal-dependent mechanism of peptidoglycan deacetylase, a streptococcal virulence factor. Proc Natl Acad Sci U S A 102: 15429–15434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caufrier F, Martinou A, Dupont C, and Bouriotis V. (2003) Carbohydrate esterase family 4 enzymes: substrate specificity. Carbohydr Res 338: 687–692. [DOI] [PubMed] [Google Scholar]
- Chibba A, Poloczek J, Little DJ, Howell PL, and Nitz M. (2012) Synthesis and evaluation of inhibitors of E. coli PgaB, a polysaccharide de-N-acetylase involved in biofilm formation. Org Biomol Chem 10: 7103–7107. [DOI] [PubMed] [Google Scholar]
- Coutinho PM and Henrissat B. (1999) Carbohydrate-active enzymes: an integrated database approach. Cambridge: The Royal Society of Chemistry. [Google Scholar]
- Cywes-Bentley C, Skurnik D, Zaidi T. et al. , (2013) Antibody to a conserved antigenic target is protective against diverse prokaryotic and eukaryotic pathogens. Proc Natl Acad Sci U S A 110: E2209–E2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby C, Hsu JW, Ghori N, and Falkow S. (2002) Caenorhabditis elegans: plague bacteria biofilm blocks food intake. Nature 417: 243–244. [DOI] [PubMed] [Google Scholar]
- DeLano WL (2002) The PyMOl User’s Manual. San Carlos, CA: Delano Scientific. [Google Scholar]
- Emsley P. and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr Sect D Biol Crystallogr 60: 2126–2132. [DOI] [PubMed] [Google Scholar]
- Fukushima T, Kitajima T, and Sekiguchi J. (2005) A polysaccharide deacetylase homologue, PdaA, in Bacillus subtilis acts as an N-acetylmuramic acid deacetylase in vitro. J Bacteriol 187: 1287–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima T, Yamamoto H, Atrih A, Foster SJ, and Sekiguchi J. (2002) A polysaccharide deacetylase gene (pdaA) is required for germination and for production of muramic delta-lactam residues in the spore cortex of Bacillus subtilis. J Bacteriol 184: 6007–6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerke C, Kraft A, Sussmuth R, Schweitzer O, and Gotz F. (1998) Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J Biol Chem 273: 18586–18593. [DOI] [PubMed] [Google Scholar]
- Haubek D, Dirienzo JM, Tinoco EM et al. , (1997) Racial tropism of a highly toxic clone of Actinobacillus actinomycetemcomitans associated with juvenile periodontitis. J Clin Microbiol 35: 3037–3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haubek D, Poulsen K, Westergaard J, Dahlen G, and Kilian M. (1996) Highly toxic clone of Actinobacillus actinomycetemcomitans in geographically widespread cases of juvenile periodontitis in adolescents of African origin. J Clin Microbiol 34: 1576–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilmann C, Schweitzer O, Gerke C, Vanittanakom N, Mack D, and Gotz F. (1996) Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol Microbiol 20: 1083–1091. [DOI] [PubMed] [Google Scholar]
- Itoh Y, Rice JD, Goller C. et al. , (2008) Roles of pgaABCD genes in synthesis, modification, and export of the Escherichia coli biofilm adhesin poly-beta-1,6-N-acetyl-D-glucosamine. J Bacteriol 190: 3670–3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izano EA, Sadovskaya I, Vinogradov E. et al. , (2007) Poly-N-acetylglucosamine mediates biofilm formation and antibiotic resistance in Actinobacillus pleuropneumoniae. Microb Pathog 43: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izano EA, Sadovskaya I, Wang H. et al. , (2008) Poly-N-acetylglucosamine mediates biofilm formation and detergent resistance in Aggregatibacter actinomycetemcomitans. Microb. Pathog 44: 52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce JG, Abeygunawardana C, Xu Q. et al. , (2003) Isolation, structural characterization, and immunological evaluation of a high-molecular-weight exopolysaccharide from Staphylococcus aureus. Carbohydr Res 338: 903–922. [DOI] [PubMed] [Google Scholar]
