Abstract
Glycoprotein B (gB) of pseudorabies virus (PrV) is essential for virus entry into target cells and direct viral cell-to-cell spread. Recently, we described a carboxy-terminally truncated derivative of PrV gB, gB-007, which was inefficiently incorporated into virions, was unable to complement infectivity, but was fully capable of restoring direct viral cell-to-cell spread of gB-negative PrV (R. Nixdorf, B. G. Klupp, and T. C. Mettenleiter, J. Virol. 74:7137–7145, 2000). Since recombinant PrV-007, which expresses gB-007 instead of wild-type gB, was able to spread directly from cell to cell, we attempted to obtain compensatory mutations leading to restoration of the entry defect by performing serial passages in cell culture. This procedure has previously been used to successfully restore entry defects in gD- or gL-deficient PrV mutants. From an initial titer of 100 PFU per ml in the supernatant, titers increased, reaching wild-type levels of up to 107 PFU after ca. 20 passages. One single-plaque isolate of the passaged mutant, designated PrV-007Pass, was further characterized. PrV-007Pass gB was efficiently incorporated into the viral envelope and restored infectivity to a gB-negative PrV mutant, PrV-gB−. Interestingly, localization of PrV-007Pass gB in the plasma membrane was similar to that of PrV-007. In contrast, wild-type gB is mainly found in intracellular vesicles. Marker rescue experiments and trans-complementation assays demonstrated the presence of compensatory mutations within the gB gene of PrV-007Pass. DNA sequencing revealed two point mutations in the gB open reading frame of PrV-007Pass, resulting in amino acid substitutions at positions 305 and 744 of gB, both of which are required for compensation of the defect in PrV-007. Our data again demonstrate the power of reversion analysis of herpesviruses and suggest that cytosolic and ectodomains play a role in incorporation of gB into virions.
Entry of herpesviruses into cells requires a cascade of events involving the function of several virus-encoded envelope glycoproteins. Primary attachment to cell surface proteoglycans carrying heparan sulfate moieties is mediated by homologs of herpes simplex virus type 1 (HSV-1) glycoprotein C (gC) (8, 22). This initial interaction is sensitive to competition by exogenous heparin but in the presence of gD converts to a more stable, heparin-resistant binding (12). Secondary binding is effected by interaction of gD with specific cellular receptors (10, 17). In the case of pseudorabies virus (PrV), these receptors belong to the immunoglobulin superfamily and encompass poliovirus receptor as well as poliovirus receptor-related proteins (4, 38, 40). In HSV-1 and bovine herpesvirus 1, gB has also been demonstrated to contribute to attachment (9, 18). For penetration, i.e., fusion of the viral envelope and the cellular plasma membrane at neutral pH, glycoproteins gD and gB and the gH-gL complex are required (21, 37). However, recently, PrV and bovine herpesvirus 1 mutants that are infectious even in the absence of gD have been isolated by serial passaging in cell culture (34–36), indicating that only gB and gH-gL are intrinsically involved in fusion. Correspondingly, only gB and gH-gL are required for direct cell-to-cell spread of PrV, whereas gD is also necessary for this process in HSV-1 (19). The central role for gB and gH-gL in the fusion process is also highlighted by the fact that these glycoproteins are conserved throughout the Herpesviridae (31).
gB homologs are the most highly conserved membrane glycoproteins within the family Herpesviridae. Most of them, including PrV gB (41), form disulfide-linked homodimers that are posttranslationally cleaved by furin protease into two subunits (27). However, proteolytic cleavage is not essential for the function of cleavable gB species (16), and HSV-1 gB is not cleaved at all (3). Thus, the importance of this prominent posttranslational modification is still unclear. PrV gB is a type I membrane protein consisting of 913 amino acids (aa), including a 58-aa putative signal peptide; three C-terminally located hydrophobic domains, of which the last one probably represents the transmembrane region; and a 93-aa cytoplasmic C-terminal tail (29). The cytoplasmic portion comprises two discrete predicted α-helical domains and putative endocytosis motifs. In our studies to analyze the function of PrV gB in more detail, we constructed mutant gB proteins lacking different portions of the carboxy terminus (24). These C-terminal truncations encompassed either one predicted α-helical domain with two putative endocytosis motifs (gB-008), two α-helical domains (gB-007), the complete cytoplasmic domain (gB-006), or the cytoplasmic domain and the membrane anchor (gB-005). As expected, in the absence of the internalization signals, gB-008 and gB-007 were mainly detected at the plasma membrane of cells either transfected with expression plasmids or infected with mutant-gB-expressing PrV recombinants. Despite the difference in subcellular localization compared to wild-type gB, which is mainly found in the cytoplasm, gB-008 was fully competent in complementing both the entry defect and the cell-to-cell spread defect of a gB-deficient PrV mutant, PrV-gB−. In contrast, gB-007 efficiently complemented direct viral cell-to-cell spread but only very poorly rescued the entry defect. This correlated with a drastically decreased level of incorporation of gB-007 into PrV virions (24).
Thus, PrV-007, a recombinant virus which expresses gB-007 instead of wild-type gB, is able to efficiently spread via direct cell-cell contact, whereas it is severely impaired in entry. Previously, we had isolated second-site revertants of PrV mutants displaying a similar phenotype by serial passage in cell cultures employing coseeding of infected and uninfected cells. By this technique, PrV mutants which are infectious in the absence of gD (PrV-gD−Pass [34] or gL (PrV-ΔgLPass [14]) have been isolated. While the exact molecular basis for gD-independent infectivity in PrV has not yet been fully elucidated but involves point mutations in gH and gB (33a), restoration of infectivity of a gL-deleted PrV mutant was due to a translocation of part of the gH gene fused in frame to most of the gD gene. The resulting gD-gH hybrid protein combines gH and gD functions and apparently does not require gL for either (14). Following these examples, we serially passaged PrV-007 in RK13 cells and isolated and characterized a second-site revertant.
