Abstract
Integration of human immunodeficiency virus type 1 (HIV-1) proviral DNA in the nuclear genome is catalyzed by the retroviral integrase (IN). In addition to IN, viral and cellular proteins associated in the high-molecular-weight preintegration complex have been suggested to be involved in this process. In an attempt to define host factors interacting with IN, we used an in vitro system to identify cellular proteins in interaction with HIV-1 IN. The yeast Saccharomyces cerevisiae was chosen since (i) its complete sequence has been established and the primary structure of all the putative proteins from this eucaryote has been deduced, (ii) there is a significant degree of homology between human and yeast proteins, and (iii) we have previously shown that the expression of HIV-1 IN in yeast induces a lethal phenotype. Strong evidences suggest that this lethality is linked to IN activity in infected human cells where integration requires the cleavage of genomic DNA. Using IN-affinity chromatography we identified four yeast proteins interacting with HIV-1 IN, including the yeast chaperonin yHSP60, which is the counterpart of human hHSP60. Yeast lethality induced by HIV-1 IN was abolished when a mutated HSP60 was coexpressed, therefore suggesting that both proteins interact in vivo. Besides interacting with HIV-1 IN, the hHSP60 was able to stimulate the in vitro processing and joining activities of IN and protected this enzyme from thermal denaturation. In addition, the functional human HSP60-HSP10 complex in the presence of ATP was able to recognize the HIV-1 IN as a substrate.
The HIV-1 integrase IN catalyzes a critical step in the infectious cycle of this retrovirus. The mechanism leading to the integration of the proviral DNA into the host genome can be divided into two separate reactions (1). HIV-1 IN catalyzes first a hydrolytic reaction of the proviral DNA substrate, termed “processing,” and second a transesterification step termed “joining” or “strand transfer,” which allows the insertion of the provirus into the nuclear genome. In the processing reaction where OH− is the nucleophilic agent, IN is able to endonucleolytically remove in vitro two nucleotides from an oligonucleotide (ODN) mimicking the retroviral LTR ends of HIV-1. In the joining or strand transfer reaction, by using the processed LTR as a nucleophile, IN catalyzes a transesterification hydrolytic reaction. The resulting double-stranded proviral DNA is part of a large nucleoprotein structure termed the PIC. This complex contains the information necessary for nuclear localization and the enzymatic machinery required to insert the proviral DNA into the host genome (for a review of the PIC, see reference 4). In addition to IN and the proviral DNA, the PIC has been reported to contain various viral and cellular proteins. However, a precise description of the viral and cellular partners involved in this complex is not yet well defined.
Although recombinant IN purified from Saccharomyces cerevisiae or bacteria can perform all the in vitro reactions using synthetic substrates (9), there is much evidence that proteins present in cytoplasmic extracts from uninfected cells are also involved in the integration process. Two proteins, the barrier-to-autointegration factor and HMG I(Y), have been identified as specific cofactors (8, 17). In addition, Kalpana et al. (21), using the yeast two-hybrid system, reported the isolation of a host factor, the integrase-interacting protein 1 (Ini1), as a binding partner of HIV-1 IN. Ini1 displays a high degree of sequence similarity to the yeast protein SNF5, a factor involved in the transcriptional activation of a number of genes.
We have previously shown that the expression of HIV-1 IN in yeast induces a lethal phenotype (5), while the expression of an inactive mutated IN does not. These results strongly suggest that the lethal phenotype could be due to cell death by DNA damage induced by the IN activity. Moreover, the inactivation of the SNF5 transcription factor gene abolished the lethal phenotype induced by the expression of HIV-1 IN in yeast, indicating that SNF5 is able to interact with HIV-1 IN in vivo (29). Therefore, the use of the yeast system should facilitate the identification of IN-interacting proteins, thus allowing further studies in a cellular context by exploring the effect of gene inactivation on the IN-induced lethal effect.
To identify yeast cellular proteins in addition to SNF5, whose human counterparts might participate in retroviral DNA integration, we developed an in vitro system to detect the interactions between HIV-1 IN and S. cerevisiae proteins. Using IN-affinity chromatography, we identified four yeast proteins which are highly homologous to their human counterparts. We focused our interest on one of these proteins corresponding to the ubiquitous chaperone HSP60. Here we describe the functional characterization of the interactions between hHSP60 and HIV-1 IN. We also show that hHSP60 stimulates both in vitro processing and joining IN activities. In addition to interacting with hHSP60, HIV-1 IN was also recognized as a substrate by the functional hHSP60-HSP10 complex.
MATERIALS AND METHODS
Abbreviations.
HIV-1, human immunodeficiency virus type 1; IN, integrase; PIC, preintegration complex; RT, reverse transcriptase; LTR, long terminal repeat; hHSP60, human HSP60; yHSP60, yeast HSP60; BSA, bovine serum albumin; CHAPS, 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate; DTT, dithiothreitol; NHS, N-hydroxysuccinimide; ODN, oligodeoxynucleotide; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis; TFA, trifluoroacetic acid ; ELISA, enzyme-linked immunosorbent assay; PBS, phosphate-buffered saline.
Yeast strains, culture media, growth conditions, and yeast transformation.
The diploid yeast strain W303 a/α (MATa/α ura3/ura3 leu2/leu2 ade3/ade3 his3/his3 trp1/trp1) was used to perform the yeast lethal assay (14) The protein extracts were obtained from yeast strain JSC310 (MATα ura3-52 leu2 trp1 prb1-1122 pep4-3 prc1–407) or JSC310 (IN) transformed with pHIV1-SF2IN (5). The W303 a/α temperature-sensitive strain expressing HSP60 G432D (35) and the W303 a/α temperature-sensitive strain (Yep351) expressing both HSP60 G432D and SCS1, derived from the diploid yeast strain W303 a/α, were used to study the in vivo effect of HSP60. Culture media used were (i) yeast complete medium YEPD (1% yeast extract, 2% Bacto Peptone, 2% glucose), (ii) yeast selective medium (YNB lacking uracil and leucine [0.67% yeast nitrogen base without amino acids, 0.1 to 8% glucose]), and (iii) YCAD lacking uracil (YNB supplemented with 0.5% Casamino Acids). Amino acids and bases (20 to 30 mg/liter) were added as required. Liquid cultures were performed in Erlenmeyer flasks filled to a fifth of their capacity and then shaken. Solid media were obtained by supplementing liquid media with 2% Bacto Agar. Yeast strains were grown at 30°C. Yeast transformation was performed using LiCl as described (7).
Yeast lethality assay.
The effect of IN and HSP60 on yeast growth was assayed using the “drop test” (5). Three-microliter droplets of plasmid-containing yeast standard suspension (about 20 000 URA+ yeast cells) were dropped on YNB solid medium lacking leucine and containing 0.1% glucose to allow high IN expression. Petri dishes were incubated for 3 days at 30°C, and then phenotypes were observed.
Bacterial strains, culture media, transformation procedure, DNA preparation, and DNA analysis.
The Escherichia coli strain DH5α was used for plasmid amplification, and the bacterial transformation was performed as described (16). BL21(DE3) was used for expression of the His-tagged IN. Bacteria were grown on standard Luria-Bertani medium containing ampicillin (50 μg/ml). Plasmids were extracted from bacteria using the boiling method (24). DNA restriction endonucleases and DNA ligase were used under conditions recommended by the supplier (Promega). DNA fragments were analyzed by electrophoresis on 1% agarose gel in the presence of ethidium bromide (0.5 μg/ml) under UV light.
