Abstract
Glioblastoma (GBM) is a highly aggressive and malignant brain tumor with limited therapeutic options and a poor prognosis. Despite current treatments, the invasive nature of GBM often leads to recurrence. A promising alternative strategy is to harness the potential of the immune system against tumor cells. Our previous data showed that the BVax (B cell–based vaccine) can induce therapeutic responses in preclinical models of GBM. In this study, we aimed to characterize the antigenic reactivity of BVax-derived Abs and evaluate their therapeutic potential. We performed immunoproteomics and functional assays in murine models and samples from patients with GBM. Our investigations revealed that BVax distributed throughout the GBM tumor microenvironment and then differentiated into Ab-producing plasmablasts. Proteomics analyses indicated that the Abs produced by BVax had unique reactivity, predominantly targeting factors associated with cell motility and the extracellular matrix. Crucially, these Abs inhibited critical processes such as GBM cell migration and invasion. These findings provide valuable insights into the therapeutic potential of BVax-derived Abs for patients with GBM, pointing toward a novel direction for GBM immunotherapy.
Keywords: Immunology, Oncology
Keywords: Brain cancer, Cancer immunotherapy, Extracellular matrix
B-cell-based therapies produce antibodies that interfere with glioma migration, invasion, and overall growth. The potential role of antibody immunity in GBM is unknown.
Introduction
Glioblastoma (GBM) is an aggressive and malignant brain tumor that arises from glial cells (1). GBM is one of the most common and deadly forms of brain cancer in adults, with a median survival of approximately 15 months after diagnosis. The current standard of care for GBM includes surgery, radiation therapy, and chemotherapy, but the overall prognosis remains poor (2). A major obstacle in treating GBM is its remarkable ability to invade and migrate into surrounding healthy brain tissues, making complete gross total surgical resection impossible and thus leading to inevitable tumor recurrence (3). As such, there is a pressing need to explore alternative therapeutic avenues to inhibit GBM progression and improve patient outcomes.
Harnessing the immune system to modulate tumor progression and remote sites of invasion is a compelling strategy for GBM treatment (4). While most immunotherapy efforts have historically focused on T cells, the role of B cells, especially in the context of GBM, remains less explored. Recent studies suggest that B cells and their secreted Abs can influence tumor growth, metastasis, and response to treatment (5–9). The presence of tertiary lymphoid structures (TLSs) in solid cancers, containing B cells undergoing somatic hypermutation, confers a favorable prognosis (10–12). As such, our laboratory is developing an immunotherapeutic approach for cancer using activated B cells as a cell-based vaccine (BVax) against GBM (13, 14). BVax is a B cell–based vaccine comprising 4-1BBL+ B cells activated through CD40 agonism and IFN-γ stimulation. Advantages of B cell therapy relative to other types of immunotherapies include its antigen-presenting capability (15–17), shared cognate antigen specificity with T cells (18), ability to generate tumor-reactive Abs (12), and circulatory mobility enabling tumor and secondary lymphoid organ infiltration (19, 20). Moreover, the relative ease as well as timely ex vivo activation and expansion from patient-derived circulating B cells reduces the cost of generating a personalized cell-based therapeutic. As such, B cell–based vaccines represent a promising, yet underinvestigated, immunotherapeutic approach (21–24) warranting further study in GBM.
In this study, we aimed to determine the humoral response induced by BVax, assess the tumor-reactive nature of BVax-derived Abs, and evaluate their therapeutic potential in preclinical models. This analysis reveals the role of BVax in the immune-tumor interplay and its therapeutic potential for patients with GBM through a blend of molecular and proteomics analyses.
Results
BVax differentiates into plasmablasts and harbors potentially tumor-reactive B cell receptors.
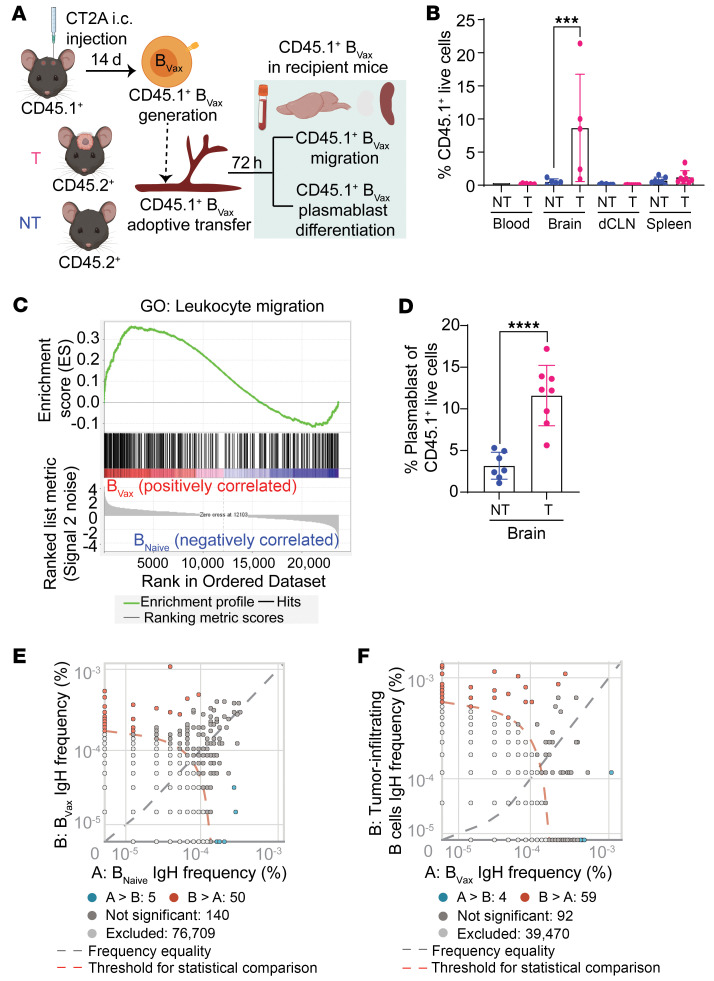
To determine the humoral responses generated by BVax, we first analyzed the potential of BVax to migrate to the tumor and differentiate into Ab-producing cells (plasmablasts). Using the CD45.1 versus CD45.2 congenic mouse model (Figure 1A), we found that, upon intravenous injection, CD45.1+ BVax preferentially migrated to the glioma-bearing brains 72 hours after injection (Figure 1B). Gene set enrichment analysis (GSEA) of BVax showed upregulation of the Gene Ontology (GO) gene set involved in leukocyte migration (Figure 1C and Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI177384DS1). The potential of BVax to differentiate into Ab-producing cells after migrating into the glioma was confirmed in vivo using CT2A tumor–bearing mice treated with BVax. Approximately 10% of the BVax showed a CD38+CD20–CD19+ plasmablast phenotype (Figure 1D). Analysis of the BVax and BNaive heavy-chain receptor repertoire (bulk IgH sequence) revealed comparable B cell receptor (BCR) repertoire diversity between BVax and BNaive cells (Supplemental Figure 1A). However, BVax might present differential reactivity compared with BNaive cells (Figure 1E, Supplemental Table 2, and Supplemental Table 3). Among the BVax BCRs, 92 were shared with glioma-infiltrating B cells (tumor-infiltrating B [TIB] cells, Figure 1F and Supplemental Table 4). Additionally, approximately 2% of BVax BCRs overlapped with glioma-infiltrating B cells but were absent in BNaive BCRs (Supplemental Figure 1B). This suggests that BVax may harbor tumor-reactive BCRs.
Figure 1. BVax differentiates into plasmablasts and generates potentially tumor-reactive B cell Igs.
(A) Schema illustrating the experimental design to investigate the potential of BVax to migrate to the glioma and differentiate into Ab-producing cells (plasmablasts). i.c., intracranial; T, tumor; NT, nontumor. (B) Percentage of BVax (CD45.1+) cells in various tissues (blood, brain, deep cervical lymph node [dCLN], and spleen) of intracranial tumor–bearing and nontumor-bearing mice via flow cytometry (n = 5 for each group). (C) GSEA of the indicated datasets comparing the transcriptional profile between BVax and BNaive. Data were pooled from 3 independent experiments. (D) Percentage of plasmablasts (CD19+CD20–CD38+) within the BVax population of the brain via flow cytometry (n = 8 for each group). (E) Representative dot plot of BCR clones from BCR IgH sequencing comparing murine BVax and BNaive. Unique clones in BVax are shown in red; unique clones in BNaive are shown in blue (n = 3 for each group). (F) Representative dot plot of BCR clones from BCR IgH sequencing comparing murine TIB cells (n = 2) and BVax (n = 3). Unique clones in TIB cells are shown in red; unique clones in BVax are shown in blue. Data are the mean ± SD. ***P < 0.001 and ****P < 0.0001, by 1-way ANOVA. NT, nontumor; T, tumor.
Characterization of murine BVax-derived immunoglobulin reactivity.