MATERIALS AND METHODS
Cells and viruses.
All virus mutants are based on PrV strain Kaplan (PrV-Ka) (11). PrV-ΔgBGFP and PrV-ΔgBβ, which are gB-deleted mutants carrying a green fluorescent protein (GFP) or lacZ expression cassette, respectively, instead of gB, have been described previously (24). PrV-007 and PrV-ΔgBGFPR were obtained after cotransfection of PrV-ΔgBGFP with appropriate plasmids, resulting in replacement of the GFP cassette by the gB-007 or full-length gB open reading frame (ORF), respectively (24).
RK13-007, a recombinant rabbit kidney cell clone which constitutively expresses gB-007 under the control of the human cytomegalovirus immediate-early promoter-enhancer, has been described previously (24).
Passaging of PrV-007.
RK13 cells were infected with PrV-007 at a multiplicity of infection (MOI) of 0.01. Cells were repeatedly split 1 to 5 until a widespread cytopathic effect (CPE) was observed, usually by 3 days postinfection (p.i.). After development of CPE, cells were trypsinized and reseeded with uninfected RK13 cells in 5 ml of medium in a 25-cm2 tissue culture flask. To accelerate the enrichment of extracellular infectivity, from passage 16 on, culture supernatant was used for infection of fresh cells after removal of cellular debris by low-speed centrifugation.
Immunodetection.
Western blotting, immunoprecipitation, and immunofluorescence microscopy were performed as described previously (20, 24). Monoclonal antibody (MAb) a80-c16 (24, 25) was used in immunoprecipitations and immunofluorescence microscopy. MAb b43-b5 was used for Western blotting. Monospecific polyclonal anti-gH serum (14) was used as a control.
Virus purification, Southern blot analysis and DNA sequencing.
Viruses were purified by sucrose gradient centrifugation as described previously (15). Sequencing of double-stranded DNA by the dideoxy chain termination method (33) was performed as previously described (13), using gB gene-specific primers. Southern blot analysis of BamHI-restricted or BamHI- and EcoRI-digested viral DNA was done by standard procedures (15, 32).
Plaque assay and replication kinetics.
Plaque assays and determination of replication kinetics were performed essentially as previously described (1).
PCR amplification of gB-007Pass.
The gB gene of PrV-007Pass was amplified in a two-step PCR using Pfx-Platinum DNA polymerase (Life Technologies, Karlsruhe, Germany) with upstream primer 5′-TAACGGATCCATGCCCGCTGGTGGCGG-3′ and downstream primer 5′-CCGAATTCCTAGGCCTCGTCCACGTCGCCTTC-3′. BamHI and EcoRI restriction sites, introduced to allow convenient cloning, are printed in italics.
Construction of cell lines expressing mutated gB proteins.
For construction of cell lines expressing gB molecules carrying either mutation, the gB-007Pass and gB-007 ORFs were recloned in pcDNA3, using an internal SphI site which is located between the two mutations (see Fig. 8). gB-079 contained only the carboxy-terminal mutation, and gB-080 contained only the amino-terminal mutation. After transfection of the plasmids into RK13 cells, stably expressing cell lines were selected by using immunofluorescence analysis with a gB-specific MAb. One cell line from each transfection with comparable immunofluorescence staining was selected for further analysis.
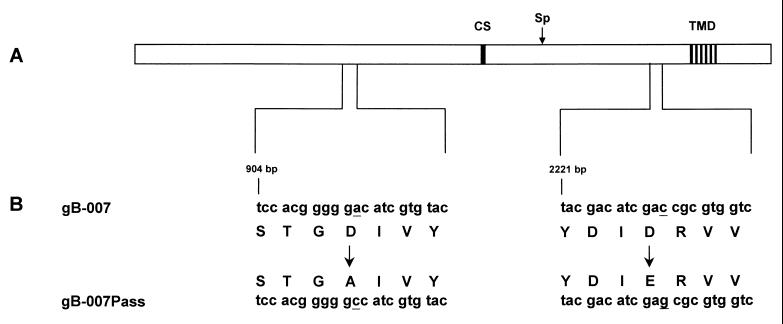
FIG. 8.
Summary of sequencing results. (A) Schematic diagram of the gB polypeptide. The locations of the transmembrane domain (TMD) and the furin protease cleavage site (CS) are indicated for orientation. The location of the SphI site (Sp) within the gB ORF, used for construction of gB molecules containing either mutation, is indicated by a vertical arrow. (B) Relevant sequences of the ORFs of gB-007 and gB-007Pass are shown in addition to the translated amino acid sequence. Single-base mutations are underlined, and amino acid substitutions are marked by arrows.
RESULTS
Isolation of PrV-007Pass.
