Abstract
Hypersensitivity pneumonitis (HP) is an immune-mediated inflammatory interstitial lung disease that may evolve to pulmonary fibrosis, a progressive disorder with a poor prognosis characterized by fibroblast activation and extracellular matrix accumulation. In HP lung fibroblasts, the gene expression of proteins involved in the interaction with the immune response, their isoforms, and how they influence their phenotype have yet to be elucidated. We analyzed the expression and splicing variants of 16 target genes involved in the interaction between HP fibroblasts and immune signaling and evaluated possible correlations with clinical data. The comparison of HP and control fibroblasts revealed distinct gene expression patterns. HP lung fibroblasts displayed an increased expression of IFI27 and PDFGRA and a downregulation of IL17RC and TGFBR3. IFI27 immunoreactive protein was markedly increased in HP lung tissues and normal fibroblasts treated with TGF-β. Furthermore, IFI27 overexpression in normal fibroblasts increased α-SMA and decreased cell number over time. The isoform analysis showed similar expression patterns for most genes, except for the AGER receptor with increased soluble variants relative to full-length AGER in HP fibroblasts. These findings indicate important differences in the expression of genes related to the immune response by HP fibroblasts, highlighting their unique characteristics and providing further insight into a possible profibrotic role of IFI27 in the disease.
Keywords: Hypersensitivity pneumonitis, mRNA splicing isoforms, IFI27, PDGFRA, AGER, α-SMA, Fibroblasts
Subject terms: Mechanisms of disease, Respiratory tract diseases
Introduction
Hypersensitivity pneumonitis (HP) is a complex and multifactorial lung disease caused by exposure to various antigens in occupational, home, or recreational environments that triggers an exaggerated immune response in predisposed individuals1. HP is classified into non-fibrotic and fibrotic based on the clinical, radiologic, and pathologic characteristics. The fibrotic form of the disease is progressive and often fatal2–4. Difficulty in identifying the causative antigen contributes to poor prognosis and development of the fibrotic form of this condition5. HP is a lung disorder that is frequently misdiagnosed6 or diagnosed at advanced stages7. Therefore, studying the cellular features that can influence its progression is crucial for better understanding the disease and identifying putative therapeutic targets.
The inflammatory response in HP is characterized by the infiltration of macrophages and lymphocytes, followed by cytokine and chemokine secretions1. The recruitment of neutrophils to the lung may cause epithelial damage and the production of damage-associated molecular patterns like Tenascin C (TNC)1,8. If the antigenic stimulation continues, adaptive immunity promotes the differentiation of lymphocytes to the Th1- and Th17-phenotypes and the production of IFN-γ and IL-17, respectively9. In addition, the induction of a Th2 environment characterized by a high output of IL-4/IL-13 and secretion of molecules like IL-17, PDGF, and TGF-β contributes to the development and progression of fibrosis through activation and differentiation of fibroblasts to myofibroblasts10. Fibroblasts possess receptors and express proteins that can impact their transformation toward a more aggressive phenotype. However, the possible splicing isoforms and their functional response in HP fibroblasts are still unknown.
Humans have over 100,000 transcripts derived from only 20,000 genes11,12. When translated, mRNA isoforms may code for proteins with variable domains and contrasting functions12. For example, long isoforms of the secreted protein TNC containing extra domains promote tissue remodeling during development and under pathological conditions. In contrast, short variants with no additional domains favor tissue stability8. Previous reports have described the presence and relevance of splicing isoforms in pulmonary diseases13,14, including interstitial diseases like idiopathic pulmonary fibrosis (IPF). So far, however, no studies have been conducted on HP.
In this study, we used fibroblasts derived from patients with HP to investigate the expression of sixteen genes implicated in the immunopathogenesis of HP and their isoform variants. Relative to controls, HP fibroblasts expressed four genes with contrasting levels, showing an increase of IFI27 (Interferon I-Inducible Protein 27), and PDGFRA (Platelet Derived Growth Factor Receptor Alpha), and a decrease of IL17RC (IL-17 receptor C), and TGFBR3 (Transforming growth factor beta type III receptor, also known as betaglycan)]. Likewise, the soluble splicing isoforms of the AGER receptor were also differentially expressed. Finally, the phenotypic characterization of lung fibroblasts overexpressing IFI27 demonstrated its role in establishing a profibrotic environment.
Results
Study population
Twenty primary HP fibroblast lines and four controls were used. Table 1 indicates the demographic and clinical characteristics of HP donor patients. Seven patients were classified as non-fibrotic and thirteen as fibrotic HP according to ATS/JRS/ALAT guidelines2. Most of the patients were women (75%). Seven patients reported being active smokers, and there were no differences in age or forced vital capacity (FVC1% predicted) and diffusing lung capacity for carbon monoxide (DLCO % predicted) parameters. However, patients with fibrotic HP showed a significant decrease in the % of oxygen saturation (SpO2) at rest compared with the non-fibrotic group. Seven patients with fibrotic HP presented honeycombing in the high-resolution computed tomography (HRCT), a hallmark of usual interstitial pneumonia (UIP). The control group consisted of commercially acquired cell lines from nonsmoking individuals and did not show age differences with HP patients (57 ± 12.5 vs. 59.7 ± 8.5 years, respectively).
Table 1.
Demographic and clinical characteristics of HP donors.
| Variable | Non-fibrotic HP n = 7 | Fibrotic HP n = 13 | p-value |
|---|---|---|---|
| Age (years) | 61 ± 10 | 59 ± 8 | 0.48 |
| Gender (M/F) | 1 / 6 | 4 / 9 | 0.61 |
| Active smoker | 1 | 6 | 0.328 |
| FVC % predicted (SD) | 62 ± 19 | 63 ± 17 | 0.697 |
| DLCO % predicted (SD) | 48 ± 18 | 55 ± 17 | 0.812 |
| SpO2 (SD) | 94.5 ± 0.03 | 89 ± 7 | < 0.0001 |
Patients were classified as fibrotic or non-fibrotic following the Official ATS/JRS/ALAT Clinical Practice Guideline2. Tests were performed at 2440mts of altitude. Numbers in age and functional tests are presented as average ± standard deviation.
FVC forced vital capacity, DLCO diffusing lung capacity for carbon monoxide, SpO2 oxygen saturation at rest, HP Hypersensitivity pneumonitis, M male, F female, SD standard deviation.
Expression of immune-associated genes in primary HP lung fibroblasts
The fibroblast phenotype in HP is still being determined, and the information on molecules and receptors that modulate their biological response is scarce. We focused on establishing differences in the expression of sixteen genes associated with the immune response in fibroblasts derived from HP patients and controls. These were selected based on global expression databases for HP and other ILDs15 (Table 2).
Table 2.
