Abstract
Rabies virus (RV) is a highly neurotropic virus that migrates from the portal of entry to the central nervous system (CNS). The cytoplasmic dynein light chain (LC8), which is involved in a variety of intracellular motile events, was shown to interact with RV phosphoprotein (P). In order to determine the functional significance of this interaction, P residues 143 to 149 or 139 to 149 encompassing a conserved LC8-interacting motif (K/RXTQT) were deleted from recombinant viruses SAD-L16 and SAD-D29. These viruses are identical except for a replacement of the arginine at position 333 (R333) of the RV glycoprotein by an aspartic acid in SAD-D29. SAD-L16 virus is fully pathogenic for mice, whereas SAD-D29 is nonpathogenic for adult mice but retained pathogenicity for suckling mice. The deletions introduced into the LC8 binding site abolished the P-LC8 interaction and blocked LC8 incorporation into virions. All the mutants propagated in cell culture as efficiently as the parent strains. The pathogenicity of the mutants was then compared with that of the parent viruses by inoculating suckling mice. SAD-L16 derivatives were as pathogenic as their parent virus after intramuscular inoculation, indicating that LC8 is dispensable for the spread of a pathogenic RV from a peripheral site to the CNS. In contrast, SAD-D29-derived deletion mutants were attenuated by as much as 30-fold after intramuscular inoculation but remained as pathogenic as the parent virus when inoculated directly into the brain. This remarkable attenuation after intramuscular but not after intracranial inoculation suggested that abolishing the P-LC8 interaction reduces the efficiency of peripheral spread of the more attenuated SAD-D29 strain. These results demonstrate that elimination of the LC8 ligand and simultaneous substitution of R333 considerably attenuate RV pathogenicity and may be helpful in designing and developing highly safe live-RV-based vaccines.
Rabies remains one of the most dreadful neurotropic infectious diseases affecting humans and animals. Rabies virus (RV) multiplies first at the site of infection and is transported by retrograde flow through peripheral nerves to the central nervous system (CNS), where it causes encephalomyelitis. The mechanism by which RV is transported retrograde from peripheral sites to the neurons is poorly understood. What has recently been described is that the cytoplasmic dynein light chain (LC8), which is involved in the retrograde intracellular transport of organelles, interacts with the phosphoprotein (P) of RV and is incorporated into mature virions (13, 25). The interaction of RV P with LC8 was shown by coimmunoprecipitation analysis, and mapping studies have demonstrated that LC8 interacts in the region between residues 138 and 172 of the P protein. Using biochemical and mutational analysis of selected LC8 binding proteins, Lo et al. (18) identified a consensus sequence containing a K/RXTQT motif that interacts with LC8. The involvement of LC8 in various intracellular transport mechanisms and other cellular functions led to the suggestion that the P-LC8 interaction might be the driving force for the retrograde transport and pathogenesis of RV. However, the functional significance of this interaction in the neuroinvasive processes of RV has not yet been elucidated.
RV is a nonsegmented negative-stranded RNA virus of the Rhabdoviridae family. The viral proteins of RV are associated either with the core component, called the ribonucleoprotein (RNP) complex, or with the viral envelope. The RNP complex consists of the RNA genome encapsidated by the nucleocapsid (N) protein in combination with polymerase (L) and the P protein. The RNP complex serves as a template for virus transcription and replication. The P protein interacts with the N and L proteins (4, 5, 12) and is believed to work as a noncatalytic cofactor of the viral RNA polymerase. P protein is multifunctional, binding to other viral proteins to act as a chaperone to help viral genome replication, and also interacting with cellular factors, possibly to participate in RV spread and pathogenesis (13, 25).
The viral envelope component is composed of a transmembrane glycoprotein (G) and a matrix (M) protein. The M protein is localized on the inner surface of the viral envelope surrounding the RNP and is involved in the virus assembly and budding process (20). In addition to M protein, the spike G protein also participates in the virus release process by increasing budding efficiency (22). G protein is responsible for cell attachment and membrane fusion and is the main protein responsible for the induction of virus-neutralizing antibodies. Moreover, the G protein is known to play a role in virulence. In particular, the arginine (R) residue at position 333 (R333) of the G protein has been shown to be responsible for RV pathogenicity (11, 23, 29). Among several neutralization-resistant RV mutants generated under selection pressure with monoclonal antibodies, only mutants that possess an amino acid differing from R333 of the G protein were found to have reduced pathogenicity for adult mice (29). This finding confirmed the presence of a direct correlation between RV pathogenicity and R333 and helped in developing attenuated RV vaccines for oral immunization of animals (17, 19). Such live rabies vaccine viruses possessing an amino acid other than R333 in the glycoprotein are nonpathogenic to immunocompetent adult mice. However, they are still pathogenic when inoculated into baby mice (33), demonstrating the existence of residual pathogenicity and the potential risk to immunocompromised animals and humans.
Despite significant scientific advances in rabies prevention and control, the disease remains a major threat to public health and continues to cause numerous human deaths in the tropics. Canine rabies is still epizootic in most countries of Africa, Asia, and South America, and infection via dogs is responsible for most human deaths from the disease. The presence of large numbers of ownerless dogs in these countries necessitates introduction of new control strategies in addition to the existing parenteral vaccination programs. Due to the success in control of wildlife rabies by oral immunization (19, 35), several developing countries are presently volunteering to make use of oral vaccination of dogs. However, compared to the live oral vaccine strains used for immunization of wildlife, vaccine strains for oral immunization of dogs have to be highly safe due to the much closer relationship between dogs and humans than between wild animals and humans. Furthermore, there is a growing interest in exploiting the potential of recombinant RV-based vectors to protect humans and animals from other infectious agents. Recently it has been shown that a recombinant RV expressing human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein might serve as a potential vector for an HIV-1 vaccine (28). The success of such RV-based vaccines is, however, largely dependent on the safety of the RV strain, particularly in immunocompromised patients.
