Abstract
Purpose
Microglia-like cells derived from stem cells (iMG) provide a plentiful cell source for studying the functions of microglia in both normal and pathological conditions. Our goal is to establish a simplified and effective method for generating iMG in a precisely defined system. Additionally, we aim to achieve functional maturation of iMG through coculture with retinal organoids.
Methods
In this study, iMG were produced under precisely defined conditions. They were subjected to LPS and poly IC stimulation. Additionally, we examined distinct phenotypic and functional variances between iMG and HMC3, a commonly used human microglia cell line. To investigate how the retinal cell interaction enhances microglial properties, iMG were cocultured with retinal organoids, producing CC-iMG. We performed RNA sequencing, electrophysiological analysis, and transmission electron microscope (TEM) to examine the maturation of CC-iMG compared to iMG.
Results
Our results demonstrated that iMG performed immune-responsive profiles closely resembling those of primary human microglia. Compared to HMC3, iMG expressed a higher level of typical microglial markers and exhibited enhanced phagocytic activity. The transcriptomic analysis uncovered notable alterations in the ion channel profile of CC-iMG compared to iMG. Electrophysiological examination demonstrated a heightened intensity of inward- and outward-rectifying K+ currents in CC-iMG. Furthermore, CC-iMG displayed elevated numbers of lysosomes and mitochondria, coupled with increased phagocytic activity.
Conclusions
These findings contribute to advancing our understanding of human microglial biology, specifically in characterizing and elucidating the functions of CC-iMG, thereby offering an in vitro microglial model for future scientific research and potential clinical applications in cell therapy.
Keywords: hiPSC-derived microglia, electrophysiology, K+ channel, proinflammatory response, lysosome and mitochondrion
Microglia, the resident immune cells of the central nervous system (CNS), play crucial roles in neuronal development, immune defense, and maintaining homeostasis, as well as contributing to neurological disorders.1 They have been prominently implicated in the initiation and progression of neurodegenerative diseases. Recent advancements in molecular biology have unveiled alterations in microglial functions and their roles in aging and various neural disorders.2 Microglia express numerous genes identified in human genome-wide association studies related to neurological disorders, such as Alzheimer's disease (AD)3 and age-related macular degeneration (AMD).4 They also play a pivotal role in the pathogenesis of various retinal diseases, including glaucoma, uveitis, diabetic retinopathy, and optic nerve damage.5–9 Consequently, the impact of microglia on neural function is evident, because both ablation and overactivation of microglia can initiate neural dysfunction.10,11 Efficient replacement of retinal microglia holds promise as a therapeutic approach for AMD,5–9 positioning them as emerging therapeutic targets for retinal disorders.
Microglial activation and function have been extensively studied in rodent experimental models; however, these models often fail to mirror human microglial properties accurately.12 As the primary immune cells in the CNS, microglia continuously survey the local microenvironmental changes, and microglial activation can be rapidly evoked by various noxious stimuli. This underscores the need for robust cell-culture models to understand their functioning and role in disease progression. HMC3, the most acceptable immortalized human microglial cell line, retains most properties of primary microglia but still has certain shortcomings.13 However, isolating primary human microglia is challenging, and the microglial transcriptional profile is quite sensitive to enzymatic digestion procedures.14 Additionally, the limited availability of postmortem human tissue hampers pathological research related to microglia. Thus there is an urgent demand for a progressive source of human microglia for both basic and clinical research.
Recently, several protocols have been established to differentiate human pluripotent stem cells (hPSC) into microglia-like cells (iMG) through yolk-sac–derived hematopoietic progenitor cells15–18 or forced expression of transcription factors crucial for microglia development.19,20 These approaches enable the production of substantial quantities of human-derived cells exhibiting a typical fetal microglial signature. Consequently, it allows for the generation of patient-specific microglia carrying gene mutation and enables the modification of microglial phenotype using CRISPR gene editing.21,22 Considering environmental discrepancies, scientists use experimental models with different microglial features, such as in vitro neuron-or astrocyte- and iMG coculture, three-dimensional (3D) organoids coculture, and in vivo chimeric mouse brain and retina.23–27 Notably, these studies have suggested that the biology of iMG is highly dependent on their environment. The extent to which iMG can mimic primary microglia biology in immune response and the difference between microglial cell line and iMG are still unclear. Additionally, it is critical to investigate the influence of neuronal interaction on microglial biology.
In the present study, we used a chemically defined system to generate iMG and evaluate their immune-responsive function by analyzing transcriptional and functional profiling after LPS and poly IC stimulation. A comparative study between iMG and HMC3 was conducted, demonstrating that iMG exhibited higher expression of microglial markers and phagocytic activity than HMC3. To further enhance their properties, iMG were cocultured with stem cell–derived retinal organoids, yielding CC-iMG under an in vivo-like microenvironment. RNA-sequencing results indicated potential changes in the ion channel properties and organelles of CC-iMG. Consequently, we performed electrophysiological analysis and transition transmission electron microscope (TEM), revealing enhanced K+ channel expression and organelle maturation in CC-iMG compared to iMG. To our knowledge, this is the first study to compare iMG and a microglial cell line. Additionally, our findings indicate that coculturing with retinal organoids can significantly enhance the function and organelle maturation of iMG, thereby bolstering its credibility as an in vitro model and a dependable cell source for cell transplantation replacement.
Material and Methods
Maintenance of Human IPSC
The human iPSC cell lines with or without nuclear green fluorescent reporter used in this study were purchased from Nuwacell (RC01010, RC01001-A; Hefei, China). Briefly, iPSCs were maintained in chemical-defined Nuwacell ncEpic hPSC medium (RP01001; Nuwacell) on Vitronectin-coated (RP01002; Nuwacell) plates at 37°C and 5% CO2 in a humidified incubator. Cells were passaged with 0.5 µM EDTA (AM9260G, Invitrogen) at a ratio of 1:10 and the medium was changed daily. Cells were regularly tested for mycoplasma contamination every three months.
Human Microglia Cell Culture
Human microglial cell line HMC3 (ATCC) was maintained in RPMI1640 medium supplemented with 10% fetal bovine serum (FBS). Cells were detached with 0.25% Trypsin-EDTA (25200-056, Gibco) and passaged with a ratio of 1:6.
Generation of HiPSC-Derived iMG
The protocol for generating iMG from GFP-expressing human-induced pluripotent stem cells (HiPSCs) involved several key steps with minor modifications from previous methods.16 Briefly, HiPSCs colonies were detached with 0.5 mM EDTA in calcium/magnesium-free PBS containing 0.9% (V/V) sodium chloride, and then transferred into a six-well suspension culture plate (657185; Greiner Bio-One, Monroe, NC, USA) in embryonic body formation medium (DMEM/F12 medium supplemented with 20% [V/V] CTS KnockOut SR XenoFree [12618012; ThermoFisher, St. Louis, MO, USA], 1% [V/V] penicillin/streptomycin, and 10 µM Y-27632 [HY-10071; MedChemExpress, Monmouth Junction, NJ, USA]). Embryonic bodies were formed automatically after incubation overnight in a humidified incubator containing 5% CO2 at 37°C. Fresh medium with the removal of Y-27632 was changed. Until the fourth day, the embryonic bodies were transferred into a gelatin-coated (G7041; Sigma-Aldrich Corp., St. Louis, MO, USA) cell culture dish with myeloid differentiation medium (X-vivo 15 plus medium [04-418Q; Lonza Group, Basel, Switzerland] supplied with 25ng/mL IL-3 [CX90; Novoprotein, Beijing, China] and 25 ng/mL M-CSF [C417; Novoprotein]). The embryonic bodies gradually attached after about seven to 10 days, and the myeloid differentiation medium was changed every week until hematopoietic progenitor cells (HPs) appeared in the suspension. HPs were collected and subjected to microglial induction medium (DMEM/F12 medium supplemented with 0.5% chemically defined N-2 supplement [17502048; ThermoFisher], 1% B-27 supplement XenoFree [A5047501, ThermoFisher], 100 ng/mL IL-34 [200-34; PeproTech, Cranbury, NJ, USA) and 50 ng/mL M-CSF. HPs were plated in six-well plates at a density of 2.5 × 104 cells/cm2. Generated iMG showed ramified morphology after seven to 14 days of induction.
HiPSC-Derived Retinal Organoids
For retinal organoid (RO) generation, HiPSCs of 80% to 90% confluency were first digested with Dispase (07923; Stemcell Technologies, Vancouver, BC, Canada), and then separated into small pieces with a syringe needle followed by detachment with a cell scraper. Cell aggregates were collected and wrapped with Matrigel (354234; Corning Inc., Corning, NY, USA) within N2B27 medium. After five days of suspension culture, cell aggregates transformed into attachment culture for six more days. Then the colonies were elevated with dispase and transferred into B27 medium, containing DMEM/F-12 medium supplemented with 2% B-27 supplement. One week after culture in B27 medium, the medium was changed to serum medium, containing DMEM/F-12 medium supplemented with 2% B-27 supplement, 8% fetal bovine serum (10270106; Thermo Fisher Scientific, Waltham, MA, USA), 0.5 µM retinal acid (R2625; Sigma-Aldrich Corp., St. Louis, MO, USA) and 100 mM Taurine (T8691; Sigma-Aldrich Corp.). Retinal organoids were cultured in the serum medium and the medium was changed weekly. Neural epithelial layer appears around differentiation day 25–30, and retinal cell types can be identified around day 40.
