Abstract
Introduction
Antimicrobial resistance is a critical global issue, and medicinal plants, as a key source of therapeutic agents, offer potential solutions by offering new antibacterial agents. Acacia polyacantha tree, known as Al Kakamout in Sudan, is a significant source of Gum Arabic and has been traditionally used to treat bacterial diseases. This study aimed to investigate a hydro-ethanol extract of Kakamout stem bark through GC-MS analysis, evaluate its antibacterial activity against two standard bacterial strains, and conduct molecular docking and ADME studies.
Methods
The stem bark of the plant was extracted by maceration using a hydro-ethanol solvent and analyzed via GC-MS. The antibacterial activity of the extract was evaluated against Staphylococcus aureus ATCC 25923 and Pseudomonas aeruginosa ATCC 27853 using the well diffusion method. The identified compounds were studied in silico to investigate their binding affinities with the target bacterial proteins. The ADMET properties were predicted for the top scoring compounds.
Results
GC-MS analysis revealed the presence of 11 compounds, with the major ones being dopamine, N, N-dimethyl-, dimethyl ether (43.76%), 4-O-methylmannose (23.27%), sucrose (8.09%), 1,4,7-triazacyclononane, 1-benzoyl- (5.41%), and lupeol, trifluoroacetate (5.24%). The extract demonstrated significant effectiveness against both bacterial strains, even at a low concentration of 50 mg/mL. Molecular docking showed that compounds 1, 3, 4, and 6 had the best docking scores with enoyl-acyl carrier protein reductase (FabI) (PDB ID: 3GR6) from S. aureus (−6.142, −10.843, −6.218 and −7.14 Kcal/mol). Similarly, compounds 1–6 exhibited favorable binding energies with LasR-TP4 complex (PDB ID: 3JPU) from P. aeruginosa (−10.025, −9.127, −8.623, −7.092, −7.722, and −6.019 Kcal/mol).
Conclusion
This study provides the first GC-MS analysis of Acacia polyacantha stem bark, identifying potential antibacterial compounds. Molecular docking and ADMET predictions suggest several promising compounds for further investigation as antibacterial agents.
Keywords: Acacia polyacantha, antibacterial, GC-MS, ADME, molecular docking
Introduction
Medicinal plants have been utilized since ancient times as important sources of therapeutic agents for the prevention and treatment of diseases. They not only provide beneficial compounds for medicinal purposes but also offer valuable structures that can be used to develop new therapeutic agents.1 Herbal medicine is widely used as primary healthcare by 80% of the global population, according to the World Health Organization (WHO), due to its perceived safety, affordability, and accessibility.2 However, modern drugs are becoming increasingly resistant to bacterial infections, posing challenges and escalating costs, particularly for impoverished communities in Africa. This necessitates the exploration of alternative treatment strategies, with a focus on the development of antimicrobial agents.3 The WHO recognizes the potential of medicinal plants as an alternative source for a diverse range of drugs.4 Therefore, exploring the untapped potential of phytomedicine in Sudan is crucial, given its unique blend of African, Arabic, and Islamic cultures, as well as its diverse climate that supports a wide variety of plant species.5 One such plant is Acacia polyacantha Willd. (Mimosaceae), commonly known as AL-Kakamout in Sudan,3 is widely distributed in tropical Africa and considered a key source of Gum Arabic.6,7 The tree commonly grows in the moist, subtropical bushveld regions, usually favoring alluvial soils close to rivers, which is its preferred habitat. It has twice compound leaves, with 14–35 pairs of pinnae and 20–60 leaflets per pinna. They are large, arranged singly along the shoots, and have darker upper surfaces with marginal and stalk hairs.8 A. polyacantha has been traditionally used in Sudanese folk medicine for snakebite management, livestock diseases such as Salmonellosis and gastrointestinal disorders,9 venereal diseases,3 and even for soothing restless children through bathing with its infusion.10 It is also known for its antimalarial properties.11 Extracts and compounds derived from A. polyacantha have exhibited various pharmacological activities, including antibacterial,9,10,12,13 larvicidal,3 anti-leishmaniasis,14 hypoglycemic,15 and radical-scavenging activities.6,9 Phytochemical analysis of the stem bark and leaves of the plant has revealed the presence of three flavonoids (epicatechin, quercetin-3-O-galactoside, and methyl gallate), two triterpenoids (lupeol and β-amyrin), one sterol (stigmasterol), two saponins (3-O- [β-D-xylopyranosyl-(1→4)-β-D-galactopyranosyl] oleanolic acid and 3-O-[β-D-galactopyranosyl- (1→4)-β-D-galactopyranosyl]-oleanolic acid), and one sugar (3-O-methyl-D-chiro-inositol).16 Given the traditional use of A. polyacantha in Sudan for the treatment of bacterial infections, this study aimed to validate its antibacterial properties and identify the constituents responsible for its antibacterial action in the hydro-ethanol extract of the stem bark. Additionally, the study aimed to analyze the binding modes of these components and evaluate their ADMET (absorption, distribution, metabolism, excretion, and toxicity) properties.
Materials and Methods
Chemicals and Reagents
The antibiotics used in the experiment were Ceftriaxone (30 μg) and Vancomycin (30 μg), both from HIMEDIA (Mumbai, India). The solvents used, ethanol (99.9% purity) and methanol (99.9% purity), were obtained from SDFCL India. All other chemicals used were of analytical grade. The filter papers used in the experiment were Whatman No.1.
Microorganisms
In this study, standard strains of Gram-negative bacteria, Pseudomonas aeruginosa (ATCC 27853), and Gram-positive bacteria, Staphylococcus aureus (ATCC 25923), were utilized.
Plant Materials and Extraction
The stem bark of A. polyacantha was collected from the Bazoora region, located in the South of Algadaref State in Sudan. The identification and authentication of the plant material were performed by the botanist Fatima Abdelrahman Yassin at the Medicinal & Aromatic Plants and Traditional Medicine Research Institute (MAPRI) in Khartoum, Sudan in February 2022. A voucher specimen with the code number A-1975-17-MAPTRI-H was deposited at the institute for reference.
The collected stem bark was then prepared by chopping it into smaller pieces, followed by drying it in the shade. Subsequently, an electric mill was used to grind the dried stem bark into a coarse powder. For extraction, 100g of the coarsely ground material was macerated with 1 liter of 70% ethanol for a duration of 72 hours. The resulting mixture was then filtered using a Buchner device to separate the extract from the plant material.
After the filtration process, the solvent was allowed to evaporate at 60°C under reduced pressure using a rotary evaporator, and the obtained extract was subjected to freeze-drying. The freeze-dried extract was stored in a refrigerator until ready for use in subsequent experiments.