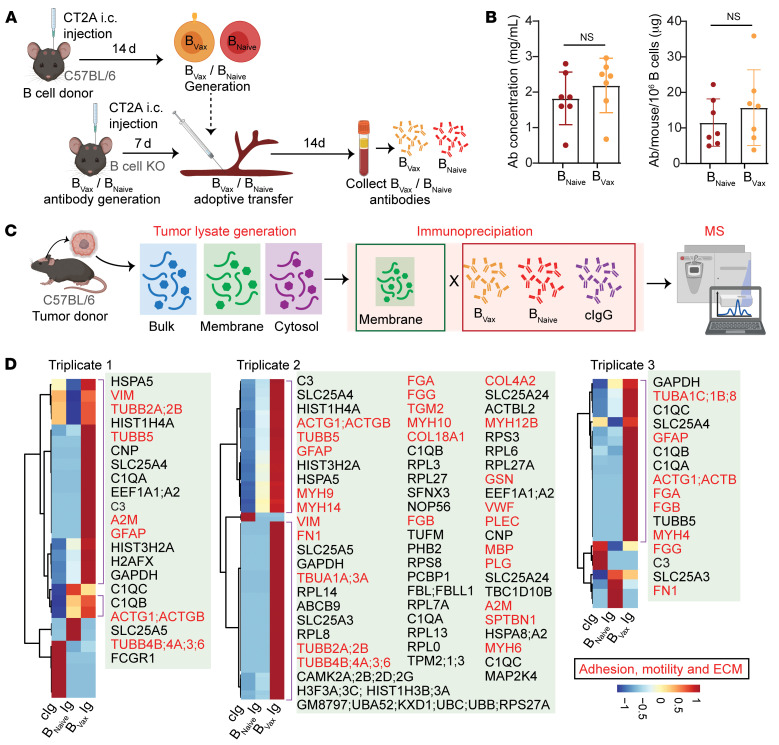
To examine BVax antigenic reactivity, BVax- and BNaive-derived immunoglobulins (Igs) were produced in vivo by adoptive transfer of BVax or BNaive into CT2A tumor–bearing, B cell–deficient mice (Figure 2A). After 2 weeks, blood from the experimental groups (BVax and BNaive) was collected, and Igs were isolated using a Protein A/G Spin Column. Comparable amounts of Igs were obtained for both groups (Figure 2B). To analyze the reactivity of these Igs, we performed a immunoproteomics study using immunoprecipitation–mass spectrometry (IP-MS). Intracranial glioma homogenates were produced and fractionated into the membrane and cytosolic fractions (Figure 2C). Proteins enriched in the membrane fraction were then immunoprecipitated with the BVax- or BNaive-derived Igs. Commercially available mouse IgG (cIgG) was used as a control. The MS analysis revealed that BVax Igs had unique reactivity compared with the BNaive-derived Igs or control Igs (Figure 2D and Supplemental Table 5). More specifically, the BVax produced Igs with preferential reactivity to factors involved in cell motility, extracellular matrix (ECM), and membrane organization, such as fibrinogen, fibronectin, and myosin, as shown by the GO pathway analysis (Supplemental Figure 2). To determine whether the BVax Abs specifically target ECM and membrane proteins when isolated from a more controlled environment, we conducted additional immunoprecipitation experiments using conditioned medium and cell membrane fractions from CT2A tumor cells cultured in vitro. Consistent with our initial findings, BVax Abs specifically targeted membrane (such as collagen receptor, integrin-linked kinase [ILK] complex, tight junction proteins, Rho GTPases, vinculin, and nischarin) and ECM proteins (such as gelsolin, collagens, fibrinogen, EGF-containing fibulin-like ECM protein 2, SPARC-like protein 1, matrix metalloproteinases, caldesmon, and asporin) involved in cell adhesion and motility (25–39) (Supplemental Figure 3 and Supplemental Table 6), confirming their preferential reactivity.
Figure 2. Characterization of murine BVax-derived Ig reactivity.
(A) Schema demonstrating how in vivo BVax-derived Igs are produced from mice bearing CT2A gliomas. (B) Amount of BVax-derived Igs generated from mice bearing GBM tumors ( n = 7 for each group). (C) Schema depicting the protocol for the murine IP-MS experiments used to identify tumor-specific antigens recognized by BVax-derived Igs. (D) Heatmap revealing hierarchical clustering of GBM tumor antigens recognized by BVax-derived Igs. Each triplicate corresponds to an independent IP-MS experiment. In triplicate 1, BVax-derived Igs were pooled from 10 mice, and BNaive-derived Igs were pooled from 11 mice. In triplicate 2, BVax-derived Igs were pooled from 11 mice, and BNaive-derived Igs were pooled from 10 mice. Triplicate 3 involved BVax-derived Igs pooled from 10 mice and BNaive-derived Igs pooled from 12 mice. Data in B are the mean ± SD and were analyzed by 2-tailed Student’s t test.
Characterization of patient BVax–derived Ig reactivity.
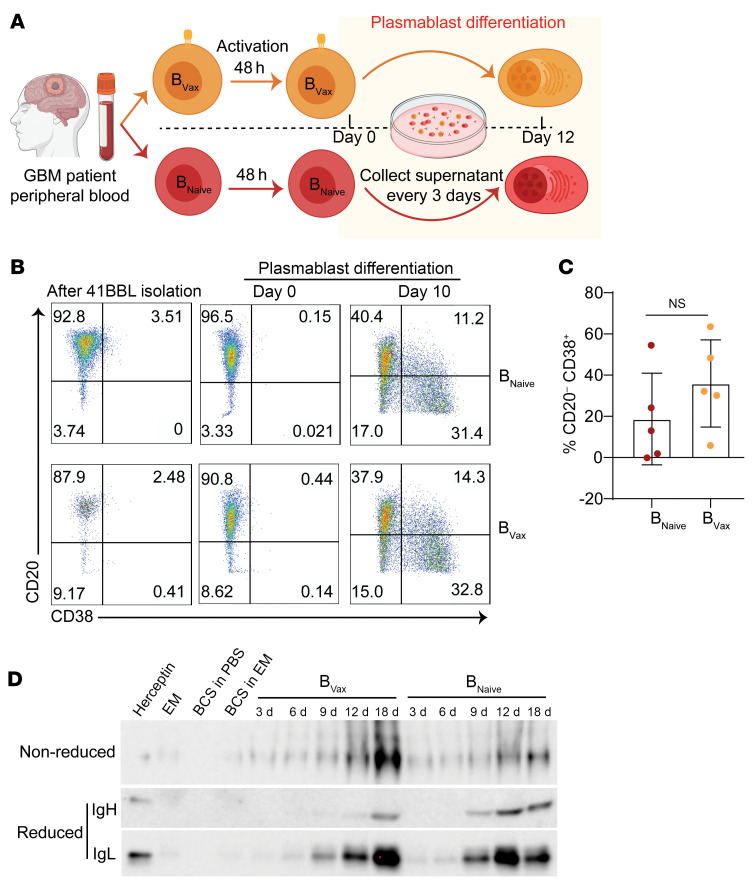
To examine the biological relevance of the murine model, we evaluated the antigenic reactivity of BVax from patients with GBM. BVax from patients newly diagnosed with GBM were differentiated into plasmablasts in vitro (Figure 3A). Flow cytometric analysis confirmed that 10 days after activation, approximately 30% of cells generated from either BNaive or BVax showed a CD19+CD20–CD38+ plasmablast phenotype (Figure 3, B and C), suggesting that both B cell types had a similar in vitro polyclonal potential to differentiate into plasmablasts. Supernatants were then collected every 3 days, and the presence of secreted Igs was confirmed by Western blotting (Figure 3D) and ELISA (Supplemental Figure 4). We detected the production of IgG (both IgH and IgL chains) in both BVax and BNaive conditions, with clear bands appearing from day 6 onward, confirming the efficacy of the ex vivo generation method in producing GBM patient–derived Abs.
Figure 3. Production of BVax-derived Igs in patients with GBM.
(A) Schema of the ex vivo generation of GBM patient BVax-derived Abs. (B) Dot plots of flow cytometric analysis of CD20 and CD38 expression during different stages in the BVax activation protocol to generate GBM patient–derived Abs ex vivo. (C) Box-and-whisker plot of the percentage of plasmablasts generated at day 10 of BVax/BNaive activation in patients with GBM (n = 5 for each group). (D) Western blot confirming the presence of Abs in the media during various stages of the ex vivo BVax activation protocol for patients with GBM. EM, expansion medium; BCS, B cell supplement. Data in C are the mean ± SD and were analyzed by 2-tailed Student’s t test.
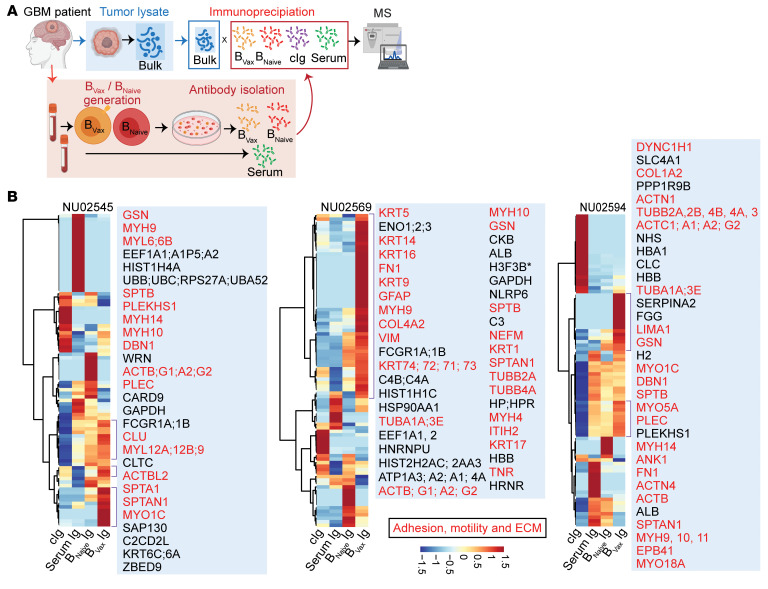
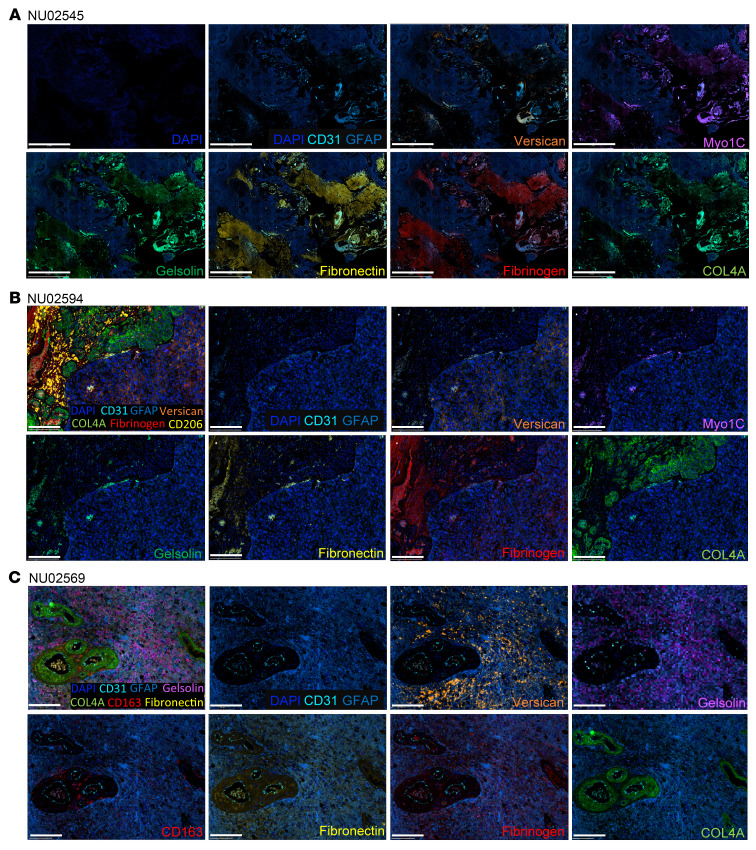
In parallel to the murine glioma analysis, Igs from supernatants of the B cell cultures harvested every 3 days were isolated using a Protein A/G Spin Column. Given the limited amount of Igs, the IP-MS was performed using autologous bulk glioma protein homogenates without fractionation (Figure 4A). The patient’s serum Igs were included to evaluate the level of peripheral baseline reactivity to the autologous tumor. The IP-MS data (Figure 4B and Supplemental Table 7) showed heterogeneity across patients (NU02545, NU0569, and NU02594). However, we found several regulators of the ECM (such as fibrinogen, versican core protein, and collagens) and cell motility (such as myosin and actin) across all patients’ BVax Igs. Furthermore, GO pathway analysis confirmed that BVax had a preferential reactivity toward biological processes involved in cell migration and motility (Supplemental Figure 5). To corroborate these findings, we conducted additional immunoprecipitation experiments using conditioned medium and cell membrane fractions from GBM43 tumor cells cultured in vitro. These experiments confirmed that BVax Abs specifically targeted ECM proteins (such as gelsolin, collagens, matrix metalloproteinases, thrombospondins, and laminin) and membrane proteins (such as ILK complex, EGFR, flotillin-2 [FLOT2], and Rho GTPases) enriched in these fractions (Supplemental Figure 6 and Supplemental Table 8), confirming their preferential reactivity. Spatial proteomics and histopathological analysis of tumors from these patients illustrate the spatial distribution of BVax-derived, Ig–recognized antigens within the ECM of GBM (Figure 5). Gelsolin (GSN), fibronectin (FN1), versican (VCAN), fibrinogen (FGB), myosin type 1 C (MYO1C), and collagen type IV α (COL4A) corresponded to perivascular clotting and hemorrhagic areas. The diverse staining patterns across patients NU02545 (Figure 5A and Supplemental Figure 7), NU02594 (Figure 5B, Supplemental Figure 8, and Supplemental Video 1), and NU02569 (Figure 5C, Supplemental Figure 9, and Supplemental Video 2) indicated patient-specific variations in ECM composition and the immune response. Such heterogeneity could explain the inconsistency of BVax-derived Ig reactivity in different patients with GBM. Additionally, we observed B cells in regions near BVax-derived, Ig-recognized antigens (Supplemental Figure 10), and peritumoral brain had low expression of these ECM proteins compared the GBM microenvironment itself (Supplemental Figure 11), indicating therapeutic potential for BVax-derived Igs.
