Abstract
An adenovirus previously isolated from a mesenteric lymph node from a chimpanzee was fully sequenced and found to be similar in overall structure to human adenoviruses. The genome of this virus, called C68, is 36,521 bp in length and is most similar to subgroup E of human adenovirus, with 90% identity in most adenovirus type 4 open reading frames that have been sequenced. Substantial differences in the hexon hypervariable regions were noted between C68 and other known adenoviruses, including adenovirus type 4. Neutralizing antibodies to C68 were highly prevalent in sera from a population of chimpanzees, while sera from humans and rhesus monkeys failed to neutralize C68. Furthermore, infection with C68 was not neutralized from sera of mice immunized with human adenovirus serotypes 2, 4, 5, 7, and 12. A replication-defective version of C68 was created by replacing the E1a and E1b genes with a minigene cassette; this vector was efficiently transcomplemented by the E1 region of human adenovirus type 5. C68 vector transduced a number of human and murine cell lines. This nonhuman adenoviral vector is sufficiently similar to human serotypes to allow growth in 293 cells and transduction of cells expressing the coxsackievirus and adenovirus receptor. As it is dissimilar in regions such as the hexon hypervariable domains, C68 vector avoids significant cross-neutralization by sera directed against human serotypes.
Vectors based on human adenovirus subgroup C (i.e., types 2 and 5) have realized widespread application in preclinical and clinical models of gene therapy (34). The viruses are rendered replication defective by deletion of E1 sequences. Multiple essential genes are disabled in more advanced versions of adenovirus vectors (7, 10, 17, 31). An important limitation of the use of adenovirus type 2- and adenovirus type 5-based vectors for human applications is that many individuals are immune to the virus as the result of a previous natural infection (6). A manifestation of existing immunity to the virus is B-cell activation, leading to persistent neutralizing antibodies that block vector uptake in vivo and diminish transduction.
One approach to accomplish immunologic distinction is to engineer the capsid of an adenovirus type 5- or adenovirus type 2-based vector. Several studies have attempted to accomplish this by exchanging the gene encoding fiber, since the protein is directly involved in receptor binding. While this has been successful in redirecting uptake of vector via a pathway distinct from that directed by the coxsackievirus and adenovirus (CAR) receptor, such chimeric viruses are still cross-neutralized due to blocking antibodies directed against hexon epitopes in the hypervariable regions (11, 14, 19, 28, 31). Recent attempts to engineer hexon proteins in chimeric viruses have been complicated by serotype-specific constraints in the hexon structure, which compromise the formation of stable chimeras. Selective modification of the hypervariable regions of hexon have diminished type-specific cross-neutralization in vitro without preventing blocking in vivo (8, 15, 25–27).
As an alternative, we decided it best to isolate a vector based solely on a nonhuman adenovirus to circumvent problems of existing immunity and have developed a vector from an adenovirus isolated from a chimpanzee. The adenovirus, called C68, was originally isolated from a mesenteric lymph node of a chimpanzee and shown to replicate in a number of primate-derived cell lines (3). Detailed restriction endonuclease mapping demonstrated similarities of C68 to human adenovirus serotype 4 (subgroup E) (21, 32). However, cross-reactivity of type-specific antisera between C68 and adenovirus type 4 is absent or diminished.
This report describes the full sequence analysis of C68 and its development as a gene transfer vector or vaccine carrier.
MATERIALS AND METHODS
Virus stocks and propagation.
The C68 virus stock was obtained from the American Type Culture Collection (ATCC; Manassas, Va.) and propagated in 293 cells (ATCC) cultured in Dulbecco's modified Eagle's medium (DMEM; Sigma, St. Louis, Mo.) supplemented with 10% fetal calf serum (FCS; Sigma or HyClone [Logan, Utah]) and 1% penicillin-streptomycin (Sigma). Infection of 293 cells was carried out in DMEM supplemented with 2% FCS for the first 24 h, after which FCS was added to bring the final concentration to 10%. Infected cells were harvested when 100% of the cells exhibited virus-induced cytopathic effect (CPE), collected, and concentrated by centrifugation. Cell pellets were resuspended in 10 mM Tris (pH 8.0) and lysed by three cycles of freezing and thawing. Virus preparations were obtained following two ultracentrifugation steps on cesium chloride density gradients, and stocks of virus were diluted to 1012 particles/ml in 10 mM Tris–100 mM NaCl–50% glycerol and stored at −70°C.
Cloning and sequencing of viral genomic DNA.
Genomic DNA was isolated from the purified virus preparation following standard methods and digested with a panel of 16 restriction enzymes following the manufacturer's recommendations. Except as noted, all restriction and modifying enzymes were obtained from Boehringer Mannheim, Indianapolis, Ind. Genomic DNA was digested with BamHI, PstI, SalI, HindIII, or XbaI, and the fragments were subcloned into plasmids (4). After deproteination, synthetic 10-bp PacI linkers (New England Biolabs, Beverly, Mass.) were ligated to the genomic DNA. Fragments containing the genomic termini were cloned into pNEB; this was followed by digestion with PacI and BamHI or PstI.
The PstI, BamHI, and HindIII clones generated from C68 are illustrated in Fig. 1B, C, and D, respectively. The fragments indicated by the shaded boxes were not cloned, but the sequence of the entire genome has been determined through sequencing overlapping clones and viral DNA directly (unshaded boxes). The cloned fragments are described in Table 1. The complete nucleotide sequence (36,521 bp) of C68 was determined by Commonwealth Biotechnologies Incorporated, Richmond, Va.
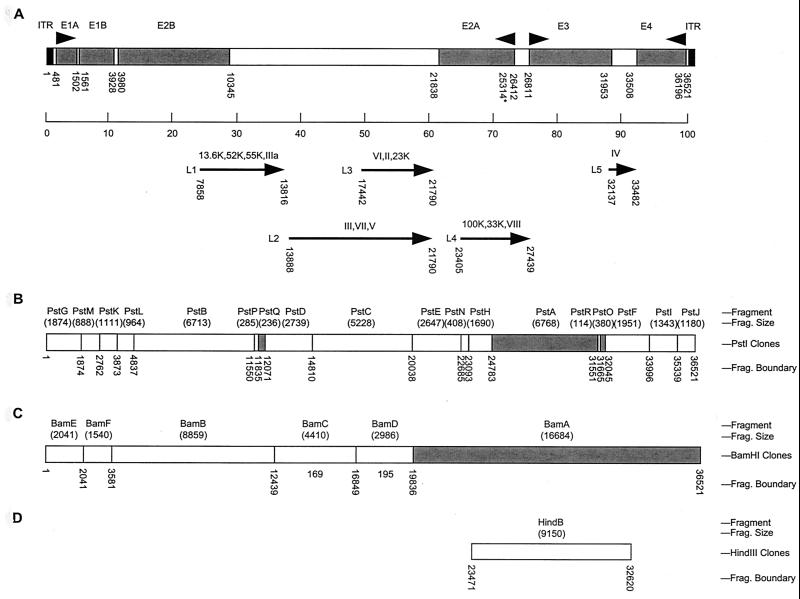
FIG. 1.
Genetic organization of the C68 genome. (A) The genome of the C68 chimpanzee adenovirus is schematically represented by the box at the top. The inverted terminal repeats are shaded black, and the early regions are shaded gray. The arrowheads above the box indicate the direction of expression of the early genes. The line below the box represents the division of the genome into 100 m.u. The arrows below the line represent the five late gene regions and the proteins encoded in each region. The numbers below the box or arrows indicate the start (promoter or initiation codon) and end (canonical polyadenylation signal) for each region. ∗ represents the E2A late promoter. (B) PstI clones. (C) BamHI clones. (D) HindIII clones. The unshaded regions indicate that a fragment was cloned into a plasmid vector, as listed in Table 1, while the shaded regions indicate that the restriction fragment was not cloned. For each section, the fragment name, alphabetical with A being the largest fragment, and the fragment size are listed above the box, and the fragment end points are listed below the box.
