Abstract
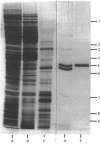
A procedure has been developed for the purification of 3-dehydroquinase from Escherichia coli. Homogeneous enzyme with specific activity 163 units/mg of protein was obtained in 19% overall yield. The subunit Mr estimated from polyacrylamide-gel electrophoresis in the presence of sodium dodecyl sulphate was 29,000. The native Mr, estimated by gel permeation chromatography on Sephacryl S-200 (superfine) and on TSK G3000SW, was in the range 52,000-58,000, indicating that the enzyme is dimeric. The catalytic properties of the enzyme have been determined and shown to be very similar to those of the biosynthetic 3-dehydroquinase component of the arom multifunctional enzyme of Neurospora crassa.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berlyn M. B., Giles N. H. Organization of enzymes in the polyaromatic synthetic pathway: separability in bacteria. J Bacteriol. 1969 Jul;99(1):222–230. doi: 10.1128/jb.99.1.222-230.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chaudhuri S., Coggins J. R. The purification of shikimate dehydrogenase from Escherichia coli. Biochem J. 1985 Feb 15;226(1):217–223. doi: 10.1042/bj2260217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coggins J. R., Boocock M. R., Campbell M. S., Chaudhuri S., Lambert J. M., Lewendon A., Mousdale D. M., Smith D. D. Functional domains involved in aromatic amino acid biosynthesis. Biochem Soc Trans. 1985 Apr;13(2):299–303. doi: 10.1042/bst0130299. [DOI] [PubMed] [Google Scholar]
- Eschenbruch M., Bürk R. R. Experimentally improved reliability of ultrasensitive silver staining of protein in polyacrylamide gels. Anal Biochem. 1982 Sep 1;125(1):96–99. doi: 10.1016/0003-2697(82)90387-6. [DOI] [PubMed] [Google Scholar]
- Frost J. W., Bender J. L., Kadonaga J. T., Knowles J. R. Dehydroquinate synthase from Escherichia coli: purification, cloning, and construction of overproducers of the enzyme. Biochemistry. 1984 Sep 11;23(19):4470–4475. doi: 10.1021/bi00314a036. [DOI] [PubMed] [Google Scholar]
- Gaertner F. H., Cole K. W. A cluster-gene: evidence for one gene, one polypeptide, five enzymes. Biochem Biophys Res Commun. 1977 Mar 21;75(2):259–264. doi: 10.1016/0006-291x(77)91037-3. [DOI] [PubMed] [Google Scholar]
- Gibson F., Pittard J. Pathways of biosynthesis of aromatic amino acids and vitamins and their control in microorganisms. Bacteriol Rev. 1968 Dec;32(4 Pt 2):465–492. [PMC free article] [PubMed] [Google Scholar]
- Kinghorn J. R., Hawkins A. R. Cloning and expression in Escherichia coli K-12 of the biosynthetic dehydroquinase function of the arom cluster gene from the eucaryote, Aspergillus nidulans. Mol Gen Genet. 1982;186(1):145–152. doi: 10.1007/BF00422927. [DOI] [PubMed] [Google Scholar]
- Kinghorn J. R., Schweizer M., Giles N. H., Kushner S. R. The cloning and analysis of the aroD gene of E. coli K-12. Gene. 1981 Jun-Jul;14(1-2):73–80. doi: 10.1016/0378-1119(81)90149-9. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lambert J. M., Boocock M. R., Coggins J. R. The 3-dehydroquinate synthase activity of the pentafunctional arom enzyme complex of Neurospora crassa is Zn2+-dependent. Biochem J. 1985 Mar 15;226(3):817–829. doi: 10.1042/bj2260817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larimer F. W., Morse C. C., Beck A. K., Cole K. W., Gaertner F. H. Isolation of the ARO1 cluster gene of Saccharomyces cerevisiae. Mol Cell Biol. 1983 Sep;3(9):1609–1614. doi: 10.1128/mcb.3.9.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarus N. R., Derechin M., Barnard E. A. Yeast hexokinase. 3. Sulfhydryl groups and protein dissociation. Biochemistry. 1968 Jun;7(6):2390–2400. doi: 10.1021/bi00846a049. [DOI] [PubMed] [Google Scholar]
- Lewendon A., Coggins J. R. Purification of 5-enolpyruvylshikimate 3-phosphate synthase from Escherichia coli. Biochem J. 1983 Jul 1;213(1):187–191. doi: 10.1042/bj2130187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumsden J., Coggins J. R. The subunit structure of the arom multienzyme complex of Neurospora crassa. A possible pentafunctional polypeptide chain. Biochem J. 1977 Mar 1;161(3):599–607. doi: 10.1042/bj1610599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumsden J., Coggins J. R. The subunit structure of the arom multienzyme complex of Neurospora crassa. Evidence from peptide 'maps' for the identity of the subunits. Biochem J. 1978 Feb 1;169(2):441–444. doi: 10.1042/bj1690441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARGOLIASH E., SMITH E. L., KREIL G., TUPPY H. Amino-acid sequence of horse heart cytochrome c. Nature. 1961 Dec 23;192:1125–1127. doi: 10.1038/1921125a0. [DOI] [PubMed] [Google Scholar]
- MITSUHASHI S., DAVIS B. D. Aromatic biosynthesis. XII. Conversion of 5-dehydroquinic acid to 5-dehydroshikimic acid dy 5-dehydroquinase. Biochim Biophys Acta. 1954 Sep;15(1):54–61. doi: 10.1016/0006-3002(54)90093-1. [DOI] [PubMed] [Google Scholar]
- Millar G., Lewendon A., Hunter M. G., Coggins J. R. The cloning and expression of the aroL gene from Escherichia coli K12. Purification and complete amino acid sequence of shikimate kinase II, the aroL-gene product. Biochem J. 1986 Jul 15;237(2):427–437. doi: 10.1042/bj2370427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi N., Yamamoto M. Analysis of the structure and transcription of the aro3 cluster gene in Schizosaccharomyces pombe. Mol Gen Genet. 1984;195(1-2):164–169. doi: 10.1007/BF00332740. [DOI] [PubMed] [Google Scholar]
- Smith D. D., Coggins J. R. Isolation of a bifunctional domain from the pentafunctional arom enzyme complex of Neurospora crassa. Biochem J. 1983 Aug 1;213(2):405–415. doi: 10.1042/bj2130405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz M. A., Deal W. C., Jr Metabolic control and structure of glycolytic enzymes. 3. Dissociation and subunit structure of rabbit muscle pyruvate kinase. Biochemistry. 1966 Apr;5(4):1399–1405. doi: 10.1021/bi00868a038. [DOI] [PubMed] [Google Scholar]
- Weber K., Osborn M. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J Biol Chem. 1969 Aug 25;244(16):4406–4412. [PubMed] [Google Scholar]