Abstract
Background
Generative artificial intelligence (GenAI) shows promise in automating key tasks involved in conducting systematic literature reviews (SLRs), including screening, bias assessment and data extraction. This potential automation is increasingly relevant as pharmaceutical developers face challenging requirements for timely and precise SLRs using the population, intervention, comparator and outcome (PICO) framework, such as those under the impending European Union (EU) Health Technology Assessment Regulation 2021/2282 (HTAR). This proof-of-concept study aimed to evaluate the feasibility, accuracy and efficiency of using GenAI for mass extraction of PICOs from PubMed abstracts.
Methods
Abstracts were retrieved from PubMed using a search string targeting randomised controlled trials. A PubMed clinical study ‘specific/narrow’ filter was also applied. Retrieved abstracts were processed using the OpenAI Batch application programming interface (API), which allowed parallel processing and interaction with Generative Pre-trained Transformer 4 Omni (GPT-4o) via custom Python scripts. PICO elements were extracted using a zero-shot prompting strategy. Results were stored in CSV files and subsequently imported into a PostgreSQL database.
Results
The PubMed search returned 682,667 abstracts. PICOs from all abstracts were extracted in < 3 h, with an average processing time of 200 s per 1000 abstracts. A total of 395,992,770 tokens were processed, with an average of 580 tokens per abstract. The total cost was $3390. On the basis of a random sample of 350 abstracts, human verification confirmed that GPT-4o accurately and comprehensively extracted 342 (98%) of all PICOs, with only outcome elements rarely missed.
Conclusions
Using GenAI to extract PICOs from clinical study abstracts could fundamentally transform the way SLRs are conducted. By enabling pharmaceutical developers to anticipate PICO requirements, this approach allows for proactive preparation for the EU HTAR process, or other health technology assessments (HTAs), streamlining efficiency and reducing the burden of meeting these requirements.
Plain Language Summary
This study explored how artificial intelligence (AI) can help automate a key part of systematic literature reviews (SLRs), which are used to summarise research in healthcare. AI was used to extract specific information referred to as population, intervention, comparator and outcome (PICOs) from nearly 700,000 abstracts of clinical studies from the PubMed database. AI extracted the PICO information from all the abstracts in under 3 h, which is significantly faster than a human could do. This demonstrates that AI could save a lot of time and effort compared with using human reviewers to do the same task. The study also found that AI performed accurately in identifying the PICO elements in the abstracts, although further human verification is still required. The use of AI in this way could help pharmaceutical developers meet upcoming requirements from the European Union, which require timely and thorough reviews of clinical evidence. AI can speed up the process, reducing the burden on pharmaceutical developers and allowing them to prepare more efficiently for these assessments. However, further testing and validation is needed before using AI in this way becomes common.
Key Points
| This study contributes to the growing body of literature demonstrating that GenAI has the potential to transform the conduct of systematic literature reviews by automating key repetitive tasks. |
| This proof-of-concept study has shown that GenAI can successfully process and extract PICO elements from over 680,000 PubMed abstracts in under 3 h, significantly reducing the time and cost compared with using human review. |
| Using GenAI for rapid initial extraction of PICO data can assist pharmaceutical developers in shaping their clinical development programs to better align with market access requirements. |
Introduction
Recent advancements in generative artificial intelligence (GenAI), particularly large language models (LLMs), have demonstrated the potential of this technology to automate key tasks involved in conducting systematic literature reviews (SLRs), widely regarded as the gold standard for synthesizing evidence in healthcare [1–3]. LLMs can automate critical tasks for SLRs, including screening abstracts and full texts, assessing risk of bias and extracting data for use in network meta-analyses with high accuracy, all while significantly reducing the time and costs required by using human review [2, 4–7]. These capabilities are especially relevant as the healthcare industry faces increasingly complex demands for evidence synthesis.
