Abstract
The Rous sarcoma virus (RSV) transmembrane (TM) glycoprotein is modified by the addition of palmitic acid. To identify whether conserved cysteines within the hydrophobic anchor region are the site(s) of palmitoylation, and to determine the role of acylation in glycoprotein function, cysteines at residues 164 and 167 of the TM protein were mutated to glycine (C164G, C167G, and C164G/C167G). In CV-1 cells, palmitate was added to env gene products containing single mutations but was absent in the double-mutant Env. Although mutant Pr95 Env precursors were synthesized with wild-type kinetics, the phenotypes of the mutants differed markedly. Env-C164G had properties similar to those of the wild type, while Env-C167G was degraded faster, and Env containing the double mutant C164G/C167G was very rapidly degraded. Degradation occurred after transient plasma membrane expression. The decrease in steady-state surface expression and increased rate of internalization into endosomes and lysosomes paralleled the decrease in palmitoylation observed for the mutants. The phenotypes of mutant viruses were assessed in avian cells in the context of the pATV8R proviral genome. Virus containing the C164G mutation replicated with wild-type kinetics but exhibited reduced peak reverse transcriptase levels. In contrast, viruses containing either the C167G or the C164G/C167G mutation were poorly infectious or noninfectious, respectively. These phenotypes correlated with different degrees of glycoprotein incorporation into virions. Infectious revertants of the double mutant demonstrated the importance of cysteine-167 for efficient plasma membrane expression and Env incorporation. The observation that both cysteines within the membrane-spanning domain are accessible for acylation has implications for the topology of this region, and a model is proposed.
The viral envelope glycoproteins of the Rous sarcoma virus (RSV) are encoded in the env region of the retroviral genome and are initially translated as a precursor protein, Pr95 (4, 25). Properly oligomerized Pr95 exits the endoplasmic reticulum (11) and undergoes complex glycosylation, as well as proteolytic cleavage, in the Golgi compartments to the final disulfide-linked products gp85 (SU) and gp37 (TM) (12, 20, 40, 43). Once SU-TM oligomers reach the cell surface, they can be incorporated into the virus particle during budding (26).
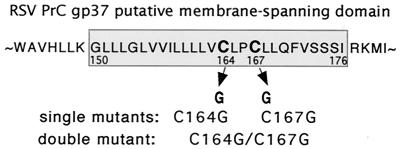
The subgroup C RSV gp37 (TM) protein is a bitopic 198-amino-acid protein consisting of a 149-amino-acid extracellular region, a membrane-spanning anchor domain, and a short C-terminal cytoplasmic tail (25, 52). Based on the hydrophobicity of the amino acid sequence, the transmembrane region had been suggested to encompass 27 amino acids, as outlined in Fig. 1. Earlier experiments in this laboratory, aimed at identifying regions of the TM protein that direct incorporation of the viral glycoproteins into virions, demonstrated that the C-terminal 22 cytoplasmic amino acids were dispensable and that loss of this cytoplasmic domain did not significantly affect glycoprotein transport, processing, or incorporation into the virion (42). Additionally, virions containing these cytoplasmic-domain-negative mutants retained infectivity. These studies suggested that the crucial (structural) information for incorporation was instead contained within the hydrophobic membrane-spanning domain (MSD). Preliminary studies with point and deletion mutants in the designated transmembrane region show that these mutations often severely depress or completely abrogate viral infectivity (D. Miller, G. L. Davis, and E. Hunter, unpublished data).
FIG. 1.
Locations of mutations within RSV PrC gp37 (TM). The previously suggested boundaries of the MSD are indicated by the shaded box. Oligonucleotide mutagenesis was used to change the TGC codon for cysteine to a GGC codon for glycine to investigate the potential thioester linkage of palmitate at these sites. Mutants were named C164G, C167G, and C164G/C167G to denote the location and type of substitution at the cysteine codon in each mutant.
Addition of palmitic acid has been shown to be a posttranslational modification of many viral glycoproteins (for a review, see references 46 and 51). The exact role of palmitoylation in transmembrane proteins is unknown, but it may serve to efficiently target the glycoprotein to the cell membrane or to distinct microdomains within it. In particular, palmitoylated transmembrane proteins, like hemagglutinin (HA) and neuraminidase of influenza A virus, have been found to associate with so-called membrane raft or detergent-resistant glycolipid domains (18, 36, 53) (see reference 46 for a review). However, this correlation is not absolute (36), as the acylated vesicular stomatitis virus (VSV) G glycoprotein does not sequester into rafts (6), nor did we find evidence for RSV Env being present in detergent-resistant glycolipid domains (39). Palmitate may help anchor the glycoprotein in the membrane, and by influencing Env topology in the plasma membrane, it could play a role in incorporation of the glycoprotein into the virion.
Cysteines were identified as sites of palmitic acid addition to membrane-spanning cellular and viral proteins, which include Sindbis virus E2 (27), measles virus F protein (7), VSV G (33, 34, 47), Semliki Forest virus E1 (50), and influenza A virus HA (37, 55, 57). No consensus motif directing this modification has been identified, but palmitoylated cysteines are generally located in the cytoplasmic tail, close to the membrane boundary, or within the membrane anchor region itself (19, 21, 50, 58, 61) (for a review, see reference 46). The importance of a complex conformational context has been suggested (45). The effects of abrogating palmitoylation on viral glycoprotein function have been analyzed for various viruses, and these effects range from insignificant to detrimental for viral replication. For VSV G, the results vary (32–34, 47, 59), and influenza virus HA displays subtype specificity with regard to the phenotypes of palmitoylation mutants (37, 44, 55).
The C-type retroviral glycoproteins of spleen focus-forming virus (54), RSV (16, 60), murine leukemia virus (MuLV) (21, 61), and human immunodeficiency virus (62) were found to be palmitoylated. Most retroviral TM proteins lack cysteines within the cytoplasmic domain. However, they are unusual in that they show a conservation of a proline-cysteine pair within the transmembrane region (41), which is a site for palmitoylation in both Friend and Moloney MuLVs (21, 61). In contrast, human immunodeficiency virus Env does contain cysteines in its unusually long cytoplasmic tail which are the sites of palmitate addition (62). The RSV TM membrane-spanning region is highly conserved among different subtypes and contains two cysteines at amino acid positions 164 and 167. This prompted us to investigate whether substituting glycine residues for each of these cysteines affected palmitoylation and the biological function of this prototype retroviral glycoprotein. We report here that both C164 and C167 are sites for palmitic acid addition and show that the lack of palmitoylation affects glycoprotein function and virus infectivity. Palmitoylation of C167, which is part of the conserved proline-cysteine pair, proved particularly critical, since molecules lacking cysteine, and thus palmitate, at this site showed significantly reduced steady-state surface expression, were inefficiently incorporated into virions, and exhibited greatly diminished virus infectivity. The accessibility of both cysteines for acylation, furthermore, prompted us to reevaluate the current model for the MSD boundaries.
MATERIALS AND METHODS
Cells and viruses.
Primary turkey embryo cells (TEC) used in infectivity studies were prepared as described by Hunter (23). The Df-1 avian fibroblast cell line was obtained from D. Foster (22, 48) and cultured in Dulbecco's modified Eagle's medium (10% fetal calf serum). African green monkey kidney (CV-1) cells used in the simian virus 40 (SV40) expression system were obtained from the American Type Culture Collection (Manassas, Va.). Bacterial plasmids were propagated in the DH-1 strain of Escherichia coli, and the M13 bacteriophage was grown in TG-1 cells.
DNA expression vectors.
The derivation of the RSV env gene from the RSV Prague C molecular clone, pATV8, was described by Davis and Hunter (9). Infectivity studies were done by inserting the mutated env genes into a modified, nonpermuted version of the pATV8 vector, pATV8R. This vector contains, between two long terminal repeats, a nonpermuted RSV genome in which a unique ClaI site has been inserted at the XbaI site near the C terminus of the env gene; modified env genes can thus be inserted via unique KpnI and ClaI sites that flank the env coding region. Details of the construction of this vector are available on request. The recombinant SV40 expression system for RSV Env has been described previously (9, 39).
Cloning and mutagenesis of env gene.
Oligonucleotide-directed single-strand DNA mutagenesis was carried out on a 717-bp EcoRI-XbaI subfragment, containing the coding region for TM, which had been inserted into M13mp18 as described by Perez and Hunter (43). Three oligonucleotide primers were employed to create the three mutations C164G, C167G, and C164G/C167G. In the first, the TGC coding for cysteine 164 was changed to the glycine codon GGC (mutant C164G); in the second, the TGC coding for cysteine 167 was changed to glycine codon GGC (mutant C167G); and in the third, both cysteine codons 164 and 167 were changed to glycine codons (mutant C164G/C167G). Insertion of the mutated sequences into the SV40 expression vector was done through unique EcoRI and XbaI restriction sites as originally described by Davis and Hunter (9).
