Abstract
Two mechanisms have been proposed for the intracellular movement of enveloped vaccinia virus virions: rapid actin polymerization and microtubule association. The first mechanism is used by the intracellular pathogens Listeria and Shigella, and the second is used by cellular vesicles transiting from the Golgi network to the plasma membrane. To distinguish between these models, two recombinant vaccinia viruses that express the B5R membrane protein fused to enhanced green fluorescent protein (GFP) were constructed. One had Tyr112 and Tyr132 of the A36R membrane protein, which are required for phosphorylation and the nucleation of actin tails, conservatively changed to Phe residues; the other had the A36R open reading frame deleted. Although the Tyr mutant was impaired in Tyr phosphorylation and actin tail formation, digital video and time-lapse confocal microscopy demonstrated that virion movement from the juxtanuclear region to the periphery was saltatory with maximal speeds of >2 μm/s and was inhibited by the microtubule-depolymerizing drug nocodazole. Moreover, this actin tail-independent movement was indistinguishable from that of a control virus with an unmutated A36R gene and closely resembled the movement of vesicles on microtubules. However, in the absence of actin tails, the Tyr mutant did not induce the formation of motile, virus-tipped microvilli and had a reduced ability to spread from cell to cell. The deletion mutant was more severely impaired, suggesting that the A36R protein has additional roles. Optical sections of unpermeabilized, B5R antibody-stained cells that expressed GFP-actin and were infected with wild-type vaccinia virus revealed that all actin tails were associated with virions on the cell surface. We concluded that the intracellular movement of intracellular enveloped virions occurs on microtubules and that the motile actin tails enhance extracellular virus spread to neighboring cells.
The infectious forms of some enveloped viruses are assembled at the plasma membrane, providing direct access to the extracellular environment. Other viruses, however, are assembled in the nucleus or internal regions of the cytoplasm and require locomotion to reach the periphery of the cell. Such movement is likely to involve the actin or microtubule cytoskeleton (28). Infectious intracellular mature vaccinia virus virions (IMV) form in juxtanuclear factory regions of the cytoplasm (5, 19) and are wrapped by a double membrane derived from trans-Golgi or endosomal cisternae to become intracellular enveloped virions (IEV), which are then translocated to the periphery of the cell where the outer IEV and plasma membranes fuse (12, 15, 19, 26, 31). The plasma membrane adherent and the released extracellular virions are called cell-associated enveloped virions (CEV) and extracellular enveloped virions (EEV), respectively. The association of actin filaments with CEV at the tips of specialized microvilli has been appreciated since the early electron and immunofluorescence microscopic studies of Stokes (29) and Hiller et al. (13). Using video microscopy, Cudmore et al. (3) reported that IEV were propelled through the cytoplasm on the tips of actin tails, much like the intracellular pathogens Listeria, Shigella, and Rickettsia (2). In a series of elegant experiments, Way and coworkers (9, 18) identified viral and cellular proteins involved in actin nucleation. Nevertheless, actin tail formation cannot entirely account for the movement of IEV, since deletion of any one of three IEV membrane proteins (A33R, A34R, or A36R) prevented the formation of actin tails and specialized microvilli but did not block the formation of extracellular virions (22, 24, 36, 38). Moreover, by visualizing the movement of a recombinant vaccinia virus (vB5R-GFP) that expressed the IEV membrane protein B5R fused to enhanced green fluorescent protein (GFP), we provided evidence that IEV were transported from the juxtanuclear region to the periphery via microtubules (34). This conclusion was based on the maximal speed (2.5 μm/s) and saltatory motion of the IEV, which was reversibly halted by the microtubule-depolymerizing drug nocodazole.
The present study was designed to directly visualize IEV movement under conditions in which actin tails could not form. Because actin filaments are intimately involved with microtubule function (11), we avoided the use of drugs, which might have broad effects. Instead, we constructed a novel vaccinia virus mutant with a specific block in actin tail nucleation. The target of the mutation was the A36R gene, which encodes a type Ib integral membrane protein component of the outer IEV membrane that is required for actin tails (20, 23, 24, 32, 38, 39). Transfection and in vitro binding studies indicated that nucleation of actin is regulated by phosphorylation of Tyr112 alone or in conjunction with Tyr132 located in the cytoplasmic domain of the A36R protein (9). We therefore constructed two vaccinia virus mutants, both of which expressed B5R-GFP to visualize IEV movement. In one mutant (vB5R-GFP/A36R-YdF), the codons for Tyr112 and Tyr132 of A36R were both conservatively changed to Phe. The other mutant (vB5R-GFP/ΔA36R) had a deletion of nearly the entire A36R gene. Both mutants were characterized with regard to A36R expression, Tyr phosphorylation, virus assembly, intracellular movement, and cell-to-cell spread. Despite the absence of Tyr phosphorylation and an inability to form actin tails or specialized microvilli, confocal and digital video microscopy revealed that the intracellular movement of vB5R-GFP/A36R-YdF was unimpaired. Nevertheless, the plaques formed by vB5R-GFP/A36R-YdF were smaller than those of vB5R-GFP. These results, and our demonstration that virions associated with actin tails and microvilli are external to the plasma membrane, support the idea that the primary role of actin tails is to facilitate virus spread. vB5R-GFP/ΔA36R exhibited a more impaired phenotype than vB5R-GFP/A36R-YdF, suggesting that the A36R protein may have an additional structural or functional role.
MATERIALS AND METHODS
Construction of B5R-GFP viruses.
Construction of vaccinia viruses vB5R-GFP and vΔA36R and the plasmid pB5R-GFP, containing the B5R open reading frame (ORF) fused to GFP sequences and approximately 500 bp of flanking sequence on each side, have been previously described (20, 33, 34). pB5R-GFP was transfected with Lipofectamine (Invitrogen) into HeLa cells that had been infected with vΔA36R (35). Recombinant viruses that formed green fluorescent foci were plaque purified three times. Proper insertion of the ORF for B5R-GFP into the final recombinant (vB5R-GFP/ΔA36R) was confirmed by PCR.
