Abstract
The infection outcome of the Parvoviridae largely relies on poorly characterized intracellular factors modulated by proliferation, differentiation, and transformation of host cells. We have studied the interactions displayed by the highly homologous p and i strains of the murine parvovirus minute virus of mice (MVM), with a series of transformed cells of rat (C6) and human (U373, U87, SW1088, SK-N-SH) nervous system origin, seeking for molecular mechanisms governing parvovirus host range. The MVMp infection of C6 and U373 cells was cytotoxic and productive, whereas the other nervous cells behaved essentially as resistant to this virus. In contrast, MVMi did not complete its life cycle in any of the human nervous cells, though it efficiently killed the astrocytic tumor cells by two types of nonproductive infections: (i) normal synthesis of all viral macromolecules with a late defect in infectious virion maturation and release to the medium in U373; and (ii) high levels of accumulation of the full set of viral messenger RNAs and of both nonstructural (NS-1) and structural (VP-1 and VP-2) proteins, under a very low viral DNA amplification, in U87 and SW1088 cells. Further analyses showed that U87 was permissive for nuclear transport of MVMi proteins, leading to efficient assembly of empty viral capsids with a normal phosphorylation and VP1-to-VP2 ratio. The DNA amplification blockade in U87 occurred after conversion of the incoming MVMi genome to the monomeric replicative form, and it operated independently of the delivery pathway used by the viral particle, since it could not be overcome by transfection with cloned infectious viral DNA. Significantly, a chimeric MVMi virus harboring the coding region of the nonstructural (NS) gene replaced with that of MVMp showed a similar pattern of restriction in U87 cells as the parental MVMi virus, and it attained in U373 cultures an infectious titer above 100-fold higher under equal levels of DNA amplification and genome encapsidation. The results suggest that the activity of complexes formed by the NS polypeptides and recruited cellular factors restrict parvovirus DNA amplification in a cell type-dependent manner and that NS functions may in addition determine MVM host range acting at postencapsidation steps of viral maturation. These data are relevant for understanding the increased multiplication of autonomous parvovirus in some transformed cells and the transduction efficacy of nonreplicative parvoviral vectors, as well as a general remark on the mechanisms by which NS genes may regulate viral tropism and pathogenesis.
Viral infection begins with the specific recognition of cell surface receptors by virus structural components followed by additional interactions leading to irreversible internalization of the particle. In order to be productive, the infection must successfully proceed through several processes, such as uncoating and genome expression, replication, and maturation. The family Parvoviridae, a large group of small viruses containing a linear single-stranded (ss) DNA genome of approximately 5,000 nucleotides (23, 77), constitutes an interesting and useful system to dissect molecular mechanisms of virus-host interactions due to the genetic simplicity of its members and the multiplicity of cellular functions on which parvovirus multiplication relies. Indeed, in addition to the cellular surface molecules acting as specific receptors for some members of the three genera of the family, as for the adeno-associated virus (AAV) (68, 81, 82), the human erythrovirus B19 (11), and the autonomous parvovirus Aleutian Disease Virus (ADV) (38) and Canine Parvovirus (CPV) (64), the multiplication of parvoviruses requires functions expressed during the S phase of the cell cycle (5, 83, 87) as well as factors expressed at certain differentiation (55, 80, 84) and transformation (56, 86) stages, the nature of which remains largely unknown.
The genome of the autonomously replicating members of the parvovirus genus is organized into two overlapping transcription units that are timely regulated (18). The left gene, driven by the P4 promoter, encodes the NS1 and NS2 nonstructural proteins, and the right gene, driven by the P38 promoter, encodes the VP1 and VP2 structural proteins. Sixty VP protein subunits assemble to form the parvoviral capsid, which structure at atomic resolution has been resolved for the canine parvovirus (CPV) (89) and the immunosuppressive strain of the minute virus of mice (MVMi) (1), among others. The two NS polypeptides play diverse roles in virus multiplication. The smaller NS2 protein (28 kDa) contains three isoforms arising from alternate splicings (26) that can bind the cell cycle regulator 14-3-3 protein family (10) and shuttle from the nucleus to the cytoplasm via the CRM1 export pathway (8). A multiplicity of functions has been assigned to NS2, such as capsid assembly (29), messenger translation (59), and DNA replication and virus production in a cell type-specific manner (48, 58), though the NS2 mode of action remains unclear. The larger NS1, the main viral replicator protein (23), is a multifunctional nuclear phosphoprotein (19) that performs crucial steps of the MVM's unique rolling-hairpin mode of DNA synthesis (reviewed in reference 28). Viral DNA replication starts with the conversion reaction, the so-called synthesis of the complementary strand of the ss virion genome by cellular factors (5) generating a double-stranded (ds) monomer replicative form (mRF) DNA. The DNA binding, nicking, and helicase activities of NS1 (27, 60), together with the concourse of additional cellular factors (15–17), are strictly required for the accomplishment of the subsequent replication steps, including the introduction of an ss nick, the amplification of mRF DNA to multimeric intermediates, and the resolution and packaging of virus genomes (27, 72). Moreover, NS1 is a potent transactivator of the viral promoters (32) by the recognition of DNA elements (51, 71) and mediates cytotoxic effects that may be modulated by the cell physiological stage (14).
Determinants of parvovirus tropism have been identified in different regions of the genome. Sequences within the NS gene were found to be important for the extension of murine parvovirus MVM (44) and porcine parvovirus (PPV) (90) host ranges, although the mechanism of action was not further explored. More data are available for the cell tropism determinants laying in the capsid gene, demonstrated for MVM (2, 44), CPV (65), PPV (6), and ADV (7, 39). Two strains of MVM, the prototype (MVMp) and the immunosuppressive (MVMi) strains isolated from cell cultures (9, 30), which differ in just a few amino acids of their coding sequences (3) but show characteristic tropism to mouse lymphohemopoietic cells in vitro (35, 54) and distinct pathogenicity in newborn mice (12, 47, 69) and SCID mice (76), were used to delineate tropism determinants for the infection of fibroblast and lymphoid cell lines in vitro (85). The main capsid determinant for MVM productive infection (2, 44), the so-called allotropic determinant, was mapped to two amino acids of the threefold spike of the capsid which, functioning coordinately (4) via the VP-2 protein (53), mediates the interaction between the incoming particle (43) and intracellular factors (78). In the nonfunctional conformation of this determinant, the MVM infection is restricted prior to transcription and gene expression (2, 43) by a proposed altered decapsidation after nuclear transport of the virions (67) that delays the initiation of viral DNA synthesis and prevents DNA amplification (67, 78).
The complexity of the cellular factors involved in parvovirus multiplication is best illustrated by the privileged interaction of these viruses with transformed cells (reviewed in reference 73). In the genus of the autonomous parvovirus, this phenomenon, called parvoviral oncosuppression, has been mainly evaluated for the H-1 and MVMp viruses infecting human and mouse fibroblasts and epithelial cells transformed by a variety of agents that lower intracellular restrictions to parvovirus multiplication (21, 56), and in which some oncogenes were particularly efficient for this effect (57, 74). The facilitated multiplication of parvoviruses in transformed cells (or oncotropism) was in correspondence with an increased gene expression and transcription activity from the P4 promoter (20, 22). In addition, some neoplastic cells seemed to be particularly sensitive to the cytotoxicity of parvovirus NS proteins (14, 57), accounting for their favored killing by these viruses under in vitro (86) and in vivo (34, 88) experimental conditions.
The potential use of parvoviruses as anticancer biological reagents demands a comprehensive understanding of their tropism toward transformed cells. For this purpose, we have explored a system formed by the homologous MVMp and MVMi strains interacting with a collection of reference cell lines derived from rat and human central nervous system tumors of neural and glial origin, at graded malignant stages. Cytopathic effect (CPE) of the cultures, virus growth, cytotoxicity, and viral macromolecular biosynthesis were monitored for each virus-cell interaction. Thereby, we found two distinct nonproductive MVMi infections internally restricted in human astrocytic tumor cells, in which either viral full gene expression and empty capsids production proceeded in the absence of DNA amplification or the DNA-full virions produced to normal levels showed a low specific infectivity. The features of these infections may underlie novel molecular mechanisms of parvovirus host range determination by nonstructural functions.
MATERIALS AND METHODS
Cells.
Cells isolated from the nervous system were purchased from the American Type Culture Collection. The U87 MG human glioblastoma (HTB-14), U373 MG human glioblastoma (HTB-17), SW1088 human astrocytoma (HTB-12), SK-N-SH human neuroblastoma (HTB-11), and C6 rat glioma (CCL-107) cells were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum (FCS), except that SW1088 cells were cultured in DMEM without sodium bicarbonate in closed tissue-culture flasks with 30 mM HEPES buffer. All the cells were maintained for a minimal number of passages.
Viruses.
