Abstract
We have identified a lytic origin of DNA replication (oriLyt) for rhesus macaque rhadinovirus (RRV), the rhesus macaque homolog of human herpesvirus 8 (HHV-8), also known as Kaposi's sarcoma-associated herpesvirus. RRV oriLyt maps to the region of the genome between open reading frame 69 (ORF69) and ORF71 (vFLIP) and is composed of an upstream A+T-rich region followed by a short (300-bp) downstream G+C-rich DNA sequence. A set of overlapping cosmids corresponding to the entire genome of RRV was capable of complementing oriLyt-dependent DNA replication only when additional ORF50 was supplied as an expression plasmid in the transfection mixture, suggesting that the level of ORF50 protein originating from input cosmid DNA was insufficient. The requirement of RRV ORF50 in the cotransfection replication assay may also suggest a direct role for this protein in DNA replication. RRV oriLyt shares a high degree of nucleotide sequence and G+C base distribution with the corresponding loci in HHV-8.
Recently, rhesus macaque rhadinovirus (RRV), the rhesus macaque homolog of human herpesvirus 8 (HHV-8), or Kaposi's sarcoma-associated herpesvirus, was isolated from a simian immunodeficiency virus-infected animal with a lymphoproliferative disorder (25, 29). The RRV genome was shown to contain many of the same genes as HHV-8, and these genes have a high degree of sequence similarity to HHV-8 (1, 25). Like HHV-8, RRV encodes many cellular homologs, and the two viruses share a similar genomic arrangement (1, 25). One defining feature of RRV, in contrast to HHV-8, is that although it has been shown that RRV can establish a latent infection in vivo, a lytic infection occurs in primary rhesus macaque fibroblasts (RFs) (1, 4, 25). The ability of RRV to infect cells in culture and produce infectious virus is an important attribute that makes RRV a very attractive model for lytic replication studies.
HHV-8 is a gamma-2 herpesvirus that is the probable cause of Kaposi's sarcoma (5, 7–9). In tissue culture, HHV-8 is present primarily in a latent form in human B cell lines (15, 16, 20). The lytic replication cycle is induced in a small number of cells by incubation with the phorbol ester tetradecanoyl phorbol acetate or n-butyrate (20). It was also shown that the viral lytic cycle could be initiated by transfection of the virus-encoded open reading frame (ORF) product ORF50 into latently infected B cells (22, 28). ORF50 is the putative homolog of EBV Rta and appears to have transactivating properties (13). These data implicated ORF50 as the key protein involved in the transition from the latent to the lytic state of viral replication. However, notwithstanding a demonstrated clear activation of the HHV-8 lytic replication cycle by either drug treatment or transfection of ORF50, it is estimated that only a small number of cells in culture undergo lytic-phase replication when induced by either method (14, 20).
Herpesvirus lytic replication initiates at distinct cis-acting regions within the genome that vary in size and complexity and are usually marked by the presence of a number of transcription factor binding sites and multiple repeat regions (2, 3, 12, 24, 30). These herpesvirus replication origins were identified using a transient-transfection replication assay in which permissive cells are transfected with plasmids containing fragments of the viral genome and then incubated with infectious virus. Viral infection supplies essential transacting factors required for efficient origin-dependent DNA replication (3, 10, 12, 26, 27). Initiation of DNA synthesis for many herpesvirus origins is via the interaction of a virus-encoded factor with a defined cis-acting element present with the lytic origin. For Epstein-Barr virus, the lytic origin is composed of four Zta binding sites that appear to act as initiator sites for replication (23). Substitutions or deletions of this region either result in a reduction of or completely abolish origin-dependent replication.
The subcloning of the entire RRV genome into cosmid and plasmid vectors, coupled with the fact that lytic infection occurs upon infection of cultured cells, allows the development of a transient-transfection replication assay for the identification of the RRV lytic origin of DNA replication (oriLyt). To this end, we transfected cosmid clones of RRV into a new cell line of telomerized RFs to identify the location of an oriLyt. The RRV oriLyt was localized to a region of the RRV genome between ORF69 and ORF71 (vFLIP). This region amplified only in the presence of infecting virus and contains an upstream A+T-rich region and a 300-bp downstream G+C repeat element. In addition, oriLyt amplification occurred in cells cotransfected with a complete set of RRV cosmids when an ORF50 expression plasmid was included in the transfection mixture, allowing the elucidation of the set of transacting factors required for origin-dependent DNA replication.
MATERIALS AND METHODS
Cells and virus.
Stock RRV (strain 17577) was propagated in primary RFs isolated as previously described (25). Telomerized RFs (telo-RFs) were used for all transfection experiments, and their development is described elsewhere (V. Kirchoff, S. Wong, S. St. Jeor, and G. S. Pari, submitted for publication).
Plasmid constructs.