The primers TGTCGACTAACGTACGCCGCCATG and TAAGCTTGAATACAGACAACGGCAAA were used to amplify the A36R ORF plus 500 bp of flanking sequence on each side and to add a HindIII site and a SalI site (underlined). The amplified product was cloned into pGEM-T to yield pBMW-32T and sequenced. The coding sequence of A36R was changed so that Phe instead of Tyr residues were encoded at residues 112 and 132, using two-stage PCR with plasmid pBMW32-T as the template. The following primer pairs were used to amplify three overlapping fragments: CAACTGAGTCAAAAATGTGTTCTGTGCTAGG and M13 reverse (Promega), CATTTTTGACTCAGTTGCTGGAAGCACGCTG and GTTCTGGAAAATAGTCTGTTCATTACGATC, and CAGACTATTTTCCAGAAACACTACAGTAGTAA and M13 forward (Promega). The resulting amplified products were purified and joined together by a fourth amplification, and the resulting product was cloned into pGEM-T and sequenced. Subsequently, the mutated A36R coding sequence plus 500 bp of flanking sequence was excised from pGEM-T by digestion with HindIII-SalI and ligated into similarly cleaved pBSgptgus (16) to yield pBMW33-BSgptgus. Recombination was used to insert the mutated A36R-coding sequence into the deleted A36R site of vB5R-GFP/ΔA36R, and the new recombinant virus was isolated by transient dominant selection, essentially as described previously (16). HeLa cells were infected at a multiplicity of 0.05 with vB5R-GFP/ΔA36R and transfected 2 h later with pBMW33-Bsgptgus as described previously (35). After 2 days, cells were harvested, frozen and thawed three times, and used to infect BS-C-1 cells that were treated with mycophenolic acid (MPA), xanthine, and hypoxanthine at concentrations of 25, 250, and 15 μg per ml, respectively. Recombinant viruses that resulted from a single crossover would have the entire plasmid integrated into the viral genome and consequently would be resistant to MPA and positive for β-glucuronidase (GUS). MPA-resistant and GUS-positive viruses were plaque purified twice in the presence of MPA before three more plaque purifications in the absence of MPA to obtain the desired double-crossover mutant. X-Gluc (5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid) staining was used to confirm the absence of the gus gene, and PCR amplification and sequencing were used to confirm the insertion of the mutated A36R ORF.
Cells.
HeLa and BS-C-1 cell monolayers were grown in Dulbecco's modified Eagle's medium (DMEM) and Earle's minimum essential medium (Quality Biologicals), respectively, supplemented with 10% fetal bovine serum. HeLa cells were transfected with pEGFP-actin (Clontech) and Lipofectamine and selected with Geneticin (Invitrogen) as suggested by the manufacturer. Isolated green fluorescent colonies were screened by staining with rhodamine-phalloidin to determine the levels of GFP-actin expression. Stable cell lines were maintained in DMEM supplemented with 10% fetal bovine serum and 50 μg of Geneticin per ml.
Virus replication and plaque assay.
All recombinant viruses were derived from the WR strain. Virus was propagated in HeLa cells, and plaque assays were carried out in BS-C-1 cells using standard procedures. Images were obtained at 3 days after infection using a Leica DMIRBE inverted fluorescence microscope with a cooled charged-coupled device (Princeton Instruments) that was controlled using Image Pro software. After imaging, cells were stained with crystal violet, rinsed with water, and allowed to air dry. Stained plates were imaged with a Kodak Image Station 440cf using Kodak 1D Image Analysis software (Kodak Digital Science), and the areas of at least 140 well-separated plaques for each virus were measured in square pixels using the same software. Average plaque sizes and standard deviations were calculated using Microsoft Excel.
To measure virus replication and spread, BS-C-1 cells were inoculated at a multiplicity of 0.01 for 2 h and then washed twice with fresh growth medium to remove unabsorbed virus. At various times after infection, the medium was removed from each well and clarified by low-speed centrifugation to remove detached cells and debris. The adherent cells were scraped into 1 ml of fresh medium and combined with the cell pellet from the clarified medium. The cells were suspended by vortexing, frozen and thawed three times, and then sonicated to release virus. Titers of virus in the cell lysate and the medium were determined in duplicate by plaque assay on BS-C-1 cells.
Virus purification.
For radioactive labeling and CsCl purification of virus, monolayers of RK13 cells were infected at a multiplicity of 10. After 2 h, the inoculum was removed and replaced with normal medium. At 4 h after infection, the medium was replaced with methionine- and cysteine-free DMEM (Invitrogen) to which 2.5% dialyzed fetal bovine serum (Invitrogen) and 500 μCi of a mixture of [35S]methionine and [35S]cysteine (Easy Tag; Amersham) had been added. At 18 h after infection, the medium was harvested and cells and large debris were removed by low-speed centrifugation. Cells were dislodged by scraping, collected by low-speed centrifugation, resuspended in swelling buffer (10 mM Tris, pH 10), incubated on ice for 10 min, disrupted by Dounce homogenization, and clarified by low-speed centrifugation. Virus from the cell lysate and from the medium was centrifuged through a 36% sucrose cushion, resuspended in swelling buffer with sonication, and banded on a preformed CsCl step gradient as previously described (7). Gradients were fractionated from the bottom of the tube, and the amount of radiation present in each fraction was determined by scintillation counting.
Syncytium formation.
HeLa cells were infected at a multiplicity of 10. After 15 h, the cells were washed with pH 7.4 phosphate-buffered saline (PBS) and then incubated for 2 min in either fusion buffer [PBS with 10 mM 2-(N-morpholino)ethanesulfonic acid and 10 mM HEPES at pH 5.5] or control buffer (PBS at pH 7.4). The buffers were replaced with DMEM growth medium and incubated for 3 h before analysis with a Leica DMIRBE inverted fluorescence microscope as described above.
Electron microscopy.
For immunoelectron microscopy, RK13 cells were grown in 60-mm-diameter dishes and infected with vaccinia virus at a multiplicity of 10. After 24 h, the cells were prepared for freezing as previously described (37) except that the final fixation step was in 8% paraformaldehyde (37). Ultrathin sections were cut, collected, immunostained, and viewed as previously described (4). The GFP polyclonal antibody (Clontech) was used at a dilution of 1:75.
For scanning electron microscopy, RK13 cells were grown on coverslips and infected at a multiplicity of 10 overnight before being fixed with 2% glutaraldehyde in 0.1 M sodium cacodylate (pH 7.4). Fixed cells were washed in cacodylate buffer, postfixed with 1% osmium tetroxide, and dehydrated in ethanol. After dehydration, samples were critical point dried, mounted on stubs, coated with gold-palladium alloy, and viewed on a Hitachi S-4700 field emission scanning electron microscope at an accelerating voltage of 3 kV.
Western blotting.