Selected target genes have altered expression in fibrosing diseases and splicing isoforms associated with other pathologies.
| Factor | Function | Genome-wide expression | Individual data | PF in KO models | Functional splicing isoforms | Diseases linked to isoforms |
|---|---|---|---|---|---|---|
| MyD88 | Adapter | bFast progression IPF (GDS4580) | aHP16, SSc PF17 | Anti-inflammatory (Myd88s)18 | B-cell lymphoma19 | |
| TNC | DAMP | bIPF (GDS1252), CF (GDS2142) | bIPF, chronic HP, SSc*17,20,21 | aPF (GDS251), SSc PF20 | Tissue remodeling (long TNC)8,22 | Chronic hepatitis22,23 |
| AGER | Receptor | aIPF (GDS1252), CF (GDS2142) | Soluble (esAGER)24 | IPF13, atherosclerosis25, anemia, cancer26 | ||
| IFI27 | Adapter | aSSc (GDS5499), IPF27–29(GDS4580)** | Altered apoptosis30, N-ter truncated31 | Cervical and breast cancer31 | ||
| PDFGRα | Receptor | Non-signaling32 | Osteosarcoma33 | |||
| PDGFRβ | Receptor | aIPF (GDS4580) | aIPF34 | aPF35 | Soluble36 | |
| IFNGR1 | Receptor | aIPF (GDS4279), SSc PF (GDS4995) | ||||
| IFNGR2 | Receptor | α | ||||
| IL4Ralfa | Receptor | bIPF (GDS4580, GDS4279) | bIIP37,38 | Soluble (sIL4R)39 | Allergic asthma14 | |
| IL13Ralfa1 | Receptor | Soluble40 | ||||
| IL13Ralfa2 | Receptor | bIPF (GDS1252) | bIIP37,38 | Soluble41 | β | |
| IL17RA | Receptor | bIPF (GDS4580) | aHP42, PF (GDS251) | Soluble43 | Prostate cancer43, γ | |
| IL17RC | Receptor | Prostate cancer44 | ||||
| TGFBR1 | Receptor | bPF bleo (GDS5078,GDS251) | bAdvanced IPF45 | Extracellular domain variation (IB)46 | Pituitary adenomas46 | |
| TGFBR2 | Receptor | aPF47 | Low or independent signaling (IIB and IIC)48,49 | Acute myeloid leukemia50, rheumatoid arthritis51 | ||
| TGFBR3 | Receptor | |||||
| aDiminished | bIncreased |
The available genome-wide (GEO profiles, NCBI) and individual expression data in indicated conditions and mouse knock-out (KO) models were summarized. a and b codes indicate increased or diminished expression relative to controls. Reported functional splicing isoforms and associated diseases were also included.
TNC tenascin C, AGER advanced glycosylation end-product specific receptor, IFI27 interferon I inducible protein 27, PDFGR platelet-derived growth factor receptor (alpha and beta), IFNGR interferon gamma receptor (R1 and R2), IL4R interleukin 4 receptor, IL13R interleukin 13 receptor, IL17R interleukin 17 receptor (A and C), TGFBR transforming growth factor beta receptor, IPF idiopathic pulmonary fibrosis, CF cystic fibrosis, IIP idiopathic interstitial pneumonia, PF pulmonary fibrosis, SSc Systemic Sclerosis.
*Several additional inflammatory and fibrotic diseases52.
**Other genome-wide assays for also showed increased [HP (GDS4587); IPF (GDS4279)] or diminished expression [IFP (GDS1012); SSc (GDS4995, GDS1012)] relative to controls. GEO profiles are indicated in parentheses.
α Mutants in splice sites have been identified in IFNGR1 and IFNGR2 genes that cause Mendelian susceptibility to Mycobacterial disease53.
β Although soluble forms of the receptor have been found during parasitic infection in humans, it has not been demonstrated that they are derived from alternative splicing54.
γ Soluble forms of IL17RA have been found in serum derived from rheumatoid arthritis patients but not demonstrated to be derived from alternative splicing84.
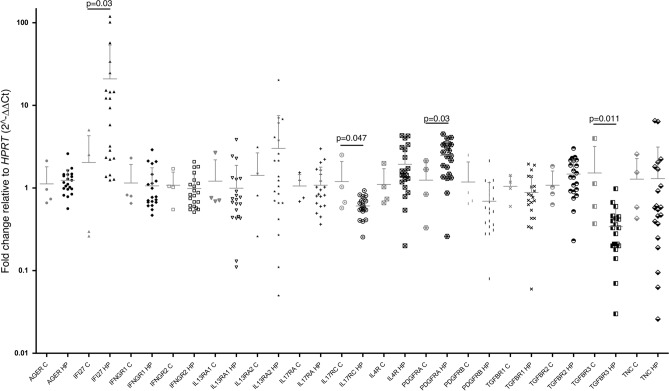
The expression of target genes in early passages of lung fibroblasts (p2-p3) was analyzed using quantitative PCR. As shown in Fig. 1, HP fibroblasts exhibited a significant decrease in the expression of membrane receptors TGFBR3 and IL17RC compared to controls (p = 0.011 and p = 0.047, respectively). Additionally, HP fibroblasts overexpressed the cell surface receptor PDGFRA and the interferon I inducible protein IFI27 (p = 0.03). No significant differences were observed for the remaining analyzed genes.
Fig. 1.
Differential mRNA expression of target genes in control and HP lung fibroblasts. Graphs of qPCR analysis of genes associated with the immune response in control (C, n = 4) and HP fibroblasts (HP, n = 20). Statistical differences between controls and HP are indicated with p values (Mann-Whitney test). The data columns include a horizontal bar indicating the mean and standard deviation. The values were adjusted by comparing the Ct values of each gene with those of HPRT using the 2∧-ΔΔCt formula.
Expression pattern of splicing variants
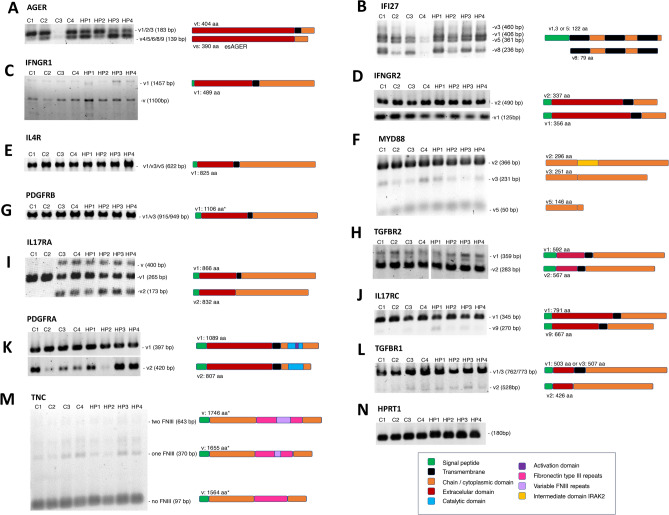
Previous reports have described the presence and relevance of splicing isoforms in pulmonary diseases (see Table 2). We, therefore, determined what splicing isoforms of the selected targets were expressed in HP and control cell lines (n = 4). The protein sequence alignments of variable regions of the splicing isoforms are shown in Figure S1. The PCR product sizes and presence or absence of bands in the initial PCR reactions established the expression of specific isoforms of most target genes in HP fibroblasts and controls (Fig. 2). The splicing isoform bands were quantified and graphed for HP and control samples (Figure S2).
Fig. 2.
Distinctive pattern of gene isoforms in HP lung fibroblasts. Alternative variants of selected genes were analyzed by end-point PCR. The splicing PCR products were resolved in 2% agarose-TBE (A-M). The expected size for each variant (V) is indicated on the right side of the gels. A schematic representation of the protein corresponding to each transcript is also included at the side. * Predicted size of complete protein. (N) HPRT amplification was used to verify the quality and loading quantities of used cDNAs. The color code guide details specific domains of the proteins represented for each target gene studied. The images were extracted from the original gels to highlight the bands of interest. For TGFBR2 (H), the image presented was generated from juxtaposed images of the same gel.
A differential isoform pattern of AGER was identified between control and HP fibroblasts. As shown in Fig. 2A and Figure S2, the full-sized AGER was increased in most control samples relative to the amplification of AGER without the transmembrane domain (esAGER). At the same time, the AGER and esAGER levels were similar in HP fibroblasts. For IFI27 mRNA isoforms, we identified four bands corresponding to three types of transcripts (Fig. 2B). However, no differences were observed between groups (Figure S2). Variant 1 of IFNGR1 was expressed along with an unknown variant (Fig. 2C). The unknown variant was not further characterized because it was found in both control and HP samples.
