Abstract
An immunodominant envelope glycoprotein is encoded by the human herpesvirus 8 (HHV-8) (also termed Kaposi's sarcoma-associated herpesvirus) K8.1 gene. The functional role of glycoprotein K8.1 is unknown, and recognizable sequence homology to K8.1 is not detectable in the genomes of most other closely related gammaherpesviruses, such as herpesvirus saimiri or Epstein-Barr virus. In search for a possible function for K8.1, we expressed the ectodomain of K8.1 fused to the Fc part of human immunoglobulin G1 (K8.1ΔTMFc). K8.1ΔTMFc specifically bound to the surface of cells expressing glycosaminoglycans but not to mutant cell lines negative for the expression of heparan sulfate proteoglycans. Binding of K8.1ΔTMFc to mammalian cells could be blocked by heparin. Interestingly, the infection of primary human endothelial cells by HHV-8 could also be blocked by similar concentrations of heparin. The specificity and affinity of these interactions were then determined by surface plasmon resonance measurements using immobilized heparin and soluble K8.1. This revealed that K8.1 binds to heparin with an affinity comparable to that of glycoproteins B and C of herpes simplex virus, which are known to be involved in target cell recognition by binding to cell surface proteoglycans, especially heparan sulfate. We conclude that cell surface glycosaminoglycans play a crucial role in HHV-8 target cell recognition and that HHV-8 envelope protein K8.1 is at least one of the proteins involved.
Human herpesvirus 8 (HHV-8), also termed Kaposi's sarcoma (KS)-associated herpesvirus, is the most recently discovered human herpesvirus (11). HHV-8 DNA is regularly present in all epidemiological forms of KS (2, 4, 7, 12, 15). In addition, HHV-8 DNA is also consistently found in primary effusion lymphomas (8, 9) and certain forms of multifocal Castleman's disease (47). A remarkably tight epidemiological relationship clearly suggests a pathogenetic role of HHV-8 in these malignant disorders. The nearly complete nucleotide sequence of this first human rhadinovirus has been determined from both a primary effusion lymphoma cell line (43) and a KS biopsy specimen (GenBank accession no. KSU75698). This showed that HHV-8 is a rhadinovirus or gamma-2 herpesvirus. Several animal rhadinoviruses are highly pathogenic upon infection of nonnatural hosts (18). In vivo, HHV-8 has been found in B cells and in KS spindle cells. The latter are derived from endothelial cells. Beyond this, the cell tropism of HHV-8 is not well characterized, and in cell culture the spectrum of cells that support lytic replication of HHV-8 appears to be rather limited. It is not clear whether this is due to restricted entry or to an intracellular block in replication at later stages of the infectious cycle. The cellular receptors and their viral ligands involved in target cell recognition by HHV-8 are unknown.
In terms of target cell recognition, the more distantly related gammaherpesvirus Epstein-Barr virus (EBV) is a much-better-studied example. Like in other viruses, target cell recognition by EBV can be separated into two sequential steps. The primary attachment of EBV to B lymphocytes is mediated by binding of the envelope glycoprotein gp350/220 to complement receptor 2 (CD21) (39, 52). Although EBV and HHV-8 belong to the same genus (gammaherpesviruses) and share most structural and many nonstructural genes, a homologue to the EBV glycoprotein gp350/220 has not been identified in the HHV-8 genome (37, 43; GenBank accession no. KSU75698). However, a nonconserved glycoprotein gene is present in all rhadinovirus genomes sequenced so far; this gene maps to a genomic position comparable to EBV open reading frame BZLF2 or BLLF1a/b, encoding glycoproteins gp42 and gp350/220, respectively. It is termed ORF51 in herpesvirus saimiri (3) or K8.1 in HHV-8 (40).
The HHV-8 glycoprotein K8.1 exists in two forms, termed K8.1α and K8.1β (40) or K8.1B and K8.1A (10), encoded by differentially spliced transcripts, with the larger one (K8.1β [K8.1A]) being predominant. It has been shown that the transmembrane glycoprotein K8.1 is part of the viral envelope (27). K8.1 is highly immunogenic in the natural host (40) and is frequently used in HHV-8 serologic assays (26, 49, 60). The physiological function of K8.1 or the other rhadinoviral glycoproteins encoded at comparable genomic positions has not been identified so far. Since K8.1 is a nonconserved virion glycoprotein and its genomic position hints at a distant relationship to glycoproteins of EBV involved in target cell recognition, we expressed soluble K8.1 and examined its binding to cultured mammalian cells. This article provides evidence that K8.1 binds with high affinity to cell surface heparan sulfate and that infection of endothelial cells by HHV-8 can be blocked by soluble heparin. In summary, we show that heparin-like moieties function as a receptor for HHV-8 and that K8.1 is at least one of the viral envelope proteins involved in this interaction.
MATERIALS AND METHODS
Cell lines and virus.
LM(tk−) murine fibroblasts (referred to here as mouse L cells) were obtained from the American Type Culture Collection (ATCC CCL-1.3). Mouse L cells are the parental cells line for mutants gro2C and sog9 (5) and gro2C-EXT1 and sog9-EXT1 (34), which were kindly provided by Frank Tufaro (University of British Columbia, Vancouver, Canada). The adherent mouse L, gro2C, and sog9 cell lines were cultured in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum, 350 mg of l-glutamine per ml, and 100 mg of gentamicin per ml. G418 was added at 700 or 500 μg/ml for the cultivation of gro2C-EXT1 or HEK 293T cells, respectively. Primary human dermal microvascular endothelial cells (HMVEC-d) were obtained from Clonetics Inc. (Walkerville, Md.) and propagated in EGM-2 MV bullet kit medium (Clonetics Inc.) according to the manufacturer's instructions. BCBL-1 cells were obtained from the AIDS Research and Reference Reagent Repository (Bethesda, Md.) and maintained in RPMI 1640 medium supplemented with 10% fetal calf serum, 350 mg of l-glutamine per ml, 100 μg of gentamicin per ml, 0.05 mM β-mercaptoethanol, and 1 mM sodium pyruvate.
Production of HHV-8 virions from the latently infected BCBL-1 cell line was induced with 12-O-tetradecanoylphorbol-13-acetate (TPA) (Sigma Chemical Co., St. Louis, Mo.) (25 ng/ml) and sodium butyrate (Sigma) for 4 days. Expression of late viral gene products was verified by indirect immunofluorescence using monoclonal antibody BS555 directed against the lytic HHV-8 antigen K8.1 (25). After 4 days of stimulation with TPA and sodium butyrate, cells and debris were removed by centrifugation for 10 min at 270 × g followed by 30 min at 2,820 × g. The supernatant was then sedimented at 12,500 × g for 3 h to pellet virions. The sediment was resuspended in OptiMEM (Gibco BRL, Rockville, Md.) to obtain a 200-fold concentration relative to the cell culture supernatant and stored at −80°C.
Construction, expression, and purification of recombinant proteins.
The pSecTag2/HygroB (Invitrogen Inc.)-based expression plasmid pAB68 contains sequences coding for the predicted extracellular domain of K8.1β (amino acids 26 to 196) (40) fused to the carboxy terminus of the 21-amino-acid murine immunoglobulin G kappa subunit ([IgG(κ)] signal peptide (PIR locus KVMS32) and the amino terminus of the Fc part from human IgG1 (GenBank accession no. S72664, amino acids 146 to 374). This plasmid was used to express the soluble ectodomain of K8.1 fused to the IgG1 Fc part (K8.1ΔTMFc), including a C-terminal Myc epitope. For cloning of the K8.1 cDNA fragment, RNA was extracted from TPA-induced BCBL-1 cells and reverse transcription-PCR was performed as described previously (40). Primers K8.1-Bg1 (GATCAGATCTTAACCATGAGTTCCACACAGATTC) and K8.1-Bg2 (GATCAGATCTATGGGTCCGTATTTCTGCATTG) were then used to amplify the sequences coding for the extracellular domains of K8.1. The abundant K8.1β form was cloned into the vector pVL1392-Fc (kindly provided by C. Ware) (32) via BglII restriction sites (underlined). The resulting K8.1-Fc fusion product was reamplified using primers K81rt-5 (CAGTGGATCCAATTGTCCACGTATCGTTC) and Fc-Xh1r (GATCCTCGAGATTTACCCGGAGACAGGGAG) and ligated via BamHI and XhoI restriction sites into the pSecTag2/HygroB plasmid (Invitrogen) in frame with the amino-terminal murine IgG(κ) signal peptide and the carboxy-terminal Myc/HIS epitope, present in the pSecTag2/HygroB vector. Plasmid pAB61 was used for the eukaryotic expression of the Fc part of human IgG1 fused to the Myc epitope sequence. To obtain pAB61, a DNA fragment coding for the Fc part was amplified from pVL1392-Fc using oligonucleotides Fc-N1 (GATCGCGGCCGCTGTGACAAAACTCACACATG) and Fc-Xh1r and cloned into plasmid pSecTag2/HygroB via NotI and XhoI restriction sites (underlined).