HeLa cells were infected with 5 PFU of virus per cell and incubated overnight. At 18 h after infection, cells were harvested by replacing the medium with lysis buffer (100 mM Tris [pH 8.0], 2% sodium dodecyl sulfate [SDS], 0.4 mM sodium orthovanadate, and 0.2 mM phenylmethylsulfonyl fluoride) followed by several passages through a syringe to shear DNA. Lysates were mixed with protein loading buffer, resolved by electrophoresis on an SDS–10 to 20% polyacrylamide gel (Invitrogen), and transferred to a nitrocellulose membrane. Membranes were incubated with anti-P-Tyr monoclonal antibody (MAb) clone 4G10 (Upstate Biotechnology) followed by horseradish peroxidase-conjugated sheep anti-mouse antibody (Amersham). Bound antibodies were detected with chemiluminescence reagents (Pierce) as directed by the manufacturer. Following detection, the blot was stripped with 10 mM 2-mercaptoethanol–2% SDS in 62.5 mM Tris (pH 6.9), blocked, and incubated with polyclonal antibody to the A36R protein followed by horseradish peroxidase-conjugated donkey anti-rabbit antibody (Amersham). Bound antibodies were detected as described above.
Fluorescence microscopy of fixed cells.
HeLa cells were grown to confluence on coverslips and infected at a multiplicity of 1. Infected cells were fixed with 4% paraformaldehyde and permeabilized with Triton X-100, both in PBS. Actin filaments were stained with rhodamine-conjugated phalloidin (Molecular Probes), and coverslips were mounted in mowoil which contained 1 μg of 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) (Molecular Probes) per ml to visualize DNA in the nucleus and viral factories. Images were collected on a Leica TCS-NT/SP inverted confocal microscope system with an attached argon ion laser (Coherent Inc.). Images were overlaid using Adobe PhotoShop version 6.0. In order to determine the percentage of cells containing actin tails, at least 200 infected cells from each of four separate experiments were scored for the presence of one or more actin tails.
For analysis of the association of vaccinia virus with actin tails, HeLa cells stably expressing GFP-actin were infected and fixed as described above but without permeabilization. Fixed cells were stained with anti-B5R MAb 19C2 (26) followed by indodicarbocyanine (Cy5)-conjugated goat anti-rat secondary antibody (Jackson ImmunoResearch Laboratories). Hoechst dye was included in the first of three washes with PBS before mounting coverslips and sealing with rubber cement. Cells were imaged by confocal microscopy as described above, and Z-series were processed using Imaris 3.0.2 (Bitplane AG) and Adobe Photoshop.
Fluorescence microscopy of live cells.
HeLa cells were plated at ≈80% confluence onto ΔTC3 dishes (Bioptechs, Inc.), and on the next day they were infected with 0.2 PFU of virus per cell. On the following day, cells were imaged by digital video microscopy using a Hammumatsu C5985 camera and controller that was attached to a Leica DMIRBE inverted fluorescence microscope. Images were digitized using an ICPCI video capture card controlled by Image Pro Plus software. During imaging, cells were maintained on a heated ΔTC3 stage (Bioptechs) with the temperature set at 35°C. Fresh medium supplemented with 2.5% fetal calf serum and 25 mM HEPES was perfused onto the dish at a rate of 0.1 ml/min throughout the experiment using a P720 peristaltic pump (Instech Laboratories). Velocities of virion movement were calculated using the public-domain software NIH Image 1.62 developed at the National Institutes of Health (available at http://rsb.info.nih.gov/nih-image/).
RESULTS
Construction of recombinant vaccinia viruses, with a deleted or mutated A36R gene, that express B5R-GFP.
We constructed two new A36R vaccinia virus mutants, both of which expressed B5R-GFP. Having shown that B5R-GFP could functionally replace the endogenous copy of B5R (34), we similarly modified a previously characterized A36R deletion mutant (vΔA36R) (20, 24, 38). HeLa cells were infected with the vΔA36R virus and transfected with a plasmid containing B5R-GFP and B5R flanking sequences including the natural B5R promoter. Recombinant viruses forming green fluorescent plaques were easily identified using an inverted fluorescence microscope without the need for selection. Individual recombinants were further plaque purified and screened by PCR to confirm the presence of the ORF encoding the B5R fusion protein and the absence of the wild-type B5R ORF (data not shown). The resulting virus was named vB5R-GFP/ΔA36R.
Transient dominant selection (8) was used to insert the mutated A36R ORF with Phe codons substituted for Tyr112 and Tyr132 codons into the A36R deletion site of vB5R-GFP/ΔA36R. There were several advantages to this approach. Starting with a small-plaque-forming A36R deletion mutant was preferable to starting with one that formed large plaques, which might obscure those of the new mutant. In addition, because the selection and marker genes used for transient dominant selection were not stably integrated into the viral genome, the final recombinant virus had the mutated A36R gene in its usual position under the regulation of its natural promoter with no adjacent reporter genes. HeLa cells were infected with vB5R-GFP/ΔA36R and then transfected with pBMW33-BSgptgus, which contained the gpt and gus genes under the control of vaccinia virus promoters adjacent to the mutated A36R ORF and flanking sequences. Plaques were picked in the presence of MPA to select for single-crossover MPA-resistant, GUS-positive recombinant viruses and then in the absence of MPA to isolate viruses that had lost the marker genes by a second recombination event. GUS-negative recombinant viruses were screened by PCR for the presence of the mutated A36R-coding sequence, and one, vB5R-GFP/A36R-YdF, was amplified and further characterized.
Plaque phenotypes of mutant viruses.