Purified stocks of the immunosuppressive strain of the parvovirus MVM (MVMi) and of the prototype strain (MVMp) were prepared from their respective infectious molecular clones, pMVMi and pMM984 (44). The viral plasmids, amplified in Escherichia coli strain JC8111 cells that preserve the viral hairpins and enriched in supercoiled forms by chromatography (Qiagen), were electroporated into the cells in which these viruses were originally isolated, the EL-4 mouse C57BL T-cell lymphoma (9) and the A9 mouse fibroblast (30) cell lines, respectively. Cells were cultured for 48 h, and the intracellular virus was harvested and used to infect at a low multiplicity of infection (MOI) the same permissive cell lines. Large viral stocks devoid of empty capsids were subsequently prepared by density gradient equilibrium centrifugation and stored at −70°C (76). Infectious virus was quantified by plaque assay on monolayers of the NB324K simian virus 40-transformed human newborn kidney cells as previously described (85).
To obtain the recombinant virus MVMi-NSp, we followed the previously described methodology that allows the recovery of MVMi/MVMp chimeric viruses upon transfection of NB324K cells (44). An NcoI-XhoI (nucleotide [nt] 261 to 2071) restriction fragment from the pMM984 plasmid encompassing most of the coding sequence of the nonstructural proteins (NS) of MVMp (3) was isolated and ligated into the MVMi genome substituting the equivalent fragment of the pMVMi plasmid. Ligation products were used to transform the E. coli JC8111 strain and colonies screened by DNA sequencing of the plasmids across the MVMi/MVMp boundary regions. An MVMi-NSp chimera plasmid was selected and transfected into NB324K by electroporation to prepare stocks of purified recombinant virus as described above for the wild-type strains, and the titer was estimated by plaque assay on the same cells. For best comparison of MVMi-NSp properties with those of its parental virus MVMi and to rule out any specific effect mediated by the host cell used to prepare the viral stocks, the experiments shown in Fig. 6 were also performed with purified MVMi grown in NB324K cells.
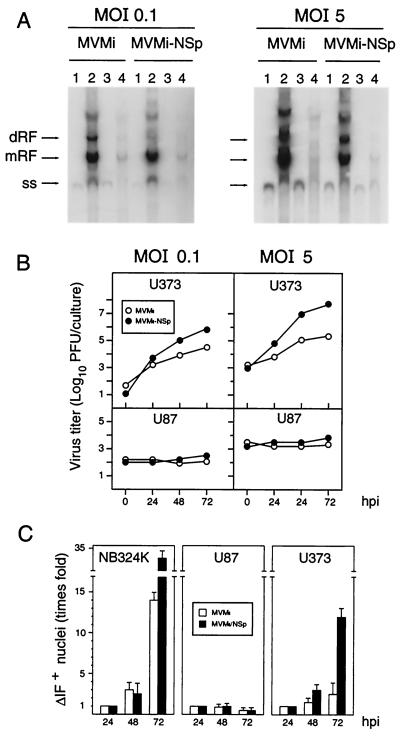
FIG. 6.
Role of NS gene in MVM infection of human astrocytic tumor cells. (A) Southern blot analysis of MVMi and MVMi-NSp viruses DNA synthesis in U373 (lanes 1 and 2) and U87 (lanes 3 and 4) cells. Samples from 2 × 105 cells infected at the indicated MOI and proceeded at 2 hpi (lanes 1 and 3) or 24 hpi (lanes 2 and 4) were loaded per gel slot, and filters were hybridized with a full-length MVM probe labeled to high specific activity. Exposure was for 48 h (MOI of 0.1) or 8 h (MOI of 5) with intensifying screen at −70°C. Markers are as described for previous figures. (B) Production of infectious MVM virus in culture of human nervous cells. Cells were scraped into the medium at the indicated postinfection times, and the total MVMi or MVMi-NSp virus produced in the cultures was determined by a plaque assay in NB3234K cells. (C) IF analysis of MVMi and MVMi-NSp virus progression in culture. Monolayers of human cells growing on coverslips were inoculated at an MOI of 0.01 PFU/cell and analyzed by IF for VP protein synthesis at the indicated times postinfection. The plots show a quantitative average of IF+ cells for VP nuclear staining from four independent experiments. Values are represented as the normalized fold increase with respect to the number of IF+ nuclei determined at 24 hpi.
Measurement of cell viability.
The effect of MVM infection on the viability of the cell lines was quantified by a clonogenic assay based in a previously described method (20). Cells seeded at a density of 5,000/cm2 were infected at increasing MOI (0.2 to 10 PFU/cell), and at 4 h postinoculation (hpi), cells were trypsinized and plated at various densities (2 × 102 to 104 cells per 60-mm-diameter dish) in triplicate. Cells were incubated for 10 days in the proper medium supplemented with a neutralizing dilution of MVM capsid antiserum to block reinfections, and arising colonies were fixed in absolute methanol and stained with 1% crystal violet. Survival is expressed as the percentage of colonies in the infected cultures with respect to the uninfected cultures normalized for a similar number of plated cells.
Blot analyses of MVM nucleic acids.
For viral transcription analysis, total RNA of infected cells was prepared as previously described (70), electrophoresed in agarose gels with 6% formaldehyde, and blotted onto nylon membranes (Gene Screen Plus; Dupont) by capillary transfer. The amount of loaded RNA was controlled by visualizing in the membranes the ribosomal RNAs stained with methylene blue. For Southern analysis of virus DNA replication, infected or transfected cells were extensively washed with phosphate-buffered saline (PBS) and processed for low-molecular-weight DNA extraction by a modified Hirt procedure (54) with carrier tRNA to ensure quantitative recoveries. DNA was fractionated by agarose gel electrophoresis and alkali blotted onto nylon membranes. Both types of membrane-bound samples were hybridized to a gel-purified MVM full-length DNA probe 32P labeled by random priming. When indicated, a 32P-labeled negative-sense riboprobe was synthesized with T7 bacteriophage RNA polymerase by using a PstI (nt 2129)-NcoI (nt 3121) restriction fragment of the MVMp genome cloned in a pGEM vector as previously described (70). Hybridizations were carried out at 42°C overnight in 50% deionized formamide, 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 5× Denhardt's solution, 10% dextran sulfate (Pharmacia), 0.5% sodium dodecyl sulfate (SDS), and 200 μg of denatured salmon sperm DNA per ml. Membranes were washed at 56°C in 0.1× SSC–0.5% SDS and exposed for autoradiography to Kodak X-Omat films for the indicated time periods.
Viral protein synthesis and subcellular distribution.
Cell lines seeded at 10,000 cells/cm2 were infected at an MOI of 5 PFU/cell with MVMp or MVMi and labeled 10 to 24 hpi in methionine-free DMEM supplemented with 10% normal medium, 10% dialyzed FCS, and 100 μCi of [35S]Met-Cys (Amersham AGQ 0080)/ml. At the end of the labeling period, cells were washed in PBS and disrupted in 50 mM Tris (pH 8.0)–0.15 M NaCl–0.2% SDS and a mixture of protease inhibitors by flushing through a 25-gauge needle. The level of synthesis of the VP1 and VP2 capsid proteins and of the NS1 nonstructural protein in the cell extracts was determined by immunoprecipitation with specific antibodies raised against NS1 and MVM capsid in rabbits (75), and the immunocomplexes were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) (10% polyacrylamide) analysis. The subcellular localization of the MVM capsid and NS1 proteins in the infected cells was determined with the same antibodies by indirect immunofluorescence (IF) as previously described (50).
Transfection.
Cells were transfected by the conventional calcium phosphate method as previously described (70). In brief, 10 μg of supercoiled plasmid was spotted as precipitated on monolayers of 106 cells growing in a 90-mm-diameter petri dish. After overnight incubation, cells were extensively washed with PBS and samples were taken either immediately (time zero posttransfection) or 48 h afterwards. To monitor that the MVMi plasmid was internalized into the nucleus of the NB324K and U87 cells, parallel cultures were similarly transfected with the addition of 2 μg of a reporter plasmid expressing the E. coli β-galactosidase enzyme under control of the MVM P4 promoter (to be described elsewhere) and stained accordingly 48 h posttransfection.
Analysis of MVM particle formation.
Infected cellular monolayers (106 cells) 35S labeled 10 to 24 hpi as above were washed and scrapped in PBS, 0.2% SDS was added, and DNA was sheared by flushing through a 25-gauge needle. The homogenates were brought to 9 ml in PBS–0.2% SDS and centrifuged for 18 h at 16,000 rpm and 15°C in a Beckman SW40 rotor (30,000 × g) through a 3-ml sucrose cushion (20% sucrose, 50 mM Tris [pH 8.0], 0.1 M NaCl, 1 mM EDTA, 0.2% SDS) to select for viral proteins assembled in particles. Pellets were resuspended and brought to 10 ml of 20 mM Tris (pH 8.0)–1 mM EDTA–0.2% Sarcosyl, adjusted to a density of 1.38 g/ml in CsCl by refractometry (ni = 1.370), and centrifuged to equilibrium for 42 h at 48,000 rpm and 15°C in a Beckman Ty65 rotor (150,000 × g). Gradients were fractionated from the top, and the positions of the 35S label-empty MVM capsids (1.32 g/ml) and DNA-full virions (1.41 to 1.46 g/ml) were determined by scintillation counting. Our method to prepare purified 32P-labeled MVM empty capsids and to perform two-dimensional analysis of VP2 tryptic phosphopeptides has been recently described (52).
