Abstract
Infection by hepatitis B virus (HBV) is mainly restricted to humans. This species specificity is likely determined at the early phase of the viral life cycle. Since the envelope proteins are the first viral factors to interact with the cell, they represent attractive candidates for controlling the HBV host range. To investigate this assumption, we took advantage of the recent discovery of a second virus belonging to the primate Orthohepadnavirus genus, the woolly monkey HBV (WMHBV). A recombinant plasmid was constructed for the expression of all WMHBV envelope proteins. In additional constructs, N-terminal sequences of the WMHBV large envelope protein were substituted for their homologous HBV counterparts. All wild-type and chimeric WMHBV surface proteins were properly synthesized by transfected human hepatoma cells, and they were competent to replace the original HBV proteins for the production of complete viral particles. The resulting pseudotyped virions were evaluated for their infectious capacity on human hepatocytes in primary culture. Virions pseudotyped with wild-type WMHBV envelope proteins showed a significant loss of infectivity. By contrast, infectivity was completely restored when the first 30 residues of the large protein originated from HBV. Analysis of smaller substitutions within this domain limited the most important region to a stretch of only nine amino acids. Reciprocally, replacement of this motif by WMHBV residues in the context of the HBV L protein significantly reduced infectivity of HBV. Hence this short region of the L protein contributes to the host range of HBV.
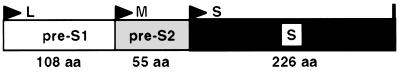
While many molecular mechanisms governing the replication of human hepatitis B virus (HBV) have been elucidated in detail (28, 29), the way in which this hepatotropic virus infects host cells still remains unclear. The surface proteins present on enveloped viruses, such as HBV, play a critical role in the initial events of viral infection. They mediate virion attachment to the cells and allow fusion between the viral envelope and cellular membranes. These early steps in the life cycle of a virus lead to the introduction of its genome into target cells and permit the initiation of viral replication. For HBV, three different types of surface glycoproteins, embedded in the viral envelope, are potential candidates to initiate viral infection. These proteins are encoded by a single open reading frame (ORF) within the HBV genome (Fig. 1). This ORF contains three in-frame initiator codons and one stop codon. Consequently, the three translated proteins share an identical C-terminal region of 226 amino acids (aa), namely, the S domain. Initiation at the third start site leads to the synthesis of the small (S) protein, containing solely the S domain. When the intermediate initiator codon is used, the middle (M) protein is translated. Compared to the S protein, it contains an additional N-terminal domain of 55 aa, known as pre-S2. Finally, utilization of the most 5′-proximal start site generates the large (L) protein, which includes the pre-S1 domain (108 aa in the ayw subtype) as well as the pre-S2 and S domains. Like the majority of polypeptides synthesized in a cell, the HBV envelope proteins are subjected to posttranslational modifications. Thus, approximately one-half of the L, M, and S protein populations are modified by the attachment of a carbohydrate group to the Asn 146 of their S domain. The M protein undergoes additional N- and O-linked glycosylations on aa 4 and 37 of its pre-S2 region, respectively. Unlike the O-linked oligosaccharides, this N-linked chain is always added to the M protein (38). Finally, the N terminus of the L protein is altered by a covalent linkage of a myristate group (33). These diverse alterations could contribute to the biological activity of the proteins. For example, the acylation has been reported to be required for the in vitro infectivity of HBV (4, 15).
FIG. 1.
Organization of the HBV gene coding for the envelope proteins. The L, M, and S proteins are encoded by a single ORF. Triangles and vertical bar, positions of the three different in-frame initiation sites and the common stop codon, respectively, which divide the gene into the pre-S1, pre-S2, and S domains (boxes). Sizes in amino acids of the regions are specified at the bottom.
One of the salient characteristics of HBV is its narrow host range. The virus efficiently infects only humans, and closely related primates, such as the chimpanzee (36). As suggested by experimental data, the initial steps of infection play a decisive role in the species specificity of HBV. Indeed, hepatocytes from refractory species are able to support viral replication only if the normal pathway of viral entry is artificially bypassed. Thus, it has been shown that primary rat hepatocytes or a rat hepatoma cell line could support HBV replication upon transient transfection of cloned HBV DNA (8, 39). Complete virions are also produced in hepatocytes of HBV transgenic mice (10, 17, 43), while there is no evidence for reinfection by the circulating virus. Hence, these observations collectively indicate that the foremost control of HBV species specificity occurs before viral transcription, i.e., at an early stage of the viral life cycle. Potentially decisive, and not mutually exclusive, critical steps include virus binding to the cell surface, the fusion process leading to the delivery of the nucleocapsid in the cytosol, its migration to the nucleus, and the release of the genome and its conversion into covalently closed circular DNA. Since the envelope proteins are implied in the binding and fusion events, they are attractive candidates which may take part in the determination of the HBV host range by interacting specifically with one or more receptors present only on cells of susceptible species. Alterations in the structural features of such cellular receptor(s) from one species to another could prevent the interactions required for HBV infection.
In this work, we investigated the envelope-mediated species specificity of HBV. For this purpose, pseudotyped HBVs were created by using the envelope protein gene of a newly isolated primate hepadnavirus, the woolly monkey HBV (WMHBV) (22). The resulting pseudotyped virus consisted of an HBV nucleocapsid enwrapped in an envelope containing the WMHBV surface proteins. Two main reasons dictated our choice on the WMHBV. First, although the origin of some nonhuman primate hepadnaviruses is still controversial (12, 23, 27, 42), it is accepted that the WMHBV is phylogenetically distinct from all the different genotypes of HBV. Second, there is suspicion that WMHBV, like all members of the Hepadnaviridae, exhibits a narrow host range. Thus, an inoculum from an infected woolly monkey failed to efficiently infect a chimpanzee (22). The aim of our strategy was first to check that the pseudotyped virus was unable to infect human hepatocytes in primary culture and then to restore its infectivity by replacing short amino acid regions in the WMHBV envelope protein by their HBV counterparts. Published data suggested that this substitution experiment might be focused on the N-terminal extremity of the L protein. Indeed, the corresponding region in the L protein of the heron HBV governs the species specificity of this avian hepadnavirus (20).