The impending implementation of the European Union (EU) Health Technology Assessment Regulation 2021/2282 (HTAR) in January 2025, for instance, will place high demands on pharmaceutical developers for precise and timely SLRs [8–10]. Under the HTAR, a dossier for Joint Clinical Assessment (JCA) must be submitted after filing for market authorization approval (MAA) with the European Medicines Agency (EMA) [9, 10]. The JCA will evaluate the health problem(s) addressed by the technology, its technical characterisation and, most critically, its relative clinical effectiveness and safety, which will be standardised using the population, intervention, comparator and outcome (PICO) framework [8–10].
The PICO framework is essential for defining study characteristics to address specific clinical research questions in SLRs [3, 11]. It guides researchers in systematically identifying, extracting, evaluating and integrating evidence from multiple studies, as well as in developing search and screening criteria for eligibility [3, 11]. However, the PICO scoping process for the HTAR introduces significant challenges, as input from all EU member states is required to ensure diverse perspectives are considered [8, 12]. Variations in national healthcare systems and standards of care may lead to uncertainty and unanticipated analyses. For example, in a hypothetical submission for a common lung cancer indication, the authors suggest the proposed PICO process could necessitate between 10 and 14 PICOs, depending on specific country and health technology assessment (HTA) requirements, leading to a minimum of 280 and up to 840 requested analyses [13]. With only around 100 days to submit a JCA dossier after the PICO scope is confirmed, pharmaceutical developers face significant pressure to meet stringent deadlines, potentially impacting the quality and comprehensiveness of the submissions [8, 10].
Given these challenging requirements, the potential of GenAI to automate SLR tasks, including the extraction and analysis of study characteristics for PICOs, offers a transformative solution. We conducted a proof-of-concept study to evaluate the feasibility, accuracy and efficiency of using GenAI for mass extraction of PICOs from PubMed abstracts of clinical trials.
Methods
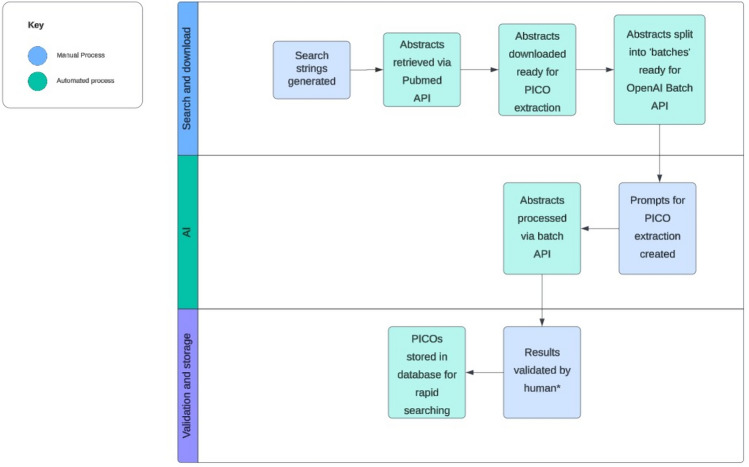
This was a proof-of-concept study designed to evaluate the feasibility, accuracy and speed of using GenAI to extract PICO data from abstracts of randomised clinical trials. Figure 1 provides an overview of the full methodology.
Fig. 1.
Summary of the PICO extraction process using GPT-4o. PICO, population, intervention, comparator, outcome; GPT-4o, Generative Pre-trained Transformer 4 Omni. *A random sample of 350 abstracts was selected for human validation
Search Strategy
Relevant abstracts were identified using the PubMed database, which was chosen due to its freely available application programming interface (API). An API is a set of rules or protocols that allows different software applications and systems to exchange data, features and functionality with each other [14, 15]. On 11 June 2024, the search API was used to retrieve a list of unique IDs [PubMed identifiers (PMIDs)] for clinical trial abstracts using the following search string: (“randomized controlled trial”[Publication Type] OR (“randomized”[Title/Abstract] AND “controlled”[Title/Abstract] AND “trial”[Title/Abstract])) NOT (“animals”[MeSH Terms] NOT “humans”[MeSH Terms]). The PubMed ‘specific/narrow’ therapy category filter, which has a sensitivity of 93% and specificity of 97% for capturing all clinical trials, was applied to streamline the results and make them more manageable (i.e. ≈700K versus 6 million results) [16].
