Abstract
The role of glycoprotein E (gE) and gI of Marek's disease virus serotype 1 (MDV-1) for growth in cultured cells was investigated. MDV-1 mutants lacking either gE (20ΔgE), gI (20ΔgI), or both gE and gI (20ΔgEI) were constructed by recE/T-mediated mutagenesis of a recently established infectious bacterial artificial chromosome (BAC) clone of MDV-1 (D. Schumacher, B. K. Tischer, W. Fuchs, and N. Osterrieder, J. Virol. 74:11088–11098, 2000). Deletion of either gE or gI, which form a complex in MDV-1-infected cells, resulted in the production of virus progeny that were unable to spread from cell to cell in either chicken embryo fibroblasts or quail muscle cells. This was reflected by the absence of virus plaques and the detection of only single infected cells after transfection, even after coseeding of transfected cells with uninfected cells. In contrast, growth of rescuant viruses, in which the deleted glycoprotein genes were reinserted by homologous recombination, was indistinguishable from that of parental BAC20 virus. In addition, the 20ΔgE mutant virus was able to spread from cell to cell when cotransfected into chicken embryo fibroblasts with an expression plasmid encoding MDV-1 gE, and the 20ΔgI mutant virus exhibited cell-to-cell spread capability after cotransfection with a gI expression plasmid. The 20ΔgEI mutant virus, however, was not able to spread in the presence of either a gE or gI expression plasmid, and only single infected cells were detected by indirect immunofluorescence. The results reported here demonstrate for the first time that both gE and gI are absolutely essential for cell-to-cell spread of a member of the Alphaherpesvirinae.
Marek's disease virus (MDV) is a member of the Alphaherpesvirinae subfamily of the Herpesviridae (59). Serotype 1 MDV (MDV-1), also referred to as gallid herpesvirus 2, induces T-cell lymphomas in chickens, whereas MDV-2 (also referred to as gallid herpesvirus 3) and MDV-3 are less pathogenic and do not induce tumors (8, 42, 47). MDV-3 represents the herpesvirus of turkeys, which is now classified as meleagrid herpesvirus 1 and which has been widely used for vaccination against Marek's disease (59).
MDV-1 and also MDV-2 exhibit unusual growth properties compared to other members of the virus subfamily, inasmuch as virtually no free virus is released into the supernatants of cultured cells, irrespective of the cell culture system used. Free infectious virus is released from the feather follicle epithelium of naturally or experimentally infected birds only (9). In this respect, MDV-1 closely resembles another alphaherpesvirus, varicella-zoster virus (VZV), which produces only small amounts of free infectious virus in cultured cells (20).
Complete sequence analysis of two strains has revealed that the MDV-1 genome harbors the alphaherpesvirus-specific repertoire of glycoprotein genes with the exception of a glycoprotein G (gG) gene, i.e., genes encoding gB, gC, gD, gE, gH, gI, gK, gL, and gM. In addition, a UL49.5 homologous open reading frame (ORF), the product of which is glycosylated in pseudorabies virus (PrV) is present (25, 57). Expression of MDV-1 gB, gC, gE, gI, gH, gL, and gK has been demonstrated (5, 43, 44, 51, 63, 64), whereas MDV-1 gD expression is absent in cultured cells due to the lack of production of gD-specific transcripts (51). gE and gI form a disulfide-linked heterodimer in all alphaherpesviruses investigated to date, which is nonessential for growth of herpes simplex virus type 1 (HSV-1), PrV, bovine herpesvirus 1 (BHV-1), and feline herpesvirus (4, 35, 65, 68). HSV-1 and PrV gE and gI have been studied in great detail (7, 10, 13–15, 21–23, 31, 16, 52–56, 61), and it could be shown that deletion of HSV-1 gE or gI results in a virus that lacks Fc receptor activity but is viable in nonpolarized cultured cells (22). Spread of HSV-1 gE and gI deletion mutants in vivo or in polarized cells which form extensive cell junctions, however, is severely impaired (4, 13–15, 23, 31). This defect of the viral mutants is caused by a missorting of the glycoprotein-deficient viruses. While wild-type viruses are primarily sorted to epithelial cell junctions, mutant viruses are not. The correct sorting of HSV-1 to tight junctions is apparently dependent on the cytoplasmic domain of gE (23). Similar to the situation in HSV-1, the PrV gE-gI complex is nonessential for growth in vitro (68), but PrV is impaired in neuropathogenicity after deletion of gE or gI (10, 24, 61, 52, 54, 56). It has also been shown that deletion of gM in addition to gE and gI results in PrV or equine herpesvirus 1 (EHV-1) progeny that are severely compromised in virus release and cell-to-cell spread of infectivity (6, 50). The inability of the PrV and EHV-1 triple mutants to efficiently spread from cell to cell as well as the defect in virus egress appears to be caused by an inefficient secondary envelopment of virions at vesicles of the Golgi apparatus. It has additionally been shown that PrV egress from cultured cells and efficient cell-to-cell spread in vivo are mediated by the cytoplasmic tails of gE and gI (56).
The growth properties of VZV in cultured cells closely resemble those of MDV-1, and it was demonstrated that VZV gE and gI form a noncovalently linked complex (1–3, 20). The roles of gE and gI in VZV replication have been studied by using mutant viruses reconstituted from overlapping cosmid clones. The effects of a deletion of either of the proteins appear to be, at least to a certain extent, cell type specific. It was reported that deletion of gI in the Oka strain resulted in a virus that was unable to grow in Vero cells and exhibited reduced replication in other cells (11). Mallory et al. (28, 29) reported that gE but not gI is essential for growth of the virus in cultured cells. Deletion of gI resulted in incomplete processing of gE as reflected by an altered mobility of the glycoprotein in sodium dodecyl sulfate (SDS)-polyacrylamide gels. Further experiments showed that gE and the gE-gI complex facilitate cell-to-cell contacts in epithelial cells and thus promote viral spread (36) and that the cytoplasmic tail of VZV gI interacts with tegument proteins (60).
In the case of MDV-1, gE and gI, which contain a total of seven and five consensus N-glycosylation sites, respectively, are expressed from a bicistronic mRNA and an mRNA that spans a large portion of the unique short (US) region of the genome from the US3 ORF to that of gE (US8). In addition, a gE-specific monocistronic mRNA was detected. This transcriptional organization is unusual for an alphaherpesvirus and may be caused by the lack of MDV-1 gD transcription in cultured cells (25, 51, 57).
The aim of this study was to explore the function of the gE-gI complex in MDV-1 by analyzing the effects of the deletion of these two glycoproteins alone or in combination. By using a recently established infectious BAC clone of MDV-1 strain 584A and recE/T-based mutagenesis (38, 39, 67), three mutant viruses were constructed. The gE and gI single mutants as well as the gE-gI double mutant were unable to grow in cultured cells. However, growth of the single mutants could be restored by cotransfection of an expression plasmid harboring the respective glycoprotein, demonstrating that both gE and gI are essential for growth of MDV-1 in cell culture. This is the first example of an alphaherpesvirus for which both gE and gI play absolutely essential roles in virus growth in cultured cells.
MATERIALS AND METHODS
Virus and cells.
Primary or secondary chicken embryo fibroblasts (CEF) or quail muscle cells (QM7; ATCC CRL-1962) were maintained in Dulbecco's modified essential medium (DMEM) supplemented with 5 to 10% fetal calf serum. MDV-1 strain 584Ap80C reconstituted from infectious BAC20 (49) was used in this study. BAC20 virus was recovered at day 5 after transfection of 1 μg of BAC20 DNA into CEF by calcium phosphate precipitation (49) unless otherwise stated. Transfections of mutant BAC clones were performed accordingly using 1 to 5 μg of BAC DNA.
Plasmids and PCR.