TABLE 1.
C68 plasmid clones and insert sizesa
| Construct | Insert size (bp) | Fragment 5′ end (nt) | Fragment 3′ end (nt) | 5′ end m.u. (%) | 3′ end m.u. (%) | Comments |
|---|---|---|---|---|---|---|
| PstI fragments | ||||||
| C68-Pst-A | 6,768 | 24784 | 31551 | 67.9 | 86.4 | Not cloned |
| pBS:C68-Pst-B | 6,713 | 4838 | 11550 | 13.2 | 31.6 | |
| pBS:C68-Pst-C | 5,228 | 14811 | 20038 | 40.6 | 54.9 | |
| pBS:C68-Pst-D | 2,739 | 12072 | 14810 | 33.1 | 40.6 | |
| pBS:C68-Pst-E | 2,647 | 20039 | 22685 | 54.9 | 32.1 | |
| pBS:C68-Pst-F | 1,951 | 32046 | 33996 | 87.8 | 93.1 | |
| pNEB:C68-Pst-G | 1,874 | 1 | 1874 | 0.0 | 5.1 | Left end/PacI linker |
| pBS:C68-Pst-H | 1,690 | 23094 | 24783 | 63.2 | 67.9 | |
| pBS:C68-Pst-I | 1,343 | 33997 | 35339 | 93.1 | 96.8 | |
| pNEB:C68-Pst-J | 1,180 | 35340 | 36519 | 96.8 | 100.0 | Right end/PacI linker |
| pBS:C68-Pst-K | 1,111 | 2763 | 3873 | 7.6 | 10.6 | |
| pBS:C68-Pst-L | 964 | 3874 | 4837 | 10.6 | 13.2 | |
| pBS:C68-Pst-M | 888 | 1875 | 2762 | 5.1 | 7.6 | |
| pBS:C68-Pst-N | 408 | 22686 | 23093 | 62.1 | 63.2 | |
| C68-Pst-O | 380 | 31666 | 32045 | 86.7 | 87.7 | Not cloned |
| pBS:C68-Pst-P | 285 | 11551 | 11835 | 31.6 | 32.4 | |
| C68-Pst-Q | 236 | 11836 | 12071 | 32.4 | 33.1 | Not cloned |
| pBS:C68-Pst-R | 114 | 31552 | 31665 | 86.4 | 86.7 | |
| BamHI fragments | ||||||
| C68-Bam-A | 16,684 | 19836 | 36519 | 54.3 | 100.0 | Right end/not cloned |
| pBS:C68-Bam-B | 8,858 | 3582 | 12439 | 9.8 | 34.1 | |
| pBS:C68-Bam-C | 4,410 | 12440 | 16849 | 34.1 | 46.1 | |
| pBS:C68-Bam-D | 2,986 | 16850 | 19835 | 46.1 | 54.3 | |
| pNEB:C68-Bam-E | 2,041 | 1 | 2041 | 0.0 | 5.6 | Left end/PacI linker |
| pBS:C68-Bam-F | 1,540 | 2042 | 3581 | 5.6 | 9.8 | |
| HindIII fragment | ||||||
| pBR:C68-Hind-B | 9,150 | 23471 | 32620 | 64.3 | 89.3 |
The name of each plasmid clone of the C68 genome is given in the first column. pBS, pBluescript SK + clone; pNEB, pNEB 193 clone; pBR, pBR322 clone; no prefix, fragment not cloned. %, fraction of genome demarcated.
Analysis of C68 sequence.
The complete nucleotide (nt) sequences of every member of the Mastadenovirus genus accessible from GenBank, including isolates from different species, were screened for identity to C68. The adenovirus type 4 minigenome was assembled from the following GenBank sequences: left-hand inverted terminal repeat (ITR) (J01964); E1A region (M14918); DNA polymerase and pTP (X74508 and 74672); VA RNA-I and -II (U10682); 52K and 55K (U52535); pVII (U70921); hexon (X84646); endoprotease (M16692); DNA-binding protein (M12407); fiber (X76547); and right-hand ITR (J01965). The adenovirus type 7 composite genome was created from the following sequence data: map units (m.u.) 3 to 21 (X03000); VA RNA-I and -II, pTP, and 52K and 55K (U52574); penton (AD001675); pVI, hexon, and endoprotease (AF065065); DNA-binding protein (K02530); E3 and fiber region (AF104384); and right-hand ITR (V00037).
The amino acid sequence alignment was generated with Clustal X, edited with Jalview (http://www.ebi.ac.uk/∼michele/jalview/), and analyzed with Boxshade (http://www.ch.embnet.org/software/BOX_form.html). Publicly available hexon protein sequences from all human adenovirus serotypes were initially aligned to identify the set showing the highest homology to C68.
Creation of an E1-deleted vector based on C68.
To construct a plasmid shuttle vector for creation of recombinant C68 virus, plasmid pSP72 (Promega, Madison, Wis.) was modified by digestion with BglII followed by filling-in of the ends with Klenow enzyme (Boehringer Mannheim, Indianapolis, Ind.) and ligation with a synthetic 12-bp PacI linker (New England Biolabs, Beverly, Mass.) to yield pSP72-Pac. A 475-bp PacI/SnaBI fragment spanning m.u. 0 to 1.3 (1 to 475 bp) of the C68 genome was isolated from the pNEB-BamE plasmid containing BamHI E fragment (Fig. 1C) of the C68 genome and cloned into PacI- and EcoRV-treated pSP72-Pac to yield pSP-C68-MU0–1.3. A minigene cassette consisting of the cytomegalovirus (CMV) early promoter driving lacZ with a simian virus 40 (SV40) polyadenylation signal was separated from pCMVβ (Clontech, Palo Alto, Calif.) as a 4.5-kb EcoRI/SalI fragment and ligated to pSP-C68-MU0–1.3 restricted with the same set of enzymes, resulting in pSP-C68-MU0–1.3-CMVLacZ.
For the initial step in the isolation of m.u. 9 to 16.7 (bp 3287 to 6099) region of C68, both pGEM-3Z (Promega, Madison, Wis.) and pBS-C68-BamF were double-digested with BamHI and SphI enzymes. Then the 293-bp fragment from pBS-C68-BamF was ligated with pGEM-3Z backbone to form pGEM-C68-MU9–9.8. A 2.4-kb fragment including C68 m.u. 9.8 to 16.7 (bp 3579 to 6099) was obtained from the pBS-C68 BamHB clone after XbaI digestion, filling-in reaction, and subsequent BamHI treatment and cloned into BamHI/SmaI double-digested pGEM-C68-MU9–9.8 to generate pGEM-C68-MU9–16.7. The C68 m.u. 9 to 16.7 region 7 (bp 3287 to 6099) was isolated from pGEM-C68-MU9–16.7 by digestion with EcoRI, filling in of the ends with Klenow enzyme (Boehringer Mannheim, Indianapolis, Ind.), ligation of a synthetic 12-bp HindIII linker (NEB), and then digestion with HindIII. This 2.7-kb fragment spanning C68 m.u. 9 to 16.7 (bp 3287 to 6099) was cloned into the HindIII site of pSP-C68-MU0–1.3-CMVlacZ to form the final shuttle plasmid pC68-CMV-LacZ. In addition, a 1.56-kb alkaline phosphatase cDNA fragment was isolated from pAdCMVALP (12) and exchanged for lacZ at NotI sites of pC68-CMV-lacZ, resulting in pC68-CMV-AP.