Construction of cosmid clones used for transfection and cotransfection experiments (cosmids 8, 9, 28a, and 44) was previously described (25). The initial plasmid subclone encoding oriLyt was made from cosmid clone 8 as follows: the XbaI 6.4 subclone, pRRVO, was constructed by cleaving cosmid 8 with XbaI and isolating the 6.4-kb fragment from nucleotide (nt) 112979 to 119470 and ligating it into XbaI-cleaved pGEM7zf(−). All other subclones were made from the plasmid pRRVO. pRRVL1 was constructed by cleaving pRRVL with BamHI, releasing the 3.2-kb fragment (genomic coordinates 112979 to 116210), and ligating it into BamHI-cleaved pGEM7zf(−). pRRVR1 was constructed by cleaving pRRVO with BamHI, removing the BamHI fragment from nt 112979 to 116472, and religating the linearized plasmid carrying nt 116210 to 119470. pRRVM1 was constructed by cleaving pRRVO with NarI and ligating the resulting 1.5-kb fragment into ClaI-cleaved pGEM7zf(−). pRRVR2 was constructed by cleaving pRRVR1 with EcoRI and ligating the resulting 2.1-kb fragment from nt 117360 to 119470 into EcoRI-cleaved pGEM7zf(−). Plasmid pRRVL6 was made by using PCR primers corresponding to the 5′ and 3′ ends (nt 112979 to 114067) of the A+T-rich region and ligating the resulting product into pGEM T-easy.
RRV oriLyt deletion mutants were made using Erase-a-Base (Promega). Plasmid pRRVL1 was cleaved with XhoI and SacI and treated with lambda exonuclease as described by the manufacturer. Several colonies were picked and analyzed by agarose gel electrophoresis. Clones were sequenced to determine the extent of the deleted sequence.
A plasmid construct expressing RRV ORF50, pEXP50, was generated by using PCR primers complementary to the putative ORF50 ORF identified from the previously published sequence (25). The PCR product was ligated into pTargeT (Promega). This vector uses the cytomegalovirus (CMV) immediate early promoter to drive the expression of an inserted gene.
Transfection-replication assay.
Telo-RFs (5 × 105) were propagated and plated for transfection using Dulbecco's modification of Eagle's medium supplemented with 10% fetal bovine serum in 6-cm dishes 16 h prior to transfection. Culture medium was removed and replaced with 3 ml of complete medium 4 h before transfection. Cells were transfected using 8 μl of Transit-LT (Mirus) and 3 μg of plasmid or cosmid DNA. Cells were incubated with the Transit-DNA mixture for 15 h, medium was removed, and cells were infected with concentrated RRV. Concentrated RRV was obtained by harvesting infected cells from a 10-day-infected roller bottle culture and pelleting virus using an SW28 rotor (25,000 rpm, 1.5 h). The resulting virus pellet was resuspended in 5 ml of complete medium. Each dish of transfected telo-RFs was incubated with 300 μl of concentrated virus stock. Transfected cells were incubated with virus for 5 to 6 days, and then total cellular DNA was extracted using Puregene DNA isolation solutions (Gentra Systems). Five micrograms of DNA was cleaved with EcoRI (15 U) and DpnI (5 U) and separated on a 0.8% agarose gel. The gel was transferred to a nylon membrane and hybridized with 32P-labeled pGEM probe in order to detect replicated plasmid DNA.
For the cotransfection replication assay, cells were transfected using the BES [N,N-bis(2-hydroxyethyl)-2-aminoethanesulfonic acid] calcium phosphate method similar to the one used for human CMV (HCMV) (19). Cells were incubated with a mixture of the oriLyt-containing plasmid pRRVL1 (10 μg) and cosmids 9, 8, 44, and 28a (2 μg each). Total cell DNA was harvested 6 days posttransfection and treated as for the infection-replication assay.
RESULTS
RRV cosmid 8 contains a lytic origin of replication.
Recently, as part of a sequencing endeavor for RRV, the entire RRV genome was subcloned into overlapping cosmid vectors (25). It was also demonstrated that these cosmids were sufficient to recover infectious virus upon cotransfection into permissive macaque fibroblasts (S. Wong, unpublished data). These experiments indicated that (i) the RRV cloned cosmids contained a competent lytic origin and (ii) the genes necessary for replication of that origin were present in the cosmids and functional upon transfection into permissive cells. These facts allowed us to develop a transient-transfection replication-infection assay by individually transfecting each cosmid from a set of four RRV overlapping cosmids (Fig. 1). Although RRV is permissive in primary RFs, it became obvious after initial transfection experiments that the transfection efficiency in these cells was very low. We had previously developed a line of telomerized RFs, and these cells were found to be highly transfectable and maintained permissiveness for RRV.