GC-MS Analysis
The stem bark extract of the plant was subjected to GC-MS analysis at the Medicinal & Aromatic Plants and Traditional Medicine Research Institute. The analysis was conducted using a GC/MS-QP2010SE instrument from Shimadzu (Kyoto, Japan), equipped with a capillary column (Rtx-5MS–30 m × 0.25 mmI.D × 0.25 µm). The experimental conditions were as follows:
The injector operated in the split mode at a temperature of 300°C, Helium was used as the carrier gas at a flow rate of 1.6 mL/min, the oven temperature was programmed to increase from 60°C to 300°C at a rate of 10°C/min, the MS conditions were set with an interface temperature of 250°C and an ion source temperature of 200°C, the mass scan covered the m/z range of 40 to 500 and had a total runtime of 34 minutes, the obtained spectra of the compounds were compared with the database of known component spectra stored in the GC-MS library of the National Institute of Standards and Technology (NIST).13
Antibacterial Activity Test
The Agar Well diffusion method was utilized to assess the antibacterial activity of the stem bark extract. Mueller Hinton Agar (MHA) served as the nutrient media.17 Initially, colonies from sub-cultured bacteria were diluted in 0.9% normal saline to achieve a concentration of 108 cfu/mL, equivalent to the turbidity of McFarland’s standard solution (0.5). McFarland’s standard solution was prepared by combining 0.6 mL of a 1.17% w/v solution of barium chloride with 99.4 mL of a 1% v/v solution of sulfuric acid. Subsequently, 20 mL of melted MHA was poured into sterile plates.
To ensure even distribution, 200 μL of the bacterial suspension was carefully added to the agar plates and gently mixed. After the media solidified at room temperature, three wells, each with a diameter of 8 millimeters, were created using the back of sterile blue tips from a graduated pipette.18 Two of these wells were filled with 100 μL of the extract at different concentrations (500, 250, 100, and 50 mg/mL), while the third well was filled with 100 μL of 50% methanol as a negative control. On the surface of the media, the positive controls (Vancomycin 30 μg disk for S. aureus and Ceftriaxone disk 30 μg for P. aeruginosa) were placed.
Subsequently, the plates were kept at room temperature for one hour and then incubated at 37°C overnight. The test was conducted twice for each extract concentration. Finally, the mean diameters of the inhibition zones were measured in millimeters (mm) using the following classification: zones less than 9 mm were considered inactive, 9–12 mm were partially active, 13–18 mm were active, and more than 18 mm were classified as very active.19
In silico Studies
The crystal structures of Staphylococcus aureus enoyl-acyl carrier protein reductase (FabI) (PDB ID: 3GR6) and the LasR-TP4 complex (PDB ID: 3JPU) from P. aeruginosa were obtained from the Protein Data Bank. Protein preparations were performed using the Protein Preparation Wizard tool in Schrodinger. The compound structures were retrieved from the PubChem database and subjected to energy minimization using the MacroModel tool. Molecular docking studies were conducted using the XP (extra precision) mode of Glide in Schrodinger, and binding sites were determined around the bound ligands using the Receptor Grid Generation tool. The Prime module in Schrodinger was used to predict the MM-GBSA free energy calculations. The druggable properties of the compounds were assessed using the QikProp module in the Schrodinger suite. Pharmacokinetic properties and toxicity of the compounds were evaluated, and Lipinski’s rule of five was applied to assess their drug-likeness.20
Results and Discussion
Yield Percent of A. Polyacantha Stem Bark Extract
Hydro-ethanol, specifically 70% ethanol, was employed as the solvent for extraction due to its environmentally friendly nature and lower cost compared to other solvents.6 Additionally, hydro-alcoholic mixtures containing a higher concentration of ethanol (70–90%) are commonly preferred for extraction processes as they have the ability to extract a wide range of compounds with varying polarities.21 In this study, hydro-ethanolic maceration was performed on the stem bark of A. polyacantha, resulting in a yield of 14.96%. This yield closely aligns with the findings of Koudoro et al,9 who achieved a yield of 15.2% using hydro-ethanol (50%) as the solvent for maceration extraction of A. polyacantha stem bark. Notably, the yield obtained using hydro-ethanol was higher than that achieved with water alone (14%) or ethanol alone (6.8%).9
GC-MS Analysis
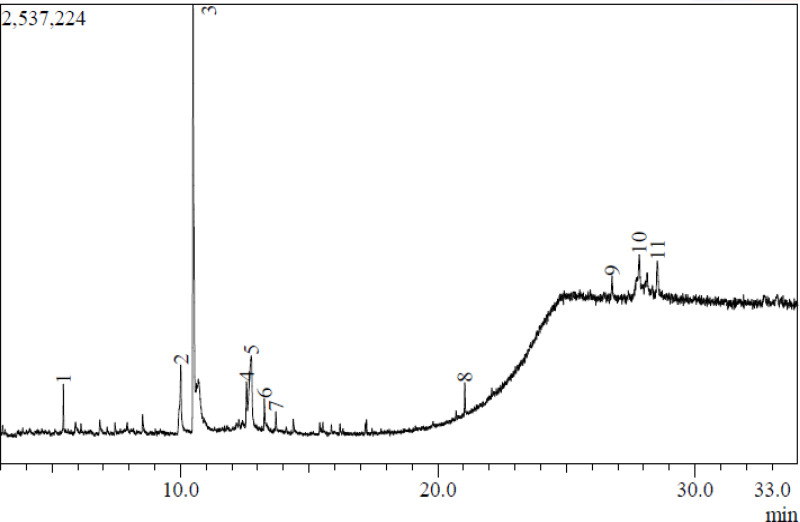
This study presents the first report on the GC-MS analysis of A. polyacantha stem bark extract (Figure 1 and Table 1), identifying a total of 11 compounds. Among these compounds, the major constituents were dopamine, N, N-dimethyl-, dimethyl ether (43.76%), 4-O-methylmannose (23.27%), sucrose (8.09%), 1,4,7-triazacyclononane, 1-benzoyl- (5.41%), lupeol, trifluoroacetate (5.24%), and stigmasterol (2.08%).
Figure 1.
GC Chromatogram of A. polyacantha stem bark extract.
Table 1.