To create the E1-deleted recombinant C68-CMVEGFP vector, a pC68-CMVEGFP shuttle plasmid was first constructed by replacing the lacZ transgene in pC68-CMV-lacZ with the enhanced green fluorescent protein (EGFP) gene. The replacement cloning process was carried out as follows. An additional NotI restriction site was introduced into the 5′ end of the EGFP coding sequence in pEGFP-1 (Clontech, Palo Alto, Calif.) by BamHI digestion, filling-in reaction, and ligation of an 8-bp synthetic NotI linker (NEB). After NotI restriction of both constructs, the EGFP sequence was isolated from the modified pEGFP-1 and used to replace the lacZ gene in pC68-CMV-lacZ. The pC68-CMVEGFP construct (3 μg) was cotransfected with SspI-digested C68 genomic DNA (1 μg) into 293 cells for homologous recombination as previously described (16). Green plaques visualized by fluorescent microscopy were isolated for two rounds of plaque purification, expansion, and purification by CsCl gradient sedimentation (16).
In an attempt to apply the convenient green/white selection process (9) to construct recombinant C68 vectors, a 7.1-kb fragment spanning m.u. 11 to 32 (bp 4012 to 11710) was isolated from the pBSC68-BamB plasmid by treatment with AgeI and BsiWI restriction endonucleases and cloned into Asp718 and AgeI sites of pC68-CMV-alkaline phosphatase shuttle plasmid, resulting in a new plasmid called pC68CMV-alkaline phosphatase-mu32. A further modification was made to remove m.u. 26 to 30 (bp 9350 to 11070) from pC68CMV-AP-MU32 by Eco47III and NruI digestions. The new shuttle plasmid, called pC68CMV-AP-MU26, has a shorter region for homologous recombination (m.u. 16.7 to 26) 3′ to the minigene.
To make a recombinant C68 vector, alkaline phosphatase is replaced with the gene of interest. The resulting pC68CMV-Nugene-mu26 construct is cotransfected with XbaI (m.u. 16.5 or bp 6026)-restricted C68-CMVGFP viral DNA into 293 cells, followed by top agar overlay. The recombinant virus plaques (white) are generated through the homologous recombination in the region of 16.7 to 26 m.u. (bp 6099 to 9350), which is shared between the pC68CMV-Nugene construct and the C68 viral backbone; the recombinants which form white plaques are selected from green plaques of uncut C68-CMVGFP virus.
The green/white selection mechanism was also introduced to the process of cloning the gene of interest into the pC68 shuttle plasmid. The alkaline phosphatase gene in both pC68CMV-AP-MU36 and pC68CMV-AP-MU26 was replaced with a cassette of the prokaryotic GFP gene driven by the lacZ promoter isolated from pGFPmu31 (Clontech, Palo Alto, Calif.). Thus, white colonies of bacterial transformants will contain the recombinant plasmid. This green/white selection process for bacterial colonies circumvented the need for making and characterizing large numbers of miniprepped DNAs and so further enhanced the efficiency in creating recombinant C68 vectors.
Virus-neutralizing antibody assays.
The neutralizing activity of sera was tested as follows. Sera collected from individual humans, rhesus monkeys, or chimpanzees were inactivated at 56°C for 30 min. A serial dilution of each sample (1:10, 1:20, 1:40, 1:80, 1:160, and 1:320 in 100 μl of DMEM containing 10% FCS) was added to equal amounts of H5.010CMVEGFP (1,000 PFU/well) or C68CMVEGFP virus and incubated at 4°C for 2 h. One hundred and fifty microliters of the mixture was transferred onto 2 × 104 293 cells in 96-well flat-bottomed plates. Control wells were infected with equal amounts of virus without addition of serum. Samples were incubated at 37°C in 5% CO2 for 48 h and examined under a fluorescent microscope. Sample dilutions that showed >50% reduction of green-fluorescent foci compared to infected controls were scored positive for neutralizing antibodies.
Structural analysis of hexon proteins.
The X-ray crystal structures of adenovirus type 5 hexon (Protein Data Bank identifier 1RUX) (27) and adenovirus type 2 hexon (1) have been further refined to yield the current hexon models (Rux and Burnett, unpublished data). Models of the homologous C68 and adenovirus type 4 hexons were initially produced using the Swiss-PdbViewer protein-modeling environment (18). Its automated procedure was used to align the C68 and adenovirus type 4 hexon amino acid sequences to those of the adenovirus type 2 and adenovirus type 5 hexon crystal structures. The sequence alignments were used to guide the threading of the model sequences onto the known molecular structures. The side chain positions of residues not seen in the known structures were selected from a library of side chain rotomers.
These initial molecular models were then manually adjusted to improve the automated alignment by moving gaps to exposed variable regions and by optimizing the packing of side chains. The positions of external loop segments not observed in the adenovirus type 2 and adenovirus type 5 template structures were either selected from a library of known loop structures or fitted manually. The conformation of each model was further refined by energy minimization using the molecular mechanics program CHARMM (5). The structures of these C68 and adenovirus type 4 hexon models were then aligned, and a new sequence alignment was calculated. The differences between the two structurally aligned hexon sequences were used to color images of the homology models. Graphic images prepared with the Swiss-PdbViewer program were exported and rendered with the Persistence of Vision Ray Tracer program (POV-Ray 2000, version 3.1g).
RESULTS
Cloning and sequence analysis of C68 genome.
Chimpanzee adenovirus C68 was obtained from the ATCC and propagated in human 293 cells. Viral genomic DNA was isolated from purified virions using established procedures (9) and digested with a panel of restriction enzymes; the data were consistent with previous studies (data not shown) (21, 23, 32). Restriction fragments spanning the entire genome of C68 were subcloned into plasmids. A schematic drawing of the C68 genome is shown in Fig. 1A, and the PstI, BamHI, and HindIII fragments that were cloned into plasmid vectors are indicated by the unshaded boxes in Fig. 1B, 1C, and 1D, respectively. The cloned fragments, fragment sizes, and genomic positions are also listed in Table 1. Both plasmid clones and genomic DNA were used as templates for sequencing. The genome was sequenced by primer walking in both directions, and each base was included in an average of approximately four reactions.
The C68 genome is 36,521 bp in length (GenBank accession no. AF394196). Preliminary comparison with GenBank sequences indicated various degrees of similarity with other human and animal adenoviruses along the entire length of the viral genome. Regions with homology to all of the previously described adenoviral genetic units, early regions 1 to 4, and the major late genes were found in the C68 genome (Fig. 1A). Nucleic acid sequence similarity between C68 and the human adenoviruses that have been completely sequenced, adenovirus types 2 (NC001405), 5 (NC001405), 12 (NC001460), 17 (NC002067), and 40 (NC01464), was used to order the clones. The open reading frames were determined and the genes were identified based on similarity to other human adenoviruses. All of the major adenoviral early and late genes are present in C68. The ITRs are 130 bp in length.
Comparison of C68 to human adenoviruses and characterization of C68 genome.
The nucleotide sequence and predicted amino acid sequences of all significant open reading frames in the C68 genome were compared to known DNA and protein sequences. The nucleotide sequence of C68 is compared to sequences of adenovirus types 2, 4, 5, 7, 12, 17, and 40 in Fig. 2. In agreement with previous restriction analysis (21, 23) C68 is most similar to human adenovirus type 4 (subgroup E).