To define the role of the P-LC8 interaction in virus transport and virus pathogenesis, it was necessary to construct mutants in which the LC8 binding domain was modified. The present paper describes the effects of deletions of either 7 or 11 residues from the LC8 binding site in the P protein with or without an R333 substitution in the G protein. Surprisingly, when deletions are introduced into the LC8 binding site of an RV that possesses an amino acid differing from R333, a dramatic reduction in pathogenicity for 1- to 2-day-old suckling mice was observed after peripheral inoculation. The profound advantage of this extensive attenuation of RV in developing live oral rabies vaccines or RV-based vectors with eliminated or diminished risks associated with retrograde neuronal spread is discussed.
MATERIALS AND METHODS
Cells and viruses.
A recombinant RV, SAD-L16, possessing the authentic sequence of the attenuated SAD-B19 (6) vaccine strain (a derivative of the Street Alabama Dufferin strain [SAD]), was generated from a full-length cDNA clone (27). BSR-T7/5 cells expressing phage T7 RNA polymerase in a stable manner (3) were used to recover infectious RV from cDNA.
Construction of cDNA clones.
In order to introduce deletions into the LC8 binding site of the P protein, a 2.2-kb BstBI fragment comprising nucleotides 1497 to 3738 of the SAD B19 sequence was first cloned into the pSK vector. Site-directed mutagenesis was carried out using the QuikChange site-directed mutagenesis kit according to the manufacturer's (Stratagene) instructions. Primer pairs 142 (5′-GGAAAGTCTTCAGAGGGCCGAGAGCTCAAG-3′) and 143 (5′-CTTGAGCTCTCGGCCCTCTGAAGACTTTCC-3′) were used to delete nucleotides in positions 1940 to 1960, corresponding to amino acids 143 to 149 of the RV P protein. A larger deletion encompassing nucleotides 1928 to 1960, which corresponds to amino acids 139 to 149 of RV P protein, was also introduced using oligonucleotides 144 (5′-CCCAACCCTCCAGGAGGCCGAGAGCTCAAG-3′) and 145 (5′-CTTGAGCTCTCGGCCTCCTGGAGGGTTGGG-3′). The accuracy of the introduced deletions was confirmed by sequencing the modified region. Clones containing the desired deletions (7 or 11 amino acids) were digested with NcoI and SnaBI, and the respective ≈0.8-kb fragments were introduced into the full-length clone SAD-L16 after removing the corresponding fragment. The resulting plasmids encoding deletions of 7 or 11 residues in the LC8 binding site of P protein were named L-ΔP7 and L-ΔP11, respectively (Fig. 1A).
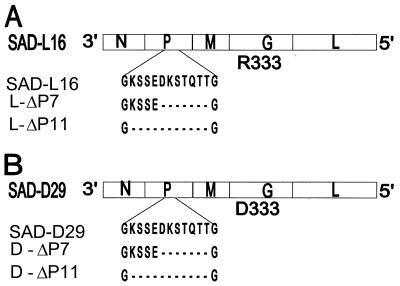
FIG. 1.
Schematic representation of the RV gene order in the negative-strand genomic RNA. (A) Recombinant RV SAD-L16, possessing the authentic sequence of SAD-B19 (6), which has an arginine (R333) at position 333 of the G protein. (B) Recombinant RV SAD-D29, possessing an aspartic acid (D333) at position 333 of the G protein. Amino acid sequences around the LC8 dynein light chain-binding site of the P protein (positions 138 to 150) are presented. The P protein sequence KSTQT (positions 144 to 148) is identical to the K/RXTQT motif found in a variety of proteins interacting with LC8. The ΔP7 and ΔP11 mutants constructed in the backbone of SAD-L16 and SAD-D29 contain the indicated deletions of 7 or 11 amino acids at the LC8 binding site of the respective P proteins.
To generate viruses possessing deletions at the LC8 binding site and simultaneous substitution of R333 (SAD B19 nucleotide positions 4370 to 4372) of the mature RV G protein, the following manipulations were carried out. First, site-directed mutagenesis was performed with 21-mer oligonucleotides (5′-ATTCCAAGTGTCGACTGACTT-3′) using pT7T-G as a template (7). The resulting plasmid coded for a modified RV G protein in which the arginine (R333), encoded by AGA, was replaced by aspartic acid (D333), encoded by GAC. A full-length cDNA clone, SAD-D29, was then generated by replacing an StuI/PpuMI cDNA fragment (SAD B19 nucleotides 4015 to 4470) of SAD-L16 with the corresponding region of the modified G protein. Finally, the above NcoI/SnaBI ≈0.8-kb DNA fragments possessing deletions in the LC8 binding site were used to replace the corresponding fragment of SAD-D29. The resulting full-length clones having D333 instead of R333 in the G protein and simultaneously a deletion of 7 or 11 amino acids at the LC8 binding site of the P protein were named D-ΔP7 and D-ΔP11, respectively (Fig. 1B).
Recovery of recombinant viruses.
Transfection experiments were carried out as described previously (27). Approximately 1.5 × 106 BSR-T7/5 cells were transfected with a plasmid mixture containing 5 μg of pT7T-N, 2.5 μg of pT7T-P, 2.5 μg of pT7T-L, and 10 μg of one of the full-length plasmids using the Stratagene mammalian transfection kit (CaPO4 protocol). Supernatants from transfected cells were passaged, and infection of cells was monitored by direct immunofluorescence with an anti-RV nucleoprotein conjugate (Centocor). The recombinant viruses were further passaged two to three times, and the resulting virus stocks were titrated by endpoint dilutions.
Replication of recombinant viruses in vitro.
