Abstract
FBXO protein family plays an essential role in the ubiquitination process acting as E3 ligases, which may contribute to the progression of cancers. However, the molecular functions of FBXOs in hepatocellular carcinoma (HCC) remain incompletely understood. Here, we investigated the overlapping genes between the FBXOs and differentially expressed genes (DEGs) of HCC identified by utilizing The Cancer Genome Atlas (TCGA) dataset, then, a prognostic model with effective predictive capacity was constructed based on the uni-cox and LASSO regression analyses. To elucidate the underlying mechanism of the FBXO model genes, KEGG analysis was carried out. Drug metabolism-cytochrome P450 and retinol metabolism were revealed as the potential pathway, which Increased the credibility of subsequent drug prediction research. Meanwhile, patients divided by the prognostic model showed a different immune infiltrating status and we also found FBXO model genes may ubiquitinate P53, inducing TP53 more prone to mutations, thereby promoting the occurrence and development of tumors. Consistent with these findings, the result of immunohistochemistry (IHC) validated an elevated expression of these model genes in HCC tissues than in the adjacent tissues. The primary aim of this investigation is to formulate a prognostic model while exploring the underlying mechanisms associated with FBXO genes in HCC. These findings offer initial research perspectives on the involvement of FBXO genes in HCC and contribute to the discovery of dependable biomarkers for the management, prognostication, and early detection of HCC in patients.
Keywords: FBXO protein family genes, Hepatocellular carcinoma (HCC), Ubiquitination, P53
Introduction
Liver cancer ranks as the fourth most prevalent reason of cancer-related mortality (Villanueva 2019). Approximately only 18% of liver cancer patients survive beyond 5 years, and in 2030, over one million patients are estimated to die from liver cancer (Wang et al. 2022a, b). Furthermore, hepatocellular carcinoma (HCC) is the main form of primary liver malignancies which exhibits a markedly unfavorable prognosis primarily attributed to the postoperative recurrence and primary tumor metastasis (Budhu et al. 2006). Nevertheless, due to the absence of effective biomarkers, the clinical diagnosis of HCC remains limited. Therefore, the early identification and timely intervention are important for enhancing the prognosis of HCC.
Nowadays, an increasing number of literatures have substantiated the association between the FBXO genes and HCC. F-box is one of the E3 ligase which commonly referred to as ubiquitin ligases and FBXO protein family is a sort of F-box only with uncharacterized domains (Tekcham et al. 2020). Moreover, E3 ubiquitin ligases are a fundamental component of the ubiquitination cascade (Tekcham et al. 2020), and they are particularly essential in the ultimate stage of ubiquitin conjugation, as they facilitate the transfer of ubiquitin molecules from ubiquitin-conjugating enzymes (E2s) to the specific substrates they target (Buetow and Huang 2016). The ubiquitination protein degradation pathway is a crucial molecular process in the human body, exhibiting significant selectivity (Liu et al. 2021). Current evidence supports its occurrence in nearly all biological mechanisms, including oncogenesis and the pathological processes of carcinoma (Tekcham et al. 2020). However, dysregulation of and E3 ubiquitin ligases has been observed in HCC, resulting in altered ubiquitination dynamics, and contributing to tumor initiation, progression, and metastasis (Lin et al. 2021). From the above content, it is concluded that protein ubiquitination is an crutial determinant of cellular homeostasis, and ubiquitin-specific enzymes, particularly deubiquitinates (DUBs), are emerging as promising targets for drug development. DUBs are composed of seven different subfamilies, among which the ubiquitin-specific protease (USPs) family includes USP1, whose deregulated expression and activity have been found in several human cancers. Although more studies are needed to verify the molecular mechanism of USP1 in carcinogenesis to promote its clinical application in the future, this still indicates that targeting USP1 is a feasible anticancer therapy (Huang et al. 2023). Besides, an article has showed FBXO45 exhibited an augmenting effect on the ubiquitination process at specific amino acid residues and the core role of FBXO45-mediated ubiquitination in regulating the onset of liver tumorigenesis (Lin et al. 2021). Moreover, there are some articles showed that FBXO31 maybe as a tumor suppressor in HCC (Huang et al. 2010) and FBXO22 could potentially manage cell proliferation in HCC by directing the degradation of the p53/p21 complex (Zhang et al. 2019). Interestingly, so far for prognostic influence and the specific function of FBXO family genes in HCC are largely unknown. To date, only a few articles have examined the relationship between the FBXO protein family and other cancers (Liu et al. 2021). Therefore, it is reasonable to explore whether most of the FBXO family genes are associated with the pathological progression and prognostic outcomes of HCC.
In these years, numerous articles have consistently demonstrated a robust association between immunity and the prognosis as well as the progress of HCC. In the existing studied, it has been demonstrated that immune checkpoint therapy confers survival benefit to a larger number of patients afflicted with HCC (Xu et al. 2018a, b; Ho et al. 2020). Among them, immune T cells have also been shown to be associated with HCC (Liu et al. 2020; Zhang et al. 2021). For example, the depletion of CD8 + T cells due to exhaustion has a significant impact on both tumor progression and the effectiveness of cancer immunotherapy (Liu et al. 2020) and an increased accumulation of CD8 + Ai-TCR-T cells within tumors has been observed to be positively correlated with tumor shrinkage (Zhang et al. 2021). Moreover, The FBXO protein family is also linked to immunological processes (Zhang et al. 2021; Yang et al. 2020; Cheng et al. 2022). According to research the presence of some of the FBXO protein family and their activated states correlate with immune infiltrations include T cells and macrophage in pancreatic cancer (Cheng et al. 2022). Lately, emerging therapeutic approaches such as cancer immunosuppressive therapy have been proved potentially to extend the survival of patients (Oura et al. 2021). Importantly, the incorporation of immune into HCC research exhibits as a substantial progression in the discipline, presenting the opportunity to augment prognostic outcomes. Consequently, our forthcoming investigation will probe the potential link of immunity and FBXO family genes in HCC patients.
In this study (flowchart shown in Fig. 1), the initial patient dataset afflicted with HCC was acquired from The Cancer Genome Atlas (TCGA) database. Subsequently, a thorough selection process was applied to discern the overlapping genes between the FBXOs and differentially expressed genes. Following this, we developed a prognostic model associated with the FBXOs by utilizing a LASSO COX regression analysis. Furthermore, the effect of the risk model was assessed through univariate and multivariate COX regression analyses, Kaplan-Meier (KM) survival curve assessment, receiver operating characteristic (ROC) curve examination, and the formulation of a nomogram. In addition, immunological effects, and ubiquitin substrate prediction of FBXO in HCC is also involved in the article. Ultimately, IHC was used to corroborate these genes expression situation in HCC samples. In conclusion, our study provides potentially effective markers for early detection and prognostic assessment of HCC.
Fig. 1.
Flowchart of construction and analysis of the prognostic model
Materials and methods
Data collection and processing
The datasets encompassing HCC were obtained by retrieving comprehensive information from the TCGA database (https://portal.gdc.cancer.gov/), specifically TCGA-LIHC. These datasets included survival information, gene expression profiles, and clinicopathological data, all of them were essential for our research analysis. Furthermore, a systematic screening process was conducted to download the gene expression data specifically pertaining to six members of the FBXO protein family from the GeneCards database. (https://www.genecards.org/) and 26 genes of the FBXO protein family from PubMed (https://pubmed.ncbi.nlm.nih.gov/).
String
The String website (https://cn.string-db.org/) which constituted a search instrument facilitating the exploration of established and anticipatory protein interrelationships was used to build the Protein-Protein Interaction (PPI) networks of the 32 genes. Its repository includes empirical datasets, outcomes derived from the computational scrutiny of abstracts within the PubMed database, amalgamated information sourced from diverse repositories, as well as prognostic inferences generated by means of bioinformatic methodologies. In this study, we delineated protein-protein interaction (PPI) networks inherent to the FBXO gene family employing the resources made available by the String database.
Establishment of risk prognostic model
TCGA database was used to validate the FBXO protein family’s differential expressions. In addition, univariate Cox regression analysis was conducted to discover underlying FBXO protein family genes related to overall survival (OS) with a threshold standard of P < 0.05. Among the 32 genes examined, twelve (FBXO4, FBXO5, FBXO6, FBXO10, FBXO11, FBXO16, FBXO30, FBXO32, FBXO41, FBXO42, FBXO43, FBXO45) demonstrated significant prognostic value. Subsequently, we utilized the LASSO regression model, implemented through the R package glmnet, to refine the selection of potential genes and construct a prognostic model. Employing Least Absolute Shrinkage and Selection Operator (LASSO) Cox regression analysis with the penalty parameter estimated through 10-fold cross-validation was to diminish the pool of candidate FBXO genes and enhance their practical relevance in a clinical setting. The computation of the risk score was accomplished via a formulation encompassing gene expression levels and their corresponding regression coefficients inherent to the signature. (Risk score = (expression of Gene 1 * β1) + (expression of Gene 2 * β2) + … + (expression of Gene n * βn). Here, β represents the regression coefficient assigned to each gene within the prognostic signature).