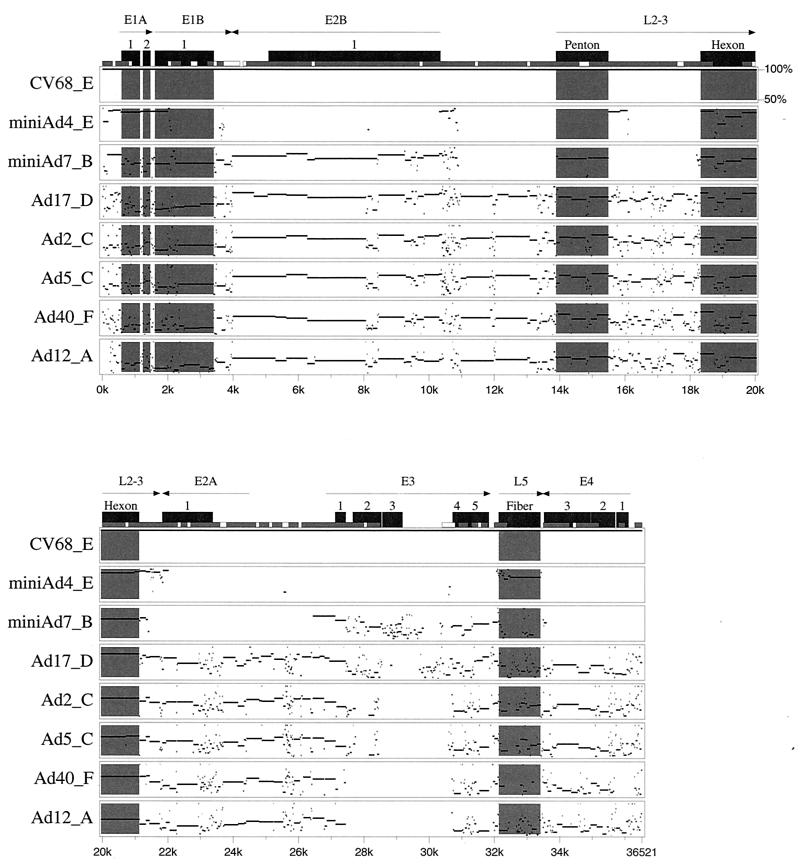
FIG. 2.
Pairwise genomic sequence comparisons between C68 and selected adenoviruses from various subgroups. The genome of each human serotype was aligned to C68, and percent identity plots (pips) were generated with PipMaker. Since only partial sequence information was available for adenovirus types 4 (Ad4) and 7, minigenomes were created and used in the analysis. Pips are arranged from top to bottom in order of decreasing identity. Regions of interest are highlighted in gray. Only segments showing greater than 50% identity to C68 are shown in the plots. In the case of adenovirus types 4 and 7, gaps of less than 50% identity represent regions for which an alignment was not generated due to insufficient input data. Arrows indicate the orientation, size, and location of genes of interest. Black boxes designate coding sequences, and numbers indicate interruptions in those sequences. Short gray/white boxes illustrate CpG island composition (CpG/GpC ratio), with white indicating 0.60 to 0.75 and gray greater than 0.75. The following GenBank documents were used in the alignment: NC001405 (adenovirus type 2), NC001406 (adenovirus type 5), NC002067 (adenovirus type 17), NC001460 (adenovirus type 12), and NC001464 (adenovirus type 40).
The E1A region of C68 extends from the TATA box at nt 480 to the poly(A) addition site at nt 1521. The consensus splice donor and acceptor sites are in the analogous positions of the human adenovirus counterparts, and the 28.2K and 24.8K proteins are similar in size to the human adenovirus proteins. The open reading frame for the smallest E1A protein of C68 is predicted to encode 101 residues, as opposed to approximately 60 amino acids for other adenoviruses. There is a TTA codon at residue 60 for C68, where other adenoviruses often have a TGA stop codon. The first 60 residues of C68 E1A 100R protein have 85% identity to the adenovirus type 4 homolog.
The C68 genome carries genes for the four E1B proteins, 20.5K, 54.7K, 10.1K, and 18.5K, as well as pIX. All five C68-encoded proteins are similar in size to other adenovirus E1B and pIX proteins. The adenovirus type 4 homolog of the E1B 21K protein has only 142 amino acids, where C68 has 186 residues and other human adenoviruses have 163 to 178 residues. The C68 and adenovirus type 4 proteins show 95% identity over the first 134 amino acids, then the similarity ends, and the adenovirus 4 protein terminates at 142 amino acids.
The C68 genome encodes homologs of the E2A 55K DNA-binding protein and the Iva2 maturation protein as well as the E2B terminal protein and the DNA polymerase. All of the E2 region proteins are similar in size to their human adenovirus counterparts, and the E2B proteins are particularly well conserved. The C68 E2B 123.6K DNA polymerase is predicted to be 1,124 residues, while adenovirus type 4 is predicted to have 1,193, although the other human adenoviruses have smaller polymerases. Residues 1 to 71 of the adenovirus type 4 polymerase have no similarity to any other adenovirus polymerase, and it is possible that this protein actually initiates at an internal ATG codon. From amino acids 72 to 1193, the adenovirus type 4 and C68 polymerases have 96% amino acid identity.
The E3 regions of human adenoviruses sequenced so far exhibit considerable sequence and coding capacity variability. Adenovirus type 40 has five E3 region genes, adenovirus type 12 has six, C68 and adenovirus type 5 have seven, adenovirus type 38 has eight, and adenovirus types 3 and 7 (subgroup B human adenoviruses) have nine putative E3 region genes. The adenovirus type 4 E3 region has not yet been sequenced. In comparison with the E3 region of adenovirus type 35, homologs to all of the eight E3 genes were identified in the C68 genome (2).
The C68 E4 region has six open reading frames, and each is homologous to proteins in the human adenovirus types 5, 12, and 40 E4 region. The E4 nomenclature is confusing because the open reading frame 2 homologs of C68, adenovirus type 12, and adenovirus type 40 are approximately 130 residues, while in adenovirus type 5 there are two open reading frames encoding proteins of 64 and 67 residues with homology, respectively, to the amino- and carboxy-terminal ends of the larger open reading frame 2 proteins. We omitted open reading frame 5 in our nomenclature because the fifth open reading frame in the E4 region is homologous to the widely studied open reading frame 6 protein of human adenovirus type 5.
We were able to locate the major late promoter and the tripartite leader sequences of the C68 genome based on homology to human adenovirus type 5 sequence. Open reading frames with the potential to encode the 15 major late proteins were located. All of the C68 late proteins are similar in size to their human adenovirus counterparts. The percent amino acid identity between chimpanzee and human adenovirus late proteins varies considerably. The C68 fiber protein is predicted to have 90% amino acid identity with the adenovirus type 4 protein, but much less similarity to the other human adenovirus fiber proteins. The CAR binding site in the fiber knob is present in C68. For a more detailed explanation of the role of CAR in C68 uptake, refer to Cohen et al. (submitted for publication).
Creation of a vector based on C68.
A replication-defective version of C68 was isolated for use in gene transfer. The classic strategy of creating a recombinant with E1 deleted by homologous recombination in an E1-expressing cell line was pursued. The first step was creation of a plasmid containing m.u. 0 through 1.3 (bp 1 to 475) followed by addition of a minigene expressing EGFP from a CMV promoter and C68 sequence spanning m.u. 9 to 16.7 (bp 3287 to 6099). This linearized plasmid was cotransfected into an E1-expressing cell line with SspI-digested C68 genomic DNA (SspI cuts at 3.6 m.u., providing 2,812 bp for homologous recombination between the shuttle vector and truncated viral backbone). Experiments were initially conducted with 293 cells, which harbor E1 from human adenovirus type 5, with the hope that this would suffice for transcomplementation. Indeed, plaques formed which represented the desired recombinant. The resulting vector was called C68-CMV-GFP.