MATERIALS AND METHODS
Nomenclature.
Names given to pseudotyped viruses refer to the identities of their surface proteins. Indeed, they differ from HBV only by their envelope proteins. Subscripts H and W are used to indicate HBV or WMHBV origin, respectively, of an envelope polypeptide or an internal domain. Thus, chimeric L proteins and pseudotyped viruses bearing these mutant polypeptides are named according to the substitution: (i) LW x-yH, when surface proteins have a WMHBV origin and amino acids from positions x to y within the pre-S1 region have been replaced by the corresponding HBV amino acids, or (ii) LH x-yW, when the envelope proteins are derived from HBV and when amino acids of the pre-S1 region, from positions x to y, have been substituted for their WMHBV counterparts. The numeration is established on the ayw subtype of the HBV L protein sequence.
Plasmid engineering.
Plasmid pHBV L- env-, designed to initiate a viral replication only when cotransfected with a helper vector expressing all viral surface proteins, derives from the previously described plasmid pHBV L- (25). Expression of the envelope proteins is prevented by the presence of two point mutations creating stop codons in the pre-S1–pre-S2–S ORF. Codon 90 of the pre-S1 region and codon 199 of the S domain are replaced by opal and amber termination signals, respectively. Both mutations were chosen because they remain silent in the overlapping polymerase gene. The additional stop codon in the S region was introduced by PCR-mediated mutagenesis (19).
Helper vectors pSV12SXH and pSV12SXW were used for the expression of the HBV or WMHBV surface polypeptides, respectively, in their wild-type (WT) state. Construct pSV12SXH corresponds to plasmid pSV12SX described in a previous work (24). Plasmid pSV12SXW contains the 2,258-bp BssHII-BssHII fragment of a cloned WMHBV genome (M. Nassal and A. Wahl-Feuerstein, unpublished data), which bears the entire WT pre-S–S coding regions. This fragment was cloned in plasmid pSV-SPORT 1 (Life Technologies) by using the compatible MluI site, located just downstream of the simian virus 40 early promoter-origin region.
PCR-mediated mutagenesis (7, 19) was used to create another set of helper vectors deriving from plasmid pSV12SXH or pSV12SXW, into which single substitutions were independently introduced to express different types of chimeric L proteins (LH x-yW and LW x-yH) without modifying the sequences of the synthesized M and S polypeptides.
All mutant subcloned fragments inserted in the final constructs were sequenced to confirm their identities.
Cell line and transfection.
To produce viral proteins or virions, the HepG2 human hepatoma cell line (1, 41) was transfected with viral DNA by electroporation. HepG2 cells were cultured in H medium (75% minimum essential medium, 25% medium 199 [Eurobio], 5 mg of insulin per liter, 4.5 mg of penicillin per liter, 50 mg of streptomycin per liter) supplemented with 3.5 × 10−7 M hydrocortisone hemisuccinate, 2 mM l-glutamine, and 10% fetal calf serum (FCS).
Virus purification.
Between 4 and 7 days after transfection of HepG2 cells, culture supernatant was harvested daily and pooled. HBV particles were precipitated with 6% polyethylene glycol 8000 (PEG 8000; Sigma) for 12 h at 4°C. The precipitates were recovered by centrifugation (5,000 × g for 45 min at 4°C) and concentrated 100-fold in phosphate-buffered saline (PBS) with 25% FCS.
Primary cell culture and infection.
Fragments of normal adult human liver were obtained from patients undergoing hepatic resection for liver metastases (the fragments were taken at a distance from the metastasis in macroscopically normal liver tissue). Access to this biopsy material was in agreement with French laws and satisfied the requirements of the French National Ethics Committee. Hepatocytes were isolated by the procedure of Guguen-Guillouzo and Guillouzo (16) and cultured in H medium supplemented with 3.5 × 10−6 M hydrocortisone hemisuccinate, 2 mM L-glutamine, 50 mg of gentamicin per liter, 2% dimethyl sulfoxide, 5% adult human serum, and 5% FCS. Three days after seeding, the cells were infected as described previously (13). The hepatocytes (1.5 × 106 per 10-cm2 petri dish) were covered with 1 ml of serum-free culture medium containing 5% PEG 8000 and 100 μl of inoculum. Infection was performed for 12 h at 37°C. Cells were then washed three times with culture medium and subjected to a further culture.
Intra- and extracellular protein analysis.
Transfected cells and their supernatants were harvested 7 days after transfection. HepG2 cells were lysed, and nuclei were removed before immunoblotting analysis by the method described in a previous work (25). Released viral particles were precipitated with 6% PEG 8000. Proteins were analyzed by electrophoresis through 12.5% polyacrylamide-sodium dodecyl sulfate gels and transferred onto a nitrocellulose filter (Amersham). Immunoblotting was performed by using enhanced chemiluminescence (Amersham) with primary monoclonal antibody F376. The epitope recognized by this antibody is located inside the pre-S2H domain (30). This allowed the detection of the L and M proteins.
Assays for HBV-specific proteins.
HBV surface antigen (HBsAg) was measured with a radioimmunoassay kit (Pasteur-Sanofi) using two monoclonal anti-HBsAg antibodies. The detection method conformed to the protocol recommended by the manufacturer.
Viral DNA extraction and analysis.
Complete viral particles were immunoprecipitated from the concentrated supernatant of transfected HepG2 cells with a polyclonal anti-HBsAg antibody (Dako). Viruses were then disrupted by the addition of a lysis buffer containing 0.5% sodium dodecyl sulfate, 10 mM Tris-HCl (pH 8), 10 mM EDTA, 10 mM NaCl, 200 μg of proteinase K per ml, and 40 μg of tRNA per ml. The lysis was performed overnight at 37°C.
The intracellular replicative forms of viral DNA were selectively extracted from human adult hepatocytes after incubating cells with 0.5 mg of trypsin per ml–0.5 mM EDTA in PBS for 10 min at 37°C in order to eliminate adsorbed virions. The reaction was stopped by adding 20% FCS. Cells were recovered by centrifugation for 3 min at 200 × g and washed once with PBS. Hepatocytes were then incubated overnight at 37°C in lysis buffer differing from the one described above only by the lack of tRNA. Subsequently, chromosomal DNA was precipitated overnight at 4°C with 1 M NaCl (18) and eliminated by centrifugation at 12,000 × g for 15 min at 4°C.