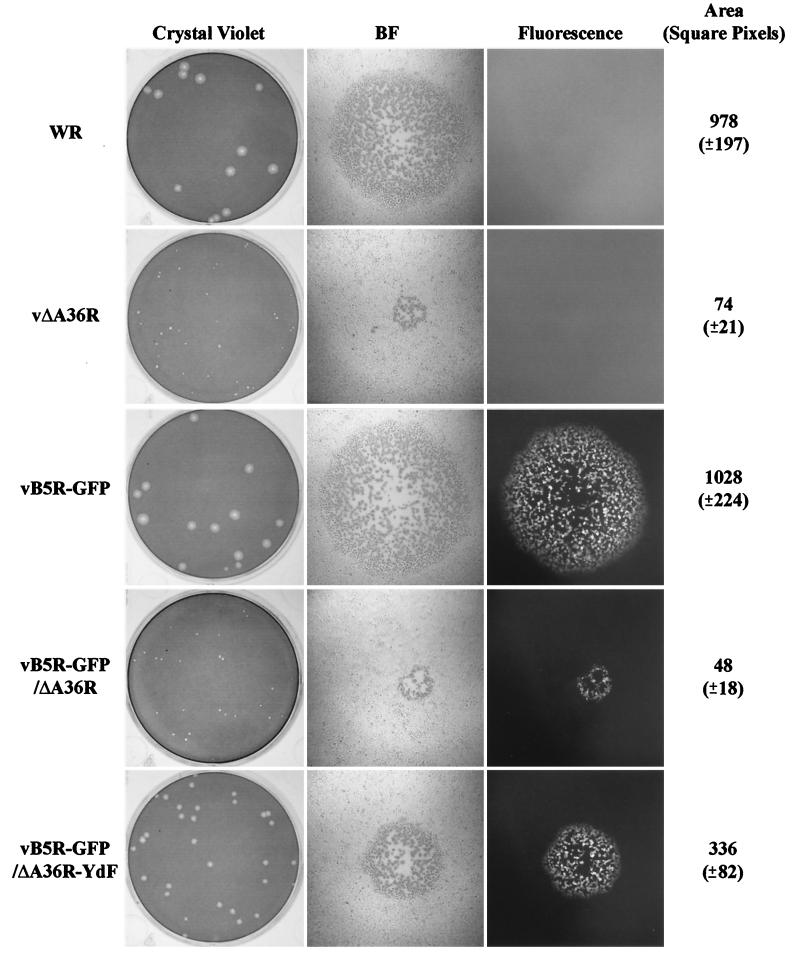
Plaques of vaccinia virus strains WR, vΔA36R, vB5R-GFP, vB5R-GFP/ΔA36R, and vB5R-GFP/A36R-YdF were compared using a methylcellulose overlay to prevent the formation of satellites. Under these conditions, plaque size is determined by direct cell-to-cell spread. As previously shown (34), standard vaccinia virus strain WR and vB5R-GFP plaques were of similar size and appearance, consistent with the normal function of the B5R-GFP fusion protein (Fig. 1). Likewise, the recombinant vB5R-GFP/ΔA36R formed tiny plaques like those of the parental vΔA36R (Fig. 1). The vB5R-GFP/A36R-YdF plaques, however, were intermediate between those containing an intact A36R ORF and those with A36R deleted (Fig. 1). These differences were quantified by measuring the areas of at least 140 plaques formed by each of the five different viruses. The vB5R-GFP/A36R-YdF plaques were about one-third of the size of those formed by viruses having an intact A36R ORF but were sevenfold larger than those formed by the A36R deletion mutant (Fig. 1). The larger plaque size of the Tyr mutant relative to the deletion mutant indicated a greater impairment of virus lacking the entire A36R ORF. As expected, the plaques of the three recombinant viruses expressing B5R-GFP were fluorescent (Fig. 1).
FIG. 1.
Plaque phenotypes. The indicated viruses were plated on monolayers of BS-C-1 cells. After 3 days, plaques were imaged using light and fluorescence microscopy and then stained with crystal violet. Areas of at least 140 individual plaques for each of the five viruses were determined, and the averages and standard deviations (in parentheses) were calculated. BF, bright field.
Effects of A36R mutations on low-pH-induced syncytium formation.
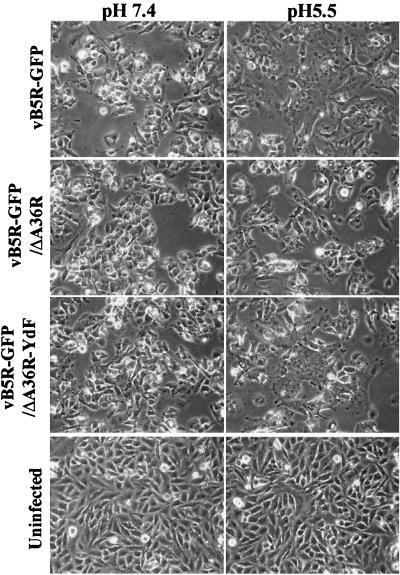
The ability of vaccinia virus-infected cells to undergo fusion by a brief exposure to low pH is due to the presence of virions on the cell surface and frequently correlates with plaque size (1, 6, 10, 35). We tested the ability of the three recombinant B5R-GFP viruses to mediate the induction of low-pH-induced fusion. HeLa cells were infected for 15 h and then incubated for 2 min at either pH 5.5 or 7.4 and for an additional 3 h in standard medium. Uninfected cells and infected cells treated with pH 7.4 buffer showed no signs of cell fusion (Fig. 2). In contrast, cells infected with either vB5R-GFP or vB5R-GFP/A36R-YdF showed extensive cell fusion and formed large syncytia when treated with pH 5.5 buffer (Fig. 2). Cells infected with vB5R-GFP/ΔA36R, however, showed less extensive fusion with few or no large polykaryons (Fig. 2), in agreement with previous analyses of an A36R deletion mutant without the B5R-GFP gene (24, 38). Thus, the difference in plaque sizes of vB5R-GFP/ΔA36R and vB5R-GFP/A36R-YdF correlated with their efficiency in forming syncytia.
FIG. 2.
Induction of syncytia. Uninfected or infected HeLa cells were incubated for 15 h, briefly treated with buffer at pH 7.4 or 5.5, and then incubated in regular medium for 3 h. The cells were examined by phase-contrast microscopy. Recombinant viruses used for infection are indicated on the left.
Replication of recombinant viruses.
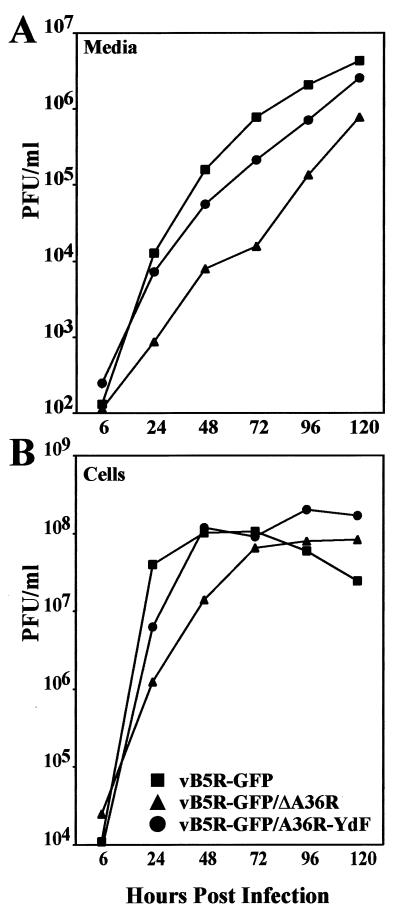
Replication of the mutant viruses was compared over a 5-day period following a low-multiplicity infection chosen to emphasize virus spread. As is typical of viruses derived from the WR strain, much of the infectivity remained cell associated even after the long period. As seen in the log plots, vB5R-GFP/ΔA36R replicated much more slowly than the others, while vB5R-GFP/A36R-YdF lagged slightly behind vB5R-GFP under these conditions (Fig. 3).
FIG. 3.
Virus replication. BS-C-1 cells were infected with 0.01 PFU of the indicated viruses per cell. Virus titers in the medium (A) and cell lysates (B) were determined by plaque assay.