Similarly, two variants of IFNGR2 (Fig. 2D), one variant of IL4R (Fig. 2E), three variants of MYD88 (Fig. 2F), one variant of PDGFRB (Fig. 2G), and two variants of TGFBR2 (Fig. 2H) were equally expressed in control and HP fibroblasts (Figure S2). Related to IL17RA, HP fibroblasts expressed three splicing variants. This pattern was observed in two control samples, while the others had only variant 1 (Fig. 2I). The soluble isoform of IL17RC, found to be scarce compared to the complete form, was detected in both groups (Fig. 2J and S2). Truncated variants of PDGFRA and TGFBR1 were expressed in control and HP fibroblasts with minimal levels compared to full-length ones (Fig. 2K,L, and S2). However, differences in expression levels in some isoforms were observed, which may be relevant to future studies. Finally, the mRNA isoforms of TNC expressed in lung fibroblasts were the same in control and HP samples and were shorter in length than the expected most extended 2 kb amplicon in the PCR reaction (Fig. 2M). The HPRT1 expression was used as a load control and did not reveal differences (Fig. 2N).
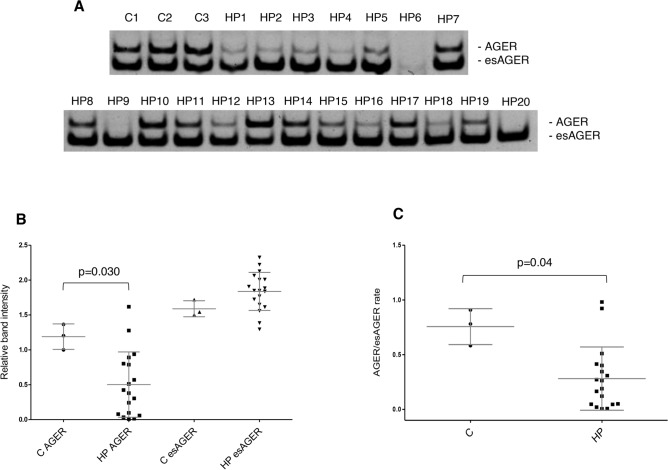
A soluble variant of AGER is overexpressed in HP fibroblasts
AGER was the only gene that showed differential expression between control and HP fibroblast splicing variants. Further analysis of the variant expression in all HP fibroblast cultures was conducted. Figure 3A,B shows a significant reduction in the AGER full-length variant expression and an increase in the soluble isoform esAGER in HP fibroblasts versus control cells. Accordingly, HP lung fibroblasts had a significantly reduced AGER/esAGER rate (Fig. 3C). These results indicate a higher presence of transcripts coding for the soluble form relative to the full-length form in these cells.
Fig. 3.
AGER isoforms are altered in HP lung fibroblast. The full-length (AGER) or soluble isoform (esAGER) expression patterns in control (C, n = 3) or HP lung fibroblasts (HP, n = 20) were analyzed by PCR. (A) Image of amplification products resolved and visualized in acrylamide gels. (B-C) Graphs of densitometric analysis of bands corresponding to amplification products.
Clinical classifications show the relevance of PDGFRA and IFI27 in HP
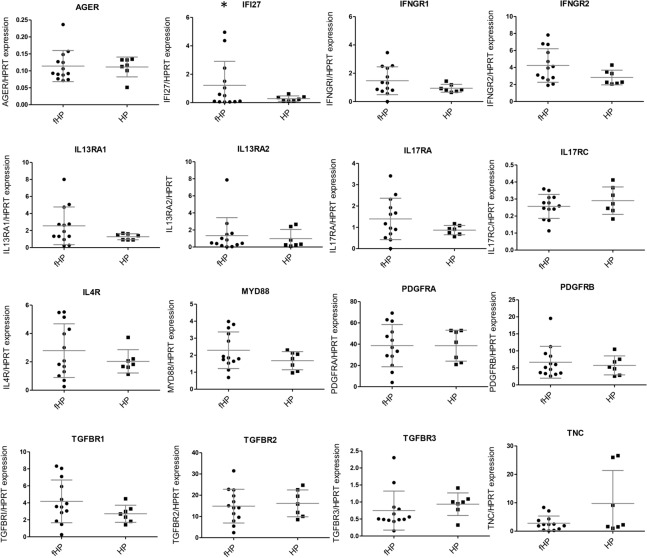
The gene expression values were correlated with clinical data to determine whether they were associated with clinical features of HP patients, including age, smoking status, percentage of lymphocytes in bronchoalveolar lavage (BAL), fibrotic or non-fibrotic HP, and the presence of honeycombing pattern. No differences in age, smoking status, or cell profile in BAL with gene expression were found.
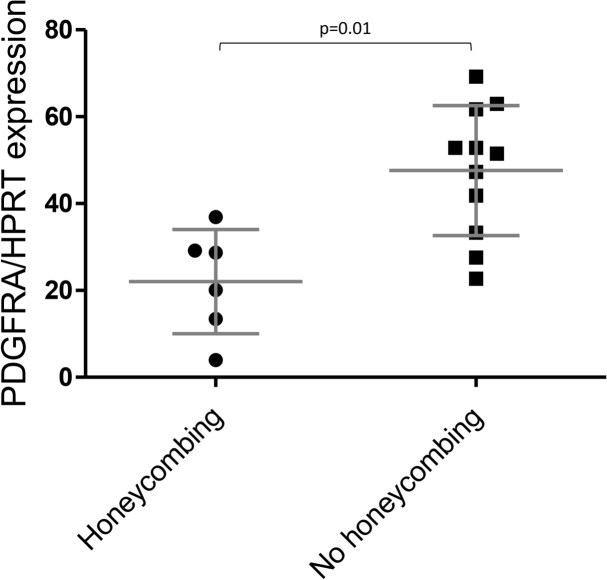
Also, no significant associations were found between fibrotic and non-fibrotic HP (Fig. 4). However, the highest levels of IFI27 were observed in the fibroblasts of patients diagnosed with fibrotic HP (fHP) (Fig. 4). Patients with fHP were also grouped according to the presence or absence of honeycombing analyzed by HRCT, and a possible association with gene expression data was determined. The honeycombing pattern represents the destruction and fibrosis of the pulmonary tissue with complete loss of the acinar architecture, containing numerous air-trapping areas with thick and fibrous walls55. As shown in Fig. 5, only PDGFRA showed a significant difference, with lower expression levels associated with a honeycombing pattern.
Fig. 4.
Gene expression associated with fibrotic or non-fibrotic HP diagnosis. Statistical differences were not found when comparing data using the Mann–Whitney test. HP (n = 7); fHP (n = 13). * fHP fibroblasts expressed higher IFI27 mRNA levels.
Fig. 5.
The honeycomb pattern correlates with HP lung fibroblast gene expression. Relationship of PDGFRA expression and honeycombing pattern. Statistical difference was assessed with the Mann-Whitney test.
IFI27 overexpression induced some myofibroblast characteristics
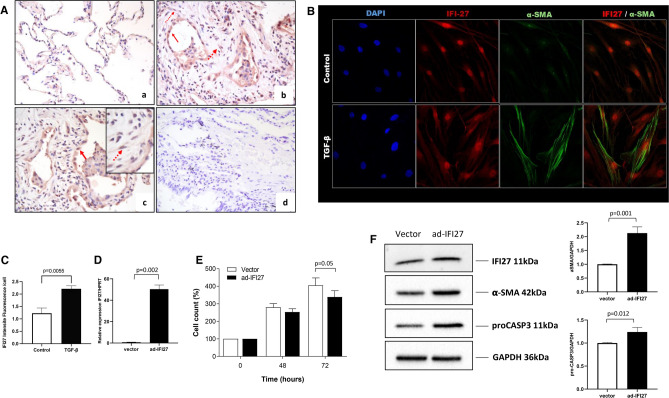
Of the genes that exhibited differential expression in HP fibroblasts compared with control fibroblasts, IFI27 had the greatest difference between HP and controls, with the highest expression values found in HP (Fig. 1). IFI27 is a molecule associated with the immune response that affects cellular proliferation and cell death56. Its role in lung fibroblast biology remained unknown and required further research. Consequently, we determined its impact on lung fibroblast phenotype. First, to evaluate if the elevated expression of IFI27 in isolated fibroblasts also occurs in lung tissue, we analyzed the presence of IFI27 by immunohistochemistry in the lungs of fibrotic HP patients. Compared to the control tissue, IFI27 was overexpressed in tissues derived from fibrotic HP lungs; interestingly, IFI27 was detected in cells with stromal and epithelial morphologies (Fig. 6A). The profibrotic cytokine TGF-β induces differentiation of fibroblasts to myofibroblasts57, promoting the elevated expression of aSMA. Thus, in search for the colocalization of IFI27 and aSMA, normal human lung fibroblasts were treated with TGF-β. Figure 6B shows that the treatment induced a-SMA overexpression and increased IFI27 protein levels (Fig. 6B,C).