Coculture of IMG and ROs
Fourteen days after iMG induction, attached ramified iMG were transferred into serum medium supplemented with 100 ng/mL IL-34 and 50 ng/mL M-CSF. ROs at differentiation days of 40–80 were added into the plates with attached iMG at a density of 2857 iMG/organoids. After five days of coculture, the attached iMG was used for subsequent analysis. Occasionally, ROs might attach to the surface of the plate, which could be lifted with a gentle shake or blow with a pipette. Undirect contact coculture was based on the transwell culture system using Millicell cell culture inserts (pore size 0.4 µm) (PIHP01250; Merck Millipore, Burlington, MA, USA).
Electrophysiological Recordings
Passive membrane properties and K current of iMG on glass slices were measured using the patch-clamp technique in whole-cell configuration following a previously published method.28 For pulse application, a patch-clamp amplifier 700B (Molecular Devices, San Jose, CA, USA) was connected to a computer, and data were recorded using the program 1550B (Molecular Devices) analog-to-digital converter. Borosilicate glass (BF150-86-10; Sutter Instrument Company, Novato, CA, USA) was used for patch electrode fabrication on a two-stage puller P97 (Sutter Instrument Company). The K+ current blockade was achieved using 20 mM TEA-Cl in a nonspecific manner.
Immunofluorescence
Cells or ROs were fixed with 4% paraformaldehyde (P0099; Beyotime Institute of Biotechnology, Jiangsu, China) in PBS buffer. Cryo-sections of ROs (25 µm in thickness) were prepared with a cryostat (CM1950; Leica, Wetzlar, Germany). The cells or sections were blocked with AB solution containing 5% donkey serum and 0.1 or 0.3% Triton X-100 in PBS for cells or sections for one hour at room temperature (RT). Primary antibodies, including anti-Iba1 (1:400, 019-19741; Wako Chemicals USA, Richmond, VA, USA), anti-hIBA1 (1:400, MAB7308-sp; R&D Systems, Minneapolis, MN, USA), anti-hP2RY12 (1:500, HPA014518; Sigma-Aldrich Corp.), anti-Calretinin (1:1000, CR7697; Swant, Bellinzona, Switzerland), anti-Cone-arrestin (1:500, AB15282; Merck Millipore), anti-Ku80 (1:1000; Cell Signaling Technology, Danvers, MA, USA) and anti-RHO (1:10000, MABN15; Merck Millipore), were diluted with indicated ratio in AB solution and incubated overnight at 4°C. Then the samples were incubated with secondary antibodies, such as AlexaFluor donkey anti-rabbit IgG 488 (1:1000, 711-585-152; Jackson Laboratories, Bar Harbor, ME, USA), Donkey anti-rabbit- IgG Cy5 (711175152, Jackson lab), and AlexaFluor donkey anti-mouse IgG 594 (715-585-151; Jackson Laboratories), for two hours at RT. The samples were observed under a confocal microscope (LSM900; Carl Zeiss, Inc., White Plains, NY, USA).
Lysosome Labeling and Quantification
For lysosome staining, Lysotracker Deep Red (L12492; ThermoFisher) was used to study lysosomal formation according to the manufacturer's instructions. Lysotracker labeled cells were mounted with ProLong Diamond Antifade Mountant (P36971; Invitrogen, Carlsbad, CA, USA). The signal of lysosome in each cell of iMG and CC-iMG were compared to assess the effect of ROs coculture.
To quantify functional lysosomes, cells were stained with Lysotracker Deep Red and then treated with 50 µM glycyl-L-phenylalanine-b-naphthylamide (GPN) for 15 minutes, or DMSO as control. GPN can be degraded by lysosomal protease cathepsin C resulting lysis of lysosomes. Thus the degree of fluorescence reduction of Lysotracker Deep Red signal before and after GPN treatment is proportional to the amount of lysosome. Note that the degree of fluorescence reduction of Lysotracker Deep Red signal before and after DMSO treatment was used as background.
Mitochondrial Staining and Quantification
The mitochondria of iMG were labeled with MitoTracker Red FM (M22425; ThermoFisher) following the manufacturer's instructions. To evaluate the influence of ROs coculture, cells were observed under confocal microscopy (LSM900; Carl Zeiss, Inc.) using iMG in serum medium without ROs as control. To quantify the mitochondrial signal, eight to 10 images were acquired.
Measurement of Cell Viability
For cell viability measurement, iMG were plated in 96-well plates for seven days and then transferred the RO into each well for five more days in serum medium. The cells in serum medium without ROs were set as control. The ATP-based method, CellTiter-Glo Luminescent Cell Viability Assay (G7571; Promega Corporation, Madison, WI, USA) was used to detect the cell viability of iMG after coculture with ROs. Relative cell viability was normalized to iMG control.
For poly IC treatment, iMG were cultured in 96-well plates for seven days, and then 1 µg/mL poly IC was added in the medium. After 24 hours’ stimulation, cell survival rates were measured using CCK-8 assay (C6005; New Cell & Molecular Biotech, Newcastle upon Tyne, UK).
RNA Isolation and Quantitative Real-Time PCR
Cells were lysed in 1 mL of TRIzol reagent (15596026CN; ThermoFisher), and total RNA was isolated according to the manufacturer's instructions. The purity and quantity of the isolated RNAs were measured with nanodrop NanoDrop, 1000 (ThermoFisher). For quantitative real-time PCR, 1 µg RNA was reverse transcribed into cDNA using HiScript III RT SuperMix for qPCR (+gDNA wiper) (R323-01; Vazyme Biotech Co., Ltd., Nanjing, China). Prepared cDNA was subjected to quantitative real-time PCR using Taq Pro Universal SYBR qPCR Master Mix (Q712-03; Vazyme Biotech Co., Ltd.) with Real-Time PCR Instrument (A28132; ThermoFisher). Primer sets for qPCR are listed in the Table.
Table.
Primer Sets for qCPR
| Gene name | Forward | Reverse |
|---|---|---|
| RPL13A | CAA GCG GAT GAA CAC CAA C | TGT GGG GCA GCA TAC CTC |
| CD68 | CCT CAG CTT TGG ATT CAT GC | GAG CCG AGA ATG TCC ACT GT |
| MPO | CGT CAA CTG CGA GAC CAG | GTC ATT GGG CGG GAT CTT |
| TNF-α | CAG CCT CTT CTC CTT CCT GAT | GCC AGA GGG CTG ATT AGA GA |
| TMEM119 | AGT CCT GTA CGC CAA GGA AC | AGG AGC AGC AAC AGA AGG AT |
| P2RY12 | TGA CAA AAA TCC AGG GTA GTG A | CGTCAG TAA AGTCTTGAGTGC TCT T |
| CX3CR1 | CCC TGG AAG GTG CTG TTA TC | TCC ATG AGA TTG GAC TGG AA |
| CYCT1 | TCC AGA ACT TCC AGT GTT GC | TGC TTC TGG GAA ATA AAT GC |
RNA Sequencing and Data Analysis
Bulk RNA sequencing was performed by BioMarker (Beijing, China) as described previously. RNA sequencing was performed on the Illumina NovaSeq 6000 platform (Illumina, San Diego, CA, USA) as PE150. A minimum of 6 G clean data yield was guaranteed for each sample. Part of the data analysis was completed by R software version 4.2.0. Differentially expressed genes were analyzed using the R package “DESeq2.” Gene oncology (GO) enrichment analysis was performed by the R package “clusterProfiler.” Data visualization was achieved with R packages “heatmap,” “ggplot2,” and “ggnewscale.”
Phagocytosis Assays
IMG were grown on glass slides and poly(methylmethacrylate) fluorescent nano-beads were added in the medium for one or two hours in a humidified incubator containing 5% CO2 at 37°C. Then cells were fixed with 4% paraformaldehyde for 10 minutes at RT and stained with anti-Iba1 as described previously. Images were captured using a Carl Zeiss, Inc. confocal microscopy (LSM900). To calculate the phagocytic activity, the intensity of embedded beads was measured using ZEN software within each cell. Cell outline was identified with IBA1 staining.
Preparation of TEM
TEM were performed following a previously published method.29 Briefly, iMG grown on a 12-well plate were fixed in fixative containing 2% paraformaldehyde (157-8; Electron Microscopy Sciences, Hatfield, PA, USA) and 2% glutaraldehyde (16020; Electron Microscopy Sciences) in 0.1 M PB, pH 7.4, incubated overnight at 4°C. After being thoroughly washed with PBS, cells were post-fixated in 1% osmic acid anhydride (20816-12-0; Electron Microscopy Sciences) for one hour at RT and stained with uranyl acetate solution. After dehydrating in a graded series of acetone, cells were embedded in freshly prepared Epon-812. Because the samples were monolayer cells, ultrathin sections of 80 nm were prepared carefully for TEM and then collected on formvar-coated grids. Images were acquired with a Hitachi-7500 electron microscope (Hitachi, Tokyo, Japan).
Graphics and Statistical Analysis
Schematic figures were generated with BioRender (https://biorender.com/). Quantitative analysis of confocal microscopy images was performed using the Histo model of ZEN software from Carl Zeiss, Inc. Data visualization and statistical analysis were performed using GraphPad Prism software version 9.0. Unpaired Student's t-test was performed for comparisons of two groups, and two-way ANOVA was used for multiple comparisons. For all analyses, a P value < 0.05 was considered statistically significant, and error bars represent standard deviation (SD) or standard error of the mean (SEM). The number of biological replicates (n) is indicated in each figure.