Figure 4. Characterization of BVax-derived Ig reactivity in patients with GBM.
(A) Schema depicting the protocol for the human IP-MS experiments used to identify tumor-specific antigens recognized by BVax-derived Abs. (B) Heatmap revealing hierarchical clustering of GBM tumor antigens recognized by BVax-derived Igs (n = 3). Targets related to adhesion, motility, or the ECM are shown in red.
Figure 5. BVax-derived, Ig-recognized antigens are part of ECM.
Representative spatial multiplex IF images generated using the COMET system (Lunaphore Technologies) from paired GBM patients (n = 3) showing that BVax-derived, Ig-recognized antigens are part of the GBM ECM (including versican, fibronectin, and COL4A), ECM modulators (gelsolin), and proteins involved in cell adhesion and motility (MYO1C and fibrinogen). (A) Images for patient NU02545. Scale bars: 2 mm. (B) Images for patient NU02594. Scale bars: 200 μm. (C) Images for patient NU02569. Scale bars: 200 μm. CD31 (endothelial cells), GFAP (glioma tumor cells and astrocytes), CD163 (macrophage scavenger receptor), CD206 (immunosuppressive macrophages).
BVax-Igs inhibit GBM invasion and migration.
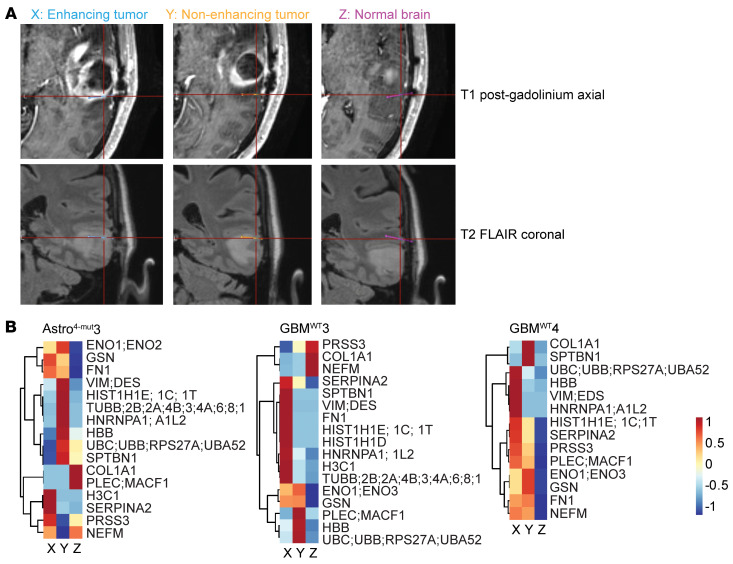
To determine whether patient BVax-derived, Ig-recognized antigens are accessible to the immune system, we performed intraoperative high-molecular-weight microdialysis, which collects proteins secreted into the interstitial fluid space (40), in the tumor and brain tissue adjacent to the tumors of patients with GBM (GBMWT3, and GBMWT4) and in 1 patient with a grade 4 isocitrate dehydrogenase–mutant (IDH-mutant) astrocytoma (Astro4-mut3), followed by MS analysis (Figure 6A). High-molecular-weight catheters (100 kDa) were applied to maximize the volume and diversity of the microdialysis (40). During the resection, catheters were placed in radiographically enhancing tumor (X) and nonenhancing tumor (Y), and in relatively normal brain adjacent to the tumor (Z). MS of the microdialysates revealed that most BVax-derived, Ig-recognized antigens were more abundantly secreted in the enhancing tumors than in normal brains, although 1 patient showed secretion of more BVax-derived, Ig-recognized antigens in the nonenhancing tumors (Figure 6B and Supplemental Table 9). Some BVax-derived, Ig-recognized antigens, such as myosin (MYH) (Figure 4B), were not identified in the microdialysates (Figure 6B). One possible explanation could be that their size surpassed the molecular weight cutoff of the catheters (>100 kDa). Overall, these findings demonstrate the presence of diverse antigenic profiles across heterogeneous zones within each tumor and the potential accessibility of these antigens to the immune system. The presence of these BVax-derived, Ig-recognized antigens in the extracellular fluid reinforces the notion that the tumor microenvironment (TME) is a potential reservoir of significant therapeutic targets.
Figure 6. BVax-derived, Ig-recognized antigens are detected in the extracellular fluid with brain microdialysis.
(A) T1 post-gadolinium axial and T2 fluid-attenuated inversion recovery (FLAIR) coronal MRIs demonstrating the stereotactic target location of each catheter in enhancing tumor, nonenhancing tumor, and normal brain (MRI for patient GBMWT3 is shown). (B) Heatmap revealed the relative intensity of high-grade glioma antigens recognized by BVax-derived Igs in the micro dialysate. Samples were collected from 3 distinct patients, different from those in Figure 5.
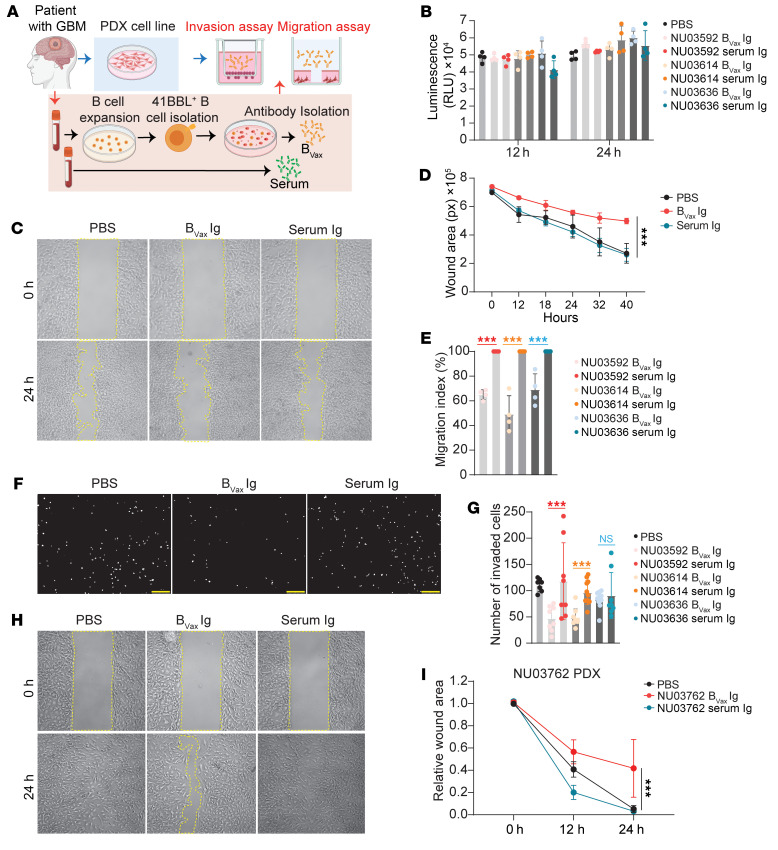
The ECM and cell motility are key biological processes controlling tumor cell migration, invasion, and epithelial-mesenchymal transition, which are hallmarks of GBM malignant behavior (41–44). Thus, we hypothesized that BVax Igs could inhibit ECM and cell motility processes by recognizing these components or regulators. To test this hypothesis, we used a patient-derived xenograft (PDX) cell line (GBM43) and BVax Igs from 3 patients with GBM (NU03592, NU03614, and NU03636). Igs from autologous plasma samples were used as a control. We performed an ex vivo PDX functional assay including invasion and migration assessment using a commercially available matrix that recapitulates mammalian ECM (Matrigel) (Figure 7A). BVax Igs did not affect PDX cell viability (assessed by ATP activity, Figure 7B) compared with paired serum Igs. However, BVax Igs significantly inhibited the PDX cells from migrating (Figure 7, C–E) and invading (Figure 7, F and G). Additionally, we conducted a migration assay using BVax Igs from patient NU03762 and the corresponding PDX cell line (NU03762 PDX). Consistent with our initial findings, BVax Igs significantly inhibited migration of the paired PDX cell line (Figure 7, H and I). These results support the potential of BVax Igs to interfere with key processes involved in GBM progression.