Fig. 6.
IFI27 overexpression reduces the cell count over time and induces activation of lung fibroblasts. (A) IFI27 immunostaining in lung tissues. (a) Lung tissue from control donor. (b-c) The positive signal for the IFI27 protein was in cells with epithelial (solid arrow) and mesenchymal morphology (dotted arrow) in HP. (d) Immunostaining of HP lung tissue where the primary antibody was omitted. (B) Immunofluorescent localization of IFI27 and αSMA. (C) Quantification of IFI27 means fluorescence intensity in (B). (D) qPCR analysis of IFI27 expression in transduced fibroblasts (E) CyQuant assay in normal lung fibroblasts (NHLF and HPF) transduced with the empty adenoX-DS-Red-Express (vector) and vector coding the full-length IFI27 (ad-IFI27). Two independent experiments in quadruplicate were done in two control cell lines. (F) Representative immunoblots of IFI27, pro-caspase 3, and αSMA. The side graph represents the densitometric analysis of three independent experiments of relative values of target proteins compared to GAPDH as a loading control. The images were extracted from the original blots to show the bands of interest. RNA and protein samples were collected 48 h after transduction.
To evaluate the role of IFI27 in the lung fibroblast phenotype, IFI27 was overexpressed using an adenoviral vector coding for the whole gene in normal primary lung fibroblasts (Fig. 6D). Then, we analyzed the effects on fibroblast cell number over time, pro-caspase 3, and α-SMA expression. Quantified with an assay based on DNA content, upregulation of IFI27 provoked a decrease in fibroblast cell number after 72 h, compared with the empty vector (Fig. 6E). To establish if this reduction in cell count was linked to apoptosis, we assessed pro-caspase-3 levels. Our findings indicate elevated pro-caspase-3 protein levels in fibroblasts overexpressing IFI27 (Fig. 6F). Since the activation and differentiation of fibroblasts into myofibroblasts is a crucial process in the progression of lung fibrosis, we also examined the effect of IFI27 on the expression of α-smooth muscle actin (αSMA). Our results showed that the IFI27 overexpression caused significantly increased αSMA protein levels, suggesting that IFI27 can regulate the fibroblast phenotype (Fig. 6F).
Discussion
Despite the importance of fibroblasts in HP, their biology, altered expression of molecules and receptors implicated in the immunopathogenesis, and possible repercussions in its phenotype remain unclear. In this study, the expression of sixteen genes involved in the immunopathogenesis of HP was investigated in primary lung fibroblasts derived from HP patients relative to controls. Likewise, we determined for the first time the expression of the splicing variants of these genes.
Gene expression analysis revealed considerable variability for most studied genes, particularly in HP fibroblasts. This result is unsurprising because primary cultures are a heterogeneous mix of fibroblast subtypes, like lipofibroblasts or myofibroblasts, whose role and importance in lung fibrosis have already been described58. Nevertheless, we found a marked overexpression of PDGFRA and IFI27, while IL17RC and TGFBR3 were downregulated in HP relative to controls.
The IL-17 pathway has been linked to the development of pulmonary fibrosis59. We observed low levels of IL17RC in HP fibroblasts and did not identify any differences in IL17RA expression, which forms the functional heterodimer for IL-17A/F signaling59. We hypothesized that the downregulation of IL17RC is insufficient to have a functional effect. However, further studies are required to determine the significance of this difference. On the other hand, TGFBR3 encodes the transmembrane glycoprotein betaglycan, a receptor involved in the signaling pathway of TGF-β. Some studies have indicated that TGFBR3 may act as an antifibrotic molecule, as it can bind to receptors TGFBRI and TGFBRII, thereby preventing the canonical signaling through the Smad2/3 cascade60. Consequently, the observed reduction of this receptor in HP fibroblasts could be associated with the development of fibrosis.
The expression of protein variants or isoforms may result from genetic mutation, changes in the cellular environment, transcription, or alternative splicing, and it has been associated with multiple diseases61. Each cell has a unique repertoire of splicing regulatory factors that can be adjusted or altered according to its needs12. We found that fibroblasts express isoforms encoding proteins with variations in the extracellular, transmembrane, or cytoplasmic domain. In some cases, PCR products corresponding to unreported variants were also amplified. Most of these variants presented similar expression levels in HP and control fibroblasts. However, our analysis showed reduced levels of full-length AGER in HP fibroblasts. In contrast, the esAGER transcripts, which lack the exon coding for the transmembrane domain, were increased in the fibroblasts of HP lungs relative to AGER.
A healthy lung is characterized by high levels of AGER expression, mainly in type I alveolar epithelial cells62,63. In contrast, AGER is diminished in some pathological states, such as IPF and lung tumors13,64. The AGER expression was reported in lung fibroblasts and fibroblastic foci in usual interstitial pneumonia65. Low soluble AGER levels were also associated with greater IPF severity and a higher rate of death66. Contrastingly, elevated serum sAGER levels characterized tuberculosis, pneumonia, acute respiratory distress syndrome, and systemic sclerosis67. Its absence is protective in the bleomycin model of pulmonary fibrosis68, indicating that a general imbalance in AGER expression can contribute to diseases.
No differences in AGER were observed in the initial gene expression analysis, which is consistent with a previous report that found no changes in AGER expression when comparing normal and IPF lung fibroblasts69. However, it is possible that the imbalance between full-length AGER and esAGER in HP fibroblasts may alter their microenvironment by sequestering ligands. Unfortunately, no studies have investigated the relationship between both variants in the same cell, and further investigation is required to evaluate the effect of the low mRNA AGER/esAGER ratio on the behavior of HP fibroblasts. As mentioned earlier, our results showed increased expression of PDGFRA in HP fibroblasts compared with control fibroblasts. Interestingly, we found that in HP samples, PDGFRA is differentially expressed with high mRNA levels in fibroblasts derived from patients without a honeycombing pattern. PDGF and its receptors have been extensively investigated and are critical in pulmonary fibrosis70. Differential expression of PDGFRA has already been documented in lung fibroblasts. Matrix resident fibroblasts associated with proliferation and damage repair without fibrosis express high levels of PDGFRA. In contrast, myofibroblasts expressing αSMA with reduced proliferative capacity have lower receptor levels71. Thus, our findings suggest that lung fibroblasts derived from fHP with honeycombing, with low PDGFRA levels, would have a higher composition of myofibroblasts than fibroblasts derived from fHP patients with no honeycombing pattern.