Assessment of risk prognostic model
The risk score pertaining to each TCGA-LIHC sample was computed employing a dedicated formula, subsequently segregating patients into high-risk and low-risk groups in accordance with the median. KM survival curves were constructed utilizing the log-rank test to contrast the overall survival (OS) outcomes across the two groups. The viability of the prognostic risk core prediction model was further affirmed through ROC analysis, PCA, and t-SNE assessments conducted within the HCC patient population. Univariate Cox proportional hazard models assessed the dependency between the prognostic model and OS, while its potential as an independent prognostic predictor was evaluated by multivariate Cox regression analysis. Additionally, a nomogram incorporating clinicopathological information and risk score was constructed using the “rms” R package. The Complex Heatmap package in R was used to make heatmaps which was visualized the relationship between clinical parameters and prognostic model genes. These analyses contribute to a comprehensive understanding of the prognostic model’s performance in liver cancer patients.
Functional enrichment analysis
Enrichment analysis of the model genes in HCC was carried out by the Cluster Profiler R package. Besides, there were two patterns being examined: co-expression analysis of 6 genes related to the model genes, and differential expression analysis between the high- and low-risk groups. Moreover, GO and KEGG databases were used to elucidate gene pathways and functions. Statistical significance was determined by adjusted p-values (p < 0.05). This analysis revealed the pathological pathways and biological functions associated with the model genes in HCC.
Immune analysis
KEGG pathway analysis also highlighted their involvement in retinol metabolism which indicated a close relation between FBXO gene family and immunity in HCC. The GSVA R package was employed by Immune infiltration analysis which further explored the impact of these prognostic model genes on immunity, shedding light on their potential implications in HCC. Additionally, ssGSEA was used to evaluate immune infiltration between the high-risk and low-risk groups.
Immunohistochemistry (IHC) staining
The research methodology was granted approval by the Institutional Research Ethics Committee of Chongqing Medical University, and all enlisted participants were informed consent. Specimens of both HCC and adjacent tissues were procured for analysis and subsequently preserved utilizing formalin fixation. The fixed tissue samples were then embedded within paraffin wax, and subsequently, they were meticulously sectioned into slices measuring 4 μm in thickness. After degumming and rehydration through a graded series of ethanol and xylene solutions, the paraffin-embedded sections underwent an overnight incubation at 4 °C with six primary antibodies of the FBXO module genes. Then, it incubated with secondary antibodies for a duration of 30 min at 37 °C. After the washing procedures, the tissue samples were counterstained with hematoxylin, subjected to progressive dehydration through a series of ethanol-xylene solutions, and ultimately sealed by neutral balsam. In the end the ensuing staining patterns were meticulously evaluated and scored by a proficient pathologist who maintained a state of blinding regarding the underlying study. The assessment was performed employing a scoring framework that ranged from 0 to 3.
Ubiquitin substrate prediction
UbiBrowser 2.0 2022b provides a database of predicted E3-substrate interactions for the entire human proteome, and it was applied to predict the ubiquitin substrate of FBXOs. UbiBrowser is the first system for predicting and displaying interactions between human ubiquitin ligases and substrates. Protein ubiquitination is a common type of post-translational modification, where ubiquitin, a small protein molecule, can bind to specific target proteins under the action of a series of specialized enzymes (ubiquitin-activating enzyme E1, conjugating enzyme E2, and ubiquitin ligase E3), leading to ubiquitination modifications. These ubiquitination modifications often result in the degradation of the target protein, thereby playing a role in regulating protein expression.
Statistical analysis
All data are presented as means ± SD (standard errors). In addition, GraphPad Prism software (version 6.0) (San Diego, CA, USA) was utilized for generating the figures. Student’s t-test was employed for conducting group comparisons. A p-value of < 0.05 was taken for statistically significant, indicating the presence of a meaningful difference between the compared groups.
Results
32 FBXO genes were discerned based on TCGA database
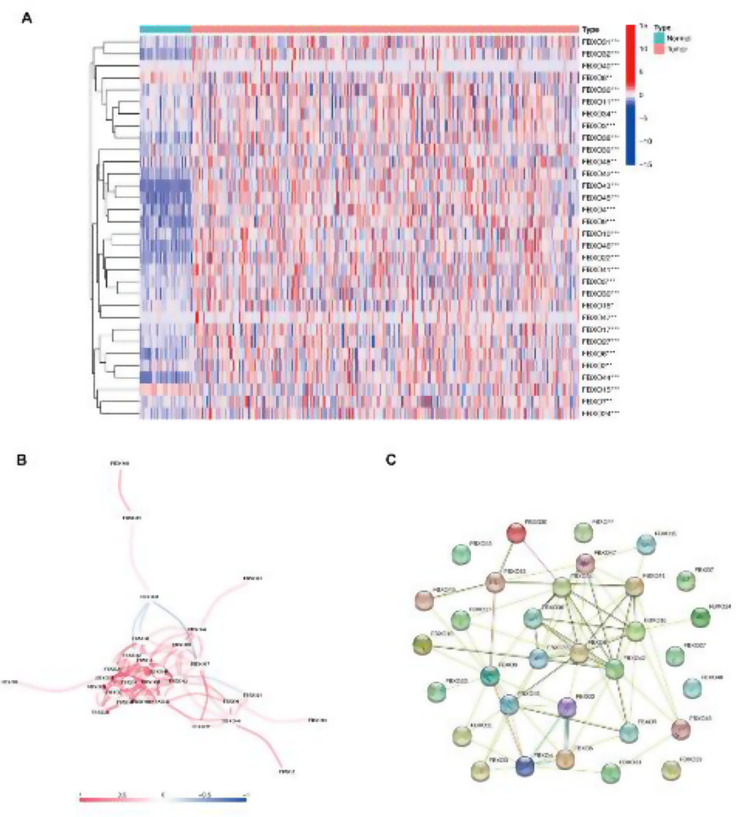
Initially, the differential expressed genes (DEGs) obtained from TCGA database. The intersection of the above DEGs and FBXO protein family 32 genes was established to get gens concerned to DEGs (NRDEGs) and FBXO protein family. After that, the heatmap was used to identify 32 FBXO family genes which were statistically significant in low- and high - risk groups in HCC (Fig. 2A). Additionally, we also showed the mRNA correlation of the 12 FBXO genes and the PPI networks of them in the String database (Fig. 2B, C).
Fig. 2.