Figure 7. BVax-derived Igs inhibit tumor invasion and migration.
(A) Schema depicting the protocol for the ex vivo functional assay. (B) Cell viability of GBM43 cells after treated with serum- or BVax-derived Igs from patients with GBM (NU03592, NU03614, and NU03636). n = 3. (C) Representative images of wound areas (marked by yellow lines) on confluent monolayers of GBM43 cells at 0 hours and 24 hours; cells were treated with serum- or BVax-derived Igs from a patient with GBM (NU03592). Original magnification, ×4 (C, F, and H). (D) Quantification of the wound area at different time points of GBM43 cells treated with serum- or BVax-derived Igs from a patient with GBM (NU03592). (E) Quantification of the migration index of GBM43 cells at 24 hours that were treated with serum- or BVax-derived Igs from patients with GBM (NU03592, NU03614, and NU03636). n = 3. (F) Representative images and (G) quantification of invading GBM43 cells (DAPI+) at 24 hours; cells had been treated with serum- or BVax-derived Igs from patients with GBM (NU03592, NU03614, and NU03636). n = 3. Each white dot represents a single invaded cell. Scale bars: 250 μm. (H) Representative images of wound areas (marked by yellow lines) on confluent monolayers of PDX cells at 0 hours and 24 hours; cells had been treated with serum- or BVax-derived Igs from the same patient (NU03762). (I) Quantification of the wound area of PDX cells at different time points; cells had been treated with serum- or BVax-derived Igs from the same patient (NU03762). Data are the mean ± SD. ***P < 0.001, by 1-way ANOVA (B, E, and G) or 2-way ANOVA (D and I).
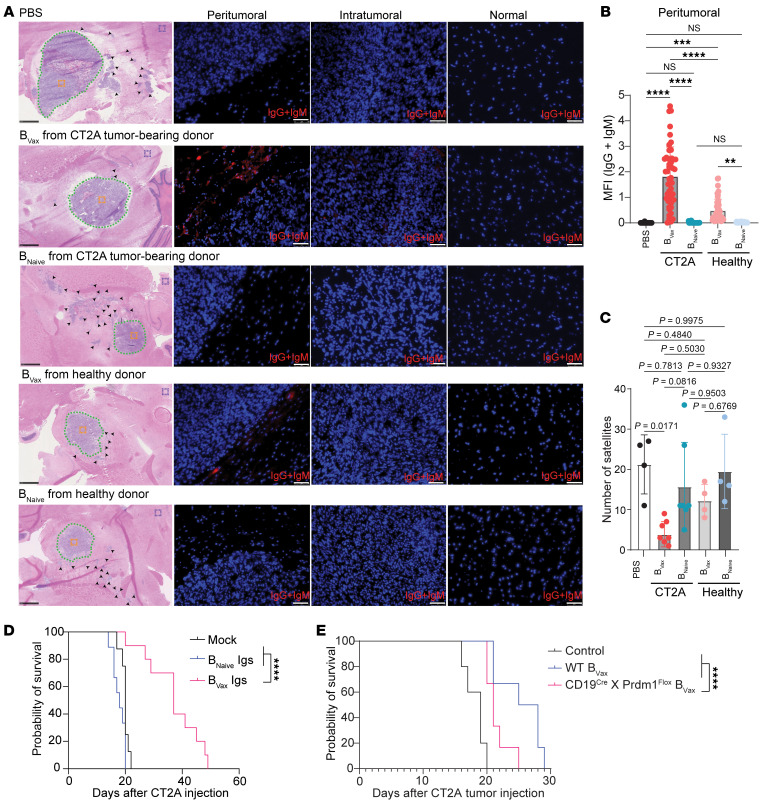
To provide direct evidence of BVax-induced Igs in tumors, we conducted additional experiments in which BVax cells from healthy or CT2A tumor–bearing C57BL/6 mice were adoptively transferred into CT2A tumor–bearing muMT (B cell–KO) mice. Following treatment, we harvested brain tissues from recipient mice and performed immunofluorescence (IF) staining for anti–mouse IgG and IgM to assess the presence and localization of BVax-derived Abs. Our results demonstrated that mice receiving BVax cells from CT2A tumor–bearing donors had significant IgG and IgM staining in the peritumoral region compared with those treated with BVax cells derived from healthy donors or with BNaive cells (Figure 8, A and B). Similar results were observed in the GL261 model, in which BVax or BNaive cells from GL261 tumor–bearing C57BL/6 mice were adoptively transferred into GL261 tumor–bearing muMT (B cell–KO) mice (Supplemental Figure 12). Histological analysis of muMT mice treated with BVax cells from CT2A tumor–bearing donors showed a decrease in satellite formation away from the tumor core, indicating the disruption of cell invasion (Figure 8, A and C). Consistent with our previously reported results (13), only mice with orthotopically implanted CT2A tumors treated with BVax Igs had a significant increase in median survival (Figure 8D). To further assess the importance of the role of BVax Igs, we conducted experiments in which we treated CT2A tumor–bearing mice with BVax generated from WT mice or mice with B cells deficient in Prdm1 (Cd19Cre Prdm1fl mice, provided by Nicole Baumgarth, Johns Hopkins Medicine, Baltimore, Maryland, USA). Prdm1 encodes Blimp1, a key factor for the development of Ig-secreting plasma cells (45, 46). Notably, the survival of Prdm1-deficient, BVax-treated mice was further reduced (Figure 8E), underscoring the essential role of BVax-induced Igs in mediating the survival benefits observed with BVax treatment.
Figure 8. BVax cells from tumor-bearing mice have a superior ability to produce Abs that localize to the peritumoral region and promote survival of GBM-bearing mice survival.
(A) Representative images of H&E and IF staining for anti–mouse IgG and IgM to assess the presence and localization of BVax-derived Igs. BVax or BNaive cells from healthy or CT2A tumor–bearing C57BL/6 mice were adoptively transferred into CT2A tumor–bearing muMT mice. Following treatment, brain tissues were harvested from recipient mice and stained for anti–mouse IgG and IgM (red). H&E-stained images show the organization of the tumors and the locations where the IF images were taken: peritumoral region (dotted green line), intratumoral region (orange box), and relatively normal brain (purple box). (B) Quantification of the relative intensity of BVax-Igs in the peritumoral region. A total of 10–15 images were taken around the peritumoral region in each mouse (dotted green line). The MFI of anti–mouse IgG and IgM (red) in each image was quantified using ImageJ as described previously (68, 69). PBS group: n = 3; BVaxCT2A group: n = 5; BNaiveCT2A group: n = 5; BVaxhealthy group: n = 3; BNaivehealthy group: n = 3. Data are representative of 2 independent experiments. (C) Quantification of satellites (black arrowhead) away from the CT2A tumor core based on H&E images from each mouse. PBS group: n = 3; BVax-CT2A group: n = 5; BNaiveCT2A group: n = 5; BVax healthy group: n = 3; BNaivehealthy group: n = 3. The data are representative of 2 independent experiments. (D) Survival of CT2A tumor–bearing mice was evaluated in 3 groups: mock-treated (n = 8), BNaive Ig–treated (n = 9), and BVax Ig–treated (n = 10). (E) Survival of CT2A tumor–bearing muMT mice was assessed according to the 3 treatment groups: PBS control (n = 5), WT BVax (n = 6), and Prdm1-deficient BVax (n = 6). Data are presented as the mean ± SD. **P < 0.01, ***P < 0.001, and ****P < 0.0001, by 1-way ANOVA (B and C) or log-rank test (D and E).
On the basis of these results, we conclude that BVax has the potential to produce Abs reactive to the tumor ECM and components of the cell motility and could interfere with the ability of the tumor to migrate and invade nontumor tissue and to ultimately affect overall tumor growth.
Discussion
Here, we show that BVax elicited antitumor reactivity, as evidenced by selective migration to glioma-bearing brains, differentiation into plasmablasts, and secretion of specific Igs. BVax-derived Igs bind to factors predominantly involved in cell motility and the ECM, essential for GBM invasion and motility (42, 47–49). These Abs are also functionally active against the key processes of cancer progression, revealing a strategy for developing novel immunotherapeutic strategies against GBM.
The BVax-derived Ig recognition of specific ECM components, such as gelsolin, fibronectin, fibrinogen, versican, and collagens, is particularly intriguing. Traditionally seen as a physical scaffold for cells, the ECM is now increasingly recognized for its role in modulating tumor behavior, progression, and response to therapy (43, 44). The ECM and the hypoxic microenvironment orchestrate the mesenchymal transition, a biological process associated with the aggressive pathological properties of GBM and therapeutic resistance (49–51). Recently, the finding of collagen 1 α1–abundant (COL1A1-abundant) oncostreams (52) and cancer-associated fibroblasts (53) in GBM and their protumor effects reinforce the concept that components of the ECM might not be mere bystanders but could actively participate in the progression of GBM. The preferential reactivity of BVax Abs observed in murine models and human GBM samples highlights its translational potential.
To date, relatively few antigens recognized by TIB cell–derived Abs have been identified in other cancer models, partially limited by the low TIB numbers from fresh tumors (54–60). Here, by expanding and differentiating BVax into Ab-secreting plasmablasts, we were able to identify potential antigens recognized by BVax-derived Abs in mouse and human GBM models. This method can also be used in other cancer models, expanding the repertoire of TIB target antigens.
Most TIB target antigens identified in patient tissue samples span across the nuclear, cytoplasmic, and extracellular compartments (24). Our data demonstrated that cytoplasmic and extracellular proteins could both be recognized by BVax Abs. While it is expected that B cell–derived Igs could recognize extracellular proteins, the detection of intracellular cytoplasmic proteins was surprising. However, the analysis of the GBM secretome obtained from microdialysis of enhancing tumor, nonenhancing tumor, and normal brain showed that cytoplasmic proteins can be detected in the extracellular fluid, hinting that cytoplasmic proteins are also accessible to the immune system. However, the mechanism behind this is still not clear. One could hypothesize that soluble antigens from the interstitial fluid could travel to regional lymph nodes, such as the deep cervical lymph nodes (61), and potentially activate B cells.