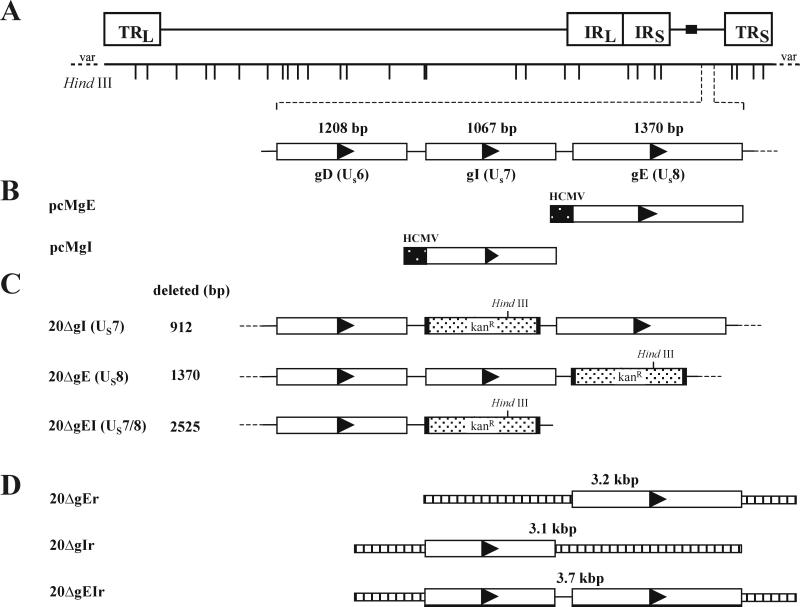
MDV-1 gE and gI ORFs were amplified from strain 584Ap80C by standard PCR as described previously (49) and using primers containing appropriate restriction enzyme sites (Table 1). The resulting amplification products were cleaved with restriction enzymes and cloned into plasmid pcDNA3 (Invitrogen). Correct insertion of the genes in recombinant plasmids pcMgE and pcMgI (Fig. 1) was determined by cycle sequencing (41). For transfections, 10 μg of purified pcMgE, pcMgI, or pcMgM (40) was used. PCR amplification of fragments containing the kanamycin resistance gene for recE/T cloning was obtained by using plasmid pACYC177 (MBI Fermentas) as a template. The primers contained 20 nucleotides each of kanamycin resistance gene-specific sequences and 50 nucleotides of MDV-1-specific sequences to allow homologous recombination (Table 1). PCR products were then used for production of gE-, gI-, or gE-gI-negative BAC20 (Fig. 1).
TABLE 1.
Primers used for generation of expression plasmids and for construction of MDV-1 mutants and rescuant viruses
| Primer | Sequencea | Fragment (plasmid generated) |
|---|---|---|
| GST-gE1 | 5′-ggattcGTAGGAGACGACGTCGCACG-3′ | GST-gE fusion protein |
| GST-gE2 | 5′-ctcgagTCAGTGGTATAAATCTAAGCG-3 | GST-gE fusion protein |
| gI-a | 5′-CAAtctagaACGGCGTGTGTGATTGCGATG-3′ | 1.5-kbp gI ORF (pcMgI) |
| gI-b | 5′-CAAgggcccCCACCTACCTATAATAGTTTC-3′ | 1.5-kbp gI ORF (pcMgI) |
| gE-a | 5′-CATAAgcatgcGAGTCAGCGTCATAATGTG-3′ | 1.1-kbp gE ORF (pcMgE) |
| gE-b | 5′-CAAgggcccATCAGTGGTATAAATCTAAGC-3′ | 1.1-kbp gE ORF (pcMgE) |
| gI-kanR-a | 5′-TCTATAGTTTATACTGGAACATCTGTTACGTTATCAACGGACCAATCTG CGATTTATTCAACAAAGCCACG-3′b | Kanamycin resistance gene for gI and gEI deletion |
| gI-kanR-b | 5′-CACATTCTTCTCTTTCCAACAATTCGACTTTCTTCTTCAGTTTTTCCATC GCCAGTGTTACAACCAATTAACC-3′b | Kanamycin resistance gene for gI deletion |
| gE-kanR-a | 5′-ATGTGTGTTTTCCAAATCCTGATAATAGTGACGACGATCAAAGTAGCTGG CGATTTATTCAACAAAGCCACG-3′b | Kanamycin resistance gene for gE deletion |
| gE-kanR-b | 5′-CTAAGCGTTTCCTAATTTTCGGCATATCATTTTTTAGCCAAGCGGTATAAG CCAGTGTTACAACCAATTAACC-3′b | Kanamycin resistance gene for gE and gEI deletion |
| gI-rev-a | 5′-TTCAAATCACCTGACGACGA-3′ | Fragment for gE rescuant |
| gE-rev-b | 5′-TAACTCTTCCAACTCCAGGG-3′ | Fragment for gI rescuant |
Restriction enzyme sites are given in lowercase boldface type; sequences in italics indicate additional bases which are not present in the MDV-1 sequence.
For primers gI-kanR-a, gI-kanR-b, gE-kanR-a, and gE-kanR-b, underlined sequences indicate sequences from pACYC177 used to amplify the kan resistance gene. Sequences printed in boldface italic type represent gI and gE sequences which allowed homologous recombination for recE/T-mediated deletion of the genes.
FIG. 1.
Schematic illustration of the procedure to delete the gE, the gI, or both the gE and gI ORFs from BAC20. (A) Shown is the organization of the approximately 190-kbp BAC20 genome and the HindIII-restriction map. TRL and TRS, long and short terminal repeats, respectively; IRL and IRS, long and short inverted repeats, respectively. (B) The locations of the gD, gI, and gE genes in the unique short region (US) as well as the constructed gE and gI expression plasmids (pcMgE and pcMgI) are shown. (C) The genomic organization of the mutant BAC clones harboring a deletion in gI (20ΔgI), gE (20ΔgE), or both gE and gI (20ΔgEI) and carrying the kanamycin resistance gene is given. (D) The construction of rescuant viruses and that of the used PCR products is outlined.
Mutagenesis of BAC20.
For mutagenesis of BAC20 DNA, a homologous recombination system was used which is performed in Escherichia coli (38, 39, 49, 67). Electrocompetent DH10B cells (GIBCO-BRL) carrying both pGETrec (39) and MDV-1 BAC20 were prepared by inoculating a fresh overnight culture into 250 ml of Luria-Bertani (LB) medium containing ampicillin (100 μg/ml) and chloramphenicol (30 μg/ml) until an optical density at 600 nm of 0.4 was reached. Expression of recE, recT, and λ gam was then induced by addition of l-arabinose to a final concentration of 0.2% and further incubation for 20 min. Cells were harvested and made electrocompetent by a standard protocol (38, 39). For recombination of a linear fragment into BAC20, 300 ng of a purified PCR product to delete the target sequences (Fig. 1; Table 1) was electroporated into 40 μl of electrocompetent pGETrec-containing BAC20 cells using standard electroporation parameters (1.25 kV/cm, 200 Ω, and 25 μF). After electroporation, cells were grown in 1 ml of LB for 60 min and moved onto LB agar plates containing chloramphenicol (30 μg/ml) and kanamycin (30 μg/ml). Double-resistant colonies were picked into liquid LB medium, and small-scale preparations of mutant BAC20 DNA were performed by alkaline lysis of E. coli (46). Large-scale preparations of mutant BAC DNAs were done using commercially available kits (Qiagen; Macherey & Nagel).
DNA analyses.
BAC DNA was cleaved with restriction enzymes (Roche Biochemicals) and separated on 0.8% agarose gels. DNA fragments were transferred to positively charged nylon membranes (Pharmacia-Amersham), and Southern blot hybridization was performed using digoxigenin-labeled gE, gI, or kanamycin resistance gene probes. Chemiluminescence detection of DNA hybrids using CSPD [disodium 3-(4-meth-oxyspiro{1,2-dioxetane-3,2′-(5′-chloro)tricyclo[3.3.1.13.7]decan}-4-yl)phenyl] was done according to the supplier's instructions (Roche Biochemicals).
Generation of an MDV-1 gE-specific antiserum.
To generate a gE-specific antiserum, the carboxy-terminal region of MDV-1 gE, from codons 420 to 497, was amplified from viral DNA by PCR using the primers listed in Table 1. The first primer contained a BamHI site followed by MDV-1 nucleotides 163650 to 163669, and the second primer contained an XhoI site followed by MDV-1 nucleotides 163862 to 163882. The amplified product was digested with BamHI and XhoI and cloned into plasmid vector pGEX-5X-3 (Pharmacia-Amersham), which contains glutathione S-transferase (GST) gene. The junction sequence between gE and pGEX-5X-3 was confirmed by DNA sequencing. The fusion protein was expressed in E. coli, which was lysed by sonication, and the GST-gE fusion protein was purified with glutathione-Sepharose according to the manufacturer's instructions. A rabbit was immunized four times at 4-week intervals with 150 μg of GST-gE fusion protein suspended in complete (first immunization) or incomplete Freund's adjuvant. Antiserum was obtained 2 weeks after the final boost and adsorbed twice with lysates of uninfected CEF.
Protein analyses.