32 DEGs related to FBXO protein family expression patterns in tumor and adjacent normal liver tissues based on TCGA- LIHC dataset. (A) Heatmap of 32 Differentially Expressed Genes (DEGs) associated with the expression patterns of FBXO protein family were identified in tumor tissues compared to adjacent normal liver tissues based on TCGA-LIHC database. (B) The mRNA correlation of the genes. (C) PPI networks of FBXO family genes in the String database
Successfully established a risk prognostic model related to FBXO protein family
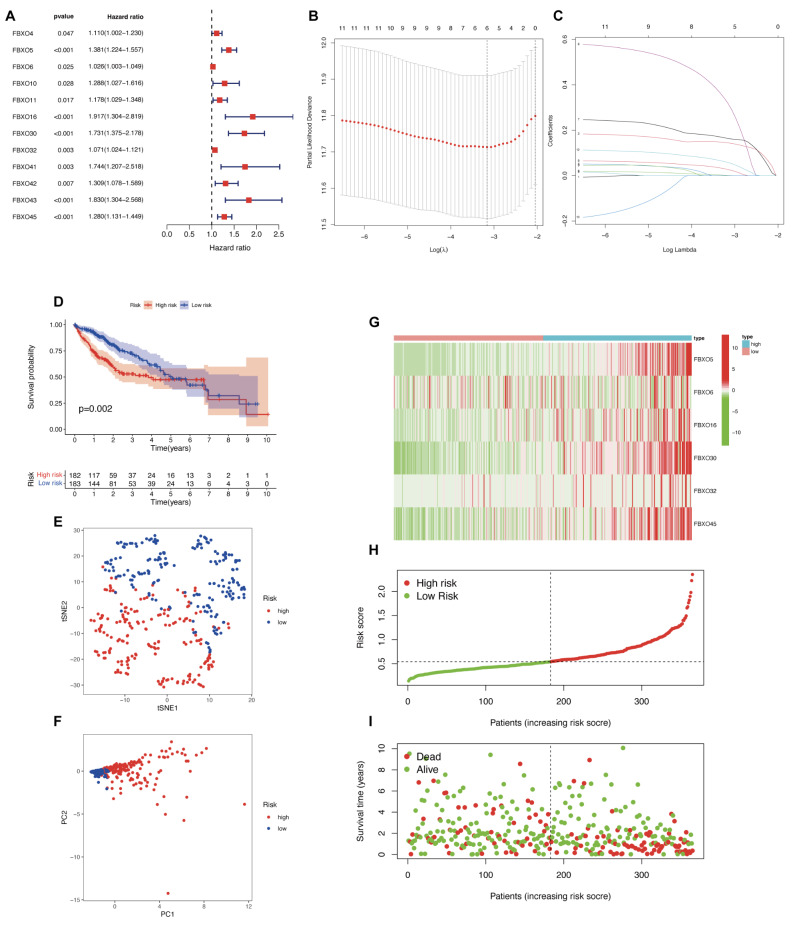
Initially, the survminer package (version 0.4.9) and survival package (version 3.2–10) were used to paint the Univariate Cox regression analysis of 12 genes which had prognostic significances including FBXO4, FBXO5, FBXO6, FBXO10, FBXO11, FBXO16, FBXO30, FBXO32, FBXO41, FBXO42, FBXO43, FBXO45. (Fig. 3A). After that, we conducted Lasso analysis to select 6 FBXO genes (FBXO5, FBXO6, FBXO16, FBXO30, FBXO32, FBXO45) from the 12 genes which have prognostic significance for the construction of an optimized risk prognosis model, utilizing a penalty coefficient for these genes (Fig. 3B-C). Respectively, the regression coefficients β1-β6 were derived as 0.144, 0.0006, 0.319, 0.164, 0.027, and 0.058. The risk score assigned to each patient was calculated through the application of the expression levels of the six genes, along with their corresponding regression coefficients, as per the subsequent formula: Risk score = EXP FBXO5*0.144 + EXP FBXO6*0.0006 + EXP FBXO16*0.318 + EXP FBXO30*0.319 + EXP FBXO32*0.027 + EXP FBXO45*0.058. On the base of risk scores and following a median division, the cohort of patients was categorized into high-risk and low-risk subsets. In addition, KM survival curves were subsequently employed to depict that the low-risk group exhibited a more favorable prognostic outcome in comparison to the high-risk group. (Fig. 3D). Notably, we successfully stratified HCC patients into distinct risk groups, exhibiting a discernible separation through principal component analysis (PCA) and t-distributed stochastic neighbor embedding (t-SNE) analysis (Fig. 3E, F). Furthermore, the differences in the expression of the six FBXO family genes across low- and high-risk groups of HCC patients within the TCGA dataset were elucidated by the heatmap (Fig. 3G) and subsequent analyses encompassed the assessment of risk curve scores as well as the distribution of risk scores for individual patients. (Figs. 3H-I).
Fig. 3.
Establishment of a prognostic model based on the FBXO model genes in HCC. (A) The univariable Cox HR regression of the 12 FBXO genes showed the HR and p-value in HCC (Criteria: p-value < 0.05). (B) Ten-time cross-validation for tuning parameter selection in the LASSO model. (C) LASSO coefficient profiles. (D) Kaplan-Meier curves showed that low-risk group had a better prognosis than the high-risk group. (E) A dot plot was generated to visualize the distinct clusters of low- and high-risk groups identified through the t-distributed Stochastic. (F) PCA plot for HCC patient. (G) Heatmap of the expression patterns of the six FBXO genes across the low- and high-risk groups. (H) The risk curve of each sample rearranged by risk score of each HCC patients. (I) The scatter plot of each HCC patient samples ‘survival status
Evaluation of the prognostic model
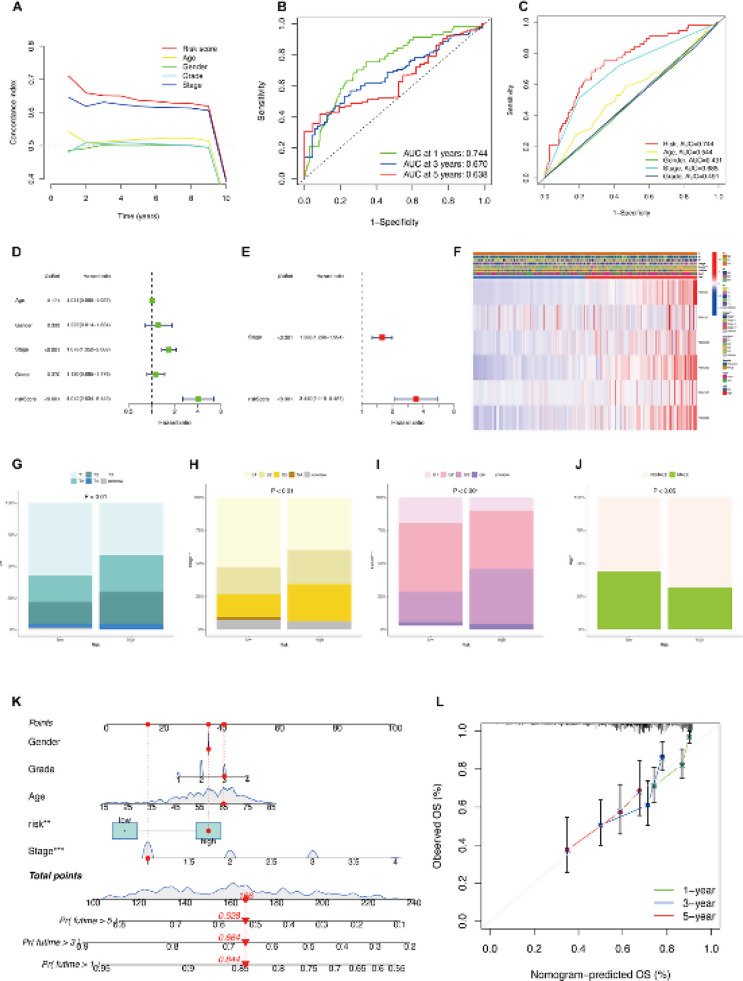
The application of the concordance index (C-index) facilitated the evaluation of correlation between anticipated survival probabilities and actual survival outcomes pertaining to each clinical parameter and it was observed that risk score and stage exhibited greater significance (Fig. 4A). Time-dependent Receiver Operating Characteristic (ROC) curves were employed to assess the predictive accuracy of the established models for overall survival (OS) in hepatocellular carcinoma (HCC) patients. The analysis revealed area under the curve (AUC) values of 0.744, 0.670, and 0.638 for 1-year, 3-year, and 5-year predictions, respectively (Fig. 4B). Comparatively, the Receiver Operating Characteristic (ROC) curves derived from models integrating clinical parameters alongside the risk score revealed that variables such as age, gender, stage, and grade manifested heightened levels of sensitivity and specificity in predictive capability when contrasted with the sole utilization of the risk score (Fig. 4C).
Fig. 4.