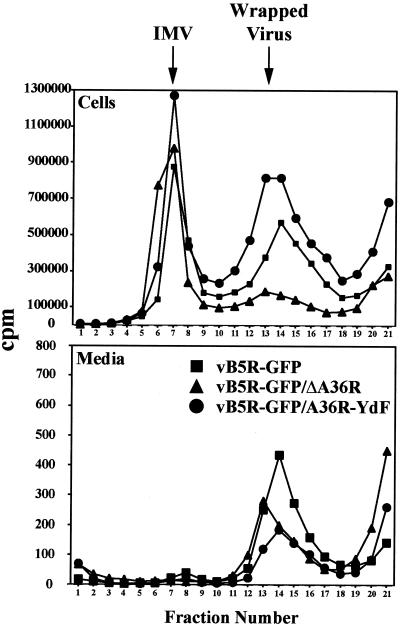
To further examine particle formation, cells infected with recombinant viruses were incubated in medium containing [35S]methionine and [35S]cysteine from 4 h on, when host protein synthesis is largely inhibited. Particles present in the medium and extracted from lysed cells were analyzed by CsCl density gradient centrifugation to distinguish IMV from wrapped virions (IEV, CEV, or EEV). When vB5R-GFP and vB5R-GFP/A36R-YdF were compared, there was little difference between the ratios of IMV to wrapped virus extracted from cells (Fig. 4). In contrast, there was much less wrapped vB5R-GFP/ΔA36R (Fig. 4). Virtually no IMV and relatively small numbers of wrapped virus particles were detected in the medium, with those from vB5R-GFP being the most abundant (Fig. 4).
FIG. 4.
Isolation of IMV and wrapped viruses. RK13 cells were infected at a multiplicity of 10 with the indicated recombinant viruses. From 4 to 18 h after infection, the cells were labeled with 500 μCi of a mixture of [35S]methionine and [35S]cysteine. Particles in the medium and cell lysates were concentrated by sedimentation through a sucrose cushion, applied to CsCl density gradients, and centrifuged. The amounts of radioactive material in the fractions were determined by scintillation counting.
Effects of A36R mutations on the formation of virus particles determined by transmission and scanning electron microcopy.
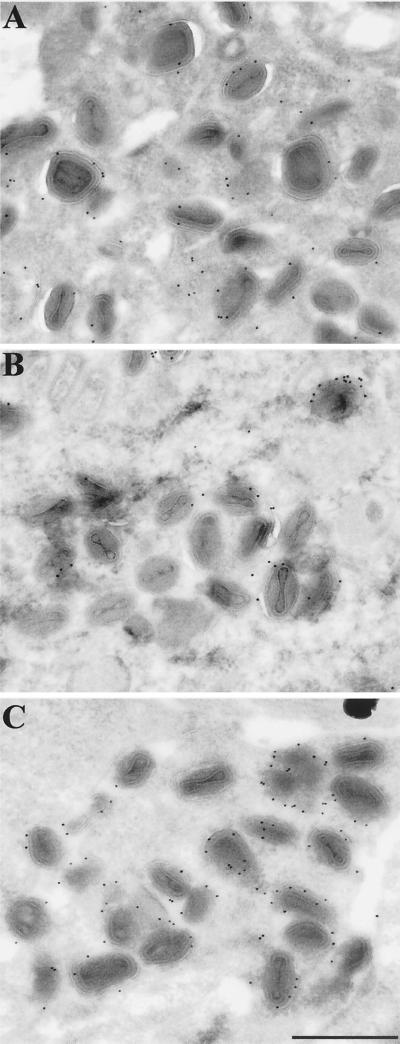
Cells infected with recombinant viruses were examined by transmission electron microscopy to determine the effects of the mutations on the intracellular forms of virus. All stages of virus assembly were detected in each case, although there were noticeably fewer IEV with vB5R-GFP/ΔA36R. The IEV membranes were specifically labeled using an antibody to GFP followed by protein A-conjugated gold particles, which demonstrated the presence of the B5R-GFP fusion protein in these membranes (Fig. 5).
FIG. 5.
Immunoelectron microscopy of infected cells. RK13 cells were infected with 10 PFU of vB5R-GFP (A), vB5R-GFP/ΔA36R (B), or vB5R-GFP/A36R-YdF (C) per cell. After 24 h, the cells were fixed, cryosectioned, and incubated successively with polyclonal anti-GFP antibody and 10-nm-diameter gold particles conjugated to protein A. Bar, 500 nm.
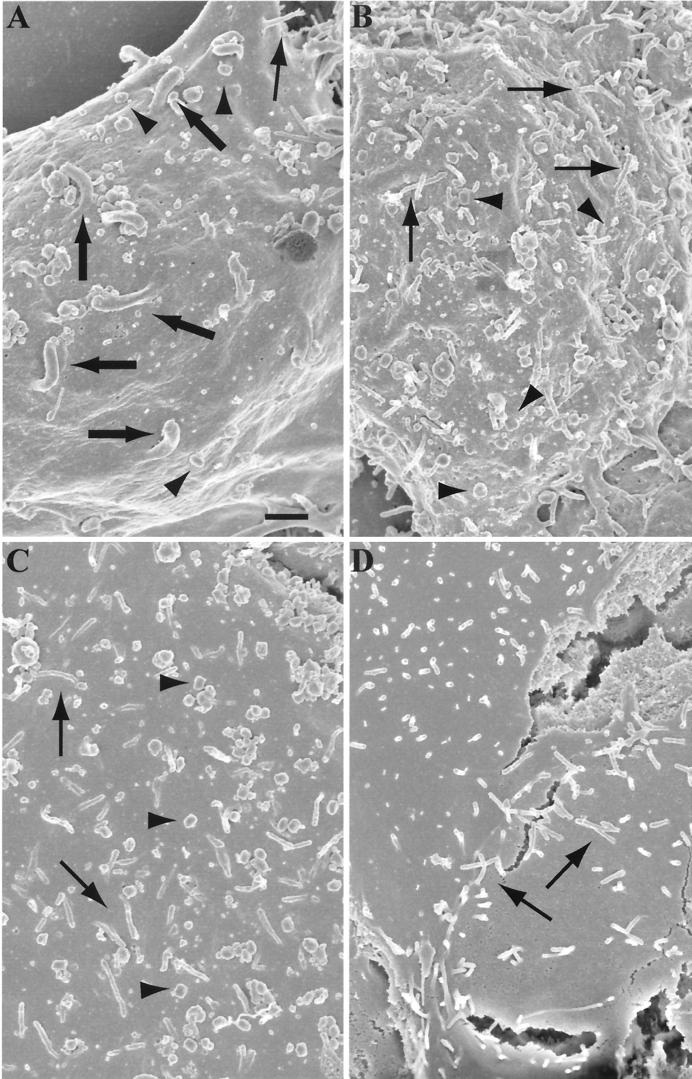
Cells infected with vaccinia virus produce thick specialized microvilli projecting from their cell surface (13, 29). These projections are tipped with virions and can be visualized by scanning electron microscopy. Using this technique, virus-tipped projections of about 0.3 μm in diameter were seen on the surfaces of cells infected with vB5R-GFP (Fig. 6A). In contrast, only slender microvilli of about 0.1 μm were present on the surfaces of uninfected cells or cells infected with vB5R-GFP/ΔA36R or vB5R-GFP/A36R-YdF (Fig. 6B, C, and D). Nevertheless, the surfaces of cells infected with each of the viruses contained brick-shaped virions, which were absent from the uninfected cells. Thus, both vB5R-GFP/ΔA36R and vB5R-GFP/A36R-YdF exhibited a specific defect in the formation of specialized microvilli.