For one-step growth curve analysis, 106 BSR-T7/5 cells were grown in 3.2-cm-diameter dishes overnight and infected in duplicate at a multiplicity of infection (MOI) of 10 with the various recombinant viruses. After 2 h of incubation at 37°C, the inoculum was removed, and the cells were rinsed three times with phosphate-buffered saline (PBS). Cells were supplied with 2.5 ml of fresh medium and incubated further at 37°C. At 4, 18, 24, and 48 h after infection, 100 μl of culture supernatants was removed and counted in duplicate on BSR-T7/5 cells. A multicycle growth analysis was done as above except that the infection of BSR-T7/5 cells was done at an MOI of 0.01.
RT-PCR and sequence analysis.
To determine whether the recombinant viruses maintained the introduced deletions, total RNA was isolated from BSR-T7/5 cells infected with passage level 4 of the respective stock viruses. Reverse transcription (RT)-PCR was performed on 1 μg of total RNA isolated from infected cells. The PCR products were analyzed on 1% agarose gels and used directly for sequencing.
Protein composition of mutant viruses and coimmunoprecipitation.
To analyze the protein composition of recombinant viruses, ≈107 BSR-T7/5 cells were infected at an MOI of 0.02 and incubated for 2 days. Virions in the supernatant were then purified and concentrated by centrifugation through a 20% sucrose cushion in a Beckman SW28 rotor at 25,000 rpm for 90 min. Pellets were resuspended and mixed with protein sample buffer to disrupt the virions. Viral proteins from purified virions were then resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes (Millipore). After incubation with a blocking solution, membranes were incubated with rabbit serum raised against RV ribonucleoprotein S50 (22) or a rabbit polyclonal anti-LC8 antibody, R4058 (15). Membranes were then incubated with peroxidase-conjugated goat anti-rabbit immunoglobulin G. Proteins were visualized after incubation with peroxidase substrate (Vector). Pulldown experiments were performed by immunoprecipitating samples of cytoplasmic extracts of infected BSR-T7/5 cells with the anti-RV RNP serum S50. Coimmunoprecipitation of LC8 was then analyzed by Western blotting after incubating blots with the anti-LC8 antibody.
Pathogenicity of recombinant viruses in mice.
Groups of six 1- to 2-day-old or 1-week-old suckling BALB/c mice were inoculated by the intramuscular (i.m.) route in the hind thigh region or by the intracranial (i.c.) route with a volume of 0.03 ml of virus suspensions at various concentrations. Mice were observed for rabies symptoms for 21 days. A 20% brain suspension was prepared from dead mice, and the presence of RV in the brain was confirmed by virus isolation in cell culture. At the end of the 21-day observation period, brain samples from surviving mice were also processed for virus isolation. The lethal dose that kills 50% of the animals (LD50) was calculated using the method of Reed and Muench (26).
RESULTS
Generation of mutant RVs from cDNA.
The finding of an interaction between the P protein of RV and the LC8 dynein light chain led to the suggestion that the retrograde transport of RV from the portal of entry might be mediated by LC8. RV P residues 138 to 172 have been mapped as domains responsible for binding to the LC8 dynein light chain (25). Moreover, comparison of P protein sequences of various RV strains reveals the presence of a conserved KSTQT sequence (residues 144 to 148), which is identical to the consensus LC8 binding motif (K/RXTQT) of various LC8 binding proteins (18).
To obtain insight into the functional significance of the LC8-P interaction, RV mutants possessing 7 or 11 amino acid deletions in the binding site encompassing residues 143 to 149 (Δ7 mutants) or residues 139 to 149 (Δ11 mutants) were constructed (Fig. 1). The deletions were first introduced into the backbone of recombinant RV SAD-L16, which possesses the authentic sequence of the attenuated RV strain SAD-B19 (6, 27).
For the purpose of developing genetically stable attenuated live oral rabies vaccine candidates and RV-based vaccine vectors, RV mutants possessing replacements of R333 were generated and characterized (not shown). Among the various R333 mutants, a mutant (SAD-D29) possessing an aspartic acid (D333) instead of R333 was chosen for further modification. The 7 or 11 amino acid deletions in the LC8 binding domain were then introduced into the backbone of SAD-D29 (Fig. 1). Full-length cDNA clones representing all the mutants were transfected as described in Materials and Methods. For all the mutants, the presence of infectious virus was detected after one passage of the transfection supernatants into fresh BSR-T7/5 cells. Stock viruses for further analysis were prepared after two to three passages in BSR-T7/5 cells. The identity of the recombinant viruses SAD-D29, D-ΔP7, D-ΔP11, L-ΔP7, and L-ΔP11 (Fig. 1) was verified by RT-PCR and by sequencing of PCR products. For all mutants, the sequences obtained corresponded exactly to the alterations introduced into the cDNA clones.
Propagation of recombinant viruses in cell culture.
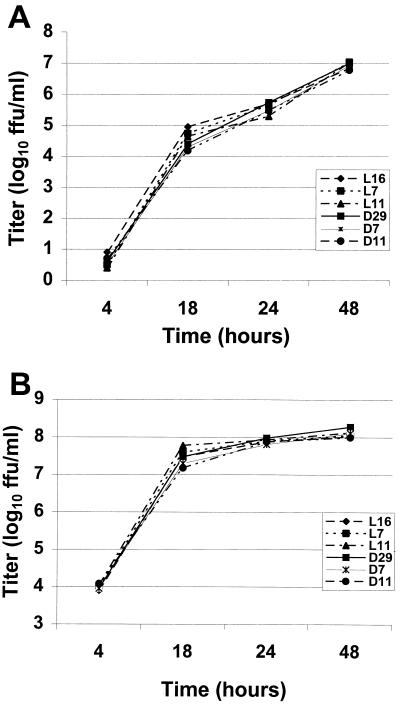
First, the titers of the stock viruses were determined by endpoint dilutions. To examine the efficiency of virus propagation at different time points, a multicycle growth curve analysis was performed by infecting BSR-T7/5 cells at an MOI of 0.01. In the titers determined during the first 18 h, SAD-L16 gave slightly higher titers than the rest of the recombinants, but this pattern disappeared at 24 h postinfection. At 48 h of infection, the titers of all the recombinants were very similar (Fig. 2A). To allow synchronous infection of all cells, BSR-T7/5 cells were also infected at an MOI of 10 and aliquots of cell culture supernatants were collected for titer determination at the indicated time points (Fig. 2B). At 18 h postinfection, the virus with the highest titer was the mutant L-ΔP11, and at 24 h postinfection all viruses yielded very similar titers. No specific pattern of growth impairment was seen for any of the mutants, indicating that as many as 11 amino acids at the LC8 binding site in the P protein are dispensable for RV replication in cell culture.