Plasmid vectors.
The HIV-1 IN gene was from a cloned genomic provirus obtained from a San Francisco isolate (SF2) (33). The expression vectors were derived from the yeast-E. coli shuttle plasmid pBS24.1, previously described (5). This plasmid, which, in addition to the 2μm sequence for autonomous replication in yeast, contained the yeast selection marker genes leu2-d (13) and URA3, was used to transform yeast strains mutated in the LEU2 and URA3 genes. Plasmid maintenance was selected either on a medium lacking uracil or on a medium lacking uracil and leucine. The latter medium allows the selection of yeast cells containing a high plasmid copy number since the plasmid leu2-d is poorly expressed. The expression of recombinant IN was under the control of the alcohol dehydrogenase 2/glyceraldehyde-3-phosphate dehydrogenase-inducible hybrid promoter (11). pET-15b(His-IN1–288), pET-15b(His-IN50–212), and pET-15b(His-IN50–288) expression vectors encoding His-tagged truncated INs were used to express His tagged INs (a generous gift of J. F. Mouscadet, UMR-8532 CNRS, Villejuif, France).
Recombinant HIV-1 IN. (i) From yeast.
JSC310 (IN) yeast cells were grown for 3 days on a YCAD selective medium, precultured in YNB liquid medium, and then incubated in YPDA complete medium for 3 days. Cells were harvested for 10 min at 4,000 × g and broken with glass beads in 5 mM HEPES (pH 7.6)–5 mM EDTA–1 mM DTT. The lysate was centrifuged at 12,000 × g for 20 min, and the pellet was solubilized in 1 M NaCl–10 mM CHAPS. The same procedure was performed using the JSC310 strain to obtain yeast extracts without IN. The final NaCl concentration of the extracts was attained by addition of salt or after dialysis, according to the affinity column elution procedure used in the subsequent purification steps.
(ii) From bacteria.
Different forms of His-tagged HIV-1 IN were overexpressed in E. coli and obtained as described by Leh et al. (22).
Purification of IN. (i) Yeast-expressed untagged IN.
Purification was performed essentially as previously described (6). The soluble fraction containing the HIV-1 IN obtained from JSC 310 (IN) was loaded on a Hitrap butyl-Sepharose 4B column (1 ml; Pharmacia-LKB), washed with LSC buffer (50 mM HEPES, pH 7.6; 0.2 M NaCl; 0.1 M EDTA; 1 mM DTT; 7 mM CHAPS; 10% glycerol) and equilibrated with 5 volumes of HSC buffer (50 mM HEPES, pH 7.6; 0.2 M NaCl; 1 M ammonium sulfate; 0.1 mM EDTA; 1 mM DTT; 7 mM CHAPS). Proteins were eluted by a decreasing ammonium sulfate gradient (1 to 0 M). Fractions containing IN activity were pooled, and 7 mM CHAPS was added. Pooled fractions were diluted 1/3 with 50 mM HEPES (pH 7.6), 0.1 M EDTA, 1 mM DTT, 10% glycerol, and 7 mM CHAPS and loaded on a Hitrap heparin Sepharose CL-4B column (1 ml; Pharmacia-LKB), washed with 5 volumes of HS buffer (50 mM HEPES, pH 7.6; 1 M NaCl; 0.1 mM EDTA; 1 mM DTT; 10% glycerol; 7 mM CHAPS), and equilibrated with a linear increasing NaCl gradient (0 to 1 M NaCl). Fractions containing IN activity were pooled and concentrated by ultrafiltration (Centricon Millipore), followed by addition of 7 mM CHAPS. Purified IN solutions were kept at −80°C. Proteins were analyzed by SDS–12% PAGE.
(ii) His-tagged IN.
His-INs were purified as described by Leh et al. (22) with minor modifications. All buffers contained 7 mM CHAPS, and ZnSO4 was omitted during elution. Proteins were dialyzed overnight against a solution containing 20 mM Tris-HCl (pH 8), 0.5 M NaCl, 2 mM DTT, and 10% glycerol. CHAPS (7 mM) was added to fractions containing IN. After elution, the His-IN fusion proteins were cleaved using biotinylated thrombin (Novagen) and dialyzed as described above. Thrombin was then captured by incubation with streptavidin magnesphere paramagnetic particles (Promega).
Selection of yeast proteins interacting with HIV-1 IN.
Purified IN was dissolved to a final concentration of 500 ng/ml in a coupling buffer (0.2 M NaHCO3, 0.5 M NaCl [pH 8.3]). An affinity chromatography column containing NHS groups covalently linked to the column was washed with 1 ml of 1 mM HCl. The IN solution was injected into the column (Pharmacia-LBK), and the coupling step was performed for 30 min at 25°C. NHS noncoupled groups were inactivated by injecting 2 ml of buffer A (0.5 M ethanolamine, 0.5 M NaCl [pH 8.3]) and 2 ml of buffer B (0.1 M acetate, 0.5 M NaCl [pH 4]) according to the procedure described by the supplier. Under these conditions, 70% of the IN was coupled to the column. Four milliliters of the JSC310 yeast protein extract (about 4 μg of protein) was loaded on the IN-coupled column in the absence of NaCl and eluted by a gradient of NaCl linearly increasing from 0 to 1 M, followed by a 2 M NaCl step to elute proteins associated to IN by ionic bonds. Fractions were analyzed by SDS–12% PAGE. Protein staining was performed either with the amido black or the silver nitrate methods.
Identification of IN-interacting proteins.
A sample of 100 ng of proteins eluted from the IN-affinity column was subjected to SDS-PAGE and stained with amido black. The selected bands were excised from the gel, destained, and cleaved in situ with the endoproteinase Lys-C (Boehringer Mannheim). Cleavage was performed in 10% (vol/vol) acetonitrile in 25 mM Tris-HCl (pH 8.6) for 16 h at 37°C, and the resulting peptides were extracted (31) and applied to a reverse-phase C18 column (Applied Biosystems). Peptides were separated by a linear gradient of solvent B (80% acetonitrile in 0.08% [vol/vol] TFA) in solvent A (0.1% [vol/vol] TFA in water). The N-terminal amino acid sequence of the isolated peptides was obtained by Edman degradation in an Applied Biosystems protein sequencer (model 491).
Proteins and antibodies.
Human proteins, HSP60, HSP10, and HSP70 and LK2 monoclonal antibodies directed against the epitope between amino acid residues 383 to 419 of hHSP60 were purchased from Sigma. HIV-1 RT was a kind gift from M. L. Andreola (UMR 5097 CNRS, Bordeaux France). Monoclonal antibodies directed against the C-terminal domain of HIV-1 IN were a kind gift from A. Leavitt (School of Medicine. University of California, San Francisco).
In vitro IN activity assays.
All assays were performed in 20 mM HEPES (pH 8)–10 mM DTT–7.5 mM MnCl2–0.05% NP-40. HIV-1 IN (1 to 5 pmol) and radiolabeled oligonucleotides (1 pmol) in a total volume of 20 μl were used. The reaction mixture was incubated at 37°C for 1 h, and the incubation was stopped by adding 10 μl of loading buffer (95% formamide, 20 mM EDTA, 0.05% bromophenol blue) and heating at 90°C for 5 min. The reaction products were analyzed by electrophoresis on 15% polyacrylamide gels with 7 M urea in Tris-borate-EDTA, pH 7.6, and autoradiographed.