The strategy for generating recombinants was modified to enable efficient and rapid isolation of recombinants. First, the alkaline phosphatase DNA in the initial shuttle vector was replaced with a prokaryotic GFP gene driven by the prokaryotic promoter from lacZ. This allowed efficient screening of bacterial transformations when attempting to incorporate a desired eukaryotic RNA polymerase II transcriptional unit into the shuttle vector. The resulting transformation can be screened for expression of GFP; white colonies are recombinants, while green colonies are residual parental plasmid.
A green-white selection has been used to screen the products of cotransfection for the isolation of human adenovirus type 5 recombinants (9); this was adapted to the C68 system. The initial shuttle vector was revised to include extended 3′ sequences from m.u. 9 to 26 (bp 3287 to 9350). This vector was cotransfected with viral DNA from the original C68-CMV-GFP isolate that had been restricted with XbaI, which cuts at m.u. 16.5 (bp 6026), allowing 3.3 kb of overlap for homologous recombination. The resulting plaques were screened under a phase contrast fluorescent microscope for nonfluorescing isolates, which represent the desired recombinants. This greatly simplified screening in comparison to the standard methods based on structure or transgene expression. A schematic description of our generic vector construction for recombinant C68 viruses is illustrated in Fig. 3.
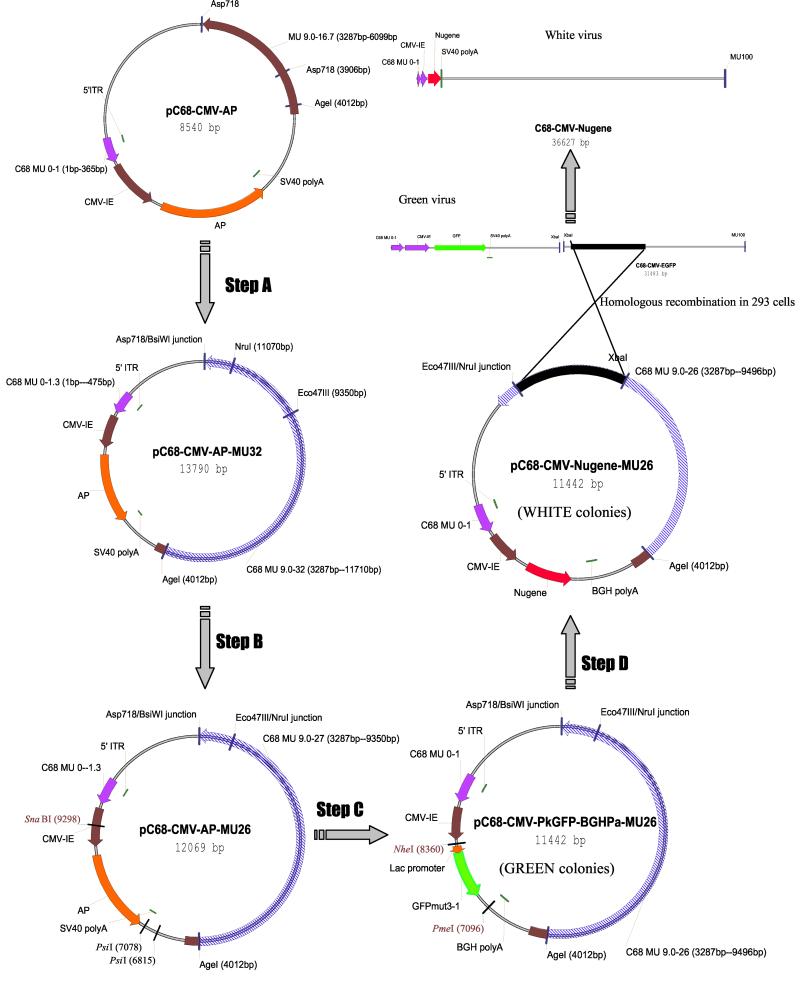
FIG. 3.
Schematic description of a generic vector construction process for replication-defective C68 viruses. Step A, C68 fragment spanning bp 4012 to 6099 was removed from pC68-CMV-AP by Asp718 and AgeI endonuclease treatments and replaced by an AgeI/BsiWI fragment. This fragment, containing bp 4012 to 11710 of C68 DNA, was isolated from pBS-C68 Bam-B. Step B, pC68-CMV-AP-MU32 was restricted with Eco47III and NruI endonucleases to delete a 1.72-kb fragment from C68 DNA and self-ligated. Step C, a part of the CMV promoter, alkaline phosphatase cDNA, and SV40 poly(A) addition signal was removed from pC68-CMV-AP-MU26 by SnaBI and PsiI digestions and substituted with a SnaBI-EcoRI fragment. This fragment, consisting of a part of the CMV promoter, prokaryotic GFP expression cassette, and bovine growth hormone poly(A) addition signal was obtained from pShut-GFPmut3-1. Step D, the prokaryotic GFP expression cassette was released from pC68-CMV-PkGFP-BGHPa-MU26 after NheI and PmeI digestions and replaced with the cDNA of the gene of interest.
Serology of C68.
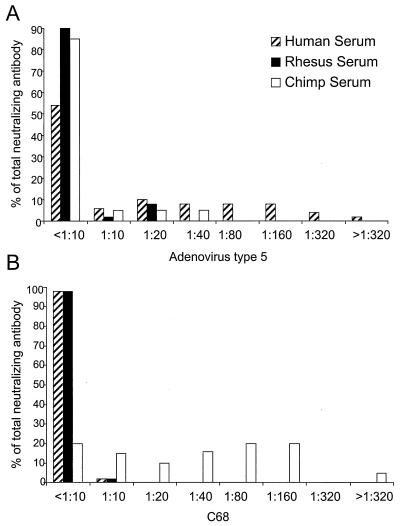
Several studies were performed to determine if there is cross-reactivity between type-specific antisera of C68 and human adenovirus. Panels of sera from 50 normal human subjects, 52 rhesus monkeys, and 20 chimpanzees were evaluated for neutralizing antibodies against adenovirus type 5- and C68-based vectors using 293 cells as an indicator cell line (Fig. 4). As expected, approximately 35% of normal human subjects demonstrated neutralizing antibody against adenovirus type 5, a frequency much higher than observed in sera of rhesus monkeys and chimpanzees (Fig. 4A). Neutralizing antibody to C68 was observed in 80% of chimpanzees and only 2% of normal human subjects or rhesus monkeys (Fig. 4B). Titers of neutralizing antibodies in the nontarget species were generally low.
FIG. 4.
Neutralizing antibodies against adenovirus type 5 and C68. Sera from 50 normal human subjects, 52 rhesus monkeys, and 20 chimpanzees were analyzed for neutralizing antibody against human adenovirus type 5 (A) or C68 (B). The percentage of total serum samples that titrated at the indicated dilutions is presented.
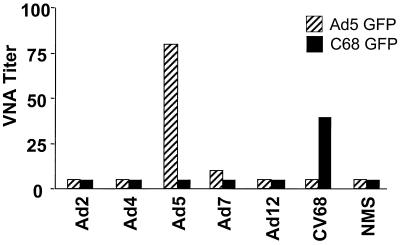
To further evaluate cross-reactivity of C68 with human adenovirus vectors, mice were immunized with 2 × 107 PFU of adenovirus types 2, 4, 5, 7, and 12 as well as C68. Sera were harvested 2 weeks later and tested for antibodies that neutralized either adenovirus type 5 or C68 vectors (Fig. 5). Neutralizing antibody to adenovirus type 5 vector was only detected in animals immunized with adenovirus type 5. Importantly, the only animals with neutralizing antibody to C68 vector were those immunized with C68 vector; none of the human serotypes tested, including adenovirus type 4, generated antibodies in mice that neutralized C68 in vitro.