Both constructs were transiently transfected in HEK 293T cells (American Type Culture Collection) with Lipofectamine PLUS (Life Technologies) as recommended by the manufacturer. Cell culture supernatant was then collected daily for 7 days. Protein expression was confirmed by Western blot analysis using antibodies directed against the Fc part of human IgG1 (DAKO). After removal of cells and debris by centrifugation, K8.1ΔTMFc and Fc proteins were purified by affinity chromatography using HiTrap protein A columns (Pharmacia) as specified by the manufacturer.
IFA and binding studies.
Adherent cells (HMVEC-d, mouse L cells, and mouse L mutant cell lines) were seeded on glass coverslips and incubated at 37°C with 5% CO2 until 90% confluence was reached. Coverslips with adherent cells were washed twice with phosphate-buffered saline (PBS) and fixed for 30 min in PBS containing 3% paraformaldehyde. After fixation, cells were washed three times with PBS. Glycine at 100 mM was added for the second washing step. Suspension cells (BCBL-1 and BJAB cells) were fixed on immunofluorescence assay (IFA) slides using a mixture of 75% acetone and 25% methanol for 10 min at −20°C.
Prior to incubation with Fc fusion proteins or antibodies, fixed cells were incubated with PBS containing 1% bovine serum albumin for 30 min at room temperature. Incubation with the first antibody was performed for 30 min at room temperature and was followed by washing three times for 5 min each in PBS. As a first antibody, either mouse monoclonal antibody BS555 directed against HHV-8 protein K8.1 (25) or monoclonal antibody 9E10 directed against an epitope of the human c-myc proto-oncogene (ATCC CRL-1729) (17) was used. For the detection of mouse monoclonal antibodies, cells were then incubated with a sheep anti-mouse IgG–Cy3 conjugate (Sigma catalog no. C2181) diluted 1:300 in PBS, followed by three washing steps in PBS as described above.
To detect binding of K8.1ΔTMFc or Fc proteins alone, adherent cells were fixed as described above. In addition, cells were incubated with Cohn fraction II of human plasma (ICN Biochemicals) at 2 mg/ml for 30 min at room temperature prior to incubation with Fc/Fc fusion proteins to avoid nonspecific binding to cellular Fc receptor molecules. Incubation with Fc/Fc fusion proteins purified from transfected cells by protein A affinity chromatography was then performed for 3 h at 4°C. The purified proteins were used at a concentration of 50 μg/ml, followed by three washing steps at room temperature for 5 min each. Washing and incubation with primary antibody (mouse anti-Myc antibody 9E10; ATCC CRL-1729) (17) and secondary antibody (anti-mouse IgG–Cy3 conjugate; Sigma no. C2181) were then performed as described above. For competitive inhibition experiments, purified K8.1ΔTMFc or Fc proteins alone were incubated with heparin (heparin sodium salt from bovine intestinal mucosa; Sigma no. H-0777) and/or chondroitin sulfate A (sodium salt from bovine trachea; Sigma no. C-9891) at 0.1 or 1 mg/ml in PBS for 15 min at room temperature prior to incubation with the fixed cells.
Infection assays.
To infect HMVEC-d with HHV-8, the cells were seeded on glass coverslips in 12-well plates. Adherent cells were inoculated with 350 μl of 100-fold-concentrated supernatant of TPA- and butyrate-induced BCBL-1 cells for 30 min at 37°C. The cells were washed three time in PBS and incubated in the appropriate medium with 5% CO2 at 37°C. Medium was exchanged after 24 h. At 2 days postinfection, cells were harvested and IFA was performed as described above using monoclonal antibody BS555 directed against K8.1 (25). For blocking experiments with glycosaminoglycans, concentrated virus stock was incubated with heparin or chondroitin sulfate A prior to infection for 30 min at 37°C. For blocking of the infection with purified K8.1ΔTMFc or Fc proteins, the cells were incubated with the purified proteins dissolved in OptiMEM (Gibco Life Technologies) at 25, 50, and 100 μg/ml for 30 min at 37°C prior to infection with concentrated BCBL-1 supernatant. For quantitation of the infection, the number of K8.1-positive plaques per view field was counted at a 400-fold magnification. The mean and standard deviation of the number of plaques per field were calculated from three fields selected at random in each of three independent assays.
SPR measurement.
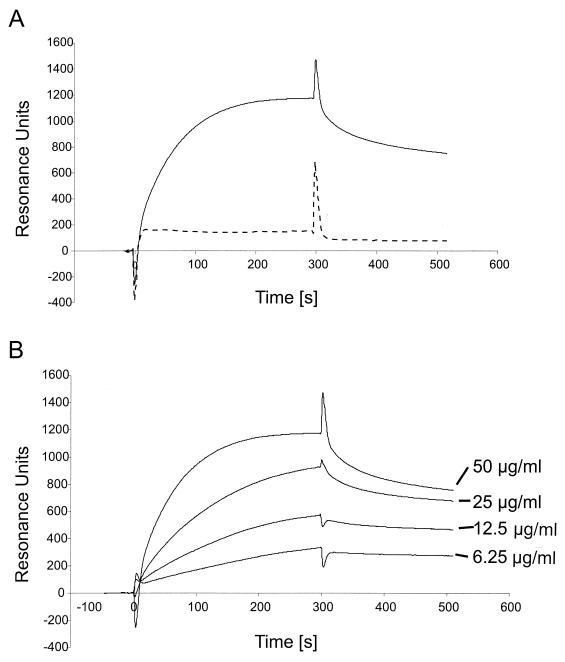
Surface plasmon resonance (SPR) experiments were performed on a BIAcore biosensor system using an SA (streptavidin-coated) biosensor chip (BIAcore AB). HBS running buffer consisted of 10 mM HEPES (pH 7.5), 0.15 M NaCl, 3.4 mM EDTA, and 0.005% Tween 20. Heparin was biotinylated and immobilized on the biosensor surface as described elsewhere (31). Briefly, heparin (bovine intestinal mucosa; Sigma) was dissolved in PBS at 20 mg/ml and mixed with a threefold molar excess of d-biotin-N-hydroxylsuccimide (Roche). After incubation for 60 min at room temperature, free biotin was removed on a NAP-5 column (Pharmacia). The biotinylated heparin was then coupled to flow cell 2 (Fc2) of the SA sensor chip by injecting 40 μl of a 25-μg/ml solution in PBS at a flow rate of 5 μl/min. This resulted in 160 resonance units (RU) of immobilized material. Flow cell 1 (Fc1) was used as reference to correct for changes in buffer composition and nonspecific binding to the sensor chip surface. For SPR measurements, 20 μl of either K8.1ΔTMFc or Fc alone diluted in PBS at various concentrations (see below) was injected at a flow rate of 4 μl/min. Following injection of the protein solution, the biosensor was rinsed with running buffer at the same flow rate for 200 s. The flow rate was then increased to 20 μl/min, and 10 μl of a 0.1 M NaOH–0.1% sodium dodecyl sulfate (SDS) solution was injected to regenerate the chip surface. For competitive binding assays, proteins (K8.1ΔTMFc or Fc alone, 25 μg/ml in PBS) were incubated with soluble glycosaminoglycans prior to injection into the biosensor chip. SPR data were analyzed with BIAevaluation 3.0 software (Biacore, Inc.). Briefly, for estimation of kon, the middle portion of the association curves (40 to 190 s in Fig. 5B) was used. For estimation of koff, the first part of the dissociation phase of the curve (315 to 415 s in Fig. 5B) was used. These kinetic data were fit most adequately by assuming a simple bimolecular reaction model for interaction between soluble analyte and immobilized ligand (Langmuir model). The goodness of fit was estimated by calculating χ2 values and inspecting residuals (difference between observed and calculated values).
FIG. 5.
(A) Binding of soluble K8.1 to a heparin-coated biosensor measured by SPR. The bindings of K8.1ΔTMFc (solid line) and Fc alone (dotted line) to a heparin biosensor are compared. Binding of K8.1 and Fc is shown as RU versus time in seconds. Both proteins were used at 50 μg/ml and injected at 4 μl/min onto the heparin-coated surface. The protein solution was injected for 300 s, followed by injection of running buffer at 4 μl/min for 250 s. The peak visible at 300 s is due to the change of the refractory index caused by replacing the buffer on the sensor chip. Due to the purification process, the buffer used to apply K8.1ΔTMFc differed slightly from the running buffer. (B) Binding of soluble K8.1 at concentrations of 6.25 to 50 μg/ml to a heparin-coated biosensor chip. K8.1ΔTMFc was injected at 4 μl/min for 300 s and reached a new equilibrium value with each higher concentration. Data from multiple runs without baseline subtraction are given as RU versus time in seconds. The peak at 300 s is due to the change of the refractory index when the protein solution was replaced with running buffer. The kinetic data shown in this figure were used to calculate the dissociation constants shown in Table 1.