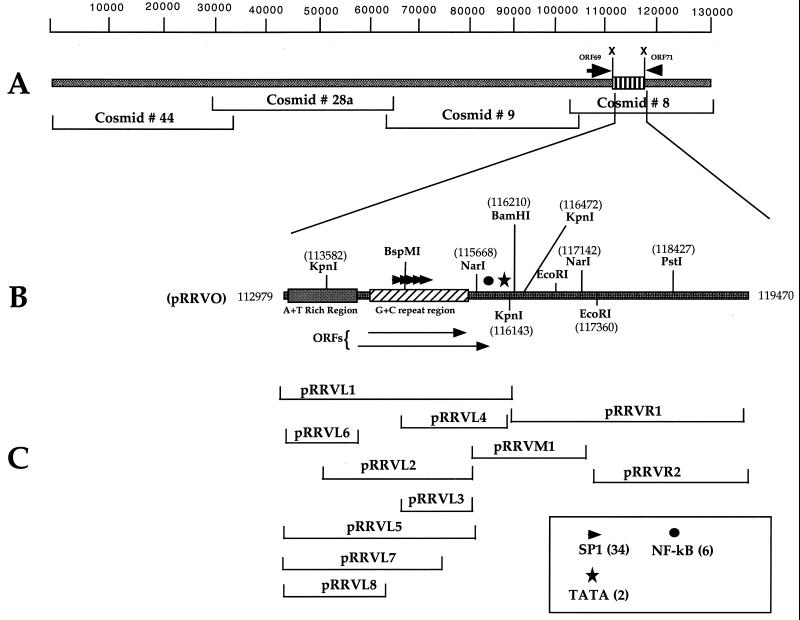
FIG. 1.
Schematic representation of the RRV genome and oriLyt. (A) Location of oriLyt mapping between ORF69 and ORF70 contained within cosmid 8. (B) Plasmid construct pRRVO containing the 6.4-kb XbaI fragment. Relative positions of some restriction endonuclease sites, A+T- and G+C-rich regions, and the positions of two putative ORFs are shown. (C) Plasmid constructs used in the transfection-infection assay. The box in the bottom right hand corner defines the symbols used for transcription factor binding sites identified within oriLyt.
To determine the location of oriLyt within cosmid subclones, we transfected individual cosmid clones into telo-RFs and subsequently infected these cells with high-titer concentrated RRV (approximate titer, 107 PFU per ml). Total cellular DNA was harvested and cleaved with EcoRI and DpnI, which cleaves only input unreplicated cosmid DNA that has been propagated in a Dam+ bacterial host. Cosmids 8, 9, 28a, and 44 were individually transfected into telo-RFs. Of the four RRV cosmids transfected, only cosmid 8 amplified in the presence of infecting virus, as demonstrated by the presence of a DpnI-resistant band (Fig. 2A, lane 1). Cosmid 8 spans nt 107105 to 130000 of the RRV genome. This sequence is located at the right end of the genome. Present within this region are a series of repeats referred to as rDL-E 1 and 2 (25). The rDL-E region, spanning approximately nt 114000 to 117000, encodes a highly repetitive G+C-rich DNA sequence. According to the published map of RRV ORFs, the rDL-E loci appeared to be devoid of any recognizable ORFs (25). We chose to focus on this region as the possible location of an oriLyt contained with cosmid 8.
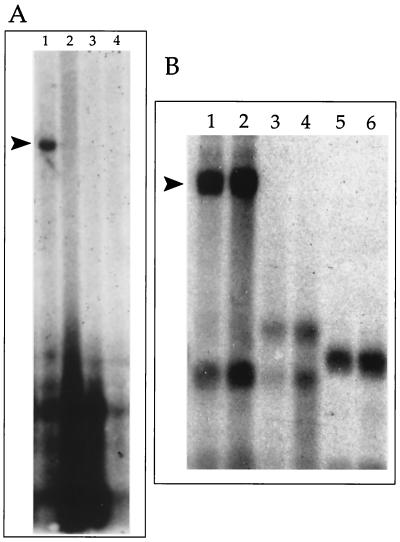
FIG. 2.
Identification of RRV oriLyt using the transient-transfection infection-replication assay. Total cellular DNA from transfected cells was cleaved with EcoRI and DpnI, separated through a 0.8% agarose gel, transferred to a nylon membrane, and probed with SuperCos (Stratagene). Arrowheads indicate replication products. (A) Autoradiogram of a Southern blot of total telo-RF DNA from cells transfected with RRV cosmids and subsequently infected with RRV. Lanes: 1, cosmid 8; 2, cosmid 9; 3, cosmid 28a; 4, cosmid 44. (B) RRV oriLyt is present within a 6.4-kb XbaI I fragment subcloned from cosmid 8. DNA was treated as for panel A except that blots were probed with pGEM7zf(−). Lanes: 1 and 2, pRRVO-transfected infected telo-RF DNA; 3 and 4, as lanes 1 and 2 except that cells were treated with PFA; 5 and 6, pRRVR1-transfected infected telo-RF DNA.