The infectious RSV proviral DNA clone pATV8R described above allowed mutant env genes to be transferred directly from the SV40 vector via the unique KpnI and ClaI restriction sites that flank the env gene. Each mutant was sequenced in both the SV40 and RSV vectors to ensure the presence of the mutation throughout the subcloning.
Generation of recombinant SV40 stocks and infection of CV-1 cells.
The SV40 slot-in vector, pSVenvKX, was cut with KpnI, self-ligated at low DNA concentration to allow circularization and juxtaposition of the late SV40 promoter upstream of env, and cotransfected with helper virus (dl1055) genomic DNA into CV-1 cells, using the DEAE-dextran method (60). At 5 to 7 days posttransfection, the CV-1 cells were freeze-thawed three times and sonicated for 30 s to release the complemented recombinant SV40 virions. Subconfluent CV-1 cells were then infected in two consecutive rounds with the sonicated lysate to make a high-titer recombinant SV40 stock. High-titer lysates were then used at appropriate dilutions to infect CV-1 cells for the experiments described, which were performed at 72 to 96 or 48 h postinfection (p.i.), as indicated.
Transfection of primary TEC and Df-1 cells.
Primary TEC were transfected by the polybrene method described by Kawai and Nishizawa (30) with the following modifications. Polybrene (30 μg) was added to 5 to 10 μg of DNA in a final volume of 1 ml of serum-free (SF) medium. This mixture was added to subconfluent TEC on 60-mm-diameter plates, which were rocked at 37°C in a humidified incubator for 6 h. The DNA solution was removed, and the cells were washed twice with SF medium and then shocked for 4 min with 25% dimethyl sulfoxide in SF medium. After the cells were gently washed twice in SF medium, growth medium containing 10% serum was added to the cells. At 24 h posttransfection, the cells were passaged to 100-mm-diameter plates. Culture medium was harvested for reverse transcriptase (RT) assays at 4-day intervals, and the cells were split at a 1:3 ratio immediately after being harvested.
Avian fibroblast cell line Df-1 cells were transfected with proviral DNA using the FuGene-6 reagent (Roche), with 6 μg of DNA and 18 μl of FuGene-6 per 1.5 × 106 cells on 100-mm-diameter plates.
Antisera used.
Polyclonal rb#2 anti-gp85 antiserum specifically interacts with RSV SU (gp85) protein. rb-anti-RSV-TM-Cpep, which was raised against a C-terminal peptide (Cys plus amino acids 184 to 198) of RSV PrC TM, was a gift from Judy White. Chicken anti-RSV PrC neutralizing antibody has been described previously (29, 39). rb-B77 serum was raised against RSV Gag protein.
Secondary antibodies for immunofluorescence studies were gt-anti-rb-Alexa488 and rb-anti-chiAlexa488, which were obtained from Molecular Probes (Eugene, Oreg.).
Radioactive labeling of viral proteins and immunoprecipitations.
[3H]palmitic acid (54 Ci/mM) and [3H]leucine (50 Ci/mM) were obtained from Amersham Corp., Arlington Heights, Ill.; [35S]methionine-cysteine (>1,150 Ci/mM) was obtained from NEN, Boston, Mass. Protocols for labeling with [3H]leucine (100 μCi/60-mm-diameter plate), immunoprecipitation, and electrophoresis were for the most part as described previously (60). Labeling with [35S]methionine-cysteine (50 μCi/100 μl of deficient medium) was performed essentially as described previously (29, 39).
To analyze fatty acid modification of the glycoproteins, recombinant-SV40-infected CV-1 cells were starved 72 to 96 h p.i. for 1 h in SF medium. [3H]palmitate was evaporated to dryness, dissolved in 4 μl of ethanol, and then mixed with 500 μl of SF medium and placed on the cells (100 μCi/60-mm-diameter plate). After being incubated for 4 h at 37°C, the cells were washed with phosphate-buffered saline (PBS) and lysed in buffer containing 1% Triton X-100 and 1% deoxycholate.
Cells pulse- or pulse-chase-labeled with [3H]leucine or [35S]methionine were lysed the same way. Cell lysates were spun to pellet the nuclei, adjusted to 0.1% sodium dodecyl sulfate (SDS), and subjected to preclearing with normal rabbit serum and Staphylococcus aureus.
Viral Env and Gag proteins were immunoprecipitated with rabbit sera and then separated by SDS-polyacrylamide gel electrophoresis (PAGE) and visualized for 3H labeling by the dimethyl sulfoxide-2,5-diphenyloxasole fluorography method of Bonner and Laskey (3) or for 35S labeling by autoradiography and OptiQuant phosphorimaging.
When incorporation of Env into virions was analyzed, Df-1 cells were [35S]methionine pulse-labeled 44 h posttransfection with proviral DNA and chased in complete medium for 6 h. The culture supernatant was precleared by low-speed centrifugation, and then virions from 1.2 ml were pelleted through a 25% (wt/vol) sucrose cushion (750 μl) in a TLS55 rotor (Beckman) for 30 min at 50,000 rpm (≈260,000 × g) and 20°C. The viral pellet was resuspended in lysis buffer and subjected to immunoprecipitation as described for cell lysates.
Indirect immunofluorescence analysis.
All analyses were done essentially as previously described (29, 39). To visualize the intracellular steady-state distributions of wild-type and mutant Env proteins, protein-expressing CV-1 cells grown on glass coverslips were fixed in cold acetone and then probed with rb-anti-RSV-TM-Cpep, with gt-anti-rb-Alexa488 as the secondary antibody. For surface immunofluorescence, unfixed cells on coverslips were incubated with chi-anti-RSV PrC antibody on ice for 30 min, washed in PBS, fixed with methanol-acetic acid, and stained with rb-anti-chiAlexa488. In order to compare clearing of wild-type and mutant PrC Env from the cell surface, CV-1 cells grown on coverslips were washed in PBS and then incubated with chi-anti-RSV PrC in the presence of 100 μM chloroquine (to prevent liposomal degradation) for 2 h at 37°C. The cells were then washed, fixed, and stained as for surface immunofluorescence. All samples were observed and photographed with a Zeiss fluorescence microscope.
RT assay.
A modification of a microtiter RT assay (10) was employed to determine the infectivity of mutated viral genomes. In this assay, 25 μl of culture supernatant was assayed directly for enzyme activity following low-speed centrifugation to remove cell debris. After a 1-h reaction, the amounts of incorporated 35S-dTTP were determined in a radioimager (Ambis Inc., San Diego, Calif.) following transfer to NA45 nitrocellulose.
RT-PCR amplification of virion RNA sequences.
Culture medium was harvested from RSV ATV8R-infected TEC at either 12 or 24 days posttransfection and precleared at 2,000 × g for 15 min. Virus was pelleted from 5.0 ml of clarified culture medium by centrifugation through a 25% (wt/vol) sucrose cushion in a Beckman SW41 rotor at 40,000 rpm (≈270,000 × g) for 1.5 h. RNA was extracted from the pelleted virions according to the method of Gilman (17) except that 1% Triton X-100 was used in place of 0.5% NP-40 and RNasin (500 U/ml) and dithiothreitol (1 mM) were added to the lysis buffer just before use.
The viral RNA pellet was resuspended in diethyl pyrocarbonate-treated H2O. RNasin (20 U), PCR amplification buffer, and 50 pmol of the 3′ minus-strand primer (5′-CGTTACAGCCCGATTCGCAGG-3′) were added, and the sample was heated to 95°C for 1 min and then chilled on ice. RNasin (20 U), deoxynucleoside triphosphates (1 mM), MgCl2 (2.5 mM), and avian myeloblastosis virus RT (25 U; Boehringer Mannheim) were added, and the reaction mixture was incubated for 10 min at room temperature for primer annealing, at 42°C for 55 min for cDNA synthesis, and at 95°C for 10 min to inactivate the enzyme. The cDNA generated was stored at −20°C or used directly in a PCR.
cDNA (60 to 100 ng), together with 50 pmol of 5′ (plus-strand) primer (5′-GATCGCGCATGGCAAGGAATTCCC-3′), was subjected to PCR using Taq polymerase (Promega). The 775-bp PCR product was agarose gel purified, and DNA was extracted using Geneclean (Bio 101) and stored at −20°C.
In order to analyze the sequences of individually cloned PCR products, DNA was cleaved with EcoRI and XbaI and cloned into the plasmid pTZ18R for amplification.
Direct DNA sequencing of PCR product.
The DNA-sequencing protocol used was based on that of Keulen and Schuurman (31) and employed Sequenase version 2.0 (United States Biochemical). Sixty to 100 ng (1 μl) of the double-stranded DNA PCR product and 20 pmol of primer C (internal primer to the PCR product; 5′-ATGGGCCGTTCATCTGC-3′) were used in the reaction. The samples were separated on an 8% urea polyacrylamide gel.