All samples were subjected to a phenol-chloroform-isoamyl alcohol extraction followed by a chloroform-isoamyl alcohol extraction. Afterwards, in the absence of salt, 0.3 M sodium acetate (pH 5.5) was added to the aqueous phase and nucleic acids were precipitated with 2 volumes of ethanol. DNA was analyzed on 1.5% agarose gels. These gels were soaked in 0.25 N HCl for 15 min, and DNA was denatured in situ in 0.4 M NaOH and transferred onto positively charged nylon membranes (Amersham) by the Southern blotting method (37). Hybridization was performed at 65°C with linearized HBV genomic α-32P-labeled DNA as a probe.
Viral RNA extraction and analysis.
Total RNA present in infected hepatocytes was extracted with the SV Total RNA Isolation System kit (Promega) and fractionated on a 1.5% agarose gel and viral RNAs were analyzed by a standard Northern blotting procedure (37). Hybridization was achieved as described for DNA analysis.
RESULTS
To evaluate the role of the surface proteins in determining the species specificity of the HBV infection, we chose a strategy based on the use of pseudotyped HBV, at the surface of which the original envelope proteins were replaced by the corresponding WMHBV polypeptides. The envelope protein gene of WMHBV was cloned in vector pSV12SXW, designed for the expression of the SW, MW, and LW proteins. Several related constructs were engineered to translate mutant LW proteins in which blocks of 10 to 30 aa were substituted for the HBV counterparts. These mutants are named LW 1-10H, LW 11-20H, LW 21-30H, and LW 1-30H, where the first and second numbers define the N- and C-terminal extremities of the substituted region, respectively.
Expression and secretion of the envelope proteins.
To check that the WT and the genetically modified WMHBV envelope proteins were efficiently expressed by a human hepatoma cell line, HepG2 cells were transiently transfected with the corresponding expression vectors. In parallel, the plasmid expressing the WT HBV envelope proteins was used as a positive control. Seven days posttransfection (dpt), intra- and extracellular viral proteins were analyzed by Western blotting with a primary monoclonal anti-pre-S2H antibody (F376) (30) (Fig. 2A and B, respectively). Since the epitope recognized by F376 is well conserved between the proteins of WMHBV and the adw2 subtype HBV (85.7% homology), against which the antibody was raised, it was anticipated that the F376 antibody would bind LW and MW proteins; however, whether its affinity would be impaired could not be predicted.
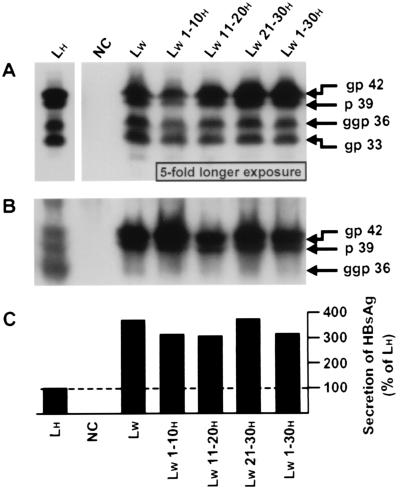
FIG. 2.
Expression and secretion of the WT and chimeric WMHBV envelope proteins by a human hepatoma cell line. HepG2 cells were transfected with 20 μg of vectors designed to express viral surface proteins: LH, expression plasmid driving the synthesis of the WT HBV proteins; NC, plasmid without any HBV insert (negative control); LW and LW x-yH, expression plasmids driving the translation of WT and chimeric WMHBV proteins, respectively. Analyses were performed 7 dpt. Proteins were extracted from cells (A) or were precipitated from the culture medium (B) and studied by Western blotting. The primary monoclonal antibody was directed against the pre-S2H region. gp 42, p 39, ggp 36, and gp 33, migration positions of the glycosylated and unglycosylated LH and LW proteins and the diglycosylated and glycosylated MH and MW proteins, respectively. In panel A, the exposure time of the autoradiogram was 5-fold longer for samples NC, LW, and LW x-yH than for the control, LH. (C) Assessment of the secreted HBsAg by radioimmunoassay. Data are percentages of the positive-control (LH) value.
As expected, a typical pattern of four main bands was revealed in the lysate of cells transfected with the vector expressing HBV envelope proteins (Fig. 2A, lane LH). The LH protein appeared as 39- and 42-kDa polypeptides, corresponding to the unglycosylated (p 39) and glycosylated (gp 42) forms, respectively. The glycosylated (gp 33) and diglycosylated (ggp 36) MH proteins were also detected at the appropriate apparent molecular masses of 33 and 36 kDa, respectively. The relatively high similarity between the amino acid sequences of the HBV and WMHBV envelope proteins led us to assume that an identical pattern would be obtained for the WMHBV proteins. For instance, the theoretical molecular weights of the proteins are equal and the analysis of their primary sequences showed that posttranslational modification motifs, such as the myristylation and N-glycosylation sites, are preserved. Bands of identical mobility were indeed observed (Fig. 2, lane LW), and a comparable glycosylation state of proteins was also confirmed by a deglycosylation experiment (data not shown). Because the use of the N-glycosylation signals in the pre-S regions is determined by their relative membrane orientations, these data suggested that the WMHBV surface proteins have transmembrane topologies equivalent to those of the corresponding HBV proteins (5, 9, 31, 35).
Results obtained for cells transfected with the chimeric constructs showed that none of the substitutions inserted in the LW protein affected the apparent sizes of, or the ratios between, the individual envelope proteins compared to those for the WT WMHBV polypeptides. However, a 5-fold-longer exposure of the WT and chimeric WMHBV samples was necessary to obtain signals equivalent in intensity to those detected for the WT HBV proteins. This may reflect a weaker affinity of the F376 antibody for the WMHBV polypeptides or/and an actually smaller amount of intracellular viral proteins due to an enhanced particle release (see below).