Phytochemical Components Identified with GC-MS of A. Polyacantha Stem Bark Hydroethanolic Extract
| Peak No. | Compound Name | Structure | Retention Time | Area % |
|---|---|---|---|---|
| 1 | Undecane |  |
5.442 | 2.51 |
| 2 | Sucrose | 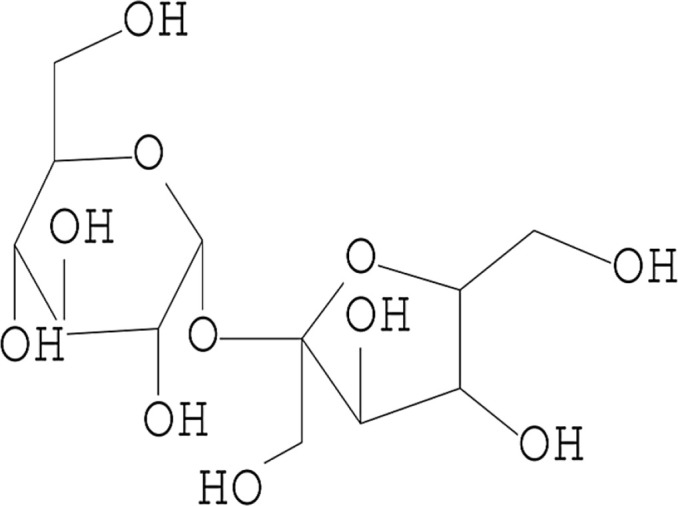 |
10.012 | 8.09 |
| 3 | Dopamine, N, N-dimethyl-, dimethyl ether | 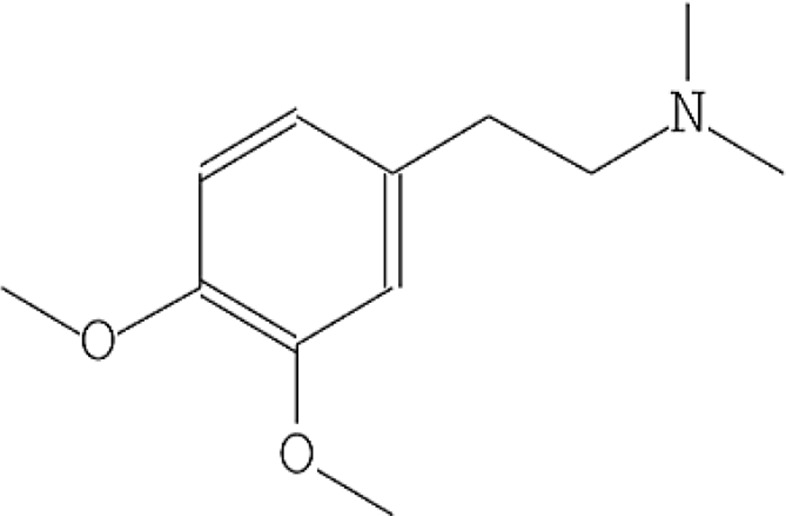 |
10.485 | 43.76 |
| 4 | 1,4,7-Triazacyclononane, 1-benzoyl- | 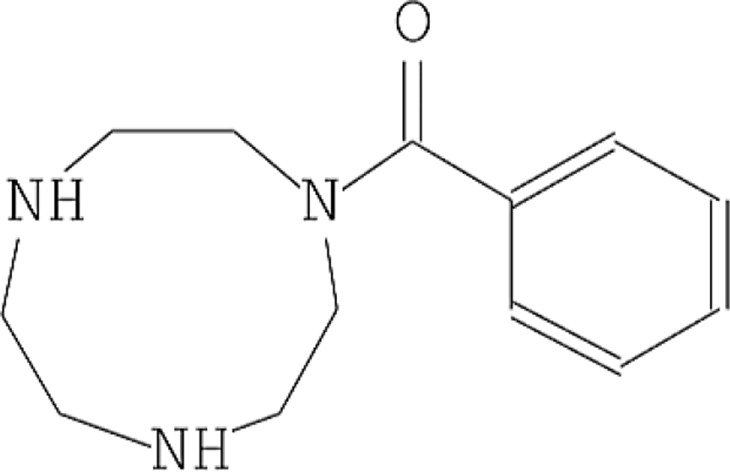 |
12.568 | 5.41 |
| 5 | 4-O-Methylmannose | 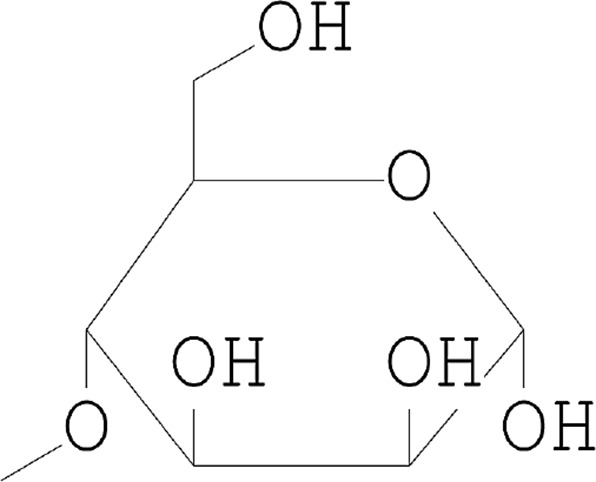 |
12.746 | 23.72 |
| 6 | 2-Propenoic acid, 3-(4 methoxyphenyl) | 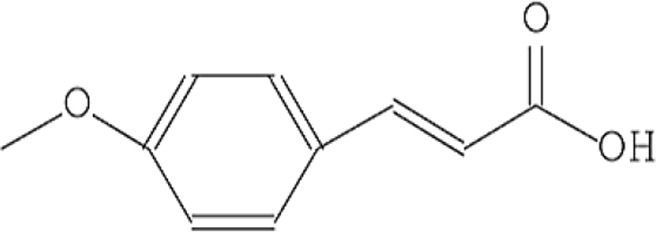 |
13.259 | 1.82 |
| 7 | 4-((1E)-3-Hydroxy-1-propenyl)-2-methoxy phenol | 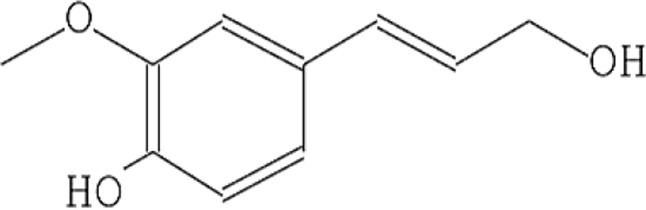 |
13.702 | 1.41 |
| 8 | Diisooctyl phthalate | 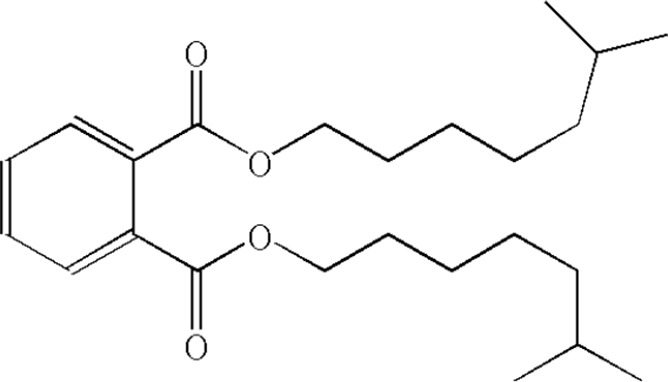 |
21.050 | 2.39 |
| 9 | Stigmasterol |  |
26.774 | 2.08 |
| 10 | 9,19-Cyclolanostan-3-ol, acetate |  |
27.829 | 4.02 |
| 11 | Lupeol, trifluoroacetate |  |
28.533 | 5.24 |
| 100.00 |
GC-MS analysis was chosen as the most suitable method for the chemical analysis of the A. polyacantha extract due to its effectiveness in identifying bioactive compounds.22
Furthermore, previous studies on ethanolic extracts of Acacia species aerial parts10,16,23,24 have also reported the presence of 4-O-methylmannose, stigmasterol, lupeol, and other triterpenes, which are known for their antibacterial properties. The identification of these compounds in the GC-MS analysis of A. polyacantha stem bark extract suggests that they could potentially contribute to the antibacterial activity observed against S. aureus and P. aeruginosa in this study.