For indirect immunofluorescence analyses (IIF), cells were grown on six-well plates (Greiner) or on glass coverslips. Cells were fixed with 90% acetone at various times after infection, IIF was done exactly as described, and samples were analyzed by conventional fluorescence microscopy (32, 49). The antibodies used were anti-gB monoclonal antibody (MAb) 2K11, anti-gE (see above), or anti-gI polyclonal rabbit antiserum (51) (kindly provided by Lucy Lee, Avian Disease and Oncology Laboratory, East Lansing, Mich.) or a convalescent-phase serum from a chicken infected with MDV-1 (anti-MDVI) (49). Radioimmunoprecipitation assays (RIPA) were done following a published protocol with slight modifications (51). Briefly, 107 CEF were infected with 105 BAC20 virus-infected CEF. At various times after infection, infected cells were overlaid with DMEM without methionine and cysteine for 30 min. Subsequently, 35S-labeled methionine and cysteine (300 μCi/ml; Tran35S-label [ICN Biochemicals]) in DMEM were added for 2 h. Cell lysates were prepared (50), precleared using 3 μl of an irrelevant rabbit antibody and a Staphylococcus aureus lysate (Pansorbin; Calbiochem), and finally incubated with 3 μl of the gE- or gI-specific rabbit antiserum for 1 h before immunocomplexes were precipitated using Pansorbin. After four washes using RIPA wash buffer (46), immunocomplexes were resuspended in 90 μl of deglycosylation buffer {50 mM K3PO4, pH 7.2; 50 mM EDTA; 0.6% [vol/vol] 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate [CHAPS]; 0.1% SDS}. Deglycosylation was performed by addition of PNGase F (0.4 U) or Endo H (2 mU) (Roche Biochemicals) for 16 h at 37°C. Samples were then analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) after addition of sample buffer (32). To test for Fc receptor binding of the gE-gI complex, purified immunoglobulin Y (IgY) from egg yolk was added to preadsorbed lysates and precipitated with a mouse anti-IgY-specific antibody (IgY and mouse anti-IgY antibody were kindly provided by B. Kaspers, University of Munich, Munich, Germany). Western blot analyses of purified IgY or IgY immunoprecipitates were done exactly as described earlier (50). The secondary antibody used for visualization was anti-chicken IgG peroxidase conjugate (Sigma).
RESULTS
MDV-1 strain 584Ap80C gE and gI form a complex which does not bind to chicken IgY.
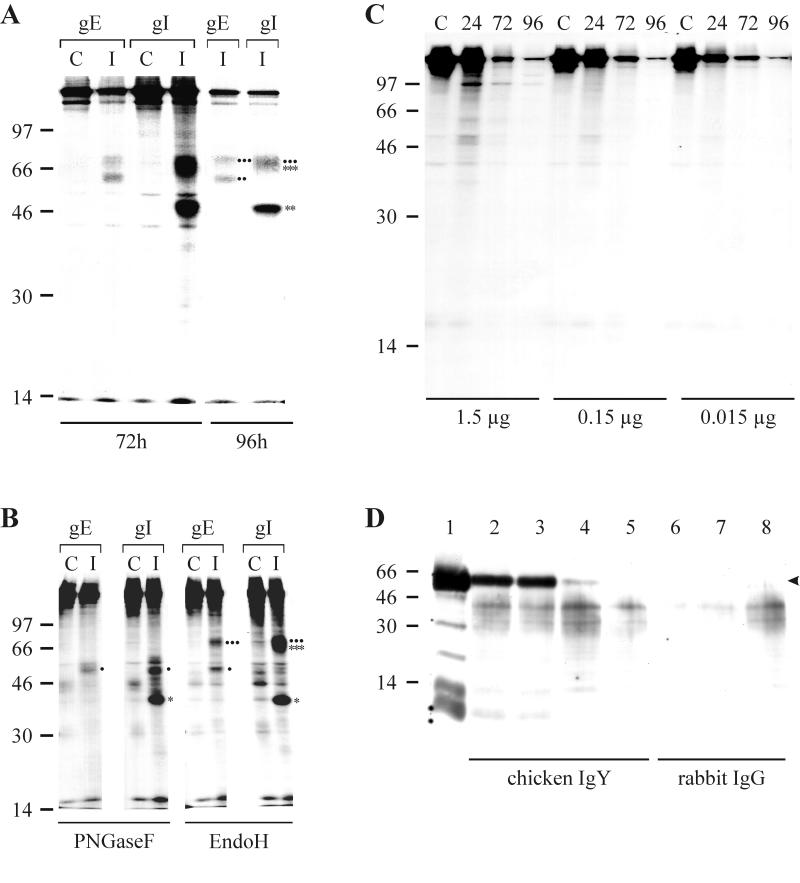
Previous work with MDV-1 strain RB1B had demonstrated that gE was coprecipitated when infected cell lysates were incubated with an anti-gI antibody (51). To verify expression of the glycoproteins in the 584Ap80C background and to analyze the putative gE-gI complex of MDV-1 in more detail, a gE-specific antiserum (anti-gE) directed against amino acid residues 420 to 497 of gE was generated after injection of a GST-gE fusion protein into a rabbit. Using the anti-gE and an anti-gI antibody (51) it could be shown by IIF that both gE and gI were expressed in BAC20-infected cells from 24 h postinfection (hpi) and throughout the observation period until 96 hpi (data not shown). The results of the IIF experiments were confirmed by performing RIPA using radiolabeled BAC20-infected cell lysates and anti-gE and anti-gI antibodies (Fig. 2A; results are shown for 72 and 96 hpi). SDS–10% PAGE of RIPA with the anti-gE antibody demonstrated that gE of MDV-1 strain 584Ap80C is expressed as two N-glycosylated moieties with molecular masses of approximately 60 to 62 kDa and 67 to 72 kDa (Fig. 2A). Treatment of the anti-gE-specific immunoprecipitates with PNGase F resulted in the appearance of a single 48-kDa protein band after SDS–10% PAGE (Fig. 2B). The higher-molecular-mass gE moiety (67 to 72 kDa) represented the Endo H-resistant mature form of the protein, because it was sensitive to treatment with PNGase F but not Endo H, whereas the faster-migrating form of gE contained high-mannose sugar side chains as reflected by its sensitivity to both PNGase F and Endo H (Fig. 2B). Radioimmunoprecipitates of BAC20-infected cell lysates using the anti-gI antibody contained proteins with apparent molecular masses of 45 to 47 kDa and 64 to 72 kDa after SDS–10% PAGE (Fig. 2A). Deglycosylation experiments with the immunoprecipitates obtained with the anti-gI antibody revealed the presence of 37- and 48-kDa bands after PNGase F digestion and protein bands of 37 kDa and 65 to 67 kDa after Endo H treatment (Fig. 2B). These results indicated that gI is expressed as a 37-kDa polypeptide which is primarily glycosylated to a high-mannose-containing glycoprotein of 45 to 47 kDa (Fig. 2B). The origin of the 65- to 67-kDa protein band which was resistant to Endo H treatment and specifically precipitated with the anti-gI but not the anti-gE antibody (Fig. 2B) is not entirely clear. We concluded that this band represents the fully glycosylated form of gI that partially comigrates with the Endo H-resistant mature form of gE. Alternatively, different forms of gE which are resistant to Endo H treatment may have been coprecipitated with the gI antibody (Fig. 2B). The observed reductions in apparent molecular masses of both gE and gI after PNGase F treatment exactly correspond to the calculated values for the gE or gI polypeptide backbone and also to sizes of the in vitro transcription-translation products (51). In addition, the detection of the gE precursor from precipitates using the anti-gI antibody after PNGase F but not after Endo H treatment strongly suggested that only mature gE forms a complex with gI (Fig. 2B).
FIG. 2.
(A to C) Digitally scanned images of radioimmunoprecipitates separated by SDS–10% PAGE. (A) Lysates of CEF infected with BAC20 virus or uninfected CEF were labeled with [35S]methionine and [35S]cysteine at 72 or 96 hpi. Cell lysates were prepared and reacted with rabbit antisera directed against gE or gI. The precipitated proteins are indicated. The mature (∗∗∗) and immature (∗∗) forms of gI as well as the mature (●●●) and immature (●●) forms of gE are given. (B) Precipitates of the anti-gE or anti-gI antibody obtained at 72 hpi were treated with PNGase F or Endo H. The various forms of gE and gI (unglycosylated precursors, Endo H-resistant and -sensitive forms) are indicated. The mature (∗∗∗) and precursor (∗) forms of gI as well as the mature (●●●) and precursor (●) forms of gE are given. Abbreviations in panels A and B: C, mock-infected cells; I, infected cells. (C) Seventy microliters of radiolabeled BAC20-infected or noninfected cell lysates were incubated with the indicated amounts of purified soluble IgY for 2 h on ice and precipitated with an IgY-specific monoclonal antibody. In panels A to C, sizes of a molecular mass marker ([14C] marker; Gibco-BRL) are given in kilodaltons. (D) Digitally scanned image of a Western blot of soluble chicken IgY or rabbit IgG (control) precipitated with the anti-IgY antibody. After immunoprecipitation using the anti-IgY antibody, precipitates were separated by SDS–10% PAGE, transferred to nitrocellulose, and probed with anti-chicken IgY peroxidase conjugate (Sigma). Lane 1 contains 1.0 μg of IgY detected with the conjugate. In lanes 2 to 5 and 6 to 8 immunoprecipitates of IgY or rabbit IgG, respectively, with the anti-IgY antibody were separated. Tenfold dilutions of IgY or IgG were used for the immunoprecipitations (lanes 2 and 6, 2 μg; lanes 3 and 7, 0.2 μg; lanes 4 and 8, 0.02 μg; lane 5, 0.002 μg). The arrowhead indicates the IgY γ chain with an apparent molecular mass of approximately 60 to 65 kDa. Sizes of a molecular mass marker (Seablue; Novex) are given in kilodaltons.