RESULTS
Palmitoylation of wild-type and mutant RSV env gene products.
As mentioned above, RSV Env is among the retroviral glycoproteins that are posttranslationally modified by palmitate addition but lack potential cysteine palmitoylation sites in their cytoplasmic tails. This prompted us to analyze the role of cysteines within the putative MSD of the subgroup C RSV gp37 (TM) protein and to determine whether they are the sites of palmitoylation. The MSDs of different avian leukosis and sarcoma viruses are highly conserved (identical amino acid sequences in the cases of, e.g., subtypes PrC, Sr-A, and SR-D) and have been suggested to encompass 27 amino acids (Fig. 1). The cysteine codons were mutated alone or in combination to code for glycine, resulting in the mutant proteins Env-C164G, -C167G, and -C164G/C167G (previously referred to as palm#6, palm#7, and palm#6+7, respectively). These changes were verified by DNA sequencing of the M13mp18 mutagenesis vector and after insertion of mutant env into both the SV40 and RSV vectors as described in Materials and Methods.
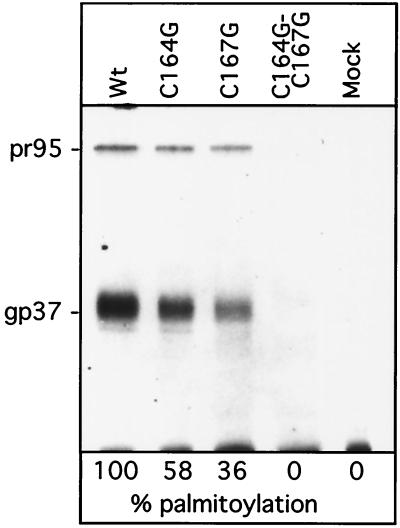
To determine whether the cysteine mutations affected palmitic acid addition, CV-1 cells infected with recombinant SV40 coding for wild-type or mutant RSV Env were labeled with [3H]palmitate at 72 h p.i. Env glycoproteins were immunoprecipitated from detergent-lysed cells and subjected to SDS-PAGE, and palmitate-labeled Env was visualized by fluorography. Wild-type Env, as well as both the Env-C164G and Env-C167G single mutants, can be labeled with [3H]palmitate (Fig. 2), with the level of palmitoylation being reduced for the mutants. In contrast, the double mutant Env-C164G/C167G was not labeled with [3H]palmitate, suggesting that both cysteines within the putative MSD are posttranslationally modified by palmitoylation. Interestingly, Env-C167G displays a more pronounced reduction of acylation than Env-C164G, which suggests that C167 is the preferred palmitoylation site. Parallel experiments in which wild-type and mutant Env-expressing CV-1 cells were pulse-labeled with [3H]leucine indicated that all four glycoproteins were expressed to similar levels (not shown).
FIG. 2.
Labeling of RSV glycoproteins with [3 H]palmitic acid. Wild-type and mutant glycoproteins expressed in CV-1 cells after infection with the recombinant SV40 vector were labeled for 4 h with [3H]palmitic acid. Immunoprecipitates were separated on SDS–12% PAGE and then analyzed by fluorography. Lane 1, wild type (wt); lane 2, C164G; lane 3, C167G; lane 4, C164G/C167G; lane 5, mock infected.
Turnover of mutant env gene products.
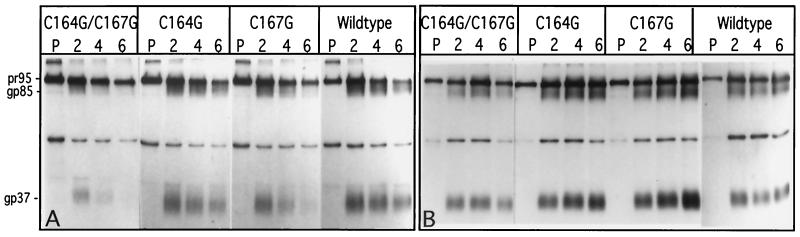
Expression of the three mutant glycoproteins was further analyzed to determine the role of palmitoylated cysteines in glycoprotein biosynthesis, processing, and stability. Four 60-mm-diameter plates of CV-1 cells were infected with each of the recombinant SV40 stocks encoding wild-type and mutant glycoproteins. At 72 to 96 h p.i., the cells were pulse-labeled for 15 min with [3H]leucine and then lysed immediately or supplemented with complete medium and chased at 37°C for 2, 4, and 6 h prior to lysis. Metabolically labeled RSV glycoproteins were visualized after immunoprecipitation and SDS-PAGE (Fig. 3A). Pulse-labeling of wild-type-expressing CV-1 cells results in a single Env-specific band, the precursor Pr95. After a 2-h chase, two additional, more diffuse bands of 85 and 37 kDa appear, representing the mature Env cleavage products, gp85 (SU) and gp37 (TM). Prolonged chase times of 4 and 6 h result in a gradual decrease in the intensity of Pr95, but also of SU and TM. The fact that no significant accumulation of the mature Env products occurs is consistent with our previous observations (9). Essentially identical results were obtained with the single mutant Env-C164G, indicating that this mutation did not affect the processing and stability of the env gene product. In contrast, while the precursor Pr95 was synthesized in amounts similar to those of the wild type for mutants Env-C167G and Env-C164G/C167G (Fig. 3A, lanes P) and showed a similar decrease over time, the levels of Env cleavage products detectable during the chase periods were clearly reduced, though to different extents. This is particularly clear when the TM (gp37) bands are compared. Env containing only the single mutation C167G was processed during the 2-h chase, and levels of gp85 and gp37 similar to that observed for wild-type Env accumulated. However, over the subsequent 4- and 6-h chase periods, the intensity of the gp85 and gp37 bands declined significantly faster. Even more dramatic reductions were observed for the double mutant, Env-C164G/C167G. In this case degradation of gp85 and gp37 appeared to occur during the 2-h chase, since both bands were significantly less intense than those observed for the wild type and had been lost almost completely by 6 h. These results indicate that the Env cleavage products of the C167G and C164G/C167G mutant precursors were being turned over more rapidly after exiting the Golgi compartments.
FIG. 3.
Kinetics of glycoprotein processing and stability in the presence and absence of chloroquine. (A) Wild-type and mutant viral glycoproteins expressed from the recombinant SV40 expression vector system were pulse-labeled with [3H]leucine for 15 min (lanes P) and then chased for 2, 4, or 6 h (lanes 2, 4, and 6, respectively). (B) The wild-type and mutant glycoproteins were pulse-labeled and chased as for panel A, but the cells were preincubated in 100 μM chloroquine for 1 h prior to the pulse and chloroquine was present in the medium throughout the chase periods. Lane designations are as in panel A.
In a previous study (9), we showed that substitution of an arginine residue for a leucine residue within the membrane-spanning region of TM (L165R; previously called μ26) resulted in rapid targeting of the Env complex to the lysosome after it had traversed the secretory pathway and had been transiently expressed on the plasma membrane. Addition of chloroquine, which prevents fusion of endosomal vesicles with the lysosome, could prevent this rapid degradation of Env-L165R, leading to the accumulation of SU and TM in prelysosomal vesicles. To determine whether the cysteine mutants were also directed into the lysosomal degradation pathway, we performed a pulse-chase experiment in the presence of 100 mM chloroquine. Under these conditions, there was no significant decrease in the intensities of the bands representing the terminally glycosylated SU and TM products for any of the constructs (Fig. 3B). These results indicate that the exchange of glycine for C167 triggered a more rapid uptake of the glycoprotein into the endocytic pathway and delivery to the lysosome. In fact, as we have described recently, in CV-1 cells Env-C167G displays an endocytosis rate of 4.7% ± 0.9% per min, which is significantly higher than the wild-type rate (1.1% ± 0.2% per min), while the rate of Env-C164G (1.3% ± 0.7% per min) is not significantly different (39).
Reduced surface expression of mutant Env proteins.