The sequences of the ODNs used to perform the processing and strand transfer assays were the following: ODN 70, 5′GTGTGGAAAATCTCTAGCAGT3′; ODN 71, 5′GTGTGGAAAATCTCTAGCA3′; and ODN 72, 5′ACTGCTAGAGATTTTCCACAC3′.
To perform the 3′ processing assay, the 5′ radiolabeled ODN 70 hybridized to ODN 72 was used as a substrate, while the 5′ radiolabeled ODN 71 hybridized to ODN 72 was used as a substrate in the strand transfer reaction.
All IN activities were quantified by scanning of the bands after gel electrophoresis and autoradiography using the NIH software.
In vitro ATPase activity assay of the HSP60-HSP10 complex.
hHSP60-HSP10 complex (0.5 pmol; 1:1) was incubated for 1 h with 6.6 pmol of [α-32P]ATP and 10 to 15 pmol of HIV-1 IN. Bilayer polyethyleneimine-cellulose chromatography plates were loaded with the reaction products. After migration in the presence of 10 mM Tris-HCl (pH 7.6)–1 M LiCl, plates were autoradiographed. The ATP consumption was determined by scanning the corresponding spots.
ELISA.
Plate wells were coated overnight at 4°C with 30 pmol of the proteins diluted in a 0.1 M carbonate solution. Wells were washed with a solution of PBS containing 0.05% Tween (PBS–0.05% Tween), saturated for 1 h with BSA (10 mg/ml) and washed three times with PBS–0.05% Tween solution. The second partner (30 pmol) was added and incubated for 1 h at 37°C. After washing three times with PBS–0.05% Tween, primary antibodies were incubated for 2 h at 37°C. After three washings, secondary antibodies were incubated for 1 h at 37°C, and the interaction was revealed with ortho-phenylenediamine after washing with PBS. The absorbance was determined at 492 nm.
RESULTS
Selection of yeast proteins interacting in vitro with HIV-1 IN.
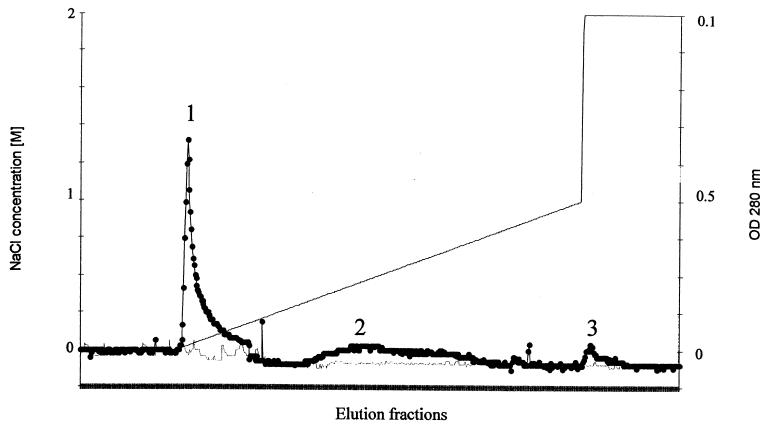
Yeast proteins able to interact with HIV-1 IN were selected by using an affinity chromatography column carrying the immobilized retroviral enzyme. The NHS chemical groups of the column were previously coupled to the NH2 groups of highly purified HIV-1 IN. After inactivation of the unlinked NHS groups, a protein extract from the JSC310 yeast strain was loaded onto the column. The protease-deficient strain JSC310 was used to minimize yeast protein digestion, as well as degradation of the IN coupled to the column. After washing out the unbound proteins (corresponding to about 90% of total protein), a linear gradient from 0 to 1 M NaCl was used to elute the IN-bound material (Fig. 1). A final step of 2 M NaCl was applied to elute proteins still bound to the immobilized IN. The profile of the three peaks of retained proteins (called peaks 1, 2, and 3) was analyzed by SDS-PAGE after silver nitrate staining (Fig. 2A). Most IN-bound proteins were eluted between 0 and 300 mM NaCl. The analysis of this fraction showed the presence of 15 to 20 proteins (Fig. 2A, lane 1). No retained proteins were detected when using an IN-free column as a control.
FIG. 1.
Elution profile of yeast proteins interacting with HIV-1 IN. Yeast proteins retained on the IN-affinity column were eluted by an increasing gradient of NaCl. Peaks 1, 2, and 3 correspond to protein-containing pooled fractions retained in the column. In parallel an IN-free column was used and was eluted similarly. Symbols: filled circle, absorbance at 280 nm of proteins eluted from the IN-coupled column; thin line, absorbance at 280 nm of proteins eluted from the IN-free column; thick line, NaCl concentration.
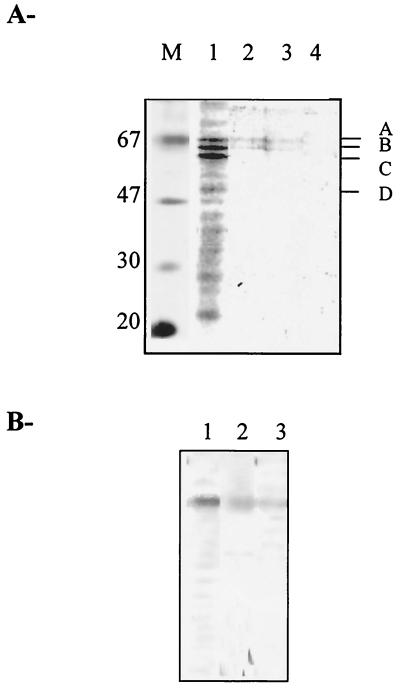
FIG. 2.
(A) SDS-PAGE analysis of the yeast fractions containing IN-interacting proteins. Fractions corresponding to peaks 1, 2, and 3 described in Fig. 1 were subjected to SDS–12% PAGE analysis and stained with silver nitrate. M: Molecular mass markers (in kilodaltons). Lanes 1, 2, and 3: fractions corresponding, respectively, to peaks 1, 2, and 3; lane 4, control corresponding to the fractions obtained with the noncoupled column. A, B, C, and D indicate proteins submitted for further identification. (B) Western blot analysis. The detection of the samples shown in Fig. 2A (lanes 1, 2, and 3) was performed with monoclonal antibodies directed against hHSP60 (see Materials and Methods).
As the protein concentration corresponding to bands A, B, C, and D was high enough to identify them by microsequencing, these fractions were pooled and the proteins were separated by SDS-PAGE. After amido black staining the corresponding bands were cut for identification.
Identification of proteins interacting in vitro with HIV-1 IN.