All retrieved PMIDs were used to locate the corresponding abstracts within a local copy of the PubMed database, which is available for download. All abstracts were included in the analysis; no further filters were applied. Once retrieved, the abstracts were stored on a local hard drive for interaction with the LLM.
LLM and API Implementation for PICO Extraction
Generative Pre-trained Transformer 4 Omni (GPT-4o) was selected as the LLM engine for the analysis. In June 2024, GPT-4o was recognised as a leading LLM, consistently ranking at the top of prominent leaderboards [17, 18]. Although these leaderboards do not specifically evaluate PICO extraction capabilities, they assess LLMs across a range of metrics that can be considered proxies for human intelligence. GPT-4o was chosen not only for its status as a frontrunner in simulating intelligence but also for its accessibility via both API and batch API, which was needed for handling the volume of the analysis.
PICOs were extracted using the OpenAI Batch API, which enabled parallel processing of the abstracts by breaking them into “chunks” [19]. This method was ideal because the extraction of PICOs was identified as an “embarrassingly parallel” problem. In computing, “embarrassingly parallel” refers to a type of problem or algorithm that can be easily separated into many discrete, independent tasks that can be executed simultaneously with minimal or no need for communication between the tasks [20]. In this case, just as multiple humans could independently review many abstracts using the same criteria without needing to coordinate with each other, multiple processes running on multiple computers and/or servers can follow the same procedure simultaneously, scaling up easily and cheaply to thousands of instances.
In addition to speeding up the process through parallel processing, the batch API significantly reduced code complexity by greatly mitigating the need to manage timeouts and API rate-limiting compared with processing abstracts individually [19, 21]. Timeouts occur when an API takes too long to respond [21], and rate limiting restricts the number of requests that can be made in a given time period [22]; both of these can disrupt processing workflows. While each batch is guaranteed to finish within 24 h, there is a possibility that some individual elements might fail or be skipped [21]. However, in practice, all of our batches completed without issue, typically in less than 2 h, even when many were launched simultaneously. The batch API’s status report tracks errors and skipped queries, which would have allowed us to identify and retry an operation or batch if necessary. However, in our case, every operation in every batch was successfully processed [19, 21]. A final advantage of using the batch API was a 50% cost reduction compared with processing abstracts individually [19, 21].
Prompting Strategy
Interaction with GPT-4o was facilitated via API calls executed through a Python script, using custom prompts developed specifically to guide extraction of the necessary PICO data from the collected abstracts. The process began with simple input/output (I/O) zero-shot prompting [23], where the LLM was provided with an abstract and asked to extract each PICO element. Zero-shot prompting is a technique where the LLM is asked to perform a task or solve a problem that it has not been explicitly trained on, without any illustrative examples or guidance [24–26]. I/O zero-shot prompting was chosen because no complex logic or instructions were required for the LLM to complete the extraction. In our experience, extracting and summarising information that is already present in text is a task that LLMs excel at [7]. For example, to extract the population data, the prompt might read: “From the medical abstract below, please extract information pertaining to the patients being studied, including disease, line of therapy and disease severity; be detailed in your answer” (Fig. 2).
Fig. 2.
Example Python code for extraction of the population of interest for one abstract
The outputs from initial prompts were reviewed, and the prompts were then refined and resubmitted for further evaluation. This iterative process of reviewing outputs and refining prompts continued until no further improvements were identified, resulting in the final, optimised set of prompts.