A FBXO-related Risk Prognostic Model was successfully constructed. (A) The concordance index (C-index) measures the risk score, age, gender, grade, and stage with time. (B) Calculate the AUC for 1-year, 3-year, and 5-year predicted OS of the total survival risk score according to the ROC curve. (C) Calculate the AUC for risk score, age, gender, stage, and grade of the total survival risk score according to the ROC curve. (D, E) Univariate and multivariate Cox regression analysis of clinicopathological features of HCC associated with OS. (F) Heatmap of the expressions of 6 FBXO genes and clinicopathologic characters of the low- and high-risk groups. (G-I) Relationship between FBXOs expression and tumor stage in HCC patients. (G) T stage, (H) S stage, (I) N stage. (J) Relationship between FBXOs expression and gander in HCC patients. (K) Clinical characteristics and prognostic model were used to establish a predictive nomogram. (L) Predictive nomogram showed the OS
Univariate and multivariate Cox regression analysis showed that risk score, and clinical stage were prognostic predictors in TCGA-LIHC instead of age, gender, and pathology grade (Fig. 4D-E). A heatmap was generated to depict the relationship between the genes of the clinical and model parameters (Fig. 4F). Moreover, the utilization of bar charts was employed to depict the association between clinical staging (T stage, S stage, G stage) and high-risk and low-risk groups (Fig. 4G-I). The analysis revealed significant associations between all stages and the respective risk groups. Additionally, a bar chart was constructed to investigate the correlation between age and the risk groups (Fig. 4J). Furthermore, we applied the prognostic model and clinical characteristics to develop a predictive nomogram aimed at setting up the survival probability of 1, 3, and 5 years in HCC patients. (Fig. 4K)
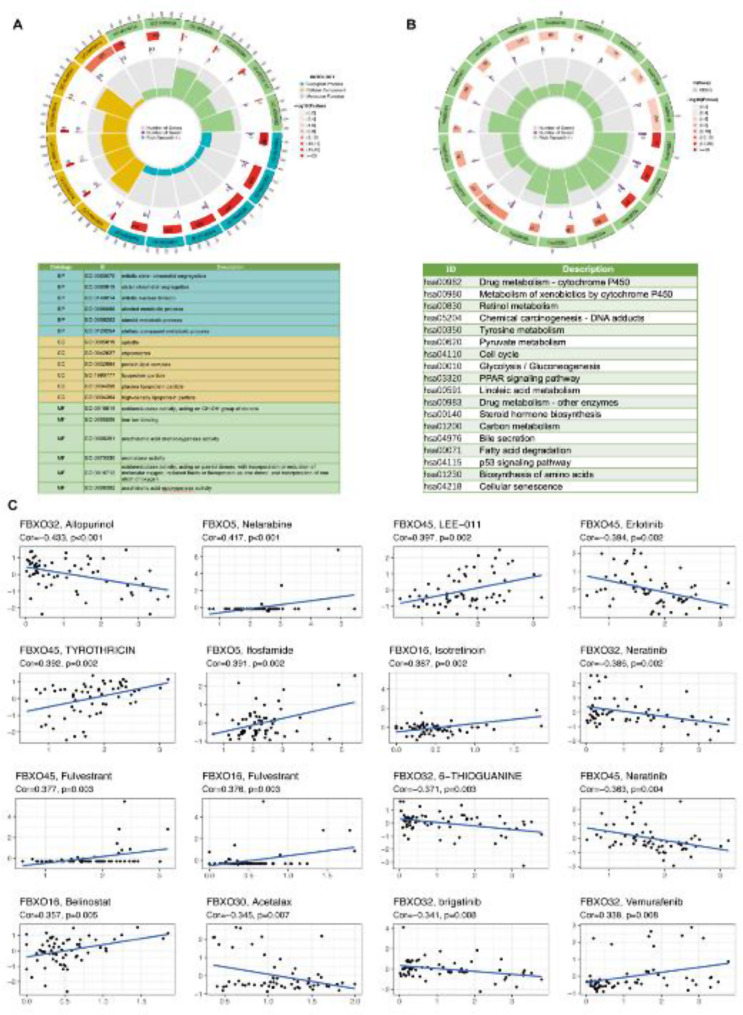
GO and KEGG analysis advised that model genes were related to metabolism and immunity
To elucidate the molecular functionalities and pathway mechanisms associated with module genes, an initial investigation encompassed the application of GO/KEGG analysis to discern disparities in gene expression profiles between the high and low cohorts characterized by heightened and diminished risk proclivities. Moreover, the outcomes underscored noteworthy involvement of specific biological processes within the ambit of KEGG enrichment analysis, including protein processing in drug metabolism-cytochrome P450, xenobiotic metabolism by cytochrome P450, and retinol metabolism. Concurrently, the GO enrichment results show great relevance to the function of molecules such as mitotic sister chromatid segregation and mitotic nuclear division. (Figure 5A and B). Intriguingly, despite the distinct analytical approaches used in GO and KEGG analyses, they both converged on the consistent finding that our model genes were closely related to the cell biochemical activity of immune cells in HCC. In addition, scatter plots were employed to illustrate the associations between five out of the six module FBXO genes (FBXO5, FBXO16, FBXO30, FBXO32, FBXO45) and several drugs. Specifically, the significant relationships were examined between FBXO5 and nelarabine and ifosfamide; FBXO16 and isotretinoin, fulvestrant, and belinostat; FBXO30 and acetalax; FBXO32 and allopurinol, neratinib, 6-THIOGUANINE, brigatinib, and vemurafenib; as well as FBXO45 and LEE-011, erlotinib, TYROTHRICIN, fulvestrant, and neratinib (Fig. 5C).
Fig. 5.
GO and KEGG analysis showed that the six model genes were related to cell metabolisms and immunity. (A) GO analysis on the differential genes between the high and low risk groups. (B) KEGG analysis on the differential genes between the high and low risk groups. (C) Drug activity prediction of the 5 of the six FBXO-related prognostic genes
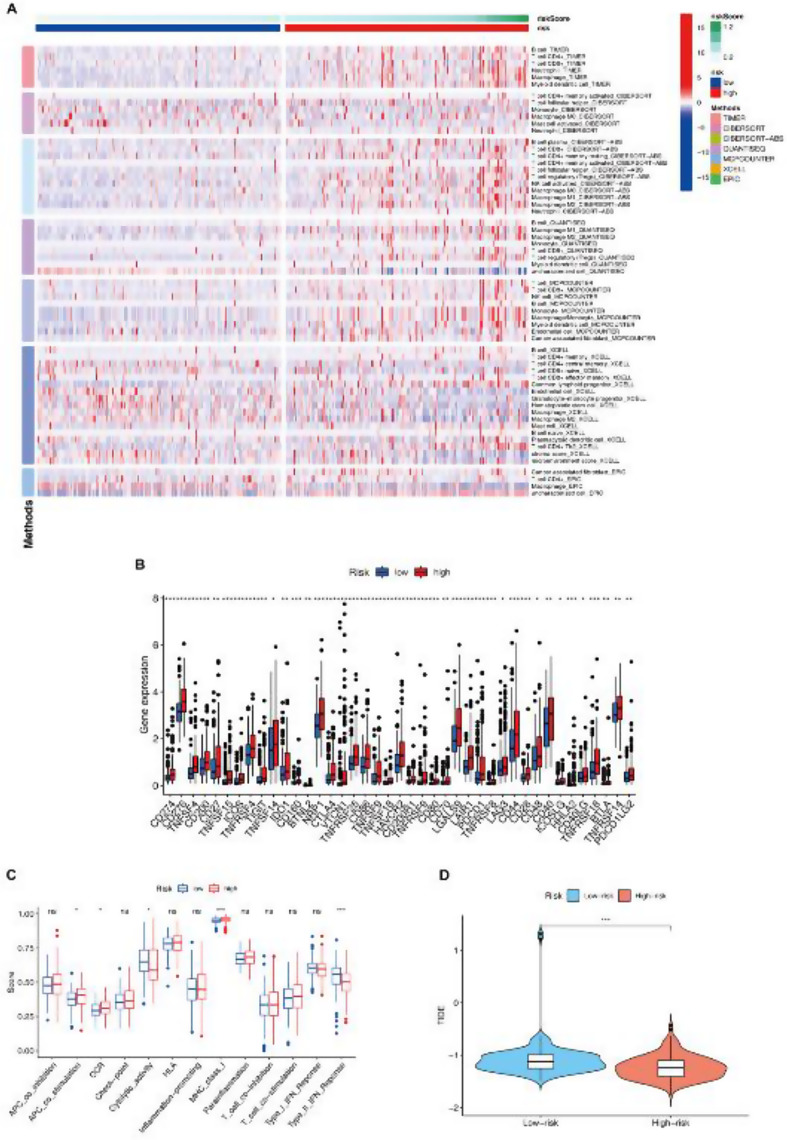
Model genes regulated immunity of HCC
Based on the results of enrichment analysis, the model genes were also associated with cellular immunity. Therefore, immunological analyses were used to evaluate the role of the model genes on immunity. Furthermore, the single-sample gene set enrichment analysis (ssGSEA) algorithms were applied to generate a heatmap illustrating the variances in immune responses between the high-risk and low-risk groups (Fig. 6A). Additionally, the comparative analysis of immune checkpoints between the high-risk and low-risk groups showed a significant correlation of Type_II_IFN_Response and MHC_class_I on our prognostic model genes (Fig. 6C). Besides, an analysis of immune checkpoints within the high-risk and low-risk cohorts demonstrated a robust positive correlation between CTLA-4, PDCD-1, LAG3 and the FBXO genes encompassed by our prognostic model. (Fig. 6B). These findings suggest a potential regulatory role of the model genes in immune processes, which could influence the prognosis of HCC. Moreover, the boxplot exhibited a statistically significant disparity in TIDE (Tumor Immune Dysfunction and Exclusion) between the two groups, as indicated by the statistical analysis (Fig. 6D).