The amido black-stained gel bands A, B, C, and D were eluted, and the proteins were digested with the endoproteinase Lys-C. The resulting peptides were extracted and separated by high-performance liquid chromatography as described in Materials and Methods section. One or two peptides of each protein were submitted to sequencing. The sequences obtained were compared to the full yeast genome at the SGD website (Saccharomyces Genome Database [http://genome-www.stanford.edu/Saccharomyces]) using BLAST software to identify the corresponding proteins. Results are shown in Table 1. The following yeast proteins were identified: yHSP60, PCK1, CCT4, and TEF1α, which correspond, respectively, to the A, B, C, and D bands shown in Fig. 2A. All these yeast proteins share a high degree of sequence similarity with their human counterparts. By using monoclonal antibodies directed against hHSP60, we confirmed the presence of the yeast homolog in the eluted fractions (Fig. 2B). Lower but significant levels of yHSP60 were also detected in the elution fractions obtained with high salt concentration (peak 3), suggesting a strong interaction between yHSP60 and IN.
TABLE 1.
Identification of IN-interacting proteinsa
| SDS-PAGE band | Peptide sequence obtained | Identified yeast protein | Known function |
|---|---|---|---|
| A | LPYDDALGAT | HSP60 | Chaperone |
| B | QVNFNLNQLIAYRGQK | PCK1 | Neoglycogenesis enzyme |
| C | QFNNXPRS | CCT4 | Tubulin chaperone |
| D | QGMVQTFASADVS | TEF1α | Translation elongation factor |
| TEF2 | Translation elongation factor |
Proteins obtained from SDS-PAGE-excised bands were digested with endoproteinase Lys-C. The sequences of the peptides obtained after digestion are indicated, as are the corresponding yeast proteins with their known functions.
On the basis of previous evidence showing that hHSP60 copurified with HIV-1 virions (2) we focused our interest on this chaperone in order to characterize the functional role(s) of the interaction with IN.
hHSP60 interacts with HIV-1 IN in vitro.
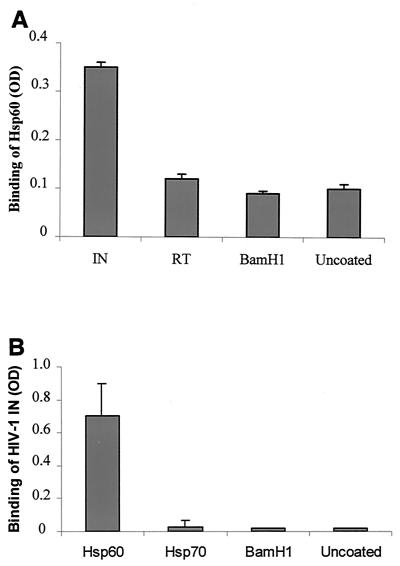
Having shown that yHSP60 interacted with HIV-1 IN, we performed ELISA experiments to verify whether the hHSP60 was also able to interact with IN. ELISA plates were coated with the enzyme and incubated with the hHSP60 protein as described in Materials and Methods. The binding of hHSP60 was detected using the mouse monoclonal anti-hHSP60 antibody LK2 described above. Results unambiguously showed the binding of hHSP60 to IN (Fig. 3A). In parallel, control experiments were performed using various proteins: BSA, the bacterial endonuclease BamHI, or recombinant HIV-1 RT also expressed and purified from yeast (32). A low level of binding was obtained with all these proteins.
FIG. 3.
Binding of hHSP60 to HIV-1 IN determined by ELISAs. (A) hHSP60 (30 pmol) was incubated in ELISA wells coated with either 30 pmol of HIV-1 IN (IN), HIV-1 RT (RT), or bacterial restriction enzyme BamHI (BamHI) or in the absence of protein (Uncoated). The amount of bound hHSP60 was determined by using the monoclonal anti-hHSP60 antibody. Values are the mean ± SD of three independent experiments. (B) HIV-1 IN (30 pmol) was incubated in ELISA wells coated with 30 pmol of hHSP60 (Hsp60), hHSP70 (Hsp70), or restriction enzyme BamHI (BamHI) or without protein (Uncoated). The amount of IN bound was determined by using a monoclonal antibody directed against HIV-1 IN. Values are the means + standard deviations (error bars) of three independent experiments. OD, optical density.
To ascertain the specificity of the interaction, ELISA plates were coated with hHSP60, BamHI, and another heat shock protein, the hHSP70, and incubated with HIV-1 IN. In this experiment the binding was monitored by using monoclonal antibodies against HIV-1 IN. Figure 3B shows that HIV-1 IN bound efficiently to hHSP60. In contrast, no binding was observed in the presence of BamHI or hHSP70, demonstrating once again the specificity of the interaction between HIV-1 IN and hHSP60. These results confirm the interaction between yHSP60 and HIV-1 IN detected by IN-affinity chromatography.
Mapping the interaction site for hHSP60 within IN.
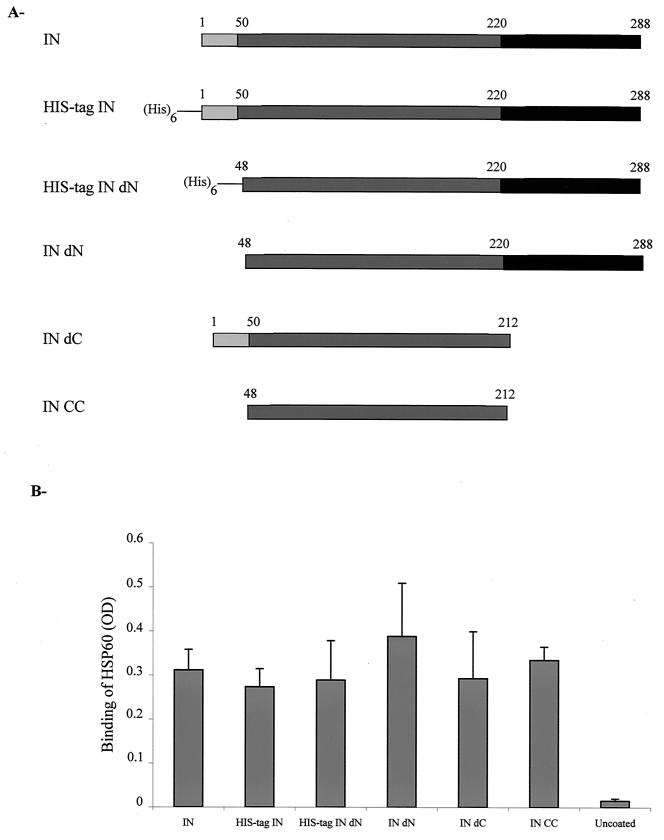
To obtain additional information concerning the IN binding site involved in the interaction with hHSP60, we performed ELISA experiments using six forms of recombinant HIV-1 IN expressed and purified from bacteria. As in all the experiments reported above, we used an HIV-1 IN expressed in yeast; it was important to verify whether the different forms of recombinant IN expressed in bacteria were also able to interact with HSP60 (Fig. 4A). Besides the two complete forms of IN, we tested three fragments of IN, one containing the N-terminal and the catalytic core domains, another carrying the C-terminal and the catalytic core domains, and another with only the catalytic core domain. hHSP60 was able to interact with all the bacterially expressed integrases (Fig. 4B). Therefore, the catalytic core domain of HIV-1 IN is clearly involved in the interaction between hHSP60 and the retroviral enzyme.
FIG. 4.