Antibacterial Activity
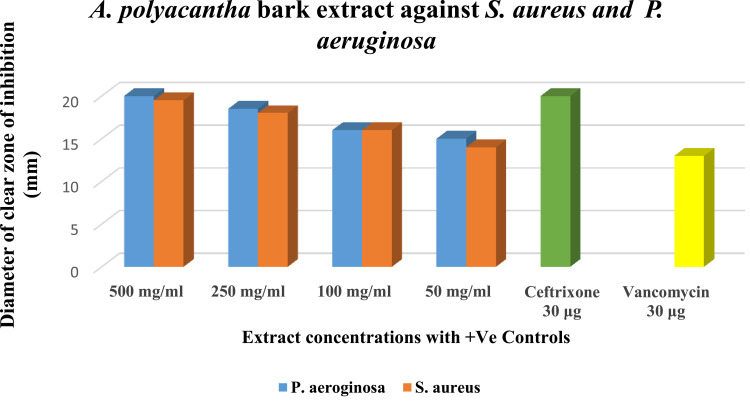
The agar well diffusion method was employed to assess the antibacterial activity of A. polyacantha stem bark extract against clinically important microorganisms, including Gram-positive Staphylococcus aureus (ATCC 25923) and Gram-negative Pseudomonas aeruginosa (ATCC 27853). Remarkably, the extract exhibited significant activity in a dose-dependent manner against both bacterial strains, even at a concentration as low as 50 mg/mL (Figure 2). These findings align with some previous studies9,10,16; however, they differ from the observations of Daffalla et al,3 who reported that the methanolic extract of the plant stem bark to be inactive against S. aureus at a concentration of 50 mg/mL. In contrast, our study demonstrated that the 50 mg/mL hydro-ethanolic extract was indeed active, producing a 14 mm inhibition zone, which is nearly comparable to the 13.6 mm zone observed with their 100 mg/mL concentration.
Figure 2.
The Susceptibility of Pseudomonas aeruginosa (ATCC 27853) and Staphylococcus aureus (ATCC 25923) to the plant extract at different concentrations.
Notes: The diameter of the clear zone of inhibition <9 mm zone was considered inactive, 9–12mm as partially active, 13–18mm as active and > 18mm as very active.19
The presence of phytoconstituents in A. polyacantha aerial parts extracts, which are known to have antibacterial activity alone or in synergism, includes steroidal compounds,25–27 triterpenoids,10,16,24,28,29 alkaloid of dopamine type,30,31 phthalates,32 phenolic compounds,33,34 phenyl propanoids35 and methyl pyranoside derivatives.23,36,16
In silico Studies
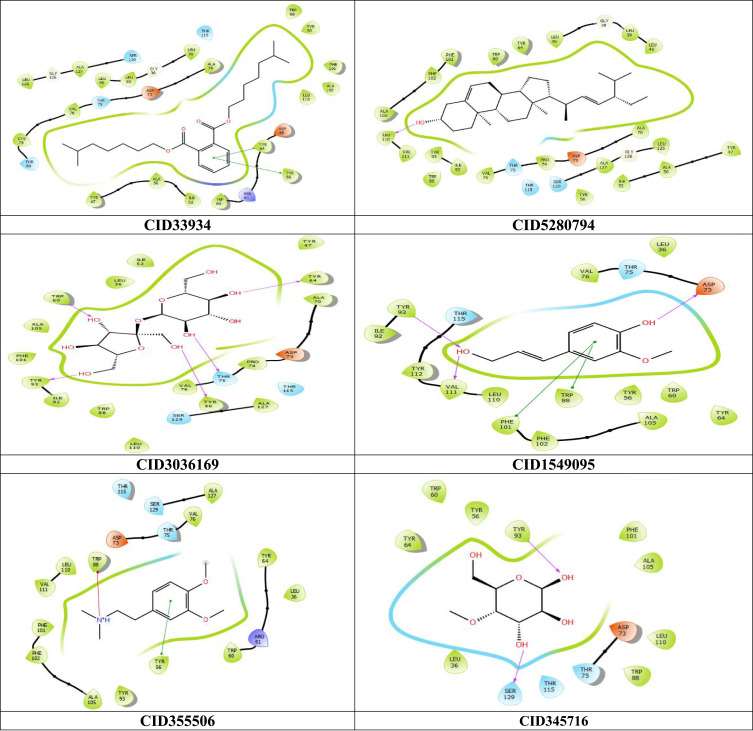
To explore the hypothetical binding modes of the compounds present in the plant stem bark extract, identified through GC-MS analysis, molecular docking was performed using two protein targets. The first target was S. aureus enoyl-acyl carrier protein reductase (FabI) (PDB ID: 3GR6), a crucial component involved in fatty acid synthesis in S. aureus,37 with an RMSD value of 2.83 Å, indicating a valid docking result. The second target was the transcriptional activator protein LasR-TP4 complex (PDB ID: 3JPU), responsible for biofilm formation and virulence factors in Pseudomonas aeruginosa,38 with an RMSD value of 0.1883 Å, demonstrating high accuracy in the docking prediction. The docking results are summarized in Table 2 and Figures 3 and 4. The binding cavities for docking were determined using the Receptor Grid Generation tool in Schrodinger, with the co-crystallized ligands serving as references.
Table 2.