FIG. 2.
(A) Multicycle replication of recombinant RVs. BSR-T7/5 cells were infected with the recombinant RVs SAD-L16 (L16), L-ΔP7 (L7), L-ΔP11 (L11), SAD-D29 (D29), D-ΔP7 (D7), and D-ΔP11 (D11) at an MOI of 0.01. (B) One-step growth curves of the same recombinant RVs after infection of BSR-T7/5 cells at an MOI of 10. Aliquots of cell culture supernatants were collected at the indicated time points, and virus titers were determined in duplicate by serial dilutions.
Protein composition of mutants.
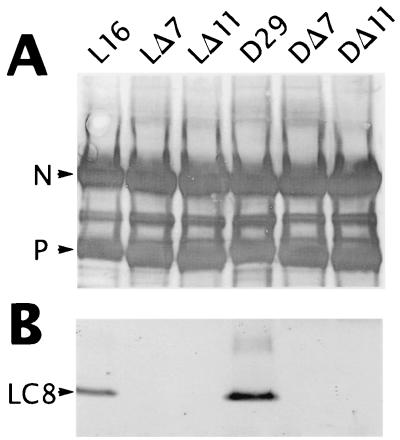
To determine the expression level of mutant P proteins and to elucidate whether LC8 was incorporated into virions, proteins from purified viruses were analyzed by Western immunoblotting. The blots were incubated with an anti-RV RNP rabbit serum (S50) or an anti-LC8 polyclonal antibody (R4058). The amounts of P protein and the ratios between P and N proteins of the mutant viruses were indistinguishable from those of the parent viruses, indicating that the deletion of 7 or 11 amino acids from the LC8 binding site had no influence on expression of mutated P proteins. In contrast to the parent viruses, no protein corresponding to LC8 could be detected in mutant viruses with a deletion at the LC8 binding site (Fig. 3). This result demonstrates that the introduced deletions were sufficient to entirely block the incorporation of LC8 into virions.
FIG. 3.
Protein composition of recombinant RVs. Approximately 107 BSR-T7/5 cells were infected with the recombinant RVs SAD-L16 (L16), L-ΔP7 (LΔ7), L-ΔP11 (LΔ11), SAD-D29 (D29), D-ΔP7 (DΔ7), and D-ΔP11 (DΔ11) at an MOI of 0.02. Two days after infection, virions from supernatants were purified over a 20% sucrose cushion, and virus pellets were analyzed by Western blotting. Using a protein marker as an indicator, the same blot was cut into two parts at approximately the 20-kDa position. The upper part of the blot was incubated with anti-RV RNP serum (A), and the lower part of the blot was incubated with a rabbit polyclonal LC8 antibody (B). Incorporation of LC8 could be detected only in the parent viruses SAD-L16 and SAD-D29, but not in the recombinant viruses possessing deletions at the LC8 binding site.
To directly determine the effects of the introduced deletions on the P-LC8 interaction, lysates of infected cells were also analyzed in a pulldown experiment. Proteins immunoprecipitated with an anti-RV RNP serum were analyzed in Western blots by incubating blots with the anti-LC8 antibody. Similar to the results obtained for purified virions, LC8 was not detected in any of the lysates from cells infected with the deletion mutants (not shown). This finding shows that the deletions abrogated the interaction between LC8 and P protein. Moreover, the results indicate that the incorporation of LC8 into virions membranes is driven by a specific interaction between P and LC8 and not by a nonspecific trapping process.
Pathogenicity of recombinant viruses in mice.
The parent SAD-L16 virus causes 100% mortality after i.c. inoculation into adult mice with a dose as low as 30 focus-forming units (FFU)/mouse. In contrast, SAD-D29 was completely nonpathogenic for adult mice even at a dose higher than 106 FFU/mouse. However, it retained full pathogenicity for 1- to 2-day-old suckling mice. Due to the suggestion that LC8 might be involved in the axonal transport of RV, it was logical to first use a peripheral route of inoculation to compare LC8 binding site mutants with their parents. In the first experiment, 1-week-old suckling mice were inoculated by i.m. injection at doses of 102 or 105 FFU/mouse. Unexpectedly, SAD-L16, L-ΔP7, and L-ΔP11 were equally pathogenic for 1-week-old suckling mice, indicating that LC8 is not required for the spread of SAD-L16 from a peripheral site of inoculation in this model (Fig. 4). In contrast, SAD-D29, D-ΔP7, and D-ΔP11 were all nonpathogenic at the doses administered (Fig. 4).
FIG. 4.
Pathogenicity of recombinant RVs. One-week-old suckling mice were inoculated with recombinant RVs SAD-L16 (L16), L-ΔP7 (L7), L-ΔP11 (L11), SAD-D29 (D29), D-ΔP7 (D7), and D-ΔP11 (D11) at a dose of 100 or 100,000 FFU/mouse i.m. Mice were observed daily for 21 days, and results recorded at the end of the experiment are presented. No rabies symptoms were observed in any group of mice inoculated with recombinant RVs possessing an aspartic acid at position 333 of the G protein.