Results
Generation of Immune-Responsive IMG Under Well-Defined Conditions
To generate iMG under well-defined conditions, HiPSCs were cultured on vitronectin-coated plates using a chemically defined medium. Modified from our previously described method,16 we generated iMG with a differentiation medium containing DMEM/F12, N-2, B27 supplement, IL-3, M-CSF, and IL-34, excluding FBS previously used. Validation of iMG was performed by assessing the expression of microglial markers IBA1 and P2RY12 (Supplementary Figs. S1A, S1B). Additionally, we evaluated the phagocytic capability of iMG through fluorescent bead engraftment (Supplementary Fig. S1C). Co-localization analysis indicated that the fluorescent nano-beads were indeed incorporated into the iMG cytoplasm (Supplementary Fig. S1D). These results confirm the successful generation of iMG from HiPSCs under chemically defined conditions.
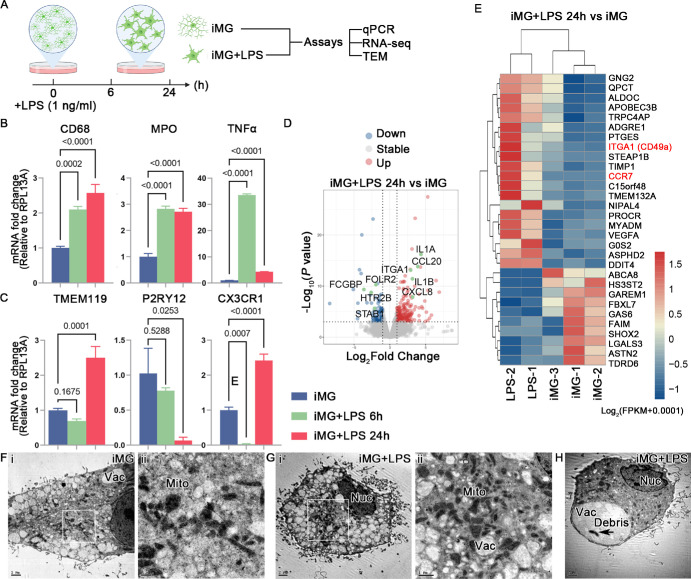
To investigate the responses of iMG to pro-inflammatory stimuli, we exposed them to bacterial endotoxin LPS or dsRNA poly IC (Figs. 1A, 2A). To simulate chronic inflammation, we opted for a relatively low concentration of LPS (1 ng/mL) and treated iMG for six and 24 hours (Fig. 1A). Initially, we assessed the expression of homeostatic and activated microglial markers, including TMEM119, P2RY12, CX3CR1, CD68, MPO, and TNFα by qPCR. The results revealed that the expression of activated microglia genes (CD68, MPO, and TNFα) significantly increased after six and 24 hours of LPS treatment, indicating that this low concentration of LPS induces iMG activation within six hours, sustaining for 24 hours (Fig. 1B). Notably, mRNA expression of TNFα peaked at a significantly higher level and declined after 24 hours of stimulation yet remained significantly upregulated compared with untreated iMG. Regarding homeostatic microglial markers, TMEM119, P2RY12, and CX3CR1 showed downregulation after six hours of LPS treatment, although the differences were not statistically significant for TMEM119 and P2RY12. Importantly, after 24 hours of treatment, the mRNA levels of TMEM119 and CX3CR1 were significantly higher in LPS-treated iMG compared with untreated iMG (Fig. 1C).
Figure 1.
LPS-triggered transcriptional response in iMG. (A) Schematic of LPS treatment and sample collection for subsequent analysis. Created with www.biorender.com. Quantitative PCR analysis for the expression of activated (B) and homeostasis (C) microglial markers. Data are represented as means ± SD; n = 3. (D) Volcano plot shows DEGs in iMG after low concentration of LPS treatment. (E) Heatmap visualization of the normalized gene expression levels for DEGs after LPS treatment for 24 hours. (F–H) Representative TEM images of iMG and LPS-treated iMG after 24 hours treatment. Nuc, nuclear, Vac, vacuole, Mito, mitochondria. Scale bar: 2 µm (F-i, G-i, H). Scale bar: 1 µm (G-ii). Scale bar: 0.5 µm (F-ii).
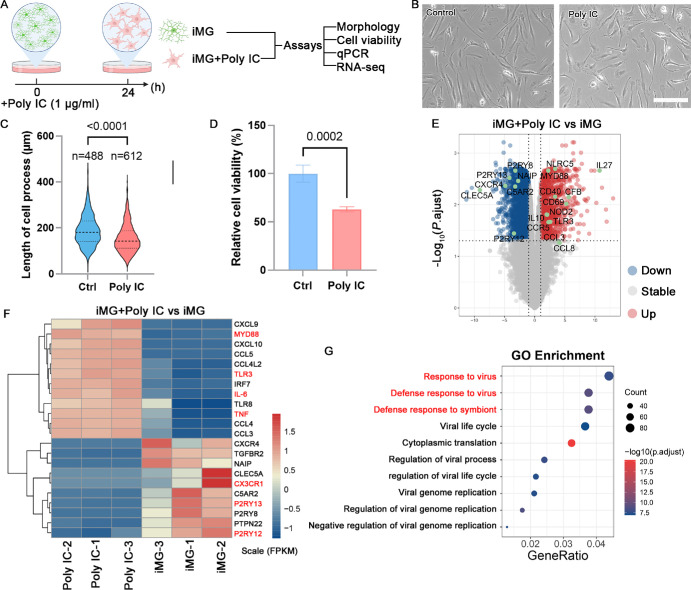
Figure 2.
Poly IC treatment evoked viral infectious-like transcriptional response in iMG. (A) Schematic of cell culture and sample collection for subsequent analysis. Created with www.biorender.com. (B) The phage contact photograph of iMG treated with or without poly IC. Scale bar: 200 µm. (C) Quantitative analysis of the length of cell process of iMG. Data are represented as means ± SD. (D) Cell viability were measured after poly IC treatment using CCK-8 kit. Data are represented as means ± SD; n = 4. (E) Volcano plot shows DEGs in comparison of poly IC treatment versus untreated iMG control. (F) Heatmap visualization of the normalized gene expression levels for DEGs after poly IC treatment for 24 hours. (G) The most significantly enriched Gene ontology of DEG.
To analyze transcriptomic changes after LPS treatment, bulk RNA-seq was performed on both untreated iMG and iMG exposed to LPS for 24 hours. LPS treatment induced differential expression in 102 genes compared to untreated iMG, with 71 genes upregulated and 31 genes downregulated. Notably, secreted cytokines CCL20, IL1A, IL1B, and CXCL8 (IL8) were induced by LPS in iMG (Fig. 1D). Additionally, the expression of ITGA1 (CD49a), a key player in cell adhesion, significantly increased after LPS exposure. Enhanced expression of CC-chemokine receptor 7 (CCR7), which may impact cell migration, was also observed in LPS-treated iMG (Fig. 1E). Furthermore, TEM analysis revealed the ultrastructural of iMG and LPS-treated iMG (Figs. 1F–H). LPS stimulation did not induce obvious alterations in mitochondria after 24 hours of treatment, with occasional observation of cell debris within the vacuoles (Fig. 1F).
To simulate viral infection in iMG, we investigated the effects of poly IC (a synthetic double-stranded RNA), a TLR3 agonist (Fig. 2A). Poly IC treatment induced noticeable changes in microglial morphology and reduced cell viability (Figs. 2B–D). Bulk RNA-seq analysis revealed that high concentrations of poly IC (1 µg/mL) treatment induced differential expression in 1923 genes, including 465 upregulated genes and 1458 downregulated genes (Fig. 2E). Among these, cytokines (CXCL10 and CCL5) and pro-inflammatory factors (IL-6 and TNF) were significantly upregulated, while homeostatic markers were markedly downregulated, such as P2RY12, P2RY13 and CX3CR1 (Fig. 3F). GO analysis indicated enrichment in response to virus and defense response to virus, suggesting that poly IC treatment triggered a robust proinflammatory response in human iPSC-derived iMG (Fig. 2G). These findings underscore iMG as a reliable cellular model, offering a platform for in vitro inflammatory studies of human microglia.
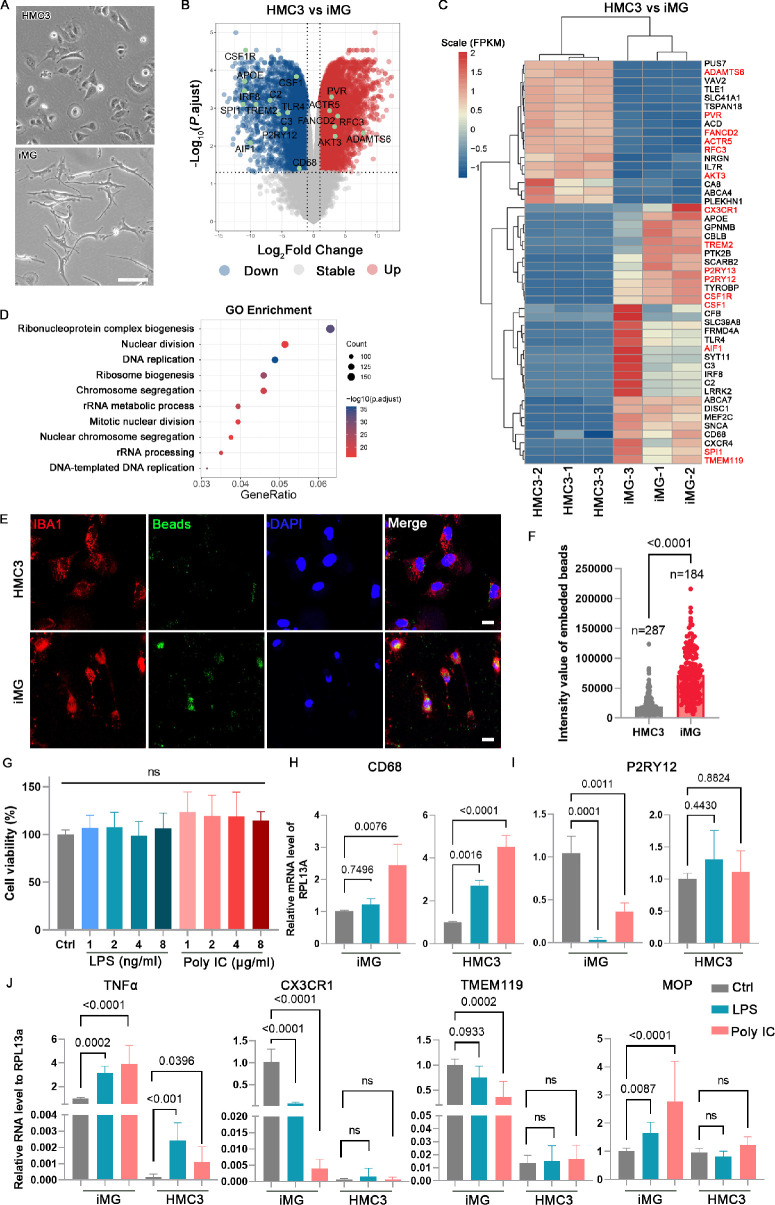
Figure 3.