Analysis of subviral particle secretion showed that, as already observed (24), the unglycosylated and glycosylated forms of the LH protein, as well as the diglycosylated form of the MH protein, were secreted in the supernatant of transfected cells (Fig. 2B, lane LH). This release indicated that amounts of the SH protein synthesized were sufficient to partly overcome the known retention property of the LH polypeptide (6, 32, 40). In fact, when overexpressed, the LH protein can inhibit its own secretion as well as the export of the other viral surface proteins. The corresponding WT proteins of WMHBV were also exported in the medium. Remarkably, despite a potentially lower affinity of the anti-pre-S2H antibody for the WMHBV proteins, larger amounts of both unglycosylated and glycosylated forms of LW were detected in the extracellular medium (Fig. 2B, lane LW). This increased secretion was also confirmed by measuring secreted HBsAg by a conventional radioimmunoassay which makes use of two monoclonal antibodies directed against the SH domain (Fig. 2C, lane LW). This efficient release of WMHBV envelope proteins might explain the low signals detected in the cell lysates (Fig. 2A). For an unknown reason, only a small amount of the diglycosylated MW protein was revealed by Western blotting (Fig. 2B, lane LW). Even if small quantitative variations were observed, similar results were obtained for all the chimeric constructions. Altogether, these data indicated that the WT and chimeric WMHBV proteins were properly synthesized under our experimental conditions.
Pseudotyped virus production.
For the generation of pseudotyped viruses, we exchanged the original surface proteins of complete HBV particles for the corresponding entities from WMHBV, either in a WT or mutant state. To this end, HepG2 cells were transiently cotransfected with a plasmid (pHBV L- env-) containing a replication-competent HBV genome but defective for the expression of envelope proteins, together with a helper vector expressing the WT or chimeric surface proteins of WMHBV. For a positive control, HepG2 cells were transfected with the same type of plasmid containing a replication-competent HBV genome lacking mutations that prevent translation of envelope proteins (pHBV-ΔEcoRI). Between 4 and 7 dpt, supernatants were harvested daily and pooled. Viral particles were then concentrated from the supernatants. The presence of virus was investigated by immunoprecipitating viral particles with a polyclonal anti-HBsAg antibody followed by the detection of viral genomes by a standard Southern blotting procedure (Fig. 3A). As already described (15), transfection of HepG2 cells with the cloned WT replication-competent HBV genome resulted in the release of virions (lane PC). As expected, no complete virion was produced from the envelope protein-defective genome in the absence of a helper vector (lane NC), because the expression of both the SH and LH peptides is required for the release of mature virions (3). Trans complementation with a helper vector expressing the WT envelope proteins of WMHBV restored the export of complete viral particles (lane LW). Extracellular virus titers comparable to those for the positive control were obtained when plasmid pHBV L- env- and the LW expression vector were used in a 4:1 molar ratio, respectively. Under the same conditions, cotransfection experiments using constructs producing chimeric L proteins also led to the secretion of similar quantities of pseudotyped virions (lanes LW x-yH).
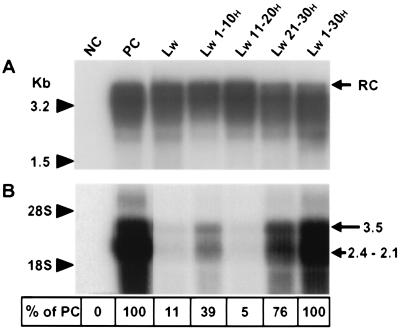
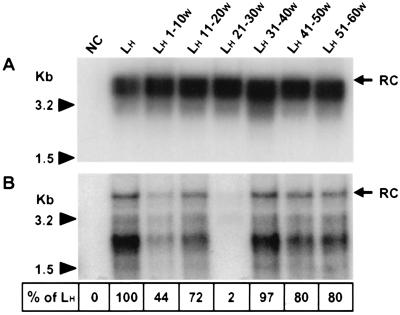
FIG. 3.
Production and infectivity of pseudotyped viruses. (A) HepG2 cells were either transfected with a plasmid (pHBV L- env-) containing a WT replication-competent HBV genome (PC for positive control) or cotransfected with an envelope protein-defective genome complemented with vectors expressing different envelope proteins: NC, plasmid without any HBV insert (negative control); LW and LW x-yH, expression plasmids driving the translation of WT and chimeric WMHBV proteins, respectively. Complete viral particles were immunoprecipitated with a polyclonal anti-HBsAg antibody from HepG2 cell supernatants collected between 4 and 7 dpt. DNA was extracted from the immunoprecipitates and analyzed by Southern blotting. Molecular size markers are on the left, and the position of the relaxed circular DNA (RC) is shown on the right. (B) Adult human hepatocytes in primary culture were incubated with concentrated supernatants obtained from HepG2 cells transfected or cotransfected as described for panel A. Total RNAs of inoculated cells were extracted 7 dpi and analyzed by Northern blotting. For each sample, 10 μg of total RNA was analyzed. The migration positions of 18S and 28S rRNAs and viral RNAs are indicated on the left and right, respectively. Shown at the bottom are the percentages of detected signal intensities relative to that revealed for PC.
To verify that the pre-S1 domain, as in WT HBV, was presented on the outer surfaces of pseudotyped virions, an immunoprecipitation was performed with a polyclonal anti-pre-S1H antibody. All mutant virions were efficiently immunoprecipitated (data not shown). This proved that the pre-S1 domain was also displayed at the surface of all pseudotyped viruses and, therefore, was available for potential interactions with cellular receptor(s).
Infectivity of the pseudotyped virions.
Progeny viruses were tested for their infectious capacity by using a previously established in vitro model of HBV infection based on normal human hepatocytes in primary culture (13, 14). The different inocula were prepared by concentrating viral particles from supernatants of HepG2 cells transfected as described above. It is important to note that all pseudotyped viruses contain the same replication-competent genome, defective for the expression of the envelope proteins. Therefore, if this modified genome is delivered into hepatocytes by an infectious pseudotyped virus, viral replication will be initiated but no complete or subviral particle will be secreted. Thus, HBsAg secretion into the culture medium could not be used to monitor infection. However, after the fusion of the viral envelope with a cellular membrane, the nucleocapsid is released into the cytosol and delivers the relaxed circular DNA genome to the nucleus for conversion into supercoiled DNA. This covalently closed circular DNA serves as a template for viral transcription. Detection of newly synthesized HBV RNA therefore constitutes a reliable proof of viral infectivity. Accordingly, while almost no viral RNA could be detected in hepatocytes immediately after in vitro infection, strong signals progressively appeared with time (data not shown).