Docking Scores and MM-GBSA Binding Free Energy of the Top Scoring Compounds with Enoyl-Acyl Carrier Protein Reductase (FabI) (PDB ID: 3GR6) and the Transcriptional Activator Enzyme LasR-TP4 Complex (PDB ID: 3JPU)
| Compound Character |
Diisooctyl Phthalate | Stigmasterol | Sucrose | Hydroxy-propenyl-methoxy-phenol | Dopamine, N, N dimethyl-dimethyl Ether | 4-O-Methylmanose | |
|---|---|---|---|---|---|---|---|
| Designated number | 1 | 2 | 3 | 4 | 5 | 6 | |
| PubChem ID | 33934 | 5,280,794 | 3,036,169 | 1,549,095 | 355,506 | 345716 | |
| Area% | 2.39 | 2.08 | 8.09 | 1.41 | 43.76 | 23.72 | |
| 3GR6 | Docking scores | −6.142 | −1.872 | −10.843 | −6.218 | −3.279 | −7.14 |
| MMGBSA dG Bind | −56.46 | −42.41 | −59.54 | −39.53 | −39.11 | −27.42 | |
| 3JPU | Docking scores | −10.025 | −9.127 | −8.623 | −7.092 | −7.722 | −6.019 |
| MMGBSA dG Bind | −70.16 | 10.34 | −66.26 | −40.67 | −59.26 | −18.67 | |
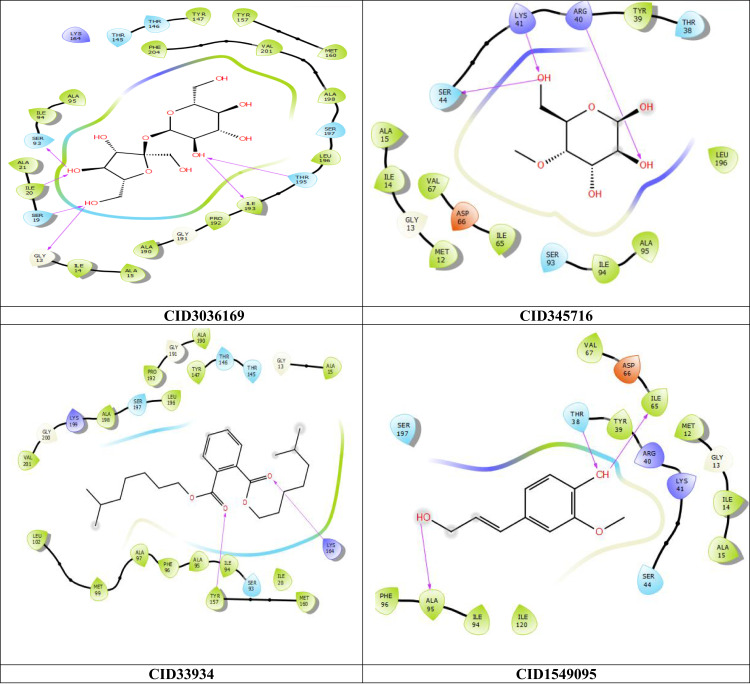
Figure 3.
2D interaction of the top four scoring compounds in complex with 3GR6 using XP docking mode of Glide software. The hydrophobic residues are in green color. The H-bond interactions with residues are illustrated by a purple dashed arrow oriented toward the electron donor.
Figure 4.
2D interaction of the top six scoring compounds in complex with 3JPU using XP docking mode of Glide software. The hydrophobic residues are in green color. The H-bond interactions with residues are illustrated by a purple dashed arrow oriented toward the electron donor.
The compounds diisooctyl phthalate, stigmasterol, sucrose, 4-((1E)-3-hydroxy-1-propenyl)-2-methoxy phenol, dopamine-N- N-dimethyl- dimethyl ether and 4-O-methylmannose were designated as compounds 1,2,3,4,5 and 6, respectively, in this manuscript.
Among the 11 compounds identified through GC-MS analysis, 6 compounds (1 to 6) exhibited the best docking scores with the 3JPU P. aeruginosa target, while 4 compounds (1,3,4 and 6) showed excellent binding energies against 3GR6 S. aureus target. Detailed docking scores and binding affinities are provided in Table 2.
Molecular docking is a valuable tool in computational drug discovery for predicting ligand–protein interactions. However, it cannot alone accurately measure the binding affinity and stability of complexes. To validate the docking results and avoid false-positive outcomes, a post-docking investigation was performed. The post-docking analysis involved calculating the binding affinity of the top ligand-protein complexes using the Molecular Mechanics Generalized Born and surface area (MM-GBSA) method. MM-GBSA is a reliable technique that considers the influence of the solvent and calculates the binding affinity based on post-docking free binding energy.20 Higher negative energy values obtained from MM-GBSA indicate stronger complex affinity. Notably, the results obtained from MM-GBSA were consistent with the docking scores, reinforcing the reliability of the docking predictions (Table 2).
Regarding the interaction between ligands and their respective target proteins, the top-scoring compounds demonstrated hydrophobic contacts, as shown in Table 3 and Figures 3 and 4. The interactions of the top ligands with target protein 3GR6 are as follows: Compound 3 formed six hydrogen bonds with THR195, ILE20, ILE193, SER19, SER93, and GLY13; Compound 6 formed three hydrogen bonds with ARG40, LYS41, and SER44; Compound 1 formed two hydrogen bonds with TYR157 and LYS164; and compound 4 formed three hydrogen bonds with ILE65, THR38, and ALA95. The residues ALA15 and ALA95 contributed to most of the hydrophobic interactions (Table 3 and Figure 3).
Table 3.