RESULTS
Distinct types of MVM infections in transformed cells of nervous system origin.
As a preliminary test of permissiveness, growing cultures of human (U373, U87, SW1088, and SK-N-SH) and rat (C6) transformed cells of the nervous system were inoculated at a low MOI (0.1 PFU/cell) with the parvovirus strains MVMp and MVMi, and the progression of the CPE arising in the cultures, manifested by detached rounded cells and a lower density of the monolayers, was inspected daily under microscope (Fig. 1A). The NB324K cell line used as a productive system to grow these viruses was the only cell type highly susceptible to both strains. MVMp caused extensive CPE in C6 and U373 cultures and undetectable CPE in U87, SW1088, and SK-N-SH cultures (not shown). MVMi instead caused at this low MOI a moderate CPE in C6 cultures, but it failed to produce any CPE in the assayed human cell cultures. However, when the infections were performed at a high MOI (5 PFU/cell), the extent of the CPE at 3 days postinoculation caused by each of these viruses further varied, since MVMi was able to cause a pronounced CPE in the three human astrocytic tumor cells (U87, U373, and SW1088), whereas the CPE of the MVMp infections remained unchanged (not shown). These results were consistent in several independent inoculations.
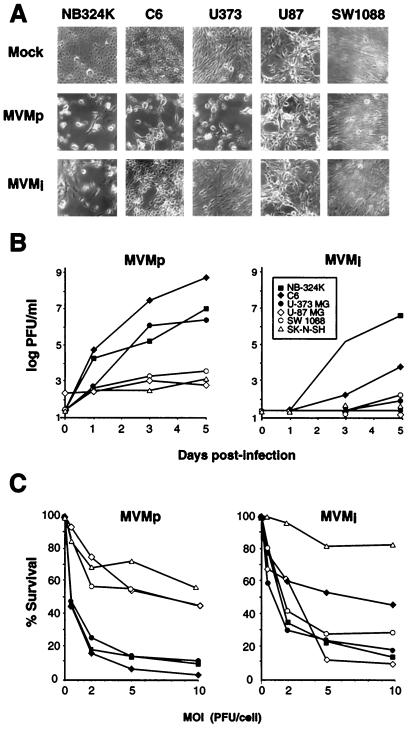
FIG. 1.
Types of MVM infection in transformed cells of nervous system origin. (A) CPE caused by MVM strains in cultured cells. Monolayers were inoculated at low MOI (0.1 PFU/cell), and micrographs were taken when the corresponding mock-infected cultures reached confluence (4 to 6 days p.i.). (B) Time course of infectious MVMp and MVMi production. Cultures were inoculated at 0.1 PFU/cell, and the release of infectious particles in the media over the time was determined by a plaque assay on NB324K cells. Each point is the average value from two independent experiments. (C) Susceptibility to MVM infections. Cells were infected with the MVM strains at the indicated MOI, and their viability was scored by a clonogenic assay. Represented are the mean values from at least two experiments for each cell line.
To get insights into the characteristic CPE provoked by each MVM strain, we determined the titer of infectious virus released in cultures infected at a low MOI (0.1) and the cytotoxicity of the interactions. MVMp grew to very high titers in C6 cultures, as efficient in U373 cells as in the reference NB324K cells, and to a much lower extent in the other cells (Fig. 1B, left). For MVMi, only the reference NB324K cells released infectious virus to high levels in the medium, whereas the yield was substantially less in C6 cells and not significant or undetectable in the rest of the cell lines (Fig. 1B, right). Thus, these rates of MVMp and MVMi infectious virus production correlated with the CPE arising in the respective cultures days after inoculation at a low MOI (Fig. 1A). A second parameter studied was the susceptibility of the cells to the virus interaction at different MOI, measured in a clonogenic assay. The percent survival of the cells to MVMp (Fig. 1C, left) was inversely proportional to the level of MVMp production in the respective cultures (Fig. 1B, left), allowing the establishment of two categories of cells for this virus: productive and sensitive versus poorly productive and resistant. For MVMi, a correlation between virus production and cytotoxic interaction was only found for the susceptible NB324K and C6 cells and the resistant SK-N-SH cells (Fig. 1B and C, right). In contrast, the U373, U87, and SW1088 astrocytic tumor cells showed a high sensitivity to MVMi (Fig. 1C, right), even though they behaved as poorly productive or nonproductive cells (Fig. 1B, right). Therefore, these experiments showed a differential capacity of MVMp and MVMi to multiply in and to kill transformed nervous cells and indicated that the MVMi life cycle undergoes a sharply abortive course in the human astrocytoma and glioblastomas, since a highly cytotoxic interaction was not accompanied by significant virus production in these cells.
Uneven macromolecular biosynthesis of parvovirus MVM in nervous tumor cells.
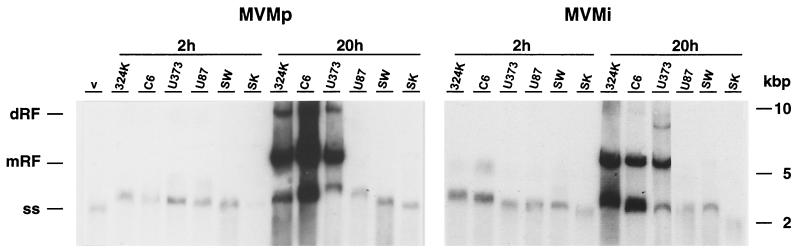
To investigate the molecular bases of the distinct MVM infections, we performed a quantitative analysis of the biosynthesis of viral macromolecular components in a single round of infection. Viral DNA synthesis analyzed by Southern blotting showed early in the virus-cell interactions (2 hpi) a similar amount of single-stranded input viral genome associated to the entire set of cell lines used (Fig. 2), an indication that virions of both MVM strains can attach to them. At a later time (20 hpi), only the NB324K, C6, and U373 cells denoted permissiveness for a high level of synthesis of MVMp and MVMi DNA replicative intermediates and genomic forms (Fig. 2). This result was in agreement with the productive character of these cells both for MVMp and, with the exception of U373, for MVMi as well (Fig. 1B). However, MVMi failed to amplify DNA to easily detectable levels in any of the other human cell lines (Fig. 2, right), in spite of the fact that it efficiently killed the U87 and SW1088 astrocytic tumor cells (Fig. 1C).
FIG. 2.
DNA amplification of MVM strains in transformed nervous cells. The cell lines were infected at 5 PFU/cell with MVMp or MVMi, and low-molecular-weight DNA was isolated at the indicated postinfection times. Samples from 105 infected cells were resolved in 1% agarose gels, blotted onto a membrane, hybridized to a 32P-labeled MVM probe, and exposed overnight to autoradiography at room temperature. dRF and mRF, MVM dimeric and monomeric replicative intermediates, respectively; ss, genomic ssDNA; v, DNA isolated from purified virions. 324K, NB324K; U373, U-373 MG; U87, U-87 MG; SW, SW1088; SK, SK-N-SH.
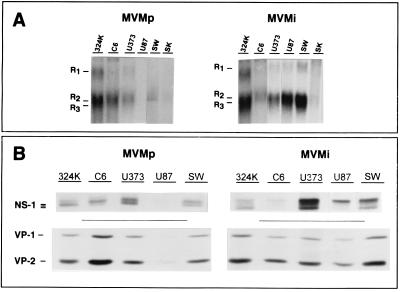
The extent of expression of the MVMp and MVMi genomes was next explored. The transcription of the MVMp genes, measured as the accumulation of viral messenger RNAs (R1, 4.8 kb; R2, 3.3 kb; R3, 3.0 kb), reached high levels in its three highly productive cell lines, reached very low levels in SW1088 and SK-N-SH cells, and was undetectable in U87 cells (Fig. 3A, left). MVMi mRNA species accumulated to high levels in all the cell lines tested except in the SK-N-SH neuroblastoma (Fig. 3A, right), in good correlation with the cytotoxic character of those interactions (Fig. 1C, right). The analysis of viral proteins synthesis performed with specific antisera showed several species of the NS1 protein, as well as the structural (VP1 and VP2) proteins, in all the cells in which transcription was evidenced (Fig. 3B). Thus, MVMp proteins were not detected in U87 cells and SK-N-SH was the only cell line negative for protein synthesis of both viral strains (not shown). There was a general correspondence between mRNA accumulation and the level of protein synthesis, though a relatively higher level of MVMp and MVMi protein synthesis was denoted in U373 cells. Altogether, this study demonstrated a good correlation between cell permissiveness to MVMp macromolecular biosynthesis and virus production, and for MVMi it evidenced the nature of the two types of nonproductive cytotoxic infections of human astrocytic tumor cells: normal levels of synthesis of all the viral macromolecules in U373 cells and high levels of viral messengers accumulation and protein synthesis in the absence of a proportional level of DNA amplification in U87 and SW1088 cells.
FIG. 3.