Western blot analysis.
TPA-stimulated and nonstimulated BCBL-1 and BJAB cells were harvested by centrifugation (10 min, 400 × g), washed twice in PBS, and lysed in 2× SDS sample buffer (4% SDS, 10% β-mercaptoethanol, 20% glycerol, 2 mM EDTA, 120 mM Tris-HCl [pH 6.8], 0.1 mg of bromphenol blue/ml). An equivalent of 105 cells was loaded per lane. Cell culture supernatant from cells transfected with either pAB68 or pAB61 expression plasmids as well as proteins purified by protein A affinity chromatography were mixed directly with 5× SDS sample buffer. Proteins were separated on 10% (wt/vol) discontinuous SDS-polyacrylamide gels containing methylenebisacrylamide and acrylamide at a ratio of 1:29. Western blot analyses were carried out as described previously (38). Briefly, proteins were transferred from 10% discontinuous SDS-polyacrylamide gels onto nitrocellulose membranes using the Hoefer SemiPhor TE70 blotting apparatus as described by the manufacturer (Pharmacia Biotech, Uppsala, Sweden). The membranes were first blocked for 2 h at 20°C in blocking buffer (10 mM Tris [pH 7.5], 150 mM NaCl, 0.5% Tween, 5% low-fat milk). Membranes were then incubated for 2 h with monoclonal antibody BS555 (25) or 9E10 (17), directed against K8.1 or the Myc epitope, respectively. This was followed by three washes in TBS-Tween (10 mM Tris [pH 7.5], 150 mM NaCl, 0.5% Tween) and 1 h of incubation with horseradish peroxidase-conjugated anti-mouse IgG antibody diluted 1:2,000 in PBS (PO447; Dako Diagnostika GmbH, Hamburg, Germany). After washing three times in PBS, peroxidase activity was detected by electrochemiluminescence. For electrochemiluminescence, 100 ml of solution A (100 mM Tris-HCl [pH 8.6], 25 mg of Luminol [Sigma no. A4685], 31 μl of 30% H2O2) was mixed with 1% solution B (1.1 mg of para-hydroxycoumaric acid [Sigma no. C9008] dissolved in 1 ml of dimethyl sulfoxide) and the solution was immediately applied to the membranes, followed by exposure for 1 to 2 min. All steps were carried out at room temperature.
RESULTS
K8.1 binds to the surface of mammalian cells.
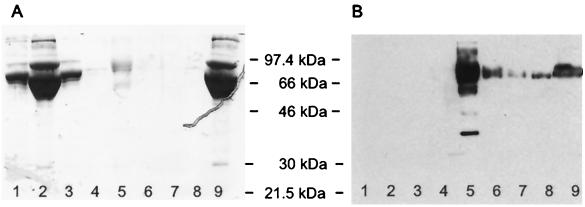
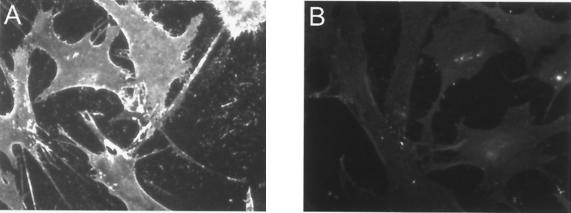
In EBV, glycoproteins gp350/220 and gp42 have been shown to be involved in binding to receptor molecules on the cell surface. Both proteins are not conserved in HHV-8 or other rhadinoviruses. Instead, the glycoprotein K8.1 is encoded at a genomic position that can be seen as being analogous to that for gp350/220. We therefore started our analysis of HHV-8 proteins involved in receptor binding with the more abundant of two K8.1 variants generated by alternative splicing, termed K8.1β (40) or K8.1A (10). The ectodomain domain of K8.1β (amino acids 27 to 196) was expressed fused to the amino terminus of human IgG1 Fc and a Myc epitope sequence. Without the 21 amino acids of the murine IgG(κ) signal peptide, the resulting protein (K8.1ΔTMFc) is 425 amino acids in length and has a calculated molecular mass of 47.1 kDa. As can be seen in Fig. 1, K8.1ΔTMFc was efficiently secreted into the supernatant of transfected 293T cells and could be purified by affinity chromatography with protein A-Sepharose (Fig. 1B, lane 5). The apparent molecular mass of approximately 70 kDa as observed in denaturing SDS-polyacrylamide gel electrophoresis is in good agreement with the expected molecular mass, given the high degree of glycosylation observed with the mature protein found in viral particles (25, 59). Protein preparations like the one shown in Fig. 1, lanes 5, were used to investigate the binding of K8.1 on the cell surface. As a control, the Fc part of human IgG was expressed and purified in the same way. HMVEC-d were used, as these cells are clearly susceptible to infection by HHV-8 both in KS lesions and in cell culture (6, 35, 41). The cells were grown on glass slides and fixed briefly with 3% paraformaldehyde to prevent internalization of bound proteins. Binding to Fc receptor molecules, which are known to be expressed on endothelial cells (21), was prevented by preincubation with Cohn fraction II from human plasma. Incubation with K8.1ΔTMFc or Fc alone was done at 4°C for 3 h. As can be seen in Fig. 2, binding of K8.1ΔTMFc (Fig. 2A) but not of the Fc fragment alone (Fig. 2B) was clearly detectable. An evenly bright staining could be observed along the plasma membrane of all endothelial cells with the Myc-tagged K8.1ΔTMFc. Similar experiments were repeated with a variety of mammalian cells, including 293 cells, BCBL-1 cells, baby hamster kidney (BHK-21) cells, rat mesangioma cells, and primary human fibroblasts. Clear and strong binding of K8.1ΔTMFc, but not of Fc alone, could invariably be detected (data not shown).
FIG. 1.
Purification of recombinant K8.1ΔTMFc fusion protein. Soluble K8.1 was expressed in a C-terminal fusion to the Fc part of human IgG1 containing a C-terminal Myc epitope (K8.1ΔTMFc). Both K8.1ΔTMFc and Fc alone (data not shown) were expressed in transiently transfected 293T cells and purified from the cell culture supernatant by affinity chromatography on protein A-Sepharose columns. An SDS-polyacrylamide gel stained with Coomassie brilliant blue (A) and a Western blot using a monoclonal antibody against K8.1 (BS555) (25) and a horseradish peroxidase-labeled secondary antibody against murine IgG (B) are shown. Lanes: 1, supernatant from 293T cells transfected with a negative control plasmid; 2, flowthrough supernatant after adsorption; 3 and 4, washing steps; 5 to 8, elution of K8.1ΔTMFc; 9, supernatant from 293T cells transfected with K8.1ΔTMFc expression plasmid prior to absorption.
FIG. 2.
Binding of K8.1 on the surface of HMVEC-d. HMVEC-d were fixed with paraformaldehyde and preincubated with Cohn fraction II to avoid binding of K8.1ΔTMFc to cellular Fc receptors. Cells were then incubated with either K8.1ΔTMFc (A) or Fc alone (B), followed by incubation with a monoclonal antibody directed to the C-terminal Myc epitope. Using an anti-mouse IgG antibody labeled with the fluorescent dye Cy3, binding of K8.1ΔTMFc but not of Fc alone could then be detected.
K8.1 binds to the surface of cells expressing heparan sulfate.