Determination of the HSP interacting IN domains. (A) Schematic representation of different recombinant forms of HIV-1 IN. All IN constructions were expressed in bacteria. IN, complete protein; His-tag IN, complete IN carrying a histidine tag; His-tag IN dN, His-tagged IN deleted from the N-terminal domain; IN dN, IN deleted from the N-terminal domain; IN dC, IN deleted from the C-terminal domain; IN CC, IN deleted from both the N-terminal and the C-terminal domains. (B) Interaction between IN constructions and hHSP60. hHSP60 (30 pmol) was incubated in ELISA wells coated either with 30 pmol of IN, His-tag IN, His-tag IN dN, IN dN, IN dC, IN CC, or no protein (Uncoated) as indicated. The amount of bound hHSP60 was determined by using the monoclonal anti-HSP60 antibody. OD, optical density. Values are the means + standard deviations (error bars) of three independent experiments.
We conclude that (i) the interaction between the hHSP60 and HIV-1 IN does not depend on the expression system used to obtain the recombinant enzyme, (ii) the same type of interaction was obtained with INs possessing or not possessing a His tag, and (iii) the interaction between hHSP60 and the IN involves at least the catalytic core domain of HIV-1 IN.
HIV-1 IN is a substrate for the hHSP60-HSP10 complex.
The hHSP60 protein performs its function in the cell in association with the HSP10 cofactor in the presence of ATP (3). This complex, which is composed of two rings of seven HSP60 molecules, binds newly synthesized peptides. The folded active state of the substrate is reached by conformational changes in the hHSP60 chaperonin induced by the ATP binding. To know whether the functional complex hHSP60-HSP10 is able to bind IN and induce conformational changes in the viral enzyme, we tested the ATPase activity of the hHSP60-HSP10-IN complex in the in vitro specific assay described in Materials and Methods.
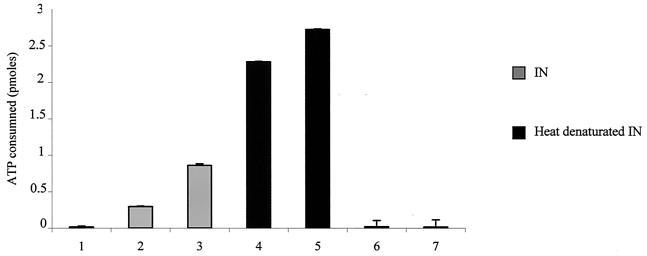
ATP consumption was observed when IN was added to the hHSP60-HSP10 complex, but not in the absence of protein (Fig. 5). Furthermore, ATP consumption was highly increased when IN preincubated at 72°C was used, suggesting that the preferential form of IN recognized by the functional chaperonin complex is the denatured form of IN.
FIG. 5.
ATPase activity assay of the hHSP60-HSP10 complex. The hHSP60-HSP10 complex (0.5 pmol in a 1:1 ratio) was incubated for 1 h at 37°C with 6.6 pmol of [α-32P]ATP in the absence of protein (bar 1), or in the presence of 10 pmol of IN (bar 2), 15 pmol of IN (bar 3), 10 pmol of IN preincubated at 72°C (bar 4), or 15 pmol IN preincubated at 72°C (bar 5). In control experiments without the HSP60-HSP10 complex, [α-32P]ATP was incubated with 10 pmol of IN (bar 6) or 15 pmol of IN (bar 7). The reaction products were chromatographed on bilayer polyethyleneimine-cellulose plates. The ATP consumption was determined by scanning the corresponding spots on the autoradiograms. Error bars, standard deviations.
hHSP60 stabilizes the IN active form.
The physiological role of chaperones involves protein conformational changes allowing these macromolecules to acquire their active form. It also protects them from denaturation (3). To further investigate the effect of hHSP60 on IN, we tested protection against heat denaturation in the presence or absence of the chaperonin.
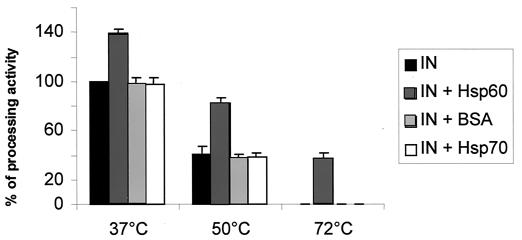
After preincubating IN at different temperatures in the presence or absence of hHSP60, BSA, or hHSP70, we tested the in vitro IN processing activity. Under these conditions the processing activity was stimulated by the presence of hHSP60, suggesting that the interaction between IN and hHSP60 is able to improve the in vitro activity of the retroviral enzyme.
As seen in Fig. 6, the presence of hHSP60 reduced the thermal denaturation of the enzyme. After preincubation with hHSP60 at 50°C, about 80% of the IN activity was retained, while 40% of the activity remained after heating at 72°C. It is important to note the striking effect observed at 72°C since no IN activity was observed in the absence of the chaperone. In contrast, no protection was observed when HIV-1 IN was preincubated in the presence of either hHSP70 or BSA. The protection of HIV-1 IN against heat denaturation by hHSP60 strongly suggests the formation of a stable complex between both proteins, which is probably mediated by the stabilization of the active form of HIV-1 IN. Such a stabilizing effect could be in part responsible for the stimulation of the IN activity (see below) by hHSP60.
FIG. 6.
Effect of HSP60 on the thermal denaturation of HIV-1 IN. HIV-1 IN (5 pmol) was preincubated at 37, 55, and 72°C for 10 min in the presence or absence of 0.50 pmol of hHSP60, hHSP70, or BSA. The processing activity assay was performed as described in Materials and Methods.
To determine whether hHSP60 was also able to induce conformational changes leading to the renaturation of HIV-1 IN, we studied the effect of the chaperone after enzyme denaturation. IN was preincubated at the same temperatures as reported above; then, hHSP60 was added for 30 min at 37°C and the in vitro IN activity was assayed. Under these conditions, no restoration of the activity was observed, indicating that denaturation at high temperatures seems to be irreversible.
hHSP60 stimulates in vitro IN activity.
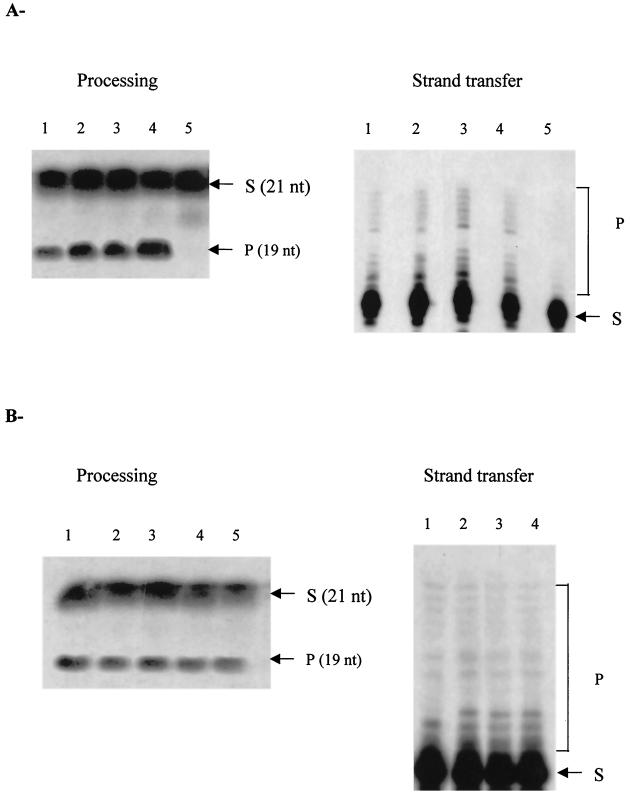
Experiments were designed to study the in vitro effect of hHSP60 on the processing and strand transfer activities catalyzed by HIV-1 IN (Fig. 7A). Preincubation with increasing amounts of hHSP60 resulted in a two- to fourfold dose-dependent stimulation of both processing and strand transfer activities. In contrast, no stimulation was observed with human HSP70 (Fig. 7B). The addition of high concentrations of hHSP60 resulted in inhibition of both IN activities (Fig. 7A, lane 5).