Fig. 6.
Model genes regulated immunity in HCC. (A) Heatmap for the variances in immune responses between the low- and high-risk groups by utilizing the single-sample gene set enrichment analysis (ssGSEA) algorithms. (B) Expressions of immune checkpoints between the high and low risk groups. (C) Single-sample gene set enrichment analysis (ssGSEA) of the correlation between immune cell subpopulations and their associated functions within the two groups. (D) The boxplot showed that there was a statistical difference in TIDE (Tumor Immune Dysfunction and Exclusion) between the two groups (p < 0.05)
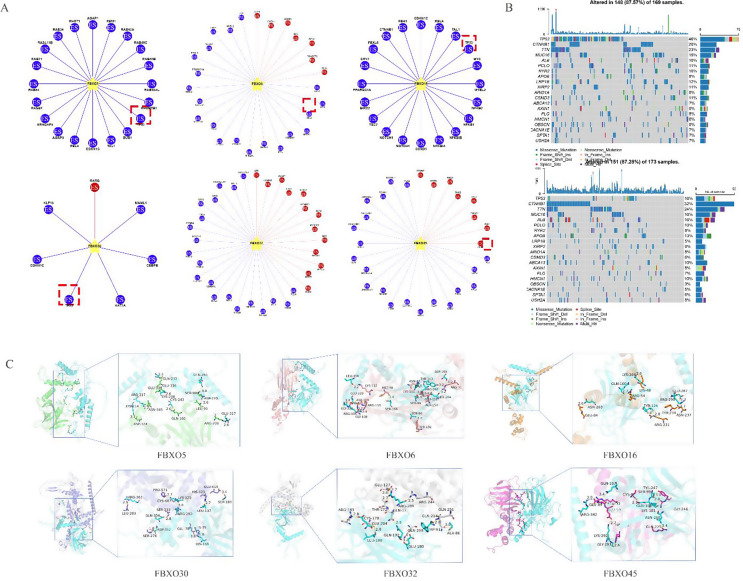
FBXO family genes promotes HCC via ubiquitination of p53
This study investigated the ubiquitin substrate of FBXO family members in HCC using the UbiBrowser. As shown in Fig. 7A, five out of six members of the FBXO family (FBXO5, FBXO6, FBXO16, FBXO30, FBXO45) acted on the same substrate, TP53. In fact, FBXO32 also ubiquitinatedd TP53, but it did not enter the top 20 in the ranking. These results indicate that the FBXO family can ubiquitinate TP53, making TP53 more prone to mutations, thereby promoting the occurrence and development of tumors (Wang et al. 2022a, b). In addition, we further explore the top 20 driver genes with the highest alteration frequency between the high and low-risk subgroups (Fig. 7B). The result showed that 46% in the high-risk group, TP53 exhibits a mutation frequency of 46%. More importantly, molecular docking results suggest that FBXO family proteins bindings with p53.
Fig. 7.
FBXO Family Genes Promotes HCC via Ubiquitination of p53. (A) Ubiquitin Substrate Prediction by UbiBrowser. (B) The driver genes with the highest alteration frequency between the high and low-risk subgroups. (C) Molecular Docking of FBXO Family Genes with p53
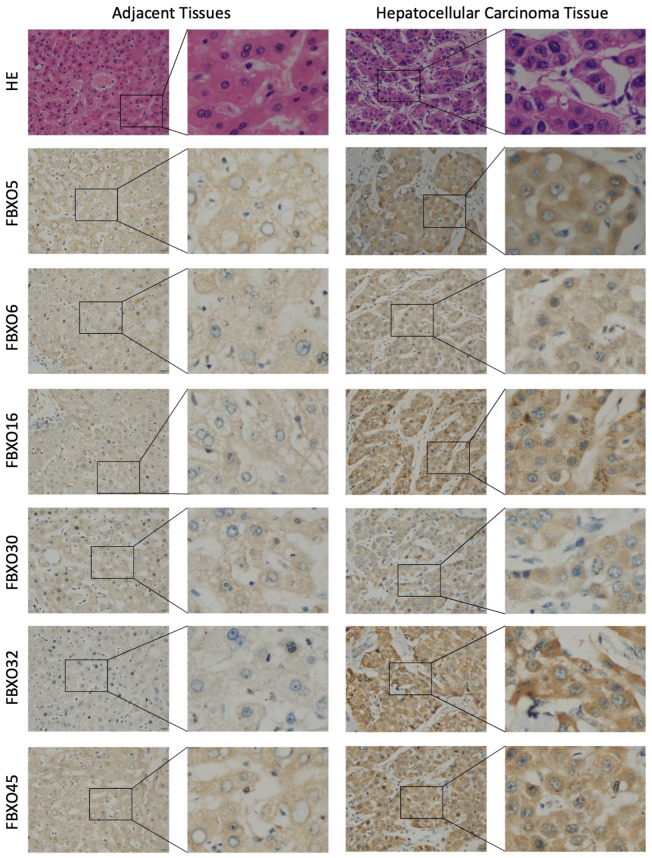
Immunohistochemistry (IHC) staining
To substantiate the dependability of the model genes, we collected clinical HCC tissues and their adjacent tissues from the First Affiliated Hospital of Chongqing Medical University. Initially, a meticulous evaluation of the histological attributes of the tumor tissues was executed through the implementation of hematoxylin and eosin (H&E) staining (Fig. 8). The outcomes of the staining procedure conspicuously manifested that the tumor tissue has more accentuated fibrotic reaction than the adjacent tissue. Furthermore, an immunohistochemical (IHC) investigation was undertaken to evaluate the expression patterns of the six model genes, namely FBXO5, FBXO6, FBXO16, FBXO30, FBXO32, and FBXO45, within both tumor tissue and adjacent tissues. (Fig. 8). The results of IHC staining undeniably demonstrated heightened levels of expression for all model genes within tumor tissues in comparison to adjacent tissues. These observed patterns in gene expression harmonize cohesively with the findings extrapolated from our bioinformatics analyses. To encapsulate, the comprehensive analyses of IHC staining has augmented the substantiation of the established model genes’ dependability within the context of HCC.
Fig. 8.
The 6 model genes were up regulated in clinical HCC samples. H&E staining and representative images of protein expression of six model genes in adjacent tissue and hepatocellular carcinoma tissue tissues
Discussion
Hepatocellular carcinoma (HCC) which accounting for 80–90% of primary liver cancers is responsible for a significant proportion of cancer-related deaths all over the world (Christopher 2006). Due to the absence of early diagnostic markers and limited efficacy of conventional treatments, prognosis of HCC remains exceedingly poor. Consequently, it is imperative to expeditiously identify highly sensitive and specific biomarkers to facilitate prognosis prediction and early detection of HCC patients. Over the recent years, an increasing amount of research has highlighted the importance of the FBXO protein family in relation to HCC, including members like FBXO31 and FBXO43 Huang et al. 2010, Wu et al. 2023. However, there was still no research article has yet explored the potential connection between the FBXO protein family and the prognosis of HCC.
In our study, amid to find out the feasible relationship between FBXO family genes and HCC, the HCC original data were retrieved from the TCGA database. Subsequently, the intersecting set of FBXO protein genes and differential expressed genes was identified by an analysis. Then, a prognostic model pertaining to FBXO family genes was constructed utilizing LASSO COX regression analysis. The model encompassed six specific genes: FBXO5, FBXO6, FBXO16, FBXO30, FBXO32, and FBXO45. To evaluate the efficacy of the risk model, various techniques including univariate analysis, multivariate COX regression, KM survival curve analysis, ROC curve analysis, and nomogram construction were employed. Subsequently, IHC was performed to verify the expression of six genes (FBXO5, FBXO6, FBXO16, FBXO30, FBXO32, and FBXO45) in HCC tissues and their homologous adjacent tissues. All the experimental findings above collectively indicate that the FBXO family genes have the potential to impact the prognostic profile of HCC.