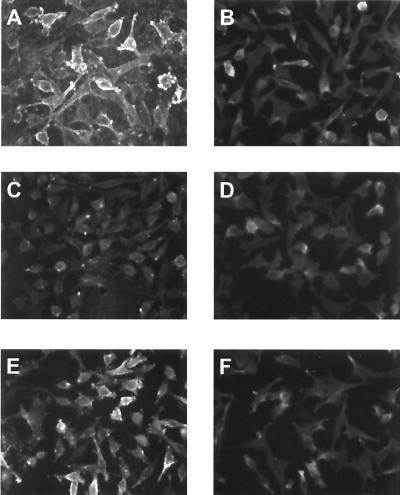
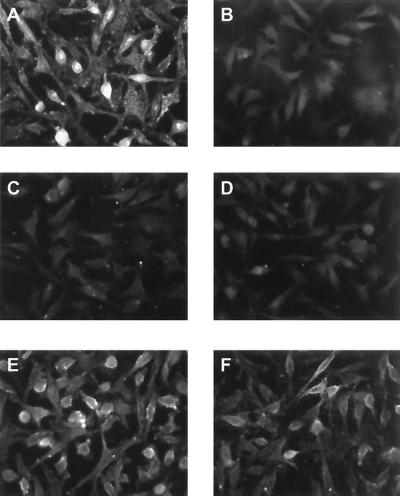
Obviously, K8.1 was able to bind to a wide spectrum of cells. The broad spectrum and the strong and relatively even staining pointed to a molecule with a wide expression pattern. Glycosaminoglycans like heparan sulfate and chondroitin sulfate are molecules fulfilling these criteria. They are involved in cell-cell adhesion and play an important role in the attachment of viruses as diverse as herpes simplex virus (HSV), dengue virus, and vaccinia virus. We thus examined whether K8.1ΔTMFc binding to cells lacking the expression of various glycosaminoglycans was also possible. The cell lines gro2C and sog9 are mutants of the mouse fibroblast L-cell line. Whereas mouse L cells express heparan sulfate and chondroitin sulfate, the mutant gro2C cells lack heparan sulfate expression (22), and neither heparan sulfate nor chondroitin sulfate is present on the surface of sog9 cells (5). The absence of heparan sulfate on these cells is due to a defect in the EXT1 gene resulting in the lack of d-glucuronic acid transferase activity (33). K8.1ΔTMFc binding assays on paraformaldehyde-fixed mouse L cells and their derivates were performed as described above after preincubation with Cohn fraction II. As can be seen in Fig. 3, K8.1ΔTMFc (Fig. 3A) but not Fc alone (Fig. 3B) clearly bound to the surface of mouse L cells. In contrast, none of these proteins could bind to the heparan sulfate-negative mutant gro2C (Fig. 3C and D). Similar data were obtained with the glycosaminoglycan-negative mutant cell line sog9 (data not shown). EXT1 is a glycosyltransferase that is required for the biosynthesis of heparan sulfate (28), and only a truncated, nonfunctional form of EXT1 is expressed in sog9 cells (33). Overexpression of EXT1 in gro2C and sog9 cells restores the biosynthesis of heparan sulfate but does not alter the expression of chondroitin sulfate (34). As shown in Fig. 3E and F, binding of K8.1ΔTMFc but not of the control protein (Fc only) (Fig. 3F) is also restored in gro2C-EXT1 cells. Similar results were obtained with the EXT1-overexpressing cell line sog9-EXT1. In summary, the ectodomain of the virion envelope protein K8.1 efficiently bound to a variety of cells, given that heparan sulfate was expressed. We conclude that the glycosaminoglycan heparan sulfate, but not chondroitin sulfate, is required for this interaction.
FIG. 3.
K8.1ΔTMFc binds on the surface of mouse L cells (A) but not on the gro2C mutant (C) lacking heparan sulfate. Cells were fixed with paraformaldehyde and treated with Cohn fraction II derived from human plasma. Cells were then incubated with K8.1ΔTMFc (A, C, and E) or Fc alone (B, D, and F). Binding was detected with a mouse monoclonal antibody against the Myc epitope which was present at the C termini of both K8.1ΔTMFc and Fc, followed by incubation with a Cy3 labeled anti-mouse IgG antibody. Specific binding of K8.1ΔTMFc (A), but not of Fc alone (B), could be detected on mouse L cells. In contrast, K8.1ΔTMFc could not bind to the gro2C mutant of mouse L cells (22) lacking heparan sulfate expression (C and D). However, when cell line gro2C-EXT1, which is a derivative of gro2C with partially restored expression of heparan sulfate (28), was used, specific binding of K8.1ΔTMFc could again be observed (E and F).
K8.1 binding to the cell surface is blocked by heparin.
To further prove the specificity of the interaction of K8.1β with cell surface heparan sulfate, competitive inhibition experiments were performed. We used heparin instead of heparan sulfate, as heparin has regularly been found to be a competitor of higher activity than heparan sulfate due to a different degree of acetylation and sulfatation (23). In addition, the composition of heparan sulfate is variable and depends on the source of isolation. Mouse L cells grown on glass slides were fixed with 3% paraformaldehyde, preincubated with Cohn fraction II to block binding to cell surface Fc receptors, and then incubated with either K8.1ΔTMFc (Fig. 4A, C, D, E, and F) or Fc alone (Fig. 4B). An experiment without competition by heparin or chondroitin sulfate is shown Fig. 4A and B. Whereas soluble K8.1 clearly bound to mouse L cells (Fig. 4A), cells incubated with Fc only remained negative (Fig. 4B). However, when the soluble K8.1 was preincubated with 0.1 or 1.0 mg of heparin per ml, K8.1ΔTMFc no longer bound to the surface of mouse L fibroblasts (Fig. 4C and D, respectively), indicating competitive inhibition of the K8.1 interaction by heparin. When chondroitin sulfate was used instead of heparin at 0.1 or 1.0 mg/ml, binding of K8.1ΔTMFc to the mouse L cells was still possible even at 1.0 mg/ml (Fig. 4E and F, respectively). These results are in good agreement with the data described above obtained using mouse L-cell mutants gro2C and sog9: binding of soluble K8.1 to these cells was not observed but could be rescued by overexpression of EXT1. The latter partially restores expression of undersulfated heparan sulfate but not chondroitin sulfate at the cell surface (34).
FIG. 4.
Binding of K8.1ΔTMFc to mouse L cells is blocked by heparin. Mouse L cells were fixed with paraformaldehyde and treated with Cohn fraction II from human blood. Prior to incubation with the cells, K8.1ΔTMFc (A, C, D, E, and F) or soluble Fc (B) was preincubated with either PBS (A and B), 0.1 mg of heparin per ml (C), 1.0 mg of heparin per ml (D), 0.1 mg of chondroitin sulfate per ml (E), or 1.0 mg of chondroitin sulfate per ml (F). Whereas heparin at both 0.1 and 1.0 mg/ml efficiently blocks binding of K8.1ΔTMFc to mouse L cells (C and D), binding is still possible in the presence of chondroitin sulfate A at up to 1.0 mg/ml (E and F).
Kinetics of K8.1 binding to heparin measured by SPR.
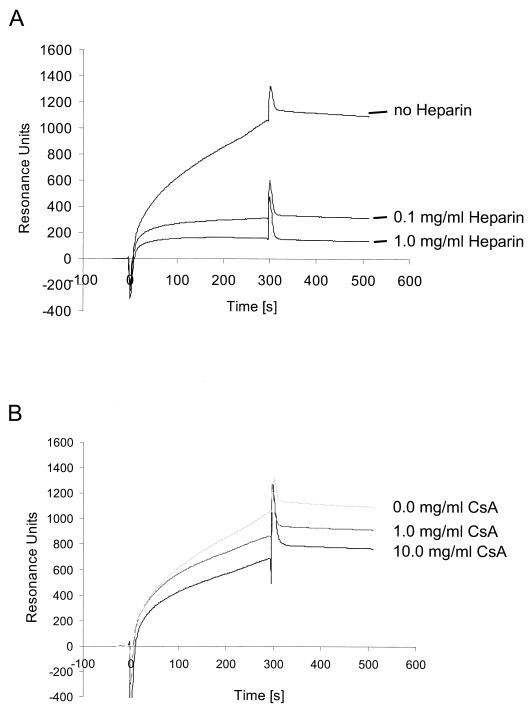
We used an SPR system to further analyze the binding of soluble K8.1 to glycosaminoglycans and calculate dissociation constants. Biotinylated heparin was immobilized on the surface of a streptavidin-coated biosensor chip and tested for the binding of soluble K8.1 (K8.1ΔTMFc) or Fc alone. A typical sensorgram is shown in Fig. 5A. Both proteins were applied at a concentration of 50 μg/ml with a flow rate of 4 μl/min. The sensorgram obtained with soluble K8.1 (Fig. 5A, continuous line) can be separated into three phases. From before the protein solution was injected at time zero on, the chip was under constant flow with running buffer. The change from running buffer to protein solution is marked by a short, sharp peak due to a change in refractory index caused by differences in buffer composition. The phase of association of K8.1 to the chip surface then takes place over the next 300 s, marked by a continuous increase of RU during the first 200 s until a maximum value of approximately 1,100 RU is reached. After injection of the protein solution, it was replaced by running buffer at 300 s, again with a sharp peak due to the change of buffering conditions. This is followed by a slow dissociation phase of 200 s with a moderate decrease of K8.1 binding resulting in a value of 800 RU. A completely different pattern is observed when soluble Fc is used instead of K8.1ΔTMFc (Fig. 5A, dotted line). The sharp peaks at 0 and 300 s, which are due to changes in buffer composition, are again visible. However, after a short initial increase to 180 RU due to increased refraction of the protein solution, no further increment of resonance is seen, indicating that the soluble Fc fragment alone does not bind to the heparin-coated surface. The results of a similar experiment are shown in Fig. 5B. Again, K8.1ΔTMFc solution was applied to a heparin-coated biosensor chip at a flow rate of 4 μg/ml for 300 s and then replaced by running buffer allowing for the dissociation of K8.1. Affinity constants were calculated from kinetic data (affinity constant [KD] = koff/kon), assuming a one-to-one interaction between the immobilized ligand (heparin) and soluble analyte (K8.1ΔTMFc). Using Biacore 3.0 evaluation software (see Materials and Methods), association and dissociation rates were calculated using the data from each of the four experiments shown in Fig. 5B. The maximum resonance signal increased in a dose-dependent manner from approximately 330 RU at 6.25 μg/ml to 1,180 RU at 50 μg/ml. For the ectodomain of K8.1β fused to the Fc part of human IgG1 used here, a mean KD of 4.8 × 10−8 M (range, from 2.3 × 10−8 M at 6.25 μg/ml to 7.7 × 10−8 M at 50 μg/ml) was calculated. The low χ2 values (0.118 to 0.168) and small residuals randomly distributed around zero indicated good agreement between the data and the Langmuir reaction model assumed here. The KD value of 4.8 × 10−8 M is well within the range of affinity constants observed for the binding of other viral ligands to their respective receptors, e.g., those of HSV type 2 (HSV-2) glycoprotein B to heparin (56), of HSV-1 glycoprotein D to the entry mediator (57), or of human immunodeficiency virus gp120 to heparin (36) (Table 1). Similar to the data obtained with mouse L cells and various mutants of glycosaminoglycan biosynthesis pathways, binding of K8.1 to a heparin-coated biosensor chip was efficiently competed by heparin at 0.1 mg/ml (Fig. 6A) but not by chondroitin sulfate A (Fig. 6B) or chondroitin sulfate B (data not shown) even at concentrations as high as 10 mg/ml.