FIG. 5.
Analysis of sera from immunized mice for neutralization of human adenovirus type 5 and C68. Groups of C3H/He mice were immunized subcutaneously with 4 × 1010 replication-competent adenovirus particles per mouse. Mice were bled 18 days later. Sera as well as a control serum from naive mice (NMS) were tested for virus-neutralizing activity (VNA) against adenovirus type 5 (Ad5)-GFP (striped bars) and C68-GFP (solid bars) virus.
Structural analysis of hexon protein of C68.
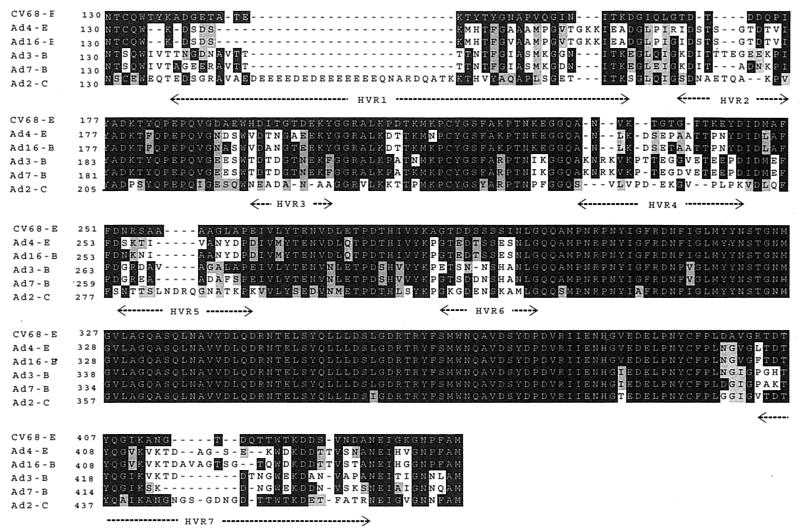
The absence of neutralizing antibodies between C68 and human serotypes compelled us to evaluate more carefully structural differences in the regions of hexon presumed to harbor type-specific epitopes. Previous studies have suggested that these epitopes are located within the seven hypervariable regions of hexon determined by Crawford-Miksza and Schnurr (8). A comparison of the amino acid sequences of hexon proteins between C68 and several human adenoviruses is shown in Fig. 6. This shows that C68 is substantially dissimilar in significant regions of these hypervariable sequences. More detailed modeling of the three-dimensional structure of hexon of C68 was performed to map the unique sequences. Models of hexon structures from C68 and adenovirus type 4 were generated based on the X-ray crystal structures of hexons for adenovirus type 2 and adenovirus type 5.
FIG. 6.
Multiple sequence alignment of hexon proteins. The deduced amino acid sequences of highly similar human adenovirus hexons were compared with the chimpanzee adenovirus using Clustal X. Serotypes and subgroups are indicated on the left margin, followed by the residue number. The numbering refers to the amino acid position with respect to the start of translation. Amino acids are shaded with respect to C68 to highlight sequence similarities (gray) and identities (black). The seven hypervariable regions within loop domains DE1 and FG1 are labeled along the bottom and correspond to the following adenovirus type 2 sequences in the alignment: hypervariable region 1, 137 to 188; hypervariable region 2, 194 to 204; hypervariable region 3, 222 to 229; hypervariable region 4, 258 to 271; hypervariable region 5, 278 to 294; hypervariable region 6, 316 to 327; and hypervariable region 7, 433 to 465. The GenBank accession numbers for the sequences shown are as follows: AAD03657 (adenovirus type 4), S37216 (adenovirus type 16), S39298 (adenovirus type 3), AAD03663 (adenovirus type 7), and NP040525 (adenovirus type 2).
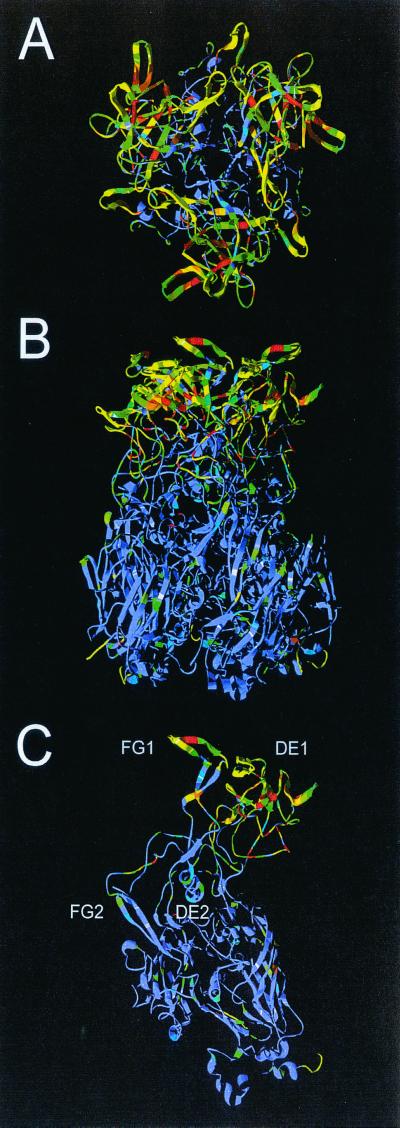
While the overall C68 sequence is very similar to that of adenovirus type 4 hexon, Fig. 7C shows that the differences between the two sequences are primarily focused in the DE1 and FG1 loops, and these contain all seven hypervariable regions. It is the DE1, FG1, and FG2 loops, each from a different subunit, that intimately associate to form the tower domains at the top of the trimeric molecule (Fig. 7A and B). The hexon towers form much of the exterior surface of the virion and are the sites of antibody attachment. As the sides and base of the hexons pack together within the capsid, these regions are shielded from antibody binding, and their sequences are conserved. In contrast, the sequences of C68 and adenovirus type 4 are quite different in the hexon towers. This immediately explains why antibodies raised to either of these viruses do not cross-react.
FIG. 7.
Predicted structure of C68 hexon. Ribbon representation of the C68 hexon homology model structure. (A) Trimer viewed from the top (exterior surface of virion); (B) trimer viewed from the side; (C) monomer. The structure is colored to indicate the sequence diversity between C68 and human adenovirus type 4. The colors range from blue (conserved) to yellow (conservative) to red (nonconservative). The four upper loops (DE1, FG1, DE2, and FG2) are labeled. DE1 contains hypervariable regions 1 to 6, and FG1 contains hypervariable region 7.
DISCUSSION
The primary impetus for creating a vector for human gene therapy based on a chimpanzee adenovirus was to circumvent problems that may arise because of existing immunity as a result of a naturally acquired adenovirus infection. Generation of virus-specific B and T cells following an infection could impact the performance of a subsequent vector administration by blocking vector uptake via neutralizing antibodies and influencing toxicity through a number of potential mechanisms (13, 20, 22, 24, 29, 35–39). Vector-specific antibodies could potentially diminish toxicity in the target organ by blocking transduction or enhance toxicity by facilitating uptake via Fc receptors on antigen-presenting cells (13, 20, 36, 37).
Complete sequence analysis of the C68 isolate revealed a structure similar to most human adenoviruses, although its sequence was distinct. The closest known homolog is the subgroup E virus adenovirus type 4. Modeling of the hypervariable regions of hexon, which presumably harbor the type-specific epitopes, revealed significant differences between adenovirus type 4 and C68.