Among predicted model genes, FBXO5, FBXO6, FBXO45 and FBXO32 have been identified as oncogenes in HCC (Lin et al. 2021; Cen et al. 2023; Sahu et al. 2017; Gao et al. 2022; Xu et al. 2018a, b). Recent research findings suggest that FBXO5 contributes to the proliferation of liver cancer cells (Gao et al. 2022; Xu et al. 2018a, 2018b) and the process is regulated through the suppression of APC/C, which hinders the stability of Skp2 while facilitating the degradation of p27kip1 Zhao et al. 2013. The FBXO5 expression level correlates with the stage and unfavorable prognosis of HCC (Gao et al. 2022). Furthermore, Chen et al. reported an overexpression of FBXO5 in HCC, suggesting its potential role in controlling the proliferation of HCC cells (Xu et al. 2018a, b). In view that FBXO6 interacts with and promotes the proteasomal degradation of NLRX1, leading to the reduced effectiveness of HCC inhibition (Cen et al. 2023) and plays a role in facilitating the ubiquitination process and promoting the degradation of the protein Chk1 to enhancing the resistance of tumor cells to specific drugs (Liu et al. 2021), suggesting that FBXO6 functions as an oncogene and has detrimental effects on the prognosis of HCC. Additionally, the identification of FBXO32 as a noteworthy contributor to the upregulation of epithelial-to-mesenchymal transition (EMT) genes’ expression substantiates its role in the metastatic progression of human tumors (Sahu et al. 2017). Moreover, the upregulation of FBXO45 has been associated with poor prognostic outcomes in HCC and plays a crucial role in activating PLK1 through IGF2BD1 mediation which further substantiating its oncogenic role in HCC progression (Lin et al. 2021). In our experiment, higher expression of FBXO5, FBXO6, FBXO32 and FBXO45 in invasive tumor specimens further underscores its role as an oncogene. Nerveless, the precise role of FBXO16 in HCC is still unclear. Additionally, the recent research documented elevated levels of FBXO30 potentially improve the prognostic outcomes in HCC because it reduced levels of EG5 who participate in distinct ubiquitination processes (Tang et al. 2022). But in our research, FBXO30 exhibits oncogenic characteristics in HCC which contradicts existing literature. Our conclusions show successful risk modelling by LASSO analysis, which was further confirmed having independent prognostic significance by univariate and multivariate COX analyses. The KM survival curves indicate a statistically significant increase in overall survival (OS) within the low-risk group compared to the high-risk group. Furthermore, the ROC analysis yielded area under the curve (AUC) values of 0.744, 0.670, and 0.638 for the 1-year, 3-year, and 5-year time points, respectively. These outcomes collectively prove the remarkable prognostic effect of the six FBXO genes in HCC patients.
To explore the specific pathological processes of the six FBXO family genes in HCC, we analyzed the role of them in the high and low-risk groups by KEGG. Moreover, the most pertinent outcome of the current investigation is the enrichment of cytochrome P450 which is associated with the processing of drugs in metabolism pathways (Patel et al. 2020). Notably, the existing literature highlights the association between CYP enzyme activity and the susceptibility to various forms of cancer development and with consistent evidence establishing a connection between CYP polymorphisms and the occurrence of liver cancer (Yeo et al. 2012). In addition, another study has presented Cytochrome P450-based cancer gene therapy as a potentially effective approach for activating prodrugs in cancer treatment, showcasing significant prospects for improving the safety and effectiveness of cancer chemotherapeutics (Agundez 2004). These studies underscore the crucial role played by cytochrome P450 enzymes in both the advancement and control of cancer, thereby drawing attention to the potential therapeutic strategies that could be developed by targeting this pathway. Hence, we examined the relationship between our six FBXOs gene-related drugs in our article. Belinostat, a histone deacetylation inhibitor which warrants consideration for enhancing its therapeutic efficacy in HCC by leveraging potential role of FBXO16 because its clinical anticancer activity and good tolerability in the treatment of HCC (Waxman et al. 1999). The drug prediction analysis presented in Fig. 5C reveals a potential negative correlation between FBXO30 and acetalax. Additionally, a prior study has provided evidence that cancer cell lines expressing higher levels of TRPM4 display heightened susceptibility to acetalax (Li et al. 2021). Furthermore, acetalax has exhibited its capacity to induce the expression of tumor necrosis factor (TNF) α and the degradation of TNFR1, thereby demonstrating its anti-tumor effects in mouse xenograft models (Yang et al. 2022). The drug predictions of FBXO32 reveal associations with allopurinol in HCC whose unique capacity to inhibit first-pass metabolism in cancer chemotherapy may enhance drug bioavailability and action in HCC treatment (Zimm et al. 1983). Regarding FBXO45, drug prediction analysis associates it with erlotinib. Encouragingly, studies have indicated that the bound of erlotinib and Lenvatinib yields significant synergistic effects both in vivo and vitro, thus offering a promising therapeutic approach for HCC (Hu et al. 2022). In the current experiment, a significant relation between the FBXO gene family and drug metabolism CYP450 was observed. Consequently, it is plausible to hypothesize that FBXO may have a potential role in drug therapy for HCC. However, additional investigation is required to validate these findings in future experiments.
The KEGG analyses also revealed a correlation between the FBXO genes and Retinol, a vital micronutrient, as known as vitamin A. This micronutrient assumes a pivotal role in the regulation of stem cell functions, cellular differentiation, and metabolism in various cell types (Gudas 2022). In various tissues, retinol ultimately transforms into retinoic acid through a sequence of biochemical reactions and leads to its immunomodulatory impacts by attaching to the retinoic acid receptor (RAR) (Mora et al. 2008). Moreover, the liver serves as a key immune-related organ situated at the center, consistently encountering blood rich in antigens from the intestines via the portal vein (Ruf et al. 2021). In the summary, our empirical inquiry has unveiled a robust association between the FBXO gene family and retinol metabolism, thereby suggesting a plausible connection between the FBXO family and immune responses. In this study, we used ssGSEA which found the tumor samples has notable disparities between low-risk and high-risk groups of different immune cell infiltrations in HCC patients. Interestingly, our analysis indicated significant variations in the levels of MHC-class I and Type II-IFN among these groups. MHC class I is a pivotal cell surface protein in the immune system (Reynisson et al. 2020) which not only restrains the evasion of the immune system by tumor cells (Lin et al. 2023) but also intensifies the engulfment of tumor cells, making it a promising target for anti-cancer immunotherapy (Barkal et al. 2018). Our findings revealed a substantially higher expression of MHC class I in the high-risk group than the low-risk group in HCC. This suggests that MHC class I could emerge as a novel potential focus for HCC immunotherapy, offering the possibility of improving the unfavorable prognosis of HCC. Type II-IFN (also known as Interferon γ) is a cytokine that plays diverse roles in enhancing protective immune responses and immunopathological processes, thereby playing a crucial role in safeguarding against tumor development (Ikeda et al. 2002). Intriguingly, Type II-IFN prompting the upregulation of MHC class I in cells and stimulating the delivery of the antigens of MHC class I are the pivotal element in anti-tumor immune responses (Ren et al. 2022). However, our results revealed that the expression of Type II-IFN was noticeably higher in the low-risk group compared with the high-risk group. This finding opens a new avenue for exploring Type II-IFN as a prospective immunotherapeutic strategy during the low-risk phase, aiming to preempt the early deterioration and migration of HCC, or as a target for immunotherapy in the context of early-stage tumors.
Furthermore, the utilization of immune checkpoint inhibitors has demonstrated considerable promise in the field of HCC tumor immunotherapy (Huang et al. 2021). Our findings revealed a significant positive correlation between our predictive model and immune checkpoints, specifically encompassing CTLA-4, PDCD1, and LAG3. PDCD1 (as known as PD-1) not only plays a key role in down-regulating immune system function and proving tolerance (Li et al. 2022) but also has surfaced as a potential effective immune treatment strategy for patients dealing with advanced HCC (Shi et al. 2022). Besides, CTLA-4 functions as an inhibitory co-receptor which exerting its influence by impeding the activation and proliferation of T cells (Sangro et al. 2013). In recent research, it has been established that CTLA-4 has the capability to dampen immune responses against tumors by impeding T cell reactions and PD-1 with CD8 + T cells interacting with PD-L1 + Kupffer cells contributes to the malfunction of effector T cells within HCC, which both are associated with less favorable prognosis in individuals with HCC (Guo et al. 2021). In addition, LAG-3 belongs to the group of immune checkpoint receptors that diminish the multiplication and stimulation of T-cells by binding with its ligand (Guo et al. 2021). Additionally, LAG-3 could also potentially serve as a marker for exhaustion alongside PD-1, arising from repetitive stimulation by antigens in cancer contexts (Wang et al. 2019). Nowadays, immunotherapy has the capability to enhance the immune system by inhibiting immune checkpoints and this advancement is widely seen as a significant breakthrough in how malignancies are treated (Guo et al. 2021). Therefore, these findings in our research underscore the potential significance of these immune checkpoints in influencing the advancement of HCC.