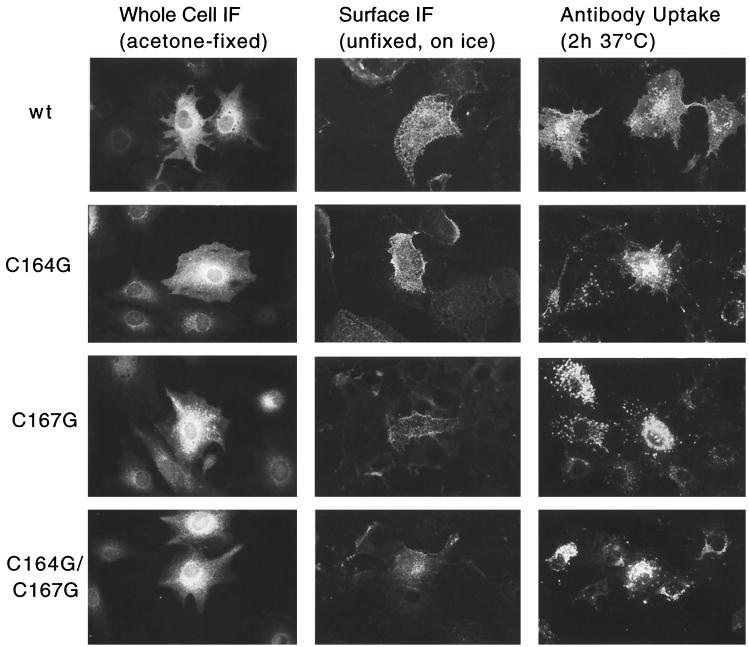
To corroborate the pulse-chase studies and to demonstrate that the viral glycoproteins are in fact reaching the cell surface, we carried out immunofluorescence analyses. CV-1 cells expressing the respective Env proteins were fixed and permeabilized for whole-cell rabbit anti-Env staining. Unfixed cells were incubated with chicken anti-RSV antisera, either on ice to detect steady-state Env surface expression or at 37°C in the presence of chloroquine to demonstrate internalization of surface-bound antibodies (Fig. 4). In fixed and permeabilized cells, the wild type and the C164G, C167G, and C164G/C167G mutant glycoproteins showed similar perinuclear staining patterns, typical for RSV Env, as described previously (39, 60). However, the contours of the plasma membrane appear stained to a somewhat lesser extent for both glycoproteins containing the C167G mutation. In accordance with this observation, the levels of steady-state surface staining of the mutant glycoproteins are reduced to various degrees compared to that in the wild type (wild type ≥ C164G > C167G >C164G/C167G). In a reciprocal manner, internalization and accumulation in intracellular prelysosomal vesicles of Env-antibody complexes is most pronounced in the double mutant and least pronounced in the wild-type Env. These observations are similar to those obtained with PrC Env-L165R, which is also transiently expressed on the cell surface but internalized rapidly (7.4% per min) (39). It should also be mentioned that, as previously described (39), the binding of the chicken-anti-RSV antibody per se does not trigger enhanced PrC Env endocytosis.
FIG. 4.
Indirect immunofluorescence of wild-type (wt) and palmitoylation mutant glycoproteins in CV-1 cells reveals different steady-state distributions. The intracellular distributions of wild-type and mutant env gene products were probed after acetone fixation and permeabilization of recombinant-SV40-infected CV-1 cells grown on glass coverslips using rb-anti-RSV-TM-Cpep (Whole Cell IF). Steady-state surface expression of wild-type and mutant env gene products was detected after incubation of SV40 vector-infected unfixed CV-1 cells on ice with neutralizing chi-anti-RSV PrC antibody followed by ethanol-acetic acid fixation and secondary antibody incubation (Surface IF). Internalization of wild-type and mutant Env proteins from the plasma membrane into prelysosomal vesicles was visualized by incubating Env-expressing cells with chi-anti-RSV PrC antibody in the presence of chloroquine at 37°C for 2 h; the cells were then fixed and stained as for surface IF (Antibody Uptake). The brightly stained vesicles represent endocytic prelysosomal vesicles, most likely late endosomes (29, 39).
The results obtained here thus demonstrate that the mutant glycoproteins, even though palmitoylated to different extents, are transported to the plasma membrane with similar kinetics. However, the steady-state distributions of these proteins are altered due to enhanced internalization from the cell surface, possibly in combination with altered efficiencies of recycling and lysosomal targeting.
Infectivity and reversion of mutant RSV genomes.
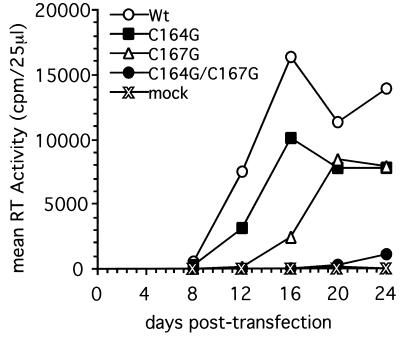
The three mutant genes were engineered into the proviral pATV8R RSV expression vector to assess the effects of the mutations on virus assembly and infectivity. Wild-type and env mutant proviral DNAs were transfected into primary TEC in duplicate. Productive replication of virions was monitored by determining RT activity in supernatants harvested on days 4, 8, 12, 16, 20, and 24 posttransfection. In a representative experiment (Fig. 5), in cell cultures transfected with the wild-type proviral DNA, RT activity could first be detected at 8 days posttransfection, rapidly increasing to a maximal level at day 16. The pATV8R containing the C164G mutation was able to replicate and spread through the culture with kinetics similar to those of the wild-type pATV8R, although the peak of RT activity was reduced by approximately 40%. pATV8R-C167G displayed a similar reduction in peak RT activity, but additionally, the peak of infectivity was delayed by about 4 days compared to that in the wild type, indicating that this virus was much less able to establish infection in the turkey cell cultures. The double mutant pATV8R-C164G/C167G showed no measurable RT activity until day 24 posttransfection, indicating that its ability to replicate was crippled by the mutation of both MSD cysteines. This might be due to the complete loss of palmitoylation per se or more indirectly to conformational changes resulting from the lack of acylation, impairing the stability of the glycoprotein and possibly its functions and/or incorporation into virions.
FIG. 5.
Infectivity of wild-type (Wt) and mutant RSV genomes in avian cells. Primary TEC were transfected in duplicate with the respective pATV8R proviral DNA or mock transfected and passaged over 24 days. Supernatants were harvested at 4-day intervals. Virus spread was assayed by the RT assay as described in Materials and Methods. Mean RT values from a representative experiment are plotted. mock, mock-infected control.
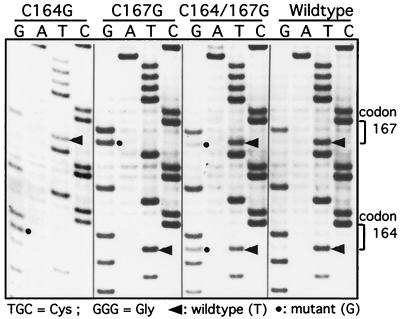
For pATV8R-C164G/C167G, we repeatedly observed a slow increase in RT activity beyond 21 days posttransfection. Thus, replication of the double mutant is much delayed and reduced even when compared to that of the C167G mutant. These observations prompted us to investigate whether the RT activity observed after 20 days resulted from a low level of residual infectivity or whether it corresponded to the appearance of revertant viruses in the culture. Thus, in addition to monitoring of RT activity, virions were pelleted from the culture supernatants on days 12 and 24 for the double mutant or on day 24 for the C164G, C167G, and wild-type viruses. A sequence corresponding to the TM coding region was amplified from the viral RNA genomes by a combination RT-PCR method. The 750-bp PCR products obtained for the wild type and each of the mutants were sequenced directly, as described in Materials and Methods (Fig. 6). The sequences amplified from the wild-type and single-mutant virions at day 24 were identical to the originally transfected wild-type and mutant sequences, and no evidence of subpopulations of revertant sequences was found by this direct sequencing approach (Fig. 6, lanes C164G, C167G, and wild type). In contrast, sequencing of the PCR product from C164G/C167G virions harvested on day 24 (Fig. 6, lane C164G/C167G) resulted in a mixture of mutant and wild-type sequences at codons 164 and 167. Interestingly, the proportion of reversion from G to T (wild type) was higher at codon 167 than at codon 164. In contrast, the sequence of the PCR product amplified from virions released from the double-mutant-infected cells at day 12 still retained both T-to-G mutations and showed no evidence of reversion (data not shown). Sequencing of the entire PCR product provides qualitative information only on the population of DNA molecules present at that time point. To further delineate the genotypes of the viruses present at days 12 and 24 in the originally double-mutant culture, and at day 24 in the single-mutant cultures, the PCR-amplified sequences were subcloned into pTZ18R. Twelve clones were isolated and sequenced for each time point. Again, we obtained no evidence of reversion during infections resulting from transfections with singly mutated proviruses (i.e., 12 of 12 clones displayed only the originally mutant sequence [data not shown]). The results for the double mutant pATV8R-C164G/C167G are summarized in Table 1. Twelve days after transfection, 12 out of 12 clones retained the double-mutant genotype, whereas at day 24, three distinct genotypes could be found: wild-type (4 of 11), C164G/C167wt (3 of 11), and C164G/C167G (4 of 11). As might be anticipated from the delayed infection kinetics of pATV8R-C167G (Fig. 5), no revertant clones corresponding to a C164wt/C167G genotype were observed in this limited analysis. These results indicate that at 24 days posttransfection a significant proportion of the replicating virus present in the double-mutant-transfected culture did revert to a more favorable, i.e., faster-replicating genotype. Furthermore, a more extensive sequence analysis of the entire TM coding region of each of the mutants revealed no additional nucleotide changes other than those observed at day 24 in the double mutant (not shown).