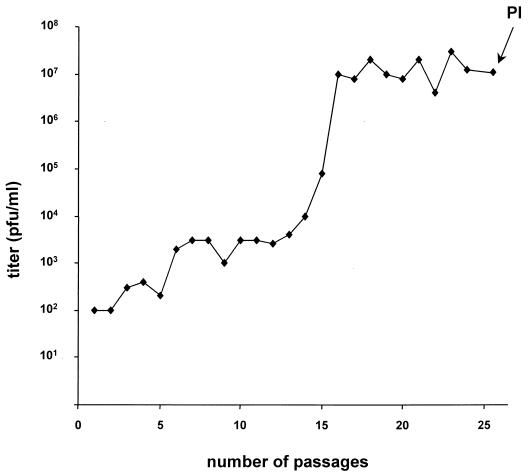
The defect in entry and direct viral cell-to-cell spread of gB-deleted PrV could be corrected by expression of wild-type gB or gB-008 either in trans or in cis. However, gB-007 complemented direct viral cell-to-cell spread efficiently but was only marginally able to mediate infection of free virions. The latter effect correlated with a decreased incorporation of gB-007 into PrV virions compared to that of wild-type gB or gB-008. Based on our results for serial passaging of PrV mutants with a similar phenotype, i.e., deficient in entry but capable of direct viral cell-to-cell spread, we passaged PrV-007 in RK13 cells by repeatedly coseeding infected and uninfected cells. The supernatants were assayed for the presence of extracellular infectious virus. As demonstrated in Fig. 1, within the first 15 passages, infectivity rose from an initial level of 102 to 104 PFU per ml. To accelerate enrichment of extracellular infectious virions, passages 16 to 25 were performed with cleared supernatant of infected cells after development of complete CPE. As anticipated, infectivity in the supernatants rose sharply, reaching ca. 107 PFU/ml after two additional passages and remaining at that plateau level henceforth. Since no further increase in viral titers was observed, six single-plaque isolates from the supernatant of passage 25 (Fig. 1) were picked and viral DNA was analyzed after cleavage with different restriction enzymes. No difference in the restriction patterns of the isolates was observed (data not shown). Thus, one plaque isolate, designated PrV-007Pass, was randomly chosen for further analysis.
FIG. 1.
Serial passaging of PrV-007 in RK13 cells. PrV-007, which expresses a C-terminally truncated gB (24), was serially passaged by repeated coseeding of infected and uninfected cells. From passage 16 on, cleared cell culture supernatant was passaged. Infectivity in the supernatant was analyzed by titration on RK13 cells. At passage 25, single-plaque isolates (PI) were picked for further analysis. Titers are indicated in PFU per milliliter of supernatant.
Genomic analysis of PrV-007Pass.
To detect gross differences in the genomic organizations of PrV-007 and PrV-007Pass, as have been observed, e.g., for PrV-ΔgLPass (14), viral DNA was isolated and cleaved with either BamHI, KpnI, or BamHI plus EcoRI. Fragments were separated in a 0.8% agarose gel and hybridized to different radiolabeled genomic fragments. In all of these tests, no difference between PrV-007 and PrV-007Pass was detected, arguing against the loss or translocation of larger genomic fragments (data not shown).
Growth kinetics of PrV-007Pass.
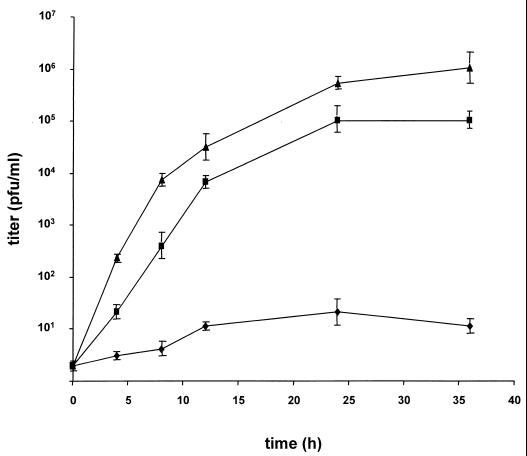
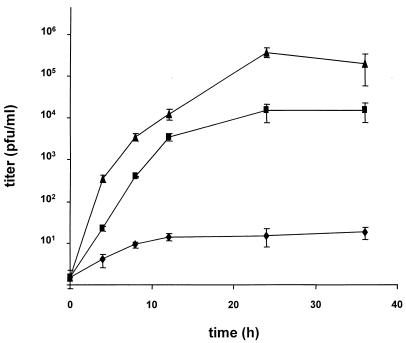
To compare PrV-007 and PrV-007Pass in terms of their replication, RK13 cells were infected at an MOI of 0.1 with PrV-007, PrV-007Pass, or PrV-ΔgBGFPR, which is a full-length-gB rescuant of the GFP-expressing gB deletion mutant PrV-ΔgBGFP (24). Although a higher MOI is generally used to explore one-step growth kinetics, this was not possible because of the low infectivity of PrV-007 due to the entry defect. As shown in Fig. 2, infection with PrV-007 resulted in only very low levels of extracellular virus (ca. 10 PFU per ml). In contrast, after infection by PrV-007Pass or PrV-ΔgBGFPR, a significant increase in extracellular infectivity was observed, with titers of ca. 105 for PrV-007Pass and 106 for PrV-ΔgBGFPR. Although the final titers of PrV-007Pass did not reach those of PrV-ΔgBGFPR, they were ca. 104-fold higher than those of PrV-007. It should be noted that this is the result of a low-multiplicity infection, which explains the lower titers compared with the passaged viruses (Fig. 1).
FIG. 2.
Replication kinetics. Growth kinetics of PrV-007 (⧫), PrV-007Pass (▪), and PrV-ΔgBGFPR (▴) in RK13 cells were determined after infection at an MOI of 0.1. Titers, indicated in PFU per milliliter, are averages of values from three independent experiments. Error bars indicate standard deviations.