In a series of experiments using BAC20 lysates which had been radiolabeled and harvested at various times after infection, the putative binding of the gE-gI complex to soluble IgY was investigated. Radiolabeled infected-cell lysates (100 μl) were incubated with 1.5, 0.15, or 0.015 μg of purified soluble IgY which was subsequently precipitated using a mouse-anti-chicken IgY antibody (17). In none of the precipitates separated by SDS–10% PAGE could a precipitation of either gE or gI be observed (Fig. 2C). The precipitation of soluble chicken IgY but not rabbit IgG by the mouse anti-IgY antibody was shown by Western blot analyses of immunoprecipitates using the mouse anti-IgY antibody (Fig. 2D). Similar to the result obtained after immunoprecipitation using radiolabeled cell lysates, purified IgY did not specifically bind to BAC20-infected cultured cells before or after fixation (data not shown). Taken together, these results strongly suggested that the MDV-1 gE-gI complex does not have Fc receptor binding activity in vitro.
Characterization of MDV-1 BACs with deletions of the gE, gI, or both gE and gI genes.
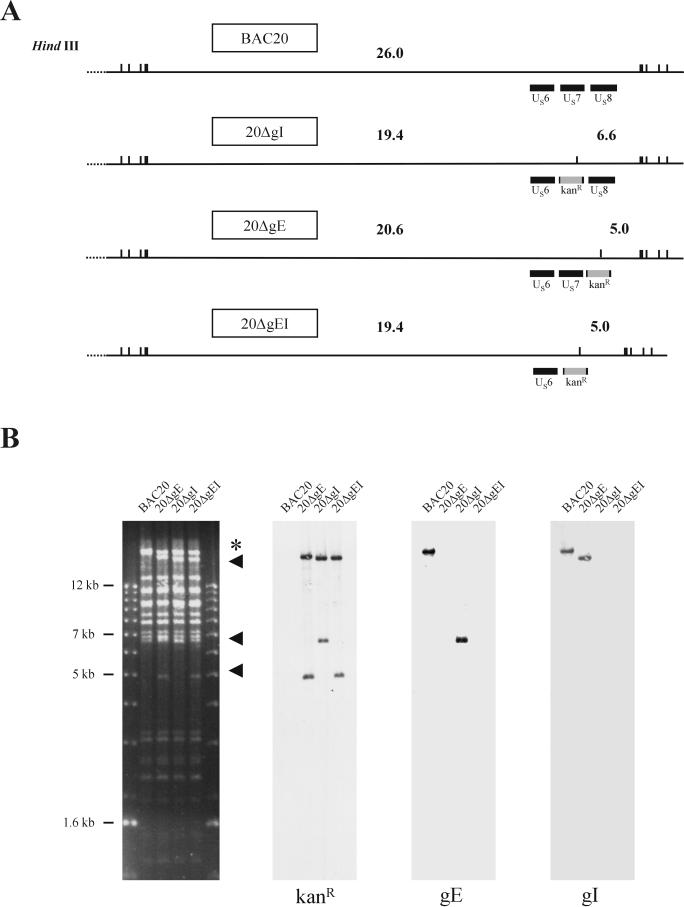
RecE/T mutagenesis was applied to remove the gI (US7) or gE (US8) or both ORFs in BAC20 DNA. Linear PCR fragments encoding the kanamycin resistance gene and 50-bp flanking sequences to allow homologous recombination were electroporated into BAC20-containing DH10B cells harboring plasmid pGETrec (39, 49). Approximately 50 BAC20 colonies which exhibited kanamycin resistance were obtained for each of the three mutants. Individual colonies were picked and analyzed by restriction enzyme analysis, Southern blotting, and cycle sequencing (41, 49). One colony from each transformation harboring a mutant BAC20 clone was chosen and termed 20ΔgE, 20ΔgI, or 20ΔgEI, respectively (Fig. 1). BAC DNA from colonies harboring the kanamycin resistance gene was prepared, digested with HindIII, and separated by 0.8% agarose gel electrophoresis (Fig. 3). In DNA cleaved with HindIII, alterations of the restriction enzyme fragments were detectable in mutant BAC clones (Fig. 3), because the insertion of the kanamycin resistance gene results in the introduction of an additional HindIII site in mutant BACs. Whereas a 26.0-kbp fragment encompassing the gE and gI ORFs was present in BAC20 (Fig. 3), fragments of 20.6 and 5.0 kbp in 20ΔgE, 19.4 and 6.6 kbp in 20ΔgI, and 19.4 and 5.0 kbp in 20ΔgEI were readily visible (Fig. 3). The genotypes of mutant BAC clones were confirmed by Southern blot analysis using kanamycin resistance gene-specific, gE, or gI probes (Fig. 3). As expected, the kanamycin resistance gene-specific probe only hybridized to fragments of mutant BAC DNA, whereas the gE and gI probes detected the 26.0-kbp fragment in BAC20 DNA only. Cycle sequencing of the junction regions between the kanamycin resistance gene and viral DNA corroborated the correct insertion of the antibiotic resistance gene and the absence of the respective ORF(s) in 20ΔgE, 20ΔgI, and 20ΔgEI, respectively (data not shown).
FIG. 3.
Calculated sizes of fragments of BAC20 and mutant BAC genomes after HindIII digestion (A) and digitally scanned images of Southern blots to analyze size variations in the various mutants (B). DNA from BAC20, 20ΔgE, 20ΔgI, or 20ΔgEI was cleaved with HindIII and transferred to nylon membranes. Sheets were incubated with digoxigenin-labeled kanamycin resistance gene-specific, gE, or gI probes. The 1-kb ladder (Gibco-BRL) was used as a size standard. Fragments that are altered in the mutant BAC genomes when compared to the BAC20 genome (5.0, 6.6, 19.4, and 20.6 kbp) are indicated (arrowheads); the 26.0-kbp fragment of BAC20 containing the gE and gI ORFs is marked with an asterisk.
Growth characteristics of 20ΔgE, 20ΔgI, and 20ΔgEI and rescuant viruses.
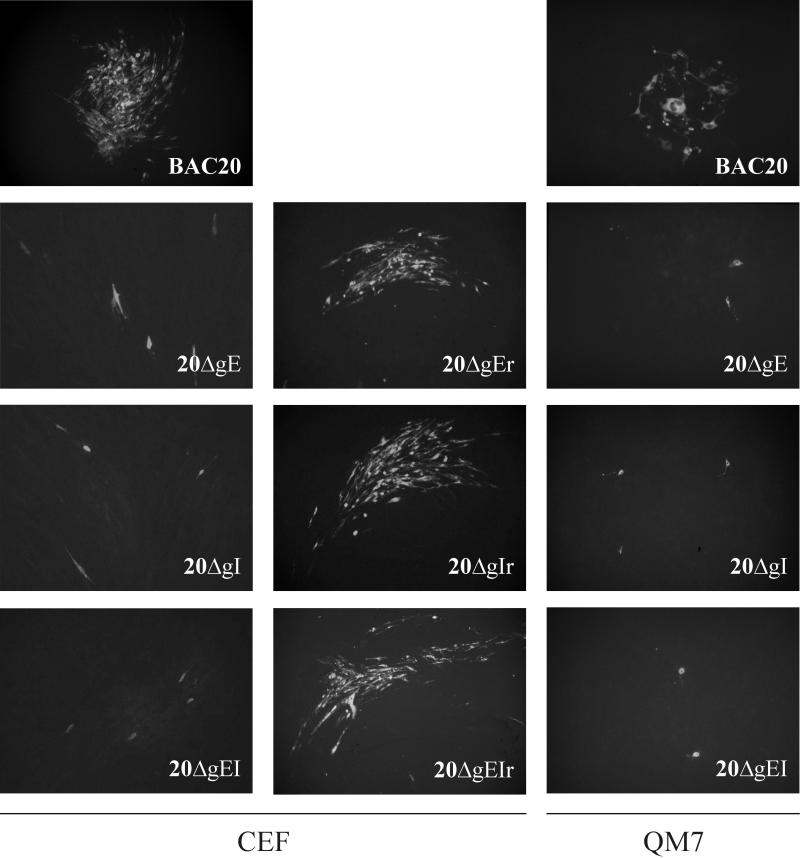
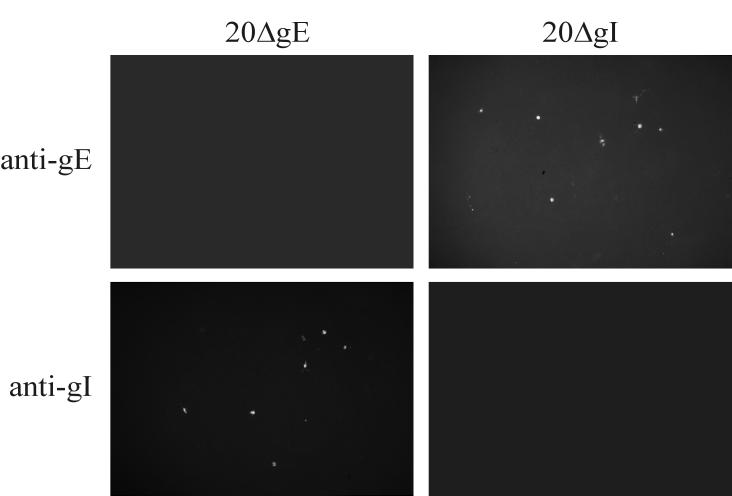
To analyze the growth properties of mutant MDV-1, DNA from mutant BAC clones was transfected into primary CEF or QM7 cells and analyzed for the appearance of virus plaques. Whereas MDV-1-specific plaques were visible from day 2 after transfection of parental BAC20 DNA into CEF, no virus plaques were obtained after transfection of either 20ΔgE, 20ΔgI, or 20ΔgEI DNA even after 7 days of incubation. These results were confirmed by IIF using the MDVI chicken antiserum or anti-gB MAb 2K11. Whereas the expected reactivity of viral plaques with the MDV-1-specific antibodies was observed in CEF transfected with BAC20, single cells only were reactive with either antibody after transfection of DNA obtained from any of the mutant BACs (data not shown). Similarly, no plaque formation was observed when cells transfected with 20ΔgE, 20ΔgI, or 20ΔgEI were coseeded with freshly prepared CEF at days 2 to 5 after infection, and