Finally, of the 16 targets initially evaluated, IFI27 was the gene with the highest mRNA levels in HP fibroblasts relative to controls, and we hypothesized that aberrant expression of this gene could affect the fibrotic phenotype of lung fibroblasts. Consequently, we decided to perform immunohistochemistry to determine if the increase in IFI27 observed in culture was also reflected in lung tissue. Interestingly, we found that IFI27 was strongly expressed in fibrotic areas of HP lungs, including cells with epithelial and interstitial morphologies. Supporting this concept, we found that normal fibroblasts stimulated with TGF-β, a well-documented promoter of myofibroblast differentiation by a-SMA induction57, increased the expression of IFI27. This supports our results and suggests that IFI27 may be an essential molecule in the pathology of HP. To further establish the influence of IFI27 on lung fibroblast behavior and phenotype, IFI27 was overexpressed in normal lung fibroblasts, and its effects on proliferation, pro-caspase 3, and αSMA protein production were evaluated. We found that IFI27 diminished the cell count after 72 h and increased levels of pro-caspase3 and αSMA proteins. This suggests that this protein may play a profibrotic role in lung fibroblasts, mainly by promoting some myofibroblasts characteristics. IFI27 may promote a pro-fibrotic phenotype by inhibiting NR4A1 activity. NR4A1 is a transcription factor that usually antagonizes TGF-β’s pro-fibrotic effects by recruiting co-repressor complexes72,73. IFI27 physically interacts with NR4A1, promoting its nuclear export and reducing its transcriptional activity72,74. Additionally, related to our pro-caspase 3 data, mitochondrial located NR4A1 exposes the BH3 domains of Bcl-2, converting it to a cytotoxic molecule75. Nevertheless, additional work is required to confirm this proposed mechanism. The possible profibrotic role of IFI27 has also been proposed as part of the mechanisms involved in mitochondrial dysfunction associated with liver cirrhosis76. Furthermore, IFI27 overexpression exacerbates the fibrotic response to unilateral ureteral obstruction in mice kidneys and, intriguingly, also enhances the expression of caspase 3 and αSMA77.
Thus, HP fibroblasts exhibit increased expression of IFI27, PDGFRA, and esAGER relative to AGER; and decreased expression of IL17RC and TGFBR3. The IFI27 expression pattern in HP fibroblasts and lung tissue suggests a profibrotic role in the pathogenesis of HP, influencing the fibroblast cell count and differentiation, leading to some of the myofibroblast characteristics of fibrotic areas.
Methods
Ethics statement and human material
The study was conducted according to the international guidelines regarding ethical and biomedical research in human subjects under the WMA Declaration of Helsinki. The protocol was approved by the Ethics and Research Committee at INER and registered with the internal number B23-19. We used cryopreserved primary lung fibroblasts from patients diagnosed with HP (Table 1) according to the ATS/JRS/ALAT guidelines2 (n = 20): 15 females (F) and 5 males (M). All patients signed informed consent. Control lung fibroblasts were purchased from Lonza (NHLF, Walkerside, Inc.) or PromoCell (HPF, USA) (n = 4; 3 F and 1 M). Samples were identified with a number, and all data were kept anonymous and protected.
Cell culture and fibroblast isolation
Primary human lung fibroblasts were achieved from a fragment of lung tissue of HP patients by open lung biopsy that was used for clinical diagnosis. Lung tissue was cut into small fragments (~ 1 × 1 mm) that were incubated with 1X Trypsin–EDTA for 15 min at 37℃. Then, Ham’s F-12 medium (GIBCO) with 10% fetal bovine serum (FBS) (GIBCO BRL) was added, and homogenates were centrifuged for 10 min at 1500 g. Pellets and supernatants were seeded into a cell culture dish with F-12 medium supplemented with 10% FBS, 100 U/ml penicillin, and 100 g/ml streptomycin. Cells were cultured at 37 °C in 5% CO2/95% air. The mesenchymal phenotype of the cells was demonstrated in previous work analyzing the expression of cell surface antigen CD9078.
The cells were grown in Ham’s F12 (Gibco, Thermo) with 10% Fetal Bovine Serum (FBS) or complete FGM2 media (PromoCell) and supplemented with penicillin (100u/ml), streptomycin (100 µg/ml) and amphotericin B (0.25 µg/ml) (Gibco, Thermo) at 37℃ and 5% CO2. Before starting the experiments, the cells between 70 and 80% confluency were incubated for 24 h with Ham’s F-12 without FBS. TGF-β1 (Biolegend) stimuli were applied for 24 h at 5 ng/ml in Ham´s F12.
RNA extraction, primers design, and verification
RNA was obtained from early passage cultures (p2-p3) to avoid loss of disease-specific characteristics of the fibroblasts due to prolonged in vitro growth79. RNA was extracted using the Trizol reagent, and the cDNA was synthesized using 800 ng of DNase-treated RNA and the RevertAid H Minus First Strand cDNA Synthesis Kit (ThermoFisher Scientific), according to the manufacturer’s instructions. Based on global gene expression data15, sixteen genes associated with the HP immune response were selected (Table 2). Primers were designed to have similar melting temperatures and annealed to different contiguous exons with the aid of the DNA-MAN program by Lynnon Biosoft Bioinformatic Solutions. Verified Refseq gene sequences were retrieved from the NCBI’s GeneBank80. The PCR amplification efficiency and melting curves for each pair were analyzed before use (Supplementary Figure S3). All the oligonucleotides used in this work showed 100% (± 15%) efficiencies and are listed in Table S1.
mRNA expression by qPCR assays
Expression of target genes in cDNA from HP and control fibroblast was analyzed by qPCR in 10 µL reactions containing 5 µL of the 2 × master mix Maxima SYBR Green/Fluorescein (Thermo Scientific), 0.5 µL of each primer [10 µM] and 8.86 ng of cDNA. Triplicate reactions were run in a LightCycler® 480 Instrument II (Roche Molecular Systems) for 40 cycles at 95℃ for 15 s and 60℃ for 1 min. The mean from triplicates was calculated after obtaining the CT value for each reaction and is relative to the HPRT Ct values utilizing the 2-∆∆CT method81. HPRT was used as a normalizer as determined by Bestkeeper V1 software82 in our conditions. Graphics and statistical analysis were performed with GraphPad Prism 6 (San Diego, CA, USA). Further details on the PCR experiments according to the MIQE guidelines are included in Figure S3 and Table S183.
Clinical correlations
The honeycombing or diagnosis of fibrotic or non-fibrotic forms of HP were correlated with the target gene expression levels. Statistically relevant clinical correlations with the expression data were analyzed as described below.
Isoform expression detection
The splicing variants of the target genes were studied using end-point PCR with two types of primer pairs. Type 1 pairs distinguished variants by obtaining different PCR amplification products with a single reaction. The forward primer annealed with a conserved exon upstream of the variable region. In contrast, the reverse primer annealed with a conserved region downstream of the variable zone. Type II pairs were variant-specific, only annealing with sequences characterizing a single variant of interest (Table S1). The amplifications were done in 25 µl reactions containing 50 ng of cDNA, 200 µM dNTPs, 2.5 mM MgCl2, 0.4 µM primers (forward and reverse), and 0.3 U of DNA Taq Polymerase (Thermo Scientific) in 1X KCl buffer. Reactions were run for 40 cycles, annealing at 60℃ for 15 s, followed by 30 s extensions at 72℃. Amplicons were detected in 1–2% agarose-Tris-Borate-EDTA gels and stained with ethidium bromide. Visualization and photographs were made using a ChemiDoc XR imaging system (BioRad) described below in equipment and configurations. For IFNGR1 variants 2 and 3, the elongation steps at 72℃ were outstretched to 45 s due to bigger expected amplicons (1,453 and 1,459 bp, respectively). cDNA from Jurkat cells was used as a positive control. Quantification of PCR products was made with the Image J program relative to the HPRT band, graphed comparing control and HP samples, and included as Figure S2.