Several subclones of cosmid 8 were transfected into telo-RFs and evaluated for their ability to amplify in the presence of infecting virus (data not shown). Of these subclones, a 6.4-kb XbaI subclone made from cosmid 8 replicated when transfected into cells that were subsequently infected with RRV (Fig. 2B, lanes 1 and 2) but not in cells treated with phosphonoformic acid (PFA) at the time of infection (Fig. 2B, lanes 3 and 4). These data indicated that the 6.4-kb XbaI fragment contained a lytic origin of replication.
RRV oriLyt is composed of an A+T-rich and a short G+C-rich sequence.
The XbaI subclone pRRVO was cleaved with BamHI, which essentially cut the 6.4-kb XbaI fragment in half (Fig. 1). The resulting two BamHI fragments were subcloned. A plasmid clone, pRRVR1, corresponding to the right half of pRRVO from nt 116210 to 119470 failed to replicate in infected telo-RF cells (Fig. 2B, lanes 5 and 6). The failure of this fragment to amplify may indicate that RRV oriLyt was localized in the right half of pRRVO. To test this hypothesis, we transfected the subclone pRRVL1, which carried the left half of pRRVO. Transfection of pRRVL1 resulted in the detection of a DpnI-resistant band and indicated that RRV oriLyt was contained in pRRVL (nt 112979 to 116210).
Now that it had been demonstrated that a replication origin was present within the plasmid subclone pRRVL1, we wanted to identify essential DNA elements contributing to origin function and further localize the boundaries of oriLyt. The right half of pRRVO contains a highly repetitive G+C-rich region spanning approximately 1.7 kb (Fig. 1). Immediately upstream of this G+C repeat region is an A+T-rich region spanning approximately 900 nt (Fig. 1). We constructed a series of subclones with portions of the repeat regions eliminated. These plasmid subclones and their location are illustrated in Fig. 1. The plasmid subclone, pRRVL1, encoding the 3-kb region from nt 112979 to 116210 was fully replication competent (Fig. 3A, lane 2). This clone contained both A+T and G+C repeat regions. When the DNA sequence in pRRVL1 was subjected to analysis using software designed to search for putative ORFs, two ORFs not previously identified in the original published sequence of either RRV strain were identified (1, 25); the locations of these ORFs are shown in Fig. 1.
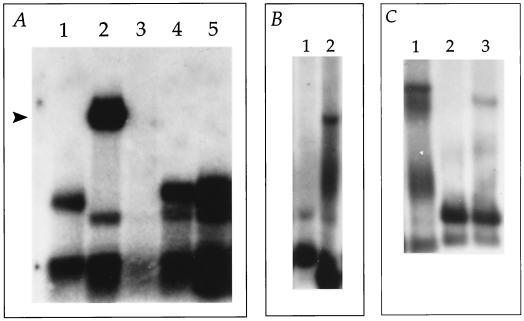
FIG. 3.
RRV oriLyt contains two essential repeat elements. Autoradiograms of Southern blots of DNA from transfected infected cells treated as described in Materials and Methods are shown. Arrowheads indicate replication products. (A) DNA from cells transfected with pRRVL3 (lane 1), pRRVL1 (lane 2), pRRVM1 (lane 3), pRRVL4 (lane 4), or pRRVL2 (lane 5). (B) Replication of a plasmid construct containing both intact G+C- and A+T-rich genomic sequences. Lanes 1 and 2, transfection with plasmid pRRVL5 in mock-infected cells and RRV-infected cells, respectively. (C) The A+T-rich region plus 200 bp of the G+C-rich genomic sequence is required for plasmid replication. Cells were transfected with oriLyt plasmids containing variable amounts of the G+C-rich region or the A+T sequence alone. Lanes: 1, pRRVL7 (nt 112979 to 115016); 2, pRRVL6 (nt 112979 to 114067); 3, pRRVL8 (nt 112979 to 114371).
