Abstract
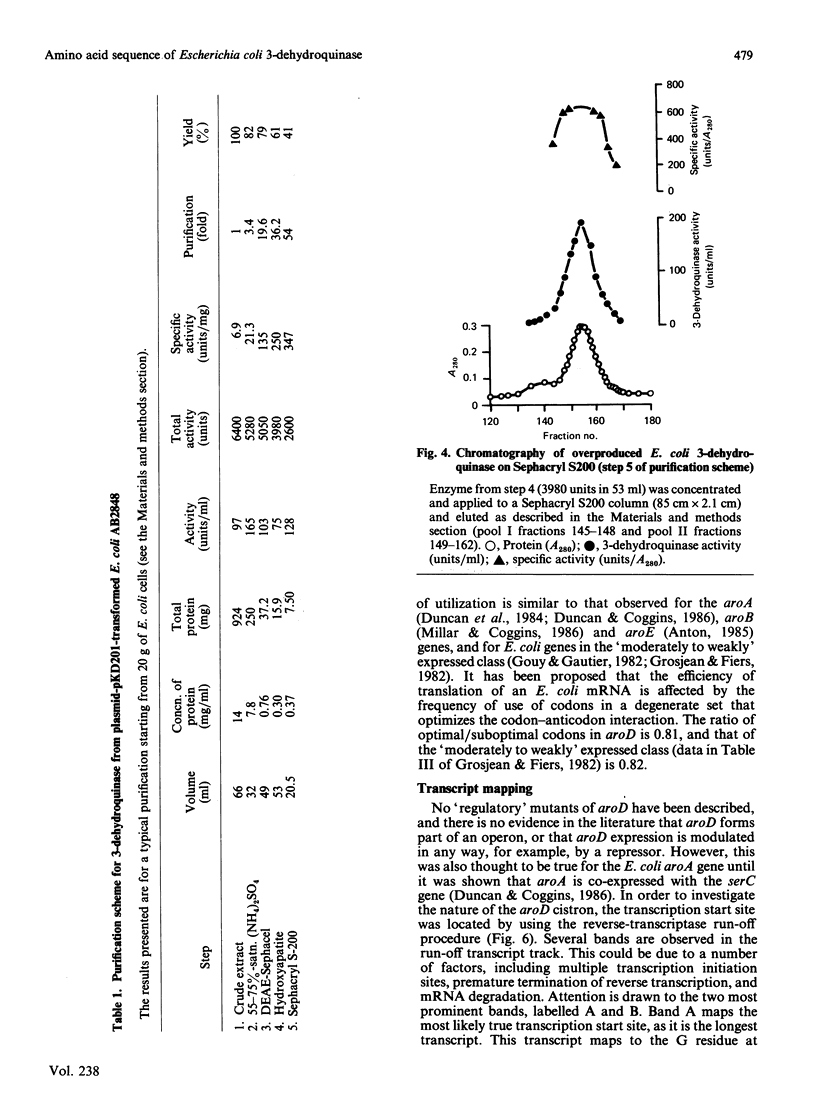
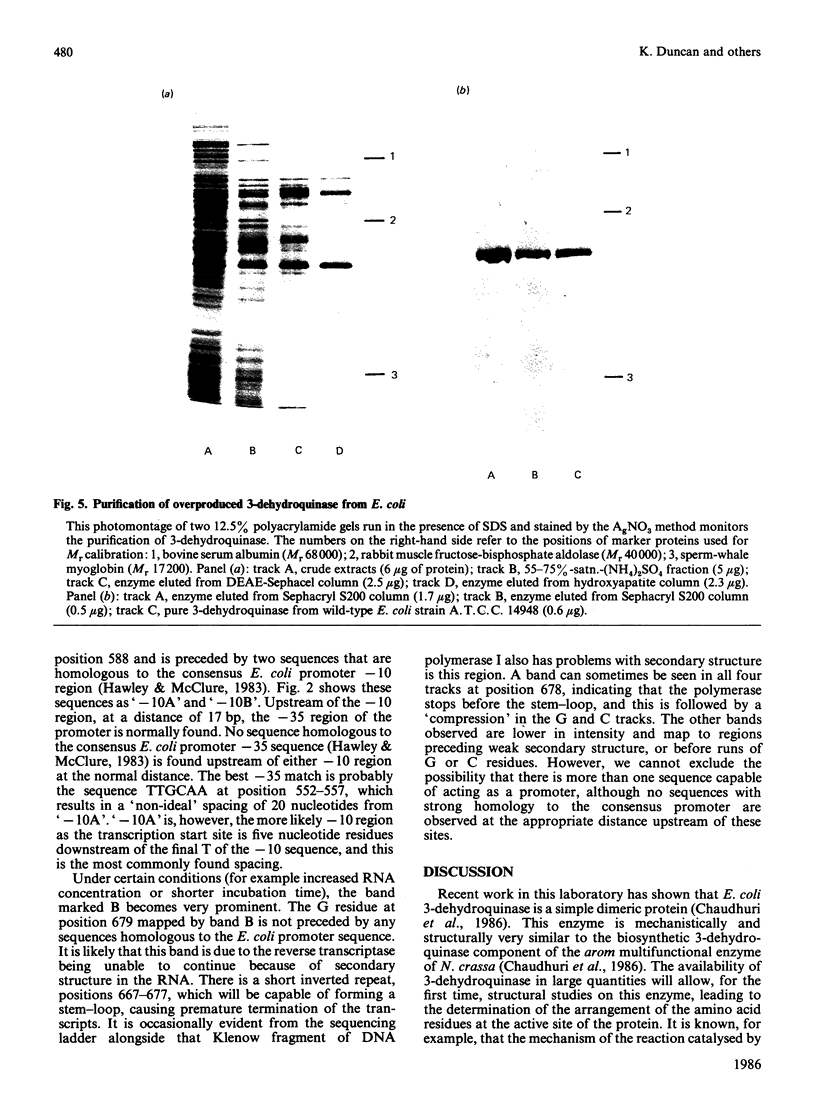
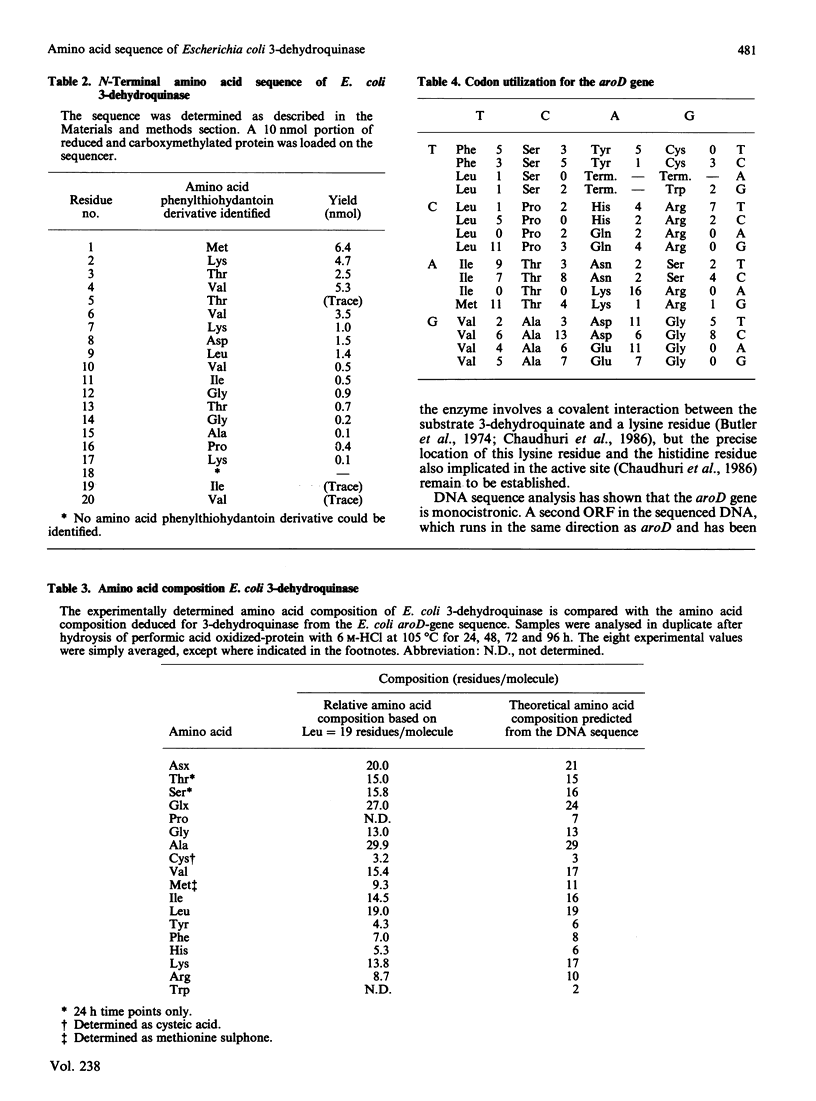
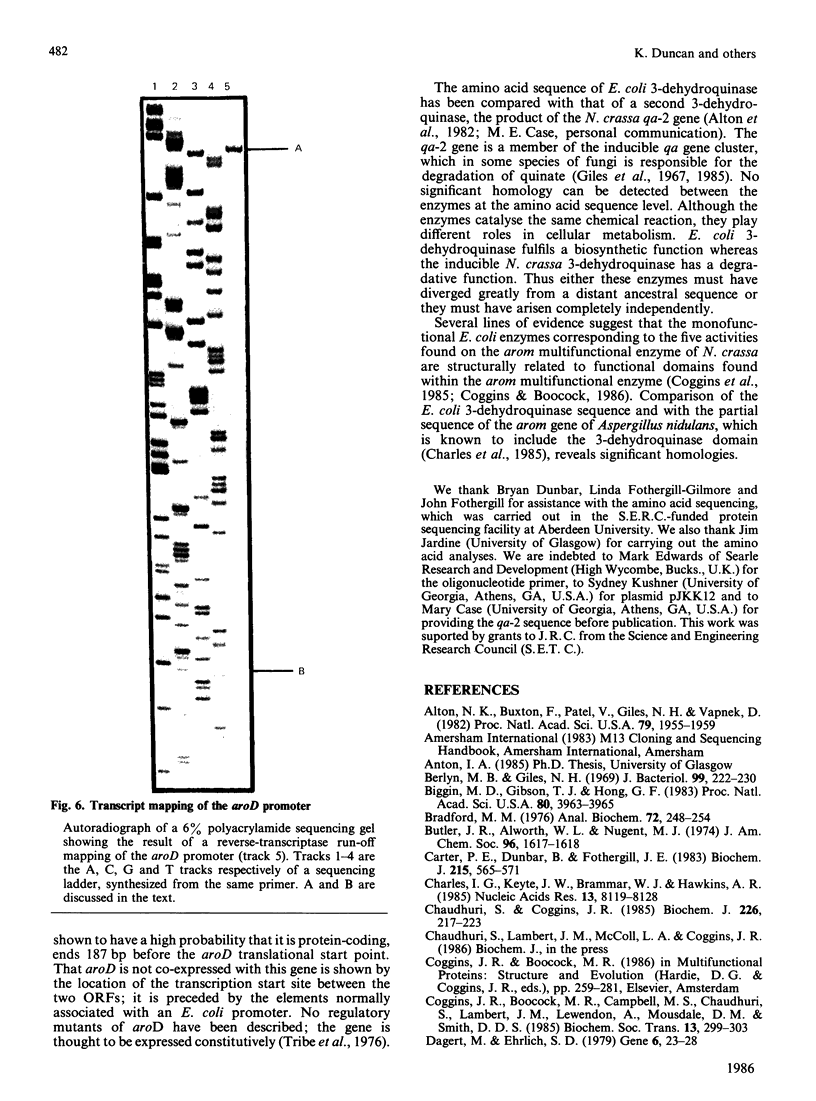
The enzyme 3-dehydroquinase was purified in milligram quantities from an overproducing strain of Escherichia coli. The amino acid sequence was deduced from the nucleotide sequence of the aroD gene and confirmed by determining the amino acid composition of the overproduced enzyme and its N-terminal amino acid sequence. The complete polypeptide chain consists of 240 amino acid residues and has a calculated subunit Mr of 26,377. Transcript mapping revealed that aroD is a typical monocistronic gene.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alton N. K., Buxton F., Patel V., Giles N. H., Vapnek D. 5'-Untranslated sequences of two structural genes in the qa gene cluster of Neurospora crassa. Proc Natl Acad Sci U S A. 1982 Mar;79(6):1955–1959. doi: 10.1073/pnas.79.6.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlyn M. B., Giles N. H. Organization of enzymes in the polyaromatic synthetic pathway: separability in bacteria. J Bacteriol. 1969 Jul;99(1):222–230. doi: 10.1128/jb.99.1.222-230.