TABLE 1.
Comparison of dissociation constants for several receptor-ligand pairsa
| Analyte | Immobilized receptor | KD (M) | Reference |
|---|---|---|---|
| HHV-8 K8.1-Fc | Heparin | 4.8 × 10−8 | |
| HIV-1 soluble gp120 | Heparin | 5.5 × 10−8 | 36 |
| HSV-2 soluble gB | Heparin | 7.7 × 10−7 | 56 |
| HSV-1 soluble gD | HveA | 3.2 × 10−6 | 57 |
| aFGF | Heparin | 9.5 × 10−8 | 31 |
| Biotin | Streptavidin | 10−15 | BIAcore |
The KD of soluble K8.1 and heparin was calculated from kinetic data (KD = koff/kon) obtained in SPR experiments using heparin-coated biosensor chips. A simple one-to-one reaction (Langmuir model) was used for the calculation. This model fitted sufficiently to the data, as can be concluded from the low χ2 values (0.118 to 0.168). HIV-1, human immunodeficiency virus type 1; aFGF, acidic fibroblast growth factor; HveA, herpesvirus entry mediator A.
FIG. 6.
Competitive inhibition of K8.1ΔTMFc binding to a heparin-coated biosensor surface by heparin (A) or chondroitin sulfate A (CsA) (B). Soluble heparin at concentrations of 0 to 1.0 mg/ml (A) or soluble chondroitin sulfate A at concentrations of 0 to 10.0 mg/ml (B) was mixed with soluble K8.1 before application to the heparin-coated biosensor chip. Whereas heparin efficiently blocked K8.1 binding at concentrations as low as 0.1 mg/ml, hardly any effect was seen with chondroitin sulfate A even at the highest concentration.
Inhibition of HHV-8 infection by heparin.
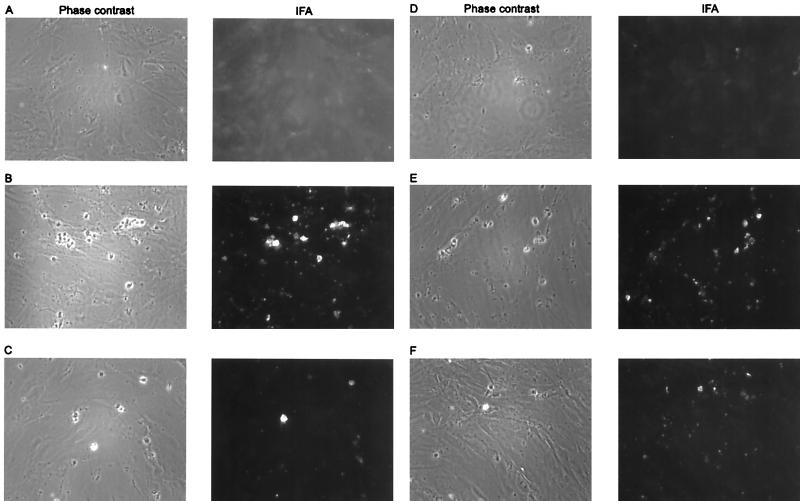
Having shown that the virion envelope protein K8.1β specifically binds to cell surface heparan sulfate with high affinity, we did experiments to evaluate the relevance of interaction with cell surface glycosaminoglycans for HHV-8 infection. Primary endothelial cells have been shown to be susceptible to HHV-8 infection. HMVEC-d were inoculated with HHV-8 concentrated from the supernatant of BCBL-1 cells induced with TPA and sodium butyrate. As can be seen in Fig. 7B and C, this resulted in lytic infection as evident from plaque formation (Fig. 7B, left) or from expression of the late viral protein(s) detected by immunofluorescence using a monoclonal antibody against K8.1 (Fig. 7B, right) (25). Recently, latent infection and spindle cell conversion in HMVEC-d upon infection with HHV-8 virions have been described (13). Although we used essentially the same type of HMVEC-d and the same culture conditions, we never observed this type of latent infection and associated morphological changes. In contrast, expression of lytic proteins was predominant in our hands.
FIG. 7.
HHV-8 infection of human endothelial cells is blocked by heparin. Concentrated supernatant (200-fold) from BCBL-1 cells stimulated with TPA was used to infect HMVEC-d. Prior to application on the HMVEC-d, the concentrated supernatant was mixed with PBS (B), 0.2 mg of heparin per ml (C), 2.0 mg of heparin per ml (D), 0.2 mg of chondroitin sulfate A per ml (E), or 2.0 mg of chondroitin sulfate A per ml (F). Uninfected cells are shown in panel A. Cells were fixed with paraformaldehyde at 2 days postinfection, and IFA was performed using a mouse monoclonal antibody against K8.1 and a Cy3-labeled secondary antibody against mouse immunoglobulin. Immunofluorescence pictures are shown on the left, and phase-contrast pictures of the same cells are shown on the right. Expression of the lytic K8.1 protein and a typical cytopathogenic effect were seen when the inoculum was treated with PBS (B) or chondroitin sulfate (E and F) prior to infection. However, when preincubation was done with heparin at either 0.2 mg/ml (C) or 2.0 mg/ml (D), the number and size of the plaques were clearly reduced.
However, when the inoculum was preincubated with heparin at 0.1 or 1.0 mg/ml, a marked decrease of plaque formation, by 69 and 87%, respectively, was observed by both phase-contrast microscopy (Fig. 7C [0.1 mg/ml] and D [1.0 mg/ml], left panels) and immunofluorescence using a monoclonal antibody against the lytically expressed K8.1 protein (Fig. 7C and D, right panels; Table 2). Formation of plaques was almost completely inhibited by heparin at 1.0 mg/ml. The specificity of this effect was again shown by using chondroitin sulfate A instead of heparin. At 0.1 mg/ml, no significant inhibition of plaque formation by chondroitin sulfate A was seen (Fig. 7E; Table 2). Even preincubation of the virus with chondroitin sulfate A at concentrations as high as 1 mg/ml resulted in only a moderate reduction of the number of plaques (Fig. 7F). Thus, at a concentration of 0.1 mg/ml, both infection of endothelial cells and binding of the virion envelope protein K8.1 are inhibited by heparin but not by chondroitin sulfate A. This points to a role of K8.1 in binding or entry of HHV-8 in the target cell. We thus examined whether infection of HMVEC-d by HHV-8 can also be blocked by soluble K8.1. When HMVEC-d were incubated with K8.1ΔTMFc at concentrations of 25, 50, and 100 μg/ml, no inhibition of the subsequent infection by HHV-8 could be seen even at the highest concentration of K8.1ΔTMFc (Table 2). Protein concentrations of above 100 μg/ml were not used, as nonspecific reduction of plaque formation by bovine serum albumin when used at higher concentrations has been observed for HHV-1 (51).
TABLE 2.