Although our data indicate that BVax Igs contributed significantly to improved survival, it is essential to consider the potential contributions of other immune mechanisms of BVax, such as CD8+ T cell activation, which also plays an important role in the BVax-conferred therapeutic effect (13, 62). Further studies are needed to delineate the interplay between BVax Igs and other immune mechanisms of BVax and how these mechanisms collectively enhance patient outcomes.
The heterogeneity observed in the BVax antigenic reactivity across different patients with GBM reminds us of the complex and diverse nature of GBM tumors. Although certain regulators like fibrinogen, myosin, and collagens emerged consistently, future research should aim at identifying and characterizing other potential tumor antigens bound by BVax Igs using a larger cohort of patient samples. In conclusion, our research highlights the therapeutic potential of BVax-derived Igs in GBM therapy. Through their unique tumor-reactive nature, BVax and the Abs they produce offer a promising strategy against this formidable malignancy. Future studies should focus on how to unleash the full potential of BVax in an immune-suppressive GBM TME.
Methods
Sex as a biological variable.
In this study, both male and female mice were used in the animal model experiments to investigate the effects of B cell–based therapy on GBM growth. However, sex was not considered as a biological variable in any of the analyses. For studies involving human samples, sex was also not considered as a biological variable because of the limitations in sample availability and the specific focus of the study.
Human specimens.
The Nervous System Tumor Bank collected all human tissue samples at Northwestern University and involved 9 patients with IDH-WT GBM. The Mayo Clinic Cancer Center collected all human tissue samples at the Mayo Clinic. Samples collected from patients with high-grade glioma included freshly resected tumors, peripheral blood, frozen tumors, and paraffin-embedded tissue sections. A neuropathologist reviewed all H&E sections to confirm that the sample included the presence of least 50% tumor cells based on cellularity.
Cell lines.
CT2A cells were obtained from MilliporeSigma, GL261 cells were obtained from the National Cancer Institute (NCI), NIH, and GBM43 PDX glioma cell lines were obtained from David James (Northwestern University, Chicago, Illinois, USA). NU03762 PDX cells were obtained from Craig M. Horbinski (Northwestern University). NU03762 PDX cells were maintained in complete RPMI, consisting of RPMI supplemented with 10% FBS (Hyclone), 100 U/mL penicillin (Corning), 100 mg/mL streptomycin (Corning), 0.1% 2-mercaptoethanol (MilliporeSigma), 2 mM l-glutamine (Invitrogen, Thermo Fisher Scientific), 25 mM HEPES (Invitrogen), and 1 mM sodium pyruvate (Invitrogen, Thermo Fisher Scientific). The other cell lines (CT2A, GL261, GBM43) were maintained in DMEM (Corning) supplemented with 10% FBS (Hyclone), 100 U/mL penicillin (Corning), and 100 mg/mL streptomycin (Corning) and incubated at 37°C in 5% CO2. Every 2 months, the cell lines were tested for mycoplasma contamination using the Universal Mycoplasma Detection Kit (American Type Culture Collection [ATCC], 30-1012K).
Murine models.
Mice used in this study included C57BL/6 and muMT mice purchased from The Jackson Laboratory and bred for use in the experiments. Cd19Cre Prdm1fl mice were provided by Nicole Baumgarth (Johns Hopkins Medicine, Baltimore, Maryland, USA). Animal experiments were initiated when the mice were 6–8 weeks old. All animals were housed at the Simpson Querry Center for Comparative Medicine in a dedicated pathogen-free animal facility with 12-hour light/12-hour dark cycles and ad libitum access to food and water. For in vivo studies, the sample size for each experiment is indicated in the figure legend. The investigators were not blinded to the groups for any experiments. This study incorporated sex as a biological variable by including both male and female mice.
Intracranial tumor implantation.
Each mouse was implanted with 1 × 105 glioma cells in a total volume of 2.5 μL PBS. Mice were anesthetized with ketamine (100 mg/kg) and xylazine (10 mg/kg) via intraperitoneal injection. After shaving the surgical site and disinfecting with povidone-iodine and 70% ethanol, an incision was made at the midline to access the skull. A 1 mm diameter burr hole was drilled 2 mm posterior to the coronal suture and 2 mm lateral to the sagittal suture. Injections were performed using a Hamilton syringe fitted with a 26 gauge blunt needle at a depth of 3.5 mm. The injection site was then sutured closed.
BVax/BNaive generation.
Cells were generated as described previously (13, 14). Briefly, 4-1BBL+ B cells were isolated and activated with anti-CD40 Ab (CD40 agonism), supplemented with BAFF (B cell survival factor), and stimulated with IFN-γ in complete RPMI media (cRPMI) (RPMI 1640 supplemented with 10% FBS, sodium pyruvate, MEM amino acids, HEPES, 2-mercaptoethanol, and penicillin/streptomycin) to generate BVax. The 4-1BBL– B cells were cultured with BAFF in cRPMI to generate BNaive.
GBM patient–derived BVax/BNaive Ig generation.
BVax/BNaive cells were cultured using the ImmunoCult Human B Cell Expansion Kit (100-0645, STEMCELL Technologies). Cells were seeded at a density of 5 × 105 cells/mL in B cell expansion medium and incubated at 37°C in a CO2 incubator. Every 3 days, supernatants were harvested and quantified for IgG using the IgG (total) Human Uncoated ELISA Kit (88-50550-22, Invitrogen). The supernatants were then stored at –80°C, and cells were replenished with fresh expansion medium. After several rounds of supernatant collection, all supernatants were pooled, and the Igs were purified using a NAb Protein A/G Spin Column (89962, Thermo Fisher Scientific).
Murine BVax/BNaive Ig generation.
The generation of murine Igs is depicted in Figure 2A. Briefly, BVax/BNaive cells were adoptively transferred into muMT mice. After 2 weeks, blood from BVax/BNaive recipient mice was collected, and Igs were isolated using a Protein A/G Spin Column.
GBM patient–derived tumor lysate for IP-MS.
Fresh tumors and paired blood samples from patients NU02545, NU02569, and NU02594 were collected by the Northwestern Brain Tumor Bank. Tumors were homogenized in NP40 buffer (40 mM HEPES, 120 mM NaCl, 1 mM EDTA, 10 mM NaPP, 50 mM NaF, 0.5% NP40, and 10 mM β-glycerophosphate) to which protease and phosphatase inhibitors were added (MilliporeSigma). Protein quantification was performed using a Bradford Assay (Bio-Rad, catalog 500-0006) according to the manufacturer’s guidelines.
Murine GBM tumor lysate for IP-MS using cellular fractionation.
CT2A cells were implanted intracranially into C57BL/6 mice. After 21 days, the brains were collected, and the tumors were dissected and flash-frozen. As per the manufacturer’s guidelines, cellular fractionation was performed with the Thermo Fisher Mem-PER Plus Kit (catalog 89842Y). Protein quantification was performed using a Bradford Assay (Bio-Rad, catalog 500-0006) according to the manufacturer’s guidelines.
Immunoprecipitation.
Cell/tissue lysates were incubated and tumbled overnight at 4°C with either BVax-derived Igs or IgG control. BVax-derived Igs were generated in vivo for the murine samples and ex vivo for patient-derived samples as described in Figure 2A and Figure 3A. Pulldown experiments were then performed with magnetic Dynabeads Protein G (Thermo Fisher Scientific). For GBM patient–derived samples, 1–2 mg tumor lysate was used with 20–60 μg Igs. For murine intracranial GBM tumor samples, 1.5 mg tumor lysate was used with 30 μg Igs. For GBM43 cell–derived samples, 2–4 mg membrane or conditioned medium proteins were used with 40–120 μg Igs. For CT2A cell–derived samples, 8 mg membrane or conditioned medium proteins were used with 160 μg Igs. Bead-protein complexes were isolated and then washed 3 times with lysis buffer and 2 times with Triton X buffer (40 mM HEPES, 120 mM NaCl, 1 mM EDTA, 10 mM NaPP, 50 mM NaF, 0.5% Triton X, and 10 mM β-glycerophosphate). The beads were then boiled for 10 minutes at 95°C in 2X Laemmli sample buffer, 20% of each sample was resolved by SDS-PAGE, and 80% was submitted to the proteomics core for MS analysis.
Proteomics immunoprecipitation analysis using LC-MS/MS.
Liquid chromatography tandem MS (LC-MS/MS) was performed as previously described (63). For visualization of protein targets identified through IP followed by MS, R programming language was used, leveraging specialized packages such as “ggplot2” and “pheatmap.” Our datasets encompassed protein measurements derived from 3 tumor samples from patients with GBM, in vitro cultures, and mouse samples, which included protein targets extracted from control Ig, BNaive or serum Ig, and BVax Ig groups. For heatmap visualization, the Ward’s method (64) was adopted to cluster samples, ensuring a more coherent and intuitive presentation.
Flow cytometry.
Flow cytometry was performed as described previously (13). The following anti-human Abs were used (all from BioLegend): 4-1BBL PerCP-Cy5.5 (5F4) (catalog 311518), CD19 Pacific blue (HIB19) (catalog 302224), CD20 BV510 (2H7) (catalog 302340), and CD38 Alexa Fluor 700 (HB-7) (catalog 356623). All samples were analyzed on a BD Symphony flow cytometry analyzer with proper single-color controls and compensation. All final analysis and data output were performed using FlowJo software (BD).
Western blot analysis.
BVax was generated ex vivo from peripheral blood samples from patients with GBM and subsequently activated as described previously (13). Supernatants were collected every few days during the activation protocol and prepared for Western blotting in nonreduced and reduced fractions. Reduced samples were prepared using 4× Laemmli Sample Buffer (Bio-Rad). Equal volumes of supernatant were loaded onto a gel. Supernatants were used as the primary Ab. The secondary Ab used was anti–human IgG-peroxidase (MilliporeSigma, A0293).
BCR sequencing.
BVax and BNaive were generated as previously described (14). Then, cells were washed twice with PBS. TIB cells were magnetically isolated using CD19-biotin (clone 6D5, BioLegend, 115504) and anti-biotin Microbeads (Miltenyi Biotec, 130-090-485). RNA isolation from BVax, BNaive, and TIB cells was performed using TRIzol (Invitrogen). BCR sequencing and bioinformatics analysis were performed by Adaptive Biotechnologies using the ImmunoSEQ platform.
Sequential IF multiplex staining.