Comparison of iMG and HMC3 in transcriptome and phagocytic function. (A) The phage contact photograph of HMC3 and iMG. Scale bar: 100 µm. (B) Volcano plot shows DEGs in comparison of HMC3 versus iMG. (C) Heatmap visualization of the normalized gene expression levels demonstrating that microglial marker genes are highly expressed in iMG and cell proliferation-related genes are highly expressed in HMC3. (D) Enrichment analysis showed the top ten Gene ontology associated with cell proliferation. (E) Phagocytosis of iMG and HMC3 were evaluated after treatment with poly(methylmethacrylate) fluorescent nano-beads for two hours. Scale bar: 20 µm. (F) Quantification of the intensity of fluorescent nano-beads in each cell. Data are represented as means ± SD. (G) Cell viability was measured after LPS and poly IC treatment using CCK-8 kit. Data are represented as means ± SD; n = 5. (H–J) Quantitative PCR analysis of the expression of activated and homeostasis microglial markers. Data are represented as means ± SD; n = 3.
Advantages of IMG Compared to HMC3
The iMG generated in this study exhibits superior bioactivities and immunological characteristics compared to HMC3, as supported by several key observations. Firstly, morphological differences between iMG and HMC3 were evident, with iMG displaying a ramified form indicative of maturation and resting states (Fig. 3A). Furthermore, RNA-seq analysis revealed distinct transcriptome profiles between iMG and HMC3 (Fig. 3B). Specifically, genes associated with cell proliferation, such as ADAMTS6, PVR, and FANCD2, were significantly upregulated in HMC3, whereas microglial homeostatic genes including P2RY12, CR3CR1, TREM2, TMEM119, and SPI1, showed notable downregulation in HMC3 compared to iMG. Moreover, the expression levels of genes involved in immune activation were significantly lower in HMC3 than iMG (Fig. 3C). GO analysis further highlighted enrichment in differentially expressed genes associated with DNA replication and mitotic nuclear division in HMC3 (Fig. 3D). Additionally, the phagocytic activity of HMC3 was significantly lower than that of iMG (Figs. 3E, 3F). Neither LPS nor poly IC treatment significantly affected HMC3 cell viability (Fig. 3G). Furthermore, qPCR results revealed that LPS and poly IC treatments had distinct effects on iMG and HMC3 cells. The expression of CD68 and TNF-α was significantly upregulated in both iMG and HMC3 cells. However, the expression of microglial homeostatic genes, including P2RY12, CX3CR1, TMEM119, and MOP, was markedly lower in HMC3 cells compared to iMG. These genes were significantly upregulated in iMG following LPS and poly IC treatment, while they remained unchanged in HMC3 cells (Figs. 3H–J). Taken together, these findings suggest iMG as a more reliable cellular model for investigating microglial biology.
Generation of ROs
Prior research has shown that coculturing with astrocytes or neural cells can enhance iMG maturation.15,26 To further explore the impact of the retinal context on iMG function, we differentiated HiPSCs into ROs (Supplementary Figs. S2A, S2B), which faithfully recapitulate retinal structure and function.30,31 The generated ROs were confirmed to contain calretinin-positive amarine cells at the early stage (D40), and cone-arrestin-positive cone and rhodopsin-positive rod photoreceptor cells at a later stage (D232; Supplementary Fig. S2B). These results confirm the suitability of ROs for establishing a retinal-microglia interaction model.
Retinal Organoid Microenvironment Induced Changes in the Ion Channel Profile of IMG
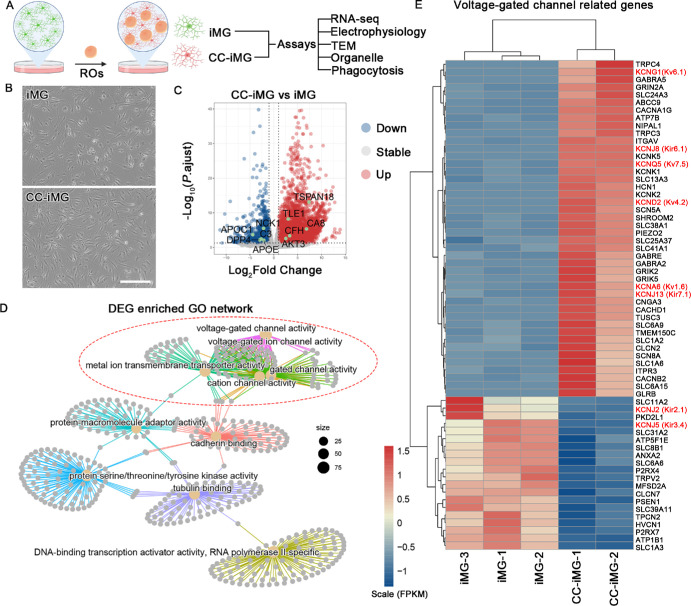
To investigate the impact of ROs on iMG function, we employed ROs at their mid-differentiation stage (D40-80) as neural organoids for coculture with iMG (Fig. 4A). After five days of coculture (CC-iMG), the morphology of CC-iMG exhibited no discernible differences compared to iMG cultured alone (Fig. 4B). However, bulk RNA-seq analysis unveiled significant alterations in the expression of 4865 genes, comprising 1367 down-regulated and 3498 up-regulated genes (Fig. 4C). Notably, this dataset encompasses transcription factors (TFs) pivotal for microglial development, including RUNX2, FOSB, and BHLHE41 (Supplementary Fig. S3). These TFs potentially underlie the transcriptional changes in CC-iMG. GO network analysis highlighted the enrichment of differentially expressed genes (DEG) in pathways associated with channel activity, such as voltage-gated channel activity and ion channel activity (Figs. 4D, 4E; Supplementary Fig. S4A). These transcriptomic shifts suggest that retinal cell interaction might influence ion channel activity in iMG.
Figure 4.
Transcriptomic changes of iMG after coculture with retinal organoids. (A) Experimental design. Retinal organoids at differentiation days 40-70 were switched to dishes containing iMG at differentiation day 7 and maintained for five days becoming CC-iMG. iMG cultured in the same medium without organoids were designated as control. IMG and CC-iMG were subjected to RNA-seq, electrophysiology, TEM, organelle, and phagocytosis analysis. Created with www.biorender.com. (B) The phage contact photograph of iMG and CC-iMG. Scale bar: 300 µm. (C) Volcano plot shows DEGs in iMG after coculture with retinal organoids. (D) GO term network analysis of DEG. (E) Heatmap visualization of the normalized gene expression levels for DEG related to voltage-gated channels.