Seven days postinfection (dpi), we investigated the presence of viral RNAs in hepatocytes by analyzing total cellular RNA by Northern blotting. A typical pattern of transcripts, 3.5 and 2.4 to 2.1 kb in size, was revealed in the extract of hepatocytes infected with the WT HBV (Fig. 3B, lane PC). The 3.5-kb band represents the pregenome and precore RNAs, whereas the large 2.1- to 2.4-kb band corresponds to messengers for the envelope proteins. Conversely, a small quantity of viral RNAs (11% of the signal detected in the positive control) was detected in cells incubated with the pseudotyped virus whose envelope contained WT WMHBV surface proteins (lane LW). Substituting amino acids located between residues 11 and 20 of the pre-S1W region for the corresponding HBV amino acids did not increase the RNA signals (5% of the signal detected in the positive control) and thus was not sufficient to restore the infectivity of the pseudotyped virus (lane LW 11-20H). By contrast, infectivity was partially or mostly rescued (39 and 76% of the signal detected in the positive control, respectively) when aa 1 to 10 or aa 21 to 30 of the LW chimeric protein had an HBV origin (lanes LW 1-10H and LW 21-30H, respectively). Finally, the infectious capacity was completely restored at a level identical to that for WT HBV when the first 30 residues of the pre-S1W domain were replaced by the corresponding HBV amino acids (lane LW 1-30H). This result is the first direct evidence demonstrating that the N-terminal part of the L protein plays an essential role in the species-specific infectivity of orthohepadnaviruses.
Reciprocal experiments.
To further corroborate the above-described results, a series of reciprocal experiments was performed; in these, six contiguous stretches of 10 aa within the 60 N-terminal residues of the HBV L protein were individually substituted for the corresponding WMHBV residues (LHx-yW). To ascertain that the mutations did not affect the synthesis of the protein, HepG2 cells were separately transfected with each of the vectors expressing the different mutant LH x-yW proteins. A Western blot analysis of the intracellular envelope proteins using the F376 antibody did not reveal any quantitative or qualitative differences from the WT HBV polypeptides whatever the introduced mutation (data not shown). Production of mutant viruses was achieved by cotransfecting HepG2 cells with a recombinant plasmid (pHBV L-) (25) containing a replication-competent L-defective HBV genome together with one of the chimeric LH x-yW protein expression vectors. To analyze the production of complete viral particles released in the supernatant of transfected cells, we applied the procedure previously described for the first series of protein chimeras (Fig. 4A). Whatever the substitution, the amounts of secreted mutant virions (lanes LH 1-10W to LH 51-60W) were almost equivalent to those of WT viruses produced when the helper vector encodes the WT form of the LH protein (lane LH). Subsequently, the infectious capacity of these mutant viruses on primary cultures of human hepatocytes was examined. Infection was assessed by the detection of replicative viral DNA forms in the cytoplasm of infected hepatocytes collected 15 dpi (Fig. 4B). Similar amounts of viral DNA were detected in hepatocytes infected by either WT HBV (lane LH) or by mutant viral particles bearing LH x-yW proteins, with substitutions either between aa 11 and 20 or between aa 31 and 60 (lanes LH 11-20W, LH 31-40W, LH 41-50W, and LH 51-60W). Indeed, as determined by densitometry, signal intensities revealed for these mutants oscillate between 72 and 97% of that detected in cells infected with the WT virus. Conversely, the amount of intracellular viral replicative intermediates was slightly reduced in hepatocytes infected by the LH 1-10W mutant virus (44% of the signal detected in WT HBV-infected cells). Importantly, the substitution LH 21-30W strongly inhibited viral infectivity as shown by the detection of only traces of HBV DNA (lane LH 21-30W) (2% of the signal detected for the WT HBV). This result is fully consistent with a crucial role of the corresponding residues in species-specific HBV infectivity.
FIG. 4.
Production and infectivity of reciprocal mutants. (A) HepG2 cells were cotransfected with the L-defective HBV genome (pHBV L-) complemented with vectors expressing different envelope proteins: NC, plasmid without any HBV insert (negative control); LH and LH x-yW, expression plasmids driving the translation of WT and mutant HBV surface proteins, respectively. Complete viral particles were immunoprecipitated with a polyclonal anti-HBsAg antibody from HepG2 cell supernatants collected between 4 and 7 dpt. DNA was extracted from the immunoprecipitates and analyzed by Southern blotting. (B) Adult human hepatocytes in primary culture were incubated with concentrated supernatants obtained from HepG2 cells cotransfected as described for panel A. Intracellular viral DNA was selectively extracted from hepatocytes collected 15 dpi and analyzed by Southern blotting. The positions of the relaxed circular DNA (RC) are shown on the right, and molecular size markers are on the left. Shown at the bottom are the percentages of detected signal intensities relative to that revealed for LH.