A transfected subclone containing the complete G+C repeat region but only a portion of the A+T repeat region failed to replicate (Fig. 3A, lane 5). This subclone, pRRVL2, contains genomic sequences from the KpnI site at nt 113582 to the NarI site at nt 115688 (Fig. 1). pRRVL2 deletes part of the A+T repeat DNA sequence region and truncates the putative 2.0-kb ORF that goes through the G+C-rich region (Fig. 1). Similarly, plasmids pRRVL3 and pRRVL4 failed to replicate (Fig. 3A, lanes 1 and 4, respectively). pRRVL3 contains only a small portion of the G+C repeat region and truncated portions of both potential ORFs. pRRVL4 extends further 3′ than pRRVL3 but still does not include the complete G+C repeats or putative ORFs. Failure of these subclones to replicate indicated that the left boundary of oriLyt extended to the original XbaI fragment at nt 112979 (Fig. 1). Another subclone, pRRVM1, also failed to replicate, indicating that an oriLyt does not lie in the middle sequence of the 6.4-kb XbaI region (Fig. 3A, lane 3). In order to test where the right boundary of oriLyt was, we generated a subclone that extended to the NarI site at nt 115688 and tested it in the replication assay. This subclone, pRRVL5, amplified in the presence of RRV, indicating that the DNA sequence located between the NarI (115668) and the BamHI (116210) sites was dispensable (Fig. 3B, lane 2).
In order to further define the boundaries of RRV oriLyt, we generated several plasmid constructs that deleted various amounts of the G+C-rich genomic sequence (Fig. 1). In addition, we subcloned the A+T-rich region alone and tested this plasmid, pRRVL6, along with the G+C deletion constructs. The plasmid construct, pRRVL7, which removes 650 bp from the 3′ end of pRRVL5 replicated, which indicated that the entire G+C-rich region was not necessary for efficient replication (Fig. 3C, lane 1). The A+T region alone, however, did not replicate, indicating that additional sequence was required (Fig. 3C, lane 2). Several other deletion constructs were tested (data not shown), and it was determined that the smallest clone that was replication competent, pRRVL8, contained approximately 300 bp of the 3′-flanking G+C sequence (Fig. 3C, lane 3). Replication of pRRVL8 was somewhat decreased compared to that of larger clones; nevertheless, this construct defines the minimal boundaries of RRV oriLyt.
RRV cosmids contain all the genes required for origin-dependent lytic DNA replication.
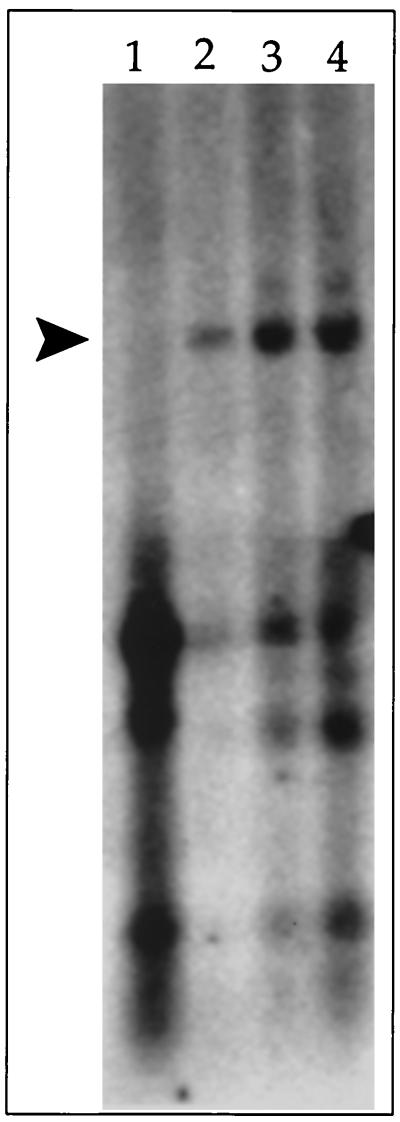
Once an oriLyt had been identified for RRV, we evaluated the ability of our overlapping cosmid library to complement origin-dependent lytic replication. These cosmids should encode all the necessary transacting factors for efficient oriLyt replication. Our initial attempts to get oriLyt-dependent DNA replication from cotransfection of four overlapping cosmids plus oriLyt (pRRVO) failed to produce a detectable replicated product (Fig. 4, lane 1). We assumed that this might be due to a low level of expression of certain viral transactivators, which then activate genes required for replication. In order to overcome this, we added increasing amounts of an expression construct encoding RRV ORF50 to the transfection mixture. For HHV-8, ORF50 was shown to activate the entire lytic cycle (11, 13). Introduction of an HHV-8 ORF50-expressing plasmid into latently infected cells resulted in the initiation of the lytic cycle of the virus, as evidenced by the appearance of viral lytic markers. Consequently, we chose to deliver additional RRV ORF50 by adding an ORF50 expression plasmid to the cotransfection mixture already containing the four overlapping cosmids and oriLyt. Transfections containing increasing amounts of pEXP50 yielded a detectable replication product (Fig. 4, lanes 2 through 4). The oriLyt band also appeared to be more intense as more pEXP50 was added, suggesting a higher degree of replication correlated to increasing levels of ORF50 protein (Fig. 4, lanes 2 through 4).
FIG. 4.