FIG. 7.
In vitro effect of hHSP60 or HSP70 on HIV-1 IN activities. (A) Increasing concentrations of hHSP60 (0, 0.14, 0.25, 0.50, and 1.41 pmol) corresponding, respectively, to lanes 1, 2, 3, 4, and 5, were preincubated for 30 min at 37°C with 5 pmol of HIV-1 IN before performing the processing or strand transfer activity assays. (B) Increasing concentrations of hHSP70 (0, 0.14, 0.25, 0.50, and 1.41 pmol) corresponding, respectively, to lanes 1, 2, 3, 4, and 5, were preincubated for 30 min at 37°C with 5 pmol of HIV-1 IN before performing processing or strand transfer activity assays. Abbreviations: S, substrate; P, product.
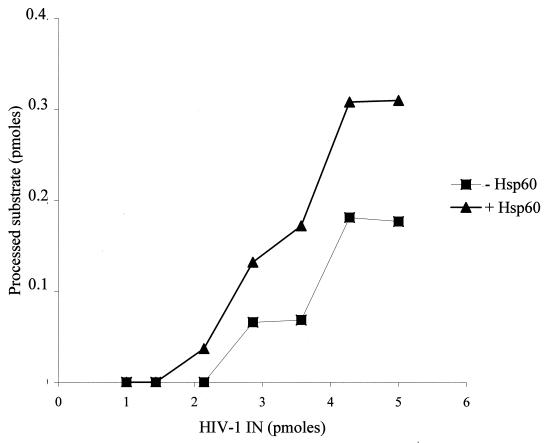
As shown in Fig. 8, a stimulation of the IN activity was also observed when using a constant concentration of hHSP60 and increasing concentrations of IN. Interestingly, in the presence of hHSP60, IN processing activity was detected even with enzyme concentrations that were too low to observe any activity under the standard conditions. Thus, in the presence of 0.5 pmol of hHSP60, a low but significant level of processing activity was observed with 2.2 pmol of IN, while no activity was detected at the same concentration of enzyme in the absence of hHSP60. The lack of stimulation of the activities of a recombinant HIV-1 RT expressed in yeast (32) or the BamHI endonuclease by the chaperone, points to the specificity of this effect on HIV-1 IN.
FIG. 8.
Effect of hHSP60 on the processing activity of HIV-1 IN. hHSP60 (0.5 pmol) was preincubated for 30 min at 37°C with different concentrations of HIV-1 IN as indicated before performing the processing assay. The IN activity was quantified by using the NIH software and was reported as indicated in the graph.
The presence of yHSP60 is necessary to obtain IN-induced lethality in S. cerevisiae
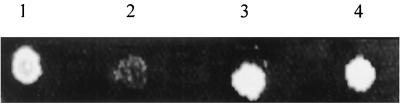
To study the effect of the chaperonin on the activity of IN within a cellular context, we used the in vivo lethal test previously described in yeast cells (5). In this system, the IN activity is detected by the emergence of a lethal phenotype. The expression of IN in a diploid yeast strain derived from W303 a/α was used, in which IN induction leads to a lethal effect (Fig. 9, spot 2). This yeast strain was identical to that reported previously (5), except for the gene encoding yHSP60, which was mutated and which thus induced the expression of a yHSP60 with an altered function at 38°C (35). No lethality was observed with the yHSP60 mutant (Fig. 9, spot 3). We verified that the yHSP60 mutation did not change the level or the activity of HIV-1 IN expressed in yeast (data not shown).
FIG. 9.
Lethality assay of yeast strains. The drop test was performed as described in Materials and Methods. Yeast cells were spotted onto YNB solid medium lacking leucine and containing 0.1% glucose for 3 days at 30°C. Spot 1, W303 a/α control yeast strain containing the pBS24.1 vector, which does not express HIV-1 IN; spot 2, W303 a/α (IN) yeast strain containing the pHIV1-SF2IN vector expressing wild-type HIV-1 IN; spot 3, W303 a/α temperature-sensitive yeast strain containing the pHIV1-SF2IN vector expressing wild type HIV-1 IN and a mutated yHSP60 product, the hps6 G432D protein; spot 4, W303 a/α temperature-sensitive yeast strain (Yep351) that expresses both the mutated HSP60 G432D and SCS1 proteins and contains the pHIV1-SF2IN vector expressing wild-type HIV-1 IN.
The use of a yeast strain able to express SCS1, a multicopy suppressor of yHSP60 temperature-sensitive mutant encoding a protein different from yHSP60 (35), did not restore the IN lethal effect (Fig. 9, spot 4). This suggests that the effect was probably due to the specific action of yHSP60 associated with the IN-induced lethality in yeast cells and that it was not related to an unspecific effect of the yHSP60 chaperone activity. This strongly supports the idea that a physiological interaction operates between HIV-1 IN and HSP60, thus suggesting the involvement of this chaperone in the lethal effect induced by IN in yeast.
DISCUSSION
In this study we have identified some yeast proteins able to interact with HIV-1 IN: PCK1, CCT4, TEF1α, and yHSP60. These proteins, which were isolated by IN-affinity chromatography, are very similar to their human counterparts. PCK1 is an enzyme involved in neoglycogenesis. A possible role of this protein in retroviral integration remains to be established. The yeast CCT4 protein is a chaperonin of tubulin. Since it has been shown that spumaretroviruses use tubulin filaments to enter the nucleus and that purified HIV-1 virions contain actin and several actin-binding proteins (28), CCT4 is a plausible candidate involved in the intracellular transfer of IN. Furthermore, using the yeast two-hybrid system, we have already identified other proteins related to tubulin and able to interact with IN (our own unpublished results). Another protein detected with the IN-affinity chromatography system was TEF1α, the yeast counterpart of the human translation elongation factor EF1α. These results can be related to those reported by Cimarelli and Luban (10). They found that EF1α interacts specifically with the HIV-1 Gag polyprotein. All this evidence suggests an interaction between EF1α and IN and the possible function of this elongation factor in the HIV-1 cycle.
We focused our attention on the characterization of the interaction between HIV-1 IN and hHSP60, the human counterpart of yHSP60. Heat shock proteins are highly conserved during evolution. These ubiquitous proteins play an essential role in cells by binding newly synthesized proteins and facilitating their folding. Here we show that hHSP60 binds in vitro to HIV-1 IN and that the catalytic core domain of IN is involved in this interaction. Although the in vitro IN-hHSP60 interactions described in this work required no additional factors as shown by ELISA and by the protection against thermal denaturation, we cannot exclude the in vivo involvement of other proteins.
We have previously shown that the induction of HIV-1 IN in yeast cells leads to the emergence of a lethal phenotype. Much genetic and molecular evidence supports this hypothesis (5). Thus, the absence of the IN-induced lethal phenotype in yeast after mutation of the yHSP60-encoding gene indicates that interaction between IN and the chaperonin operates in vivo and plays a key role in cellular lethality.