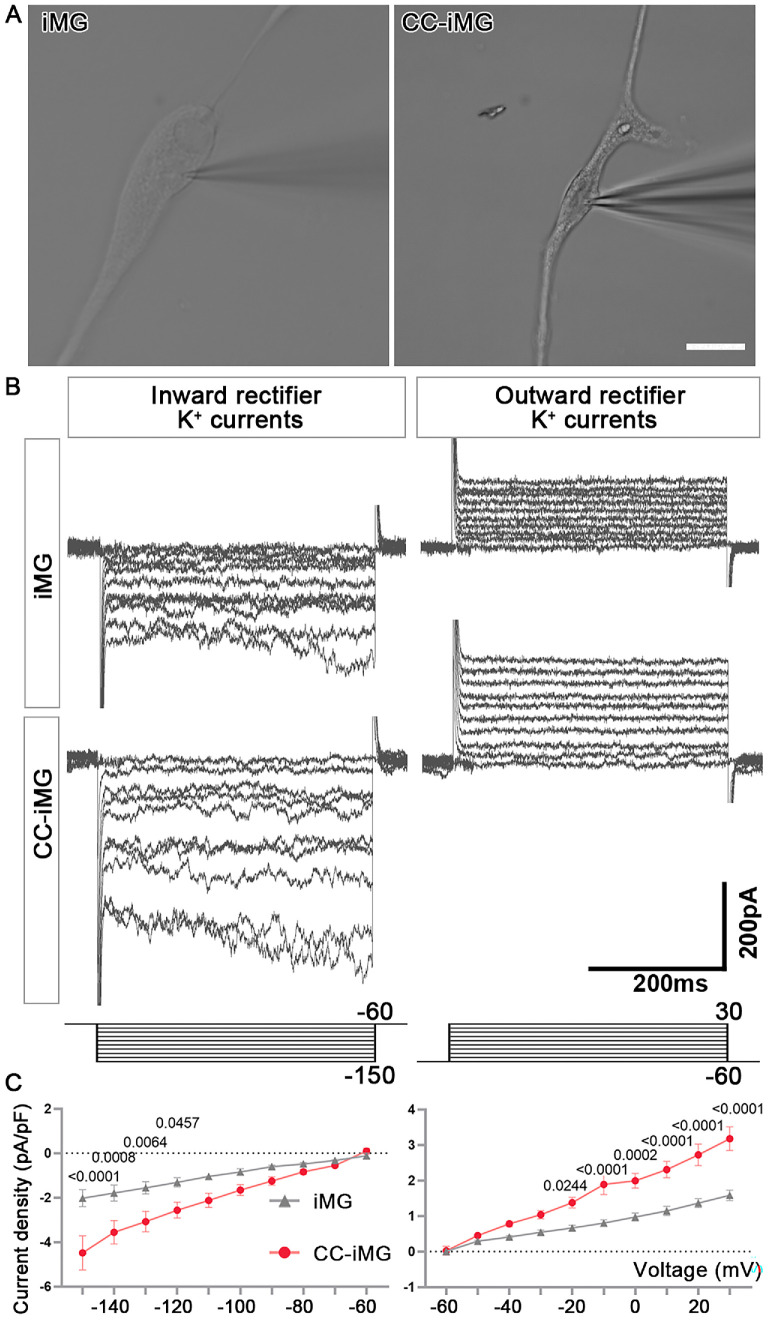
Retinal Organoid Microenvironment Enhances the Electrophysiological Function of IMG
To validate whether changes in the expression of voltage-gated channel-related genes influenced the electrophysiological properties of iMG following coculture with ROs, we utilized the patch clamp technique to assess rectifying K+ channels in both iMG and CC-iMG. Only relatively large cells with ramified processes were selected for the experiment (Fig. 5A). A total of ten cells in each group were recorded. Microglial cells exhibit both inward- and outward-rectifying K+ channels and their expression pattern is strongly influenced by the retinal cell interaction, making them a sensitive marker of microglial function.28 In this study, K+ channel currents were successfully recorded in iMG using the whole-cell configuration. Voltage pulses ranging from -150 and +30 mV, stepped by 10 mV at 10 s intervals between each stimulus, elicited inward-rectifying and outward-rectifying K+ currents from a holding potential of -60 mV (Figs. 5B, 5C; Supplementary Figs. S4B–D). Compared to iMG, the average membrane capacitance of CC-iMG was significantly lower (Supplementary Fig. S4E). However, no significant differences were found between iMG and CC-iMG in the average membrane resistance and the average resting membrane potential (Supplementary Figs. S4F, S4G). Importantly, CC-iMG exhibited a significantly larger mean inward K+ and outward K+ current density compared to iMG controls. These findings highlight the significant impact of the retinal organoid microenvironment on the electrophysiological function of iMG.
Figure 5.
Enhanced electrophysiological properties of iMG after coculture with retinal organoids. (A) Whole-cell voltage recordings obtained from iMG. Scale bar: 20 µm. (B) Example of an inward and outward rectifier K+ current elicited by voltage pulses of potentials between −150 and 30 mV. (C) The average current density of inward and outward currents normalized with respect to membrane capacitance. Data are represented as means ± SEM; n = 10 cells from iMG and CC-iMG each.
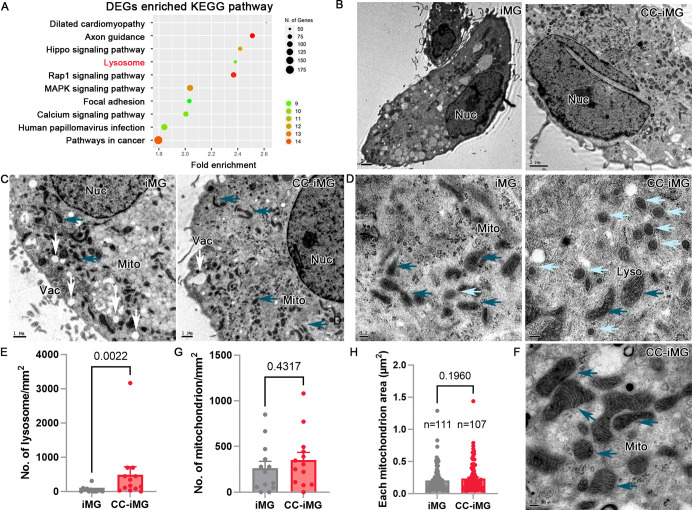
Retinal Organoid Microenvironment Promotes Organelle Maturation of iMG
Both lysosomes and mitochondria are pivotal organelles playing key roles in the development of iMG. Lysosomes, in particular, are crucial for microglial function due to their involvement in the phagocytic clearance of cell debris and pathogens. Our enriched KEGG pathway analysis revealed significant alterations in lysosomal pathways following ROs coculture (Fig. 6A). To elucidate the changes in cellular organelle in iMG, cells with or without ROs coculture underwent ultrastructural analysis using TEM (Fig. 6B). Amplified TEM images revealed vacuole-like vesicles with diameters exceeding 0.35 µm (Fig. 6C), indicative of lysosomal structures. In iMG, lysosomes typically exhibited diameters ranging from 50 to 200 nm, appearing as spheres filled with granules (Fig. 6D). Notably, lysosomes in CC-iMG displayed intact and clearly delineated membranes, a feature rarely observed in iMG lysosomes (Fig. 6D). Additionally, the number of lysosomes in CC-iMG was significantly increased compared to iMG (Fig. 6E). Mitochondria in CC-iMG exhibited prominent inner membranes and mature spines compared to those in iMG (Fig. 6F). However, the number and area of mitochondria were comparable between CC-iMG and iMG (Figs. 6G, 6H). These findings highlight the profound influence of retinal organoid interaction on lysosomal and mitochondrial morphology and abundance in iMG.
Figure 6.
Electron micrographs showing enriched organelles in iMG after coculture with retinal organoids. (A) The most significantly enriched KEGG pathway of DEGs. (B) Representative TEM images of iMG and CC-iMG. Nuc, nuclear. (C) Amplified TEM images showing more vesicles (white arrow) in iMG and more organelle, like mitochondrial (Mito, dark blue arrow) in CC-iMG. (D, F) Representative micrographs showing more mature lysosomes (Lyso, light blue arrow) and mitochondria (Mito, dark blue arrow) in CC-iMG. Scale bar: 2 µm (A, B). Scale bar: 1 µm (C). Scale bar: 0.2 µm (D, F). Quantification of number of lysosome (E), mitochondrion (G) and area of each mitochondrion (H) in iMG and CC-iMG.
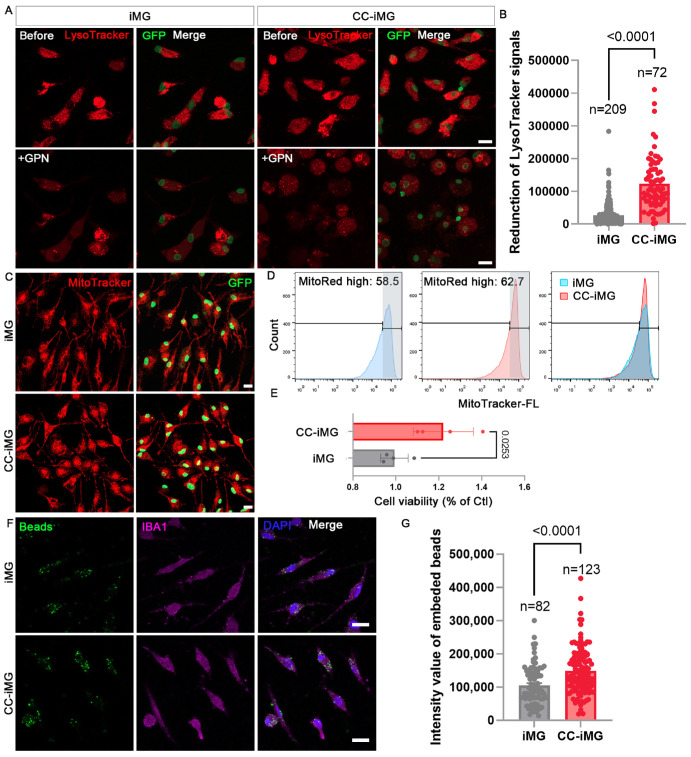
The LysoTracker probes selectively accumulate in acidic organelles, although not exclusively in lysosomes. The fluorescence intensity of the LysoTracker signal markedly increased in CC-iMG compared to that in iMG (Supplementary Figs. S5A, S5B). The percentage of cells exhibiting high fluorescence increased from 68.4% to 79.7% after coculture with ROs, and the median fluorescence intensity was also significantly elevated (Supplementary Fig. S5C). To distinguish lysosomal and prelysosomal endocytic vacuoles, Glycyl-L-phenylalanine 2-naphthylamide (GPN), a lysosome-disrupting enzyme substrate, was applied to treat iMG and CC-iMG. Consequently, the functional lysosomes can be lysed with GPN treatment (Fig. 7A). We observed a significantly higher reduction in LysoTracker-positive structures in CC-iMG compared to iMG, indicating a greater abundance of mature lysosomes in CC-iMG (Fig. 7B; Supplementary Figs. S5D, S5E).
Figure 7.