- Kaplan JB, Ragunath C, Velliyagounder K, Fine DH, and Ramasubbu N. (2004a) Enzymatic detachment of Staphylococcus epidermidis biofilms. Antimicrob Agents Chemother 48: 2633–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan JB, Velliyagounder K, Ragunath C. et al. , (2004b) Genes involved in the synthesis and degradation of matrix polysaccharide in Actinobacillus actinomycetemcomitans and Actinobacillus pleuropneumoniae biofilms. J Bacteriol 186: 8213–8220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropec A, Maira-Litran T, Jefferson KK et al. , (2005) Poly-N-acetylglucosamine production in Staphylococcus aureus is essential for irulence in murine models of systemic infection. Infect Immun 73: 6868–6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, and Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst 26: 283–291. [Google Scholar]
- Little DJ, Li G, Ing C. et al. , (2014) Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-d-glucosamine. Proc Natl Acad Sci U S A. 111: 11013–11018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little DJ, Milek S, Bamford NC et al. , (2015) The protein BpsB is a poly-beta-1,6-N-acetyl-D-glucosamine deacetylase required for biofilm formation in Bordetella bronchiseptica. J Biol Chem 290: 22827–22840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little DJ, Poloczek J, Whitney JC, Robinson H, Nitz M, and Howell PL (2012) The structure- and metal-dependent activity of Escherichia coli PgaB provides insight into the partial de-N-acetylation of poly-beta-1,6-N-acetyl-D-glucosamine. J Biol Chem 287: 31126–31137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Du Y, Wang X, and Sun L. (2004) Chitosan kills bacteria through cell membrane damage. Inter J Food Microbiol 95: 147–155. [DOI] [PubMed] [Google Scholar]
- Mack D, Fischer W, Krokotsch A. et al. , (1996) The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1,6-linked glucosaminoglycan: purification and structural analysis. J Bacteriol 178: 175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah TF and O’Toole GA (2001) Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol 9: 34–39. [DOI] [PubMed] [Google Scholar]
- Maira-Litran T, Kropec A, Abeygunawardana C. et al. , (2002) Immunochemical properties of the staphylococcal poly-N-acetylglucosamine surface polysaccharide. Infect Immun 70: 4433–4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney D, Hubner J, Muller E, Wang Y, Goldmann DA, and Pier GB (1998) The ica locus of Staphylococcus epidermidis encodes production of the capsular polysaccharide/adhesin. Infect Immun 66: 4711–4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montanier C, Flint JE, Bolam DN et al. , (2010) Circular permutation provides an evolutionary link between two families of calcium-dependent carbohydrate binding modules. J Biol Chem 285: 31742–31754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Skubak P, Lebedev AA et al. , (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr Sect D Biol Crystallogr 67: 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama T, Noguchi H, Yoshida H, Park SY, and Tame JR (2013) The structure of the deacetylase domain of Escherichia coli PgaB, an enzyme required for biofilm formation: a circularly permuted member of the carbohydrate esterase 4 family. Acta Crystallogr Sect D Biol Crystallogr 69: 44–51. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor W. (1997) Processing of X-ray Crystallographic Data in Oscillation Mode. Methods Enzymol 276: 307–326. [DOI] [PubMed] [Google Scholar]
- Parise G, Mishra M, Itoh Y, Romeo T, and Deora R. (2007) Role of a putative polysaccharide locus in Bordetella biofilm development. J Bacteriol 189: 750–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokrovskaya V, Poloczek J, Little DJ, Griffiths H, Howell PL, and Nitz M. (2013) Functional characterization of Staphylococcus epidermidis IcaB, a de-N-acetylase important for biofilm formation. Biochem 52: 5463–5471. [DOI] [PubMed] [Google Scholar]