Inhibition of HHV-8 infection by glycosaminoglycansa
| Inhibitor | Concn (mg/ml) | No. of
plaques
|
% Inhibition | |
|---|---|---|---|---|
| Mean | SD | |||
| None (PBS) | 42.3 | 1.7 | 0 | |
| Heparin | 0.1 | 13.0 | 0.7 | 69 |
| 1.0 | 5.7 | 1.7 | 87 | |
| CsA | 0.1 | 41.0 | 3.0 | 3 |
| 1.0 | 23.7 | 4.6 | 44 | |
| K8.1ΔTMFc | 0.1 | 41.0 | NAb | 3 |
Concentrated supernatant (200-fold) from BCBL-1 cells stimulated with TPA was used to infect HMVEC-d. Prior to application on the HMVEC-d, the concentrated supernatant was mixed with PBS, heparin, or chondroitin sulfate A (CsA) in various concentrations. HMVEC-d were incubated with K8.1ΔTMFc for 30 min at 37°C prior to infection. Cells were fixed with paraformaldehyde at 2 days postinfection, and IFA was performed using a mouse monoclonal antibody against K8.1 and a Cy3-labeled secondary antibody against mouse immunoglobulin. The number of K8.1-positive plaques was counted in three randomly selected fields at a magnification of × 400. With the exception of blocking by K8.1ΔTMFc, experiments were done in triplicate. At a concentration of 0.1 mg/ml, a clear reduction of HHV-8 infection was observed when heparin, but not chondroitin sulfate A or soluble K8.1 protein, was used.
NA, not applicable.
DISCUSSION
Although in theory many different cell surface molecules could serve as viral receptors, the viral receptors identified so far appear to belong to only a few families (for a review, see reference 19).These include several members of the immunoglobulin superfamily, integrins, complement receptors, and cell surface glycosaminoglycans. Thus, unrelated viruses may use closely related or even identical receptors. On the other hand, closely related viruses may also use unrelated receptors to infect the same type of cell. Examples of the latter are HHV-6 and HHV-7. While HHV-7, like human immunodeficiency virus, employs the CD4 receptor to infect T lymphocytes (30), the closely related HHV-6 does not use this protein (29) but instead binds to the complement regulatory protein CD46 (44), which also serves as receptor for measles virus (16). Thus, one cannot infer the cellular receptor used by a virus from family relationships.
Binding of a virion protein to cell surface glycosaminoglycans is part of the target cell recognition process of many viruses of different families, with the best-examined example being heparan sulfate and its role in target cell recognition by HSVs (45, 48, 58). Several other viruses have also been found to interact with glycosaminoglycans. These include the Picornaviridae (foot-and-mouth disease virus), Togaviridae (Sindbis virus and dengue virus), Parvoviridae (adeno-associated virus 2), and Poxviridae (vaccinia virus). More recently, gp120 of human immunodeficiency virus has also been shown to bind to cell surface heparan sulfate, which enhances infectivity (36).
HHV-8 infects endothelial cells (6, 20) as well as B lymphocytes in vivo and in cell culture (42). There are only a few studies that deal with the identification of the full spectrum of cells which can be infected by HHV-8 (41). The cellular receptors and viral ligands involved in target cell recognition by HHV-8 are completely unknown.
In this report we clearly demonstrate that virus binding to heparin-like moieties on the cell surface is required for HHV-8 replication in susceptible HMVEC-d. We could efficiently block infection of these cells with heparin but not with chondroitin sulfate A, a related glycosaminoglycan. In addition, we show that the viral glycoprotein K8.1 is involved in this step, as (i) soluble K8.1 which was expressed in fusion to the Fc part of human IgG1 binds to a wide variety of mammalian cells, including mouse L cells, a murine fibroblast cell line; (ii) as described above for infection of endothelial cells, this interaction could be competitively inhibited by heparin but not by chondroitin sulfate A; (iii) K8.1 binding was not observed on a mouse L mutant cell line termed gro2C lacking heparan sulfate expression, but when gro2C cells were partially reconstituted for heparan sulfate expression by overexpression of the EXT1 tumor suppressor gene (34), binding of the K8.1 ectodomain could again be observed; and (iv) the affinity constant for binding of soluble K8.1 to heparin is well within the range observed for other viral glycoproteins binding to their receptors (Table 1), e.g., HSV-2 gB binding to glycosaminoglycans (56) or HSV-1 gD binding to the herpesvirus entry mediator (57).
Very little is known about the mechanisms of target cell recognition in any rhadinovirus. It has been shown for bovine herpesvirus 4 that interaction of glycoprotein 8, also termed gp135k, with heparin-like moieties on the cell surface is involved in attachment (54). However, the gene encoding gp8/gp135k has not yet been identified. EBV is the virus most closely related to HHV-8 for which some of the cellular receptors and their viral ligands have been identified. The EBV glycoproteins involved in attachment (gp350/220) and entry (gp42) by binding to CD21 (39) or the HLA DR β-chain (50), respectively, are not conserved in the HHV-8 genome. Moreover, heparan sulfate binding has not been shown to be of importance for EBV.
However, due to its genomic position, the gene encoding the immunogenic HHV-8 K8.1 (40) may be seen as distantly related to these two envelope proteins of EBV. A nonconserved transmembrane glycoprotein gene is present at an equivalent genomic position in all rhadinovirus genomes sequenced so far. A typical serine-threonine-rich stretch is present in the ectodomains of all of these glycoproteins, and this remote similarity is also found in the gp350/220 protein of EBV, pointing to a possible functional relationship. The evidence that binding of K8.1 to heparan sulfate is involved in target cell recognition by HHV-8 is also supported by the finding that a fully glycosylated form of K8.1 has been shown to be part of the virion envelope (27).
Although we were able to show that K8.1 clearly binds to heparan sulfate and heparan sulfate binding by HHV-8 is important for efficient infection, it remains an open question at which step of the infection process this interaction is important. Generally, infection of a cell by a virus can be divided into two steps. A first interaction of viral proteins with cellular receptors mediates attachment. Usually, this juxtaposes viral proteins and cellular coreceptors, enabling binding of the viral protein to the coreceptors, which mediates entry through membrane fusion or triggers release of the nucleic acid. In most cases, the interaction of a virus with glycosaminoglycans is important for the first step, termed attachment. This holds true for the interaction of HSV glycoproteins C and B with heparan sulfate. In the case of HSVs, membrane fusion is subsequently enabled by interaction of glycoprotein D with one of several herpesvirus entry mediators that belong to the family of nectins, such as the tumor necrosis factor receptors (14, 55). It should be noted in this context that glycoproteins C and D of HSVs are not conserved in HHV-8 or other gammaherpesviruses. Additional viral glycoproteins, especially gH and gL, are then required to initiate membrane fusion (53). However, it has also been shown that binding of gD to 3-O-sulfated heparan sulfate is able to mediate entry of HSV-1 (46). The efficient binding of K8.1 to cell surface glycosaminoglycans, its localization in the virion envelope, and the ability of heparin to block both binding of K8.1 to the cell surface and infection of endothelial cells by HHV-8 make it very likely that K8.1 mediates HHV-8 attachment. This does not imply that K8.1 is required for the attachment step. Our finding that infection of endothelial cells by HHV-8 cannot be blocked by soluble K8.1 indicates that K8.1 may not be essential for this step. Even at the highest concentration of K8.1 used, no reduction in plaque formation could be observed, and antibody BS555 directed against K8.1 was not able to inhibit infection. This is surprising only at first sight. A similar situation is observed in HSV, where binding to heparan sulfate greatly enhances infection. Notably, whereas soluble gC could compete with attachment of HSV-1, plaque formation was not be inhibited by gC when used at concentrations of up to 100 μg/ml (51). This is most likely due to the fact that HSV has evolved redundant mechanisms for the initial attachment step. Both glycoproteins B and C of HSV are able to bind to glycosaminoglycans, and gC of HSV is thus not essential for this first step (24). Indeed, preliminary data from this laboratory indicate that glycoprotein H when expressed in a soluble form is also able to bind to cell surface glycosaminoglycans (unpublished data), and a recent study indicated that HHV-8 glycoprotein B may bind to heparan sulfate (1). In summary, we show that cell surface heparan sulfate is required for efficient infection by HHV-8. The data presented indicate that the virion envelope protein K8.1 plays a role in the viral life cycle that is comparable to the function of HSV glycoprotein C.
ACKNOWLEDGMENTS
This work was supported by grant SFB466 from the German Research Foundation on Lymphoproliferation and Viral Immunodeficiency, by the Ria-Freifrau-von-Fritsch Foundation, and by the European Union Concerted Action on AIDS-Associated Kaposi's Sarcoma.
We thank Matthias Peipp for helpful hints and critical discussion.