A pathogenicity experiment was then carried out in the more sensitive 1- to 2-day-old suckling mice by i.m. injection at doses of 102 or 105 FFU/mouse. As shown in Fig. 5, all mice inoculated with SAD-L16, L-ΔP7, and L-ΔP11 died of rabies within 10 days of inoculation. This again demonstrated that deletion of 7 or 11 residues from the LC8 binding site of the P protein does not affect the peripheral spread of SAD-L16 in suckling mice. However, when the LC8 binding site deletion mutations are combined with the R333 substitution mutation, a substantial degree of attenuation of SAD-D29 was achieved. Within 10 days postinoculation, all mice inoculated with both doses of the parent SAD-D29 had died of rabies. In contrast, lower mortality rates of 83 and 50% occurred in the groups of mice that received 105 FFU/mouse of D-ΔP7 or D-ΔP11, respectively (Fig. 5). Fifteen days after infection, the mortality reached 100 and 83% (five of six), respectively, and remained the same until the end of the 21-day observation period (Fig. 5).
FIG. 5.
Pathogenicity of recombinant RVs. Two-days-old suckling mice were inoculated with recombinant RVs at a dose of 100 or 100,000 FFU/mouse i.m. Results recorded at the end of the observation period of 10 or 21 days are presented. All animals inoculated with SAD-L16 (L16), L-ΔP7 (L7), L-ΔP11 (L11), and SAD-D29 (D29) died of rabies within 10 days after inoculation. At the end of 21 days of observation, the number of mice that died of rabies in the groups inoculated with 100 FFU/mouse of D-ΔP7 (D7) or D-ΔP11 (D11) was one and zero, respectively.
Surprisingly, there were no deaths in the groups of mice inoculated with D-ΔP7 or D-ΔP11 at a dose of 102 FFU/mouse until 11 days postchallenge. Total mortality during the 21-day observation period was less than 17% (one of six) for the D-ΔP7-inoculated group, whereas no mice died in the D-ΔP11-inoculated group. These results demonstrate that the combination of LC8 binding site mutation and the amino acid change at R333 of the G protein considerably attenuate RV virulence for 1- to 2-day-old suckling mice.
Next, the degree of attenuation of the recombinant D-ΔP11 was analyzed by administering graded doses into 1- to 2-day-old suckling mice by the i.m. or i.c. route. In an earlier experiment, one of six mice survived after infection with D-ΔP11 at a dose of 105 FFU/mouse (83% mortality, Fig. 5). In this independent experiment, doses of 105 (not shown) and 104 FFU/mouse killed all inoculated animals (Fig. 6). However, as in the previous experiment, D-ΔP11 administered at a dose of 102 FFU/mouse by i.m. injection was completely nonpathogenic for 1- to 2-day-old suckling mice. As shown in Fig. 6, the dose of D-ΔP11 that was required to kill 50% (LD50 per 30 μl) of the i.m.-inoculated 2-day-old suckling mice was 556 FFU, whereas the LD50 of the parent SAD-D29 virus was only 18 FFU. This demonstrates that D-ΔP11 is attenuated by as much as 30-fold when administered by the i.m. route. Interestingly, very similar LD50s of 10 and 14 FFU were obtained for SAD-D29 and D-ΔP11, respectively, when the strains were administered i.c. (Fig. 6). This remarkable attenuation after i.m. but not after i.c. administration shows that D-ΔP11 spreads inefficiently from a peripheral site of infection to the CNS compared to the parent virus.
FIG. 6.
Comparison of pathogenicity of recombinant RVs after i.c. or i.m. inoculation. Two-day-old suckling mice were inoculated with SAD-D29 (D29) or D-ΔP11 (D11) by the i.c. or i.m. route at the indicated doses. After i.c. inoculation, the LD50s of SAD-D29 and D-ΔP11 were 10 and 14 FFU/30 μl, respectively, whereas after i.m. inoculation, the LD50 of D-ΔP11 was 556 FFU, compared to only 18 FFU for SAD-D29, indicating that D-ΔP11 is attenuated by as much as 30-fold.
DISCUSSION
Neurotropism is one of the most striking biological characteristics of RV infection. RV enters the body in the bite or when virus-laden saliva from a rabid animal enters an open wound. After viral replication at the portal of entry, RV is believed to enter nerve endings and spread centripetally along peripheral nerves and eventually gain access to the CNS. However, the events of RV dissemination from the portal of entry to the CNS are among the most obscure events of the infection. The strict neurotropic characteristics of RV suggest the involvement of neuronal receptors for entry of the virus into nerves (32, 34). Nevertheless, the recent description of an interaction between LC8 and RV P is relevant to speculation about the involvement of cellular factors besides neuronal receptors (13, 25). To determine the functional significance of the LC8-P interaction in RV dissemination and pathogenesis, RV P protein mutants possessing deletions in the LC8 binding site were generated and characterized. A substantial degree of attenuation of RV for 1- to 2-day-old suckling mice was achieved by combining the LC8 binding site mutation with that of R333 substitution.
LC8 has been implicated in a wide variety of cellular functions, ranging from intracellular organelle transport to regulation of cellular functions. The finding that LC8 interacts with the P protein of RV led to suggestions that LC8 may be involved in the axonal transport and pathogenesis of RV (13, 25). In order to define the role of this interaction in RV neurotropism, it was necessary to construct mutants in which the LC8 binding domain was modified by deletion or mutagenesis. The LC8 binding domain was mapped to the central part of P, encompassing residues 138 to 172 (25). Based on biochemical and mutational analysis of selected LC8 binding proteins, Lo et al. (18) recently showed that LC8 binds to its targets via a conserved K/RXTQT motif. In addition, sequence alignment analysis of P proteins from various RV strains demonstrated the existence of a conserved KSTQT sequence (residues 144 to 148), suggesting that LC8 binds to P protein via this motif.