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Carter P. E., Dunbar B., Fothergill J. E. The serine proteinase chain of human complement component C1s. Cyanogen bromide cleavage and N-terminal sequences of the fragments. Biochem J. 1983 Dec 1;215(3):565–571. doi: 10.1042/bj2150565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles I. G., Keyte J. W., Brammar W. J., Hawkins A. R. Nucleotide sequence encoding the biosynthetic dehydroquinase function of the penta-functional arom locus of Aspergillus nidulans. Nucleic Acids Res. 1985 Nov 25;13(22):8119–8128. doi: 10.1093/nar/13.22.8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri S., Coggins J. R. The purification of shikimate dehydrogenase from Escherichia coli. Biochem J. 1985 Feb 15;226(1):217–223. doi: 10.1042/bj2260217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coggins J. R., Boocock M. R., Campbell M. S., Chaudhuri S., Lambert J. M., Lewendon A., Mousdale D. M., Smith D. D. Functional domains involved in aromatic amino acid biosynthesis. Biochem Soc Trans. 1985 Apr;13(2):299–303. doi: 10.1042/bst0130299. [DOI] [PubMed] [Google Scholar]
- Dagert M., Ehrlich S. D. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene. 1979 May;6(1):23–28. doi: 10.1016/0378-1119(79)90082-9. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan K., Coggins J. R. The serC-aro A operon of Escherichia coli. A mixed function operon encoding enzymes from two different amino acid biosynthetic pathways. Biochem J. 1986 Feb 15;234(1):49–57. doi: 10.1042/bj2340049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eschenbruch M., Bürk R. R. Experimentally improved reliability of ultrasensitive silver staining of protein in polyacrylamide gels. Anal Biochem. 1982 Sep 1;125(1):96–99. doi: 10.1016/0003-2697(82)90387-6. [DOI] [PubMed] [Google Scholar]
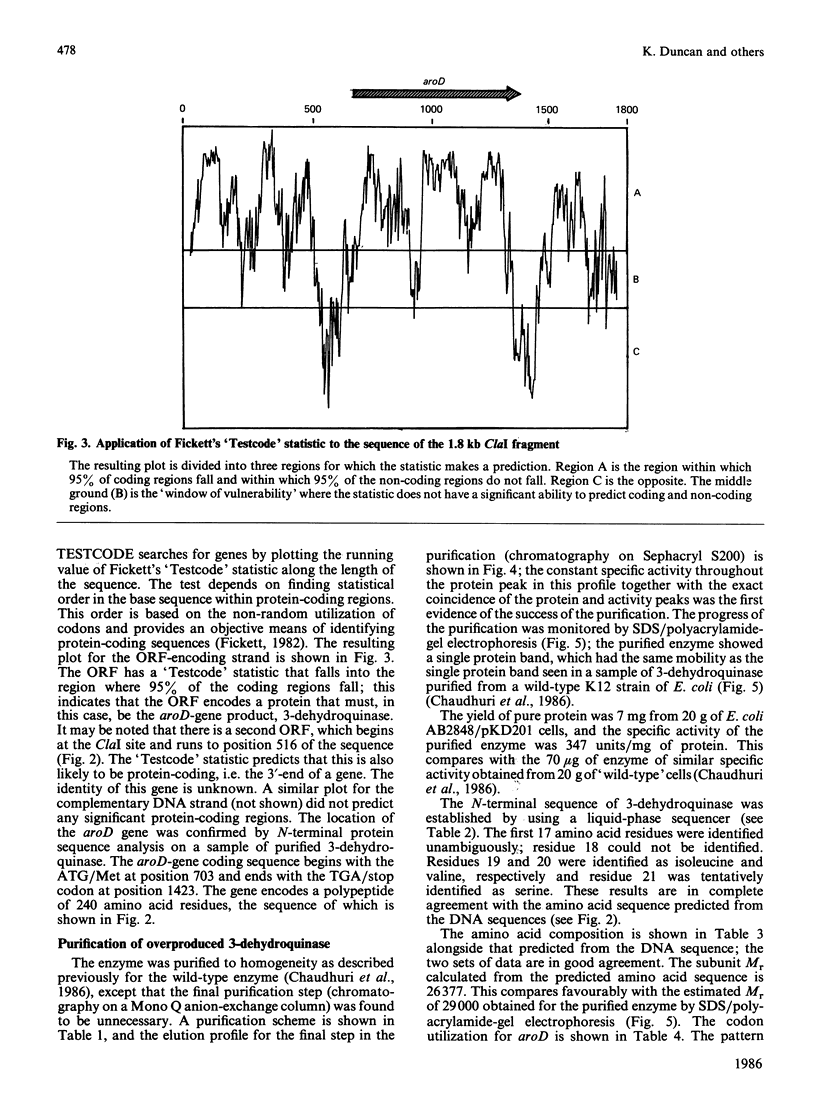
- Fickett J. W. Recognition of protein coding regions in DNA sequences. Nucleic Acids Res. 1982 Sep 11;10(17):5303–5318. doi: 10.1093/nar/10.17.5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaertner F. H., Cole K. W. A cluster-gene: evidence for one gene, one polypeptide, five enzymes. Biochem Biophys Res Commun. 1977 Mar 21;75(2):259–264. doi: 10.1016/0006-291x(77)91037-3. [DOI] [PubMed] [Google Scholar]
- Giles N. H., Case M. E., Baum J., Geever R., Huiet L., Patel V., Tyler B. Gene organization and regulation in the qa (quinic acid) gene cluster of Neurospora crassa. Microbiol Rev. 1985 Sep;49(3):338–358. doi: 10.1128/mr.49.3.338-358.