Since FBXOs serve as E3 ubiquitin ligases, in order to further explore the role of the FBXO family in HCC, we used the UbiBrowser database to search for their substrate target proteins. The database results indicated that TP53 is the substrate target protein for all six FBXO members. The TP53 gene (also referred to as p53) encodes the p53 protein which represents one of the most frequently mutated gene and is related in over 50% of all human cancers (Juan Liu et al. 2023). In addition, the progression of cancer can be attributed to the proteolytic degradation of p53 facilitated by activated E3 ubiquitination enzymes (Ming Zhou et al. 2021). Notably, there are investigations revealing that the upregulation of the ubiquitination process targeting the tumor suppressor gene TP53 is associated with enhanced growth in ovarian cancer (Ming Zhou et al. 2021), while the ubiquitination of P53 is implicated in the pathological progression of colorectal cancer (Wang et al. 2020). FBXO proteins serve as integral components of E3 ubiquitin ligase, playing crucial regulatory roles in various cellular processes (Liu et al. 2021). Moreover, the functional prediction revealed that the FBXOs were involved in ubiquitination-related pathways such as TP5352which is as the same as ours. Based on the reported literature it is known that FBXO45 expression was related to P53 mutation in PDAC (Zhang et al. 2021). In addition, FBXO45 selectively ubiquitinates and degrades p73 which is a member of the p53 family, resulting in diminished cellular apoptosis in cancers (Wang et al. 2020). However, there was no significant difference in the expression of FBXO32 between the two groups (Jing et al. 2023). Additionally, in p53-deficient cells, up-regulation of FBXO5 could facilitate genomic instability, tetra ploidy, and cell proliferation in breast cancer (Zhang et al. 2022). Interestedly, there were rare evidence to improve that FBXO6, FBXO16, and FBXO30 had relationship with the P53-mediated ubiquitination in cancers, and our research may demonstrate them. To sum up, it is reasonably to suppose that FBXO family genes could ubiquitinate TP53 which results in p53 being more prone to mutation and promotes HCC progression.
This study holds great potential for the future. By identifying the overlap between FBXO genes and DEGs in HCC, our research provides potential novel biomarkers that can be used for the early detection, prognosis assessment and management of HCC. Moreover, KEGG analysis offered strong directions for subsequent drug development research. Additionally, the discovery of different immune infiltrating statuses can inform personalized immunotherapy, advancing precision medicine. However, our study also has certain limitations. We did not elucidate the specific molecular mechanisms of FBXO proteins in HCC, nor did we specify how these proteins influence P53 and TP53 mutations through ubiquitination. The specific impact of different immune infiltrating statuses on HCC progression and treatment response also requires further investigation. In future research, functional experiments such as cell-based assays and animal models could be conducted to explore the precise mechanisms of FBXO proteins, particularly their roles in P53 ubiquitination. Combining techniques like immunohistochemistry (IHC) and flow cytometry can provide deeper insights into the effects of different immune infiltrating statuses on HCC. Our study aims to contribute to optimized personalized treatment strategies for HCC, thereby enhancing treatment efficiency and patient survival rates. As research on drug metabolism pathways progresses, new targeted drugs and therapeutic approaches are expected to emerge. Nonetheless, our study has limitations, including the constraint of sample size, potential biases in the TCGA dataset, and the fact that our conclusions are primarily based on bioinformatics analyses. These findings require further experimental data for validation and support.
Conclusion
The FBXO gene family, an isoform of ubiquitinated E3 ligase, has garnered significant attention due to its intricate association with tumor development, prognosis, and immunotherapy. Consequently, numerous articles have been conducted to explore the correlation between specific FBXO genes and various types of cancer. However, only a small number of studies have focused on the role of the FBXO gene family in HCC. In this experiment, we screened 6 module genes from the FBXO gene family as potential biomarkers which hold promise for their prognostic utility in predicting survival outcomes among HCC patients. Moreover, the prognostic model shows that FBXO family as oncogene is detrimental to the prognostic situation of HCC patients. In addition, analysis of immune infiltration and KEGG enrichment of the FBXOs in HCC provides potential immunotherapeutic directions and targets. However, the absence of experimental verification in our study serves as a limitation. Subsequently, the precise pathological mechanism of the FBXO genes in HCC was not explicitly elucidated in our study. Therefore, our research primarily offers novel research perspectives and valuable insights into the role of the FBXO gene family in HCC progression, prognosis, as well as anti-HCC potential immunotherapeutic interventions. In subsequent investigations, it is imperative to provide more precise elucidation and comprehensive exploration of the distinct pathological mechanisms associated with the FBXO gene family in HCC to enhance the credibility and robustness of our research.
Author contributions
Qingge Gong and Ning Jiang conceived and designed this study. Qingge Gong and La Zhang collected and analyzed the relative data. Qingge Gong and La Zhang wrote the paper. Jiao Guo and Wei Zhao conducted the experiments. Changhong Yang and Ning Jiang revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 82203310), Creative Research Group of CQ University (Grant No. CXQT21017), the Postdoctoral Cultivation Project of the First Affiliated Hospital of Chongqing Medical University (CYYY-BSHPYXM-202315), Chongqing full-time postdoctoral program (2109012646561514) and Program for Youth Innovation in Future Medicine from Chongqing Medical University.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Qingge Gong and La Zhang contributed equally to this work and should be considered co-first authors.
Contributor Information
Changhong Yang, Email: changhong@cqmu.edu.cn.
Ning Jiang, Email: jiangning@cqmu.edu.cn.
References
- Agundez JA (2004) Cytochrome P450 gene polymorphism and cancer. Curr Drug Metab 5(3):211–224 [DOI] [PubMed] [Google Scholar]
- Barkal AA, Weiskopf K, Kao KS et al (2018) Engagement of MHC class I by the inhibitory receptor LILRB1 suppresses macrophages and is a target of cancer immunotherapy. Nat Immunol 19(1):76–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budhu A, Forgues M, Ye QH et al (2006) Prediction of venous metastases, recurrence, and prognosis in hepatocellular carcinoma based on a unique immune response signature of the liver microenvironment. Cancer Cell 10(2):99–111 [DOI] [PubMed] [Google Scholar]
- Buetow L, Huang DT (2016) Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat Rev Mol Cell Biol 17(10):626–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cen M, Ouyang W, Lin X et al (2023) FBXO6 regulates the antiviral immune responses via mediating alveolar macrophages survival. J Med Virol 95(1):e28203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K, Cai N, Zhu J, Yang X, Liang H, Zhang W (2022) Tumor-associated macrophages in liver cancer: from mechanisms to therapy. Cancer Commun (Lond) 42(11):1112–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopher L. Brooks and Wei Gu. p53 ubiquitination: Mdm2 and Beyond. Mol Cell. (2006) 10.1016/j.molcel.2006.01.020 [DOI] [PMC free article] [PubMed]
- Gao J, Yang D, Cao R et al (2022) The role of Fbxo5 in the development of human malignant tumors. Am J Cancer Res 12(4):1456–1464 [PMC free article] [PubMed] [Google Scholar]
- Gudas LJ (2022) Retinoid metabolism: new insights. J Mol Endocrinol 69(4):T37–t49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Qi F, Rao Q et al (2021) Serum LAG-3 predicts outcome and treatment response in Hepatocellular Carcinoma patients with Transarterial Chemoembolization. Front Immunol 12:754961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho WJ, Danilova L, Lim SJ et al (2020) Viral status, immune microenvironment and immunological response to checkpoint inhibitors in hepatocellular carcinoma. J Immunother Cancer ;8(1) [DOI] [PMC free article] [PubMed]
- Hu B, Zou T, Qin W et al (2022) Inhibition of EGFR overcomes acquired Lenvatinib Resistance Driven by STAT3-ABCB1 signaling in Hepatocellular Carcinoma. Cancer Res 82(20):3845–3857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HL, Zheng WL, Zhao R, Zhang B, Ma WL (2010) FBXO31 is down-regulated and may function as a tumor suppressor in hepatocellular carcinoma. Oncol Rep 24(3):715–720 [DOI] [PubMed] [Google Scholar]