MVM gene expression in transformed nervous cells. Cells were infected at 5 PFU/cell with MVMp or MVMi and were analyzed for viral transcription and protein synthesis in the first round of infection. (A) Northern blot analysis of messengers accumulation at 15 hpi. Five micrograms of total RNA was loaded per slot, and filters were exposed for autoradiography for 2 days with an intensifying screen at −70°C. The positions of the three R1, R2, and R3 viral mature transcripts are indicated. R2 and R3 are not well resolved due to their similar size. (B) PAGE analysis of MVM nonstructural (NS1) and structural (VP1 and VP2) protein synthesis. Cells were labeled 12 to 20 hpi with [35S]Met-Cys, and proteins were immunoprecipitated with specific antibodies. An equivalent of 105 infected cells was loaded per slot. Gels were exposed for autoradiography for 4 days.
Virus particle formation in nonproductive MVMi infections.
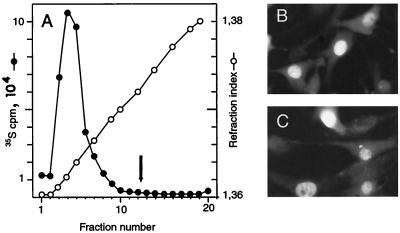
The apparent lack of infectious MVMi production in the medium of the human glioblastoma cultures prompted us to investigate the fate of the abundant viral proteins synthesized in these cells. To assess the capacity of the structural proteins to assemble in viral particles, 35S-labeled protein extracts of MVMi infected U373 and U87 cells (MOI of 5) were subjected to equilibrium centrifugation in CsCl gradients. An intracellular accumulation of empty (ρ = 1.32 g/ml) and DNA-full (ρ = 1.41 g/ml) particles at the normal rate found in MVM-infected cells (54) was obtained in U373 extracts (not shown), in agreement with the quantitative ssDNA production demonstrable in blots (Fig. 2) that is believed to result from viral genome encapsidation (23). As depicted in Fig. 4A, the U87 cells yielded also high levels of particles sedimenting at the position of empty capsids; however, no significant peak of 35S counts was detected at the banding position of the DNA-full virion. These empty particles showed all the features of the MVM capsids recovered from productively infected NB324K cells, namely hemagglutination activity of mouse erythrocytes, a complex pattern of phosphorylation demonstrable by two-dimensional tryptic analysis (52), and a protein composition of VP1 to VP2 at an approximate ratio of one to five (data not shown). The VP antigen accumulated in the nucleus of the infected U87 cells (Fig. 4B), the subcellular compartment where the MVMi assembly occurs (50). NS1, the major viral replicative polypeptide of MVM reported to translocate into the nucleus of murine and human permissive cells (22, 23), also accumulated normally into the nucleus of most U87 cells of the cultures (Fig. 4C). The intense and general nuclear staining indicated that most U87-infected cells expressed the structural and the NS1 cytototoxic protein, corresponding with the high proportion of these cells killed by the MVMi infection in clonogenic assays (Fig. 1C, left). These data showed that none of the evaluated protein properties, including level of synthesis, nuclear transport and capsid assembly, accounts for the abortive character of the MVMi infection in U87 cells.
FIG. 4.
Capsid formation and nuclear localization of MVMi proteins in U87 glioblastoma cells. (A) Equilibrium centrifugation of 35S-labeled MVMi particles produced in U87 cells. The arrow marks the main banding position of the DNA full virions (ρ = 1.41 g/ml). Subcellular localization of VP structural proteins (B) and of the NS-1 nonstructural protein (C) of MVMi in U87 cells infected at an MOI of 5 and stained by IF with specific antibodies.
MVMi replication in U87 glioblastoma is blocked after DNA conversion step.
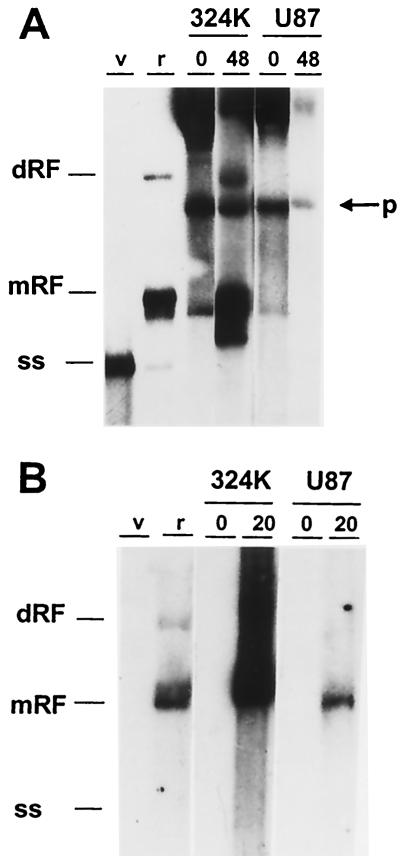
The apparent lack of MVMi DNA amplification in U87 (Fig. 2) was inconsistent with the high level of viral gene expression reached in these cells (Fig. 3), given that MVM transcription initiation requires the conversion of the incoming ss viral genome to a dsDNA template (5), allowing the onset of P4 promoter activity. To get insights into the mechanisms restricting MVMi DNA amplification in U87 cells, the synthesis of the viral intermediate replicative forms in transfected and infected U87 cells was carefully examined. The MVMi infectious plasmid clone was efficiently replicated in transfected control permissive NB324K cells, as the viral ss genomes and the replicative intermediates (mRF and dRF) accumulated by 48 h posttransfection (Fig. 5A). In contrast, none of these viral DNA molecules was resolved in transfected U87 cells, and the signal of the plasmid weakened with time (Fig. 5A).
FIG. 5.
Analysis of MVMi DNA amplification blockade in U87 glioblastoma cells. (A) Cell monolayers were transfected by the calcium phosphate procedure with an infectious MVMi plasmid, and viral DNA synthesis determined by blot hybridization with a ds full-length MVM probe to low-molecular-weight DNA isolated at the indicated posttransfection times. Autoradiography was for 2 days with intensifying screen at −70°C. p, input plasmid. (B) Cells were infected with MVMi at 5 PFU/cell, and low-molecular-weight DNA was isolated at 20 hpi and blot hybridized with a negative-sense riboprobe. Samples from 105 infected cells were loaded per slot. Filters were exposed for autoradiography for 14 h (NB324K [324K]) or for 120 h (U87) at room temperature. Note the lack of detection of the negative sense viral genome. dRF and mRF, replicative intermediates; ss; genomic ssDNA. Markers for panels A and B were used in similar amounts: v, MVMi genome isolated from purified virions; r, replicative forms isolated from MVMi-infected EL-4 lymphoma cells.
To investigate whether the input ssDNA of MVMi virions was copied into dsDNA in infected U87 cells, low-molecular-weight DNA isolated at 20 hpi was hybridized to an MVM strand-specific riboprobe labeled to high specific activity. The negative-sense probe failed to recognize the ssDNA isolated from purified virus, which is consistent with the reported preferable encapsidation of negative-sense DNA by the MVM virions (23, 28). The abundant replicative intermediates originated in the permissive NB324K cells could be easily detected with this probe in a short exposure (Fig. 5B). Interestingly enough, in the U87 cells infected by MVMi at an MOI of 5, a low copy number of mRF could be detected upon a much longer exposure of the films (Fig. 5B, right), but larger replicative forms (as the dRF intermediate) were not synthesized to a comparable level. This result showed that the input ssDNA virus genome can be converted to monomeric dsDNA forms in the nonpermissive U87 cells. The conversion reaction provides a template from which the MVMi transcription and protein expression found in U87 cells (Fig. 3) can be explained. Thus, the block hampering MVMi DNA amplification in U87 cells operated on transfected plasmid as well as on ssDNA genomes delivered by the viral particle at a postconversion step.
NS gene of MVMp enhances MVMi multiplication in U373 human glioblastoma.
The features of MVMi abortive infection in U373 and U87 cells drew our interest to the viral nonstructural (NS1 and NS2) proteins, as known mediators of many replicative and maturation functions during MVM infection. The U373 cells, in which the MVMp fully productive cycle (Fig. 1 to 3) ensures functional NS proteins and proper collaboration of cellular factors, provided a system in which a hypothetical involvement of NS proteins in MVMi abortive infections could be tested. For this purpose, an intertypic recombinant virus carrying most of the NS1 and NS2 coding sequence (NSp) of MVMp cloned into the MVMi genome background (MVMi-NSp) was constructed, grown in and purified from NB324K cells, and used for comparison of its DNA replication and maturation capacities in glioblastoma cells infected at different MOI with those of the parental MVMi. This recombinant virus showed, very much like MVMp, a larger plaque size in NB324K cells (not shown). At an MOI of 0.1, both viruses synthesized by 24 hpi abundant DNA replicative intermediates (mRF and dRF) and virus genomes (ss) in U373 cells (Fig. 6A, lane 2). In U87 cells, however (Fig. 6A, lanes 3 and 4), no significant viral DNA synthesis was detected besides the conversion of the input ss genomes into mRF forms. Similarly, at an MOI of 5, the synthesis of replicative intermediates and of ss forms corresponding to encapsidated genomes occurred to comparably high levels for both viruses only in U373 cells (Fig. 6A, right). Thus, the activities endowed by the NS coding sequence of MVMp (NSp) maintained the level of MVMi DNA replication in U373 but did not overcome the restrictions imposed by the U87 cells.