DISCUSSION
In this work, we took advantage of HBV species specificity to investigate its infection process. Our strategy was based on the creation of a pseudotyped HBV, at the surface of which the original envelope proteins were replaced by those from WMHBV. As a prerequisite of our approach, it was necessary to check the efficient expression of both WT and chimeric WMHBV envelope proteins in human cells and then their ability to replace their homologous HBV counterparts for the production of complete viral particles. Different analytical methodologies used in this work showed that the human HepG2 hepatoma cell line supports the biosynthesis of all expected WMHBV surface proteins. However, we also observed that their secretion level was higher than that of the HBV proteins. A retention signal between residues 1 to 8 in the ayw subtype LH protein has been described (21), and there are divergences in this region between the LH and LW proteins. However, these differences could not explain the higher secretion level of the WMHBV envelope proteins since the presence of the HBV retention signal in the LW 1-10H protein (Fig. 2A, lane LW 1-10H) did not modify the export of the WMHBV surface protein in the culture medium. So, whether this phenomenon is due to a reduced retention property of the WMHBV proteins per se, to an impairment of their retention out of the context of woolly monkey cells, or to an overproduction of the SW protein remains to be determined. Despite these different secretion efficiencies, the WMHBV proteins were able to efficiently substitute for the original surface proteins of HBV without affecting virion production. Moreover, the infectivity investigations were not impaired by this property specific to the WMHBV surface proteins because no correlation between the secretion capacity of the envelope proteins and the infectious ability of the generated pseudotyped viruses was observed. The pseudotyping ability was expected because a successful experimental phenotype mixing between envelope proteins from HBV and a more phylogenetically distant hepadnavirus, the woodchuck hepatitis virus (WHV), has already been reported (11). Thus, HBV morphogenesis tolerates the substitution of either the pre-S regions in the LH protein or the entire SH protein by the WHV counterparts. Successful pseudotyping, despite the general divergence between the amino acid sequences of HBV and WHV envelope polypeptides, seems to be linked to the sequence homology of two short amino acid regions. These are believed to mediate direct binding to nucleocapsid during the envelopment event. They are located at the junction of the pre-S1 and pre-S2 regions in the LH protein and in the first cytosolic loop, flanked by transmembrane domains I and II, of the SH protein (2, 25, 26, 34). The sequences of the WMHBV and HBV surface proteins exhibit even greater homologies within these two domains, which should explain the ability of the WMHBV envelope proteins to direct the envelopment of the HBV nucleocapsid and to ensure the budding of complete viral particles.
The host range determination of primate hepadnaviruses may be controlled during the infection process by several independent viral factors such as the envelope or capsid proteins. Our experimental approach provides compelling evidence that at least one viral determinant implied in the narrow host range of these viruses is located within the N-terminal region of the L protein. The pseudotyped virus with HBV nucleocapsid and WT WMHBV envelope proteins was unable to efficiently infect human hepatocytes, despite the accessibility of the pre-S1W region at its surface. The use of chimeric L proteins enabled us to identify the WMHBV amino acid motif which is unsuited for the infection of human cells. Thus, the pseudotyped virus with chimeric WMHBV envelope proteins bearing the first 30 HBV amino acids in the N-terminal part of their LW proteins had the same infectious capacity as the WT HBV. Hence, among all divergences between the sequences of the WMHBV and HBV surface proteins, only those located between aa 1 to 30 of the L protein are critically involved in the control of the envelope-mediated species specificity of the infection process. Within the relevant domain, shorter substitutions allowed us to precisely locate crucial residues between aa 1 to 10 and 21 to 30. The significance of these data was further corroborated by the totally coherent results obtained in the reciprocal experiments in which replacement of the same amino acids in the LH protein by WMHBV-specific residues decreased the infectivity of the corresponding virions. Again, the infectious capacity was most dramatically affected by the mutations between positions 21 and 30 and, to a lesser extent, by those between positions 1 and 10. Importantly, all of the four other mutants were still infectious. These results also argue against a potential influence of the different secretion properties of the WMHBV envelope proteins on the infectious capacity of the LW and LW x-yH pseudotyped viruses.
Even though a potential myristylation signal is predicted at the N terminus of the LW protein, its actual use has not been experimentally proven. Since this posttranslational modification is required for HBV infectivity (4, 15), its absence could formally explain the loss of virus infectivity. However, we consider this unlikely because the pseudotyped virus bearing the mutant LW 21-30H protein with the authentic WMHBV acylation signal was able to infect human hepatocytes. Moreover, replacement of the HBV myristylation signal by that of WMHBV in the LH 1-10W protein only partially affected HBV infectivity, whereas selective inactivation of this posttranslational signal is known to completely delete the infectious capacity of HBV (4, 15).
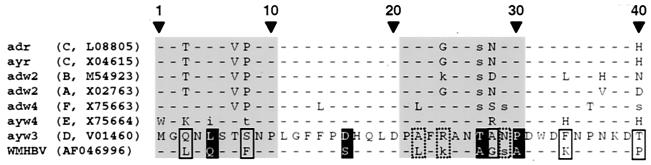
To further delineate potentially important residues within the domains identified in this study, the first 40 residues of the pre-S1 sequences from the WMHBV and HBV isolates used here were compared with those of 160 different HBV isolates (data not shown). For clarity, only six representative sequences are included in Fig. 5. Since the sequence of the WMHBV genome has been determined from an amplicon obtained by PCR, this could raise doubts as to the authenticity of the sequence. However, this part of the pre-S1 sequence appears to be highly conserved in WMHBV because identical sequences have been found in further isolates, including the genome described by Lanford et al. (22). The alignment shows that of the 20 residues in the crucial regions from aa 1 to 10 and 21 to 31, only 9 differ between the LW protein and the ayw3 subtype LH protein used in the present study. These variant residues could be classified in three different categories: (i) at positions 22, 24, and 29, amino acids identical to those present in the LW protein are found in some HBV isolates; (ii) at positions 3, 8, and 28, there is already a marked diversity in the nature of amino acids among the human viruses; (iii) at positions 5, 27, and 30, by contrast, identical or physicochemically similar residues are present in all HBV isolates but not in the WMHBV sequence. Residues from the last category are therefore prime candidates for limiting the host range specificity of WMHBV; however, this does not exclude potential contributions from residues of categories i and ii.
FIG. 5.
Comparison of WMHBV and HBV sequences in the N terminus pre-S1 domain. The WMHBV amino acid sequence is compared with the homologous sequence of HBV subtype ayw 3 used in this work. Six other human isolates, representing genotypes A to F, were also aligned. The genotype and the GenBank accession number of each sequence are in parentheses. The amino acid sequence from Met 1 to Thr 40 of pre-S1H (subtype ayw 3) is entirely shown in one-letter code. Dashes and lowercase letters, residues identical or similar to those in the corresponding positions of subtype ayw 3, respectively. Grey backgrounds indicate the two domains involved in the determination of HBV species specificity. Black box and open box, WMHBV-specific amino acids located at positions where the corresponding residues are conserved or variable among HBV strains, respectively; dotted-line box, residues different from those present in the ayw 3 subtype HBV but found in some other HBV isolates.