Proteins from the HSP60 chaperonin family recognize a well-defined set of newly translated polypeptides. The HSP60 bacterial counterpart, GroEL, interacts strongly with approximately 300 polypeptides (18). The bacterial chaperonin substrates consist preferentially of two or more domains with αβ folds, which contain α-helices and buried β-shields presenting extensive hydrophobic surfaces. Houry et al. (18) defined the typical GroEL substrate as a protein with a molecular mass between 20 and 60 kDa presenting such characteristics. Since the HIV-1 IN protein has a similar quaternary structure, this enzyme may act as a cellular substrate of this chaperonin. This hypothesis is supported by our results concerning the hHSP60-IN interaction and the functionality of the HSP60-HSP10 complex in the presence of IN.
Several hypotheses may be proposed with regard to the biological relevance of the interaction between hHSP60 and HIV-1 IN. On the basis of the well-known functions of chaperones, hHSP60 may act either by stabilizing the active form of IN or by inducing conformational changes allowing inactive forms of IN to reach a functional folding state. Inside the cell, such conformational changes involve the HSP60-HSP10 multimeric complex (3). Our results concerning the ATPase activity of this complex in the presence of IN support a cellular role of HSP60-HSP10 on the folding of HIV-1 IN.
Thermal denaturation experiments showed that hHSP60 protects HIV-1 IN from inactivation, thus indicating that hHSP60 could also act as a chaperone for HIV-1 IN in protein folding and assembly. The latter effect may be related to the fact that purified HIV-1 IN aggregates easily into insoluble multimeric complexes. Recent results from our group and others (12) indicate that the active form of the enzyme is a dimer. Thus, a possible role of hHSP60 in the cell may be to prevent aggregation of HIV-1 IN. The in vivo interaction of hHSP60 or hHSP60-like proteins with IN may be to maintain the proper conformation and/or the oligomerization state of the viral integrase.
Previous reports have shown a possible involvement of hHSP60 in the HIV-1 biological cycle. The hHSP60 has been shown to copurify with HIV-1 and simian immunodeficiency virus viral particles (2), thus suggesting that this protein is able to bind to one or more molecules encapsidated into HIV-1 particles. After submission of this work, the interaction of HSP60 and the hepatitis B virus DNA polymerase was described, leading to conclusions very similar to those described here (30). Heat shock proteins have also been found to be associated with VSV (15), vaccinia virus (20), adenovirus (23), Sindbis virus (26), and canine distemper virus (27). Moreover, the expression of hHSP60 is significantly increased in virus-infected cells (25). Taken together, these results strongly suggest that the interactions shown here between hHSP60 and HIV-1 IN may be part of a more general infection mechanism involving cell factors and virus components, which in turn supports the hypothesis of a functional role of hHSP60 in the HIV-1 biological cycle. Thus, the in vitro results obtained in this work may be related to possible functional interactions between hHSP60 and HIV-1 IN in the infected human cell. As there is no direct evidence concerning the in vivo role of the interactions described here, it remains to be confirmed that the effect of HSP60 described in vitro operates during the virus life cycle.
Even if a large amount of HSP60 was found in the mitochondria of mammalian cells, a small but significant fraction has been also detected in the nuclear and the cytoplasmic fractions (19, 34), suggesting that HIV-1 IN and the chaperonin may share the same compartments in the infected cell. Furthermore, as hHSP60 is nucleus encoded and synthesized in the cytoplasm, the interaction between IN and hHSP60 may reflect a transient step, requiring a low amount of chaperonin before the latter is transferred to the mitochondria.
Taken together, our results concerning the interactions of HIV-1 IN and HSP60, the ones described very recently in the case of hepatitis B virus DNA polymerase (30), and those described above in other viral systems may indicate a general role of this chaperonin in the conversion of some viral encoded proteins into an active state.
Work is under way to obtain further information regarding the structure-function relationship of hHSP60-IN interactions.
ACKNOWLEDGMENTS
We are deeply grateful to L. Tarrago-Litvak for precious advice and help with the manuscript. Editing of the manuscript was done with the capable help of Ray Cooke (English Department, University Bordeaux 2). We thank A. Leavitt (University of California, San Francisco) for generously providing HIV-1 IN IgG, R. L. Hallberg (Syracuse University, Syracuse, N.Y.) for the gift of yeast strains, J. F. Mouscadet (UMR-8532 CNRS, Villejuif, France) for the gift of the His-tagged truncated IN expression plasmids, and M.-L. Sallafranque-Andreola for the gift of the HIV-1 reverse transcriptase protein. We are grateful to J. H. Alix and A. El Yaagoubi (IBPC, Paris, France) for fruitful discussions.
This work was supported by the French Agence Nationale de Recherche contre le SIDA (ANRS), the Centre National de la Recherche Scientifique, and the University Victor Segalen Bordeaux 2. V.P. and V.R.D.S. benefited from a Ph.D. fellowship from the MNERT.
REFERENCES
- 1.Asante-Appiah E, Skalka A M. Molecular mechanisms in retrovirus DNA integration. Antivir Res. 1997;36:139–156. doi: 10.1016/s0166-3542(97)00046-6. [DOI] [PubMed] [Google Scholar]
- 2.Bartz S R, Pauza C D, Ivanyi J, Jindal S, Welch W J, Malkovsky M. An Hsp60 related protein is associated with purified HIV and SIV. J Med Primatol. 1994;23:151–154. doi: 10.1111/j.1600-0684.1994.tb00116.x. [DOI] [PubMed] [Google Scholar]
- 3.Bukau B, Horwich A L. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 4.Bukrinsky M, Haffar O K. HIV-1 nuclear import: in search of a leader. Front Biosci. 1999;4:772–781. doi: 10.2741/bukrinsky. [DOI] [PubMed] [Google Scholar]
- 5.Caumont A B, Jamieson G A, Pichuantes S, Nguyen A T, Litvak S, Dupont C-H. Expression of functional HIV-1 integrase in the yeast Saccharomyces cerevisiae leads to the emergence of a lethal phenotype: potential use for inhibitor screening. Curr Genet. 1996;29:503–510. doi: 10.1007/BF02426953. [DOI] [PubMed] [Google Scholar]
- 6.Caumont A B, Jamieson G A, Richard de Soultrait V, Parissi V, Fournier M, Zakharova O D, Bayandin R, Litvak S, Tarrago-Litvak L, Nevinsky G A. High affinity interaction of HIV-1 integrase with specific and non-specific single-stranded short oligonucleotides. FEBS Lett. 1999;455:154–158. doi: 10.1016/s0014-5793(99)00859-5. [DOI] [PubMed] [Google Scholar]