FIG. 6.
Sequence analysis of RT-PCR products from mutant and wild-type virions at day 24. Cells were transfected and cultured as for Fig. 5. Culture medium was harvested on day 24 posttransfection, virions were pelleted, and the extracted RNA was subjected to RT-PCR amplification. The resulting product was excised from a gel and sequenced directly. Wild-type T residues are denoted by the arrowheads, and mutant G residues are denoted by the circles.
TABLE 1.
Results of sequencing of double mutanta
| Days posttransfection with pATV8R-C164G/C167G | Genotype of PCR clones | Prevalence of genotypeb |
|---|---|---|
| 12 | C164G/C167G | 12/12 |
| 24 | C164G/C167G | 4/11 |
| C164G/C167wt | 3/11 | |
| C164wt/C167G | 0/11 | |
| C164wt/C167wt | 4/11 |
Virion RNA was extracted from virus pelleted from cell culture supernatant and reverse transcribed using a negative-strand primer located just 3′ of the env gene. PCR amplification was performed using a second (plus-strand) primer located just 5′ of the SU-TM boundary. The PCR product was cleaved with EcoRI and XbaI and cloned into the plasmid ptZ18R. Twelve clones were randomly selected from both the day 12 and day 24 time points for double-strand sequencing.
Number with genotype/number sequenced.
Incorporation of mutant glycoproteins into RSV virions.
In order to determine the nature of the replication defect in the mutant RSV virions, we examined the protein composition of virions released from avian cells transfected with the mutant proviral genomes. Performing these studies in TEC, for which transient transfection is quite inefficient, is complicated by the undetectable levels of infectivity of the double mutant. Thus, harvesting virions for analysis at the peak of viral replication proved difficult. We therefore decided to use the recently established chicken fibroblast cell line Df-1 (22, 48) to address this question.
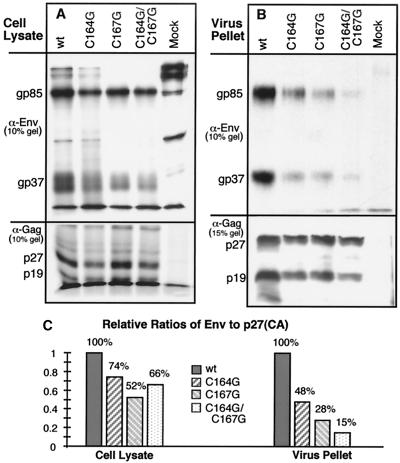
Df-1 cells were transfected with wild-type and mutant proviral pATV8R DNAs and pulse-labeled with [35S]methionine 44 h posttransfection. This was followed by a 6-h chase in complete medium. At the end of the chase period, the cells were lysed, the supernatants were harvested and cleared, and the virions were pelleted through 25% (wt/vol) sucrose and lysed in detergent buffer as described in Materials and Methods. All lysates were subjected to anti-Env immunoprecipitation (using rb-anti-RSV-TM-Cpep), followed by anti-Gag immunoprecipitation. Samples were analyzed by SDS-PAGE, autoradiography, and OptiQuant quantitation (Fig. 7). In the cell lysates, the Gag expression levels of the wild-type and mutant proviruses were comparable. The amounts of intracellular Env expression relative to that of p27 (CA) after a pulse and 6-h chase were slightly reduced for the mutants (Fig. 7A and C), as could be expected from the results shown in Fig. 3. In contrast, when the relative amounts of Env and Gag in virions were analyzed, significant differences between the wild type and the mutants became apparent, as is clearly visible in the autoradiographs in Fig. 7B. When adjusted to the wild-type ratio, the single mutants C164G and C167G and the double mutant C164G/C167G showed a progressive reduction of glycoprotein incorporated into virions which, in the experiment shown, amounted to 48, 28, and 15%, respectively, of wild-type levels (Fig. 7C). These results are in accordance with observations obtained from experiments with virions harvested from TEC 20 days after transfection with the respective proviruses (data not shown).
FIG. 7.
Incorporation of wild-type and mutant glycoproteins into virions released from avian cells. The stable chicken fibroblast cell line Df-1 was transfected with wild-type (wt) and mutant pATV8R DNAs or mock transfected; 44 h posttransfection, the cells were pulse-labeled with [35S]methionine and chased for 6 h in complete medium. Virions were pelleted from the medium as described in Materials and Methods. Lysates of viral pellets and of cells were subjected to consecutive immunoprecipitation with anti-Env and anti-Gag sera. Viral proteins from cell lysates were separated on SDS–10% PAGE gels (A), as was virion-associated Env (B). Virus-associated Gag was visualized on an SDS–15% PAGE gel (B). (C) Amounts of p27 (CA), SU, and TM were quantitated, and relative ratios, adjusted to wild type, were plotted.
DISCUSSION
In this study we demonstrate that cysteines 164 and 167 within the membrane anchor region of the TM protein of RSV are the sites of palmitic acid addition to this viral glycoprotein. We further observed that mutation of these sites to glycine, either alone or in combination, reduced the stability and steady-state plasma membrane expression of the glycoprotein to various extents. The different efficiencies with which glycoprotein was found to be incorporated into virus particles reflected the levels of surface expression and the ability of the mutant glycoprotein to confer infectivity upon RSV virions.
Previous studies by Gebhardt et al. (16) and Wills et al. (60) had demonstrated that the gp37 (TM) protein of RSV was palmitoylated. The TM MSD had been predicted to encompass amino acids 150 to 176 based on the hydrophobicity of this region, and the predominance of serines (located at positions −4, −3, −2, +6, and +7 relative to the putative inner membrane boundary) within the carboxy terminus of the TM protein prompted the former authors to predict that the fatty acid might be added to a serine residue within this region. Two cysteines are present within the putative MSD, and Rose et al. and Magee et al. (34, 47) showed that same year that VSV G protein was palmitoylated at a cytoplasmic cysteine close to the membrane boundary; this palmitic acid is linked through a thiol ester (33, 47). For RSV Env, however, it was hypothesized that the two cysteines in the MSD might be located too distant from the suggested membrane-cytoplasm interface to be modified by palmitate (49). Moreover, the proposed serine modification was supported by the work of Jing and Trowbridge (28), who observed that in avian cells palmitic acid could be added to serine residues in the context of a mutated human transferrin receptor. On the other hand, in most cellular and viral transmembrane proteins studied to date, palmitic acid is linked through a thioester bond to cysteine residues. These are usually located in the cytoplasmic tail close to the membrane boundary or within the membrane anchor region itself (19, 21, 51, 58, 61) (for a review, see reference 46).
As with the VSV G protein (33, 34, 47), in the Semliki Forest virus E1 protein palmitate is added to a cysteine on the cytoplasmic face of the lipid bilayer (50), whereas in rabies virus G protein the palmitoylated cysteines are located within or at the border of the membrane anchor domain (15). In the HA proteins (H2, H3, and H7) of different influenza viruses, three cysteines located in both the membrane-spanning and cytoplasmic domains represent addition sites for palmitic acid (37, 55, 57). Both Friend and Moloney MuLV Env proteins are palmitoylated at cysteines either within or adjacent to the MSD (21, 61).
Even though the existing model for the MSD of RSV TM suggested that the only two cysteines (C164 and C167) in this region were located too distant from the proposed membrane-cytoplasm interface to be acylated (49), we demonstrate here that both cysteines are indeed accessible sites for palmitate addition. Interestingly, the more C-terminal and thus more membrane boundary-proximal cysteine (C167) appears to be not only the preferred acylation site but also more crucial for glycoprotein function .
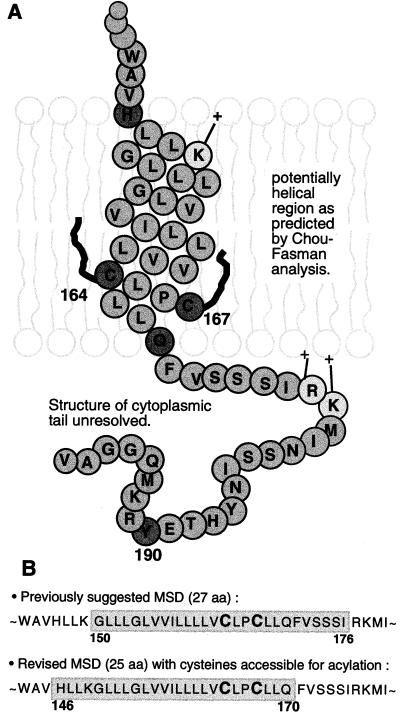
The efficient and apparently necessary palmitoylation of the cysteine residues raises the possibility that the MSD is shorter than the 27 uncharged residues between lysine 149 (K149) and arginine 177 (R177). It is possible, as has been suggested for other proteins (for a review, see references 46 and 51), that the palmitate groups insert into the bilayer and constrain the conformation of the polypeptide such that the cytoplasmic domain is held close to the membrane. Based on this possibility, a revised model for the MSD of the RSV TM protein is shown in Fig. 8. While this model predicts that only 24 (H146 to Q170) amino acids will span the membrane and act as the membrane anchor, it is supported by our previous observation (9) that a mutant TM with leucine 165 (L165) replaced by an arginine (L165R) remained firmly anchored in the membrane. Moreover, deletions within the MSDs of both the RSV Env and VSV G proteins (1, 24) and studies with synthetic anchor domains (9) have demonstrated that 14 to 16 amino acids are sufficient to act as anchor sequences. If this shorter and slightly N-terminally shifted domain does indeed act as a membrane anchor in the molecule, then our previous cytoplasmic truncation of 22 amino acids (42) would leave a residual cytoplasmic domain of 6 amino acids that could continue to interact, directly or indirectly, with capsid proteins during assembly. In our revised MSD model, the charged side chains of K149, and possibly R177 and K178, could be embedded within the polar head groups of the outer and inner layers of the membrane, respectively. Indeed, when we performed a Chou-Fasman-Rose analysis, it predicted an alpha-helical structure for a peptide corresponding to H146 to Q170, undisturbed by P166, supporting our model.