The multiplex panel included the following unconjugated Abs: CD31 (endothelial cells; Abcam, EPR3131), GFAP (glioma tumor cells and astrocytes; Abcam, EPR1034Y), CD163 (macrophage scavenger receptor; Abcam, EPR19518), CD206 (immunosuppressive macrophages; Abcam, ab64693), COL4A (collagen 4 subtypes A1/A2; Abcam, ab6586), versican (MilliporeSigma, HPA004726), fibrinogen (Abcam, ab34269), fibronectin (Abcam, ab2413), gelsolin (Cell Signaling Technology, D9W8Y), MYO1C (MilliporeSigma, HPA001768), and CD20 (Dako Agilent Technologies, L26). All Abs were validated using conventional IHC and/or IF staining in conjunction with the corresponding fluorophore and the spectral DAPI (Thermo Fisher Scientific, 62247) counterstain. For optimal concentration and the best signal/noise ratio, all Abs were tested at 3 different dilutions, starting with the manufacturer-recommended dilution (MRD), then MRD/2 and MRD/4. Secondary Alexa Fluor 555 (Thermo Fisher Scientific, A32727) and Alexa Fluor 647 (Thermo Fisher Scientific, A32733) were used at 1:200 and 1:400 dilutions, respectively. The optimizations and full runs of the multiplex panel were executed using the sequential IF (seqIF) methodology integrated into the Lunaphore COMET platform (characterization 2 and 3 protocols, and seqIF protocols, respectively; ref. 65). Staining was done on a maximum of 4 tissue slides simultaneously, where automated cycles of 2 Abs were stained at a time, followed by fully automated imaging and elution, with no sample manipulation required. All reagents were diluted in Multistaining Buffer (BU06, Lunaphore Technologies). The elution step lasted 2 minutes for each cycle and was performed with Elution Buffer (BU07-L, Lunaphore Technologies) at 37°C. Quenching lasted 30 seconds and was performed with Quenching Buffer (BU08-L, Lunaphore Technologies). Staining incubation durations were 4 minutes for all primary Abs and 2 minutes for secondary Abs. Imaging was performed in Imaging Buffer (BU09, Lunaphore Technologies) with an integrated epifluorescence microscope at ×20 magnification. Image registration was performed immediately after conclusion of the staining and imaging procedures by COMET Control Software. Each seqIF protocol resulted in a multilayer OME-TIFF file, in which the imaging outputs from each cycle were stitched and aligned. COMET OME-TIFF files contained a DAPI image, intrinsic tissue autofluorescence in TRITC and Cy5 channels, and a single fluorescent layer per marker. Markers were subsequently pseudocolored for visualization of markers in the Viewer from Lunaphore.
Intraoperative microdialysis.
Intraoperative microdialysis was performed at Mayo Clinic during 3 standard-of-care glioma resections based on previously published methods (40), including 1 for a recurrent grade 4 IDH-mutant astrocytoma (Astro4-mut3), 1 primary GBM (GBMWT3), and 1 recurrent GBM (GBMWT4). Briefly, 3 high-molecular-weight microdialysis catheters (100 kDa; M Dialysis 71 High cutoff brain microdialysis catheters) were inserted into radiographically diverse regions (enhancing, nonenhancing, and normal brain) based on stereotactic neuronavigation. Microdialysis was performed at a flow rate of 2 μL/min using the 107 microdialysis pump and perfusion fluid with 3% dextran 500 kDa to improve analyte recovery. Catheters were flushed prior to insertion to minimize dead space, and microvials were then changed every 20 minutes until the sampling area needed to be resected. Microdialysates were split into 2 aliquots and then placed on dry ice. The third aliquot after catheter insertion was sent for MS proteomics analysis at the Mayo Clinic Proteomics Core via LC-MS/MS for label-free relative quantitation by intensity-based abundance quantity (iBAQ).
Migration assay.
This assay used a silicone insert with a defined cell-free gap (80206, IBIDI), as described previously (66). Briefly, cells were seeded at 3.5 × 104 per well of culture insert chamber (80206, IBIDI). Cells were left undisturbed for approximately 12 hours in a 37°C CO2 incubator. After successful attachment, cells were then washed with Dulbecco’s PBS (DPBS), and the silicone insert was carefully lifted using sterile forceps. Culture medium was switched to cDMEM containing 100 μg/mL BVax or serum Abs. At different time points, imaging was obtained by bright-field via Leica Microscope until the visible gap closed. The distance or area between cells was measured via pixels on Fiji ImageJ. Migration index was calculated as follows: migration index (%) = (wound area at 0 hours – wound area at 24 hours)/wound area at 0 hours × 100.
Invasion assay.
For the invasion assay, a Corning’s Matrigel Invasion Chamber (354480, Corning) was used. Briefly, after rehydration of the chamber, 5 × 104 cells were suspended in DMEM and 100 μg/mL BVax or serum Abs and seeded into the invasion chamber. The bottom chambers were filled with 750 μL cDMEM, which contained DMEM, 10% FBS, 100 U/mL penicillin, and 100 mg/mL streptomycin. After a 24-hour incubation, invaded cells were fixed overnight in 4% formaldehyde and then washed and stained with DAPI (P36931, Thermo Fisher Scientific) as described previously (67). The membrane was imaged, and invading cells were counted using Fiji ImageJ.
Cell viability assay.
The CellTiter-Glo 2.0 Cell Viability Assay kit (G9242, Promega) was used to assess viability of PDX cells after treatment with BVax-derived Abs or serum Abs (100 μg/mL), as described by the manufacturer.
Direct evidence of BVax-induced Igs in tumors.
In the CT2A glioma model, BVax or BNaive cells from healthy or CT2A tumor–bearing C57BL/6 mice were adoptively transferred into CT2A tumor–bearing muMT mice 7 days after tumor injection. Each mouse received 2 × 106 cells every 3 days. Seven days after the third treatment, brain tissues were harvested from recipient mice, frozen with OCT and sectioned, and then stained for anti–mouse IgG and IgM (Cy3 AffiniPure Goat Anti–mouse IgG + IgM [H+L], 1:500, Jackson ImmunoResearch, 115-165-044). Because muMT mice do not have endogenous Igs, using anti–mouse Ig will detect only BVax- or BNaive-derived Abs, allowing for specific identification of these Abs. Nearby sections were stained for H&E to check the organization of the tumors. IF images were taken from the peritumoral region, the intratumoral region, and relatively normal brain. For quantification of Ig intensity, 10–15 images were taken around the peritumoral region in each mouse. The MFI of anti–mouse IgG and IgM (red) in each image was quantified as previously described using ImageJ (NIH) (68, 69). To evaluate the invasive feature of CT2A tumor cells after treatment, satellites away from the CT2A tumor core were quantified on the basis of H&E images from each mouse (70). In GL261 glioma model, BVax or BNaive cells from GL261 tumor–bearing C57BL/6 mice were adoptively transferred into GL261 tumor–bearing muMT mice 7 days after tumor injection. Each mouse received 2 × 106 cells every 3 days. Seven days after the second treatment, brain tissues from recipient mice were harvested, stained and quantified as in the CT2A glioma model.
Essential role of BVax induced Igs.
CT2A tumor–bearing, B cell–deficient mice were treated with purified BVax or BNaive Igs (12.5 μg/mouse/injection) as described previously (13), and their survival was monitored. Additionally, CT2A tumor–bearing, B cell–deficient mice were treated with106 BVax generated from WT or Cd19Cre Prdm1fl mice (B cells deficient in Prdm1 were provided by Nicole Baumgarth, Johns Hopkins Medicine, Baltimore, Maryland, USA), and their survival was monitored.
Statistics.
GraphPad Prism version 8 (GraphPad Software) and R version 4.2.3 (R Foundation for Statistical Computing) were used for all statistical analyses. The sample size for the experiments was 3 or more. Results are represented as the mean ± SD unless otherwise indicated. Comparisons between 2 groups were conducted using a 2-tailed Student’s t test. Comparisons between more than 2 groups were conducted using a 1-way ANOVA with Tukey’s or Dunnett’s post hoc multiple-comparison test. For animal survival experiments, Kaplan-Meier survival curves were generated, and a log-rank test was applied to compare survival distributions. All reported P values are 2 sided and were considered statistically significant at a P value of less than 0.05.
Study approval.
All animal experiments conducted in this study were reviewed and approved by the IACUC of Northwestern University under protocol number ISO16696. The study protocols adhered to the IACUC’s guidelines to ensure the ethical treatment of animals. For studies involving human samples, the research was reviewed and approved by the IRB of Northwestern University (protocol no. STU00202003) and the Mayo Clinic (protocol no. 19-004694). All patients who contributed to this study signed a written consent form, and the study was conducted according to the US Common Rule of Ethical Standards.
Data availability.
The mass spectrometric proteomics data were deposited in the ProteomeXchange Consortium database via the PRIDE (71) partner repository and are available via ProteomeXchange (accession code PXD046712; project webpage: http://www.ebi.ac.uk/pride/archive/projects/PXD046712; FTP download: https://ftp.pride.ebi.ac.uk/pride/data/archive/2024/09/PXD046712). Additionally, the supporting data values for all figures and analyses are provided in the Supporting Data Values file, which is included with the online supplemental material.
Author contributions
CLC conceived the project. BAC, SW, MF, and CLC designed the study. SW, BAC, JLK, HN, VA, GVC, HW, IEO, DH, VA, MD, LKB, TYC, CW, AR, PZ, LCP, KM, CRC, TB, MF, and CLC were involved in data acquisition. BAC, SW, and CLC performed the statistical analysis. SW, BAC, HN, JM, MSL, AUA, CRC, TB, CMH, RS, ABH, AMS, MF, and CLC were involved in the interpretation of data. AR was responsible for the mouse colonies. BAC helped with animal surgery. SW, BAC, and CLC prepared the manuscript.