only single infected cells were detectable after coseeding of CEF with cells transfected with mutant genomes (Fig. 4). MDV-1-specific plaques, however, could be easily identified after coseeding of CEF transfected with BAC20 and freshly prepared CEF (Fig. 4). A cell-type-specific essentiality of either gE or gI was excluded by performing identical transfection and infection experiments in QM7 cells. As described above, MDV-1 plaque formation was observed in cells transfected with BAC20 but not in those transfected with either 20ΔgE, 20ΔgI, or the double deletion mutant 20ΔgEI (Fig. 4). In contrast, rescuant viruses, in which the deleted gE gene (20ΔgE) or gI gene (20ΔgI) or both genes (20ΔgEI) had been reinserted, were able to produce plaques on CEF and QM7 cells which were indistinguishable from those induced by the parental BAC20 virus (Fig. 4). Rescuant viruses were isolated by cotransfection of the individual mutant BAC clones and PCR products encompassing the previously introduced deletions (Fig. 1). To verify expression of gE in the 20ΔgI and of gI in 20ΔgE mutant virus, respectively, QM7 cells were transfected with mutant MDV-1 BAC DNAs. The anti-gE antibody detected single cells after transfection of the 20ΔgI mutant, whereas the anti-gI antibody was reactive with single cells after transfection of 20ΔgE DNA (Fig. 5). No reactivity with either antibody was obtained in cells transfected or infected with the double deletion mutant 20ΔgEI (data not shown).
FIG. 4.
Growth of BAC20, mutant, and rescuant viruses on CEF or QM7 cells. Cells were infected with wild-type, mutant, or rescuant viruses by coseeding of cells transfected with the various viruses with fresh CEF or QM7 cells. At 4 days after infection, IIF using anti-gB MAb 2K11 was performed. In the case of BAC20 virus, plaque formation in CEF and QM7 cells was observed. In contrast, only single infected cells were observed after infection of CEF or QM7 cells with 20ΔgE, 20ΔgI, or 20ΔgEI. The ability to produce MDV-1-specific plaques was restored in rescuant viruses from 20ΔgE, 20ΔgI, and 20ΔgEI on both CEF and QM7 cells. Individual pictures represent views of 1,000 by 650 μm.
FIG. 5.
Detection of gE or gI expression in QM7 cells transfected with 20ΔgE or 20ΔgI DNA. At 5 days after transfection, cells were fixed and incubated with anti-gE or anti-gI antibodies. Expression of gE was readily detected in the case of 20ΔgI. Similarly, gI expression was demonstrated in cells transfected with 20ΔgE DNA. Individual pictures represent views of 1,000 by 650 μm.
Transcomplementation of growth of 20ΔgE and 20ΔgI.
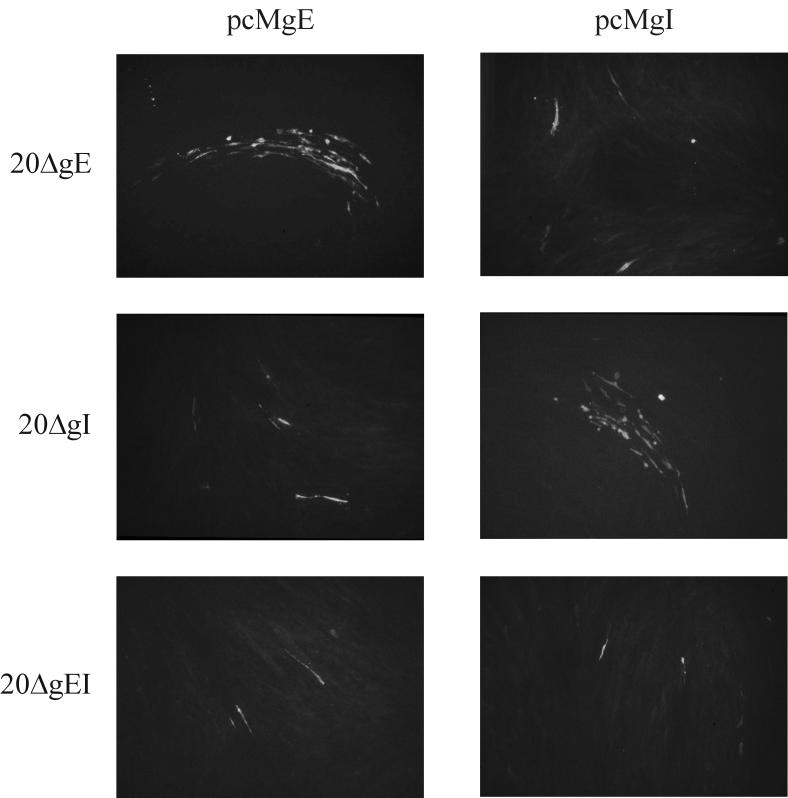
The transfection and coseeding experiments of the various mutants and the respective rescuant viruses strongly suggested that both gE and gI of MDV-1 are essential for virus growth in vitro and thus are crucially involved in cell-to-cell spread of MDV-1. To demonstrate that the two glycoproteins are indeed essential, transient transcomplementation assays were performed. Firstly, CEF were cotransfected with 20ΔgE and pcMgE, pcMgI, or pcMgM (41). Each of these plasmids expresses the respective MDV-1 ORF under the control of the HCMV IE promoter-enhancer (Fig. 1). Transfected cells were scanned for MDV-1 plaque formation from day 2 after transfection, and IIF was performed. Plaque formation was detectable from day 3 in CEF after cotransfection of 20ΔgE BAC DNA and the pcMgE expression plasmid. In contrast, no plaque formation but only single infected cells were observed in CEF cotransfected with the mutant genome and pcMgI or pcMgM (Fig. 6). Similarly, cell-to-cell spread capability of the gI-negative mutant (20ΔgI) was transcomplemented by the corresponding expression plasmid pcMgI only, and single infected cells were observed after cotransfection of the gI-negative virus and the gE or gM expression plasmid (Fig. 6). When DNA of the double deletion mutant 20ΔgEI was cotransfected with either pcMgE or pcMgI, however, no virus plaques were observed at any time after transfection. Only single infected cells were observed after staining of the monolayers with the anti-MDV-1 chicken antibody (Fig. 6). The transcomplementation of the single deletion mutants 20ΔgE and 20ΔgI could also be demonstrated in QM7 cells. After cotransfection of 20ΔgE with pcMgE as well as 20ΔgI with pcMgI, MDV-1 specific plaques were observed. In contrast, no plaque formation was observed after cotransfection of 20ΔgEI with either or both expression plasmids, or after cotransfection of DNA of single mutants with the noncorresponding expression plasmids in these cells (data not shown). From the results of the cotransfection experiments using CEF and QM7 cells it was concluded that cell-to-cell spread of MDV-1 in cultured cells is dependent on the expression of both gE and gI.
FIG. 6.