FIG. 8.
Revised schematic organization of the RSV gp37 (TM) MSD. Palmitoyl groups are shown inserted into the hydrophobic environment of the bilayer, while the amine groups of the histidine and glutamine residues are located in the polar head group region. The long positively charged side chain of the arginine allows this region of the peptide backbone to be buried in the hydrophobic region of the lipid bilayer. The leucine (residue 165) that in mutant μ26 (9) is converted to an arginine is in this model located close to the inner membrane-cytoplasm boundary. This would allow insertion of the charged residue without significantly interfering with the membrane anchor properties of the region.
No consensus sequence for palmitate addition has been described, although in some of the ras-related proteins it has been suggested that a cysteine followed by three residues of which the first two are aliphatic might act as such a signal (34). However, neither of the two target cysteines in the RSV Env protein strictly follows this rule. Moreover, a deletion mutation within the MSD of this protein indicated that a cysteine residue within one amino acid of the C terminus could act as an acceptor for palmitoylation. This mutation, which eliminated both C164 and C167, resulted in a reading frame shift and introduced a new cysteine as the penultimate amino acid (residue 169) which was efficiently palmitoylated (G. L. Davis and E. Hunter, unpublished data). These results, as well as those of other investigators (19, 21, 50, 58, 61), support the suggestion of Schmidt (51) and Resh (46) that any cysteine within a few residues of the membrane-cytoplasm junction can be the target of palmitoylation. Nevertheless, Ponimaskin and Schmidt have recently emphasized the role of the conformational context for the palmitoylation of cysteines introduced into the Sendai virus F protein (45).
The exact function of palmitic acid addition remains unresolved. It is interesting that for the human transferrin receptor, removal of palmitoylation sites resulted in the receptor being endocytosed with faster kinetics and higher levels of iron accumulated in mutant receptor-expressing cells (2, 28). The mutation of C167 of the RSV TM to a glycine residue also resulted in a higher rate of endocytosis and degradation of the mature protein complex in lysosomes (this study and reference 39). This was even more pronounced in cells expressing the double mutant. A lack of palmitoylation may thus destabilize the molecule or alter its association with the membrane sufficiently for entry into the endocytic pathway to be triggered. Previously, Collawn and colleagues (8) identified a cytoplasmic tyrosine residue in the transferrin receptor as critical for endocytic signaling. The structural context of this tyrosine (YXRF), a tight turn in the polypeptide chain, is similar to that of a similar signal (YNPX) in the low-density lipoprotein receptor (35, 56). A tyrosine in a similar context (YRKM) is located at residue 190 of the RSV PrC TM cytoplasmic domain, and as we have recently described (39), this Y190 can act as part of an efficient endocytosis motif in RSV Env. Interestingly, both the single and double mutant containing the C167G mutation display a phenotype reminiscent of the Env mutant L165R, insofar as all three mutants show rapid internalization of Env from the cell surface into the lysosomal pathway (Fig. 3B and 4) (29, 39). We have previously hypothesized that the mutations L165R and C167G are both likely to induce conformational changes in the MSD and tail domain that result in an activation or increased accessibility of the Y190RKM motif to the endocytic machinery, leading to the observed increase in endocytosis rates (RSV PrC Env wild type, 1.1%/min; L165R, 7.4%/min; C167G, 4.7%/min [39]). Since the mutation L165R does not abrogate palmitoylation (our unpublished observation), the phenotypes observed for Env-C167G and Env-C164G/C167G might not be due to the absence of palmitoylation at residue 167 per se but rather to a resulting overall conformational change within TM.
Site-directed mutagenesis studies of other viral glycoproteins have suggested that palmitoylation is not necessarily required for intracellular transport or biological activity. This was found to be the case for influenza virus HA (which contains three highly conserved cysteines which are modified by palmitate) of either the H7 or H3 subtype (55, 58) and of the H2 subtype in a study by Naim et al. (38). However, in earlier work on the H2-subtype HA, the ability to induce cell fusion was reported to be lost following mutagenesis of either of the two cytoplasmic cysteines (37). Additionally, Brewer and Roth reported that mutating C560 in HA H2 changed the polarized transport of HA from the apical to the basolateral surface in MDCK cells (5). In studies of Sindbis virus, mutations of cysteines in the E2 cytoplasmic domain and the 6K region of the glycoprotein precursor resulted in virus assembly defects (13, 14). For VSV G, different investigators obtained partially contradictory results regarding the effects of abrogating palmitate addition on glycoprotein function (32–34, 47, 59). In a recent study of MuLV Env by Yang and Compans (61), transport, processing, surface expression, and cell fusion were not altered by abrogating Env palmitoylation. However, the authors did not investigate potential effects on viral replication.
Our studies of the role of palmitoylation in subgroup C RSV Env, in both mammalian and avian cells, reveal a slightly different and more consistent picture. Since either cysteine can be mutated and residual palmitoylation is observed, this argues that both transmembrane cysteines can be palmitoylated. Palmitoylation appeared to be unnecessary for synthesis or intracellular transport of the RSV Env precursor protein. Mutation of C164 to glycine had only moderate effects on the half-life and steady-state distribution of Env. In contrast, glycoproteins containing the C167G or both mutations were progressively less stable and were internalized significantly faster than the wild type (reference 39 and this work). Even though incorporation of Env-C164G into virions was reduced approximately 50%, viral infection kinetics were not significantly affected; only peak RT values were reduced. In contrast, single- or double-mutant glycoproteins containing the C167G mutation, which leads to a further reduction in incorporation of protein into virions, resulted in a significant reduction in the ability of env mutant viruses to propagate in avian cells. The fact that this defective replication phenotype was far more pronounced when both palmitoylation sites were mutated suggests that palmitoylation of C164 does play some role in Env function. This is supported by the infectivity studies of the double mutant in which reversion to an infectious phenotype involved conversion of both glycine residues to cysteines.
The progressive reduction in glycoprotein incorporation into virions is mirrored by the defects observed in virus infectivity. However, this is not a simple linear correlation. It is conceivable that a threshold level of incorporated Env might be necessary for replication with wild-type-like kinetics, since even though Env-C164G incorporation is reduced twofold, the replication kinetics of this mutant are only moderately affected. Thus, in addition to further reducing the amounts of glycoprotein on virions, Env bearing C167G or the double mutation might also be functionally impaired. The availability of Env molecules for incorporation into virions could be reduced by the shorter surface residence time of the mutant Env-C167G and Env-C164G/C167G glycoproteins; however, these mutant proteins are transiently expressed at significant levels on the cell surface, as was shown by immunofluorescent antibody labeling experiments. On the other hand, it is possible that for Env-C167G and Env-C164G/C167G an enhanced association with the endocytic machinery directly prevents entry of the mutant glycoproteins into the assembly site and thus blocks incorporation into virus.
Interestingly, when virus replication was first detected in cultures transfected with the defective pATV8R-C164G/C167G genome (20-plus days), the virus pool comprised a mixture of mutant and revertant viruses in which either one (codon 167) or both mutant codons had reverted to cysteine codons. This is in contrast to the single mutants, for which no revertants were observed either by bulk sequencing or by cloning of RT-PCR products. It is likely that at the input multiplicities used, there was insufficient selective pressure for revertant virus to arise in the cultures infected by single-mutant virus. In contrast, because the double mutant is so highly attenuated for infectivity, selective pressures would be much greater, and so reverting mutations that facilitate replication would be selected for. It is likely that reversion of the C164G mutation would provide a replicative advantage to the virus. However, because the C167G single mutant produced would be present at such a low multiplicity, there would be a much greater selective pressure exerted on it to also revert before it could be detected in the culture. In contrast, reversion of the C167G mutation in the double mutant would yield C164G, which replicates almost as well as the wild type, thereby explaining why a mixture of C164G and wild-type revertants are seen at day 24 p.i. for the double mutant. In the limited number of PCR clones amplified from virions at 24 days posttransfection, we obtained no evidence of second-site reversion events, where for example other codons had mutated to cysteines, indicating that the positions of the palmitoylated residues are important for biological function.