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles N. H., Partridge C. W., Ahmed S. I., Case M. E. The occurrence of two dehydroquinases in Neurospora crassa, one constitutive and one inducible. Proc Natl Acad Sci U S A. 1967 Nov;58(5):1930–1937. doi: 10.1073/pnas.58.5.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M., Gautier C. Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res. 1982 Nov 25;10(22):7055–7074. doi: 10.1093/nar/10.22.7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H., Fiers W. Preferential codon usage in prokaryotic genes: the optimal codon-anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene. 1982 Jun;18(3):199–209. doi: 10.1016/0378-1119(82)90157-3. [DOI] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
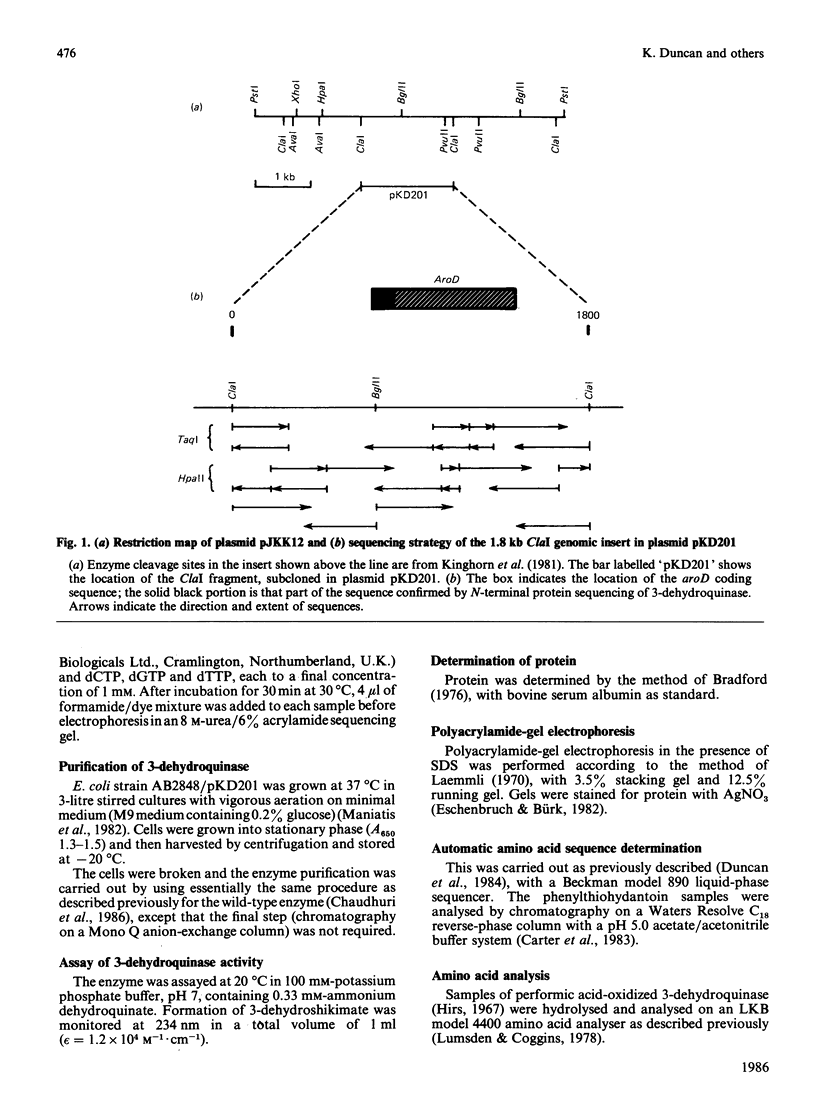
- Kinghorn J. R., Schweizer M., Giles N. H., Kushner S. R. The cloning and analysis of the aroD gene of E. coli K-12. Gene. 1981 Jun-Jul;14(1-2):73–80. doi: 10.1016/0378-1119(81)90149-9. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lambert J. M., Boocock M. R., Coggins J. R. The 3-dehydroquinate synthase activity of the pentafunctional arom enzyme complex of Neurospora crassa is Zn2+-dependent. Biochem J. 1985 Mar 15;226(3):817–829. doi: 10.1042/bj2260817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumsden J., Coggins J. R. The subunit