Transcomplementation of mutant viruses by the corresponding expression plasmids. Mutant 20ΔgE, 20ΔgI, or 20ΔgEI BAC DNA was cotransfected with pcMgE or pcMgI. Five days after transfection, cells were fixed with acetone and IIF using anti-gB MAb 2K11 was performed. Cell-to-cell spread of 20ΔgE and 20ΔgI was rescued by the corresponding expression plasmid (pcMgE and pcMgI). In contrast, growth of 20ΔgEI could not be restored by any expression plasmid. Individual pictures represent views of 1,000 by 650 μm
DISCUSSION
In this communication we have performed an analysis of the function of the gE-gI complex of MDV-1 strain 584Ap80C for virus growth. The salient findings reported here are that gE and gI form a complex that does not have chicken IgY Fc receptor binding activity and that deletion of either gE or gI even in a highly passaged MDV-1 results in virus mutants which are not viable in cultured cells. These findings demonstrate that gE and gI of MDV-1 are essential for virus growth in vitro, i.e., for direct cell-to-cell spread of infection.
gE and gI of members of the family Alphaherpesvirinae form a multifunctional complex which is involved in virus egress and cell-to-cell spread and represents an Fc receptor in HSV-1 and VZV (21, 27). The results of the RIPA reported here indicate that only the fully processed forms of gE and gI of MDV-1 form a complex and thus corroborate and extend previously reported data on RB1B gE-gI complex formation (51). Analysis of precipitates using gE- or gI-specific antibodies clearly indicates complex formation between the two glycoproteins, but the coprecipitation of mature gE with the gI antibody and very similar molecular masses of mature gE and gI make interpretation of the data difficult. At present, based on the results of previous studies (51) and the deglycosylation experiments performed here we propose that MDV-1 gE is synthesized as 48-kDa precursor protein in infected cells that is processed to a 60- to 62-kDa glycoprotein containing high-mannose sugar side chains. After entering the trans-Golgi network (TGN), gE is trimmed to the mature 67- to 72-kDa Endo H-resistant protein which complexes with gI. In case of gI, a 37-kDa protein precursor appears to be N-glycosylated to form a 45- to 47-kDa high-mannose-containing glycoprotein, which may be subsequently trimmed to a 65- to 67-kDa mature glycoprotein. The 65- to 67-kDa Endo H-resistant band was only observed after precipitation with the gI but not the gE antibody, but we cannot exclude the possibility that this band contains different forms of gE and not mature gI. It is interesting that two different polyclonal anti-gE antibodies did not precipitate gI in cells that had been infected with two different viruses (reference 51 and this study). Also, the anti-gI antibody was able to precipitate the mature form of gE only, indicating that full processing of MDV-1 gE may not be dependent on interaction with gI. The nature of gE-gI interaction and glycoprotein maturation will be addressed in detail after generation of novel MAbs and by using QM7 cell lines that constitutively express one of the glycoproteins and can be transfected with the respective complex partner, because we were not able to definitely clarify the origin of the bands obtained after immunoprecipitation with the available antibodies. The question of a putative Fc receptor activity of the MDV-1 gE-gI complex which has been reported for HSV-1, HSV-2, and VZV was examined by using purified chicken IgY (IgG) from egg yolk either in solution or on infected cells. Even with high amounts of purified IgY, we were not able to demonstrate a specific binding of MDV-1 gE or gI to the antibody preparation. This result is in good agreement with those from previous reports, inasmuch as Fc receptor activity so far has only been demonstrated for human but not animal alphaherpesviruses (20–22).
Concerning the effect on virus growth after deletion of either gE or gI in MDV-1, the experiments reported in this communication demonstrate that cell-to-cell spread of MDV-1 is essentially dependent on expression of gE and gI. Previous studies of the requirement of gE and gI for growth of VZV, a closely related virus that primarily grows by direct cell-to-cell spread in cultured cells, have shown that deletion of gE resulted in a marked reduction of virus growth, whereas a gI-negative VZV exhibited a cell type-specific growth restriction (11, 28, 29). The contributions of the gE-gI complex to direct cell-to-cell spread and the exact mechanisms by which the complex is involved in this process remain enigmatic. A number of recent studies, however, have shown that HSV-1 and also VZV gE and gI are present at tight junctions and that absence of gE or gI leads to missorting of HSV-1 virions in polarized epithelial cells (23, 31, 62). Also, interaction of HSV-1 gE with α-catenin, an F-actin binding protein, might suggest that cell-to-cell spread involves gE-gI as the viral counterpart and F-actin as the player of the host cell in this process (45). Alphaherpesvirus gE-gI complexes are thought to function in virus egress and cell-to-cell spread by virtue of interaction with tegument proteins, resulting in secondary envelopment of nucleocapsid at membranes of the Golgi network (6, 7,50). This view is substantiated by the fact that activity of the PrV gE-gI complex in vivo is impaired in the absence of the cytoplasmic tails of gE or gI (52–56). Whereas deletion of gE and/or gI can occur after serial passage of PrV and EHV-1 especially in nonhomologous cell culture systems (19, 33), simultaneous absence of the gE-gI complex and gM in PrV (6) or either gM or the UL49.5 product in EHV-1 leads to virus progeny that are severely impaired in both virus egress and direct cell-to-cell spread (50). These studies have revealed that the gE-gI and the gM-UL49.5 complexes serve overlapping but different functions in alphaherpesvirus egress and cell-to-cell spread. In the case of another alphaherpesvirus, BHV-1, absence of gE leads to an accelerated virus egress, at least early after infection and in the beginning of virus release from infected cells (58).
The apparently greater importance of gE and gI for growth of MDV-1 in cultured cells as demonstrated here may be caused by the smaller repertoire of MDV-1 glycoproteins expressed in cultured cells. In MDV-1-infected duck or chicken embryo fibroblasts, neither gD nor a gD-specific transcript is detectable (51), and, as indicated above, MDV-1 is transmitted from an infected to an uninfected cell only by virtue of direct cell-to-cell spread of virus and no free extracellular virus is produced (40, 66). In this respect, MDV-1 also closely resembles VZV, which produces plaques but only minute amounts of extracellular infectivity in cultured cells (20). It should also be noted that other members of the Alphaherpesvirinae family, such as PrV, that lack gD, gE, and gI are also severely impaired in cell-to-cell spread (34, 37). These observations emphasize the importance of gE and gI for cell-to-cell spread of alphaherpesviruses in the absence of gD. Based on the reported gE and/or gI function in various virus systems and the assumption that the glycoproteins encoded by the unique short region have arisen by virtue of gene duplication (30), it is conceivable that gD and the gE-gI complex may serve partially overlapping functions in cell-to-cell spread of members of the Alphaherpesvirinae family. In the concerted action of glycoproteins required for cell-to-cell spread, gD certainly plays an important role, although expression and function of this glycoprotein are highly variable: gD is entirely absent in the VZV genome (12), is not expressed by MDV-1 in cultured cells, is essential for virus entry only in the case of PrV, and is absolutely required for cell-to-cell spread and virus entry in HSV-1 and BHV-1 (12, 18, 26, 51). In BHV-1, the essentiality of gD in cell-to-cell spread can be overcome by serial passage of a gD-negative mutant in cultured cells (48). To further elucidate the possible interaction of unique short glycoproteins and cell-to-cell spread of members of the Alphaherpesvirinae family, future experiments will concentrate on the effect of a constitutive or inducible expression of gD in both wild-type and gE-gI-negative MDV-1, the trafficking properties of gE and gI in the presence or absence of the respective complex partner, and an assessment of the function of gM in growth of MDV-1. These studies may shed more light on the general principles of spread of alphaherpesviruses from an infected to an uninfected cell and in the functional interaction of the involved glycoproteins.
ACKNOWLEDGMENTS
We gratefully acknowledge the expert technical assistance of Kerstin Wink. Jean-Francois Vautherot, INRA, Tours, France, generously provided MAb 2K11 and Lucy Lee, Avian Disease and Oncology Laboratory, provided the anti-gI antiserum.
This work was supported by grant QLK2-CT-1999-00601 from the Commission of the European Union.