Accuracy of the PICO Extractions
To assess the accuracy of the PICO extraction, a random subsample of 350 abstracts was selected for human verification after the LLM completed the screening of all abstracts and the PICO extractions were considered final. The verification process involved confirming whether all PICOs were correctly identified. The sample of 350 abstracts, representing 0.05% of the total dataset, was considered an acceptable size within the study’s timeframe, and it exceeded the amount a reviewer could feasibly verify in a single working day. Additionally, accuracy served as a criterion for stopping. If the extraction had been highly inaccurate, verification would have continued until the reasons for inaccuracy were identified.
Time and Cost Analysis of the PICO Extractions
To evaluate the time savings of using GenAI for PICO extraction, we tracked the number of abstracts a human reviewer could process (i.e. comparing the PICOs extracted with the original abstract text) in 1 day. The total number of PICO abstract extractions processed by GenAI was then compared with human labour, assuming an 8-h working day and 250 working days per year (accounting for weekends and public holidays). The working days per year assumption is likely an underestimation, as it does not account for variations in vacation policies or potential sick leave.
The cost of using the OpenAI Batch API was compared with the cost of processing abstracts individually. Each operation performed using the OpenAI LLM incurs a token usage, which reflects the combined number of tokens in both the input query and the generated response. These token counts are provided in the metadata of each API response. OpenAI applies a token-based pricing structure, with charges varying by model. This pricing applies uniformly, whether queries are processed individually or in batches through the Batch API. For batch operations, OpenAI offers a 50% discount to promote the use of this method [19, 21]. Following batch processing, we aggregated the reported token counts to calculate the total token usage, which was then multiplied by the published model-specific token rate. The final cost was determined by applying the 50% discount to this calculated total.
PICO Extraction Data Storage
The final extracted PICO data were initially stored as CSV files and then imported into a PostgreSQL database. PostgreSQL is a leading relational database system known for its robust full-text search capabilities, offering ‘tsvector’ and ‘tsquery’ data types that optimise full-text search and query functions. It also provides a variety of other tools useful for full-text search, such as stemming, stop words and thesauri. An advantage of using PostgreSQL over a dedicated document database (e.g. Elasticsearch) is that it allows for complex queries combining traditional structured query language (SQL) database operations with full-text search. The PostgreSQL database was constructed via the following steps: (1) a PostgreSQL database was provisioned using the Amazon Web Services Relational Database Service (AWS RDS); (2) a database schema optimised for full-text search was designed; (3) data were uploaded using custom bulk insert operations written in Python.
Results
The PubMed search returned 682,667 abstracts. PICOs from all abstracts were successfully extracted using the method described above in less than 3 h, with an average processing time of 200 seconds per 1000 abstracts. In contrast, one reviewer required approximately 8 h (or one workday) to review and compare PICO data for just 274 abstracts. Therefore, at this rate, it would take one reviewer approximately 2492 working days to review the total number of abstracts, which equates to at least 10 working years. A total of 395,992,770 tokens were processed, with an average of 580 tokens per abstract. The total cost was $3390, which represents a 50% cost reduction compared with processing abstracts individually.
Accuracy of the PICO Extractions
On the basis of the random sample of 350 abstracts, human verification confirmed that GPT-4o accurately and comprehensively extracted 342 (98%) of all PICOs. In the remaining cases, the model missed some outcome elements, but consistently extracted the population, intervention and comparator correctly. If the extractions had not achieved such high levels of accuracy, review would have continued. An example PICO output based on one abstract (the same abstract referenced in Fig. 2) is presented in Table 1.
Table 1.