Molecular Interactions of the Top Compounds with Enoyl-Acyl Carrier Protein Reductase (FabI) (PDB ID: 3GR6) and the Transcriptional Activator Enzyme LasR-TP4 Complex (PDB ID: 3JPU)
| Target | Ligand | Pi-cation Interaction | Pi-pi Interaction | Hydrogen Bonding Interaction | Hydrophobic Interaction |
|---|---|---|---|---|---|
| 3GR6 | Sucrose (3) | ILE193, SER19, SER93, GLY13, THR195, ILE20 | ALA15, ALA190, ALA198, ALA21, ALA95, ILE14, ILE193, ILE20, ILE94, LEU196, MET160, PHE204, PRO192, TYR147, TYR157, VAL201 | ||
| 4-O-Methylmanose (6) | ARG40, LYS41, SER44 | ALA95, ILE14, ILE65, ILE94, LEU196, MET12, TYR39, VAL15, VAL67 | |||
| Diisooctyl phthalate (1) | LYS164, TYR157 | ALA15, ALA190, ALA198, ALA95, ALA97, ILE20, ILE94, LEU102, LEU196, MET160, MET99, PHE96, PRO192, TYR147, TYR157, VAL201 | |||
| Hydroxy-propenyl-methoxyphenol (4) | ALA95, ILE65, THR38 | ALA15, ALA95, ILE120, ILE14, ILE65, ILE94, MET12, PHE96, TYR39, VAL67 | |||
| 3JPU | Diisooctyl phthalate (1) | TYR56, TYR64 | ALA105, ALA127, ALA50, ALA70, CYS79, ILE52, LEU30, LEU39, LEU40 LEU110, LEU125, PHE101, TRO60, TRP88, TYR47, TYR56, TYR64, TYR93, VAL76 | ||
| Stigmasterol (2) | LEU110 | ALA105, ALA127, ALA50, ALA70, ILE52, ILE92, LEU110, LEU125, LEU36, LEU39, LEU40, PHE101, PHE102, PRO74, TRP60, TRP88, TYR47, TYR56, TYR64, TYR93, VAL111, VAL76 | |||
| Sucrose (3) | THR75, TRP60, TYR56, TYR64, TYR93 | ALA105, ALA127, ALA70, ILE52, ILE92, LEU110, LEU36, PHE101, PRO74, TRP60, TRP88, TYR47, TYR56, TYR64, TYR93, VAL76 | |||
| Hydroxy-propenyl-methoxyphenol (4) | PHE101, TRP88 | ASP73, TYR93, VAL111 | ALA105, ILE92, LEU110, LEU36, PHE101, PHE102, TRP60, TRP88, TYR112, TYR56, TYR64, TYR93, VAL111, VAL76, | ||
| Dopamine, N, N-dimethyl-, dimethyl ether (5) | TRP88 | TYR56 | ALA105, ALA127, LEU110, LEU36, PHE101, PHE102, TRP60, TRP88, TYR56, TYR64, TYR93, VAL111, VAL76 | ||
| 4-O-Methylmannose (6) | SER126, TYR93 | ALA105, LEU36, LEU110, PHE101, TRP60, TRP88, TYR56, TYR64, TYR93 |
In Figure 4 and Table 3, which depict the interactions of the top ligands with target protein 3JPU, the compound 1 formed pi–pi interactions with TYR64 and TYR56; Compound 2 formed a hydrogen bond with LEU110; Compound 3 formed five hydrogen bonds with TYR56, TYR64, TYR93, TRP60, and THR75; Compound 4 formed three hydrogen bonds with TYR93, VAL111, and ASP73, as well as two pi–pi interactions with TRP88 and PHE101; Compound 5 showed a pi–pi interaction with TYR56 and a pi–cation interaction with TRP88; and compound 6 formed two hydrogen bonds with TYR93 and SER126. The residue ALA105 was involved in all of the hydrophobic interactions.
Previous computational studies on target protein 3JPU39 have also reported similar hydrophobic interactions with residues LEU125, LEU110, LEU40, LEU36, PHE101, ILE52, and VAL76, as well as hydrogen bonding with TRP60, TYR56, and ASP73. Pi–pi interactions were observed with TYR64, PHE101, and TRP88. Additionally, hydrophobic interactions were found with residues ALA50, ALA70, ILE52, VAL76, LEU36, TRP60, and TYR56, while a hydrogen bond was formed with TYR64, as reported by Chakraborty et al.40
The ADME (absorption, distribution, metabolism, and excretion) profile of drug-like compounds plays a crucial role in drug discovery. In this study, the QikProp module of Maestro was utilized to predict the ADMET properties of the compounds found in the extract. Various physicochemical and pharmacokinetic characteristics were calculated and summarized in Table 4. These included human oral absorption, QPlogBB (molecule permeability to the brain), QPlogPo/w (lipophilicity profile prediction), QPlogS (aqueous solubility prediction), QPPCaco (molecule permeability to cell membranes), QPlogHERG (cardiac toxicity prediction), QPPMDCK (kidney cell permeability), and Lipinski’s rule of five, which is an important measure of drug-likeness. In Table 4, the compounds 5 and 4 exhibited high oral absorption, while compound 6 showed moderate oral absorption. On the other hand, compounds 1, 2 and 3 displayed low oral absorption. All of the compounds showed limited effects on the central nervous system, falling within the acceptable range (between −3 and +1.2) except for compound 1, which had a value of −6.079, indicating cardiac toxicity beyond the acceptable range (below −5). Furthermore, all compounds demonstrated acceptable ranges of both cell membrane permeability (QPPCaco) and Madin-Darby canine Kidney (MDCK) cell permeability, except for compound 3 which exhibited poor permeability in both categories (with values of 7.749 and 2.587, respectively). The compounds 4, 5, and 6 displayed favorable lipophilicity and aqueous solubility, whereas compound 3, exhibited good aqueous solubility but lacked sufficient lipophilicity. In contrast, compounds 1 and 2 showed poor lipophilicity and aqueous solubility profiles. Importantly, all compounds adhered to Lipinski’s rule of five, with values below the maximum threshold of 4, indicating good drug-likeness. The compounds 4, 5 and 6 (with area percentages of 1.41%, 43.76%, and 23.72%, respectively) possessed the most favorable predicted ADMET properties and drug-likeness characteristics among the tested compounds.
Table 4.
ADMET Properties of the Top-Scoring Compounds with 3GR6 and 3JPU
| Compound Character |
Diisooctyl Phthalate | Stigmasterol | Sucrose | Hydroxy-propenyl-methoxy-phenol | Dopamine, N, N dimethyl-dimethyl Ether | 4-O-Methylmanose |
|---|---|---|---|---|---|---|
| Designated number | 1 | 2 | 3 | 4 | 5 | 6 |
| Human Oral Absorption | 1 | 1 | 1 | 3 | 3 | 2 |
| QPlogBB (Brain permeability) | −1.094 | −0.288 | −2.986 | −0.695 | 0.608 | −1.214 |
| QPlogHERG (Cardiac toxixity) | −6.079 | −4.635 | −3.334 | −4.097 | −4.726 | −2.782 |
| QPlogPo/w (Lipophilicity) | 6.626 | 7.418 | −3.692 | 1.345 | 2.358 | −1.711 |
| QPlogS (Aqueous solubility) | −7.308 | −8.499 | −0.052 | −1.391 | −1.422 | −0.862 |
| QPPCaco (Cell membrane permeability) | 2075.250 | 3373.767 | 7.749 | 871.614 | 2164.817 | 190.957 |
| QPPMDCK (MDCK cell permeability) | 1089.095 | 1841.535 | 2.587 | 426.430 | 1261.190 | 82.628 |
| RuleOfFive | 1 | 1 | 2 | 0 | 0 | 0 |
Notes: Human oral absorption (1, 2, or 3 for low, medium, or high). QPlogBB: Predicted blood-brain barrier permeability (acceptable range −3–1.2). QPlogHERG Predicted IC50 value for blockage of Human ether-related gene (HERG) K+ channels which is a molecular target responsible for the cardiac toxicity (concern below −5.0). QPlogPo/w: Predicted octanol/water partition coefficient log P (acceptable range - 2.0–6.5). QPlogS: Predicted aqueous solubility in mol/L (acceptable range - 6.5–0.5). QPPCaco: Predicted caco cell membrane permeability in nm/s (acceptable range: <25 is poor and >500 is great). QPPMDCK: Predicted apparent Madin-Darby canine Kidney (MDCK) cell permeability in nm/s (acceptable range in nm/s (acceptable range: <25 is poor and >500 is great). RuleOfFive: Number of violations of Lipinski’s rule of five, which is considered a crucial measure of drug-likeness (Maximum is 4).