FIG. 6.
Scanning electron microscopy of infected cells. RK13 cells were infected with 10 PFU of vB5R-GFP (A), vB5R-GFP/ΔA36R (B), or vB5R-GFP/A36R-YdF (C) per cell or were uninfected (D). After 16 h, the cells were fixed, coated with gold-palladium alloy, and viewed with an Hitachi S-4700 field emission scanning electron microscope at an accelerating voltage of 3 kV. Thick arrows, virus-tipped specialized microvilli; thin arrows, slender cellular microvilli; arrowheads, virions on the cell surface. Bar, 1 μm.
A36R Tyr phosphorylation.
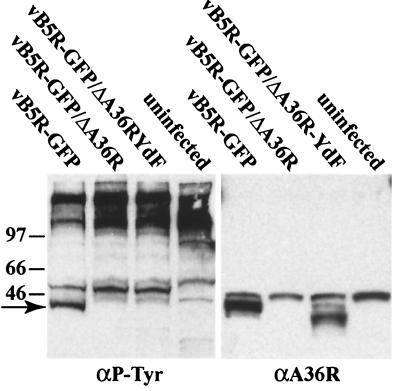
Tyr112 and Tyr132 were identified as the phosphorylation sites of the A36R protein by transfection experiments (9). The generation of a mutant vaccinia virus with these two Tyr residues mutated allowed us to investigate phosphorylation under more natural infection conditions. Proteins from lysates of uninfected cells or cells infected with recombinant viruses were resolved by SDS-polyacrylamide gel electrophoresis, blotted onto nitrocellulose, and probed with an antibody against P-Tyr. In addition to high-molecular-weight and other bands common to all samples, a prominent band below the 46-kDa marker was unique to cells infected with vB5R-GFP, the only virus with an intact, unmutated A36R gene (Fig. 7). To confirm that the unique P-Tyr band corresponded to A36R and that vB5R-GFP/A36R-YdF expressed the A36R protein, the blot was stripped and reprobed with anti-A36R antibody (Fig. 7). A background band was present at the position of the 46-kDa marker in all lanes. However, the A36R protein corresponding in position to the unique P-Tyr band was found in the lane containing the vB5R-GFP extract. The mutated A36R protein migrated slightly faster than wild-type A36R but was expressed at a similar level. These data indicated that mutation of Tyr112 and Tyr132 of the A36R protein prevented Tyr phosphorylation of the residues recognized by the 4G10 antibody.
FIG. 7.
Tyrosine phosphorylation of the A36R protein. Infected or mock-infected HeLa cells were incubated for 18 h, harvested, and analyzed by SDS-polyacrylamide gel electrophoresis and Western blotting with a P-Tyr-specific MAb (αP-Tyr). After detection of P-Tyr, the blot was stripped and reprobed with polyclonal antibody to the A36R protein (αA36R). The arrow marks the position of the Tyr-phosphorylated A36R protein. Positions and masses (in kilodaltons) of marker proteins are shown on the left.
Effects of A36R mutations on formation of actin tails.
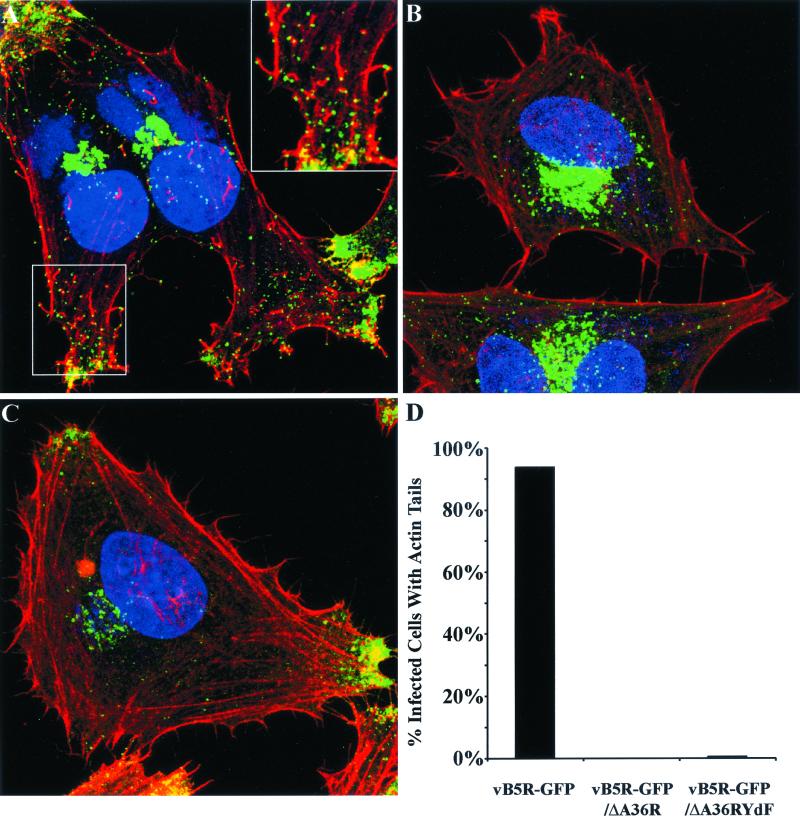
Evidence that Tyr phosphorylation was required to nucleate actin tails had previously been obtained by a complementation experiment in which plasmids containing wild-type or mutated A36R ORFs were transfected into cells infected with an A36R deletion mutant and then the fixed cells were stained with phalloidin (9). We confirmed those results by carrying out similar transfection-infection experiments in a cell line that stably expressed GFP-actin. Videos showing formation and movement of actin tails when a plasmid containing the intact A36R ORF was transfected but not after transfection with a plasmid containing a mutated A36R ORF, with Phe substituted for Tyr112 and Tyr132, are provided as Supplemental Material no. 1 at http://www.niaid.nih.gov/dir/labs/lvd/moss.htm. In Fig. 8, we show results obtained by infecting cells with vB5R-GFP, vB5R-GFP/ΔA36R, or vB5R-GFP/A36R-YdF and staining with rhodamine-phalloidin in order to visualize actin tails. The infected cells were also stained with DAPI to discern nuclei and DNA-containing viral factory areas. Examination of cells infected with vB5R-GFP revealed numerous red actin tails attached to green fluorescent particles (Fig. 8A). In contrast, actin tails were not seen in cells infected with vB5R-GFP/ΔA36R (Fig. 8B) and were rare in cells infected with vB5R-GFP/A36R-YdF (Fig. 8C). These data were quantified by determining the percentage of infected cells that contained one or more actin tails (Fig. 8D). Therefore, mutation of Tyr112 and Tyr132 of A36R was sufficient to inhibit actin tail formation by vB5R-GFP/A36R-YdF.