Critical to the success of an adenovirus vector for in vivo applications is a process for efficient production and purification of lots that are potent and free of contaminants. C68 retains enough similarity to subgroup C adenoviruses to allow high-level replication of an E1-deleted C68 vector in 293 cells containing the E1 region of adenovirus type 5 (17). This useful similarity eliminated the need to create a new packaging cell line with the C68 E1 gene. We have created and produced in 293 cells four C68-based replication-defective vectors that carry different transgenes. The average yield of viral particles of these vectors was two- to threefold higher than that of human serotype 5-based vectors. However, the ratio of viral particles to PFU measured in plaque assays in 293 cells is two- to threefold higher for C68 vectors than for those based on adenovirus type 5 (Table 2). Transducibility of HeLa and A549 cells with adenovirus type 5 and C68 vectors expressing green fluorescent protein at the same multiplicity of infection (MOI = 10 [based on PFU]) was also compared in a quantitative transduction assay. Infectivity of these vectors, based on PFU assays, correlated directly with transduction efficiency.
TABLE 2.
Comparison of preparations of recombinant C68 and human adenovirustype 5 virusesa
| Construct | Total yield (particles) | Viral particle/PFU ratio | RCA |
|---|---|---|---|
| C68-CMV-HIV-gag short | 1 × 1014 | 31 | Negative |
| C68-CMV-Rab-gp | 7.5 × 1013 | 286 | Negative |
| C68-CMV-HIV-gag full | 8 × 1013 | 178 | Negative |
| C68-CMVEGFP | 1.6 × 1014 | 570 | Negative |
| Mean ± SD | |||
| C68 vectors (n = 4) | 1.04 (±0.39) × 1014 | 266 ± 228 | Negative |
| Ad5 vectors (n = 53) | 4.5 (±4.2) × 1013 | 78 ± 88 | 24% of prepns positive |
Total viral particle yield, ratio of viral particles to PFU, and results of RCA tests from four different C68 vector preparations and 53 preparations of human adenovirus type 5 (Ad5) vectors that contain different transgenes were compared. All preparations were from 50 plates (150 mm) of 293 infections. HIV-gag short and HIV-gag full represent different open reading frames of the gag gene of HIV. Rab-gp is the gene encoding rabies virus glycoprotein.
No gross contamination with replication-competent adenovirus (RCA) has been identified in our C68 preparations, possibly due to the sequence divergence between overlapping sequences of the C68 vector and adenovirus type 5 E1 in 293 cells. These showed 40% differences 5′ to the E1 deletion and 38% differences 3′ to the E1 deletion (Table 2). The potential for decreased likelihood of RCA is an important advantage in both preclinical and clinical applications.
Important to the utility of C68 vector in human trials is the absence of neutralizing antibody in the human population. Previous studies regarding this issue provided conflicting views, especially with respect to cross-neutralization between C68 and adenovirus type 4 (3, 23, 33). In our study, a screen of 50 normal human subjects failed to detect any significant neutralizing antibodies (>1:10) using the same assay that showed neutralizing antibodies in >80% of chimpanzees. Furthermore, sera from mice immunized with multiple human adenovirus serotypes, including adenovirus type 4, did not neutralize infection with C68.
In a comparison study, it has been shown that C68 vectors are internalized via the CAR receptor (Cohen et al., submitted). The utility of the C68 vector in models of gene therapy is currently under investigation. Preliminary studies indicate that it functions as an excellent vaccine for human immunodeficiency virus (HIV) and rabies virus in murine models (Fitzgerald et al., submitted; Xiang et al., submitted).
ACKNOWLEDGMENTS
The first two authors contributed equally to this work.
The technical support of the Vector and Immunology cores of the Institute for Human Gene Therapy is greatly appreciated.
Support was provided by NIH (P30 DK47757–08 and P01 HL59407–02), CF Foundation, and Genovo, Inc., to J. Wilson. NIH (AI-17270) and the Wistar Cancer Center Core Grant (CA 09171) supported R.M. Burnett. J. Wilson owns equity in Targeted Genetics (formerly Genovo).
REFERENCES
- 1.Athappilly F K, Murali R, Rux J J, Cai Z, Burnett R M. The refined crystal structure of hexon, the major coat protein of adenovirus type 2, at 2.9 Å resolution. J Mol Biol. 1994;242:430–455. doi: 10.1006/jmbi.1994.1593. [DOI] [PubMed] [Google Scholar]
- 2.Basler C F, Horwitz M S. Subgroup B adenovirus type 35 early region 3 mRNAs differ from those of the subgroup C adenoviruses. Virology. 1996;215:165–177. doi: 10.1006/viro.1996.0019. [DOI] [PubMed] [Google Scholar]
- 3.Basnight M, Rogers N G, Gibbs C J, Gajdusek D C. Characterization of four new adenovirus serotypes isolated from chimpanzee tissue explants. Am J Epidemiol. 1971;94:166. doi: 10.1093/oxfordjournals.aje.a121308. [DOI] [PubMed] [Google Scholar]
- 4.Berkner K L, Sharp P A. Generation of adenovirus by transfection of plasmids. Nucleic Acids Res. 1983;11:6003–6020. doi: 10.1093/nar/11.17.6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brooks B R, Bruccoleri R E, Olafson B D, States D J, Swaminathan S, Karplus M. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J Comp Chem. 1983;4:187–217. [Google Scholar]
- 6.Chirmule N, Propert K J, Magosin S A, Qian Y, Qian R, Wilson J M. Immune responses to adenovirus and adeno-associated virus in humans. Gene Ther. 1996;6:1574–1583. doi: 10.1038/sj.gt.3300994. [DOI] [PubMed] [Google Scholar]
- 7.Christ M, Louis B, Stoeckel F, Dieterle A, Grave L, Dreyer D, Kintz J, Ali Hadji D, Lusky M, Mehtali M. Modulation of the inflammatory properties and hepatotoxicity of recombinant adenovirus vectors by the viral E4 gene products. Hum Gene Ther. 2000;11:415–427. doi: 10.1089/10430340050015888. [DOI] [PubMed] [Google Scholar]
- 8.Crawford-Miksza L, Schnurr D P. Analysis of 15 adenovirus hexon proteins reveals the location and structure of seven hypervariable regions containing serotype-specific residues. J Virol. 1996;70:1836–1844. doi: 10.1128/jvi.70.3.1836-1844.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis A R, Meyers K, Wilson J M. High throughput method for creating and screening recombinant adenoviruses. Gene Ther. 1998;5:1148–1152. doi: 10.1038/sj.gt.3300705. [DOI] [PubMed] [Google Scholar]
- 10.Dedieu J F, Vigne E, Torrent C, Jullien C, Mahfouz I, Caillaud J M, Aubailly N, Orsini C, Guillaume J M, Opolon P, Delaere P, Perricaudet M, Yeh P. Long-term gene delivery into the livers of immunocompetent mice with E1/E4-defective adenoviruses. J Virol. 1997;71:4626–4637. doi: 10.1128/jvi.71.6.4626-4637.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dmitriev I, Krasnykh V, Miller C R, Wang M, Kashentseva E, Mikheeva G, Belousova N, Curiel D T. An adenovirus vector with genetically modified fibers demonstrates expanded tropism via utilization of a coxsackievirus and adenovirus receptor-independent cell entry mechanism. J Virol. 1998;72:9706–9713. doi: 10.1128/jvi.72.12.9706-9713.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fisher K J, Gao G, Weitzman M D, DeMatteo R, Burda J F, Wilson J M. Transduction with recombinant adeno-associated virus for gene therapy is limited by leading-strand synthesis. J Virol. 1996;70:520–532. doi: 10.1128/jvi.70.1.520-532.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gahery-Segard H, Juillard V, Gaston J, Lengagne R, Pavirani A, Boulanger P, Guillet J G. Humoral immune response to the capsid components of recombinant adenoviruses: routes of immunization modulate virus-induced Ig subclass shifts. Eur J Immunol. 1997;27:653–659. doi: 10.1002/eji.1830270312. [DOI] [PubMed] [Google Scholar]