Rescue of PrV-gB− with gB-007Pass results in a PrV-007Pass phenotype.
To pinpoint the compensatory mutations within the genome of PrV-007Pass, genomic DNA of PrV-ΔgBGFP was cotransfected with cloned KpnI fragment C isolated from either PrV-007 or PrV-007Pass, leading to recombinants PrV-064 and PrV-065, respectively. KpnI-C encompasses, among others, the gB gene (23). As expected, growth kinetics and plaque formation of PrV-064 did not differ from those of PrV-007. However, PrV-065 showed a phenotype indistinguishable from that of PrV-007Pass, indicating that the compensatory mutations were present in KpnI fragment C (data not shown).
To analyze whether these compensatory mutations were located in the gB gene, which is contained in KpnI-C, the gB ORF of PrV-007Pass was PCR amplified from the cloned KpnI-C fragment and inserted into vector pcDNA3, which allows constitutive expression in eukaryotic cells under the control of the human cytomegalovirus immediate-early promoter-enhancer. A stably expressing cell line, designated RK13-007Pass, was established after transfection of RK13 cells.
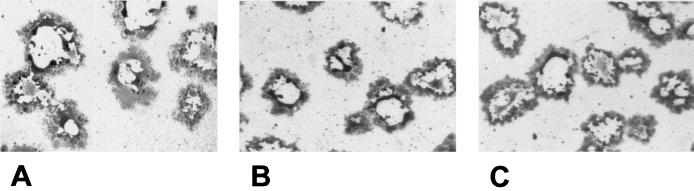
To investigate the function of gB-007Pass in direct viral cell-to-cell spread, cells expressing either wild-type gB, gB-007, or gB-007Pass were infected by phenotypically complemented PrV-ΔgBβ and plaque formation was analyzed. As shown in Fig. 3, no obvious differences in size and appearance of plaques were observed at 2 days p.i.
FIG. 3.
Plaque formation on gB-expressing cells. Recombinant RK13 cells expressing wild-type gB (A), gB-007 (B), or gB-007Pass (C) were infected with PrV-ΔgBβ and were stained with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) at 2 days p.i..
To assay the capacity of gB-007Pass to complement the entry defect of a gB-negative PrV mutant, wild-type-gB-, gB-007-, or gB-007Pass-expressing RK13 cells were infected with phenotypically complemented PrV-ΔgBβ and supernatants were titrated on wild-type gB-expressing cells. As shown in Fig. 4, cells expressing gB-007Pass produced infectious progeny in the supernatant at titers only ca. 10-fold lower than those produced on wild-type-gB-expressing cells. However, titers after replication on gB-007Pass-expressing cells were again more than 1,000-fold higher than those of gB-007-expressing cells, which produced very little cell-free infectivity (ca. 10 PFU/ml). Thus, these results strongly indicate that the compensatory mutations were located in gB-007Pass.
FIG. 4.
Complementation of infectivity. RK13 cells expressing wild-type gB (▴), gB-007 (⧫), or gB-007Pass (▪) were infected by phenotypically complemented PrV-ΔgBβ at an MOI of 10, and extracellular infectivity was determined by titration on wild-type-gB-expressing cells at the indicated time points. Titers are measured in PFU per milliliter. Data are averages of values from three independent experiments. Error bars indicate standard deviations.
Characterization of gB-007Pass.
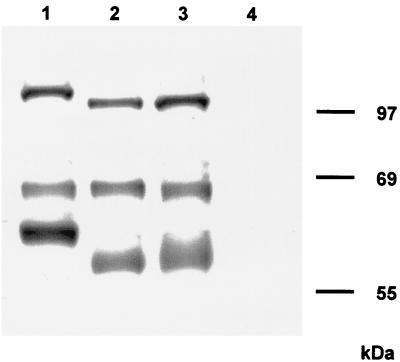
To test for differences between gB-007 and gB-007Pass, proteins were metabolically labeled with Tran-S35-Label (ICN, Eschwege, Germany), immunoprecipitated, and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; 10% polyacrylamide) under reducing conditions. As shown in Fig. 5, the two gB subunits and the uncleaved precursor were precipitated from RK13-gB (lane 1), RK13-007 (lane 2), and RK13-007Pass (lane 3) cells. As expected, the larger (69-kDa) amino-terminal subunits of all gB species migrated identically, whereas the uncleaved precursor and the smaller carboxy-terminal subunits of RK13-007 and RK13-007Pass gB migrated faster, correlating with the carboxy-terminal truncation. No difference was observed between gB-007 and gB-007Pass, arguing against induction of gross alterations in gB-007Pass during cell passage. Also, no difference between gB-007 and gB-007Pass was detected after in vitro transcription-translation (data not shown).
FIG. 5.
Immunoprecipitation of gB. Lysates from metabolically radiolabeled RK13 cells expressing gB (lane 1), gB-007 (lane 2), or gB-007Pass (lane 3) or normal RK13 cells (lane 4) were precipitated with the gB-specific MAb a80-c16. Precipitates were analyzed by SDS-PAGE under reducing conditions. Labeled proteins were visualized by autoradiography.