FIG. 8.
Visualization of actin tails by confocal microscopy. (A to C) HeLa cells were infected with vB5R-GFP (A), vB5R-GFP/ΔA36R (B), or vB5R-GFP/A36R-YdF (C) and stained with rhodamine-phalloidin (red) and DAPI (blue) to visualize F-actin and DNA, respectively. Green is GFP fluorescence. The boxed area in the lower left of panel A is enlarged in the upper right in order to more clearly see actin tails. (D) The percentages of infected cells that contained one or more actin tails were determined.
We also noted that in addition to the GFP staining in the juxtanuclear region and individual particles in the cytoplasm, there was intense GFP staining at the cell vertices. The latter staining, which appears to represent dense collections of IEV, was prominent only in cells infected with vB5R-GFP and vB5R-GFP/A36R-YdF (Fig. 8A and C). The reduction in this staining in cells infected with vB5R-GFP/ΔA36R (Fig. 8B) was consistent with the lower density of IEV seen by electron microscopy.
Intracellular movement of vB5R-GFP/A36R-YdF.
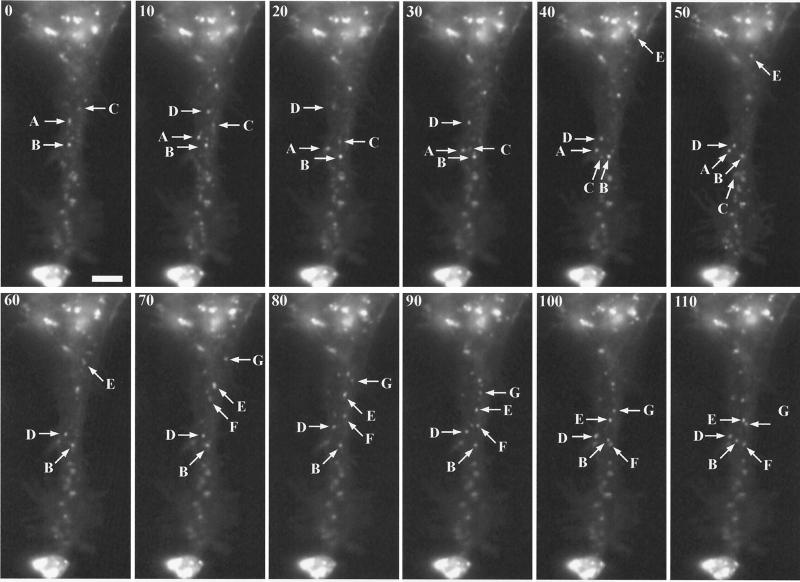
Having established that vB5R-GFP/A36R-YdF was specifically defective in the formation of actin tails, we compared the intracellular movement of this mutant with that previously described for vB5R-GFP. As with vB5R-GFP, the overall IEV movement in vB5R-GFP/A36R-YdF-infected cells was directional from the juxtanuclear region to the periphery. The net movement was parallel to the long axis of the cell, leading to the accumulation of IEV at the position expected for the plus ends of microtubules (Fig. 9). The accumulation of the fluorescent particles in the cell vertices of cells infected with either vB5R-GFP or vB5R-GFP/A36R-YdF was also seen in Fig. 8A and C and was a general feature of such infections. A movie showing movement of the particles seen in Fig. 9 is provided as Supplemental Material no. 2 at http://www.niaid.nih.gov/dir/labs /lvd/moss.htm. By capturing images at one frame per second, the speeds of individual IEV were determined. Although not shown, occasional virions displayed short intervals of movement toward the center of the cell during their progression to the periphery. The saltatory nature of the movement as well as maximal speeds of greater than 2 μm per s (Table 1) were similar to those previously reported for vB5R-GFP (34) and for microtubule-associated post-Golgi vesicles (14). Also, as previously reported for vB5R-GFP (34), vB5R-GFP/A36R-YdF movement was arrested by the microtubule-depolymerizing drug nocodazole (data not shown). Thus, intracellular IEV movement had the characteristics of microtubule association and was unaffected by the absence of actin tails.
FIG. 9.
Visualization of IEV by digital video microscopy. HeLa cells were infected with 0.2 PFU of vB5R-GFP/A36R-YdF per cell. After 12 h, images were collected at one frame per second. Selected frames are shown, with the cumulative time elapsed (in seconds) indicated in the upper left corner. The lettered arrows in each frame point to the same IEV particles moving downwards in subsequent frames. Note the intense fluorescence, at the bottom of each panel, where IEV accumulate at the vertex of the cell. Bar, 5 μm. The entire time-lapse video is provided as Supplemental Material no. 2 at http://www.niaid.nih.gov/dir/labs/lvd/moss.htm.
TABLE 1.