- Ramasubbu N, Thomas LM, Ragunath C, and Kaplan JB (2005) Structural analysis of dispersin B, a biofilm-releasing glycoside hydrolase from the periodontopathogen Actinobacillus actinomycetemcomitans. J Mol Biol 349: 475–486. [DOI] [PubMed] [Google Scholar]
- Rohde H, Frankenberger S, Zahringer U, and Mack D. (2010) Structure, function and contribution of polysaccharide intercellular adhesin (PIA) to Staphylococcus epidermidis biofilm formation and pathogenesis of biomaterial-associated infections. Eur J Cell Biol 89: 103–111. [DOI] [PubMed] [Google Scholar]
- Sadovskaya I, Chaignon P, Kogan G, Chokr A, Vinogradov E, and Jabbouri S. (2006) Carbohydrate-containing components of biofilms produced in vitro by some staphylococcal strains related to orthopaedic prosthesis infections. FEMS Immunol Med Microbiol 47: 75–82. [DOI] [PubMed] [Google Scholar]
- Shaddox L, Wiedey J, Bimstein E. et al. , (2010) Hyper-responsive phenotype in localized aggressive periodontitis. J Dent Res 89: 143–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugam M, Gopal P, El Abbar F. et al. , (2015) Role of Exopolysaccharide in Aggregatibacter actinomycetemcomitans-Induced Bone Resorption in a Rat Model for Periodontal Disease. PLoS One 10: e0117487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacy A, Everett J, Jorth P, Trivedi U, Rumbaugh KP, and Whiteley M. (2014) Bacterial fight-and-flight responses enhance virulence in a polymicrobial infection. Proc Natl Acad Sci U S A 111: 7819–7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subashchandrabose S, Smith SN, Spurbeck RR, Kole MM, and Mobley HL (2013) Genome-wide detection of fitness genes in uropathogenic Escherichia coli during systemic infection. PLoS Path 9: e1003788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland IW (2001) Exopolysaccharides in biofilms, flocs and related structures. Water Sci Technol 43: 77–86. [PubMed] [Google Scholar]
- Taylor EJ, Gloster TM, Turkenburg JP et al. , (2006) Structure and activity of two metal ion-dependent acetylxylan esterases involved in plant cell wall degradation reveals a close similarity to peptidoglycan deacetylases. J Biol Chem 281: 10968–10975. [DOI] [PubMed] [Google Scholar]
- Vollmer W. and Tomasz A. (2000) The pgdA gene encodes for a peptidoglycan N-acetylglucosamine deacetylase in Streptococcus pneumoniae. J Biol Chem 275: 20496–20501. [DOI] [PubMed] [Google Scholar]
- Vuong C, Kocianova S, Voyich JM et al. , (2004a) A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J Biol Chem 279: 54881–54886. [DOI] [PubMed] [Google Scholar]
- Vuong C, Voyich JM, Fischer ER et al. , (2004b) Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell Microbiol 6: 269–275. [DOI] [PubMed] [Google Scholar]
- Wang X, Preston JF 3rd, and Romeo T. (2004) The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J Bacteriol 186: 2724–2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watnick P. and Kolter R. (2000) Biofilm, city of microbes. J Bacteriol 182: 2675–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield GB, Marmont LS, and Howell PL (2015) Enzymatic modifications of exopolysaccharides enhance bacterial persistence. Front Microbiol 6: 471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD et al. , (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr Sect D Biol Crystallogr 67: 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Labrie J, Tremblay YD, Haine D, Mourez M, and Jacques M. (2013) Zinc as an agent for the prevention of biofilm formation by pathogenic bacteria. J Appl Microbiol 115: 30–40. [DOI] [PubMed] [Google Scholar]
- Yu Y. and Lutz S. (2011) Circular permutation: a different way to engineer enzyme structure and function. Trends Biotechnol 29: 18–25. [DOI] [PubMed] [Google Scholar]
- Zambon JJ (1985) Actinobacillus actinomycetemcomitans in human periodontal disease. J Clin Periodontol 12: 1–20. [DOI] [PubMed] [Google Scholar]