Addition of an ORF50 expression construct facilitates origin-dependent DNA replication in the cotransfection replication assay. A Southern blot of the cotransfection replication assay is shown. Telo-RFs were transfected with all four overlapping cosmids plus pRRVO alone (lane 1) or pRRVO and 0.5, 1, or 2 μg of pEXP50 (lanes 2 through 4, respectively). The arrowhead indicates the replication product.
Inspection of the oriLyt DNA sequence.
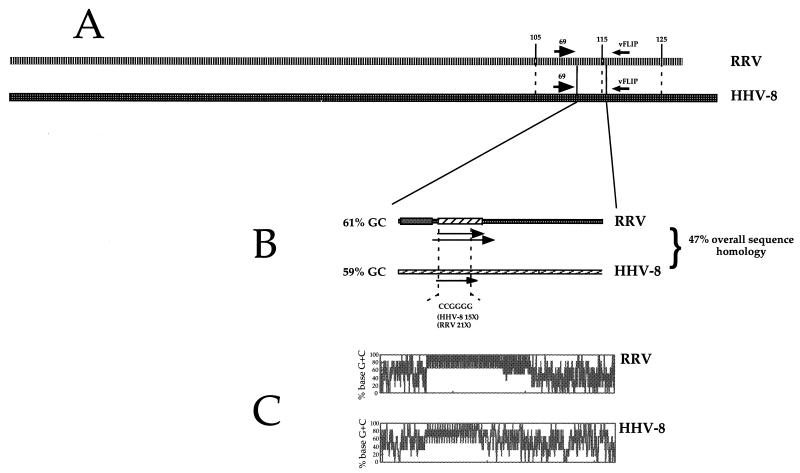
We compared the relative positions of RRV oriLyt and the same locus in HHV-8. RRV oriLyt maps to the region between ORF69 and the vFLIP (ORF71) gene locus mapping approximately between nt 110000 and 120000. Homologs of these genes are located in the same relative position in the HHV-8 genome (Fig. 5A). It was suggested that this region, in HHV-8, might contain a lytic origin (18). When the HHV-8 and RRV genomes within the genomic regions between ORF 69 and vFLIP are compared, several similarities are apparent. This region in HHV-8 is a highly repetitive G+C-rich DNA sequence and contains a number of transcription factor binding sites. In addition, except for one ORF identified by DNA sequence analysis and not described in the original report of the HHV-8 genomic sequence, this region apparently does not contain any identifiable ORFs (21).
FIG. 5.
Comparison of RRV oriLyt and the corresponding region in HHV-8. (A) Alignment of RRV and HHV-8 genomes, showing the genomic regions between ORF69 and vFLIP. (B) Comparison of the 2-kb region from RRV oriLyt and the similar region in HHV-8. The relative positions of two putative ORFs within each region are shown. (C) Schematic representation of the oriLyt region of RRV and the positional homolog of HHV-8. The G+C base pair distribution across each region is shown.
RRV oriLyt has an overall G+C content of 61%; a region of similar size and position in the HHV-8 genome was calculated to have a 59% G+C content. Between these two regions (HHV-8 putative oriLyt and RRV oriLyt) there is 47% nucleotide sequence homology. Several similar or identical repeat elements are present within RRV and HHV-8 regions; one example is the DNA sequence CCGGGG, which is repeated 21 times in RRV oriLyt and 15 times in the putative HHV-8 oriLyt region (Fig. 5B). As with RRV, we were also able to identify a small ORF (800 bp) within the G+C repeat region of the putative HHV-8 oriLyt. This putative ORF is located in a similar place in the RRV oriLyt, which is within the G+C-rich repeat regions (Fig. 5B). Distribution of G+C base pairs also had similar patterns in HHV-8 and RRV homologous regions. There appears to be the same A+T-rich region followed by a downstream G+C-rich region (Fig. 5C). Although the G+C-rich region appears to be longer in RRV, the pattern is similar in HHV-8 (Fig. 5C). Most striking is the similarity of the pattern of base distribution, where two A+T-rich regions flank a highly repetitive G+C-rich domain.
DISCUSSION
Lytic replication of HHV-8 occurs in a small number of persistently infected B cells upon treatment with tetradecanoyl phorbol myristate or sodium butyrate or transfection with the viral transactivator ORF50 (11, 13, 28). This is not the case for the rhesus macaque homolog, RRV, where a lytic infection occurs upon incubation with infectious virus in primary RFs. This feature lends itself to the development of a transient-replication assay similar to those used to identify lytic origins of other herpesviruses (3, 12, 26, 27). RRV shares most of the genes with HHV-8, including the cellular homologs for interleukin 6, cyclin D, and Bcl2. There are, however, some notable differences. For example, RRV lacks the genes for kaposin and nut1 and some of the vMIP-encoding loci. Despite these differences, RRV is still a good model for the study of gammaherpesvirus lytic replication. All of the core replication proteins along with the viral transactivators have a high degree of nucleotide sequence and positional homology between the two viruses. This may allow a hybrid replication assay where proteins from one system will be used to complement origin-dependent replication for the other.