Notably, compound 5 (Area%=43.76%) emerged as the most prominent compound in the extract, exhibiting excellent predicted ADMET properties. Additionally, it displayed high docking scores and binding energies when interacting with the 3JPU target, indicating its enhanced activity against P. aeruginosa.
Compounds 6 and 4 interacted with both 3GR6 and 3JPU targets, demonstrating good docking scores and binding energies. These compounds exhibited antibacterial activity against both strains, with compound 6 accounting for 23.72% of the extract, making it the most promising candidate among the tested compounds for antibacterial purposes.
Conclusion
This study successfully validated the antibacterial properties of the hydro-ethanol extract of Acacia polyacantha stem bark and identified 11 bioactive components through the first GC-MS analysis of this extract, with the most abundant being dopamine, N, N-dimethyl-, dimethyl ether, 4-O-methylmannose, and sucrose. In vitro antibacterial tests demonstrated significant antibacterial activity against both Staphylococcus aureus and Pseudomonas aeruginosa, even at relatively low concentrations. In silico molecular docking identified strong-binding affinities for several compounds (1–6) to key bacterial enzymes, including enoyl-acyl carrier protein reductase, which is essential for fatty acid synthesis in S. aureus and the LasR-TP4 complex, responsible for virulence factors in P. aeruginosa. ADMET predictions indicated that compounds 4, 5, and 6 possess favorable drug-likeness properties, highlighting their potential as candidates for further preclinical investigation in drug development. Overall, these findings suggest that the hydro-ethanol extract of A. polyacantha could be a promising natural source of antibacterial agents. Further studies are necessary to explore the full phytochemical profile and clinical potential of this plant.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Vashist H, Jindal A. Antimicrobial activities of medicinal plants – review. Int J Res Pharm Biomed Sci. 2012;3(1):222–230. [Google Scholar]
- 2.Ahmad Khan MS, Ahmad I. Herbal medicine: current trends and future prospects. In: New Look to Phytomedicine: Advancements in Herbal Products as Novel Drug Leads. Elsevier Inc; 2018:3–13. doi: 10.1016/B978-0-12-814619-4.00001-X [DOI] [Google Scholar]
- 3.Daffalla HM, Ali KS, Tajelser T, Hagr TE, Ahmed NS, Ahmed RH Larvicidal and antibacterial activities of methanol extract of Acacia polyacantha willd. J Adv Res Pharm Sci Pharmacol Interv. 2018;2(2):7–11. [Google Scholar]
- 4.Mickymaray S. Efficacy and mechanism of traditional medicinal plants and bioactive compounds against clinically important pathogens. Antibiotics. 2019;8(4):257. doi: 10.3390/antibiotics8040257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khalid H, Abdalla WE, Abdelgadir H, Opatz T, Efferth T. Gems from traditional north-African medicine: medicinal and aromatic plants from Sudan. Natural Products and Bioprospecting. 2012;2(3):92–103. doi: 10.1007/s13659-012-0015-2 [DOI] [Google Scholar]
- 6.Saha Tchinda JB, Mbitnkeu Fetngna Tchebe T, Tchoukoua A, et al. Fatty acid profiles, antioxidant, and phenolic contents of oils extracted from Acacia polyacantha and Azadirachta indica (Neem) seeds using green solvents. J Food Process Preserv. 2021;45(2):0–3. doi: 10.1111/jfpp.15115 [DOI] [Google Scholar]
- 7.Awadasseid A, Adam IM, Jamal M, Khameis F, Teia F. Physicochemical properties of kakamut gum (Acacia polyacantha) and physicochemical properties of kakamut gum (Acacia polyacantha) and hashab gum (Acacia senegal): a comparative analysis. International Journal of Applied Agricultural Sciences. 2019;5(6):138. doi: 10.11648/j.ijaas.20190506.12 [DOI] [Google Scholar]
- 8.Deshmukh S, Shrivastava B, Bhajipale N. A Review on acacia species of therapeutics importance. Int J Pharm Biol Sci Arch. 2018;6(4):24–34. [Google Scholar]
- 9.Pascal AD, Boniface Y, Koudoro AY, et al. Chemical characterization and biological activities of extracts from two plants (Cissus quadrangularis and Acacia polyacantha) used in veterinary medicine in Benin. J Pharmacogn Phytochem. 2015;3:91–96. [Google Scholar]
- 10.Mambe FT, Na-Iya J, Fotso GW, et al. Antibacterial and Antibiotic Modifying Potential of Crude Extracts, Fractions, and Compounds from Acacia polyacantha Willd. against MDR Gram-Negative Bacteria. Evidence-Based Complement Altern Med. 2019;2019:1–13. doi: 10.1155/2019/7507549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manya MH, Keymeulen F, Ngezahayo J, et al. Antimalarial herbal remedies of Bukavu and Uvira areas in DR Congo: an ethnobotanical survey. J Ethnopharmacol. 2020;249:112422. doi: 10.1016/j.jep.2019.112422 [DOI] [PubMed] [Google Scholar]
- 12.Shaza A, Ak M. Phytochemical screening of Sudanese Leptadenia pyrotechnica. Acacia Polyacantha Ant Activ Acacia Polyacantha. 2019;4(5):8–10. [Google Scholar]
- 13.Abdel Karim M, Hamid AM, Hagr TE. Gas chromatography / mass spectroscopy analysis, antibacterial activity of fixed oil from Acacia polyacantha (Sudanese Kakamout) Seeds. Sudan Online Res J. 2021;(2):21–27. doi: 10.51527/v2i1.4me [DOI] [Google Scholar]
- 14.“Efficacy of crude bark extract of acacia polyacantha against Leishmania Donovani in mice”. Available from: http://erepository.uoeld.ac.ke/handle/123456789/1345. Accessed March 15, 2022.