structure of the arom multienzyme complex of Neurospora crassa. A possible pentafunctional polypeptide chain. Biochem J. 1977 Mar 1;161(3):599–607. doi: 10.1042/bj1610599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumsden J., Coggins J. R. The subunit structure of the arom multienzyme complex of Neurospora crassa. Evidence from peptide 'maps' for the identity of the subunits. Biochem J. 1978 Feb 1;169(2):441–444. doi: 10.1042/bj1690441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Millar G., Coggins J. R. The complete amino acid sequence of 3-dehydroquinate synthase of Escherichia coli K12. FEBS Lett. 1986 May 5;200(1):11–17. doi: 10.1016/0014-5793(86)80501-4. [DOI] [PubMed] [Google Scholar]
- Pittard J., Wallace B. J. Distribution and function of genes concerned with aromatic biosynthesis in Escherichia coli. J Bacteriol. 1966 Apr;91(4):1494–1508. doi: 10.1128/jb.91.4.1494-1508.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polley L. D. Purification and characterization of 3-dehydroquinate hydrolase and shikmate oxidoreductase. Evidence for a bifunctional enzyme. Biochim Biophys Acta. 1978 Sep 11;526(1):259–266. doi: 10.1016/0005-2744(78)90310-8. [DOI] [PubMed] [Google Scholar]