REFERENCES
- 1.Akula S M, Pramod N P, Wang F Z, Chandran B. Human herpesvirus 8 envelope-associated glycoprotein B interacts with heparan sulfate-like moieties. Virology. 2001;284:235–249. doi: 10.1006/viro.2001.0921. [DOI] [PubMed] [Google Scholar]
- 2.Albini A, Aluigi M G, Benelli R, Berti E, Biberfeld P, Blasig C, Calabro M L, Calvo F, Chieco-Bianchi L, Corbellino M, Del Mistro A, Ekman M, Favero A, Hofschneider P H, Kaaya E, Lebbe C, Morel P, Neipel F, Noonan D M, Parravicini C, Repetto L, Schalling M, Stürzl M, Tschachler E. Oncogenesis in HIV-infection: KSHV and Kaposi's sarcoma. Int J Oncol. 1996;9:5–8. [PubMed] [Google Scholar]
- 3.Albrecht J C, Nicholas J, Biller D, Cameron K R, Biesinger B, Newman C, Wittmann S, Craxton M A, Coleman H, Fleckenstein B, Honess R W. Primary structure of the herpesvirus saimiri genome. J Virol. 1992;66:5047–5058. doi: 10.1128/jvi.66.8.5047-5058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ambroziak J A, Blackbourn D J, Herndier B G, Glogau R G, Gullett J H, McDonald A R, Lennette E T, Levy J A. Herpes-like sequences in HIV-infected and uninfected Kaposi's sarcoma patients. Science. 1995;268:582–583. doi: 10.1126/science.7725108. [DOI] [PubMed] [Google Scholar]
- 5.Banfield B W, Leduc Y, Esford L, Schubert K, Tufaro F. Sequential isolation of proteoglycan synthesis mutants by using herpes simplex virus as a selective agent: evidence for a proteoglycan-independent virus entry pathway. J Virol. 1995;69:3290–3298. doi: 10.1128/jvi.69.6.3290-3298.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boshoff C, Schulz T F, Kennedy M M, Graham A K, Fisher C, Thomas A, McGee J O, Weiss R A, O'Leary J J. Kaposi's sarcoma-associated herpesvirus infects endothelial and spindle cells. Nat Med. 1995;1:1274–1278. doi: 10.1038/nm1295-1274. [DOI] [PubMed] [Google Scholar]
- 7.Boshoff C, Whitby D, Hatziioannou T, Fisher C, van-der Walt J, Hatzakis A, Weiss R, Schulz T. Kaposi's-sarcoma-associated herpesvirus in HIV-negative Kaposi's sarcoma. Lancet. 1995;345:1043–1044. doi: 10.1016/s0140-6736(95)90780-7. [DOI] [PubMed] [Google Scholar]
- 8.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 9.Cesarman E, Moore P S, Rao P H, Inghirami G, Knowles D M, Chang Y. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood. 1995;86:2708–2714. [PubMed] [Google Scholar]
- 10.Chandran B, Bloomer C, Chan S R, Zhu L, Goldstein E, Horvat R. Human herpesvirus-8 ORF K8.1 gene encodes immunogenic glycoproteins generated by spliced transcripts. Virology. 1998;249:140–149. doi: 10.1006/viro.1998.9316. [DOI] [PubMed] [Google Scholar]
- 11.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 12.Chang Y, Ziegler J, Wabinga H, Katangole Mbidde E, Boshoff C, Schulz T, Whitby D, Maddalena D, Jaffe H W, Weiss R A, Moore P S. Kaposi's sarcoma-associated herpesvirus and Kaposi's sarcoma in Africa. Uganda Kaposi's Sarcoma Study Group Arch Intern Med. 1996;156:202–204. [PubMed] [Google Scholar]
- 13.Ciufo D M, Cannon J S, Poole L J, Wu F Y, Murray P, Ambinder R F, Hayward G S. Spindle cell conversion by Kaposi's sarcoma-associated herpesvirus: formation of colonies and plaques with mixed lytic and latent gene expression in infected primary dermal microvascular endothelial cell cultures. J Virol. 2001;75:5614–5626. doi: 10.1128/JVI.75.12.5614-5626.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Connolly S A, Whitbeck J C, Rux A H, Krummenacher C, Drunen Littel-van den Hurk S, Cohen G H, Eisenberg R J. Glycoprotein D homologs in herpes simplex virus type 1, pseudorabies virus, and bovine herpes virus type 1 bind directly to human HveC (nectin-1) with different affinities. Virology. 2001;280:7–18. doi: 10.1006/viro.2000.0747. [DOI] [PubMed] [Google Scholar]
- 15.de Lellis L, Fabris M, Cassai E, Corallini A, Giraldo G, Feo C, Monini P. Herpesvirus-like DNA sequences in non-AIDS Kaposi's sarcoma. J Infect Dis. 1995;172:1605–1607. doi: 10.1093/infdis/172.6.1605. [DOI] [PubMed] [Google Scholar]
- 16.Dörig R E, Marcil A, Chopra A, Richardson C D. The human CD46 molecule is a receptor for measles virus (Edmonston strain) Cell. 1993;75:295–305. doi: 10.1016/0092-8674(93)80071-l. [DOI] [PubMed] [Google Scholar]
- 17.Evan G I, Lewis G K, Ramsay G, Bishop J M. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fleckenstein B, Desrosiers R C. Herpesvirus saimiri and Herpesvirus ateles. In: Roizman B, editor. The herpesviruses. New York, N.Y: Plenum Press; 1982. pp. 253–331. [Google Scholar]
- 19.Flint S J, Enquist L W, Krug R M, Racaniello V R, Skalka A M. Virus attachment to host cells. In: Flint J S, Enquist L W, Krug R M, Racaniello V R, Skalka A M, editors. Principles of virology. Washington, D.C.: ASM Press; 2000. pp. 100–131. [Google Scholar]
- 20.Flore O, Rafii S, Ely S, O'Leary J J, Hyjek E M, Cesarman E. Transformation of primary human endothelial cells by Kaposi's sarcoma-associated herpesvirus. Nature. 1998;394:588–592. doi: 10.1038/29093. [DOI] [PubMed] [Google Scholar]
- 21.Groger M, Sarmay G, Fiebiger E, Wolff K, Petzelbauer P. Dermal microvascular endothelial cells express CD32 receptors in vivo and in vitro. J Immunol. 1996;156:1549–1556. [PubMed] [Google Scholar]
- 22.Gruenheid S, Gatzke L, Meadows H, Tufaro F. Herpes simplex virus infection and propagation in a mouse L cell mutant lacking heparan sulfate proteoglycans. J Virol. 1993;67:93–100. doi: 10.1128/jvi.67.1.93-100.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hallak L K, Collins P L, Knudson W, Peeples M E. Iduronic acid-containing glycosaminoglycans on target cells are required for efficient respiratory syncytial virus infection. Virology. 2000;271:264–275. doi: 10.1006/viro.2000.0293. [DOI] [PubMed] [Google Scholar]
- 24.Herold B C, WuDunn D, Soltys N, Spear P G. Glycoprotein C of herpes simplex virus type 1 plays a principal role in the adsorption of virus to cells and in infectivity. J Virol. 1991;65:1090–1098. doi: 10.1128/jvi.65.3.1090-1098.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lang D, Birkmann A, Neipel F, Hinderer W, Rothe M, Ernst M, Sonneborn H H. Generation of monoclonal antibodies directed against the immunogenic glycoprotein K8.1 of human herpesvirus 8. Hybridoma. 2000;19:287–295. doi: 10.1089/027245700429837. [DOI] [PubMed] [Google Scholar]
- 26.Lang D, Hinderer W, Rothe M, Sonneborn H H, Neipel F, Raab M S, Rabenau H, Masquelier B, Fleury H. Comparison of the immunoglobulin-G-specific seroreactivity of different recombinant antigens of the human herpesvirus 8. Virology. 1999;260:47–54. doi: 10.1006/viro.1999.9804. [DOI] [PubMed] [Google Scholar]
- 27.Li M, MacKey J, Czajak S C, Desrosiers R C, Lackner A A, Jung J U. Identification and characterization of Kaposi's sarcoma-associated herpesvirus K8.1 virion glycoprotein. J Virol. 1999;73:1341–1349. doi: 10.1128/jvi.73.2.1341-1349.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lind T, Tufaro F, McCormick C, Lindahl U, Lidholt K. The putative tumor suppressors EXT1 and EXT2 are glycosyltransferases required for the biosynthesis of heparan sulfate. J Biol Chem. 1998;273:26265–26268. doi: 10.1074/jbc.273.41.26265. [DOI] [PubMed] [Google Scholar]
- 29.Lusso P, Gallo R C, DeRocco S E, Markham P D. CD4 is not the membrane receptor for HHV-6. Lancet. 1989;1:730. doi: 10.1016/s0140-6736(89)92249-6. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 30.Lusso P, Secchiero P, Crowley R W, Garzino Demo A, Berneman Z N, Gallo R C. CD4 is a critical component of the receptor for human herpesvirus 7: interference with human immunodeficiency virus. Proc Natl Acad Sci USA. 1994;91:3872–3876. doi: 10.1073/pnas.91.9.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mach H, Volkin D B, Burke C J, Middaugh C R, Linhardt R J, Fromm J R, Loganathan D, Mattsson L. Nature of the interaction of heparin with acidic fibroblast growth factor. Biochemistry. 1993;32:5480–5489. doi: 10.1021/bi00071a026. [DOI] [PubMed] [Google Scholar]