IFI27 cloning and viral transductions
IFI27 was subcloned from the commercial clone for variant 1 OHu14279 (Genscript) to pAdenoX-DSRed-Express adenoviral vector utilizing the InFusion kit (Clontech) and the high-fidelity Phusion polymerase (Thermo Scientific). The infective particles were produced and amplified with the adenoX-293 packaging cells and purified using the adenovirus affinity purification kit (Clontech). After titration with the AdenoX-Rapid-Titer kit (Clontech), infective particles were used to transduce primary fibroblasts with an adjusted multiplicity of infection (MOI) of 100 in cultures with or without serum for 48 h.
Immunoblots
Proteins were extracted with RIPA buffer (Sigma Aldrich) containing protease inhibitors (Calbiochem V) and quantified using the Bradford protein assay (BioRad). Total proteins (20–30 µg) were separated by SDS-PAGE and transferred to nitrocellulose membranes. Blocking was made with 5% nonfat milk for 1 h. The membranes were then incubated overnight at 4℃ with the following primary antibodies at the indicated dilutions: anti-IFI27 (Abcam, ab171919) 1:1000, anti-GAPDH (Invitrogen, PA1-987) 1:2000, anti-beta actin (Sigma, A5441) 1:10000, anti-αSMA (Sigma, A2547) 1:500, and anti-Caspase3 (Sigma, C9598) 1:1000. The membranes were incubated with the corresponding HRP-conjugated mouse or rabbit secondary antibodies for 1 h. The proteins were then revealed using the SuperSignal West Pico Chemiluminescent Substrate in a ChemiDoc-XR imaging system (BioRad) described below in equipment and configurations.
Immunohistochemistry
Lung tissue derived from two patients diagnosed with fHP and one control were processed for conventional immunohistochemistry. The tissues were deparaffinized, rehydrated, and treated with 3% hydrogen peroxide. After blocking and treating with citrate buffer for antigen retrieval, the tissues were incubated overnight with the primary antibody anti-IFI27 (Abcam, ab171919) at a 1:40 dilution. The samples were incubated with a biotinylated secondary antibody and streptavidin-HRP (BioGenex), followed by the 3-amino-9-ethyl carbazole (AEC) chromogen. Afterward, the samples were contrasted with hematoxylin and photographed using a Leica DR microscope.
Cyquant assay
Cell count over time was determined following the manufacturer’s instructions for the commercial CyQUANT kit (Invitrogen) in fibroblasts overexpressing IFI27 relative to cells transduced with an empty vector. Briefly, 5 × 103 cells were seeded on 96-well plates. After 48 and 72 h of growth, 200 µL of the CyQUANT dye/cell-lysis buffer was added. Fluorescence was detected using a microplate reader (BioTek Synergy HTX) set at 485/530 nm excitation and emission wavelengths. Two independent experiments in quadruplicate were done in two control cell lines.
Immunofluorescence assay
Control fibroblasts were seeded on round coverslips (12 mm) and stimulated for 24 h with TGF-β. The cells were fixed for 20 min with 4% paraformaldehyde, washed twice with PBS, left for 10 min in a PBS-Triton X-100 (0.3%) solution, and then 30 min in a blocking solution with PBS-1% BSA. The cells were left in contact with anti-IFI27 (Abcam, ab171919; 1:40) and anti-aSMA (Clone 1A4 Sigma, A2547; 1:100) antibodies at 4 °C overnight. The secondary antibodies anti-rabbit-AlexaFluor-647 and anti-mouse-Dylight-488 (1:200) were incubated at room temperature for 1 h and with DAPI (300 nM) for 10 min. The coverslips were mounted on microscope slides with Mowiol medium. The images were captured on a Leica TCS-SP8 confocal microscope using a 60X objective. The color and merged channels were processed with the ImageJ 1.54f. Software (NIH, U.S.) without changes or adjustments to any additional parameters of the individual images. The immunofluorescent staining was quantified using ImageJ. Briefly, regions of interest (ROIs) were selected using the square selection tool following background subtraction. The mean fluorescence intensity was then calculated and normalized to the number of cells, determined by counting cell nuclei stained with DAPI.
Equipment and configurations
The images were digitized with a ChemiDoc-XRS imaging system using the ImageLab 6.0 software (BioRad). The equipment was configured to capture images of gels stained with ethidium bromide, with automatic exposure to detect faint bands. Subsequently, the images were reversed to show black bands on a white background and recorded in the TIFF format using the same software. For blots, the computer was set to take photos at a 60 s interval with the feature of highlighting saturated pixels enabled. The brightness was adjusted for images with a high background.
For immunofluorescence, 8 bits digital images at 1024 × 1024 (ScanSpeed 400) were acquired with a Confocal Laser Scanning Microscope Leica TCS SP8 (Leica, Wetzlar, Germany); objective Plan Apochromat CS2 63x/1.40 Oil, HyD detectors, and laser line 405 nm (intensity 3.9784%), laser line 488 (intensity 2.0001%), and laser line 638 nm (intensity 1.3005%).
Statistical analysis
The statistical analysis was performed using GraphPad Prism software. The normal distribution was evaluated with the Shapiro-Wilks test. Differences between two samples or conditions were assessed with the Mann-Whitney test unless the data distribution was normal. Group differences for dimensional variables were assessed using ANOVA or Kruskal Wallis tests. The data were presented as mean ± standard deviation. Values of p ≤ 0.05 were used to determine statistical significance.
Supplementary Information
Acknowledgements
We thank Karla Jocelyn Meza Negrete for her technical support. We thank LANSBIODYT-UNICUA-UNAM (CONAHCYT) for technical support in acquiring confocal images and Angeles Garcia Vicente for her technical support in analyzing IHC images.
Author contributions
Conceptualization: A.L.TM, AP, MS, JC; Methodology: A.L.TM, E.HP, IH, MM, MN, CM, F. HS, RR, I. BR; Investigation: A.L.TM., E.HP, IH, MM, MN, CM, F. HS, RR, I. BR, MG, JC; Resources: JC, CB, MS; Writing-original draft: A.L.TM; Writing-review and editing: A.L.TM, AP, MS, JC; Visualization: A.L.TM, JC, AP, MS; Supervision: ALTM, MS, JC; Project administration: JC. All authors have read and approved the final manuscript.
Data availability
Data is provided within the manuscript or supplementary information files.
Declarations
Competing interests
The authors declare no competing interests.
Institutional review board
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Bioethics Committee of Instituto Nacional de Enfermedades Respiratorias Ismael Cosío Villegas B23-19.
Informed consent
Informed consent was obtained from all subjects involved in the study, and their personal and identification data were protected.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-74267-x.