Example PICO output based on an abstract
| Reference | Choueiri TK, Owles T, Albiges L, et al. Cabozantinib plus Nivolumab and Ipilimumab in Renal-Cell Carcinoma. N Engl J Med 2023;388:1767-78. https://doi.org/10.1056/NEJMoa2212851, PMID: 37163623 |
| Study design | Phase 3, double-blind trial |
| Population | Advanced clear-cell renal-cell carcinoma, previously untreated, intermediate or poor prognostic risk according to the International Metastatic Renal-Cell Carcinoma Database Consortium categories |
| Interventions and comparators | Cabozantinib plus nivolumab and ipilimumab versus nivolumab and ipilimumab plus placebo |
| Outcomes | Progression-free survival, overall survival, adverse events |
Discussion
This proof-of-concept study highlights the potential of GenAI, specifically the LLM GPT-4o, for large-scale, rapid and accurate PICO extraction. Prior work in this area has explored the feasibility of using natural language processing (NLP) to automate PICO extraction from biomedical abstracts with limited success [5, 27, 28]. To our knowledge, this study represents the first instance of PICO extraction achieved with this level of speed, scale and accuracy.
Our LLM-based process extracted PICO data from approximately 683,000 abstracts with up to 98% accuracy, on the basis of a random sample of 350 abstracts. This level of accuracy is notable given that GPT-4o correctly extracted the population, intervention and comparator elements among all 350 verified extractions. While it did occasionally miss outcomes, particularly when there was a long list in the abstract, these occurrences were relatively rare. Since clinical outcomes would typically require human verification using the full publication, these results suggest that our process can efficiently and accurately screen thousands of abstracts for pharmaceutical developers. While the results are promising, further large-scale verification will be essential to build confidence in regularly using GenAI extraction methods in regulatory and HTA settings. It is important to note, however, that human data extraction is also not error-free; several studies have reported high error rates, often involving misidentification or placing correct data in the wrong fields [29–31]. The continued improvement of foundation models, combined with growing user experience and expertise in utilising LLMs, indicates that these technologies may not only augment but also potentially outperform manual extraction methods in tasks such as SLRs. LLMs could become a reliable tool for large-scale data extraction, gradually reducing the reliance on human validation.
Robust SLRs are essential for reducing bias, ensuring reliable results and supporting informed decision-making in healthcare [1, 3, 11]. They are widely required by national HTA agencies and will be a key component of the EU HTAR JCA process [8–10]. However, traditional SLRs are both time-consuming and costly to complete [4, 11, 32]. A study of Collaboration for Environmental Evidence (CEE) SLRs published between 2012 and 2017 highlights the substantial labour required to complete a typical SLR [32]. For example, one reviewer would need nearly 10 days to screen the titles only for 8500 records retrieved as part of a typical search strategy. The mean number of abstracts screenable in a day was estimated at only 192, and full texts at 44. Given that an estimated 1200 records typically remain after title screening, it would therefore take one reviewer an additional 6.25 days to screen abstracts. For data extraction, only around seven full-text publications could be reviewed each day, which would translate to 14 days to complete extraction of only 100 studies (i.e. the average number included in an SLR) remained at this stage.
In our study, PICO extraction for approximately 683,000 abstracts took under 3 h. We estimate that retrieving the abstracts and refining the process took an additional 10 days, due to the experimentation and learning required. While PICO extraction is just one element of an SLR, the potential time and resource savings achieved using GenAI are clearly exponential – key tasks for SLRs could be completed in hours rather than weeks. Under the new EU HTAR, developers can engage in Joint Clinical Consultation (JSC) with the aim of obtaining guidance to shape the clinical development plan of a new health technology, to generate evidence that will be appropriate for a future JCA and EMA assessment [9, 10] Conducting an analysis of PICOs using GenAI before the JSC could be highly beneficial for developers. Potential PICOs could be rapidly and pre-emptively assessed, facilitating more informed discussions with the JSC, rather than waiting until the scoping process has begun to generate the appropriate PICO framework and subsequent analyses.