The effect that the NSp sequence exerts on the completion of the MVMi life cycle was evaluated by monitoring infectious virus production and viral progression in cultures infected at different MOI. As depicted in Fig. 6B, a significant production of MVMi was detected in total homogenates of U373 cells infected at an MOI of 0.1 or 5, reaching a titer of up to a 100-fold increase with respect to that of the input virus by 72 hpi (Fig. 6B, top). Remarkably, the production of infectious MVMi-NSp virus in U373 cultures was significantly higher at any postinfection time, reaching a titer by 72 hpi of a 5-logarithmic-unit increase with respect to that of the inocula at the two MOI tested. In contrast, the U87 cells did not yield infectious particles of any type of virus at both MOI (Fig. 6B, bottom). In agreement with these results, the MVMi progression in the cultures, monitored by the number of cells showing IF nuclear staining of expressed VP proteins, was high in NB324K cells and low in U373 cells (Fig. 6C). The number of IF+ adhered U87 cells slightly decreased with the time, though a punctuated cytoplasmic capsid staining with nuclear exclusion was evident at 72 hpi in virtually all the U87 cells in the culture (not shown). The MVMi-NSp virus also synthesized capsid proteins that localized in the nucleus of the U373 and U87 cells, and like the parental virus, it progressed in NB324K and U373 cultures but not in U87 cultures. However, this recombinant virus actually progressed more efficiently than the parental virus in both cell types, and the number of cells showing nuclear VP staining increased by a factor of 12 in U373 cells by 72 hpi. These results indicated that the NS coding region of MVMp enables MVMi to enhance its multiplication and maturation capacities in the U373 human astrocytic cells.
DISCUSSION
Intracellular host range restrictions to parvovirus infections.
This report adresses a comprehensive assessment of the interaction of the parvovirus MVMp and MVMi strains with transformed cells of nervous system origin. The characteristics of the infections were remarkably diverse within the series of cells tested (Fig. 1). Indeed, both viruses completed a cytotoxic and productive cycle exclusively in the rat glioma C6 cells. In the human cells, the astrocytoma U373 was the only nervous cell line fully permissive to MVMp, and the rest behaved as resistant and nonproductive hosts for this viral strain. MVMi did not complete a productive cycle in any of the human nervous cells, but its interaction with the three human tumor astrocytic cells (U373, U87, and SW1088) led to highly cytotoxic infections demonstrable in clonogenic assays (Fig. 1C, right) noncorresponded with a significant production of infectious virus in the culture media (Fig. 1B, right). The consistent evidence on distinct types of infection caused by each strain of this murine parvovirus in the human transformed nervous cells prompted us to further analyze this system in search of novel mechanisms of parvovirus host range determination.
At the molecular level, the MVMi abortive infections of the astrocytic tumor cells were not restricted at the same step of virus life cycle. In U373 cells, the virus macromolecular synthesis was complete and occurred to high levels (Fig. 2 and 3), but the production of mature infectious virions was low (Fig. 6B) and the exit from the cells to the medium very inefficient (Fig. 1B, right), hampering the progression of the infection in the culture (Fig. 6C). In the MVMi cytotoxic and abortive interaction with the human U87 and SW1088 cells, the block of the virus cycle was at an earlier step. These cells tolerated high levels of viral transcription and protein synthesis but no significant DNA amplification (Fig. 2 and 3), as the conversion reaction of the input viral genome to mRF could only be detected in blots hybridized with high-specific-activity probes (Fig. 5B and 6A). The NS1 and the structural viral proteins translocated and accumulated efficiently in the nucleus in the cellular population, forming empty capsids (Fig. 4) with a normal VP1-to-VP2 ratio. Even in the U87 cultures infected at a low MOI (Fig. 6C), most cells showed at a late postinfection time a characteristic-punctuated cytoplasmic staining for MVMi structural proteins, presumably due to the capacity of the empty capsids produced in high amounts by the few initially infected cells to reenter uninfected ones. Thus, all the analyses focused on capsid synthesis, composition, and properties suggest that it is the nonavailability of multimeric replicative DNA forms to be encapsidated what hampers virions formation in the U87 cells.
The main features of the MVMi life cycle in the human astrocytic tumor cells set these abortive infections apart from previous reports on parvovirus host range determination, in which the virus macromolecular synthesis was also investigated (2, 43, 63, 65, 90). Particularly in MVM, the best characterized system, the MVMp and MVMi nonproductive infections of mouse fibroblast and lymphoid cells are restricted prior DNA amplification, and neither transcription nor gene expression are detectable (2, 43, 78). The viral determinant allowing a productive interaction with fibroblasts, named the allotropic determinant (44), was mapped within a region of the capsid gene that acts with the incoming particle, as the restriction could be overcome by DNA transfection (43). Similar allotropic-like determinants mapping to the VP gene and operating by a few adjacent residues have been described for the PPV, ADV, and CPV parvoviruses (6, 7, 65, 66, 90). Capsid determinants may also underlie the lack of MVMp transcription and gene expression in U87 cells (Fig. 3) since the MVMi-NSp virus, harboring the NS gene of MVMp packaged into an MVMi coat, killed U87 cells (not shown) and expressed capsid proteins in the first round of infection (Fig. 6C). In contrast, although the abortive MVMi infection of the human U87 and SW1088 cells reported here was also restricted before DNA amplification (Fig. 2), viral transcription and gene expression proceeded normally (Fig. 3), and the restriction could not be overcome by transfection with an infectious molecular clone (Fig. 5A). Thus, in these human transformed nervous cells, the MVMi infection is restricted at the intracellular level by mechanisms in which the virus particle is not involved.
The unbalanced macromolecular biosynthesis in the abortive MVMi infection of U87 cells may shed light into the course of the viral life cycle along a nonrestricted infection. Indeed, the data indicate that high levels of transcription and gene expression may be reached by using as template the limited number of mRF molecules resulting from the conversion reaction. A similar phenomenon may occur in a normal productive infection, while the destiny of the dRF and higher concatemer replicative intermediates resulting from the virus DNA amplification might be the packaging into maturing capsids. This hypothesis is supported by the data obtained from dependovirus AAV vectors in which transduction was correlated with the conversion of the vector genome to dsDNA forms (36), as well as from the lack of correlation between gene expression and viral DNA amplification in MVM-based vectors (33).
Mechanisms of host range control by NS functions.
The intertypic recombinant MVMi-NSp virus amplified DNA and yielded ss genomic forms in the astrocytic U373 cells to the same high level as the parental MVMi (Fig. 6A), although the infectious titers attained by the virus (Fig. 6B) and the progression in U373 cultures infected at a low MOI (Fig. 6C) were significantly higher. As the ss genomes result from the encapsidation process (23), the yield of virus particles in U373 is the same in both viruses, and thus the specific infectivity of the newly formed MVMi-NSp virions must be severalfold higher. Therefore, the NSp gene enables an efficient production of infectious virus in U373 cells acting at as-yet-unidentified postencapsidation steps of the MVMi maturation process. A contribution of NS functions to the ordered genome packaging within the MVMi coat (1) may increase virion stability or uncoating in the subsequent infection. The copy of the NS1 protein placed outside the infectious virion covalently linked to the encapsidated 5′ end of the ss MVM genome (25), as a possible factor mediating the quality of the packaging steps, is a tentative hypothesis to be investigated.
On the contrary, the restrictions imposed by the human U87 and SW1088 astrocytic tumor cells to MVMi DNA synthesis could not be overcome by the NSp gene (Fig. 6), and no infectious virus was produced. Here, the mRF DNA of MVMi serves as adequate template for the transcription machinery (Fig. 3A), but the NS functions necessary for DNA amplification seemed to be hampered. NS1 as well as NS2 have been described to be necessary for viral DNA amplification and efficient virus production, although the NS2 activities were dispensable in human transformed cells (48, 58) and necessary for efficient messenger translation (48, 59), a defect not seen in U87 or SW1088 cells (Fig. 3). NS1, however, is necessary in any cell type for the replication steps following the conversion reaction that leads to DNA amplification (27, 60, 72), and thus it is the main candidate polypeptide for explaining the lack of MVMi genome amplification in the tumor astrocytic cells. What is particularly intriguing is why the NS1 protein of MVMi expressed to high levels in U87 and SW1088 cells and normally translocated to the nucleus failed to amplify DNA, even though the other well-established NS1 mediated functions, such as P38 promoter transactivation (32) or cytotoxicity, which indeed can be modulated in transformed cells (14, 57), were not affected (Fig. 1 to 4). Interestingly enough, NS1 replicative functions can be modulated in vitro by phosphorylation through members of the protein kinase C family (31, 61), and some of these phosphorylations may also take place in the viral infection (19). We are presently investigating whether the NS1 phosphorylation state differ between transformed cells. Alternatively, a number of proteins have been described to assist on NS1 functions (15–17, 24) which may be altered or expressed to lower levels in certain neoplastic cells. Given the few coding differences between the NS proteins of the MVM strains (3), the human tumor astrocytic cells provide an useful system to map those residues functionally involved in the different NS1 activities, as well as to work out components of the replication machinery of transformed cells recruited for parvovirus replication.