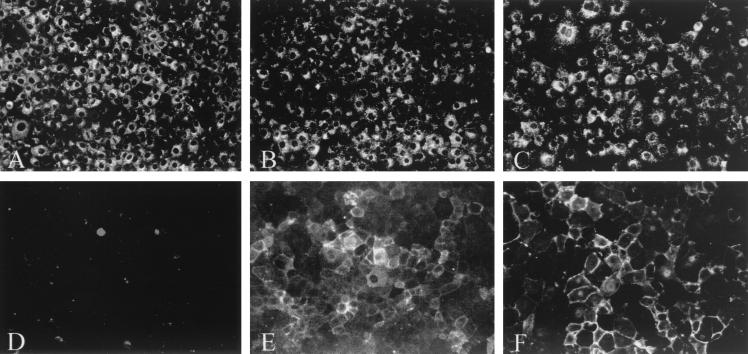
Since gB-007 was detected in the plasma membranes of infected cells, in contrast to wild-type gB (24), gB-007- or gB-007Pass-expressing cells were analyzed by indirect immunofluorescence using a gB-specific MAb. As shown in Fig. 6A to C, all cell lines exhibited similar levels of fluorescence after permeabilization of the plasma membrane with detergent. However, without permeabilization, gB-specific surface fluorescence was detected only in gB-007 (Fig. 6E)- and gB-007Pass (Fig. 6F)-expressing cells, but, as expected, not in gB-expressing cells (Fig. 6D). Thus, the cell surface localization of gB-007Pass did not differ from that of gB-007.
FIG. 6.
Subcellular localization of gB. RK13-gB (A and D), RK13-007 (B and E), and RK13-007Pass cells (C and F) were grown to confluency, fixed with 3% paraformaldehyde (A to C) or 3% paraformaldehyde–0.3% Triton X-100 (D to F), and incubated with anti-gB MAb a80-c16. Immunofluorescence microscopy was performed after incubation with fluorescein isothiocyanate-conjugated secondary antibodies.
Incorporation of gB-007Pass into virions.
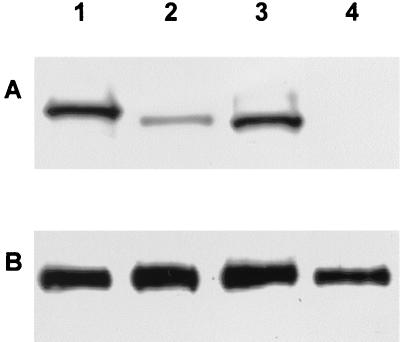
PrV gB is essential for infectivity of free virions. Virus particles lacking gB are unable to enter target cells (26, 28). We recently demonstrated the inefficiency of gB-007 incorporation into virions, which parallels its poor complementation of infectivity (24). To determine whether gB-007Pass is more efficiently incorporated into virions, normal RK13 as well as recombinant RK13-007, RK13-007Pass, and RK13-gB cells were infected at an MOI of 10 with phenotypically complemented PrV-ΔgBGFP. Two days after infection, when a complete CPE had developed, supernatants were harvested and virions were purified by sucrose gradient centrifugation and analyzed by Western blotting. As shown in Fig. 7A, full-length gB as well as gB-007 and gB-007Pass were detected in the purified virion preparations. However, whereas the signal intensity of gB-007Pass (Fig. 7A, lane 3) was similar to that of wild-type gB (Fig. 7A, lane 1), the amount of gB-007 found in purified virions (Fig. 7A, lane 2) was significantly smaller. gH, used as a control, was detectable at a similar level in the virus preparations (Fig. 7B). As expected, gB was absent from virus progeny of normal RK13 cells (Fig. 7A, lane 4). These results indicate that gB-007Pass is incorporated into virions at levels similar to wild-type gB and significantly more efficiently than gB-007.
FIG. 7.
Incorporation of gB into virions. RK13-gB (lane 1), RK13-007 (lane 2), RK13-007Pass (lane 3), and normal RK13 cells (lane 4) were infected with PrV-ΔgBβ at an MOI of 10. Samples of sucrose gradient-purified progeny virions were subjected to SDS-PAGE under nonreducing conditions followed by Western blot analysis with anti-gB MAb b43-b5 (A) or a polyclonal gH-specific antiserum (B).
Identification of mutations in gB-007Pass.
These studies of the differences in the biological functions of gB-007 and gB-007Pass point to compensatory mutation(s) within the gB-007Pass ORF. Therefore, the PCR-amplified gB-007 and gB-007Pass genes as well as the gB genes contained in the cloned genomic KpnI-C fragments of PrV-007 and PrV-007Pass were sequenced and analyzed. As outlined in Fig. 8, two single-base substitutions were detected in PrV-007Pass from both sources, compared with the two PrV-007 sequences, which were identical. The mutations result in amino acid substitutions at positions 305 and 744 of the deduced gB polypeptide, both of which are in the predicted gB ectodomain. To test whether both mutations are required for restoration of gB function, cell lines expressing gB proteins containing either mutation were constructed and assayed for functional complementation after infection with gB-trans-complemented PrV-ΔgBβ by titration of progeny viruses from the supernatant on wild-type-gB-expressing cells. Whereas titers on gB-007-expressing cells were less than 102 PFU/ml, they reached 105 PFU/ml on gB-007Pass-expressing cells. On cells expressing gB proteins with either mutation, progeny viral titers were between 102 and 103 PFU/ml, which indicates that the presence of both mutations is required for efficient functional compensation of the defect in gB-007.