References
- 1.Vasakova, M. et al. Hypersensitivity pneumonitis: Current concepts of pathogenesis and potential targets for treatment. Am. J. Respir. Crit. Care Med. 200, 301–308 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Raghu, G. et al. Diagnosis of hypersensitivity pneumonitis in adults. An official ATS/JRS/ALAT clinical practice guideline. Am. J. Respir. Crit. Care Med. 202, e36–e69 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Churg, A. Hypersensitivity pneumonitis: new concepts and classifications. Mod. Pathol. 35, 15–27 (2022). [DOI] [PubMed] [Google Scholar]
- 4.McSharry, C., Anderson, K., Bourke, S. J. & Boyd, G. Takes your breath away–the immunology of allergic alveolitis. Clin. Exp. Immunol. 128, 3–9 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaxiola, M. et al. Morphologic diversity of chronic pigeon breeder’s disease: clinical features and survival. Respir. Med. 105, 608–614 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Morell, F. et al. Chronic hypersensitivity pneumonitis in patients diagnosed with idiopathic pulmonary fibrosis: a prospective case-cohort study. Lancet Respir. Med. 1, 685–694 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Vourlekis, J. S. et al. The effect of pulmonary fibrosis on survival in patients with hypersensitivity pneumonitis. Am. J. Med. 116, 662–668 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Giblin, S. P. & Midwood, K. S. Tenascin-C: Form versus function. Cell Adhes. Migration 9, 48–82 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mohr, L. C. Hypersensitivity pneumonitis. Curr. Opinion Pulm. Med. 10, 401–411 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Barrera, L. et al. Functional diversity of T-cell subpopulations in subacute and chronic hypersensitivity pneumonitis. Am. J. Respir. Crit. Care Med. 177, 44–55 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Wang, E. T. et al. Alternative isoform regulation in human tissue transcriptomes. Nature 456, 470–476 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marasco, L. E. & Kornblihtt, A. R. The physiology of alternative splicing. Nat. Rev. Mol. Cell Biol. 24, 242–254 (2023). [DOI] [PubMed] [Google Scholar]
- 13.Yamaguchi, K. et al. Reduced endogenous secretory RAGE in blood and bronchoalveolar lavage fluid is associated with poor prognosis in idiopathic pulmonary fibrosis. Respir. Res. 21, 145 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hytonen, A. M. et al. Haplotypes of the interleukin-4 receptor alpha chain gene associate with susceptibility to and severity of atopic asthma. Clin. Exp. Allergy 34, 1570–1575 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Selman, M. et al. Gene expression profiles distinguish idiopathic pulmonary fibrosis from hypersensitivity pneumonitis. . Am. J. Respir. Crit. Care Med. 173, 188–198 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nance, S. C., Yi, A. K., Re, F. C. & Fitzpatrick, E. A. MyD88 is necessary for neutrophil recruitment in hypersensitivity pneumonitis. J. Leukoc. Biol. 83, 1207–1217 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhattacharyya, S. et al. Tenascin-C drives persistence of organ fibrosis. Nat. Commun. 7, 11703 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andrews, C. S., Matsuyama, S., Lee, B. C. & Li, J. D. Resveratrol suppresses NTHi-induced inflammation via up-regulation of the negative regulator MyD88 short. Sci. Rep. 6, 34445 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cardona Gloria, Y. et al. Absence of non-canonical, inhibitory MYD88 splice variants in B cell lymphomas correlates with sustained NF-kappaB signaling. Front. Immunol. 12, 616451 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kasprzycka, M., Hammarstrom, C. & Haraldsen, G. Tenascins in fibrotic disorders-from bench to bedside. Cell Adhes. Migration 9, 83–89 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brissett, M., Veraldi, K. L., Pilewski, J. M., Medsger, T. A. Jr. & Feghali-Bostwick, C. A. Localized expression of tenascin in systemic sclerosis-associated pulmonary fibrosis and its regulation by insulin-like growth factor binding protein 3. Arthritis Rheum. 64, 272–280 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanaka, H. et al. Circulating level of large splice variants of tenascin-C is a marker of piecemeal necrosis activity in patients with chronic hepatitis C. Liver Int. 26, 311–318 (2006). [DOI] [PubMed] [Google Scholar]
- 23.El-Karef, A. et al. Expression of large tenascin-C splice variants by hepatic stellate cells/myofibroblasts in chronic hepatitis C. J. Hepatol. 46, 664–673 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Kalea, A. Z. et al. Alternative splicing of the murine receptor for advanced glycation end-products (RAGE) gene. FASEB J. 23, 1766–1774 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalea, A. Z., Schmidt, A. M. & Hudson, B. I. Alternative splicing of RAGE: roles in biology and disease. Front. Biosci. (Landmark Ed.) 16, 2756–2770 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Yan, S. F., Ramasamy, R. & Schmidt, A. M. Soluble RAGE: therapy and biomarker in unraveling the RAGE axis in chronic disease and aging. Biochem. Pharmacol. 79, 1379–1386 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Selman, M. & Pardo, A. Role of epithelial cells in idiopathic pulmonary fibrosis: from innocent targets to serial killers. Proc. Am. Thorac. Soc. 3, 364–372 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Kim, S. Y. et al. Classification of usual interstitial pneumonia in patients with interstitial lung disease: assessment of a machine learning approach using high-dimensional transcriptional data. Lancet Respir. Med. 3, 473–482 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Hiyama, K. et al. Exploration of the genes responsible for unlimited proliferation of immortalized lung fibroblasts. Exp. Lung Res. 34, 373–390 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Gytz, H. et al. Apoptotic properties of the type 1 interferon induced family of human mitochondrial membrane ISG12 proteins. Biol. Cell 109, 94–112 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Smidt, K. C. et al. A nine-nucleotide deletion and splice variation in the coding region of the interferon induced ISG12 gene. Biochim. Biophys. Acta 1638, 227–234 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Mueller, A. A., van Velthoven, C. T., Fukumoto, K. D., Cheung, T. H. & Rando, T. A. Intronic polyadenylation of PDGFRalpha in resident stem cells attenuates muscle fibrosis. Nature 540, 276–279 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minato, Y. et al. Transcriptional regulation of a new variant of human platelet-derived growth factor receptor alpha transcript by E2F–1. Gene 403, 89–97 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Aono, Y. et al. Role of platelet-derived growth factor/platelet-derived growth factor receptor axis in the trafficking of circulating fibrocytes in pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 51, 793–801 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Kishi, M. et al. Blockade of platelet-derived growth factor receptor-beta, not receptor-alpha ameliorates bleomycin-induced pulmonary fibrosis in mice. PLoS ONE 13, e0209786 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Payne, L. B. et al. A soluble platelet-derived growth factor receptor-beta originates via pre-mRNA splicing in the healthy brain and is upregulated during hypoxia and aging. Biomolecules 13, 17 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jakubzick, C. et al. Augmented pulmonary IL-4 and IL-13 receptor subunit expression in idiopathic interstitial pneumonia. J. Clin. Pathol. 57, 477–486 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jakubzick, C. et al. Human pulmonary fibroblasts exhibit altered interleukin-4 and interleukin-13 receptor subunit expression in idiopathic interstitial pneumonia. Am. J. Pathol. 164, 1989–2001 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fitch, P. S., Brown, V., Schock, B. C., Ennis, M. & Shields, M. D. Interleukin-4 and interleukin-4 soluble receptor alpha levels in bronchoalveolar lavage from children with asthma. Ann. Allergy Asthma Immunol. 90, 429–433 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Osawa, M. et al. Characterization of the mouse interleukin-13 receptor alpha1 gene. Immunogenetics 51, 974–981 (2000). [DOI] [PubMed] [Google Scholar]
- 41.Tabata, Y. et al. Allergy-driven alternative splicing of IL-13 receptor alpha2 yields distinct membrane and soluble forms. J. Immunol. 177, 7905–7912 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Hasan, S. A. et al. Role of IL-17A and neutrophils in fibrosis in experimental hypersensitivity pneumonitis. J. Allergy Clin. Immunol. 131, 1663–1673 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Sohda, M. et al. Identification of a soluble isoform of human IL-17RA generated by alternative splicing. Cytokine 64, 642–645 (2013). [DOI] [PubMed] [Google Scholar]