Coculture with retinal organoids promotes organelle maturation in iMG. (A) Representative images of LysoTracker red staining of iMG and CC-iMG before and after GPN (50 µM) treatment. (B) Quantification of reduced fluorescent intensity of LysoTracker after GPN treatment. The values extract the mean background noise with vehicle (DMSO) treatment presented in Supplementary Figure S4E. (C) Representative images of MitoTracker red staining of iMG and CC-iMG. (D) Flow cytometry analysis of MitoTracker stained iMG and CC-iMG. (E) Cell viability was measured with an ATP-based method using CellTiter-Glo Luminescent Cell Viability Assay. Cell viability in CC-iMG was normalized with that of iMG (n = 4). (F) Phagocytosis of iMG and CC-iMG were evaluated after treatment with poly(methylmethacrylate) fluorescent nano-beads for 30 minutes. Scale bar: 10 µm. (G) Quantification of the intensity of fluorescent nano-beads in each cell. (B, E, G) Data are represented as means ± SD.
Dysfunctional mitochondria released by microglia can trigger neuronal death.32 To further understand the changes in mitochondria in CC-iMG, we performed MitoTracker staining followed by flow cytometry analysis (Figs. 7C, 7D). Flow cytometry analysis showed a slight increase in the proportion of cells with higher fluorescence intensity (Fig. 7D). We then investigated whether the function of mitochondria in CC-iMG was enhanced. To assess this, we measured cell viability using an ATP-based method, which revealed an increase in ATP production ability elicited by retinal tissue coculture (Fig. 7E). To assess the non-specific engraftment of iMG and CC-iMG, fluorescent nano-beads were used (Fig. 7F). Coculture with ROs significantly increased the phagocytic activity of iMG (Fig. 7G). These findings collectively suggest that the retinal organoid microenvironment promotes organelle maturation and phagocytic activity of iMG.
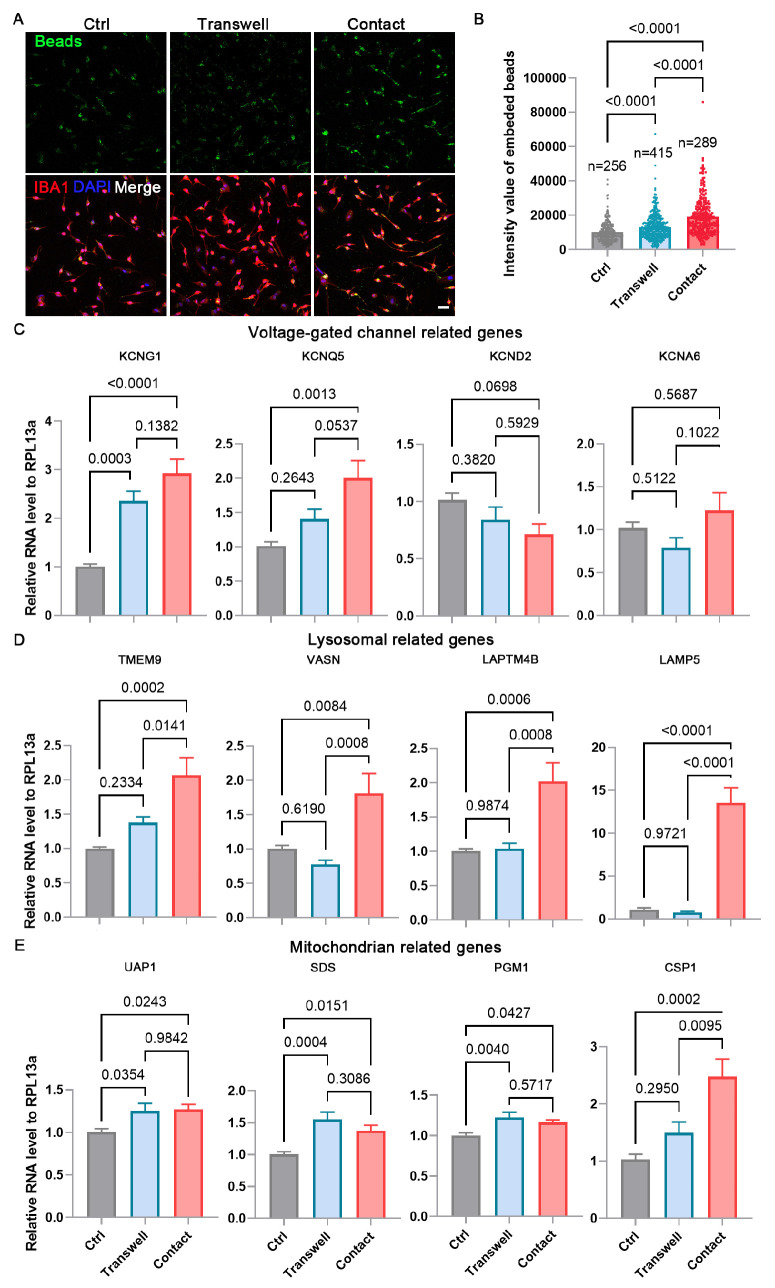
To further investigate whether direct contact or secreted factors from ROs influence the physiology of iMG, we used a transwell system to separate ROs and iMG. Phagocytic activity was significantly increased in the transwell group compared to the iMG monoculture control. However, the phagocytic activity was markedly higher in the contact coculture group than in the transwell group (Figs. 8A, 8B). Additionally, we evaluated the expression of genes related to voltage-gated channels, lysosomes, and mitochondria using qPCR (Figs. 8C–E). The expression of the voltage-gated channel gene KCNG1 was upregulated in both the transwell and contact coculture groups compared to the control group. In contrast, the expression of KCNQ5 was significantly increased in the contact coculture group but did not significantly change in the transwell group (Fig. 8C). A similar pattern was observed for all detected lysosomal genes and the mitochondrial gene CSP1 (Figs. 8D, 8E). The expression of mitochondrial genes UAP1, SDS, and PGM1 was significantly upregulated in both the transwell and contact coculture groups (Fig. 8E). These results indicate that secreted factors from ROs can influence iMG physiology, although direct contact coculture has a more pronounced effect.
Figure 8.
Direct contact coculture has a more pronounced effect on iMG than transwell. (A) Phagocytosis of iMG and CC-iMG (transwell and contact) was evaluated after treatment with poly(methylmethacrylate) fluorescent nano-beads for 30 minutes. Scale bar: 50 µm. (B) Quantification of the intensity of fluorescent nano-beads in each cell. Data are represented as means ± SD. (C–E) Quantitative PCR analysis of the expression of genes related to voltage-gated channel (C), lysosomes (D), and mitochondria (E). Data are represented as means ± SD; n = 3.
Discussion
In this study, we generated iMG from hiPSC and assessed their immune response following pro-inflammatory stimulation, which closely mirrored that of cultured primary human microglia. Additionally, we elucidated transcriptional and functional differences between iMG and the commonly used microglial cell line, HMC3, underscoring iMG's potential as a more reliable microglial model. To further refine the microglial phenotype of iMG, we cocultured them with retinal organoids for five days, yielding what we refer to as CC-iMG. Comparative analysis revealed significant alterations in the transcriptional signature, electrophysiological properties, and organelle composition of CC-iMG compared to iMG alone. This study represents the first comparative analysis of transcriptome-level differences between iMG and microglial cell lines. Additionally, we used TME and electrophysiology to investigate how retinal cell interaction influences the ion channel activity and organelles of iMG.
Microglia play a pivotal role in the pathology and progression of various neurodegenerative diseases. Therefore it is essential that in vitro models of microglia faithfully recapitulate their function to advance our understanding of these cells and explore novel drugs or therapeutic interventions for neurological disorders. Recently, protocols for generating iMG have been established in several laboratories, providing a valuable platform for studying microglial biology.15–20,26,33 These iMG, inducted by small molecule cocktails, express typical microglial signature genes, acquire phagocytic function, and respond to inflammatory stimuli. However, many current approaches rely on animal-derived Matrigel and FBS. For medical applications, it is crucial to generate cells under fully defined conditions. Truncated recombinant human vitronectin is widely used as a Matrigel substitute for producing therapeutic cells from hPSC in an xeno-free culture system, maintaining hPSC pluripotency with a chemically defined medium.34 Although the addition of FBS in microglial culture medium can mitigate the negative impacts of in vitro culture and enhance microglial phagocytic activity, it is not essential for microglial survival.35 Thus we present here the development of a simplified and highly efficient protocol for generating iMG from hPSCs in a fully defined culture system suitable for clinical application.
Microglia-mediate neuroinflammation has been recognized as a significant contributor to neurodegenerative diseases.36 To assess the immune responsiveness of iMG, we stimulated them with pro-inflammatory stimuli. Consistent with findings in primary human microglia,35,37,38 we observed a significant upregulation of TNF-α, CCR7, and IL1β with LPS treatment and an enhanced toll-like receptor 3 (TLR3) signaling pathway with poly IC treatment. The minor changes in the transcription profile induced by LPS may be attributed to two main factors: the low concentration of LPS (1 ng/mL) and the presence of components in the culture medium similar to Abud medium, which exhibits an immunosuppressive effect.15,35 Recent studies have demonstrated that exposure to neural substrates generates transcriptional heterogeneity in iMG, resembling subpopulations identified in human brain microglia.39 Additionally, iMG treated with LPS and interferon-gamma (IFNγ) exhibit similarities to AD-relevant microglia,40 suggesting that iMG can serve as a substitute for primary human microglia in relevant studies.