It is worth questioning whether the effects that the NS functions exert on MVM host range may be an exclusive phenomenon of the particular system of transformed nervous cells used in this report and are thus of poor biological interest for other viral-host interactions in the Parvoviridae. There are, however, previous genetic evidences for an involvement of NS sequences in parvovirus host range reported for PPV and MVM restricted infections (44, 90). In addition, MVMp may kill transformed fibroblasts independently of virus production (46), with gene expression uncoupled from DNA replication in some types of transformed fibroblasts (37). In the erythrovirus genus, the B19 parvovirus suppresses primary human megakaryocyte colony formation by a nonproductive interaction in which a low level of viral transcription was supported without DNA replication (79) and in nonpermissive cell lines blockades in viral transcript maturation (49) or capsid protein production (41) have been described. Furthermore, our results may provide a support of basic knowledge for the increasing evidences of the important roles that NS genes play on host range and tropism in diverse viral systems in vivo (e.g., references 13 and 62). In polyomavirus, in which the NS genes play host range functions as well (40, 42), the distribution of the initially targeted lung cells of inoculated mice showing viral DNA synthesis and proteins expression did not match (45). A dependence for NS functions and intracellular factors expressed at certain differentiation stages that restrict viral genome amplification may be a general phenomenon governing the tropism of the small DNA viruses.
ACKNOWLEDGMENTS
M.-P. Rubio and S. Guerra contributed equally to this work.
We are indebted to P. Tattersall for kindly providing the infectious molecular clones of MVMp and MVMi viruses and to J. Rommelaere and J. Cornelis for helpful comments.
This work was supported by grant SAF 98-0019 from the Comisión Interministerial de Ciencia y Tecnología and an institutional grant from Fundación Ramón Areces to the Centro de Biología Molecular “Severo Ochoa.” M.-P.R. was supported by a contract and S.G. was supported by a predoctoral fellowship, both from the Spanish Ministry of Education.
REFERENCES
- 1.Agbandje-McKenna M, LLamas-Saiz A, Wang F, Tattersall P, Rossmann M G. Functional implications of the structure of the murine parvovirus minute virus of mice. Structure. 1998;6:1369–1381. doi: 10.1016/s0969-2126(98)00137-3. [DOI] [PubMed] [Google Scholar]
- 2.Antonietti J-P, Sahli R, Beard P, Hirt B. Characterization of the cell type-specific determinant in the genome of minute virus of mice. J Virol. 1988;62:552–557. doi: 10.1128/jvi.62.2.552-557.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Astell C R, Gardiner E M, Tattersall P. DNA sequence of the lymphotropic variant of minute virus of mice, MVM(i), and comparison with the DNA sequence of the fibrotropic prototype strain. J Virol. 1986;57:656–669. doi: 10.1128/jvi.57.2.656-669.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ball-Goodrich L J, Tattersall P. Two amino acid substitutions within the capsid are coordinately required for acquisition of fibrotropism by the lymphotropic strain of minute virus of mice. J Virol. 1992;66:3415–3423. doi: 10.1128/jvi.66.6.3415-3423.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bashir T, Hörlein R, Rommelaere J, Willwand K. Cyclin A activates the DNA polymerase δ-dependent elongation machinery in vitro: a parvovirus DNA replication model. Proc Natl Acad Sci USA. 2000;97:5522–5527. doi: 10.1073/pnas.090485297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergeron J, Hébert B, Tijssen P. Genome organization of the kresse strain of porcine parvovirus: identification of the allotropic determinant and comparison with those of NADL-2 and field isolates. J Virol. 1996;70:2508–2515. doi: 10.1128/jvi.70.4.2508-2515.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bloom M E, Bradley D B, Wei W, Perryman S, Wolfinbarger J B. Characterization of chimeric full-length molecular clones of aleutian mink disease parvovirus (ADV): identification of a determinant governing replication of ADV in cell culture. J Virol. 1993;67:5976–5988. doi: 10.1128/jvi.67.10.5976-5988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bodendorf U, Cziepluch C, Jauniaux J-C, Rommelaere J, Salomé N. Nuclear export factor CRM1 interacts with nonstructural proteins NS2 from parvovirus minute virus of mice. J Virol. 1999;73:7769–7779. doi: 10.1128/jvi.73.9.7769-7779.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonnard G D, Manders E K, Campbell D A, Herberman R B, Collins M J. Immunosuppressive activity of a subline of the mouse EL-4 lymphoma. Evidence for minute virus of mice causing the inhibition. J Exp Med. 1976;143:187–205. doi: 10.1084/jem.143.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brockhaus K, Plaza S, Pintel D J, Rommelaere J, Salomé N. Nonstructural proteins NS2 of minute virus of mice associate in vivo with 14-3-3 protein family members. J Virol. 1996;70:7527–7534. doi: 10.1128/jvi.70.11.7527-7534.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown K E, Anderson S M, Young N S. Erythrocyte P antigen: cellular receptor for B19 parvovirus. Science. 1993;262:114–117. doi: 10.1126/science.8211117. [DOI] [PubMed] [Google Scholar]
- 12.Brownstein D G, Smith A L, Jacoby R O, Johnson E A, Hansen G, Tattersall P. Pathogenesis of infection with a virulent allotropic variant of minute virus of mice and regulation by host genotype. Lab Investig. 1991;65:357–363. [PubMed] [Google Scholar]
- 13.Brune W, Ménard C, Heesemann J, Koszinowski U H. A ribonucleotide reductase homolog of cytomegalovirus and endothelial cell tropism. Science. 2001;291:303–305. doi: 10.1126/science.291.5502.303. [DOI] [PubMed] [Google Scholar]
- 14.Caillet-Fauquet P, Perros M, Brandenburger A, Spegelaere P, Rommelaere J. Programmed killing of human cells by means of an inducible clone of parvoviral genes encoding non-structural proteins. EMBO J. 1990;9:2989–2995. doi: 10.1002/j.1460-2075.1990.tb07491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christensen J, Cotmore S F, Tattersall P. A novel cellular site-specific DNA-binding protein cooperates with the viral NS1 polipeptide to initiate parvovirus DNA replication. J Virol. 1997;71:1405–1416. doi: 10.1128/jvi.71.2.1405-1416.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christensen J, Cotmore S F, Tattersall P. Parvovirus initiation factor PIF: a novel human DNA-binding factor which coordinately recognizes two ACGT motifs. J Virol. 1997;71:5733–5741. doi: 10.1128/jvi.71.8.5733-5741.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christensen J, Cotmore S F, Tattersall P. Minute virus of mice initiator protein NS1 and host KDWK family transcription factor must form a precise ternary complex with origin DNA for nicking to occur. J Virol. 2001;75:7009–7017. doi: 10.1128/JVI.75.15.7009-7017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clemens K E, Pintel D J. The two transcription units of the autonomous parvovirus minute virus of mice are transcribed in a temporal order. J Virol. 1988;62:1448–1451. doi: 10.1128/jvi.62.4.1448-1451.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corbau R, Salome N, Rommelaere J, Nüesch J P F. Phosphorylation of the viral nonstructural protein NS1 during MVMp infection of A9 cells. Virology. 1999;259:402–415. doi: 10.1006/viro.1999.9786. [DOI] [PubMed] [Google Scholar]
- 20.Cornelis J J, Spruyt N, Spegelaere P, Guetta E, Darawshi T, Cotmore S F, Tal J, Rommelaere J. Sensitization of transformed rat fibroblasts to killing by parvovirus minute virus of mice correlates with an increase in viral gene expression. J Virol. 1988;62:3438–3444. doi: 10.1128/jvi.62.9.3438-3444.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cornelis J J, Becquart P, Duponchel N, Salome N, Avalosse B L, Namba M, Rommelaere J. Transformation of human fibroblasts by ionizing radiation, a chemical carcinogen, or simian virus 40 correlates with an increase in susceptibility to the autonomous parvovirus H-1 and minute virus of mice. J Virol. 1988;62:1679–1686. doi: 10.1128/jvi.62.5.1679-1686.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cornelis J J, Chen Y Q, Spruyt N, Duponchel N, Cotmore S F, Tattersall P, Rommelaere J. Susceptibility of human cells to killing by the parvovirus H-1 and minute virus of mice correlates with viral transcription. J Virol. 1990;64:2537–2544. doi: 10.1128/jvi.64.6.2537-2544.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cotmore S F, Tattersall P. The autonomously replicating parvoviruses of vertebrates. Adv Virus Res. 1987;33:91–173. doi: 10.1016/s0065-3527(08)60317-6. [DOI] [PubMed] [Google Scholar]