DISCUSSION
gB plays a central role in two important membrane fusion processes during herpesvirus infection, i.e., fusion between the virion envelope and the cellular plasma membrane during viral entry and fusion between the plasma membranes of infected cells and those of adjacent uninfected cells to allow direct viral cell-to-cell spread. We recently described a carboxy-terminally truncated PrV gB, gB-007, which was fully functional in mediating direct viral cell-to-cell spread but unable to complement the entry defect of a gB-deleted PrV mutant. This correlated with an inefficient incorporation of the mutated gB-007 into virus particles (24). Here we showed by reversion analysis that two point mutations in the gB-007 ectodomain restored efficient incorporation into virions and allowed gB-007Pass to complement the PrV-gB− entry defect. Thus, compensation for a carboxy-terminal truncation could be achieved by mutations in the ectodomain.
During maturation of herpesvirus virions, capsids assembled in the nucleus traverse the nuclear membrane by first budding at the inner leaflet, thereby acquiring a primary envelope. This envelope is then lost by fusion with the outer leaflet of the nuclear membrane and translocation of capsids into the cytoplasm. In the trans-Golgi network, these capsids gain their complete set of tegument proteins and their final envelope by budding into Golgi-derived vesicles (5). Therefore, incorporation into the envelope requires the presence of viral glycoproteins in those vesicles. The carboxy-terminal cytoplasmic domains of herpesvirus glycoproteins are supposed to play a role in intracellular localization of the proteins as well as in directing efficient incorporation into virions. We previously showed that carboxy-terminal truncation of gB, resulting in the loss of one predicted α-helix including two putative endocytosis signals, indeed results in abrogation of endocytosis of gB from the plasma membrane, leading to a primarily membrane-associated localization. Surprisingly, this gB-008 mutant protein was functional in both entry and direct viral cell-to-cell spread, similar to wild-type gB (24), indicating that endocytosis apparently does not play a role in gB function or incorporation into virions. In contrast, an additional deletion of the second predicted α-helix led to a loss of function during entry, correlating with inefficient incorporation into virions. Surprisingly, this defect could be corrected by two point mutations within the ectodomain of gB. A defect on one side of the membrane is, therefore, compensated by mutations on the other side of the membrane.
There are several explanations for this phenomenon: The cytoplasmic portions of herpesvirus glycoproteins have been shown to be involved in directing incorporation of the glycoprotein into virions (25, 39), possibly by mediating interaction with tegument proteins which in turn interact with capsid during secondary envelopment (2). In this context, mutations in the ectodomain of gB could increase the interaction between gB and other viral glycoproteins so that gB is efficiently incorporated into virions by a piggyback mechanism. However, so far, no functional interaction of gB with other glycoproteins, other than the formation of homo-oligomers, has been observed (6, 7). Mutations in the ectodomain could also somehow alter the conformation of gB-007, resulting in efficient incorporation, or affect overall processing. Since we do not have any evidence supporting the last two possibilities, and indeed we showed that gB-007 and gB-007Pass do not differ with regard to intracellular distribution or appearance of gB subunits in SDS-PAGE, we currently favor the first hypothesis. It has, for instance, been shown that gD is efficiently incorporated into virions despite deletion of the cytoplasmic domain (25). Therefore, there has to be another mechanism for glycoprotein targeting to the viral envelope besides interaction of their cytoplasmic domains with tegument proteins. This may be taken as indirect evidence for lateral interactions between different herpesvirus glycoprotein ectodomains in the viral envelope. Interactions between different envelope glycoproteins, in particular gB, gC, and gD, have been documented by cross-linking studies (6, 7, 30), although their functional importance is unclear (30). However, intermolecular interactions between different glycoproteins could also explain the dependence on more than one glycoprotein for fusion, which resulted in the suggestion of a so-called fusion complex.
In our analyses, we were not able to separate the function of C-terminally truncated gB in entry and its ability to become incorporated into virions. It seems trivial that a viral protein can only function in entry when it is present (in sufficient quantities) in the virion envelope. However, we cannot formally exclude the possibility that the function of PrV gB in entry is indeed separable from efficient incorporation into virions.
In our comparisons of gB-007 and gB-007Pass, we observed only the two point mutations described above. Our data indicate that both are required for restoration of the phenotype and that introduction of either mutation alone into gB-007 is not sufficient to restore function. However, it is conceivable that one mutation rescues virion incorporation and the other complements the functional defect in entry. This will be investigated in the future.
By in vitro serial passaging of PrV mutants which are capable of direct viral cell-to-cell spread but unable to produce free infectious virions, reversion mutants which are infectious in the absence of an otherwise essential glycoprotein, i.e., gD or gL, have been isolated (14, 34). While the molecular basis for the repaired phenotype of PrV-ΔgLPass been shown to be due to the creation of a hybrid gene encoding a hybrid gD-gH protein which exerts gD and gH function without the need for gL, the compensatory mutations resulting in gD-independent infectivity have not yet all been mapped. However, they include mutations in gH and gB (33a). Here we showed by reversion analysis that a carboxy-terminal deletion of gB which affects incorporation into virions and function during entry is compensated by two point mutations in the ectodomain. These results again prove the power of reversion analysis for uncovering unexpected compensatory mutations and document the amazing adaptability of herpesviruses.
ACKNOWLEDGMENTS
This work was supported by the Deutsche Forschungsgemeinschaft (grant Me 854/4-2).
We thank Nadine Müller and Uta Hartwig for expert technical assistance.