The results presented here indicate an important role for cysteine palmitoylation of the subgroup C RSV envelope protein for glycoprotein function and virus infectivity. The amino acid sequences of TM proteins from different avian leukosis and sarcoma virus subgroups are quite conserved. However, they display an interesting divergence in the cytoplasmic tail just C terminal of the MSD. It will be interesting to analyze the role of palmitoylation for Env proteins from other RSV subtypes and to determine how far the context of sequence and conformation influences the phenotypes of various mutants.
ACKNOWLEDGMENTS
We are very grateful to Susan R. Dubay for her valuable experimental advice, and we thank Tshana Thomas for excellent technical assistance.
This work was supported by grant CA-29884 from the National Institutes of Health and by the facilities of the DNA sequence core of the Center for AIDS Research, Program Grant P30-AI-27767, from the National Institutes of Health. David Miller was supported on Training Grant CA-09467.
REFERENCES
- 1.Adams G A, Rose J K. Structural requirements of a membrane-spanning domain for protein anchoring and cell surface transport. Cell. 1985;41:1007–1015. doi: 10.1016/s0092-8674(85)80081-7. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez E, Girones N, Davis R J. Inhibition of the receptor-mediated endocytosis of diferric transferrin is associated with the covalent modification of the transferrin receptor with palmitic acid. J Biol Chem. 1990;265:16644–16655. [PubMed] [Google Scholar]
- 3.Bonner W M, Laskey R A. A film detection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974;46:83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- 4.Bosch J V, Schwarz R T. Processing of gPr92env, the precursor to the glycoproteins of Rous sarcoma virus: use of inhibitors of oligosaccharide trimming and glycoprotein transport. Virology. 1984;132:95–109. doi: 10.1016/0042-6822(84)90094-1. [DOI] [PubMed] [Google Scholar]
- 5.Brewer C B, Roth M G. A single amino acid change in the cytoplasmic domain alters the polarized delivery of influenza virus hemagglutinin. J Cell Biol. 1991;114:413–421. doi: 10.1083/jcb.114.3.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown D A, Rose J K. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell. 1992;68:533–544. doi: 10.1016/0092-8674(92)90189-j. [DOI] [PubMed] [Google Scholar]
- 7.Caballero M, Carabana J, Ortego J, Fernandez-Munoz R, Celma M L. Measles virus fusion protein is palmitoylated on transmembrane-intracytoplasmic cysteine residues which participate in cell fusion. J Virol. 1998;72:8198–8204. doi: 10.1128/jvi.72.10.8198-8204.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collawn J F, Stangel M, Kuhn L A, Esekogwu V, Jing S Q, Trowbridge I S, Tainer J A. Transferrin receptor internalization sequence YXRF implicates a tight turn as the structural recognition motif for endocytosis. Cell. 1990;63:1061–1072. doi: 10.1016/0092-8674(90)90509-d. [DOI] [PubMed] [Google Scholar]
- 9.Davis G L, Hunter E. A charged amino acid substitution within the transmembrane anchor of the Rous sarcoma virus envelope glycoprotein affects surface expression but not intracellular transport. J Cell Biol. 1987;105:1191–1203. doi: 10.1083/jcb.105.3.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dubay J W, Roberts S J, Brody B, Hunter E. Mutations in the leucine zipper of the human immunodeficiency virus type 1 transmembrane glycoprotein affect fusion and infectivity. J Virol. 1992;66:4748–4756. doi: 10.1128/jvi.66.8.4748-4756.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Einfeld D, Hunter E. Oligomeric structure of a prototype retrovirus glycoprotein. Proc Natl Acad Sci USA. 1988;85:8688–8692. doi: 10.1073/pnas.85.22.8688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.England J M, Bolognesi D P, Dietzschold B, Halpern M S. Evidence that a precursor glycoprotein is cleaved to yield the major glycoprotein of avian tumor virus. J Virol. 1977;21:810–814. doi: 10.1128/jvi.21.2.810-814.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaedigk-Nitschko K, Ding M X, Levy M A, Schlesinger M J. Site-directed mutations in the Sindbis virus 6K protein reveal sites for fatty acylation and the underacylated protein affects virus release and virion structure. Virology. 1990;175:282–291. doi: 10.1016/0042-6822(90)90210-i. [DOI] [PubMed] [Google Scholar]
- 14.Gaedigk-Nitschko K, Schlesinger M J. Site-directed mutations in Sindbis virus E2 glycoprotein's cytoplasmic domain and the 6K protein lead to similar defects in virus assembly and budding. Virology. 1991;183:206–214. doi: 10.1016/0042-6822(91)90133-v. [DOI] [PubMed] [Google Scholar]
- 15.Gaudin Y, Tuffereau C, Benmansour A, Flamand A. Fatty acylation of rabies virus proteins. Virology. 1991;184:441–444. doi: 10.1016/0042-6822(91)90866-a. [DOI] [PubMed] [Google Scholar]
- 16.Gebhardt A, Bosch J V, Ziemiecki A, Friis R R. Rous sarcoma virus p19 and gp35 can be chemically crosslinked to high molecular weight complexes. An insight into virus assembly. J Mol Biol. 1984;174:297–317. doi: 10.1016/0022-2836(84)90340-1. [DOI] [PubMed] [Google Scholar]
- 17.Gilman M. Preparation of RNA from eukaryotic and prokaryotic cells. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current Protocols in Molecular Biology. Vol. 1. Boston, Mass: John Wiley & Sons, Inc.; 1995. pp. 4.1.1–4.1.6. [Google Scholar]
- 18.Harder T, Scheiffele P, Verkade P, Simons K. Lipid domain structure of the plasma membrane revealed by patching of membrane components. J Cell Biol. 1998;141:929–942. doi: 10.1083/jcb.141.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hausmann J, Ortmann D, Witt E, Veit M, Seidel W. Adenovirus death protein, a transmembrane protein encoded in the E3 region, is palmitoylated at the cytoplasmic tail. Virology. 1998;244:343–351. doi: 10.1006/viro.1998.9135. [DOI] [PubMed] [Google Scholar]
- 20.Hayman M. Synthesis and processing of avian sarcoma virus glycoproteins. Virology. 1978;85:475–486. doi: 10.1016/0042-6822(78)90454-3. [DOI] [PubMed] [Google Scholar]
- 21.Hensel J, Hintz M, Karas M, Linder D, Stahl B, Geyer R. Localization of the palmitoylation site in the transmembrane protein p12E of Friend murine leukaemia virus. Eur J Biochem. 1995;232:373–380. doi: 10.1111/j.1432-1033.1995.373zz.x. [DOI] [PubMed] [Google Scholar]
- 22.Himly M, Foster D N, Bottoli I, Iacovoni J S, Vogt P K. The DF-1 chicken fibroblast cell line: transformation induced by diverse oncogenes and cell death resulting from infection by avian leukosis viruses. Virology. 1998;248:295–304. doi: 10.1006/viro.1998.9290. [DOI] [PubMed] [Google Scholar]
- 23.Hunter E. Biological techniques for avian sarcoma viruses. Methods Enzymol. 1979;58:379–393. doi: 10.1016/s0076-6879(79)58153-1. [DOI] [PubMed] [Google Scholar]
- 24.Hunter E. Membrane insertion and transport of viral glycoproteins: a mutational analysis. In: Das R C, Robbins P W, editors. Protein transfer and organelle biogenesis. New York, N.Y: Academic Press; 1988. pp. 109–158. [Google Scholar]
- 25.Hunter E, Hill E, Hardwick M, Bhown A, Schwartz D E, Tizard R. Complete sequence of the Rous sarcoma virus env gene: identification of structural and functional regions of its product. J Virol. 1983;46:920–936. doi: 10.1128/jvi.46.3.920-936.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hunter E, Swanstrom R. Retrovirus envelope glycoproteins. Curr Top Microbiol Immunol. 1990;157:187–253. doi: 10.1007/978-3-642-75218-6_7. [DOI] [PubMed] [Google Scholar]
- 27.Ivanova L, Schlesinger M J. Site-directed mutations in the Sindbis virus E2 glycoprotein identify palmitoylation sites and affect virus budding. J Virol. 1993;67:2546–2551. doi: 10.1128/jvi.67.5.2546-2551.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jing S Q, Trowbridge I S. Nonacylated human transferrin receptors are rapidly internalized and mediate iron uptake. J Biol Chem. 1990;265:11555–11559. [PubMed] [Google Scholar]