Because the HBV host range is crucially determined at the viral entry level, the identification of amino acid residues involved in the viral interspecies barrier is important progress in the understanding of the molecular events which occur during the early steps of the viral life cycle. Unfortunately, our current knowledge of the HBV infection process does not allow us to firmly attribute the exact function of the amino acids underlined in this work. However, they may be directly or indirectly involved in the interaction with a cellular receptor during either the attachment of the virus to the cell surface or the penetration by fusion of the viral envelope with a cellular membrane. In these cases, the recognized cellular factors from different species should exhibit a polymorphism complementary to the one displayed by the hepadnaviral L proteins.
Not the least, our data also provide strong evidence that WMHBV is not, or at least much less, infectious to humans than HBV.
ACKNOWLEDGMENTS
P.C. and J.L.S. contributed equally to this work.
P.C. and J.L.S are, respectively, recipients of fellowships from the Ligue Nationale contre le Cancer (Comité des Côtes d'Armor) and from the Académie Nationale de Médecine. This work was supported by INSERM, the Association pour la Recherche contre le Cancer, and the Ligue Nationale contre le Cancer (Comité d'Ille et Vilaine) and by a grant to Michael Nassal from the Bundesministerium für Bildung und Forschung.
We gratefully acknowledge Agatha Budkowska for the gift of the F376 antibody.
REFERENCES
- 1.Aden D P, Fogel A, Plotkin S, Damjanov I, Knowles B B. Controlled synthesis of HBsAg in a differentiated human liver carcinoma-derived cell line. Nature (London) 1979;282:615–616. doi: 10.1038/282615a0. [DOI] [PubMed] [Google Scholar]
- 2.Bruss V. A short linear sequence in the pre-S domain of the large hepatitis B virus envelope protein required for virion formation. J Virol. 1997;71:9350–9357. doi: 10.1128/jvi.71.12.9350-9357.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruss V, Ganem D. The role of envelope proteins in hepatitis B virus assembly. Proc Natl Acad Sci USA. 1991;88:1059–1063. doi: 10.1073/pnas.88.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruss V, Hagelsten J, Gerhardt E, Galle P R. Myristylation of the large surface protein is required for hepatitis B virus in vitro infectivity. Virology. 1996;218:396–399. doi: 10.1006/viro.1996.0209. [DOI] [PubMed] [Google Scholar]
- 5.Bruss V, Lu X Y, Thomssen R, Gerlich W H. Post-translational alterations in transmembrane topology of the hepatitis B virus large envelope protein. EMBO J. 1994;13:2273–2279. doi: 10.1002/j.1460-2075.1994.tb06509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chisari F V, Filippi P, McLachlan A, Milich D R, Riggs M, Lee S, Palmiter R D, Pinkert C A, Brinster R L. Expression of hepatitis B virus large envelope polypeptide inhibits hepatitis B surface antigen secretion in transgenic mice. J Virol. 1986;60:880–887. doi: 10.1128/jvi.60.3.880-887.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deminie C A, Emerman M. Incorporation of human immunodeficiency virus type 1 Gag proteins into murine leukemia virus virions. J Virol. 1993;67:6499–6506. doi: 10.1128/jvi.67.11.6499-6506.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diot C, Gripon P, Rissel M, Guguen-Guillouzo C. Replication of hepatitis-B virus in differentiated adult rat hepatocytes transfected with cloned viral DNA. J Med Virol. 1992;36:93–100. doi: 10.1002/jmv.1890360206. [DOI] [PubMed] [Google Scholar]
- 9.Eble B E, Lingappa V R, Ganem D. The N-terminal (pre-S2) domain of a hepatitis B virus surface glycoprotein is translocated across membranes by downstream signal sequences. J Virol. 1990;64:1414–1419. doi: 10.1128/jvi.64.3.1414-1419.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farza H, Hadchouel M, Scotto J, Tiollais P, Babinet C, Pourcel C. Replication and gene expression of hepatitis B virus in a transgenic mouse that contains the complete viral genome. J Virol. 1988;62:4144–4152. doi: 10.1128/jvi.62.11.4144-4152.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerhardt E, Bruss V. Phenotypic mixing of rodent but not avian hepadnavirus surface proteins into human hepatitis B virus particles. J Virol. 1995;69:1201–1208. doi: 10.1128/jvi.69.2.1201-1208.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grethe S, Heckel J O, Rietschel W, Hufert F T. Molecular epidemiology of hepatitis B virus variants in nonhuman primates. J Virol. 2000;74:5377–5381. doi: 10.1128/jvi.74.11.5377-5381.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gripon P, Diot C, Guguen-Guillouzo C. Reproducible high level infection of cultured adult human hepatocytes by hepatitis-B virus: effect of polyethylene glycol on adsorption and penetration. Virology. 1993;192:534–540. doi: 10.1006/viro.1993.1069. [DOI] [PubMed] [Google Scholar]
- 14.Gripon P, Diot C, Theze N, Fourel I, Loreal O, Brechot C, Guguen-Guillouzo C. Hepatitis B virus infection of adult human hepatocytes cultured in the presence of dimethyl sulfoxide. J Virol. 1988;62:4136–4143. doi: 10.1128/jvi.62.11.4136-4143.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gripon P, Le Seyec J, Rumin S, Guguen-Guillouzo C. Myristylation of the hepatitis B virus large surface protein is essential for viral infectivity. Virology. 1995;213:292–299. doi: 10.1006/viro.1995.0002. [DOI] [PubMed] [Google Scholar]
- 16.Guguen-Guillouzo C, Guillouzo A. Methods for preparation of adult and fetal hepatocytes. In: Guillouzo A, Guguen-Guillouzo C, editors. Isolated and cultured hepatocytes. Les éditions INSERM Paris. London, United Kingdom: John Libbey and Co., Ltd.; 1986. pp. 1–12. [Google Scholar]