This proof-of-concept study has several limitations. Firstly, only PubMed abstracts were used for extraction of PICO data due to its freely available API. However, PubMed is not the only online database of medical literature, and this limits the generalisability of our findings. Further studies, conducted in collaboration with professional librarians who specialise in search strategies, are needed to ensure this methodology can be implemented across multiple databases. Secondly, as abstracts are often limited to around 300 words, it is possible that PICO elements for HTA analyses were missed. Abstracts present limited information; therefore, further testing and verification would be needed to apply this approach to full texts. In our previous work, we have noted the need to segment publications into chunks before interacting with the LLM (GPT-4), due to a token limit of approximately 6000 words [7, 33]. With GPT-4o, this token limit is no longer an issue, meaning that full-texts could be screened using this method. Another potential limitation of this study is that, while the proof-of-concept demonstrated strong performance with a large volume of records, its effectiveness in specific research areas remains unclear. We did not investigate whether the methodology used would successfully identify relevant abstracts in more specialised searches. Therefore, validation with a narrower set of abstracts focussed on a particular research area is necessary. Lastly, we applied an English-language-only filter to our search, so it is unknown how this method would perform with non-English-language abstracts.
The JCA clinical effectiveness assessment relies on the collection and synthesis of data using standard HTA methodologies – i.e. through SLR, meta-analyses and modelling. For decision-makers, it is crucial that all relevant clinical evidence from the published scientific literature is identified in a clear and comprehensive manner. SLRs based on GenAI methods offer promising methodologies that can contribute to this goal. To our knowledge, there has been no specific advice or commentary regarding the use of GenAI for developing JCA dossiers. However, the EMA and Heads of Medicines Agencies have recognised AI’s potential in medicines regulation [34]. Their AI workplan (2023–2028) includes an aim to identify and provide frameworks for the use of AI to increase efficiency, enhance understanding and analysis of data and support decision-making [34].
Conclusions
This promising application of GenAI to extract PICOs from clinical study abstracts holds transformative potential for the JCA process and could fundamentally enhance the way SLRs are conducted. By enabling pharmaceutical developers to anticipate member state PICO requests, this approach allows for proactive preparation for the JCA scoping process, or other HTAs, streamlining efficiency and reducing the burden of meeting these requirements. In addition, the creation of a fully searchable PICO database would be an invaluable resource for medical researchers, pharmaceutical developers and healthcare professionals involved in market access, helping to ensure that clinical development programs and analyses are strategically aligned with future needs and opportunities.
Acknowledgements
Medical writing support (including literature searching, writing the first draft, journal styling, journal submission, manuscript revisions and preparing tables) was provided by Diana Steinway, Appa Communications, Leuven, Belgium.
Declarations
Competing Interests
All authors are employees at Estima Scientific.
Funding
This research received no funding from external sources, and no funding was received for the preparation or publication of the manuscript.
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Availability of Data and Material
The data that support the findings of this study are not publicly available due to reasons of commercial sensitivity.
Code Availability
The code that supports the findings of this study is not openly available due to reasons of commercial sensitivity.
Author Contributions
The authors confirm that all contributors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship. T.R. and A.G. made significant contributions to the conception and design of the study. All authors were involved in data acquisition, analysis, or interpretation. J.L. devised and verified the search strategy. T.R. and A.G. were responsible for writing the Python script, creating the prompts and developing the database used in the study. Each author participated in drafting the work or critically revising it for important intellectual content, approved the final version for publication and agrees to be accountable for all aspects of the work, ensuring that any concerns regarding accuracy or integrity are appropriately investigated and resolved.
References
- 1.Murad MH, Asi N, Alsawas M, Alahdab F. New evidence pyramid. Evid Based Med. 2016;21:125–7. 10.1136/ebmed-2016-110401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fleurence RL, Bian J, Wang X, Xu H, Dawoud D, Fakhouri T, et al. Generative AI for health technology assessment: opportunities, challenges, and policy considerations. 2024. Available from: https://www.researchgate.net/publication/382302525_Generative_AI_for_Health_Technology_Assessment_Opportunities_Challenges_and_Policy_Considerations. Accessed 23 Aug 2024.