- 7.Chen D C, Yang B C, Kuo T T. One-step transformation of yeast in stationary phase. Curr Genet. 1992;21:83–84. doi: 10.1007/BF00318659. [DOI] [PubMed] [Google Scholar]
- 8.Chen H, Engelman A. The barrier-to-autointegration protein is a host factor for HIV type 1 integration. Proc Natl Acad Sci USA. 1998;95:15270–15274. doi: 10.1073/pnas.95.26.15270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chow, S. A. 1997. In vitro assays for activities of retroviral integrase. A companion to methods in enzymology. 12:306–317. [DOI] [PubMed]
- 10.Cimarelli A, Luban J. Translation elongation factor 1-alpha interacts specifically with the human immunodeficiency virus type 1 Gag polyprotein. J Virol. 1999;73:5388–5401. doi: 10.1128/jvi.73.7.5388-5401.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cousens L S, Shuster J R, Gallegos C, Ku L L, Stempien M M, Urdea M S, Sanchez-Pescador R, Taylor A, Tekamp-Olson P. High level expression of proinsulin in the yeast Saccharomyces cerevisiae. Gene. 1987;61:265–275. doi: 10.1016/0378-1119(87)90190-9. [DOI] [PubMed] [Google Scholar]
- 12.Deprez E, Tauc P, Leh H, Mouscadet J-F, Auclair C, Brochon J C. Oligomeric states of the HIV-1 integrase as measured by time-resolved fluorescence anisotropy. Biochemistry. 2000;39:9275–9284. doi: 10.1021/bi000397j. [DOI] [PubMed] [Google Scholar]
- 13.Erhart E, Hollenberg C P. The presence of a defective LEU2 gene on 2μ DNA recombinant plasmids of Saccharomyces cerevisiae is responsible for curing and high copy number. J Bacteriol. 1983;156:625–635. doi: 10.1128/jb.156.2.625-635.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hallberg E M, Shu Y, Hallberg R L. Loss of mitochondrial hsp60 function: nonequivalent effects on matrix-targeted and intermembrane-targeted proteins. Mol Cell Biol. 1993;13:3050–3057. doi: 10.1128/mcb.13.5.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammond C, Helenius A. Folding of VSV G protein: sequential interaction with BiP and calnexin. Science. 1994;266:456–458. doi: 10.1126/science.7939687. [DOI] [PubMed] [Google Scholar]
- 16.Hanahan D. Studies on transformation of E. coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 17.Hindmarsch P, Ridky T, Reeves R, Andrake M, Skalka A M, Leis J. HMG protein family members stimulate human immunodeficiency virus type 1 and avian sarcoma virus concerted DNA integration in vitro. J Virol. 1999;73:2994–3003. doi: 10.1128/jvi.73.4.2994-3003.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Houry W A, Dmitrij F, Eckerskorn C, Lottspeich F, Hartl U. Identification of in vivo substrates of the chaperonin GroEl. Nature. 1999;402:147–154. doi: 10.1038/45977. [DOI] [PubMed] [Google Scholar]
- 19.Itoh H, Kobayshi R, Wakui H, Komatsuda A, Ohtani H, Miura A B, Otaka M, Masamune O, Andoh H, Koyama K, Sato Y, Yashima Y. Mammalian 60-kDa stress protein (chaperonin homolog) J Biol Chem. 1995;270:13429–13435. doi: 10.1074/jbc.270.22.13429. [DOI] [PubMed] [Google Scholar]
- 20.Jindal S, Young R A. Vaccinia virus infection induces a stress response that leads to association of Hsp70 with viral proteins. J Virol. 1992;66:5357–5362. doi: 10.1128/jvi.66.9.5357-5362.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalpana G V, Marmon S, Wang W, Crabtree G R, Goff S P. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor Snf5. Science. 1994;266:2002–2006. doi: 10.1126/science.7801128. [DOI] [PubMed] [Google Scholar]
- 22.Leh H, Brodin P, Bischerour J, Deprez E, Tauc P, Brochon J C, LeCam E, Coulaud D, Auclair C, Mouscadet J F. Determinants of Mg2+-dependent activities of recombinant human immunodeficiency virus type 1 integrase. Biochemistry. 2000;39:9285–9294. doi: 10.1021/bi000398b. [DOI] [PubMed] [Google Scholar]
- 23.Macejak D V, Luftig R B. Association of Hsp70 with the adenovirus type 5 fiber protein in infected Hep-2 cells. Virology. 1991;180:120–125. doi: 10.1016/0042-6822(91)90015-4. [DOI] [PubMed] [Google Scholar]
- 24.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 25.Morimoto R I, Tissieres A, Georgopoulos G. The biology of heat-shock proteins and molecular chaperones. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 26.Mulvey M, Brown D T. Involvement of the molecular chaperone BiP in maturation of Sindbis virus envelope glycoproteins. J Virol. 1995;69:1621–1627. doi: 10.1128/jvi.69.3.1621-1627.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oglesbee M J, Liu Z, Henney H, Brooks C L. The highly inducible member of the 70 kDa family of heat shock proteins increases canine distemper virus polymerase activity. J Gen Virol. 1996;77:2125–2135. doi: 10.1099/0022-1317-77-9-2125. [DOI] [PubMed] [Google Scholar]
- 28.Ott D E, Coren L V, Kane B P, Bush L K, Johnson D G, Sowder R C N, Chertova E N, Arthur L O, Henderson L E. Cytoskeletal proteins inside human immunodeficiency virus type 1 virions. J Virol. 1996;70:7734–7743. doi: 10.1128/jvi.70.11.7734-7743.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parissi V, Caumont A B, Richard de Soultrait V, Dupont C-H, Pichuantes S, Litvak S. Inactivation of the SNF5 transcription factor gene abolishes the lethal phenotype induced by the expression of HIV-1 integrase in yeast. Gene. 2000;247:129–136. doi: 10.1016/s0378-1119(00)00108-6. [DOI] [PubMed] [Google Scholar]
- 30.Park S G, Jung G. Human hepatitis B virus polymerase interacts with the molecular chaperonin Hsp60. J Virol. 2001;75:6962–6968. doi: 10.1128/JVI.75.15.6962-6968.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosenfeld J, Capdevielle J, Guillemot J C, Ferrara P. In-gel digestion of proteins for internal sequence analysis after one-or two-dimensional gel electrophoresis. Anal Biochem. 1992;203:173–179. doi: 10.1016/0003-2697(92)90061-b. [DOI] [PubMed] [Google Scholar]
- 32.Sallafranque-Andreola M-L, Robert D, Barr J P, Fournier M, Litvak S, Sarih-Cottin L, Tarrago-Litvak L. Human immunodeficiency virus reverse transcriptase expressed in transformed yeast cells. Eur J Biochem. 1989;184:367–374. doi: 10.1111/j.1432-1033.1989.tb15028.x. [DOI] [PubMed] [Google Scholar]
- 33.Sanchez-Pescador R, Power M D, Barr P J, Steimer K S, Stempien M M, Brown-Shimer S L, Gee W W, Renard A, Randolph A, Levy J A, Dina D, Luciw P A. Nucleotide sequence and expression of an AIDS-associated retrovirus (ARV-2) Science. 1985;227:484–492. doi: 10.1126/science.2578227. [DOI] [PubMed] [Google Scholar]
- 34.Sanders B M, Nguyen J, Douglass T G, Miller S. Heat-inducible proteins that react with antibodies to chaperonin 60 are localizated in the nucleus of a fish cell line. Biochem J. 1994;297:21–25. doi: 10.1042/bj2970021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shu Y, Hallberg R L. SCS1, a multicopy suppressor of hsp60-ts mutant alleles, does not encode a mitochondrially targeted protein. Mol Cell Biol. 1995;15:5618–5626. doi: 10.1128/mcb.15.10.5618. [DOI] [PMC free article] [PubMed] [Google Scholar]