REFERENCES
- 1.Alconada A, Bauer U, Hoflack B. A tyrosine-based motif and a casein kinase II phosphorylation site regulate the intracellular trafficking of the varicella-zoster virus glycoprotein I, a protein localized in the trans-Golgi network. EMBO J. 1996;15:6096–6110. [PMC free article] [PubMed] [Google Scholar]
- 2.Alconada A, Bauer U, Baudoux L, Piette J, Hoflack B. Intracellular transport of the glycoproteins gE and gI of the varicella-zoster virus. gE accelerates the maturation of gI and determines its accumulation in the trans-Golgi network. J Biol Chem. 1998;273:13430–13436. doi: 10.1074/jbc.273.22.13430. [DOI] [PubMed] [Google Scholar]
- 3.Alconada A, Bauer U, Sodeik B, Hoflack B. Intracellular traffic of herpes simplex virus glycoprotein gE: characterization of the sorting signals required for its trans-Golgi network localization. J Virol. 1999;73:377–387. doi: 10.1128/jvi.73.1.377-387.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balan P, Davis-Poynter N, Bell S, Atkinson H, Browne H, Minson T. An analysis of the in vitro and in vivo phenotypes of mutants of herpes simplex virus type 1 lacking glycoproteins gG, gE, gI or the putative gJ. J Gen Virol. 1994;75:1245–1258. doi: 10.1099/0022-1317-75-6-1245. [DOI] [PubMed] [Google Scholar]
- 5.Binns M M, Ross N L. Nucleotide sequence of the Marek's disease virus (MDV) RB-1B A antigen gene and the identification of the MDV A antigen as the herpes simplex virus-1 glycoprotein C homologue. Virus Res. 1989;12:371–381. doi: 10.1016/0168-1702(89)90094-4. [DOI] [PubMed] [Google Scholar]
- 6.Brack A R, Dijkstra J M, Granzow H, Klupp B G, Mettenleiter T C. Inhibition of virion maturation by simultaneous deletion of glycoproteins E, I, and M of pseudorabies virus. J Virol. 1999;73:5364–5372. doi: 10.1128/jvi.73.7.5364-5372.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brack A R, Klupp B G, Granzow H, Tirabassi R, Enquist L W, Mettenleiter T C. Role of the cytoplasmic tail of pseudorabies virus glycoprotein E in virion formation. J Virol. 2000;74:4004–4016. doi: 10.1128/jvi.74.9.4004-4016.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calnek B W, Witter R L. Marek's disease. In: Calnek B W, editor. Diseases of poultry. Ames: Iowa State University Press; 1991. pp. 342–385. [Google Scholar]
- 9.Calnek B W, Adldinger H K, Kahn D E. Feather follicle epithelium: a source of enveloped and infectious cell-free herpesvirus from Marek's disease. Avian Dis. 1970;14:219–233. [PubMed] [Google Scholar]
- 10.Card J P, Whealy M E, Robbins A K, Enquist L W. Pseudorabies virus envelope glycoprotein gI influences both neurotropism and virulence during infection of the rat visual system. J Virol. 1992;66:3032–3041. doi: 10.1128/jvi.66.5.3032-3041.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen J I, Nguyen H. Varicella-zoster virus glycoprotein I is essential for growth of virus in Vero cells. J Virol. 1997;71:6913–6920. doi: 10.1128/jvi.71.9.6913-6920.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davison A J, Scott J E. The complete DNA sequence of varicella-zoster virus. J Gen Virol. 1986;67:1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- 13.Dingwell K S, Brunetti C R, Hendricks R L, Tang Q, Tang M, Rainbow A J, Johnson D C. Herpes simplex virus glycoproteins E and I facilitate cell-to-cell spread in vivo and across junctions of cultured cells. J Virol. 1994;68:834–845. doi: 10.1128/jvi.68.2.834-845.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dingwell K S, Doering L C, Johnson D C. Glycoproteins E and I facilitate neuron-to-neuron spread of herpes simplex virus. J Virol. 1995;69:7087–7098. doi: 10.1128/jvi.69.11.7087-7098.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dingwell K S, Johnson D C. The herpes simplex virus gE-gI complex facilitates cell-to-cell spread and binds to components of cell junctions. J Virol. 1998;72:8933–8942. doi: 10.1128/jvi.72.11.8933-8942.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enquist L W, Dubin J, Whealy M E, Card J P. Complementation analysis of pseudorabies virus gE and gI mutants in retinal ganglion cell neurotropism. J Virol. 1994;68:5275–5279. doi: 10.1128/jvi.68.8.5275-5279.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Erhard M H, Von Q, Schranner I, Jungling A, Kaspers B, Schmidt P, Kuhlmann R. Development of specific enzyme-linked immunosorbent antibody assay systems for the detection of chicken immunoglobulins G, M, and A using monoclonal antibodies. Poult Sci. 1992;71:302–310. doi: 10.3382/ps.0710302. [DOI] [PubMed] [Google Scholar]
- 18.Fehler F, Herrmann J M, Saalmüller A, Mettenleiter T C, Keil G M. Glycoprotein IV of bovine herpesvirus 1-expressing cell line complements and rescues a conditionally lethal viral mutant. J Virol. 1992;66:831–839. doi: 10.1128/jvi.66.2.831-839.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flowers C C, O'Callaghan D J. The equine herpesvirus type 1 (EHV-1) homolog of herpes simplex virus type 1 US9 and the nature of a major deletion within the unique short segment of the EHV-1 KyA strain genome. Virology. 1992;190:307–315. doi: 10.1016/0042-6822(92)91217-i. [DOI] [PubMed] [Google Scholar]
- 20.Gershon M D, Gershon A A. Role of glycoproteins in varicella-zoster virus infection. Contrib Microbiol. 1999;3:43–60. doi: 10.1159/000060309. [DOI] [PubMed] [Google Scholar]
- 21.Johnson D C, Feenstra V. Identification of a novel herpes simplex virus type 1-induced glycoprotein which complexes with gE and binds immunoglobulin. J Virol. 1987;61:2208–2216. doi: 10.1128/jvi.61.7.2208-2216.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson D C, Frame M C, Ligas M W, Cross A M, Stow N D. Herpes simplex virus immunoglobulin G Fc receptor activity depends on a complex of two viral glycoproteins, gE and gI. J Virol. 1988;62:1347–1354. doi: 10.1128/jvi.62.4.1347-1354.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson D C, Webb M, Wisner T W, Brunetti C. Herpes simplex virus gE/gI sorts nascent virions to epithelial cell junctions, promoting virus spread. J Virol. 2001;75:821–833. doi: 10.1128/JVI.75.2.821-833.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kritas S K, Nauwynck H J, Pensaert M B. Dissemination of wild-type and gC-, gE-and gI-deleted mutants of Aujeszky's disease virus in the maxillary nerve and trigeminal ganglion of pigs after intranasal inoculation. J Gen Virol. 1995;76:2063–2066. doi: 10.1099/0022-1317-76-8-2063. [DOI] [PubMed] [Google Scholar]
- 25.Lee L F, Wu P, Sui D, Ren D, Kamil J, Kung H J, Witter R L. The complete unique long sequence and the overall genomic organization of the GA strain of Marek's disease virus. Proc Natl Acad Sci USA. 2000;97:6091–6096. doi: 10.1073/pnas.97.11.6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ligas M W, Johnson D C. A herpes simplex virus mutant in which glycoprotein D sequences are replaced by beta-galactosidase sequences binds to but is unable to penetrate into cells. J Virol. 1988;62:1486–1494. doi: 10.1128/jvi.62.5.1486-1494.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Litwin V, Jackson W, Grose C. Receptor properties of two varicella-zoster virus glycoproteins, gpI and gpIV, homologous to herpes simplex virus gE and gI. J Virol. 1992;66:3643–3651. doi: 10.1128/jvi.66.6.3643-3651.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mallory S, Sommer M, Arvin A M. Mutational analysis of the role of glycoprotein I in varicella-zoster virus replication and its effects on glycoprotein E conformation and trafficking. J Virol. 1997;71:8279–8288. doi: 10.1128/jvi.71.11.8279-8288.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mallory S, Sommer M, Arvin A M. Analysis of the glycoproteins I and E of varicella-zoster virus (VZV) using deletional mutations of VZV cosmids. J Infect Dis. 1998;178:S22–S26. doi: 10.1086/514277. [DOI] [PubMed] [Google Scholar]
- 30.McGeoch D J. Evolutionary relationships of virion glycoprotein genes in the S regions of alphaherpesvirus genomes. J Gen Virol. 1990;71:2361–2367. doi: 10.1099/0022-1317-71-10-2361. [DOI] [PubMed] [Google Scholar]
- 31.McMillan T N, Johnson D C. Cytoplasmic domain of herpes simplex virus gE causes accumulation in the trans-Golgi network, a site of virus envelopment and sorting of virions to cell junctions. J Virol. 2001;75:1928–1940. doi: 10.1128/JVI.75.4.1928-1940.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meindl A, Osterrieder N. The equine herpesvirus 1 Us2 homolog encodes a nonessential membrane-associated virion component. J Virol. 1999;73:3430–3437. doi: 10.1128/jvi.73.4.3430-3437.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mettenleiter T C, Lomniczi B, Sugg N, Schreurs C, Ben-Porat T. Host cell-specific growth advantage of pseudorabies virus with a deletion in the genome sequences encoding a structural glycoprotein. J Virol. 1988;62:12–19. doi: 10.1128/jvi.62.1.12-19.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mettenleiter T C, Klupp B G, Weiland F, Visser N. Characterization of a quadruple glycoprotein-deleted pseudorabies virus mutant for use as a biologically safe live virus vaccine. J Gen Virol. 1994;75:4–33. doi: 10.1099/0022-1317-75-7-1723. [DOI] [PubMed] [Google Scholar]