RV mutants possessing 7 or 11 amino acid deletions in the binding site propagated in cell culture with comparable efficiency and to similar titers as the parent viruses, demonstrating that the introduced deletions have no influence on the efficiency of virus replication in cell culture (Fig. 2). As demonstrated by immunoblotting analyses of purified virions, the amount of P protein and the P-to-N ratio in the mutants were indistinguishable from those of the parent viruses. Moreover, incubation of blots with LC8 antibody clearly demonstrated the absence of LC8 in purified virions as well as in cell lysates subjected to coimmunoprecipitation experiments. Titration of aliquots of supernatants subjected to immunoblotting revealed comparable titers, ranging from 8.0 log10 FFU/ml for SAD-L16 to 8.15 log10 FFU/ml for SAD-D29. Although the levels of the more abundant P and N proteins were indistinguishable, this slight titer difference might have contributed to the different amounts of the less-abundant LC8 observed in SAD-L16 and SAD-D29 viruses (Fig. 3). Taken together, these results demonstrate that LC8 incorporation into RV virions is mediated by a specific interaction with P protein rather than a passive entrapment into the virion envelope. Moreover, deletion of residues 143 to 149 from the LC8 binding site was sufficient to completely abolish the P-LC8 interaction and to block incorporation of LC8 into mature virions. In agreement with this finding, Lo et al. (18) demonstrated that a single change in one of the four consensus amino acid residues in the K/RXTQT motif can weaken or entirely abolish the binding of a peptide to LC8. The mutational analysis also suggested that the TQT tripeptide plays a more dominant role in the interaction between LC8 and its target proteins.
Pathogenicity of RV is usually measured by inoculating mice via different routes. The virus strain, the route of inoculation, and the age of the mice are important factors in determining RV virulence. In order to determine whether the mutations introduced into the LC8 binding site block the spread of the virus from peripheral sites to the CNS, 1-week-old suckling mice were inoculated by i.m. administration. As shown in Fig. 4, SAD-L16 and mutant derivatives were equally pathogenic, whereas SAD-D29 and its derivatives were all nonpathogenic. This result raises obvious questions as to why the L16-derived mutants are as pathogenic as their parent virus in spite of the absence of the P-LC8 interaction. In experiments carried out in 1- to 2-day-old mice, the pathogenicity of SAD-L16 and its derivatives was again indistinguishable. In both models, the P-LC8 interaction seems to be not at all necessary for spread of the less attenuated SAD-L16 from a peripheral site of inoculation.
In an attempt to see differences between the attenuated SAD-D29 and its derivatives, similar experiments were carried out in more sensitive 1- to 2-day-old mice. Surprisingly, the elimination of the LC8 ligand through deletion of critical residues from the binding site and the simultaneous R333 substitution considerably reduced the pathogenicity of RV strains D-ΔP7 and D-ΔP11 after peripheral inoculation (Fig. 5). Very marginal differences between the mortality levels of D-ΔP7 and D-ΔP11 were observed (one more mouse died in the group of mice inoculated with D-ΔP7 than in the group inoculated with D-ΔP11; see Results section). However, due to deletion of four more amino acids in the P protein in D-ΔP11 than in D-ΔP7, minor impairment of D-ΔP11 in in vivo replication cannot be excluded.
Antigenic site III of RV G protein, particularly R333, has been implicated as playing an important role in RV neuroinvasive processes (11, 23, 29). Although R333 mutants were able to penetrate neuron cells as efficiently as the wild-type parent strains, their propagation was restricted after the second and third cycles, demonstrating a defect in trans-neuronal transfer (9). Moreover, the presence of R333 and a lysine at position 330 of the G protein has been shown to be important for RV to be able to bind to neuronal receptors and propagate in motoneurons (10, 34). It appears that R333 plays a dominant role in RV neuroinvasion, and the role of LC8 comes to light when the function of R333 is hampered.
To determine the degree of attenuation of the deletion mutants, graded doses of D-ΔP11 and SAD-D29 were administered to 1-to 2-day-old suckling mice i.m. or i.c. Compared to the parent virus, D-ΔP11 was attenuated by as much as 30-fold when administered by the i.m. route. Interestingly, the LD50 of both viruses was nearly identical by the i.c. route, indicating that D-ΔP11 spreads inefficiently from a peripheral site of inoculation to the CNS. The involvement of cytoplasmic dynein and the microtubular network in the retrograde transport of some viruses, including herpes simplex virus type 1, adenovirus, and pseudorabies virus, has been described in the past (14, 30, 31, 36). Furthermore, LC8 was shown to be essential for retrograde intraflagellar transport in Chlamydomonas (16, 24). These findings indicate that various organisms may utilize cytoplasmic dynein as a motor in reaching their preferred site. In the case of RV, however, abolishing the P-LC8 interaction affected only the efficiency of peripheral spread of the more attenuated strain lacking R333, but even for this recombinant (D-ΔP11), LC8 was dispensable at higher dose levels (Fig. 6). Although a substantial degree of attenuation was achieved by combining the R333 and LC8 binding site mutations, the contribution of LC8 to the peripheral spread of a fully virulent RV appears to be limited. Therefore, cellular factors other than LC8 or an entirely different mechanism should be involved in the retrograde axonal transport of RV. Future work should provide direct evidences for the contribution of LC8 to RV axonal transport and elucidate the neoroinvasive processes during RV infection.
The findings that RV infection in mice could take place in the oral mucosa (8) and that foxes could be immunized by oral administration of live attenuated RV (1, 2) led to the present-day wildlife rabies control programs of oral vaccinations (reviewed in reference 35). Two of the serious problems presented by the oral vaccine viruses were residual pathogenicity for target and nontarget species as well as the possibility of reversion to virulence. Significant advances in increasing the safety of oral rabies vaccine strains was made by selecting monoclonal antibody-resistant mutants with mutations at R333 (11, 29). Derivatives of SAD virus possessing amino acids differing from R333 have been developed for oral immunization of foxes and other animals (19).