We developed a transient-replication assay using RRV where infecting virus supplies all of the necessary transacting factors for lytic replication. For these studies we generated an RF cell line by introducing the gene expressing the catalytic subunit of telomerase (6, 17). These cells, telo-RFs, are highly transfectable and are much more robust than primary RFs. In addition to using these cells for our assay, we found it necessary to use concentrated virus for infection. Regular stock virus was not of a high enough titer to efficiently infect all of the cells in the culture dish. The replication-infection assay revealed that a lytic origin was present in RRV cosmid 8, and subsequent subcloning identified a 2.3-kb region containing an A+T-rich region followed by a downstream 300-bp G+C-rich region. Transfection data suggest that both the A+T-rich and the short (300-bp) G+C-rich regions are essential for efficient replication. This region would include any putative promoter elements required for the transcription of mRNA containing one or both of the potential ORFs found in the downstream region. This RRV oriLyt region closely resembles the corresponding HHV-8 loci. In addition, a recently identified lytic origin for the bovine rhadinovirus bovine herpesvirus 4 shows positional homology to the genomes of RRV and HHV-8 (30). The bovine herpesvirus 4 oriLyt also has a G+C-rich region flanked by an A+T-rich region (30). The conservation of these structures may reflect a distinct mechanism for the initiation of DNA replication for this subset of herpesviruses.
We also demonstrated that a set of overlapping cosmids for RRV can complement origin-dependent DNA replication. We found it necessary to add additional ORF50 protein in the form of a plasmid construct under the control of the HCMV major immediate early promoter. Additional ORF50 may be required because expression from the native ORF50 within cosmid 28a was not sufficient to activate the other replication factors. A similar scenario was observed for the cosmid cotransfection assay for HCMV. In the HCMV system, origin-dependent replication was inefficient unless additional IE2 was added to the transfection mixture in the form of a plasmid expression vector (19). Another possibility for both systems is that these transactivators may directly participate in lytic replication. The cotransfection assay should allow the elucidation of factors required for oriLyt-dependent DNA replication.
Now that the lytic origin for RRV has been identified and a cotransfection assay has been established, we can start to compare this obviously more lytic viral system to HHV-8, a predominately latent system.
ACKNOWLEDGMENT
This work was supported by a grant from the National Institutes of Health (R01 CA85164)
REFERENCES
- 1.Alexander L, Denekamp L, Knapp A, Auerbach M R, Damania B, Desrosiers R C. The primary sequence of rhesus monkey rhadinovirus isolate 26-95: sequence similarities to Kaposi's sarcoma-associated herpesvirus and rhesus monkey rhadinovirus isolate 17577. J Virol. 2000;74:3388–3398. doi: 10.1128/jvi.74.7.3388-3398.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anders D G, Kacica M A, Pari G, Punturieri S M. Boundaries and structure of human cytomegalovirus oriLyt, a complex origin for lytic-phase DNA replication. J Virol. 1992;66:3373–3384. doi: 10.1128/jvi.66.6.3373-3384.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anders D G, Punturieri S M. Multicomponent origin of cytomegalovirus lytic-phase DNA replication. J Virol. 1991;65:931–937. doi: 10.1128/jvi.65.2.931-937.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergquam E P, Avery N, Shiigi S M, Axthelm M K, Wong S W. Rhesus rhadinovirus establishes a latent infection in B lymphocytes in vivo. J Virol. 1999;73:7874–7876. doi: 10.1128/jvi.73.9.7874-7876.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birley H D, Schultz T F. Kaposi's sarcoma and the new herpesvirus. J Med Microbiol. 1997;46:433–435. doi: 10.1099/00222615-46-6-433. [DOI] [PubMed] [Google Scholar]
- 6.Bodnar A G, Ouellette M, Frolkis M, Holt S E, Chiu C P, Morin G B, Harley C B, Shay J W, Lichtsteiner S, Wright W E. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 7.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 8.Cesarman E, Moore P S, Rao P H, Inghirami G, Knowles D M, Chang Y. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood. 1995;86:2708–2714. [PubMed] [Google Scholar]
- 9.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 10.Dewhurst S, Dollard S C, Pellett P E, Dambaugh T R. Identification of a lytic-phase origin of DNA replication in human herpesvirus 6B strain Z29. J Virol. 1993;67:7680–7683. doi: 10.1128/jvi.67.12.7680-7683.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gradoville L, Gerlach J, Grogan E, Shedd D, Nikiforow S, Metroka C, Miller G. Kaposi's sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J Virol. 2000;74:6207–6212. doi: 10.1128/jvi.74.13.6207-6212.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hammerschmidt W, Sugden B. Identification and characterization of oriLyt, a lytic origin of DNA replication of Epstein-Barr virus. Cell. 1988;55:427–433. doi: 10.1016/0092-8674(88)90028-1. [DOI] [PubMed] [Google Scholar]