- Huang SL, Wang YM, Wang QY et al (2021) Mechanisms and clinical trials of Hepatocellular Carcinoma Immunotherapy. Front Genet 12:691391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Wang Y, Zhang P, Li Q (2023) Ubiquitin-specific peptidase 1: assessing its role in cancer therapy. Clin Exp Med 23(7):2953–2966 [DOI] [PubMed] [Google Scholar]
- Ikeda H, Old LJ, Schreiber RD (2002) The roles of IFN gamma in protection against tumor development and cancer immunoediting. Cytokine Growth Factor Rev 13(2):95–109 [DOI] [PubMed] [Google Scholar]
- Jing Y, Xin-Ping Wang J, Yang et al (2023) SCF-FBXL8 contributes to liver metastasis and stem-cell-like features in colorectal cancer cells by mediating ubiquitination and degradation of TP53. CLINICAL AND TRANSLATION MEDICINE. 10.1002/ctm2.1208 [DOI] [PMC free article] [PubMed]
- Juan Liu C, Zhang D, Xu et al (2023) The ubiquitin ligase TRIM21 regulates mutant p53 accumulation and gain of function in cancer. J Clin Invest 133(6):e164354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Umbach DM, Krahn JM, Shats I, Li X, Li L (2021) Predicting tumor response to drugs based on gene-expression biomarkers of sensitivity learned from cancer cell lines. BMC Genomics 22(1):272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Han J, Yang Y, Chen Y (2022) PD-1/PD-L1 checkpoint inhibitors in advanced hepatocellular carcinoma immunotherapy. Front Immunol 13:1070961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin XT, Yu HQ, Fang L et al (2021) Elevated FBXO45 promotes liver tumorigenesis through enhancing IGF2BP1 ubiquitination and subsequent PLK1 upregulation. Elife ;10 [DOI] [PMC free article] [PubMed]
- Lin W, Chen L, Zhang H et al (2023) Tumor-intrinsic YTHDF1 drives immune evasion and resistance to immune checkpoint inhibitors via promoting MHC-I degradation. Nat Commun 14(1):265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Tian Y, Li Y et al (2020) In vivo therapeutic effects of affinity-improved-TCR engineered T-cells on HBV-related hepatocellular carcinoma. J Immunother Cancer ;8(2) [DOI] [PMC free article] [PubMed]
- Liu Y, Pan B, Qu W, Cao Y, Li J, Zhao H (2021) Systematic analysis of the expression and prognosis relevance of FBXO family reveals the significance of FBXO1 in human breast cancer. Cancer Cell Int 21(1):130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming Zhou H, Cheng Y, Fu et al (2021) Long noncoding RNA DARS-AS1 regulates TP53 ubiquitination and affects ovarian cancer progression by modulation miR‐194‐5p/RBX1 axis. CLINICAL AND TRANSLATION MEDICINE.;10.1002/jbt.22865 [DOI] [PubMed]
- Mora JR, Iwata M, von Andrian UH (2008) Vitamin effects on the immune system: vitamins A and D take centre stage. Nat Rev Immunol 8(9):685–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oura K, Morishita A, Tani J, Masaki T (2021) Tumor Immune Microenvironment and Immunosuppressive Therapy in Hepatocellular Carcinoma: a review. Int J Mol Sci ;22(11) [DOI] [PMC free article] [PubMed]
- Patel R, Barker J, ElShaer A (2020) Pharmaceutical excipients and Drug Metabolism: a Mini-review. Int J Mol Sci ;21(21) [DOI] [PMC free article] [PubMed]
- Ren J, Li N, Pei S et al (2022) Histone methyltransferase WHSC1 loss dampens MHC-I antigen presentation pathway to impair IFN-γ-stimulated antitumor immunity. J Clin Invest ;132(8) [DOI] [PMC free article] [PubMed]
- Reynisson B, Alvarez B, Paul S, Peters B, Nielsen M (2020) NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res 48(W1):W449–w54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf B, Heinrich B, Greten TF (2021) Immunobiology and immunotherapy of HCC: spotlight on innate and innate-like immune cells. Cell Mol Immunol 18(1):112–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu SK, Tiwari N, Pataskar A et al (2017) FBXO32 promotes microenvironment underlying epithelial-mesenchymal transition via CtBP1 during tumour metastasis and brain development. Nat Commun 8(1):1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangro B, Gomez-Martin C, de la Mata M et al (2013) A clinical trial of CTLA-4 blockade with tremelimumab in patients with hepatocellular carcinoma and chronic hepatitis C. J Hepatol 59(1):81–88 [DOI] [PubMed] [Google Scholar]
- Shi J, Liu J, Tu X et al (2022) Single-cell immune signature for detecting early-stage HCC and early assessing anti-PD-1 immunotherapy efficacy. J Immunother Cancer;10(1) [DOI] [PMC free article] [PubMed]
- Tang L, Wang T, Li W, Yu S, Yao S, Cheng H (2022) Construction of cuproptosis-related lncRNAs/mRNAs model and prognostic prediction of hepatocellular carcinoma. Am J Cancer Res 12(10):4693–4707 [PMC free article] [PubMed] [Google Scholar]
- Tekcham DS, Chen D, Liu Y et al (2020) F-box proteins and cancer: an update from functional and regulatory mechanism to therapeutic clinical prospects. Theranostics 10(9):4150–4167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva A (2019) Hepatocellular Carcinoma. N Engl J Med 380(15):1450–1462 [DOI] [PubMed] [Google Scholar]
- Wang J, Sanmamed MF, Datar I et al (2019) Fibrinogen-like protein 1 is a major Immune Inhibitory ligand of LAG-3. Cell ;176(1–2): 334 – 47.e12. [DOI] [PMC free article] [PubMed]
- Wang Zhi-wei Lin1 et al (2020) FBXO45 is a potential therapeutic target for cancer therapy. Cell Death Discovery.; 10.1038/s41420-020-0291-2 [DOI] [PMC free article] [PubMed]
- Wang W, Yuan T, Ma L et al (2022a) Hepatobiliary tumor Organoids Reveal HLA Class I Neoantigen Landscape and Antitumoral Activity of Neoantigen peptide enhanced with Immune Checkpoint inhibitors. Adv Sci (Weinh) 9(22):e2105810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Li Y, He M et al (2022b) UbiBrowser 2.0: a comprehensive resource for proteome-wide ubiquitin ligase/deubiquitinase-substrate interactions in eukaryotic species. Nucleic Acids Res 50:D719–D728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman DJ, Chen L, Hecht JE, Jounaidi Y (1999) Cytochrome P450-based cancer gene therapy: recent advances and future prospects. Drug Metab Rev 31(2):503–522 [DOI] [PubMed] [Google Scholar]
- Wu S, Qin L, Yang J, Wang J, Shen Y (2023) Association between F-box-only protein 43 overexpression and hepatocellular carcinoma pathogenesis and prognosis. Cancer Med 12(8):10062–10076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Jin T, Zhu Y, Dai C (2018a) Immune checkpoint therapy in liver cancer. J Exp Clin Cancer Res 37(1):110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Luo L, Yu Y et al (2018b) Screening therapeutic targets of Ribavirin in hepatocellular carcinoma. Oncol Lett 15(6):9625–9632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Liu F, Liu W et al (2020) Analysis of single-cell RNAseq identifies transitional states of T cells associated with hepatocellular carcinoma. Clin Transl Med 10(3):e133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Wei S, Zhang J et al (2022) Construction of a predictive model for immunotherapy efficacy in lung squamous cell carcinoma based on the degree of tumor-infiltrating immune cells and molecular typing. J Transl Med 20(1):364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo W, Chung HC, Chan SL et al (2012) Epigenetic therapy using belinostat for patients with unresectable hepatocellular carcinoma: a multicenter phase I/II study with biomarker and pharmacokinetic analysis of tumors from patients in the Mayo Phase II Consortium and the Cancer Therapeutics Research Group. J Clin Oncol 30(27):3361–3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Chen J, Ning D et al (2019) FBXO22 promotes the development of hepatocellular carcinoma by regulating the ubiquitination and degradation of p21. J Exp Clin Cancer Res 38(1):101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu Q, Cui M et al (2021) Comprehensive Analysis of expression, Prognostic Value, and Immune Infiltration for Ubiquitination-related FBXOs in pancreatic ductal adenocarcinoma. Front Immunol 12:774435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu Q, Cui M et al (2022) Comprehensive Analysis of expression, Prognostic Value, and Immune Infiltration for Ubiquitination-related FBXOs in pancreatic ductal adenocarcinoma. Front Immunol. 10.3389/fimmu.2021.774435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Tang Q, Ni R et al (2013) Early mitotic inhibitor-1, an anaphase-promoting complex/cyclosome inhibitor, can control tumor cell proliferation in hepatocellular carcinoma: correlation with Skp2 stability and degradation of p27(Kip1). Hum Pathol 44(3):365–373 [DOI] [PubMed] [Google Scholar]
- Zimm S, Collins JM, O’Neill D, Chabner BA, Poplack DG (1983) Inhibition of first-pass metabolism in cancer chemotherapy: interaction of 6-mercaptopurine and allopurinol. Clin Pharmacol Ther 34(6):810–817 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analysed during the current study.