Unlike other attenuated mutants such as SAD-Bern and SAD-B19 (35), R333 mutants are nonpathogenic for adult mice after i.c. inoculation. However, these strains remained pathogenic for baby mice, suggesting the potential danger of such live vaccines for immunocompromised humans who may come in contact with the virus. In this work, by applying the reverse genetics technology, an extensive attenuation of R333 mutants was achieved by deleting critical residues from the LC8 binding site in the P protein. The introduction of the combined mutations into the P and G proteins did not affect in vitro replication. This makes recombinant RV mutants D-ΔP7 and D-ΔP11 potential candidates for oral immunization of animals.
Highly safe live rabies vaccine viruses can not only be used to immunize against rabies, but potentially can also serve as vaccine vectors to protect humans and animals from other infectious agents. In addition to the five viral proteins, RV has been shown to express foreign genes in a stable manner for more than 25 serial passages (21). The potential of RV-based vectors as vaccines against other viral diseases such as AIDS was also demonstrated recently (28). The introduction of the described combined G and P protein mutations into RV vectors will undoubtedly diminish the risk associated with residual pathogenicity of RV and facilitate the design of safe RV-based vectors. The substantially attenuated D-ΔP7 and D-ΔP11 viruses are not only attractive candidates for live RV based vaccines but also provide some insight into the ill-defined events of RV dissemination through peripheral nerves to the CNS.
ACKNOWLEDGMENTS
I am grateful to L. T. C. de Vaan, M. Braber, and N. de Haas for perfect technical assistance, E. Schuurmans for digitizing Fig. 3, and S. King for kindly providing the anti-LC8 antibody. I thank colleagues from the animal service department of Intervet for their assistance and A. Gray, P. van der Marel, and I. Tarpey for valuable suggestions on the manuscript.
REFERENCES
- 1.Baer G M, Abelsth M K, Debbie J G. Oral vaccination of foxes against rabies. Am J Epidemiol. 1971;93:487–490. doi: 10.1093/oxfordjournals.aje.a121283. [DOI] [PubMed] [Google Scholar]
- 2.Black J G, Lawson K F. Sylvatic rabies studies in the silver fox (Vulpes vulpes): susceptibility and immune response. Can J Comp Med. 1970;34:309–311. [PMC free article] [PubMed] [Google Scholar]
- 3.Buchholz U J, Finke S, Conzelmann K-K. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J Virol. 1999;73:251–259. doi: 10.1128/jvi.73.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chenik M, Chebli K, Gaudin Y, Blondel D. In vivo interaction of rabies virus phosphoprotein (P) and nucleoprotein (N), existence of two N binding sites on P protein. J Gen Virol. 1994;75:2889–2896. doi: 10.1099/0022-1317-75-11-2889. [DOI] [PubMed] [Google Scholar]
- 5.Chenik M, Schnell M, Conzelmann K-K, Blondel D. Mapping the interacting domains between the rabies virus polymerase and phosphoprotein. J Virol. 1998;72:1925–1930. doi: 10.1128/jvi.72.3.1925-1930.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conzelmann K-K, Cox J H, Schneider L G, Thiel H-J. Molecular cloning and complete nucleotide sequence of the attenuated rabies virus SAD B19. Virology. 1990;175:485–499. doi: 10.1016/0042-6822(90)90433-r. [DOI] [PubMed] [Google Scholar]
- 7.Conzelmann K-K, Schnell M. Rescue of synthetic genomic RNA analogs of rabies virus by plasmid-encoded proteins. J Virol. 1994;68:713–719. doi: 10.1128/jvi.68.2.713-719.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Correa-Giron E P, Allen R, Sulkin S E. The infectivity and pathogenesis of rabies virus administered orally. Am J Epidemiol. 1970;91:203–215. doi: 10.1093/oxfordjournals.aje.a121129. [DOI] [PubMed] [Google Scholar]
- 9.Coulon P, Derbin C, Kucera P, Lafay F, Prehaud C, Flamand A. Invasion of the peripheral nervous systems of adult mice by the CVS strain of rabies virus and its avirulent derivative AvO1. J Virol. 1989;63:3550–3554. doi: 10.1128/jvi.63.8.3550-3554.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coulon P, Ternaux J-P, Flamand A, Tuffereau C. An avirulent mutant of rabies virus is unable to infect motor neurons in vivo and in vitro. J Virol. 1998;72:273–278. doi: 10.1128/jvi.72.1.273-278.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dietzschold B, Wunner W H, Wiktor T J, Lopes A D, Lafon M, Smith C L, Koprowski H. Characterization of an antigenic determinant of the glycoprotein that correlates with pathogenicity of rabies virus. Proc Natl Acad Sci USA. 1983;80:70–74. doi: 10.1073/pnas.80.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu Z F, Zheng Y, Wunner W H, Koprowski H, Dietzschold B. Both the N- and the C-terminal domains of the nominal phosphoprotein of rabies virus are involved in binding to the nucleoprotein. Virology. 1994;200:590–597. doi: 10.1006/viro.1994.1222. [DOI] [PubMed] [Google Scholar]
- 13.Jacob Y, Badrane H, Ceccaldi P-E, Tordo N. Cytoplasmic dynein LC8 interacts with lyssavirus phosphoprotein. J Virol. 2000;74:10217–10222. doi: 10.1128/jvi.74.21.10217-10222.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaelin K, Dezelee S, Masse M J, Bras F, Flamand A. The UL25 protein of pseudorabies virus associates with capsids and localizes to the nucleus and to microtubules. J Virol. 2000;74:474–482. doi: 10.1128/jvi.74.1.474-482.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.King S M, Patel-King R S. The Mr = 8,000 and 11,000 outer arm dynein light chains from Chlamydomonas flagella have cytoplasmic homologues. J Biol Chem. 1995;270:11445–11452. doi: 10.1074/jbc.270.19.11445. [DOI] [PubMed] [Google Scholar]