REFERENCES
- 1.Brack A R, Dijkstra J M, Granzow H, Klupp B G, Mettenleiter T C. Inhibition of virion maturation by simultaneous deletion of glycoproteins E, I, and M of pseudorabies virus. J Virol. 1999;73:5364–5372. doi: 10.1128/jvi.73.7.5364-5372.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brack A R, Klupp B G, Granzow H, Tirabassi R, Enquist L W, Mettenleiter T C. Role of the cytoplasmic tail of pseudorabies virus glycoprotein E in virion formation. J Virol. 2000;74:4004–4016. doi: 10.1128/jvi.74.9.4004-4016.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Claesson-Welsh L, Spear P G. Oligomerization of herpes simplex virus glycoprotein B. J Virol. 1986;60:803–806. doi: 10.1128/jvi.60.2.803-806.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geraghty R J, Krummenacher C, Cohen G H, Eisenberg R J, Spear P G. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science. 1998;280:1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 5.Granzow H, Klupp B G, Fuchs W, Veits J, Osterrieder N, Mettenleiter T C. Egress of alphaherpesviruses: comparative ultrastructural study. J Virol. 2001;75:3675–3684. doi: 10.1128/JVI.75.8.3675-3684.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Handler C, Eisenberg R J, Cohen G H. Oligomeric structure of glycoproteins in herpes simplex virus type 1. J Virol. 1996;70:6067–6075. doi: 10.1128/jvi.70.9.6067-6070.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Handler C, Cohen G H, Eisenberg R J. Cross-linking of glycoprotein oligomers during herpes simplex virus type 1 entry. J Virol. 1996;70:6076–6082. doi: 10.1128/jvi.70.9.6076-6082.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herold B C, WuDunn D, Soltys N, Spear P G. Glycoprotein C of herpes simplex virus type 1 plays a principal role in the adsorption of virus to cells and in infectivity. J Virol. 1991;65:1090–1098. doi: 10.1128/jvi.65.3.1090-1098.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herold B C, Visalli R J, Susmarski N, Brandt C, Spear P G. Glycoprotein C-independent binding of herpes simplex virus to cells requires cell surface heparan sulphate and glycoprotein B. J Gen Virol. 1994;75:1211–1222. doi: 10.1099/0022-1317-75-6-1211. [DOI] [PubMed] [Google Scholar]
- 10.Johnson D C, Burke R L, Gregory T. Soluble forms of herpes simplex virus glycoprotein D bind to a limited number of cell surface receptors and inhibit virus entry into cells. J Virol. 1990;64:2569–2576. doi: 10.1128/jvi.64.6.2569-2576.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaplan A S, Vatter A. A comparison of herpes simplex and pseudorabies virus. Virology. 1959;7:394–407. doi: 10.1016/0042-6822(59)90068-6. [DOI] [PubMed] [Google Scholar]
- 12.Karger A, Mettenleiter T C. Glycoproteins gIII and gp50 play dominant roles in the biphasic attachment of pseudorabies virus. Virology. 1993;194:654–664. doi: 10.1006/viro.1993.1305. [DOI] [PubMed] [Google Scholar]
- 13.Klupp B G, Mettenleiter T C. Sequence and expression of the glycoprotein gH gene of pseudorabies virus. Virology. 1991;182:732–741. doi: 10.1016/0042-6822(91)90614-h. [DOI] [PubMed] [Google Scholar]
- 14.Klupp B G, Mettenleiter T C. Glycoprotein gL-independent infectivity of pseudorabies virus is mediated by a gD-gH fusion protein. J Virol. 1999;73:3014–3022. doi: 10.1128/jvi.73.4.3014-3022.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kopp A, Mettenleiter T C. Stable rescue of a glycoprotein gII deletion mutant of pseudorabies virus by glycoprotein gI of bovine herpesvirus 1. J Virol. 1992;66:2754–2762. doi: 10.1128/jvi.66.5.2754-2762.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kopp A, Blewett E, Misra V, Mettenleiter T C. Proteolytic cleavage of bovine herpesvirus 1 (BHV-1) glycoprotein gB is not necessary for its function in BHV-1 or pseudorabies virus. J Virol. 1994;68:1667–1674. doi: 10.1128/jvi.68.3.1667-1674.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krummenacher C, Nicola A V, Whitbeck J C, Lou H, Hou W, Lambris J D, Geraghty R J, Spear P G, Cohen G H, Eisenberg R J. Herpes simplex virus glycoprotein D can bind to poliovirus receptor-related protein 1 or herpesvirus entry mediator, two structurally unrelated mediators of virus entry. J Virol. 1998;72:7064–7074. doi: 10.1128/jvi.72.9.7064-7074.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, van Drunen Littel-van den Hurk S, Babiuk L A, Liang X. Characterization of cell-binding properties of bovine herpesvirus 1 glycoproteins B, C, and D: identification of a dual cell-binding function of gB. J Virol. 1995;69:4758–4768. doi: 10.1128/jvi.69.8.4758-4768.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ligas M W, Johnson D C. A herpes simplex virus mutant in which glycoprotein D sequences are replaced by β-galactosidase sequences binds to but is unable to penetrate into cells. J Virol. 1988;62:1486–1494. doi: 10.1128/jvi.62.5.1486-1494.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lukàcs N, Thiel H-J, Mettenleiter T C, Rziha H-J. Demonstration of three major species of pseudorabies virus glycoproteins and identification of a disulfide-linked glycoprotein complex. J Virol. 1985;53:166–173. doi: 10.1128/jvi.53.1.166-173.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mettenleiter T C. Aujeszky's disease (pseudorabies) virus: the virus and molecular pathogenesis—state of the art, June 1999. Vet Res. 2000;31:99–115. doi: 10.1051/vetres:2000110. [DOI] [PubMed] [Google Scholar]