- 17.Guidotti L G, Matzke B, Schaller H, Chisari F V. High-level hepatitis B virus replication in transgenic mice. J Virol. 1995;69:6158–6159. doi: 10.1128/jvi.69.10.6158-6169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 19.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 20.Ishikawa T, Ganem D. The pre-S domain of the large viral envelope protein determines host range in avian hepatitis B viruses. Proc Natl Acad Sci USA. 1995;92:6259–6263. doi: 10.1073/pnas.92.14.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuroki K, Russnak R, Ganem D. Novel N-terminal amino acid sequence required for retention of a hepatitis B virus glycoprotein in the endoplasmic reticulum. Mol Cell Biol. 1989;9:4459–4466. doi: 10.1128/mcb.9.10.4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lanford R E, Chavez D, Brasky K M, Burns R B, Rico-Hesse R. Isolation of a hepadnavirus from the woolly monkey, a New World primate. Proc Natl Acad Sci USA. 1998;95:5757–5761. doi: 10.1073/pnas.95.10.5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lanford R E, Chavez D, Rico-Hesse R, Mootnick A. Hepadnavirus infection in captive gibbons. J Virol. 2000;74:2955–2959. doi: 10.1128/jvi.74.6.2955-2959.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le Seyec J, Chouteau P, Cannie I, Guguen-Guillouzo C, Gripon P. Infection process of the hepatitis B virus depends on the presence of a defined sequence in the pre-S1 domain. J Virol. 1999;73:2052–2057. doi: 10.1128/jvi.73.3.2052-2057.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le Seyec J, Chouteau P, Cannie I, Guguen-Guillouzo C, Gripon P. Role of the pre-S2 domain of the large envelope protein in hepatitis B virus assembly and infectivity. J Virol. 1998;72:5573–5578. doi: 10.1128/jvi.72.7.5573-5578.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loffler-Mary H, Dumortier J, Klentsch-Zimmer C, Prange R. Hepatitis B virus assembly is sensitive to changes in the cytosolic S loop of the envelope proteins. Virology. 2000;270:358–367. doi: 10.1006/viro.2000.0268. [DOI] [PubMed] [Google Scholar]
- 27.MacDonald D M, Holmes E C, Lewis J C, Simmonds P. Detection of hepatitis B virus infection in wild-born chimpanzees (Pan troglodytes verus): phylogenetic relationships with human and other primate genotypes. J Virol. 2000;74:4253–4257. doi: 10.1128/jvi.74.9.4253-4257.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nassal M. Hepatitis B virus replication: novel roles for virus-host interactions. Intervirology. 1999;42:100–116. doi: 10.1159/000024970. [DOI] [PubMed] [Google Scholar]
- 29.Nassal M. Macromolecular interactions in hepatitis B virus replication and particle formation. In: Cann A J, editor. Frontiers in molecular biology: DNA virus replication. Vol. 26. Oxford, United Kingdom: Oxford University Press; 2000. pp. 1–40. [Google Scholar]
- 30.Neurath A R, Adamowicz P, Kent S B, Riottot M M, Strick N, Parker K, Offensperger W, Petit M A, Wahl S, Budkowska A, et al. Characterization of monoclonal antibodies specific for the pre-S2 region of the hepatitis B virus envelope protein. Mol Immunol. 1986;23:991–997. doi: 10.1016/0161-5890(86)90130-6. [DOI] [PubMed] [Google Scholar]
- 31.Ostapchuk P, Hearing P, Ganem D. A dramatic shift in the transmembrane topology of a viral envelope glycoprotein accompanies hepatitis B viral morphogenesis. EMBO J. 1994;13:1048–1057. doi: 10.1002/j.1460-2075.1994.tb06353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Persing D H, Varmus H E, Ganem D. Inhibition of secretion of hepatitis B surface antigen by a related presurface polypeptide. Science. 1986;234:1388–1391. doi: 10.1126/science.3787251. [DOI] [PubMed] [Google Scholar]
- 33.Persing D H, Varmus H E, Ganem D. The pre-S1 protein of hepatitis B virus is acylated at its amino terminus with myristic acid. J Virol. 1987;61:1672–1677. doi: 10.1128/jvi.61.5.1672-1677.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poisson F, Severac A, Hourioux C, Goudeau A, Roingeard P. Both pre-S1 and S domains of hepatitis B virus envelope proteins interact with the core particle. Virology. 1997;228:115–120. doi: 10.1006/viro.1996.8367. [DOI] [PubMed] [Google Scholar]
- 35.Prange R, Streeck R E. Novel transmembrane topology of the hepatitis B virus envelope proteins. EMBO J. 1995;14:247–256. doi: 10.1002/j.1460-2075.1995.tb06998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson W S, Marion P L, Feitelson M, Siddiqui A. The hepadna virus group: hepatitis B and related viruses. In: Szmuness W, et al., editors. Viral hepatitis. Philadelphia, Pa: Franklin Press; 1982. pp. 57–68. [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Schmitt S, Glebe D, Alving K, Tolle T K, Linder M, Geyer H, Linder D, Peter-Katalinic J, Gerlich W H, Geyer R. Analysis of the pre-S2 N- and O-linked glycans of the M surface protein from human hepatitis B virus. J Biol Chem. 1999;274:11945–11957. doi: 10.1074/jbc.274.17.11945. [DOI] [PubMed] [Google Scholar]
- 39.Shih C H, Li L S, Roychoudhury S, Ho M H. In vitro propagation of human hepatitis B virus in a rat hepatoma cell line. Proc Natl Acad Sci USA. 1989;86:6323–6327. doi: 10.1073/pnas.86.16.6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Standring D N, Ou J H, Rutter W J. Assembly of viral particles in Xenopus oocytes: pre-surface-antigens regulate secretion of the hepatitis B viral surface envelope particle. Proc Natl Acad Sci USA. 1986;83:9338–9342. doi: 10.1073/pnas.83.24.9338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sureau C, Romet-Lemonne J L, Mullins J I, Essex M. Production of hepatitis B virus by a differentiated human hepatoma cell line after transfection with cloned circular HBV DNA. Cell. 1986;47:37–47. doi: 10.1016/0092-8674(86)90364-8. [DOI] [PubMed] [Google Scholar]
- 42.Warren K S, Heeney J L, Swan R A, Heriyanto, Verschoor E J. A new group of hepadnaviruses naturally infecting orangutans (Pongo pygmaeus) J Virol. 1999;73:7860–7865. doi: 10.1128/jvi.73.9.7860-7865.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamamura K, Araki K, Hino O, Tomita N, Miyazaki J, Matsubara K. HBV production in transgenic mice. Gastroenterol Jpn. 1990;25(Suppl. 2):49–52. doi: 10.1007/BF02779928. [DOI] [PubMed] [Google Scholar]