- 3.Page MJ, Moher D, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. PRISMA 2020 explanation and elaboration: updated guidance and exemplars for reporting systematic reviews. BMJ. 2021;372: n160. 10.1136/bmj.n160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borah R, Brown AW, Capers PL, Kaiser KA. Analysis of the time and workers needed to conduct systematic reviews of medical interventions using data from the PROSPERO registry. BMJ Open. 2017;7: e012545. 10.1136/bmjopen-2016-012545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khraisha Q, Put S, Kappenberg J, Warraitch A, Hadfield K. Can large language models replace humans in systematic reviews? Evaluating GPT-4’s efficacy in screening and extracting data from peer-reviewed and grey literature in multiple languages. Res Synth Methods. 2024;15:616–26. 10.1002/jrsm.1715. [DOI] [PubMed] [Google Scholar]
- 6.Guo E, Gupta M, Deng J, Park YJ, Paget M, Naugler C. Automated paper screening for clinical reviews using large language models: data analysis study. J Med Internet Res. 2024;26: e48996. 10.2196/48996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reason T, Benbow E, Langham J, Gimblett A, Klijn SL, Malcolm B. Artificial intelligence to automate network meta-analyses: four case studies to evaluate the potential application of large language models. Pharmacoecon Open. 2024;8:205–20. 10.1007/s41669-024-00476-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schuster V. EU HTA regulation and joint clinical assessment—threat or opportunity? J Mark Access Health Policy. 2024;12:100–4. 10.3390/jmahp12020008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.European Commission. Regulation on health technology assessment. 2024. https://health.ec.europa.eu/health-technology-assessment/regulation-health-technology-assessment_en. Accessed 22 Jul 2024.
- 10.European Commission. Commission Implementing Regulation (EU) 2024/1381 of 23 May 2024 laying down, pursuant to Regulation (EU) 2021/2282 on health technology assessment, procedural rules for the interaction during, exchange of information on, and participation in, the preparation and update of joint clinical assessments of medicinal products for human use at Union level, as well as templates for those joint clinical assessments. 2024. https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=OJ%3AL_202401381. Accessed 22 Jul 2024.
- 11.UNC Health Sciences Library. Systematic reviews: home. 2024. https://guides.lib.unc.edu/systematic-reviews/overview. Accessed 16 Aug 2024.
- 12.Delaitre-Bonnin C. Navigating the challenges and opportunities of the PICO framework in the new EU HTA regulation. 2024. https://www.ppd.com/blog/navigating-challenges-opportunities-pico-framework-new-eu-hta-regulation/. Accessed 23 Jul 2024.
- 13.van Engen A, Kruger R, Ryan J, Tzelis D, Wager P. Impact of additive PICOs in a European Joint Health Technology Assessment: a hypothetical case study in lung cancer. Poster HTA97 presented at ISPOR Europe 2022.
- 14.Gordon WJ, Rudin RS. Why APIs? Anticipated value, barriers, and opportunities for standards-based application programming interfaces in healthcare: perspectives of US thought leaders. JAMIA Open. 2022. 10.1093/jamiaopen/ooac023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.IBM. What is an API (application programming interface)? 2024. https://www.ibm.com/topics/api. Accessed 9 Sep 2024.
- 16.PubMed. PubMed user guide: clinical study categories. 2024. https://pubmed.ncbi.nlm.nih.gov/help/#clinical-study-category-filters. Accessed 27 Aug 2024.
- 17.Artificial Analysis. Independent analysis of AI models and API providers. 2024. https://artificialanalysis.ai/. Accessed 13 Aug 2024.
- 18.Hugging Face. LMSYS Chatbot Arena Leaderboard. 2024. https://huggingface.co/spaces/lmsys/chatbot-arena-leaderboard. Accessed 13 Aug 2024.
- 19.OpenAI. Batch API. 2024. https://platform.openai.com/docs/guides/batch/overview. Accessed 24 Jul 2024.