Supplementary Material
Acknowledgments
BC is supported by the Neurosurgery Research and Education Foundation (NREF), the Stach Strong Foundation, and the Glioblastoma Foundation. CLC is supported by National Cancer Institute (NCI), NIH (R37CA258426 and P50CA221747) and the Cancer Research Institute (CR68036). MSL is supported by the NCI, NIH (P50CA221747 and R35CA197725) and the National Institute of Neurological Diseases (NINDS), NIH (R01NS115955) DH is supported by the American Brain Tumor Association (ABTA) Jack and Fay Netchin Medical Student Fellowship in honor of Paul Fabbri and a Northwestern RISE research fellowship award. CRC is supported by the NIH (T32 GBM145408). TCB is supported by the NCI, NIH (R37 CA276851). The authors of this project are supported by the Cancer Research Institute and the Malnati Brain Tumor Institute. The Northwestern Nervous System Tumor Bank is supported by the Specialized Program of Research Excellence (SPORE) for Translational Approaches to Brain Cancer (P50CA221747). The spatial multiplex analysis is supported by NIH grants CA120813 and NS120547. The Flow Cytometry Core Facility is supported by a NCI Cancer Center Support grant (CA060553). Proteomics services were performed by the Northwestern Proteomics Core Facility, generously supported by a grant from the NCI, NIH (CCSG P30 CA060553), awarded to the Robert H. Lurie Comprehensive Cancer Center; an instrumentation award (S10OD025194) from NIH Office of the Director; and the National Resource for Translational and Developmental Proteomics, supported by P41 GM108569. The Mayo Clinic Cancer Center is supported by the NCI, NIH (P30 CA15083).
Schematic illustrations were created using BioRender.com and Adobe illustrator. The Lunaphore COMET system for multiplex IF was enabled by a gift from the Stephen M. Coffman trust to the Northwestern Medicine Malnati Brain Tumor Institute of the Lurie Cancer Center. We give special thanks to Bella Najem, BA in Fashion Design and Media, for her media and animated video generation expertise. We also acknowledge the assistance of the Mayo Clinic Proteomics Core, a shared resource of the Mayo Clinic Cancer Center.
Version 1. 08/29/2024
In-Press Preview
Version 2. 10/15/2024
Electronic publication
Funding Statement
Catalina Lee-Chang
Catalina Lee-Chang
Maciej S. Lesniak
Maciej S. Lesniak
Cécile Riviere-Cazaux
Terry Burns
Spatial multiplex analysis
The Flow Cytometry Core Facility
Northwestern Proteomics Core Facility
Robert H Lurie Comprehensive Cancer Center instrumentation award
Mayo Clinic Cancer Center
Footnotes
BAC’s present address is: Department of Neurological Surgery, University of Illinois Chicago, Chicago, Illinois, USA.
MF’s present address is: Grove Biopharma, Inc. Chicago, Illinois, USA.
Conflict of interest: ABH serves on the advisory board of Caris Life Sciences and the WCG Oncology Advisory Board; receives royalty and milestone payments from DNAtrix for the licensing of the patent “Biomarkers and combination therapies using oncolytic virus and immunomodulation” (no. 11,065,285); is supported by research grants from Celularity, Alnylam, and AbbVie; and receives consulting fees from Novocure and Istari Oncology. She additionally has active granted patents titled “miRNA for treating cancer and for use with adoptive immunotherapies” (no. 9,675,633) and “Concurrent chemotherapy and immunotherapy” (no. 9,399,662), with a patent-pending titled “Low-intensity ultrasound combination cancer therapies” (international applications PCT/US2022/019435 and US 63/158,642). MF is currently employed as a principal scientist at Grove Biopharma Inc. CLC has pending patents titled “B cell–based immunotherapy for the treatment of glioblastoma and other cancer” (PCT/US2021/046331 and US 62/936,867) and “B cell production and expansion” (US63/590,202).
Copyright: © 2024, Wang et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: J Clin Invest. 2024;134(20):e177384. https://doi.org/10.1172/JCI177384.
Contributor Information
Si Wang, Email: si.wang@northwestern.edu.
Brandyn A. Castro, Email: brandyn.castro@northwestern.edu.
Joshua L. Katz, Email: joshua.katz2@northwestern.edu.
Hinda Najem, Email: hinda.najem@northwestern.edu.
Gustavo I. Vazquez-Cervantes, Email: guigvace@northwestern.edu.
Hanxiao Wan, Email: hanxiao.wan@northwestern.edu.
Ian E. Olson, Email: ian.olson1@northwestern.edu.
David Hou, Email: david.hou@northwestern.edu.
Mark Dapash, Email: mark-dapash@northwestern.edu.
Leah K. Billingham, Email: leah.billingham@northwestern.edu.
Chao Wei, Email: cwei7023@gmail.com.
Aida Rashidi, Email: aida.rashidi@northwestern.edu.
Leonidas C. Platanias, Email: I-platanias@northwestern.edu.
Kathleen McCortney, Email: Kathleen.mccortney@northwestern.edu.
Craig M. Horbinski, Email: craig.horbinski@northwestern.edu.
Roger Stupp, Email: roger.stupp@usz.ch.
Peng Zhang, Email: peng@northwestern.edu.
Atique U. Ahmed, Email: atique.ahmed@northwestern.edu.
Adam M. Sonabend, Email: Adam.sonabend@northwestern.edu.
Amy B. Heimberger, Email: Amy.heimberger@northwestern.edu.
Maciej S. Lesniak, Email: maciej.lesniak@northwestern.edu.
Jason Miska, Email: jason.miska@northwestern.edu.
Mariafausta Fischietti, Email: fausta.fischietti@grovebiopharma.com.
Catalina Lee-Chang, Email: catalina.leechang@northwestern.edu.
References
- 1.Laug D, et al. A glial blueprint for gliomagenesis. Nat Rev Neurosci. 2018;19(7):393–403. doi: 10.1038/s41583-018-0014-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schaff LR, Mellinghoff IK. Glioblastoma and other primary brain malignancies in adults: a review. JAMA. 2023;329(7):574–587. doi: 10.1001/jama.2023.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hambardzumyan D, Bergers G. Glioblastoma: defining tumor niches. Trends Cancer. 2015;1(4):252–265. doi: 10.1016/j.trecan.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y, Zhang Z. The history and advances in cancer immunotherapy: understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell Mol Immunol. 2020;17(8):807–821. doi: 10.1038/s41423-020-0488-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petitprez F, et al. B cells are associated with survival and immunotherapy response in sarcoma. Nature. 2020;577(7791):556–560. doi: 10.1038/s41586-019-1906-8. [DOI] [PubMed] [Google Scholar]
- 6.Helmink BA, et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature. 2020;577(7791):549–555. doi: 10.1038/s41586-019-1922-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cabrita R, et al. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature. 2020;577(7791):561–565. doi: 10.1038/s41586-019-1914-8. [DOI] [PubMed] [Google Scholar]
- 8.Sharonov GV, et al. B cells, plasma cells and antibody repertoires in the tumour microenvironment. Nat Rev Immunol. 2020;20(5):294–307. doi: 10.1038/s41577-019-0257-x. [DOI] [PubMed] [Google Scholar]
- 9.Laumont CM, Nelson BH. B cells in the tumor microenvironment: Multi-faceted organizers, regulators, and effectors of anti-tumor immunity. Cancer Cell. 2023;41(3):466–489. doi: 10.1016/j.ccell.2023.02.017. [DOI] [PubMed] [Google Scholar]
- 10.Vanhersecke L, et al. Mature tertiary lymphoid structures predict immune checkpoint inhibitor efficacy in solid tumors independently of PD-L1 expression. Nat Cancer. 2021;2(8):794–802. doi: 10.1038/s43018-021-00232-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fridman WH, et al. B cells and tertiary lymphoid structures as determinants of tumour immune contexture and clinical outcome. Nat Rev Clin Oncol. 2022;19(7):441–457. doi: 10.1038/s41571-022-00619-z. [DOI] [PubMed] [Google Scholar]
- 12.Meylan M, et al. Tertiary lymphoid structures generate and propagate anti-tumor antibody-producing plasma cells in renal cell cancer. Immunity. 2022;55(3):527–541. doi: 10.1016/j.immuni.2022.02.001. [DOI] [PubMed] [Google Scholar]
- 13.Lee-Chang C, et al. Activation of 4-1BBL+ B cells with CD40 agonism and IFNγ elicits potent immunity against glioblastoma. J Exp Med. 2021;218(1):e20200913. doi: 10.1084/jem.20200913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hou D, et al. Generation of B cell-based cellular vaccine for cancer in murine models. STAR Protoc. 2023;4(2):102219. doi: 10.1016/j.xpro.2023.102219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hua Z, Hou B. The role of B cell antigen presentation in the initiation of CD4+ T cell response. Immunol Rev. 2020;296(1):24–35. doi: 10.1111/imr.12859. [DOI] [PubMed] [Google Scholar]
- 16.Rivera A, et al. Role of B cells as antigen-presenting cells in vivo revisited: antigen-specific B cells are essential for T cell expansion in lymph nodes and for systemic T cell responses to low antigen concentrations. Int Immunol. 2001;13(12):1583–1593. doi: 10.1093/intimm/13.12.1583. [DOI] [PubMed] [Google Scholar]
- 17.Lee-Chang C, Lesniak MS. Next-generation antigen-presenting cell immune therapeutics for gliomas. J Clin Invest. 2023;133(3):e163449. doi: 10.1172/JCI163449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murata K, et al. Landscape mapping of shared antigenic epitopes and their cognate TCRs of tumor-infiltrating T lymphocytes in melanoma. Elife. 2020;9:e53244. doi: 10.7554/eLife.53244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzalez NK, et al. In vitro and in vivo imaging of initial B-T-cell interactions in the setting of B cell based cancer immunotherapy. Oncoimmunology. 2015;4(9):e1038684. doi: 10.1080/2162402X.2015.1038684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Domingues P, et al. Tumor infiltrating immune cells in gliomas and meningiomas. Brain Behav Immun. 2016;53:1–15. doi: 10.1016/j.bbi.2015.07.019. [DOI] [PubMed] [Google Scholar]
- 21.Kim EK, et al. Enhanced antitumor immunotherapeutic effect of B cell-based vaccine transduced with modified adenoviral vector containing type 35 fiber structures. Gene Ther. 2014;21(1):106–114. doi: 10.1038/gt.2013.65. [DOI] [PubMed] [Google Scholar]
- 22.Schultze JL, et al. CD40-activated human B cells: an alternative source of highly efficient antigen presenting cells to generate autologous antigen-specific T cells for adoptive immunotherapy. J Clin Invest. 1997;100(11):2757–2765. doi: 10.1172/JCI119822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khasraw M, et al. New approaches to glioblastoma. Annu Rev Med. 2022;73:279–292. doi: 10.1146/annurev-med-042420-102102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laumont CM, et al. Tumour-infiltrating B cells: immunological mechanisms, clinical impact and therapeutic opportunities. Nat Rev Cancer. 2022;22(7):414–430. doi: 10.1038/s41568-022-00466-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie J, et al. ITGB1 drives hepatocellular carcinoma progression by modulating cell cycle process through PXN/YWHAZ/AKT pathways. Front Cell Dev Biol. 2021;9:711149. doi: 10.3389/fcell.2021.711149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shu C, et al. Integrin β1 regulates proliferation, apoptosis, and migration of trophoblasts through activation of phosphoinositide 3 kinase/protein kinase B signaling. J Obstet Gynaecol Res. 2021;47(7):2406–2416. doi: 10.1111/jog.14782. [DOI] [PubMed] [Google Scholar]