- 15.Okpanchi A, Goji AD, Ezekiel I, Tanko Y, Mohammed A, Adelaiye AB Effect of aqueous-methanolic stem bark extract of Acacia polyacantha on blood glucose levels on normoglycemic Wistar rats. Int J Anim Vet Adv. 2012;4(13):163–166. [Google Scholar]
- 16.Ashu FA, Na-Iya J, Wamba BEN, et al. Antistaphylococcal activity of extracts, fractions, and compounds of Acacia polyacantha wild (Fabaceae). Evidence-Based Complement Altern Med. 2020;2020(1). doi: 10.1155/2020/2654247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abdallah MS, Mustafa M, Nallappan MA, Choi S, Paik JH, Rusea G. Determination of phenolics and flavonoids of some useful medicinal plants and bioassay-guided fractionation substances of sclerocarya birrea (a. rich) hochst stem (bark) extract and their efficacy against Salmonella typhi. Front Chem. 2021;9:1. doi: 10.3389/fchem.2021.670530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alajmi MF, Alam P, Alqasoumi SI, et al. Comparative anticancer and antimicrobial activity of aerial parts of Acacia salicina, Acacia laeta, Acacia hamulosa and Acacia tortilis grown in Saudi Arabia. Saudi Pharm J. 2017;25(8):1248–1252. doi: 10.1016/j.jsps.2017.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altayb HN, Yassin NF, Hosawi S, Kazmi I. In-vitro and in-silico antibacterial activity of Azadirachta indica (Neem), methanolic extract, and identification of Beta.d-Mannofuranoside as a promising antibacterial agent. BMC Plant Biol. 2022;22(1):1–14. doi: 10.1186/s12870-022-03650-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohamed LM, Eltigani MM, Abdallah MH, et al. Discovery of novel natural products as dual MNK/PIM inhibitors for acute myeloid leukemia treatment: pharmacophore modeling, molecular docking, and molecular dynamics studies. Front Chem. 2022;10(July). doi: 10.3389/fchem.2022.975191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacotet-Navarro M, Laguerre M, Fabiano‐Tixier A-S, et al. What is the best ethanol-water ratio for the extraction of antioxidants from rosemary? Impact of the solvent on yield, composition, and activity of the extracts. Electrophoresis. 2018;39(15):1946–1956. doi: 10.1002/elps.201700397 [DOI] [PubMed] [Google Scholar]
- 22.Huebschmann H-J. Interview for Handbook of GC-MS. GIT Lab J. 2017;01–02:33474. [Google Scholar]
- 23.EL-Kamali MA, H. H, Rabih HA, Kordofani. Chemical compositions of ethanolic extract and their fractions of Acacia ehrenbergiana aerial parts from Sudan. J Pharmacogn Phytochem. 2019;8(5):902–904. [Google Scholar]
- 24.Fotso GW, Na-Iya J, Mbaveng T. A, et al. Polyacanthoside A, a new oleanane-type triterpenoid saponin with cytotoxic effects from the leaves of Acacia polyacantha (Fabaceae). Nat Prod Res. 2019;33(24):3521–3526. doi: 10.1080/14786419.2018.1486312 [DOI] [PubMed] [Google Scholar]
- 25.Na MW, Lee E, Kang DM, et al. Identification of antibacterial sterols from Korean wild mushroom daedaleopsis confragosa via bioactivity-and LC-MS/MS profile-guided fractionation. Molecules. 2022;27(6):1–11. doi: 10.3390/molecules27061865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vollaro A, Esposito A, Antonaki E, et al. Steroid derivatives as potential antimicrobial agents against staphylococcus aureus planktonic cells. Microorganisms. 2020;8(4):1–14. doi: 10.3390/microorganisms8040468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alawode TT, Lajide L, Olaleye M, Owolabi B. Stigmasterol and β-sitosterol: antimicrobial compounds in the leaves of Icacina trichantha identified by GC–MS. Beni-Suef Univ J Basic Appl Sci. 2021;10(1). doi: 10.1186/s43088-021-00170-3 [DOI] [Google Scholar]
- 28.Compean KL, Ynalvez RA. 2014 _ Antimicrobial activity of plant secondary metabolites.pdf. Research Journal of Medicinal Plant. 2014;8(5):204–213. [Google Scholar]
- 29.Amoussa AMO, Lagnika L, Bourjot M, Vonthron-Senecheau C, Sanni A. Triterpenoids from Acacia ataxacantha DC: antimicrobial and antioxidant activities. BMC Complement Altern Med. 2016;16(1):1–8. doi: 10.1186/s12906-016-1266-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou JW, Luo HZ, Jiang H, Jian TK, Chen ZQ, Jia AQ. Hordenine: a novel quorum sensing inhibitor and antibiofilm agent against Pseudomonas aeruginosa. J Agric Food Chem. 2018;66(7):1620–1628. doi: 10.1021/acs.jafc.7b05035 [DOI] [PubMed] [Google Scholar]
- 31.Mostafa HS. Banana plant as a source of valuable antimicrobial compounds and its current applications in the food sector. J Food Sci. 2021;86(9):3778–3797. doi: 10.1111/1750-3841.15854 [DOI] [PubMed] [Google Scholar]
- 32.Salem O, E.m H, Ghazi SM, Hanna SN. Antimicrobial activity of microalgal extracts with special emphasize on Nostoc sp. Life Sci J. 2014;11(12):752–758. [Google Scholar]
- 33.Evans SM, Cowan MM. Plant products as antimicrobial agents. Cosmetic and Drug Microbiology. 2016;205–231. doi: 10.3109/9781420019919-17 [DOI] [Google Scholar]
- 34.Al-Huqail AA, Behiry SI, Salem MZM, Ali HM, Siddiqui MH, Salem AZM. Antifungal, antibacterial, and antioxidant activities of Acacia saligna (Labill.) H. L. Wendl. Flower extract: HPLC analysis of phenolic and flavonoid compounds. Molecules. 2019;24(4):700. doi: 10.3390/molecules24040700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Płowuszyńska A, Gliszczyńska A. Recent developments in therapeutic and nutraceutical applications of p-methoxycinnamic acid from plant origin. Molecules. 2021;26(13):3827. doi: 10.3390/molecules26133827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kabir A. Antibacterial and antifungal evaluation of some derivatives of methyl α -D-mannopyranoside. Int J Agric Biol. 2005;7:754–756. [Google Scholar]
- 37.Priyadarshi A, Kim EE, Hwang KY. Structural insights into Staphylococcus aureus. Proteins. 2009:480–486. doi: 10.1002/prot.22581 [DOI] [PubMed] [Google Scholar]
- 38.Yang M, Meng F, Gu W, et al. Review article effects of natural products on bacterial communication and network-quorum sensing. BioMed Res Int. 2020;2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zou Y, Nair SK. Molecular basis for the recognition of structurally distinct autoinducer mimics by the Pseudomonas aeruginosa LasR Quorum-sensing signaling receptor. Chem. Biol. 2009;16(9):961–970. doi: 10.1016/j.chembiol.2009.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chakraborty P, Daware AV, Kumari M, et al. Free tryptophan residues inhibit quorum sensing of Pseudomonas aeruginosa: a potential approach to inhibit the development of microbial biofilm. Arch. Microbiol. 2018;200(10):1419–1425. doi: 10.1007/s00203-018-1557-4 [DOI] [PubMed] [Google Scholar]