- 16.King S M, Barbarese E, Dillmann III J F, Patel-King R S, Carson J H, Pfister K K. Brain cytoplasmic and flagellar outer arm dyneins share a highly conserved Mr 8000 light chain. J Biol Chem. 1996;271:19358–19366. doi: 10.1074/jbc.271.32.19358. [DOI] [PubMed] [Google Scholar]
- 17.Lafay F, Benejean J, Tuffereau C, Flamand A, Coulon P. Vaccination against rabies: construction and characterization of SAG2, a double avirulent derivative of SADBern. Vaccine. 1994;12:317–320. doi: 10.1016/0264-410x(94)90095-7. [DOI] [PubMed] [Google Scholar]
- 18.Lo K W-H, Naisbitt S, Fan J-S, Sheng M, Zhang M. The 8-kDa dynein light chain binds to its targets via a conserved (K/RXTQT) motif. J Biol Chem. 2001;276:14059–14066. doi: 10.1074/jbc.M010320200. [DOI] [PubMed] [Google Scholar]
- 19.Masson E, Cliquet F, Aubert M, Barrat J, Aubert A, Artois M, Schumacher C L. Safety study of the SAG2 rabies virus mutant in several non-target species with a view to its future use for the immunization of foxes in Europe. Vaccine. 1996;14:1506–1510. doi: 10.1016/s0264-410x(96)00114-4. [DOI] [PubMed] [Google Scholar]
- 20.Mebatsion T, Weiland F, Conzelmann K-K. Matrix protein of rabies virus is responsible for the assembly and budding of bullet-shaped particles and interacts with the transmembrane spike glycoprotein. J Virol. 1999;73:242–250. doi: 10.1128/jvi.73.1.242-250.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mebatsion T, Schnell M J, Cox J H, Finke S, Conzelmann K-K. Highly stable expression of a foreign gene from rabies virus vectors. Proc Natl Acad Sci USA. 1996;93:7310–7314. doi: 10.1073/pnas.93.14.7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mebatsion T, Matthias K, Conzelmann K-K. Budding of rabies virus particles in the absence of the spike glycoprotein. Cell. 1996;84:941–951. doi: 10.1016/s0092-8674(00)81072-7. [DOI] [PubMed] [Google Scholar]
- 23.Morimoto K, McGettingan J P, Foley H D, Hooper D C, Dietzschold B, Schnell M J. Genetic engineering of live rabies vaccines. Vaccine. 2001;19:3543–3551. doi: 10.1016/s0264-410x(01)00064-0. [DOI] [PubMed] [Google Scholar]
- 24.Pazour G J, Wilkerson C G, Witman G B. A dynein light chain is essential for the retrograde particle movement of intraflagellar transport (IFT) J Cell Biol. 1998;141:979–992. doi: 10.1083/jcb.141.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raux H, Flamand A, Blondel D. Interaction of the rabies virus P protein with the LC8 dynein light chain. J Virol. 2000;74:10212–10216. doi: 10.1128/jvi.74.21.10212-10216.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reed L J, Muench H. A simple method of estimating fifty percent end points. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 27.Schnell M J, Mebatsion T, Conzelmann K-K. Infectious rabies viruses from cloned cDNA. EMBO J. 1994;13:4195–4203. doi: 10.1002/j.1460-2075.1994.tb06739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schnell M J, Foley H D, Siler C A, McGettigan J P, Dietzschold B, Pomerantz R J. Recombinant rabies virus as potential live-viral vaccines for HIV-1. Proc Natl Acad Sci USA. 2000;97:3544–3549. doi: 10.1073/pnas.050589197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seif I, Coulon P, Rollin P E, Flamand A. Rabies virulence: Effect on pathogenicity and sequence characterization of rabies virus mutations affecting antigenic site III of the glycoprotein. J Virol. 1985;53:926–934. doi: 10.1128/jvi.53.3.926-934.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sodeik B, Ebersold M W, Helenius A. Microtubule-mediated transport of incoming herpes simplex 1 virus capsid to the nucleus. J Cell Biol. 1997;136:1007–1021. doi: 10.1083/jcb.136.5.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suomalainen M, Nakano M Y, Keller S, Boucke K, Stidwill R P, Greber U F. Microtubule-dependent plus- and minus end-directed motilities are competing processes for nuclear targeting of adenovirus. J Cell Biol. 1999;144:657–672. doi: 10.1083/jcb.144.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thoulouze M I, Lafage M, Schachner M, Hartmann U, Cremer H, Lafon M. The neural cell adhesion molecule is a receptor for rabies virus. J Virol. 1998;72:7181–7190. doi: 10.1128/jvi.72.9.7181-7190.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tuffereau C, Leblois H, Benejean J, Coulon P, Lafay F, Flamand A. Arginine or lysine in position 333 of ERA and CVS glycoprotein is necessary for rabies virulence in mice. Virology. 1989;172:206–212. doi: 10.1016/0042-6822(89)90122-0. [DOI] [PubMed] [Google Scholar]
- 34.Tuffereau C, Benejean J, Roque Alfonso A-M, Flamand A, Fishman M C. Neuronal cell surface molecules mediate specific binding to rabies virus glycoprotein expressed by a recombinant baculovirus on the surface of lepidoptera cells. J Virol. 1998;72:1085–1091. doi: 10.1128/jvi.72.2.1085-1091.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vos A, Müller T, Schuster P, Schlüter H, Neubert A. Oral vaccination of foxes against rabies with SAD B19 in Europe, 1983–1998: a review. Vet Bull. 2000;70:1–6. [Google Scholar]
- 36.Ye G J, Vaughan K T, Roizman B. The herpes simplex virus 1 U(L)34 protein interacts with a cytoplasmic dynein intermediate chain and targets nuclear membrane. J Virol. 2000;74:1355–1363. doi: 10.1128/jvi.74.3.1355-1363.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]