The human microglial cell line, HMC3, is a widely used in vitro model for microglial studies.13 However, a previous study raised concerns about the origin of CHME-5 cells, another term for HMC3, suggesting they were rat glioblastoma cells rather than human cells.41 To confirm the human origin of HMC3 cells used in our laboratory, we conducted immunofluorescence staining using a human nuclear antibody against Ku80 and amplified human CYCT1-specific primers (Supplementary Fig. S6). These analyses confirmed the human identity of the HMC3 cells used in our experiments. Our investigation revealed a significant decrease in the expression of microglial marker genes, including P2RY12, CSF1R, SPI1, TREM2, and TMEM119, in HMC3 compared to iMG. Furthermore, the expression of these genes is barely changed following LPS and poly IC treatments in HMC3, which is completely in contrast to the responses found in iMG (Figs. 3I, 3J) and primary microglia. Additionally, poly IC treatments induced significant cell death in iMG but not in HMC3 (Figs. 2D, 3G). These observations underscore the importance of iMG as a complementary model to current microglial research, providing valuable insights into the dynamics and plasticity of microglial ion channels under neural interaction conditions.
Previous studies have highlighted the suitability of iMG for coculture with glia and neurons, leading to partial enhancements in their functions and morphology.26,42 Direct contact with niche cells significantly enhances their transcriptional resemblance to ex vivo microglia.42 Our prior research demonstrated that indirect coculture with retinal organoids affects the transcriptional signature of iMG, including the upregulation of receptors such as TLR3 and TMEM119, and the downregulation of genes associated with cytokine production.16 Conversely, coculture with cortical neurons containing Tau mutations results in a decreased degree of ramification and increased apoptosis, mitochondrial fission, and ROS production in iMG.19 Thus we have developed retinal organoids and established a direct contact coculture system to investigate how retinal cell interaction influences the functional properties of iMG, including electrophysiological function and organelle biogenesis. Our goal is to accurately recapitulate microglia in vivo to better understand their role in pathologies.
Microglia, characterized by very high membrane resistance, possess the ability to rapidly modulate their membrane potential through the function of a few ion channels, thereby regulating various associated functions such as phagocytosis, migration, cytokine secretion, shape changes, reactive oxygen species release, and proliferation.43 A previous study has identified the presence of inward rectifier K+ current in iMG, which can be blocked by Cs+.20 Similarly, we observed partial blocking of both inward and outward rectifier K+ currents in iMG by TEA-Cl (Supplementary Fig. S4D). In this study, we investigated the electrophysiological properties of iMG before and after coculturing with retinal organoids. The characteristics of both inward and outward rectifier K+ currents in iMG closely resemble those documented for cultured human microglia,44 indicating that iMG can highly recapitulate the electrophysiological function of primary human microglia.
The expression of ion channel genes in microglia is closely linked to their states and functions. Here, we observed a significant alteration in the iron channel gene expression pattern of iMG upon exposure to retinal organoids. Our comparison of the macroscopic current profile in iMG before and after exposure to retinal organoids revealed an enhanced expression of both inward and outward K+ current. The inward rectifier K+ current has been recorded in cultured microglial cells from various species, including rats, mice, bovines, and humans.44–47 Notably, a significant reduction in this current can be induced by LPS stimulation,48 suggesting that an increased inward rectifier K+ current in CC-iMG may indicate a healthier state compared to iMG alone. Although altered outward K+ current has been recognized as a hallmark of activated microglial cells in both in vitro and in vivo studies,49,50 reports have also shown increased outward K+ current in primary human microglia between days 5-7 post-plating compared to days 1-2 post-plating,44 as well in HIV-1 Tat protein-induced microglia.50 Our result showing increased outward K+ current are consistent with these findings. However, our previous study demonstrated that iMG produce similar levels of cytokines, including IL-8, IL-1β, IL-6, TNF-α, IL-10, and IL-20P70, before and after coculture with retinal organoids,16 suggesting that CC-iMG are not activated iMG. Furthermore, the enhanced expression of inward and outward K+ current density in CC-iMG is also consistent with the gene transcriptomic changes of K+ channels (Figs. 4D, 4E), where both inward rectifier Kir2.1 (KCNJ2), Kir3.4 (KCNJ5), Kir6.1 (KCNJ8), Kir7.1 (KCNJ13), and outward rectifier Kv6.1 (KCNG1), Kv7.5 (KCNQ5), Kv4.2 (KCND2), Kv1.6 (KCNA6) increased (Supplementary Fig. S4A). Currently, microglial ion channels are garnering more attention as potential targets for the discovery of new drugs or therapeutics,51 owing to their significant role in neural disorders.52 Therefore iMG are well suited for further investigations to determine the key factors leading to increased microglial ion channels, which may provide valuable insights into microglial function in neuropathologies.
Microglia cell replacement therapy has emerged as a potential treatment strategy, because microglia have the capacity to repopulate after depletion.53 Repopulated microglia have demonstrated protective effects and have been shown to positively modulate the microenvironment in the injured brain and in neurodegenerative diseases.54,55 For diseases stemming from microglial dysfunction, such as Trem2 mutation-induced AD, replacing dysfunctional microglia with healthy counterparts has been shown to restore disease phenotype.56 With advancements in techniques, the replacement of mouse microglia with iMG has been achieved in both the brain and retina.23–27 This study presents a straightforward method to generate more mature iMG through coculture with retinal organoids, offering a promising and reliable candidate cell source for microglia cell replacement therapy.
Supplementary Material
Acknowledgments
The authors thank Miss Yizhen Zhang for her significant comments and helpful discussion.
Supported in part by grants from the Natural Science Foundation of China (82070981), Photo-biological Effects of Specific Wavelength Segments of Red Light and Relevant Applications (512024Y-11446), National Key R&D Program of China (2022YFA1105502), State Key Laboratory of Neuroscience (SKLN-202103), Zhejiang Natural Science Foundation of China (LY22H120008), Wenzhou Association for Science and Technology (kjfw27), Scientific Research Program of Wenzhou (Y2023172), and the Fundamental Research Fund of China National Institute of Standardization (512023Y-10404).
Author Contributions: MLG designed and supervised the experiments. MLG, TYW, and CT performed stem cell differentiation, part immunofluorescence, and qPCR. TYW performed flow cytometry and part immunofluorescence. XL completed the electrophysiological analysis. JZ and CT completed electron microscopy. XFH, MYL, PZB, LJC, and QRK completed RNA-seq data analysis and visualization. MLG, JZ, JQC, and XFH conducted data and image analysis. ZCL, YG, SSZ, and SHP performed part immunofluorescence and quantification. MLG and TYW wrote the draft of the manuscript. JZ wrote the final manuscript. All authors approved the final manuscript.
Data Availability and Materials: The RNA sequencing datasets generated in this study has been be uploaded in the Genome Sequence Archive for human (https://ngdc.cncb.ac.cn/gsa-human/) and the accession number is HRA008070.