- 44.You, Z. et al. Differential expression of IL-17RC isoforms in androgen-dependent and androgen-independent prostate cancers. Neoplasia 9, 464–470 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khalil, N. et al. Regulation of the effects of TGF-beta 1 by activation of latent TGF-beta 1 and differential expression of TGF-beta receptors (T beta R-I and T beta R-II) in idiopathic pulmonary fibrosis. Thorax 56, 907–915 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alexander, J. M., Bikkal, H. A., Zervas, N. T., Laws, E. R. Jr. & Klibanski, A. Tumor-specific expression and alternate splicing of messenger ribonucleic acid encoding activin/transforming growth factor-beta receptors in human pituitary adenomas. J. Clin. Endocrinol. Metab. 81, 783–790 (1996). [DOI] [PubMed] [Google Scholar]
- 47.Li, M. et al. Epithelium-specific deletion of TGF-beta receptor type II protects mice from bleomycin-induced pulmonary fibrosis. J. Clin. Investig. 121, 277–287 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rotzer, D. et al. Type III TGF-beta receptor-independent signalling of TGF-beta2 via TbetaRII-B, an alternatively spliced TGF-beta type II receptor. EMBO J. 20, 480–490 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Konrad, L. et al. Alternative splicing of TGF-betas and their high-affinity receptors T beta RI, T beta RII and T beta RIII (betaglycan) reveal new variants in human prostatic cells. BMC Genomics 8, 318 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu, Y. et al. Abnormal expression of TGF-beta type II receptor isoforms contributes to acute myeloid leukemia. Oncotarget 8, 10037–10049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carrea, A., Preisegger, M. A., Velasco Zamora, J. & Dewey, R. A. The mRNA levels of TGF-beta type II receptor splice variants in monocytes are associated with disease activity in patients with rheumatoid arthritis. Clin. Exp. Rheumatol. 39, 310–318 (2021). [PubMed] [Google Scholar]
- 52.Gocheva, V. et al. Quantitative proteomics identify Tenascin-C as a promoter of lung cancer progression and contributor to a signature prognostic of patient survival. Proc. Natl. Acad. Sci. U. S. A. 114, E5625–E5634 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bandari, A. K. et al. A novel splice site mutation in IFNGR2 in patients with primary immunodeficiency exhibiting susceptibility to mycobacterial diseases. Front. Immunol. 10, 1964 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mentink-Kane, M. M. et al. IL-13 receptor alpha 2 down-modulates granulomatous inflammation and prolongs host survival in schistosomiasis. Proc. Natl. Acad. Sci. U. S. A. 101, 586–590 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hansell, D. M. et al. Fleischner society: glossary of terms for thoracic imaging. Radiology 246, 697–722 (2008). [DOI] [PubMed] [Google Scholar]
- 56.Hsieh, W. L. et al. IFI27, a novel epidermal growth factor-stabilized protein, is functionally involved in proliferation and cell cycling of human epidermal keratinocytes. Cell Prolif. 48, 187–197 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walker, E. J., Heydet, D., Veldre, T. & Ghildyal, R. Transcriptomic changes during TGF-beta-mediated differentiation of airway fibroblasts to myofibroblasts. Sci. Rep. 9, 20377 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ushakumary, M. G., Riccetti, M. & Perl, A. T. Resident interstitial lung fibroblasts and their role in alveolar stem cell niche development, homeostasis, injury, and regeneration. Stem Cells Transl. Med. 10, 1021–1032 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nie, Y. J., Wu, S. H., Xuan, Y. H. & Yan, G. Role of IL-17 family cytokines in the progression of IPF from inflammation to fibrosis. Mil. Med. Res. 9, 21 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tazat, K., Hector-Greene, M., Blobe, G. C. & Henis, Y. I. TbetaRIII independently binds type I and type II TGF-beta receptors to inhibit TGF-beta signaling. Mol. Biol. Cell 26, 3535–3545 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim, B. H., Woo, T. G., Kang, S. M., Park, S. & Park, B. J. Splicing variants, protein-protein interactions, and drug targeting in hutchinson-gilford progeria syndrome and small cell lung cancer. Genes (Basel) 13, 165 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Buckley, S. T. & Ehrhardt, C. The receptor for advanced glycation end products (RAGE) and the lung. J. Biomed. Biotechnol. 2010, 917108 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Demling, N. et al. Promotion of cell adherence and spreading: a novel function of RAGE, the highly selective differentiation marker of human alveolar epithelial type I cells. Cell Tissue Res. 323, 475–488 (2006). [DOI] [PubMed] [Google Scholar]
- 64.Sparvero, L. J. et al. RAGE (receptor for advanced glycation endproducts), RAGE ligands, and their role in cancer and inflammation. J. Transl. Med. 7, 17 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Machahua, C. et al. Increased AGE-RAGE ratio in idiopathic pulmonary fibrosis. Respir. Res. 17, 144 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Manichaikul, A. et al. Plasma soluble receptor for advanced glycation end products in idiopathic pulmonary fibrosis. Ann. Am. Thorac. Soc. 14, 628–635 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Salehi, M., Amiri, S., Ilghari, D., Hasham, L. F. A. & Piri, H. The remarkable roles of the receptor for advanced glycation end products (RAGE) and its soluble isoforms in COVID-19: The importance of RAGE pathway in the lung injuries. Indian J. Clin. Biochem. 38, 159–171 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Englert, J. M. et al. Paradoxical function for the receptor for advanced glycation end products in mouse models of pulmonary fibrosis. Int. J. Clin. Exp. Pathol. 4, 241–254 (2011). [PMC free article] [PubMed] [Google Scholar]
- 69.Queisser, M. A. et al. Loss of RAGE in pulmonary fibrosis: molecular relations to functional changes in pulmonary cell types. Am. J. Respir. Cell Mol. Biol. 39, 337–345 (2008). [DOI] [PubMed] [Google Scholar]
- 70.Grimminger, F., Gunther, A. & Vancheri, C. The role of tyrosine kinases in the pathogenesis of idiopathic pulmonary fibrosis. Eur. Respir. J. 45, 1426–1433 (2015). [DOI] [PubMed] [Google Scholar]
- 71.Green, J., Endale, M., Auer, H. & Perl, A. K. Diversity of interstitial lung fibroblasts is regulated by platelet-derived growth factor receptor alpha kinase activity. Am. J. Respir. Cell Mol. Biol. 54, 532–545 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Palumbo-Zerr, K. et al. Orphan nuclear receptor NR4A1 regulates transforming growth factor-beta signaling and fibrosis. Nat. Med. 21, 150–158 (2015). [DOI] [PubMed] [Google Scholar]
- 73.Gao, L. et al. The roles of orphan nuclear receptor 4 group A1 and A2 in fibrosis. Int. Immunopharmacol. 139, 112705 (2024). [DOI] [PubMed] [Google Scholar]
- 74.Papac-Milicevic, N. et al. The interferon stimulated gene 12 inactivates vasculoprotective functions of NR4A nuclear receptors. Circ. Res. 110, e50-63 (2012). [DOI] [PubMed] [Google Scholar]
- 75.Li, X. et al. High level expression of ISG12(1) promotes cell apoptosis via mitochondrial-dependent pathway and so as to hinder newcastle disease virus replication. Vet. Microbiol. 228, 147–156 (2019). [DOI] [PubMed] [Google Scholar]
- 76.Xiong, Z. et al. Integrated bioinformatics and validation reveal IFI27 and its related molecules as potential identifying genes in liver cirrhosis. Biomolecules 14, 13 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen, Z. et al. Single cell multi-omics of fibrotic kidney reveal epigenetic regulation of antioxidation and apoptosis within proximal tubule. Cell Mol. Life Sci. 81, 56 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Becerril, C. et al. Mesenchymal-epithelial transition in fibroblasts of human normal lungs and interstitial lung diseases. Biomolecules 11, 378 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hall, C. et al. Chronic activation of human cardiac fibroblasts in vitro attenuates the reversibility of the myofibroblast phenotype. Sci. Rep. 13, 12137 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sayers, E. W. et al. GenBank. Nucleic Acids Res. 48, D84–D86 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- 82.Pfaffl, M. W., Tichopad, A., Prgomet, C. & Neuvians, T. P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–excel-based tool using pair-wise correlations. Biotechnol. Lett. 26, 509–515 (2004). [DOI] [PubMed] [Google Scholar]
- 83.Bustin, S. A. et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622 (2009). [DOI] [PubMed] [Google Scholar]
- 84.Rodriguez-Montano, R. et al. IL-23/IL-17 axis and soluble receptors isoforms sIL-23R and sIL-17RA in patients with rheumatoid arthritis-presenting periodontitis. J. Clin. Lab. Anal. 35, e23963 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is provided within the manuscript or supplementary information files.