- 35.Mijnes J D, van der Horst L M, van Anken E, Horzinek M C, Rottier P J, de Groot R J. Biosynthesis of glycoproteins E and I of feline herpesvirus: gE-gI interaction is required for intracellular transport. J Virol. 1996;70:5466–5475. doi: 10.1128/jvi.70.8.5466-5475.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mo C, Schneeberger E E, Arvin A M. Glycoprotein E of varicella-zoster virus enhances cell-cell contact in polarized epithelial cells. J Virol. 2000;74:11377–11387. doi: 10.1128/jvi.74.23.11377-11387.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mulder W, Pol J, Kimman T, Kok G, Priem J, Peeters B. Glycoprotein D-negative pseudorabies virus can spread transneuronally via direct neuron-to-neuron transmission in its natural host, the pig, but not after additional inactivation of gE or gI. J Virol. 1996;70:2191–2200. doi: 10.1128/jvi.70.4.2191-2200.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muyrers J P, Zhang Y, Testa G, Stewart A F. Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic Acids Res. 1999;27:1555–1557. doi: 10.1093/nar/27.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Narayanan K, Williamson R, Zhang Y, Stewart A F, Ioannou P A. Efficient and precise engineering of a 200 kb beta-globin human/bacterial artificial chromosome in E. coli DH10B using an inducible homologous recombination system. Gene Ther. 1999;6:442–447. doi: 10.1038/sj.gt.3300901. [DOI] [PubMed] [Google Scholar]
- 40.Niikura M, Witter R L, Jang H K, Ono M, Mikami T, Silva R F. MDV glycoprotein D is expressed in the feather follicle epithelium of infected chickens. Acta Virol. 1999;43:159–163. [PubMed] [Google Scholar]
- 41.Osterrieder N. Sequence and initial characterization of the U(L)10 (glycoprotein M) and U(L)11 homologous genes of serotype 1 Marek's Disease Virus. Arch Virol. 1999;144:1853–1863. doi: 10.1007/s007050050710. [DOI] [PubMed] [Google Scholar]
- 42.Payne L N, editor. Marek's disease—scientific basis and methods of control. Boston, Mass: Martinus Nijhoff Publishing; 1985. pp. 43–76. [Google Scholar]
- 43.Ren D, Lee L F, Coussens P M. Identification and characterization of Marek's disease virus genes homologous to ICP27 and glycoprotein K of herpes simplex virus-1. Virology. 1994;204:242–250. doi: 10.1006/viro.1994.1528. [DOI] [PubMed] [Google Scholar]
- 44.Ross L J, Sanderson M, Scott S D, Binns M M, Doel T, Milne B. Nucleotide sequence and characterization of the Marek's disease virus homologue of glycoprotein B of herpes simplex virus. J Gen Virol. 1989;70:1789–1804. doi: 10.1099/0022-1317-70-7-1789. [DOI] [PubMed] [Google Scholar]
- 45.Sakisaka T, Taniguchi T, Nakanishi H, Takahashi K, Miyahara M, Ikeda W, Yokoyama S, Peng Y F, Yamanishi K, Takai Y. Requirement of interaction of nectin-1alpha/HveC with afadin for efficient cell-cell spread of herpes simplex virus type 1. J Virol. 2001;75:4734–4743. doi: 10.1128/JVI.75.10.4734-4743.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 47.Schat K A. Characteristics of the virus. In: Payne L N, editor. Marek's disease. Hingham, Mass: Kluwer Academic Publishers; 1985. pp. 77–112. [Google Scholar]
- 48.Schröder C, Linde G, Fehler F, Keil G M. From essential to beneficial: glycoprotein D loses importance for replication of bovine herpesvirus 1 in cell culture. J Virol. 1997;71:25–33. doi: 10.1128/jvi.71.1.25-33.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schumacher D, Tischer B K, Fuchs W, Osterrieder N. Reconstitution of Marek's disease virus serotype 1 (MDV-1) from DNA cloned as a bacterial artificial chromosome and characterization of a glycoprotein B-negative MDV-1 mutant. J Virol. 2000;74:11088–11098. doi: 10.1128/jvi.74.23.11088-11098.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seyboldt C, Granzow H, Osterrieder N. Equine herpesvirus 1 (EHV-1) glycoprotein M: effect of deletions of transmembrane domains. Virology. 2000;278:477–489. doi: 10.1006/viro.2000.0664. [DOI] [PubMed] [Google Scholar]
- 51.Tan X, Brunovskis P, Velicer L F. Transcriptional analysis of Marek's disease virus glycoprotein D, I, and E genes: gD expression is undetectable in cell culture. J Virol. 2001;75:2067–2075. doi: 10.1128/JVI.75.5.2067-2075.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tirabassi R S, Townley R A, Eldridge M G, Enquist L W. Characterization of pseudorabies virus mutants expressing carboxy-terminal truncations of gE: evidence for envelope incorporation, virulence, and neurotropism domains. J Virol. 1997;71:6455–6464. doi: 10.1128/jvi.71.9.6455-6464.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tirabassi R S, Enquist L W. Role of envelope protein gE endocytosis in the pseudorabies virus life cycle. J Virol. 1998;72:4571–4579. doi: 10.1128/jvi.72.6.4571-4579.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tirabassi R S, Townley R A, Eldridge M G, Enquist L W. Molecular mechanisms of neurotropic herpesvirus invasion and spread in the CNS. Neurosci Biobehav Rev. 1998;22:709–720. doi: 10.1016/s0149-7634(98)00009-8. [DOI] [PubMed] [Google Scholar]
- 55.Tirabassi R S, Enquist L W. Mutation of the YXXL endocytosis motif in the cytoplasmic tail of pseudorabies virus gE. J Virol. 1999;73:2717–2728. doi: 10.1128/jvi.73.4.2717-2728.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tirabassi R S, Enquist L W. Role of the pseudorabies virus gI cytoplasmic domain in neuroinvasion, virulence, and posttranslational N-linked glycosylation. J Virol. 2000;74:3505–3516. doi: 10.1128/jvi.74.8.3505-3516.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tulman E R, Afonso C L, Lu Z, Zsak L, Rock D L, Kutish G F. The genome of a very virulent Marek's disease virus. J Virol. 2000;74:7980–7988. doi: 10.1128/jvi.74.17.7980-7988.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Engelenburg F A, Kaashoek M J, Rijsewijk F A, van den Burg L, Moerman A, Gielkens A L, van Oirschot J T. A glycoprotein E deletion mutant of bovine herpesvirus 1 is avirulent in calves. J Gen Virol. 1994;75:2311–2318. doi: 10.1099/0022-1317-75-9-2311. [DOI] [PubMed] [Google Scholar]
- 59.van Regenmortel M H V, Fauquet C M, Bishop D H L, Carstens E B, Estes M K, Lemon S M, Maniloff J, Mayo M A, McGeoch D J, Pringle C R, Wickner R B, editors. Virus Taxonomy. Seventh Report of the International Committee on Taxonomy of Viruses. New York, N.Y: Academic Press; 1999. [Google Scholar]
- 60.Wang Z H, Gershon M D, Lungu O, Zhu Z, Mallory S, Arvin A M, Gershon A A. Essential role played by the C-terminal domain of glycoprotein I in envelopment of varicella-zoster virus in the trans-Golgi network: interactions of glycoproteins with tegument. J Virol. 2001;75:323–340. doi: 10.1128/JVI.75.1.323-340.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Whealy M E, Card J P, Robbins A K, Dubin J R, Rziha H J, Enquist L W. Specific pseudorabies virus infection of the rat visual system requires both gI and gp63 glycoproteins. J Virol. 1993;67:3786–3797. doi: 10.1128/jvi.67.7.3786-3797.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wisner T, Brunetti C, Dingwell K, Johnson D C. The extracellular domain of herpes simplex virus gE is sufficient for accumulation at cell junctions but not for cell-to-cell spread. J Virol. 2000;74:2278–2287. doi: 10.1128/jvi.74.5.2278-2287.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu P, Reed W M, Yoshida S, Sui D, Lee L F. Identification and characterization of glycoprotein H of MDV-1 GA strain. Acta Virol. 1999;43:152–158. [PubMed] [Google Scholar]
- 64.Yoshida S, Lee L F, Yanagida N, Nazerian K. Identification and characterization of a Marek's disease virus gene homologous to glycoprotein L of herpes simplex virus. Virology. 1994;204:414–419. doi: 10.1006/viro.1994.1546. [DOI] [PubMed] [Google Scholar]
- 65.Yoshitake N, Xuan X, Otsuka H. Identification and characterization of bovine herpesvirus-1 glycoproteins E and I. J Gen Virol. 1997;78:1399–1403. doi: 10.1099/0022-1317-78-6-1399. [DOI] [PubMed] [Google Scholar]
- 66.Zelnik V, Majerciak V, Szabova D, Geerligs H, Kopacek J, Ross L J, Pastorek J. Glycoprotein gD of MDV lacks functions typical for alpha-herpesvirus gD homologues. Acta Virol. 1999;43:164–168. [PubMed] [Google Scholar]
- 67.Zhang Y, Buchholz F, Muyrers J P, Stewart A F. A new logic for DNA engineering using recombination in Escherichia coli. Nat Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 68.Zuckermann F A, Mettenleiter T C, Schreurs C, Sugg N, Ben-Porat T. Complex between glycoproteins gI and gp63 of pseudorabies virus: its effect on virus replication. J Virol. 1988;62:4622–4626. doi: 10.1128/jvi.62.12.4622-4626.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]