Disclosure: M.-L. Gao, None; T.-Y. Wang, None; X. Lin, None; C. Tang, None; M. Li, None; Z.-P. Bai, None; Z.-C. Liu, None; L.-J. Chen, None; Q.-R. Kong, None; S.-H. Pan, None; S.-S. Zeng, None; Y. Guo, None; J.-Q. Cai, None; X.-F. Huang, None; J. Zhang, None
References
- 1. Li Q, Barres BA.. Microglia and macrophages in brain homeostasis and disease. Nat Rev Immunol. 2018; 18: 225–242. [DOI] [PubMed] [Google Scholar]
- 2. Lopes KP, Snijders GJL, Humphrey J, et al.. Genetic analysis of the human microglial transcriptome across brain regions, aging and disease pathologies. Nat Genet. 2022; 54: 4–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gosselin D, Skola D, Coufal NG, et al.. An environment-dependent transcriptional network specifies human microglia identity. Science. 2017; 356(6344): eaal3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fritsche LG, Fariss RN, Stambolian D, Abecasis GR, Curcio CA, Swaroop A.. Age-related macular degeneration: genetics and biology coming together. Ann Rev Genom Hum Genet. 2014; 15: 151–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kumar S, Akopian A, Bloomfield SA.. Neuroprotection of retinal ganglion cells suppresses microglia activation in a mouse model of glaucoma. Invest Ophthalmol Vis Sci. 2023; 64: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shu N, Zhang Z, Wang X, et al.. Apigenin alleviates autoimmune uveitis by inhibiting microglia M1 pro-inflammatory polarization. Invest Ophthalmol Vis Sci. 2023; 64: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhou J, Lin S, Hu Q, et al.. Microglial CD11b knockout contributes to axonal debris clearance and axonal degradation attenuation via IGF-1 after acute optic nerve injury. Invest Ophthalmol Vis Sci. 2023; 64: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huang JM, Zhao N, Hao XN, et al.. CX3CL1/CX3CR1 signaling mediated neuroglia activation is implicated in the retinal degeneration: a potential therapeutic target to prevent photoreceptor death. Invest Ophthalmol Vis Sci. 2024; 65: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li H, Chen D, Sun W, et al.. KATP opener attenuates diabetic-induced müller gliosis and inflammation by modulating Kir6.1 in microglia. Invest Ophthalmol Vis Sci. 2021; 62: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ryan SK, Zelic M, Han Y, et al.. Microglia ferroptosis is regulated by SEC24B and contributes to neurodegeneration. Nat Neurosci. 2023; 26: 12–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang X, Zhao L, Zhang J, et al.. Requirement for microglia for the maintenance of synaptic function and integrity in the mature retina. J Neurosci. 2016; 36: 2827–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sabogal-Guáqueta AM, Marmolejo-Garza A, Trombetta-Lima M, et al.. Species-specific metabolic reprogramming in human and mouse microglia during inflammatory pathway induction. Nat Commun. 2023; 14: 6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dello Russo C, Cappoli N, Coletta I, et al.. The human microglial HMC3 cell line: where do we stand? A systematic literature review. J Neuroinflam. 2018; 15: 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marsh SE, Walker AJ, Kamath T, et al.. Dissection of artifactual and confounding glial signatures by single-cell sequencing of mouse and human brain. Nat Neurosci. 2022; 25: 306–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Abud EM, Ramirez RN, Martinez ES, et al.. iPSC-derived human microglia-like cells to study neurological diseases. Neuron. 2017; 94: 278–293.e279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gao ML, Zhang X, Han F, et al.. Functional microglia derived from human pluripotent stem cells empower retinal organ. Sci China Life Sci. 2022; 65: 1057–1071. [DOI] [PubMed] [Google Scholar]
- 17. Muffat J, Li Y, Yuan B, et al.. Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat Med. 2016; 22: 1358–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pandya H, Shen MJ, Ichikawa DM, et al.. Differentiation of human and murine induced pluripotent stem cells to microglia-like cells. Nat Neurosci. 2017; 20: 753–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Speicher AM, Korn L, Csatári J, et al.. Deterministic programming of human pluripotent stem cells into microglia facilitates studying their role in health and disease. Proc Natl Acad Sci USA. 2022; 119: e2123476119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen SW, Hung YS, Fuh JL, et al.. Efficient conversion of human induced pluripotent stem cells into microglia by defined transcription factors. Stem Cell Rep. 2021; 16: 1363–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brownjohn PW, Smith J, Solanki R, et al.. Functional studies of missense TREM2 mutations in human stem cell-derived microglia. Stem Cell Reports. 2018; 10: 1294–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu T, Zhu B, Liu Y, et al.. Multi-omic comparison of Alzheimer's variants in human ESC-derived microglia reveals convergence at APOE. J Exp Med. 2020; 217(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mancuso R, Van Den Daele J, Fattorelli N, et al.. Stem-cell-derived human microglia transplanted in mouse brain to study human disease. Nat Neurosci. 2019; 22: 2111–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xu R, Li X, Boreland AJ, et al.. Human iPSC-derived mature microglia retain their identity and functionally integrate in the chimeric mouse brain. Nat Commun. 2020; 11: 1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hasselmann J, Coburn MA, England W, et al.. Development of a chimeric model to study and manipulate human microglia in vivo. Neuron. 2019; 103: 1016–1033.e1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Haenseler W, Sansom SN, Buchrieser J, et al.. A highly efficient human pluripotent stem cell microglia model displays a neuronal-co-culture-specific expression profile and inflammatory response. Stem Cell Rep. 2017; 8: 1727–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Svoboda DS, Barrasa MI, Shu J, et al.. Human iPSC-derived microglia assume a primary microglia-like state after transplantation into the neonatal mouse brain. Proc Natl Acad Sci USA. 2019; 116: 25293–25303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schilling T, Eder C.. Microglial K(+) channel expression in young adult and aged mice. Glia. 2015; 63: 664–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang J, Tuo J, Cao X, Shen D, Li W, Chan CC.. Early degeneration of photoreceptor synapse in Ccl2/Cx3cr1-deficient mice on Crb1(rd8) background. Synapse. 2013; 67: 515–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Deng WL, Gao ML, Lei XL, et al.. Gene correction reverses ciliopathy and photoreceptor loss in iPSC-derived retinal organoids from retinitis pigmentosa patients. Stem Cell Rep. 2018; 10: 1267–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cowan CS, Renner M, De Gennaro M, et al.. Cell types of the human retina and its organoids at single-cell resolution. Cell. 2020; 182: 1623–1640.e1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Joshi AU, Minhas PS, Liddelow SA, et al.. Fragmented mitochondria released from microglia trigger A1 astrocytic response and propagate inflammatory neurodegeneration. Nat Neurosci. 2019; 22: 1635–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hasselmann J, Blurton-Jones M.. Human iPSC-derived microglia: a growing toolset to study the brain's innate immune cells. Glia. 2020; 68: 721–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Braam SR, Zeinstra L, Litjens S, et al.. Recombinant vitronectin is a functionally defined substrate that supports human embryonic stem cell self-renewal via alphavbeta5 integrin. Stem Cells. 2008; 26: 2257–2265. [DOI] [PubMed] [Google Scholar]
- 35. Dorion MF, Yaqubi M, Murdoch HJ, et al.. Systematic comparison of culture media uncovers phenotypic shift of primary human microglia defined by reduced reliance to CSF1R signaling. Glia. 2023; 71: 1278–1293. [DOI] [PubMed] [Google Scholar]
- 36. Xu L, He D, Bai Y.. Microglia-mediated inflammation and neurodegenerative disease. Mol Neurobiol. 2016; 53: 6709–6715. [DOI] [PubMed] [Google Scholar]
- 37. Melief J, Sneeboer MA, Litjens M, et al.. Characterizing primary human microglia: a comparative study with myeloid subsets and culture models. Glia. 2016; 64: 1857–1868. [DOI] [PubMed] [Google Scholar]
- 38. Melief J, Koning N, Schuurman KG, et al.. Phenotyping primary human microglia: tight regulation of LPS responsiveness. Glia. 2012; 60: 1506–1517. [DOI] [PubMed] [Google Scholar]
- 39. Dolan MJ, Therrien M, Jereb S, et al.. Exposure of iPSC-derived human microglia to brain substrates enables the generation and manipulation of diverse transcriptional states in vitro. Nat Immunol. 2023; 24: 1382–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Monzón-Sandoval J, Burlacu E, Agarwal D, et al.. Lipopolysaccharide distinctively alters human microglia transcriptomes to resemble microglia from Alzheimer's disease mouse models. Dis Models Mech. 2022; 15(10): dmm049349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Garcia-Mesa Y, Jay TR, Checkley MA, et al.. Immortalization of primary microglia: a new platform to study HIV regulation in the central nervous system. J Neurovirol. 2017; 23: 47–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Grubman A, Vandekolk TH, Schröder J, et al.. A CX3CR1 reporter hESC line facilitates integrative analysis of in-vitro-derived microglia and improved microglia identity upon neuron-glia co-culture. Stem Cell Rep. 2020; 14: 1018–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Izquierdo P, Attwell D, Madry C.. Ion channels and receptors as determinants of microglial function. Trends Neurosci. 2019; 42: 278–292. [DOI] [PubMed] [Google Scholar]
- 44. McLarnon JG, Xu R, Lee YB, Kim SU.. Ion channels of human microglia in culture. Neuroscience. 1997; 78: 1217–1228. [DOI] [PubMed] [Google Scholar]
- 45. Kettenmann H, Hoppe D, Gottmann K, Banati R, Kreutzberg G.. Cultured microglial cells have a distinct pattern of membrane channels different from peritoneal macrophages. J Neurosci Res. 1990; 26: 278–287. [DOI] [PubMed] [Google Scholar]
- 46. McLarnon JG, Sawyer D, Kim SU.. Cation and anion unitary ion channel currents in cultured bovine microglia. Brain Res. 1995; 693: 8–20. [DOI] [PubMed] [Google Scholar]
- 47. Visentin S, Agresti C, Patrizio M, Levi G.. Ion channels in rat microglia and their different sensitivity to lipopolysaccharide and interferon-gamma. J Neurosci Res. 1995; 42: 439–451. [DOI] [PubMed] [Google Scholar]
- 48. Prinz M, Kann O, Draheim HJ, et al.. Microglial activation by components of gram-positive and -negative bacteria: distinct and common routes to the induction of ion channels and cytokines. J Neuropathol Exp Neurol. 1999; 58: 1078–1089. [DOI] [PubMed] [Google Scholar]
- 49. Nörenberg W, Gebicke-Haerter PJ, Illes P.. Voltage-dependent potassium channels in activated rat microglia. J Physiol. 1994; 475: 15–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Visentin S, Renzi M, Levi G.. Altered outward-rectifying K(+) current reveals microglial activation induced by HIV-1 Tat protein. Glia. 2001; 33: 181–190. [DOI] [PubMed] [Google Scholar]
- 51. Skaper SD. Ion channels on microglia: therapeutic targets for neuroprotection. CNS Neurol Disord Drug Targets. 2011; 10: 44–56. [DOI] [PubMed] [Google Scholar]
- 52. Plescher M, Seifert G, Hansen JN, Bedner P, Steinhäuser C, Halle A.. Plaque-dependent morphological and electrophysiological heterogeneity of microglia in an Alzheimer's disease mouse model. Glia. 2018; 66: 1464–1480. [DOI] [PubMed] [Google Scholar]
- 53. Zhang L, Wang Y, Liu T, Mao Y, Peng B.. Novel Microglia-based therapeutic approaches to neurodegenerative disorders. Neuroscience Bull. 2023; 39: 491–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Willis EF, MacDonald KPA, Nguyen QH, et al.. Repopulating microglia promote brain repair in an IL-6-dependent manner. Cell. 2020; 180: 833–846.e816. [DOI] [PubMed] [Google Scholar]
- 55. Cheng X, Gao H, Tao Z, et al.. Repopulated retinal microglia promote Müller glia reprogramming and preserve visual function in retinal degenerative mice. Theranostics. 2023; 13: 1698–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yoo Y, Neumayer G, Shibuya Y, Mader MM, Wernig M.. A cell therapy approach to restore microglial Trem2 function in a mouse model of Alzheimer's disease. Cell Stem Cell. 2023; 30: 1043–1053.e1046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.