- Sanger F. Determination of nucleotide sequences in DNA. Science. 1981 Dec 11;214(4526):1205–1210. doi: 10.1126/science.7302589. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. D., Coggins J. R. Isolation of a bifunctional domain from the pentafunctional arom enzyme complex of Neurospora crassa. Biochem J. 1983 Aug 1;213(2):405–415. doi: 10.1042/bj2130405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. O., Birnstiel M. L. A simple method for DNA restriction site mapping. Nucleic Acids Res. 1976 Sep;3(9):2387–2398. doi: 10.1093/nar/3.9.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. A new computer method for the storage and manipulation of DNA gel reading data. Nucleic Acids Res. 1980 Aug 25;8(16):3673–3694. doi: 10.1093/nar/8.16.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribe D. E., Camakaris H., Pittard J. Constitutive and repressivle enzymes of the common pathway of aromatic biosynthesis in Escherichia coli K-12: regulation of enzyme synthesis at different growth rates. J Bacteriol. 1976 Sep;127(3):1085–1097. doi: 10.1128/jb.127.3.1085-1097.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twigg A. J., Sherratt D. Trans-complementable copy-number mutants of plasmid ColE1. Nature. 1980 Jan 10;283(5743):216–218. doi: 10.1038/283216a0. [DOI] [PubMed] [Google Scholar]