- 27.Wang J-f, et al. ITGA5 promotes tumor progression through the activation of the FAK/AKT signaling pathway in human gastric cancer. Oxid Med Cell Longev. 2022;2022(1):8611306. doi: 10.1155/2022/8611306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deng Y, et al. Integrin α5/ITGA5 promotes the proliferation, migration, invasion and progression of oral squamous carcinoma by epithelial-mesenchymal transition. Cancer Manag Res. 2019;11:9609–9620. doi: 10.2147/CMAR.S223201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qian Y, et al. ILK mediates actin filament rearrangements and cell migration and invasion through PI3K/Akt/Rac1 signaling. Oncogene. 2005;24(19):3154–3165. doi: 10.1038/sj.onc.1208525. [DOI] [PubMed] [Google Scholar]
- 30.Reymond N, et al. Cdc42 promotes transendothelial migration of cancer cells through β1 integrin. J Cell Biol. 2012;199(4):653–668. doi: 10.1083/jcb.201205169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamao M, et al. Distinct predictive performance of Rac1 and Cdc42 in cell migration. Sci Rep. 2015;5(1):17527. doi: 10.1038/srep17527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang W, et al. TRPV4 activates the Cdc42/N-wasp pathway to promote glioblastoma invasion by altering cellular protrusions. Sci Rep. 2020;10(1):14151. doi: 10.1038/s41598-020-70822-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mierke CT, et al. Vinculin facilitates cell invasion into three-dimensional collagen matrices. J Biol Chem. 2010;285(17):13121–13130. doi: 10.1074/jbc.M109.087171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thievessen I, et al. Vinculin is required for cell polarization, migration, and extracellular matrix remodeling in 3D collagen. FASEB J. 2015;29(11):4555–4567. doi: 10.1096/fj.14-268235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rahman A, et al. Vinculin regulates directionality and cell polarity in 2D, 3D matrix and 3D microtrack migration. Mol Biol Cell. 2016;27(9):1431–1441. doi: 10.1091/mbc.E15-06-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cai YJ, et al. Impact of Nischarin on EMT regulators in breast cancer cell lines. Oncol Lett. 2020;20(6):291. doi: 10.3892/ol.2020.12154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alahari SK, et al. The integrin-binding protein Nischarin regulates cell migration by inhibiting PAK. EMBO J. 2004;23(14):2777–2788. doi: 10.1038/sj.emboj.7600291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo Y, et al. PRL1 promotes cell migration and invasion by increasing MMP2 and MMP9 expression through Src and ERK1/2 pathways. Biochemistry. 2009;48(8):1838–1846. doi: 10.1021/bi8020789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giannelli G, et al. Induction of cell migration by matrix metalloprotease-2 cleavage of laminin-5. Science. 1997;277(5323):225–228. doi: 10.1126/science.277.5323.225. [DOI] [PubMed] [Google Scholar]
- 40.Riviere-Cazaux C, et al. Blood-brain barrier disruption defines the extracellular metabolome of live human high-grade gliomas. Commun Biol. 2023;6(1):653. doi: 10.1038/s42003-023-05035-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Friedl P, Alexander S. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell. 2011;147(5):992–1009. doi: 10.1016/j.cell.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 42.Lu P, et al. The extracellular matrix: a dynamic niche in cancer progression. J Cell Biol. 2012;196(4):395–406. doi: 10.1083/jcb.201102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mair DB, et al. Mechanisms of invasion and motility of high-grade gliomas in the brain. Mol Biol Cell. 2018;29(21):2509–2515. doi: 10.1091/mbc.E18-02-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vollmann-Zwerenz A, et al. Tumor cell invasion in glioblastoma. Int J Mol Sci. 2020;21(6):1932. doi: 10.3390/ijms21061932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Turner CA, et al. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell. 1994;77(2):297–306. doi: 10.1016/0092-8674(94)90321-2. [DOI] [PubMed] [Google Scholar]
- 46.Shapiro-Shelef M, et al. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 2003;19(4):607–620. doi: 10.1016/S1074-7613(03)00267-X. [DOI] [PubMed] [Google Scholar]
- 47.Marino S, et al. The extracellular matrix in glioblastomas: a glance at its structural modifications in shaping the tumoral microenvironment-a systematic review. Cancers (Basel) 2023;15(6):1879. doi: 10.3390/cancers15061879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koh I, et al. The mode and dynamics of glioblastoma cell invasion into a decellularized tissue-derived extracellular matrix-based three-dimensional tumor model. Sci Rep. 2018;8(1):4608. doi: 10.1038/s41598-018-22681-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.So JS, et al. Mechanisms of invasion in glioblastoma: extracellular matrix, Ca2+ signaling, and glutamate. Front Cell Neurosci. 2021;15(2+):663092. doi: 10.3389/fncel.2021.663092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Azam Z, et al. Mesenchymal transformation: the rosetta stone of glioblastoma pathogenesis and therapy resistance. Adv Sci (Weinh) 2020;7(22):2002015. doi: 10.1002/advs.202002015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Behnan J, et al. The landscape of the mesenchymal signature in brain tumours. Brain. 2019;142(4):847–866. doi: 10.1093/brain/awz044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Comba A, et al. Spatiotemporal analysis of glioma heterogeneity reveals COL1A1 as an actionable target to disrupt tumor progression. Nat Commun. 2022;13(1):3606. doi: 10.1038/s41467-022-31340-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jain S, et al. Single-cell RNA sequencing and spatial transcriptomics reveal cancer-associated fibroblasts in glioblastoma with protumoral effects. J Clin Invest. 2023;133(5):e147087. doi: 10.1172/JCI147087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garaud S, et al. Antigen specificity and clinical significance of IgG and IgA autoantibodies produced in situ by tumor-infiltrating B cells in breast cancer. Front Immunol. 2018;9:2660. doi: 10.3389/fimmu.2018.02660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Germain C, et al. Presence of B cells in tertiary lymphoid structures is associated with a protective immunity in patients with lung cancer. Am J Respir Crit Care Med. 2014;189(7):832–844. doi: 10.1164/rccm.201309-1611OC. [DOI] [PubMed] [Google Scholar]
- 56.Wieland A, et al. Defining HPV-specific B cell responses in patients with head and neck cancer. Nature. 2021;597(7875):274–278. doi: 10.1038/s41586-020-2931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Biswas S, et al. IgA transcytosis and antigen recognition govern ovarian cancer immunity. Nature. 2021;591(7850):464–470. doi: 10.1038/s41586-020-03144-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yasuda M, et al. Antigens recognized by IgG derived from tumor-infiltrating B lymphocytes in human lung cancer. Anticancer Res. 2006;26(5a):3607–3611. [PubMed] [Google Scholar]
- 59.Yasuda M, et al. Tumor-infiltrating B lymphocytes as a potential source of identifying tumor antigen in human lung cancer. Cancer Res. 2002;62(6):1751–1756. [PubMed] [Google Scholar]
- 60.Pavoni E, et al. Tumor-infiltrating B lymphocytes as an efficient source of highly specific immunoglobulins recognizing tumor cells. BMC Biotechnol. 2007;7:70. doi: 10.1186/1472-6750-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Laman JD, Weller RO. Drainage of cells and soluble antigen from the CNS to regional lymph nodes. J Neuroimmune Pharmacol. 2013;8(4):840–856. doi: 10.1007/s11481-013-9470-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hou D, et al. Antigen-presenting B cells promote TCF-1+ PD1- stem-like CD8+ T-cell proliferation in glioblastoma. Front Immunol. 2024;14:1295218. doi: 10.3389/fimmu.2023.1295218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fischietti M, et al. Schlafen 5 as a novel therapeutic target in pancreatic ductal adenocarcinoma. Oncogene. 2021;40(18):3273–3286. doi: 10.1038/s41388-021-01761-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ward JH. Hierarchical grouping to optimize an objective function. J Am Stat Assoc. 1963;58(301):236–244. doi: 10.1080/01621459.1963.10500845. [DOI] [Google Scholar]
- 65.Rivest F, et al. Fully automated sequential immunofluorescence (seqIF) for hyperplex spatial proteomics. Sci Rep. 2023;13(1):16994. doi: 10.1038/s41598-023-43435-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pijuan J, et al. In vitro cell migration, invasion, and adhesion assays: from cell imaging to data analysis. Front Cell Dev Biol. 2019;7:107. doi: 10.3389/fcell.2019.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stoellinger HM, Alexanian AR. Modifications to the transwell migration/invasion assay method that eases assay performance and improves the accuracy. Assay Drug Dev Technol. 2022;20(2):75–82. doi: 10.1089/adt.2021.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Leite C, et al. Differentiation of human umbilical cord matrix mesenchymal stem cells into neural-like progenitor cells and maturation into an oligodendroglial-like lineage. PLoS One. 2014;9(10):e111059. doi: 10.1371/journal.pone.0111059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shihan MH, et al. A simple method for quantitating confocal fluorescent images. Biochem Biophys Rep. 2021;25:100916. doi: 10.1016/j.bbrep.2021.100916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Binello E, et al. Stemness of the CT-2A immunocompetent mouse brain tumor model: characterization in vitro. J Cancer. 2012;3:166–174. doi: 10.7150/jca.4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Perez-Riverol Y, et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2021;50(d1):D543–D552. doi: 10.1093/nar/gkab1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometric proteomics data were deposited in the ProteomeXchange Consortium database via the PRIDE (71) partner repository and are available via ProteomeXchange (accession code PXD046712; project webpage: http://www.ebi.ac.uk/pride/archive/projects/PXD046712; FTP download: https://ftp.pride.ebi.ac.uk/pride/data/archive/2024/09/PXD046712). Additionally, the supporting data values for all figures and analyses are provided in the Supporting Data Values file, which is included with the online supplemental material.