- 32.Mauri D N, Ebner R, Montgomery R I, Kochel K D, Cheung T C, Yu G L, Ruben S, Murphy M, Eisenberg R J, Cohen G H, Spear P G, Ware C F. LIGHT, a new member of the TNF superfamily, and lymphotoxin alpha are ligands for herpesvirus entry mediator. Immunity. 1998;8:21–30. doi: 10.1016/s1074-7613(00)80455-0. [DOI] [PubMed] [Google Scholar]
- 33.McCormick C, Duncan G, Goutsos K T, Tufaro F. The putative tumor suppressors EXT1 and EXT2 form a stable complex that accumulates in the Golgi apparatus and catalyzes the synthesis of heparan sulfate. Proc Natl Acad Sci USA. 2000;97:668–673. doi: 10.1073/pnas.97.2.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCormick C, Leduc Y, Martindale D, Mattison K, Esford L E, Dyer A P, Tufaro F. The putative tumour suppressor EXT1 alters the expression of cell-surface heparan sulfate. Nat Genet. 1998;19:158–161. doi: 10.1038/514. [DOI] [PubMed] [Google Scholar]
- 35.Moses A V, Fish K N, Ruhl R, Smith P P, Strussenberg J G, Zhu L, Chandran B, Nelson J A. Long-term infection and transformation of dermal microvascular endothelial cells by human herpesvirus 8. J Virol. 1999;73:6892–6902. doi: 10.1128/jvi.73.8.6892-6902.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moulard M, Lortat-Jacob H, Mondor I, Roca G, Wyatt R, Sodroski J, Zhao L, Olson W, Kwong P D, Sattentau Q J. Selective interactions of polyanions with basic surfaces on human immunodeficiency virus type 1 gp120. J Virol. 2000;74:1948–1960. doi: 10.1128/jvi.74.4.1948-1960.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neipel F, Albrecht J C, Fleckenstein B. Cell-homologous genes in the Kaposi's sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J Virol. 1997;71:4187–4192. doi: 10.1128/jvi.71.6.4187-4192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neipel F, Ellinger K, Fleckenstein B. Gene for the major antigenic structural protein (p100) of human herpesvirus type 6. J Virol. 1992;66:3918–3924. doi: 10.1128/jvi.66.6.3918-3924.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nemerow G R, Mold C, Schwend V K, Tollefson V, Cooper N R. Identification of gp350 as the viral glycoprotein mediating attachment of Epstein-Barr virus (EBV) to the EBV/C3d receptor of B cells: sequence homology of gp350 and C3 complement fragment C3d. J Virol. 1987;61:1416–1420. doi: 10.1128/jvi.61.5.1416-1420.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raab M S, Albrecht J C, Birkmann A, Yaguboglu S, Lang D, Fleckenstein B, Neipel F. The immunogenic glycoprotein gp35–37 of human herpesvirus 8 is encoded by open reading frame K8.1. J Virol. 1998;72:6725–6731. doi: 10.1128/jvi.72.8.6725-6731.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Renne R, Blackbourn D, Whitby D, Levy J, Ganem D. Limited transmission of Kaposi's sarcoma-associated herpesvirus in cultured cells. J Virol. 1998;72:5182–5188. doi: 10.1128/jvi.72.6.5182-5188.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Renne R, Zhong W, Herndier B, McGrath M S, Abbey N, Kedes D H, Ganem D E. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 43.Russo J J, Bohenzky R A, Chen M C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi's sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Santoro F, Kennedy P E, Locatelli G, Malnati M S, Berger E A, Lusso P. CD46 is a cellular receptor for human herpesvirus 6. Cell. 1999;99:817–827. doi: 10.1016/s0092-8674(00)81678-5. [DOI] [PubMed] [Google Scholar]
- 45.Shieh M T, WuDunn D, Montgomery R I, Esko J D, Spear P G. Cell surface receptors for herpes simplex virus are heparan sulfate proteoglycans. J Cell Biol. 1992;116:1273–1281. doi: 10.1083/jcb.116.5.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shukla D, Liu J, Blaiklock P, Shworak N W, Bai X, Esko J D, Cohen G H, Eisenberg R J, Rosenberg R D, Spear P G. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell. 1999;99:13–22. doi: 10.1016/s0092-8674(00)80058-6. [DOI] [PubMed] [Google Scholar]
- 47.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals Hatem D, Babinet P, d'Agay M F, Clauvel J P, Raphael M, Degos L, et al. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 48.Spear P G, Shieh M T, Herold B C, WuDunn D, Koshy T I. Heparan sulfate glycosaminoglycans as primary cell surface receptors for herpes simplex virus. Adv Exp Med Biol. 1992;313:341–353. doi: 10.1007/978-1-4899-2444-5_33. [DOI] [PubMed] [Google Scholar]
- 49.Spira T J, Lam L, Dollard S C, Meng Y X, Pau C P, Black J B, Burns D, Cooper B, Hamid M, Huong J, Kite-Powell K, Pellett P E. Comparison of serologic assays and PCR for diagnosis of human herpesvirus 8 infection. J Clin Microbiol. 2000;38:2174–2180. doi: 10.1128/jcm.38.6.2174-2180.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spriggs M K, Armitage R J, Comeau M R, Strockbine L, Farrah T, Macduff B, Ulrich D, Alderson M R, Mullberg J, Cohen J I. The extracellular domain of the Epstein-Barr virus BZLF2 protein binds the HLA-DR beta chain and inhibits antigen presentation. J Virol. 1996;70:5557–5563. doi: 10.1128/jvi.70.8.5557-5563.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tal-Singer R, Peng C, Ponce D L, Abrams W R, Banfield B W, Tufaro F, Cohen G H, Eisenberg R J. Interaction of herpes simplex virus glycoprotein gC with mammalian cell surface molecules. J Virol. 1995;69:4471–4483. doi: 10.1128/jvi.69.7.4471-4483.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tanner J, Weis J, Fearon D, Whang Y, Kieff E. Epstein-Barr virus gp350/220 binding to the B lymphocyte C3d receptor mediates adsorption, capping, and endocytosis. Cell. 1987;50:203–213. doi: 10.1016/0092-8674(87)90216-9. [DOI] [PubMed] [Google Scholar]
- 53.Turner A, Bruun B, Minson T, Browne H. Glycoproteins gB, gD, and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a Cos cell transfection system. J Virol. 1998;72:873–875. doi: 10.1128/jvi.72.1.873-875.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vanderplasschen A, Bublot M, Dubuisson J, Pastoret P P, Thiry E. Attachment of the gammaherpesvirus bovine herpesvirus 4 is mediated by the interaction of gp8 glycoprotein with heparinlike moieties on the cell surface. Virology. 1993;196:232–240. doi: 10.1006/viro.1993.1471. [DOI] [PubMed] [Google Scholar]
- 55.Whitbeck J C, Connolly S A, Willis S H, Hou W, Krummenacher C, Ponce D L, Lou H, Baribaud I, Eisenberg R J, Cohen G H. Localization of the gD-binding region of the human herpes simplex virus receptor, HveA. J Virol. 2001;75:171–180. doi: 10.1128/JVI.75.1.171-180.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Williams R K, Straus S E. Specificity and affinity of binding of herpes simplex virus type 2 glycoprotein B to glycosaminoglycans. J Virol. 1997;71:1375–1380. doi: 10.1128/jvi.71.2.1375-1380.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Willis S H, Rux A H, Peng C, Whitbeck J C, Nicola A V, Lou H, Hou W, Salvador L, Eisenberg R J, Cohen G H. Examination of the kinetics of herpes simplex virus glycoprotein D binding to the herpesvirus entry mediator, using surface plasmon resonance. J Virol. 1998;72:5937–5947. doi: 10.1128/jvi.72.7.5937-5947.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.WuDunn D, Spear P G. Initial interaction of herpes simplex virus with cells binding to heparan sulfate. J Virol. 1989;63:52–58. doi: 10.1128/jvi.63.1.52-58.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu L, Puri V, Chandran B. Characterization of human herpesvirus-8 K8.1A/B glycoproteins by monoclonal antibodies. Virology. 1999;262:237–249. doi: 10.1006/viro.1999.9900. [DOI] [PubMed] [Google Scholar]
- 60.Zhu L, Wang R, Sweat A, Goldstein E, Horvat R, Chandran B. Comparison of human sera reactivities in immunoblots with recombinant human herpesvirus (HHV)-8 proteins associated with the latent (ORF73) and lytic (ORFs 65, K8.1A, and K8.1B) replicative cycles and in immunofluorescence assays with HHV-8-infected BCBL-1 cells. Virology. 1999;256:381–392. doi: 10.1006/viro.1999.9674. [DOI] [PubMed] [Google Scholar]