- 29.Johnston P B, Dong J Y, Hunter E. Transport of a lysosomally targeted Rous sarcoma virus envelope glycoprotein involves transient expression on the cell surface. Virology. 1995;206:353–361. doi: 10.1016/s0042-6822(95)80050-6. [DOI] [PubMed] [Google Scholar]
- 30.Kawai S, Nishizawa M. New procedure for DNA transfection with polycation and dimethyl sulfoxide. Mol Cell Biol. 1984;4:1172–1174. doi: 10.1128/mcb.4.6.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keulen W, Schuurman R. Modified protocol for DNA sequence analysis using sequenase® 2.0. BioTechniques. 1991;10:185–188. [PubMed] [Google Scholar]
- 32.Kotwal G J, Ghosh H P. Role of fatty acid acylation of membrane glycoproteins. Absence of palmitic acid in glycoproteins of two serotypes of vesicular stomatitis virus. J Biol Chem. 1984;259:4699–4701. [PubMed] [Google Scholar]
- 33.Mack D, Kruppa J. Fatty acid acylation at the single cysteine residue in the cytoplasmic domain of the glycoprotein of vesicular-stomatitis virus. Biochem J. 1988;256:1021–1027. doi: 10.1042/bj2561021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Magee A I, Koyama A H, Malfer C, Wen D, Schlesinger M J. Release of fatty acids from virus glycoproteins by hydroxylamine. Biochim Biophys Acta. 1984;798:156–166. doi: 10.1016/0304-4165(84)90298-8. [DOI] [PubMed] [Google Scholar]
- 35.Matter K, Hunziker W, Mellman I. Basolateral sorting of LDL receptor in MDCK cells: the cytoplasmic domain contains two tyrosine-dependent targeting determinants. Cell. 1992;71:741–753. doi: 10.1016/0092-8674(92)90551-m. [DOI] [PubMed] [Google Scholar]
- 36.Melkonian K A, Ostermeyer A G, Chen J Z, Roth M G, Brown D A. Role of lipid modifications in targeting proteins to detergent-resistant membrane rafts. Many raft proteins are acylated, while few are prenylated. J Biol Chem. 1999;274:3910–3917. doi: 10.1074/jbc.274.6.3910. [DOI] [PubMed] [Google Scholar]
- 37.Naeve C W, Williams D. Fatty acids on the A/Japan/305/57 influenza virus hemagglutinin have a role in membrane fusion. EMBO J. 1990;9:3857–3866. doi: 10.1002/j.1460-2075.1990.tb07604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naim H Y, Amarneh B, Ktistakis N T, Roth M G. Effects of altering palmitoylation sites on biosynthesis and function of the influenza virus hemagglutinin. J Virol. 1992;66:7585–7588. doi: 10.1128/jvi.66.12.7585-7588.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ochsenbauer C, Dubay S R, Hunter E. The Rous sarcoma virus Env glycoprotein contains a highly conserved motif homologous to tyrosine-based endocytosis signals and displays an unusual internalization phenotype. Mol Cell Biol. 2000;20:249–260. doi: 10.1128/mcb.20.1.249-260.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palade G. Intracellular aspects of the process of protein synthesis. Science. 1975;189:347–358. doi: 10.1126/science.1096303. [DOI] [PubMed] [Google Scholar]
- 41.Patarca R, Haseltine W A. Similarities among retrovirus proteins. Nature. 1984;312:496. doi: 10.1038/312496a0. [DOI] [PubMed] [Google Scholar]
- 42.Perez L G, Davis G L, Hunter E. Mutants of the Rous sarcoma virus envelope glycoprotein that lack the transmembrane anchor and cytoplasmic domains: analysis of intracellular transport and assembly into virions. J Virol. 1987;61:2981–2988. doi: 10.1128/jvi.61.10.2981-2988.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perez L G, Hunter E. Mutations within the proteolytic cleavage site of the Rous sarcoma virus glycoprotein that block processing to gp85 and gp37. J Virol. 1987;61:1609–1614. doi: 10.1128/jvi.61.5.1609-1614.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Philipp H C, Schroth B, Veit M, Krumbiegel M, Herrmann A, Schmidt M F. Assessment of fusogenic properties of influenza virus hemagglutinin deacylated by site-directed mutagenesis and hydroxylamine treatment. Virology. 1995;210:20–28. doi: 10.1006/viro.1995.1313. [DOI] [PubMed] [Google Scholar]
- 45.Ponimaskin E, Schmidt M F. Domain-structure of cytoplasmic border region is main determinant for palmitoylation of influenza virus hemagglutinin (H7) Virology. 1998;249:325–335. doi: 10.1006/viro.1998.9303. [DOI] [PubMed] [Google Scholar]
- 46.Resh M D. Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim Biophys Acta. 1999;1451:1–16. doi: 10.1016/s0167-4889(99)00075-0. [DOI] [PubMed] [Google Scholar]
- 47.Rose J K, Adams G A, Gallione C J. The presence of cysteine in the cytoplasmic domain of the vesicular stomatitis virus glycoprotein is required for palmitate addition. Proc Natl Acad Sci USA. 1984;81:2050–2054. doi: 10.1073/pnas.81.7.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schaefer-Klein J, Givol I, Barsov E V, Whitcomb J M, VanBrocklin M, Foster D N, Federspiel M J, Hughes S H. The EV-O-derived cell line DF-1 supports the efficient replication of avian leukosis-sarcoma viruses and vectors. Virology. 1998;248:305–311. doi: 10.1006/viro.1998.9291. [DOI] [PubMed] [Google Scholar]
- 49.Schlesinger M J, Veit M, Schmidt M F G. Palmitoylation of cellular and viral proteins. In: Schlesinger M J, editor. Lipid modifications of proteins. Boca Raton, Fla: CRC Press; 1993. pp. 1–19. [Google Scholar]
- 50.Schmidt M, Schmidt M F, Rott R. Chemical identification of cysteine as palmitoylation site in a transmembrane protein (Semliki Forest virus E1) J Biol Chem. 1988;263:18635–18639. [PubMed] [Google Scholar]
- 51.Schmidt M F. Fatty acylation of proteins. Biochim Biophys Acta. 1989;988:411–426. doi: 10.1016/0304-4157(89)90013-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schwarz D, Tizard R, Gilbert W. Nucleotide sequence of Rous sarcoma virus. Cell. 1983;32:853–869. doi: 10.1016/0092-8674(83)90071-5. [DOI] [PubMed] [Google Scholar]
- 53.Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 54.Srinivas R V, Compans R W. Membrane association and defective transport of spleen focus-forming virus glycoproteins. J Biol Chem. 1983;258:14718–14724. [PubMed] [Google Scholar]
- 55.Steinhauer D A, Wharton S A, Wiley D C, Skehel J J. Deacylation of the hemagglutinin of influenza A/Aichi/2/68 has no effect on membrane fusion properties. Virology. 1991;184:445–448. doi: 10.1016/0042-6822(91)90867-b. [DOI] [PubMed] [Google Scholar]
- 56.Trowbridge I S, Collawn J F. Structural requirements for high efficiency endocytosis of the human transferrin receptor. J Inorg Biochem. 1992;47:209–217. doi: 10.1016/0162-0134(92)84066-v. [DOI] [PubMed] [Google Scholar]
- 57.Veit M, Klenk H D, Kendal A, Rott R. The M2 protein of influenza A virus is acylated. J Gen Virol. 1991;72:1461–1465. doi: 10.1099/0022-1317-72-6-1461. [DOI] [PubMed] [Google Scholar]
- 58.Veit M, Schmidt M F, Rott R. Different palmitoylation of paramyxovirus glycoproteins. Virology. 1989;168:173–176. doi: 10.1016/0042-6822(89)90417-0. [DOI] [PubMed] [Google Scholar]
- 59.Whitt M A, Rose J K. Fatty acid acylation is not required for membrane fusion activity or glycoprotein assembly into VSV virions. Virology. 1991;185:875–878. doi: 10.1016/0042-6822(91)90563-q. [DOI] [PubMed] [Google Scholar]
- 60.Wills J W, Srinivas R V, Hunter E. Mutations of the Rous sarcoma virus env gene that affect the transport and subcellular location of the glycoprotein products. J Cell Biol. 1984;99:2011–2023. doi: 10.1083/jcb.99.6.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang C, Compans R W. Palmitoylation of the murine leukemia virus envelope glycoprotein transmembrane subunits. Virology. 1996;221:87–97. doi: 10.1006/viro.1996.0355. [DOI] [PubMed] [Google Scholar]
- 62.Yang C, Spies C P, Compans R W. The human and simian immunodeficiency virus envelope glycoprotein transmembrane subunits are palmitoylated. Proc Natl Acad Sci USA. 1995;92:9871–9875. doi: 10.1073/pnas.92.21.9871. [DOI] [PMC free article] [PubMed] [Google Scholar]