- 24.Cotmore S F, Tattersall P. High-mobility group 1/2 proteins are essential for initiating rolling-circle type DNA replication at a parvovirus hairpin origin. J Virol. 1988;72:8477–8484. doi: 10.1128/jvi.72.11.8477-8484.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cotmore S F, Tattersall P. A genome-linked copy of the NS-1 polypeptide is located on the outside of infectious parvovirus particles. J Virol. 1989;63:3902–3911. doi: 10.1128/jvi.63.9.3902-3911.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cotmore S F, Tattersall P. Alternate splicing in a Parvoviral nonstructural gene links a common amino-terminal sequence to downstream domains which confer radically different localization and turnover characteristics. Virology. 1990;177:477–487. doi: 10.1016/0042-6822(90)90512-p. [DOI] [PubMed] [Google Scholar]
- 27.Cotmore S F, Tattersall P. In vivo resolution of circular plasmids containing concatemere junction fragments from Minute Virus of Mice DNA and their subsequent replication as linear molecules. J Virol. 1992;66:420–431. doi: 10.1128/jvi.66.1.420-431.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cotmore S F, Tattersall P. DNA replication in the autonomous parvoviruses. Semin Virol. 1995;6:271–281. [Google Scholar]
- 29.Cotmore S F, Abramo J D, Carbonell L F, Bratton J, Tattersall P. The NS2 polypeptide of parvovirus MVM is required for capsid assembly in murine cells. Virology. 1997;231:267–280. doi: 10.1006/viro.1997.8545. [DOI] [PubMed] [Google Scholar]
- 30.Crawford L V. A minute virus of mice. Virology. 1966;29:605–612. doi: 10.1016/0042-6822(66)90284-4. [DOI] [PubMed] [Google Scholar]
- 31.Dettwiler S, Rommelaere J, Nuesch J P F. DNA unwinding functions of minute virus of mice NS1 protein are modulated specifically by the lambda isoform of protein kinase C. J Virol. 1999;73:7410–7420. doi: 10.1128/jvi.73.9.7410-7420.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doerig C, Hirt B, Beard P, Antonietti J P. Minute virus of mice non-structural protein NS-1 is necessary and sufficient for trans-activation of the viral P39 promoter. J Gen Virol. 1988;69:2563–2573. doi: 10.1099/0022-1317-69-10-2563. [DOI] [PubMed] [Google Scholar]
- 33.Dupont F, Tenenbaum L, Guo L-P, Spegelaere P, Zeicher M, Rommelaere J. Use of an autonomous parvovirus vector for selective transfer of a foreign gene into transformed human cells of different tissue origins and its expression therein. J Virol. 1994;68:1397–1406. doi: 10.1128/jvi.68.3.1397-1406.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dupressoir T, Vanacker J-M, Cornelis J J, Duponchel N, Rommelaere J. Inhibition by parvovirus H-1 of the formation of tumors in nude mice and colonies in vitro by transformed human mammary epithelial cells. Cancer Res. 1989;49:3203–3208. [PubMed] [Google Scholar]
- 35.Engers H D, Louis J A, Zubler R H, Hirt B. Inhibition of T-cell mediated functions by MVM(i), a parvovirus closely related to minute virus of mice. J Immunol. 1981;127:2280–2285. [PubMed] [Google Scholar]
- 36.Fisher K J, Gao G-P, Weitzman M D, DeMatteo R, Burda J F, Wilson J M. Transduction with recombinant adeno-associated virus for gene therapy is limited by leading-strand synthesis. J Virol. 1996;70:520–532. doi: 10.1128/jvi.70.1.520-532.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fox E, Moen P T, Jr, Bodnar J W. Replication of minute virus of mice DNA in adenovirus-infected or adenovirus-transformed cells. Virology. 1990;176:403–412. doi: 10.1016/0042-6822(90)90010-o. [DOI] [PubMed] [Google Scholar]
- 38.Fox J M, Bloom M E. Identification of a cell surface protein from crandell feline kidney cells that specifically binds aleutian mink disease parvovirus. J Virol. 1999;73:3835–3842. doi: 10.1128/jvi.73.5.3835-3842.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fox M J, McCrackin Stevenson M A, Bloom M E. Replication of aleutian mink disease parvovirus in vivo is influenced by residues in the VP2 protein. J Virol. 1999;73:8713–8719. doi: 10.1128/jvi.73.10.8713-8719.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Freund R, Bauer P H, Crissman A, Bradbury E M, Benjamin T L. Host range and cell cycle activation properties of polyomavirus large T-antigen mutants defective in pRB binding. J Virol. 1994;68:7227–7234. doi: 10.1128/jvi.68.11.7227-7234.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gallinella G, Manaresi E, Zuffi E, Venturoli S, Bonsi L, Bagnara G P, Musiani M, Zerbini M. Different patterns of restriction to B19 parvovirus replication in human blast cell lines. Virology. 2000;278:361–367. doi: 10.1006/viro.2000.0673. [DOI] [PubMed] [Google Scholar]
- 42.Garcea R L, Talmage D A, Harmatz A, Freund R, Benjamin T L. Separation of host range from transformation functions of the hr-t gene of polyomavirus. Virology. 1989;168:312–319. doi: 10.1016/0042-6822(89)90271-7. [DOI] [PubMed] [Google Scholar]
- 43.Gardiner E M, Tattersall P. Evidence that developmentally regulated control of gene expression by a parvoviral allotropic determinant is particle mediated. J Virol. 1988;62:1713–1722. doi: 10.1128/jvi.62.5.1713-1722.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gardiner E M, Tattersall P. Mapping of the fibrotropic and lymphotropic host range determinants of the parvovirus minute virus of mice. J Virol. 1988;62:2605–2613. doi: 10.1128/jvi.62.8.2605-2613.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gottlieb K, Villarreal L P. The distribution and kinetics of polyomavirus in lungs of intranasally infected newborn mice. Virology. 2000;266:52–65. doi: 10.1006/viro.1999.0030. [DOI] [PubMed] [Google Scholar]
- 46.Guetta E, Mincberg M, Mousset S, Bertinchamps C, Rommelaere J, Tal J. Selective killing of transformed rat cells by minute virus of mice does not require infectious virus production. J Virol. 1990;64:458–462. doi: 10.1128/jvi.64.1.458-462.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kimsey P B, Engers H D, Hirt B, Jongeneel V. Pathogenicity of fibroblast- and lymphocyte-specific variants of minute virus of mice. J Virol. 1986;59:8–13. doi: 10.1128/jvi.59.1.8-13.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li X, Rhode S L. Nonstructural protein NS2 of parvovirus H-1 is required for efficient viral protein synthesis and virus production in rat cells in vivo and in vitro. Virology. 1991;184:117–130. doi: 10.1016/0042-6822(91)90828-y. [DOI] [PubMed] [Google Scholar]
- 49.Liu J, Green S W, Takashi T, Young N S. A block in full-length transcript maturation in cells nonpermissive for B19 parvovirus. J Virol. 1992;66:4686–4693. doi: 10.1128/jvi.66.8.4686-4692.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lombardo E, Ramírez J C, Agbandje-McKenna M, Almendral J M. A β-stranded motif drives capsid protein oligomers of the parvovirus minute virus of mice into the nucleus for viral assembly. J Virol. 2000;74:3804–3814. doi: 10.1128/jvi.74.8.3804-3814.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lorson C, Pearson J, Burguer L, Pintel D J. An Sp1-binding site and TATA element are sufficient to support full transactivation by proximally bound NS1 protein of minute virus of mice. Virology. 1998;240:326–337. doi: 10.1006/viro.1997.8940. [DOI] [PubMed] [Google Scholar]
- 52.Maroto B, Ramírez J C, Almendral J M. The phosphorylation status of the parvovirus minute virus of mice particle: mapping and biological relevance of the major phosphorylation sites. J Virol. 2000;74:10892–10902. doi: 10.1128/jvi.74.23.10892-10902.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maxwell I H, Spitzer A L, Maxwell F, Pintel D J. The capsid determinant of fibrotropism for the MVMp strain of minute virus of mice functions via VP2 and not VP1. J Virol. 1995;69:5829–5832. doi: 10.1128/jvi.69.9.5829-5832.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McMaster G K, Beard P, Engers H D, Hirt B. Characterization of an immunosuppressive parvovirus related to the minute virus of mice. J Virol. 1981;38:317–326. doi: 10.1128/jvi.38.1.317-326.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miller R A, Ward D C, Ruddle F H. Embryonal carcinoma cells (and their somatic cell hybrids) are resistant to infection by the murine parvovirus MVM, which does infect other teratocarcinoma-derived cell lines. J Cell Physiol. 1977;91:393–402. doi: 10.1002/jcp.1040910309. [DOI] [PubMed] [Google Scholar]
- 56.Mousset S, Rommelaere J. Minute virus of mice inhibits cell transformation by simian virus 40. Nature (London) 1982;300:537–539. doi: 10.1038/300537a0. [DOI] [PubMed] [Google Scholar]