- 13.Lukac D M, Kirshner J R, Ganem D. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J Virol. 1999;73:9348–9361. doi: 10.1128/jvi.73.11.9348-9361.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lukac D M, Renne R, Kirshner J R, Ganem D. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF50 transactivator, a homolog of the EBV R protein. Virology. 1998;252:304–312. doi: 10.1006/viro.1998.9486. [DOI] [PubMed] [Google Scholar]
- 15.Miller G, Heston L, Grogan E, Gradoville L, Rigsby M, Sun R, Shedd D, Kushnaryov V M, Grossberg S, Chang Y. Selective switch between latency and lytic replication of Kaposi's sarcoma herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells. J Virol. 1997;71:314–324. doi: 10.1128/jvi.71.1.314-324.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller G, Rigsby M O, Heston L, Grogan E, Sun R, Metroka C, Levy J A, Gao S J, Chang Y, Moore P. Antibodies to butyrate-inducible antigens of Kaposi's sarcoma-associated herpesvirus in patients with HIV-1 infection. N Engl J Med. 1996;334:1292–1297. doi: 10.1056/NEJM199605163342003. [DOI] [PubMed] [Google Scholar]
- 17.Nakamura T M, Morin G B, Chapman K B, Weinrich S L, Andrews W H, Lingner J, Harley C B, Cech T R. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 18.Nicholas J, Zong J C, Alcendor D J, Ciufo D M, Poole L J, Sarisky R T, Chiou C J, Zhang X, Wan X, Guo H G, Reitz M S, Hayward G S. Novel organizational features, captured cellular genes, and strain variability within the genome of KSHV/HHV8. J Natl Cancer Inst Monogr. 1998;23:79–88. doi: 10.1093/oxfordjournals.jncimonographs.a024179. [DOI] [PubMed] [Google Scholar]
- 19.Pari G S, Kacica M A, Anders D G. Open reading frames UL44, IRS1/TRS1, and UL36-38 are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA synthesis. J Virol. 1993;67:2575–2582. doi: 10.1128/jvi.67.5.2575-2582.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 21.Russo J J, Bohenzky R A, Chien M C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sarid R, Flore O, Bohenzky R A, Chang Y, Moore P S. Transcription mapping of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1) J Virol. 1998;72:1005–1012. doi: 10.1128/jvi.72.2.1005-1012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sarisky R T, Gao Z, Lieberman P M, Fixman E D, Hayward G S, Hayward S D. A replication function associated with the activation domain of the Epstein-Barr virus Zta transactivator. J Virol. 1996;70:8340–8347. doi: 10.1128/jvi.70.12.8340-8347.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schepers A D, Pich D, Mankertz J, Hammerschmidt W. Cis-acting elements in the lytic origin of DNA replication of Epstein-Barr virus. J Virol. 1993;67:4237–4245. doi: 10.1128/jvi.67.7.4237-4245.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Searles R P, Bergquam E P, Axthelm M K, Wong S W. Sequence and genomic analysis of a rhesus macaque rhadinovirus with similarity to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J Virol. 1999;73:3040–3053. doi: 10.1128/jvi.73.4.3040-3053.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stow N D. Localization of an origin of DNA replication within the TRs/IRs repeated region of the herpes simplex virus type 1 genome. EMBO J. 1982;1:863–867. doi: 10.1002/j.1460-2075.1982.tb01261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stow N D, Davidson A J. Identification of varicella-zoster virus origin of DNA replication and its activation by herpes simplex virus type 1 gene products. J Gen Virol. 1986;67:1613–1623. doi: 10.1099/0022-1317-67-8-1613. [DOI] [PubMed] [Google Scholar]
- 28.Sun R, Lin S F, Staskus K, Gradoville L, Grogan E, Haase A, Miller G. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J Virol. 1999;73:2232–2242. doi: 10.1128/jvi.73.3.2232-2242.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong S W, Bergquam E P, Swanson R M, Lee F W, Shiigi S M, Avery N A, Fanton J W, Axthelm M K. Induction of B cell hyperplasia in simian immunodeficiency virus-infected rhesus macaques with the simian homologue of Kaposi's sarcoma-associated herpesvirus. J Exp Med. 1999;190:827–840. doi: 10.1084/jem.190.6.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zimmermann W, Broll H, Ehlers B, Buhk H J, Rosenthal A, Goltz M. Genome sequence of bovine herpesvirus 4, a bovine rhadinovirus, and identification of an origin of DNA replication. J Virol. 2001;75:1186–1194. doi: 10.1128/JVI.75.3.1186-1194.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]