- 22.Mettenleiter T C, Zsak L, Zuckermann F, Sugg N, Kern H, Ben-Porat T. Interaction of glycoprotein gIII with a cellular heparinlike substance mediates adsorption of pseudorabies virus. J Virol. 1990;64:278–286. doi: 10.1128/jvi.64.1.278-286.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mettenleiter T C, Lukàcs N, Thiel H-J, Schreurs C, Rziha H-J. Location of the structural gene of pseudorabies virus glycoprotein complex gII. Virology. 1986;152:66–75. doi: 10.1016/0042-6822(86)90372-7. [DOI] [PubMed] [Google Scholar]
- 24.Nixdorf R, Klupp B G, Karger A, Mettenleiter T C. Effects of truncation of the carboxy terminus of pseudorabies virus glycoprotein B on infectivity. J Virol. 2000;74:7137–7145. doi: 10.1128/jvi.74.15.7137-7145.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nixdorf R, Klupp B G, Mettenleiter T C. Role of the cytoplasmic tails of pseudorabies virus glycoproteins B, E and M in intracellular localization and virion incorporation. J Gen Virol. 2001;82:215–226. doi: 10.1099/0022-1317-82-1-215. [DOI] [PubMed] [Google Scholar]
- 26.Peeters B, de Wind N, Hooisma M, Wagenaar F, Gielkens A, Moormann R. Pseudorabies virus envelope glycoproteins gp50 and gII are essential for virus penetration, but only gII is involved in membrane fusion. J Virol. 1992;66:894–905. doi: 10.1128/jvi.66.2.894-905.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pereira L. Function of glycoprotein B homologues of the family Herpesviridae. Infect Agents Dis. 1994;3:9–28. [PubMed] [Google Scholar]
- 28.Rauh I, Mettenleiter T C. Pseudorabies virus glycoproteins gII and gp50 are essential for virus penetration. J Virol. 1991;65:5348–5356. doi: 10.1128/jvi.65.10.5348-5356.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robbins A K, Dorney D J, Wathen M W, Whealy M E, Gold C, Watson R J, Holland L E, Weed S D, Levine M, Glorioso J, Enquist L W. The pseudorabies virus gII gene is closely related to the gB glycoprotein gene of herpes simplex virus. J Virol. 1987;61:2691–2701. doi: 10.1128/jvi.61.9.2691-2701.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodger G, Boname J, Bell S, Minson A. Assembly and organization of glycoproteins B, C, D, and H in herpes simplex virus type 1 particles lacking individual glycoproteins: no evidence for the formation of a complex of these molecules. J Virol. 2001;75:710–716. doi: 10.1128/JVI.75.2.710-716.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roizman B. Herpesviridae. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2221–2231. [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33a.Schmidt J, Gerdts V, Beyer J, Klupp B G, Mettenleiter T C. Glycoprotein D-independent infectivity of pseudorabies virus results in an alteration of in vivo host range and correlates with mutations in glycoproteins B and H. J Virol. 2001;75:10054–10064. doi: 10.1128/JVI.75.21.10054-10064.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmidt J, Klupp B G, Karger A, Mettenleiter T C. Adaptability in herpesviruses: glycoprotein D-independent infectivity of pseudorabies virus. J Virol. 1997;71:17–24. doi: 10.1128/jvi.71.1.17-24.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schröder C, Keil G M. Bovine herpesvirus 1 requires glycoprotein H for infectivity and direct spreading and glycoproteins gHw450 and gB for glycoprotein D-independent cell-to-cell spread. J Gen Virol. 1999;80:57–61. doi: 10.1099/0022-1317-80-1-57. [DOI] [PubMed] [Google Scholar]
- 36.Schröder C, Linde G, Fehler F, Keil G M. From essential to beneficial: glycoprotein D loses importance for replication of bovine herpesvirus 1 in cell culture. J Virol. 1997;71:25–33. doi: 10.1128/jvi.71.1.25-33.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spear P G. Entry of alphaherpesviruses into cells. Semin Virol. 1993;4:167–180. [Google Scholar]
- 38.Spear P G, Eisenberg R J, Cohen G H. Three classes of cell surface receptors for alphaherpesvirus entry. Virology. 2000;275:1–8. doi: 10.1006/viro.2000.0529. [DOI] [PubMed] [Google Scholar]
- 39.Tirabassi R S, Townley R A, Eldridge M G, Enquist L W. Characterization of pseudorabies virus mutants expressing carboxy-terminal truncations of gE: evidence for envelope incorporation, virulence, and neurotropism domains. J Virol. 1997;71:6455–6464. doi: 10.1128/jvi.71.9.6455-6464.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Warner M S, Geraghty R J, Martinez W M, Montgomery R I, Whitbeck J C, Xu R, Eisenberg R J, Cohen G H, Spear P G. A cell surface protein with herpesvirus entry activity (HveB) confers susceptibility to infection by mutants of herpes simplex virus type 1, herpes simplex virus type 2, and pseudorabies virus. Virology. 1998;246:179–189. doi: 10.1006/viro.1998.9218. [DOI] [PubMed] [Google Scholar]
- 41.Wölfer U, Kruft V, Sawitzky D, Hampl H, Wittmann-Liebold B, Habermehl K-O. Processing of pseudorabies virus glycoprotein gII. J Virol. 1990;64:3122–3125. doi: 10.1128/jvi.64.6.3122-3125.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]