- 20.Reilly AC, Staid A, Gao M, Guikema SD. Tutorial: parallel computing of simulation models for risk analysis. Risk Anal. 2016;36:1844–54. 10.1111/risa.12565. [DOI] [PubMed] [Google Scholar]
- 21.OpenAI. Batch API FAQ. 2024. https://help.openai.com/en/articles/9197833-batch-api-faq. Accessed 24 Jul 2024.
- 22.Serbout S, Malki AE, Pautasso C, Zdun U. API rate limit adoption—A pattern collection. Proceedings of the 28th European Conference on Pattern Languages of Programs; Irsee, Germany: Association for Computing Machinery; 2024. p. 1–20, Article 5. 10.1145/3628034.3628039.
- 23.Giray L. Prompt engineering with ChatGPT: a guide for academic writers. Ann Biomed Eng. 2023;51:2629–33. 10.1007/s10439-023-03272-4. [DOI] [PubMed] [Google Scholar]
- 24.Yim RP, Rudrapatna VA. Zero-shot prompting is the most accurate and scalable strategy for abstracting the Mayo Endoscopic Subscore from colonoscopy reports using GPT-4. medRxiv. 2024:2024.03.22.24304745. 10.1101/2024.03.22.24304745.
- 25.Xu W, Lan Y, Hu Z, Lan Y, Lee RK-W, Lim E-P. Plan-and-solve prompting: improving zero-shot chain-of-thought reasoning by large language models. arXiv:230504091. 2023. 10.48550/arXiv.2305.04091.
- 26.Gowda DD, Suneel S, Naidu PR, Ramanan S, Suneetha S. Challenges and Limitations of Few-Shot and Zero-Shot Learning. In: Advances in Bioinformatics and Biomedical Engineering: IGI Global; 2024. p. 113–137. 10.4018/979-8-3693-1822-5.ch007.
- 27.Nye B, Jessy Li J, Patel R, Yang Y, Marshall IJ, Nenkova A, et al. A corpus with multi-level annotations of patients, interventions and outcomes to support language processing for medical literature. Proc Conf Assoc Comput Linguist Meet. 2018;2018:197–207. [PMC free article] [PubMed] [Google Scholar]
- 28.Hu Y, Keloth VK, Raja K, Chen Y, Xu H. Towards precise PICO extraction from abstracts of randomized controlled trials using a section-specific learning approach. Bioinformatics. 2023. 10.1093/bioinformatics/btad542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mathes T, Klaßen P, Pieper D. Frequency of data extraction errors and methods to increase data extraction quality: a methodological review. BMC Med Res Methodol. 2017;17:152. 10.1186/s12874-017-0431-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu C, Yu T, Furuya-Kanamori L, Lin L, Zorzela L, Zhou X, et al. Validity of data extraction in evidence synthesis practice of adverse events: reproducibility study. BMJ. 2022;377:e069155. 10.1136/bmj-2021-069155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rice H, Roussi K, King E, Martin A. Frequency and type of errors in data extraction within systematic literature reviews. Session SA83 presented at ISPOR Europe, 17–20 November 2024, Barcelona, Spain.
- 32.Haddaway N, Westgate M. Predicting the time needed for environmental systematic reviews and systematic maps. Conserv Biol. 2018. 10.1111/cobi.13231. [DOI] [PubMed] [Google Scholar]
- 33.Reason T, Rawlinson W, Langham J, Gimblett A, Malcolm B, Klijn S. Artificial intelligence to automate health economic modelling: a case study to evaluate the potential application of large language models. Pharmacoecon Open. 2024;8:191–203. 10.1007/s41669-024-00477-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.EMA. Artificial intelligence workplan to guide use of AI in medicines regulation. 2023. https://www.ema.europa.eu/en/news/artificial-intelligence-workplan-guide-use-ai-medicines-regulation. Accessed 17 Sep 2024 2024.