- 14.Gall J, Kass-Eisler A, Leinwand L, Falck-Pedersen E. Adenovirus type 5 and 7 capsid chimera: fiber replacement alters receptor tropism without affecting primary immune neutralization epitopes. J Virol. 1996;70:2116–2123. doi: 10.1128/jvi.70.4.2116-2123.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gall J G D, Crystal R G, Falck-Pedersen E. Construction and characterization of hexon-chimeric adenoviruses: specification of adenovirus serotype. J Virol. 1998;72:10260–10264. doi: 10.1128/jvi.72.12.10260-10264.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao G, Yang Y, Wilson J M. Biology of adenovirus vectors with E1 and E4 deletions for liver-directed gene therapy. J Virol. 1996;70:8934–8943. doi: 10.1128/jvi.70.12.8934-8943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graham F L, Smiley J, Russell W C, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 18.Guex N, Peitsch M C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 19.Havenga M J E, Lemckert A A C, Grimbergen J M, Vogels R, Huisman L G M, Valerio D, Bout A, Quax P H A. Improved adenovirus vectors for infection of cardiovascular tissues. J Virol. 2001;75:3335–3342. doi: 10.1128/JVI.75.7.3335-3342.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Juillard V, Villefroy P, Godfrin D, Pavirani A, Venet A, Guillet J G. Long-term humoral and cellular immunity induced by a single immunization with replication-defective adenovirus recombinant vector. Eur J Immunol. 1995;25:3467–3673. doi: 10.1002/eji.1830251239. [DOI] [PubMed] [Google Scholar]
- 21.Kitchingman G R. Restriction mapping and molecular cloning of adenovirus type 4 (subgroup E) DNA. Gene. 1982;20:205–210. doi: 10.1016/0378-1119(82)90039-7. [DOI] [PubMed] [Google Scholar]
- 22.Kozarsky K F, McKinley D R, Austin L L, Raper S E, Stratford-Perricaudet L D. In vivocorrection of low density lipoprotein receptor deficiency in the Watanabe heritable hyperlipidemic rabbit with recombinant adenoviruses. J Biol Chem. 1994;269:13695–13702. [PubMed] [Google Scholar]
- 23.Li Q, Wadell G. The degree of genetic variability among adenovirus type 4 strains isolated from man and chimpanzee. Arch Virol. 1998;101:65–77. doi: 10.1007/BF01314652. [DOI] [PubMed] [Google Scholar]
- 24.Morral N, O'Neal W, Zhou H, Langston C, Beaudet A. Immune responses to reporter proteins and high viral dose limit duration of expression with adenoviral vectors: comparison of E2a wild type and E2a deleted vectors. Hum Gene Ther. 1997;8:1275–1286. doi: 10.1089/hum.1997.8.10-1275. [DOI] [PubMed] [Google Scholar]
- 25.Ostapchuk P, Hearing P. Pseudopackaging of adenovirus type 5 genomes into capsids containing the hexon proteins of adenovirus serotypes B, D, or E. J Virol. 2001;75:45–51. doi: 10.1128/JVI.75.1.45-51.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roy S, Shirley P S, McClelland A, Kaleko M. Circumvention of immunity to the adenovirus major coat protein hexon. J Virol. 1998;72:6875–6879. doi: 10.1128/jvi.72.8.6875-6879.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rux J J, Burnett R M. Type-specific epitope locations revealed by X-ray crystallographic study of adenovirus type 5 hexon. Mol Ther. 2000;1:18–30. doi: 10.1006/mthe.1999.0001. [DOI] [PubMed] [Google Scholar]
- 28.Shayakhmetov D M, Papayannopoulou T, Stamatoyannopoulos G, Lieber A. Efficient gene transfer into human CD34+cells by a retargeted adenovirus vector. J Virol. 2000;74:2567–2583. doi: 10.1128/jvi.74.6.2567-2583.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tripathy S K, Black H B, Goldwasser E, Leiden J M. Immune responses to transgene-encoded proteins limit the stability of gene expression after injection of replication-defective adenovirus vectors. Nat Med. 1996;2:545–550. doi: 10.1038/nm0596-545. [DOI] [PubMed] [Google Scholar]
- 30.Wang Q, Greenburg G, Bunch D, Finer M H. Persistent transgene expression in mouse liver following in vivogene transfer with a ΔE1/ΔE4 adenovirus vector. Gene Ther. 1997;4:393–400. doi: 10.1038/sj.gt.3300404. [DOI] [PubMed] [Google Scholar]
- 31.Wickham T J, Tzeng E, Shears L L, Roelvink P W, Li Y, Lee G M, Brough D E, Lizonova A, Kovesdi I. Increased in vitro and in vivo gene transfer by adenovirus vectors containing chimeric fiber proteins. J Virol. 1997;71:8221–8229. doi: 10.1128/jvi.71.11.8221-8229.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wigand R, Mauss M, Adrian T. Chimpanzee adenoviruses are related to four subgenera of human adenoviruses. Intervirology. 1989;30:1–9. doi: 10.1159/000150069. [DOI] [PubMed] [Google Scholar]
- 33.Willimzik H F, Kalter S S, Lester T L, Wigand R. Immunological relationship among adenoviruses of humans, simians, and nonprimates as determined by the neutralization test. Intervirology. 1981;15:28–36. doi: 10.1159/000149211. [DOI] [PubMed] [Google Scholar]
- 34.Wivel N A, Gao G, Wilson J M. Adenovirus vectors: the development of human gene therapy. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1999. [Google Scholar]
- 35.Yang Y, Ertl H C, Wilson J M. MHC class I-restricted cytotoxic T lymphocytes to viral antigens destroy hepatocytes in mice infected with E1-deleted recombinant adenoviruses. Immunity. 1994;1:433–442. doi: 10.1016/1074-7613(94)90074-4. [DOI] [PubMed] [Google Scholar]
- 36.Yang Y, Li Q, Ertl H C, Wilson J M. Cellular and humoral immune responses to viral antigens create barriers to lung-directed gene therapy with recombinant adenoviruses. J Virol. 1995;69:2004–2015. doi: 10.1128/jvi.69.4.2004-2015.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Y, Trinchieri G, Wilson J M. Recombinant IL-12 prevents formation of blocking IgA antibodies to recombinant adenovirus and allows repeated gene therapy to mouse lung. Nat Med. 1995;1:890–893. doi: 10.1038/nm0995-890. [DOI] [PubMed] [Google Scholar]
- 38.Yao S N, Farjo A, Roessler B J, Davidson B L, Kurachi K. Adenovirus-mediated transfer of human factor IX gene in immunodeficient and normal mice: evidence for prolonged stability and activity of the transgene in liver. Viral Immunol. 1996;9:141–153. doi: 10.1089/vim.1996.9.141. [DOI] [PubMed] [Google Scholar]
- 39.Zsengeller Z K, Wert S E, Hull W M, Hu X, Yei S, Trapnell B C, Whitsett J A. Persistence of replication-deficient adenovirus-mediated gene transfer in lungs of immune-deficient (nu/nu) mice. Hum Gene Ther. 1995;6:457–467. doi: 10.1089/hum.1995.6.4-457. [DOI] [PubMed] [Google Scholar]