- 57.Mousset S, Ouadrhiri Y, Caillet-Fauquet P, Rommelaere J. The cytotoxicity of the autonomous parvovirus minute virus of mice nonstructural proteins in FR3T3 rat cells depends on oncogene expression. J Virol. 1994;68:6446–6453. doi: 10.1128/jvi.68.10.6446-6453.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Naeger L K, Cater J, Pintel D J. The small nonstructural protein (NS2) of the parvovirus minute virus of mice is required for efficient DNA replication and infectious virus production in a cell-type specific manner. J Virol. 1990;64:6166–6175. doi: 10.1128/jvi.64.12.6166-6175.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Naeger L K, Salomé N, Pintel D J. NS2 is required for efficient translation of viral mRNA in minute virus of mice-infected murine cells. J Virol. 1993;67:1034–1043. doi: 10.1128/jvi.67.2.1034-1043.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nüesch J P F, Cotmore S F, Tattersall P. Sequence motifs in the replicator protein of parvovirus MVM essential for nicking and covalent attachment to the viral origin: identification of the linking tyrosine. Virology. 1995;209:122–135. doi: 10.1006/viro.1995.1236. [DOI] [PubMed] [Google Scholar]
- 61.Nuesch J P F, Dettwiler S, Corbau R, Rommelaere J. Replicative functions of minute virus of mice NS1 protein are regulated in vitro by phosphorylation through protein kinase C. J Virol. 1998;72:9966–9977. doi: 10.1128/jvi.72.12.9966-9977.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Núnez J I, Baranowsky E, Molina N, Ruiz-Jarabo C M, Sánchez C, Domingo E, Sobrino F. A single amino acid substitution in nonstructural protein 3A can mediate adaptation of foot-and-mouse disease virus to the guinea pig. J Virol. 2001;75:3977–3983. doi: 10.1128/JVI.75.8.3977-3983.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oraveerakul K, Choi C-S, Molitor T W. Restriction of porcine parvovirus replication in nonpermissive cells. J Virol. 1992;66:715–722. doi: 10.1128/jvi.66.2.715-722.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Parker J S L, Murphy W J, Wang D, O'Brien S J, Parrish C R. Canine and feline parvoviruses can use human or feline transferrin receptors to bind, enter, and infect cells. J Virol. 2001;75:3896–3902. doi: 10.1128/JVI.75.8.3896-3902.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Parrish C R. Mapping specific functions in the capsid structure of canine parvovirus and feline panleukopenia virus using infectious plasmid clones. Virology. 1991;183:195–205. doi: 10.1016/0042-6822(91)90132-u. [DOI] [PubMed] [Google Scholar]
- 66.Parrish C R, Carmichael L E. Characterization and recombination mapping of an antigenic and host range mutation of canine parvovirus. Virology. 1986;148:121–132. doi: 10.1016/0042-6822(86)90408-3. [DOI] [PubMed] [Google Scholar]
- 67.Previsani N, Fontana S, Hirt B, Beard P. Growth of the parvovirus minute virus of mice MVMp3 in EL4 lymphocytes is restricted after cell entry and before viral DNA amplification: cell-specific differences in virus uncoating in vitro. J Virol. 1997;71:7769–7780. doi: 10.1128/jvi.71.10.7769-7780.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qing K, Mah C, Hansen J, Zhou S, Dwarki V, Srivastava A. Human fibroblast growth factor receptor 1 is a co-receptor for infection by adeno-associated virus 2. Nat Med. 1999;5:71–77. doi: 10.1038/4758. [DOI] [PubMed] [Google Scholar]
- 69.Ramírez J C, Fairén A, Almendral J M. Parvovirus minute virus of mice strain i multiplication and pathogenesis in the newborn mouse brain are restricted to proliferative areas and to migratory cerebellar young neurons. J Virol. 1996;70:8109–8116. doi: 10.1128/jvi.70.11.8109-8116.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ramírez J C, Santarén J A, Almendral J M. Transcriptional inhibition of the parvovirus Minute Virus of Mice by constitutive expression of an antisense RNA targeted against the NS-1 transactivator protein. Virology. 1995;206:57–68. doi: 10.1016/s0042-6822(95)80019-0. [DOI] [PubMed] [Google Scholar]
- 71.Rhode S L, Richard S M. Characterization of the trans-activation responsive element of the parvovirus H-1 P38 promoter. J Virol. 1987;61:2807–2815. doi: 10.1128/jvi.61.9.2807-2815.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rhode S L., III Both excision and replication of cloned automous parvovirus DNA require the NS1 (rep) protein. J Virol. 1989;63:4249–4256. doi: 10.1128/jvi.63.10.4249-4256.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rommelaere J, Tattersall P. Oncosuppression by parvoviruses. In: Tjissen P, editor. Handbook of parvoviruses. II. Boca Raton, Fla: CRC Press; 1990. pp. 41–57. [Google Scholar]
- 74.Salomé N, van Hille B, Duponchel N, Meneguzzi G, Cuzin F, Rommelaere J, Cornelis J J. Sensitization of transformed rat cells to parvovirus MVMp is restricted to specific oncogenes. Oncogene. 1990;5:123–130. [PubMed] [Google Scholar]
- 75.Santarén J F, Ramírez J C, Almendral J M. Protein species of the parvovirus minute virus of mice strain MVMp: involvement of phosphorylated VP-2 subtypes in viral morphogenesis. J Virol. 1993;67:5126–5138. doi: 10.1128/jvi.67.9.5126-5138.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Segovia J C, Gallego J M, Bueren J A, Almendral J M. Severe leukopenia and dysregulated erythropoiesis in SCID mice persistently infected by the parvovirus minute virus of mice. J Virol. 1999;73:1774–1784. doi: 10.1128/jvi.73.3.1774-1784.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Siegl G, Bates R C, Berns K I, Carter B J, Kelly D C, Kurstak E, Tattersall P. Characteristics and taxonomy of Parvoviridae. Intervirology. 1985;23:61–73. doi: 10.1159/000149587. [DOI] [PubMed] [Google Scholar]
- 78.Spalholz B A, Tattersall P. Interaction of minute virus of mice with differentiated cells: strain-dependent cell specificity is mediated by intracellular factors. J Virol. 1983;46:937–943. doi: 10.1128/jvi.46.3.937-943.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Srivastava A, Bruno E, Briddell R, Cooper R, Srivastava C, Van Besien K, Hoffman R. Parvovirus B19-induced perturbation of human megakaryocytopoiesis in vitro. Blood. 1990;76:1997–2004. [PubMed] [Google Scholar]
- 80.Studdert M J. Tissue tropism of parvoviruses. In: Tijssen P, editor. Handbook of Parvoviruses. II. Boca Raton, Fla: CRC Press; 1990. pp. 3–27. [Google Scholar]
- 81.Summerford C, Samulski R J. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J Virol. 1998;72:1438–1445. doi: 10.1128/jvi.72.2.1438-1445.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Summerford C, Bartlett J S, Samulski R J. ∝Vβ5 integrin: a co-receptor for adeno-associated virus type 2 infection. Nat Med. 1999;5:78–82. doi: 10.1038/4768. [DOI] [PubMed] [Google Scholar]
- 83.Tattersall P. Replication of parvovirus minute virus of mice. Dependence of virus multiplication and plaque formation on cell growth. J Virol. 1972;10:586–590. doi: 10.1128/jvi.10.4.586-590.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tattersall P, Gardiner E M. Autonomous parvovirus host-cell interactions. In: Tjissen P, editor. Handbook of Parvoviruses. Vol. 1. Boca Raton, Fla: CRC Press; 1990. pp. 111–121. [Google Scholar]
- 85.Tattersall P, Bratton J. Reciprocal productive and restrictive virus-cell interaction of immunosuppressive and prototype strains of minute virus of mice. J Virol. 1983;46:944–955. doi: 10.1128/jvi.46.3.944-955.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Telerman A, Tuynder M, Dupressoir D, Robaye B, Sigaux F, Shaulian E, Oren M, Rommelaere J, Amson R. A model of tumor suppression using H-1 parvovirus. Proc Natl Acad Sci USA. 1993;90:8702–8706. doi: 10.1073/pnas.90.18.8702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tennant R W, Laymant K R, Hand R E., Jr Effect of cell physiological state on infection by rat virus. J Virol. 1969;4:872–878. doi: 10.1128/jvi.4.6.872-878.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Toolan H W, Ledinko N. Inhibition by H-1 virus on the incidence of tumors produced by adenovirus 12 in hamsters. Virology. 1968;35:475–478. doi: 10.1016/0042-6822(68)90226-2. [DOI] [PubMed] [Google Scholar]
- 89.Tsao J, Chapman M S, Agbandje M, Keller W, Smith K, Wu H, Luo M, Smith T J, Rossmann M G, Compans R W, Parrish C R. The three-dimensional structure of canine parvovirus and its functional implications. Science. 1991;251:1456–1464. doi: 10.1126/science.2006420. [DOI] [PubMed] [Google Scholar]
- 90.Vasudevacharya J, Compans R W. The NS and capsid genes determine the host range of porcine parvovirus. Virology. 1992;187:515–524. doi: 10.1016/0042-6822(92)90454-w. [DOI] [PubMed] [Google Scholar]