Frame-by-frame measurements for six individual virions
| Frame no.a | Distance (μm) moved by virion:
|
|||||
|---|---|---|---|---|---|---|
| A | B | C | D | E | F | |
| 1 | 2.79 | 0.67 | 0.49 | 0.42 | 0.43 | 1.23 |
| 2 | 0.00 | 0.00 | 1.79 | 0.00 | 0.00 | 0.93 |
| 3 | 0.00 | 0.60 | 0.00 | 0.00 | 0.81 | 0.81 |
| 4 | 0.00 | 0.00 | 0.91 | 0.21 | 0.76 | 0.00 |
| 5 | 0.54 | 1.73 | 0.00 | 0.39 | 0.87 | 0.95 |
| 6 | 0.47 | 0.00 | 0.00 | 1.28 | 0.41 | 0.69 |
| 7 | 0.00 | 0.00 | 1.13 | 0.30 | 0.47 | 0.00 |
| 8 | 0.57 | 0.87 | 0.00 | 0.52 | 0.84 | 0.76 |
| 9 | 0.80 | 0.00 | 0.87 | 0.00 | 0.00 | 0.78 |
| 10 | 0.67 | 1.41 | 0.34 | 0.00 | 0.00 | 1.59 |
| 11 | 0.61 | 0.57 | 1.02 | 0.67 | 0.33 | 0.00 |
| 12 | 0.48 | 1.15 | 1.50 | 0.24 | 0.48 | 0.52 |
| 13 | 0.67 | 0.00 | 0.39 | 0.20 | 0.80 | |
| 14 | 0.00 | 1.01 | 0.00 | 0.00 | 0.87 | |
| 15 | 1.76 | 0.94 | 0.00 | 0.24 | 0.89 | |
| 16 | 0.94 | 0.00 | 0.00 | 0.00 | 0.27 | |
| 17 | 1.65 | 0.00 | 0.48 | 2.36 | ||
| 18 | 0.51 | 0.00 | 0.39 | 0.43 | ||
| 19 | 0.00 | 1.07 | 1.46 | |||
| 20 | 0.49 | 0.85 | ||||
| 21 | 0.63 | |||||
| Total (μm) | 10.30 | 12.20 | 9.50 | 5.33 | 13.32 | 8.25 |
| Time (s) | 16 | 21 | 19 | 18 | 20 | 12 |
| Speed (μm/s) | 0.64 | 0.58 | 0.50 | 0.30 | 0.67 | 0.69 |
Frame rate, one frame per second.
Actin tails are associated with extracellular virions.
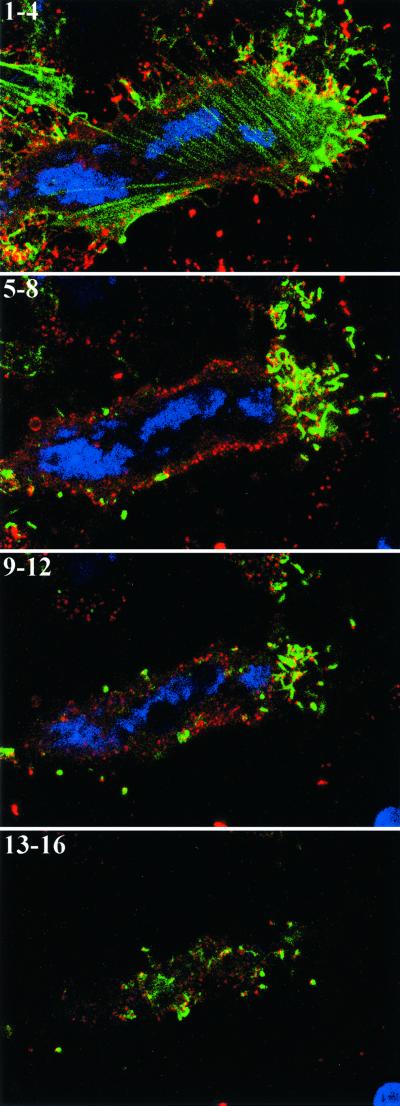
Our finding that actin tails have no role in intracellular movement appeared to conflict with images that appear to show actin tails associated with intracellular virions (see, e.g., Fig. 8A). However, such virions could be on the cell surface rather than in the cytoplasm. In previous studies, it was not possible to distinguish between intra- and extracellular virions because either the virions were labeled with GFP or the infected cells were permeabilized prior to antibody labeling. To overcome this technical problem, a HeLa cell line that stably expresses GFP-actin was constructed and then infected with wild-type vaccinia virus. The unpermeabilized cells were then stained with a MAb to the B5R membrane protein and examined by confocal microscopy. The absence of juxtanuclear staining, which is intense in permeabilized cells, provided evidence that only extracellular virions could be labeled with antibody under these conditions. Maximum-intensity projections for consecutive portions of a single Z-stack from a representative cell are shown in Fig. 10. By reconstructing the Z-series, we determined that all of the actin tails were associated with labeled virions on the cell surface. A movie showing rotation of a three-dimensional reconstruction is available as Supplemental Material no. 4 at http://www.niaid.nih.gov/dir/labs/lvd/moss.htm. This result was consistent with the formation of actin tails following fusion of the IEV with the plasma membrane and has important implications regarding the mechanism of virus spread and the potential susceptibility of these virions to neutralization with antibody.
FIG. 10.
Association of extracellular virus with actin tails. HeLa cells, which stably express GFP-actin (green), were infected with wild-type vaccinia virus. Unpermeabilized cells were stained with MAb 19C2 to the B5R membrane protein followed by Cy5-conjugated donkey anti-rat antibody (red) and Hoechst dye to stain DNA (blue). Entire cells were imaged by confocal microscopy as a series of optical sections. Starting from the bottom of one cell, every four sequential sections (1 to 4, 5 to 8, 9 to 12, and 13 to 16) were summed, and the set of four maximum-intensity projections is shown. The extracellular virions appear red at the tips of green actin tails and yellow if overlying an actin tail.
DISCUSSION
Following wrapping with trans-Golgi or endosomal cisternae, vaccinia virus virions must travel from the interior of the cell to the plasma membrane. In a recent review, Sodeik (28) pointed out that molecular crowding due to the presence of organelles, cytoskeleton, and high protein concentrations effectively restricts free diffusion of molecules larger than 500 kDa. For IEV, she calculated that it would take 5.7 h to travel 10 μm. A possible transport mechanism, used by a number of intracellular pathogens, involves rapid actin polymerization (2). Indeed, an actin polymerization mechanism to explain vaccinia virus virion movement was proposed by Cudmore et al. (3) based on video microscopic studies. However, our video microscopy experiments carried out with a recombinant vaccinia virus expressing GFP fused to the B5R membrane protein revealed movement that was similar to vesicle transport along microtubules and which could be inhibited by a microtubule-depolymerizing agent (34). Because interpretations based on drug inhibition are always risky, we have now extended these observations by specifically preventing the nucleation of actin tails on vaccinia virus particles. To accomplish this, we used data of Frischknecht et al. (9) indicating that actin nucleation occurs by phosphorylation of the A36R protein on Tyr112 alone or with Tyr132. These residues had been identified by transcomplementation, rather than by constructing a vaccinia virus mutant with point mutations of the Tyr residues. Thus, our objectives were to construct and use recombinant vaccinia viruses to (i) confirm the site of A36R tyrosine phosphorylation, (ii) confirm the role of A36R Tyr112 and Tyr132 in actin nucleation, (iii) analyze the effects of the Tyr mutations on the assembly and movement of IEV and cell-to-cell virus spread, (iv) compare the phenotype of the A36R Tyr mutant with that of an A36R deletion mutant, and (v) determine whether intra- or extracellular virions are associated with actin tails.
The recombinant viruses containing a deleted or mutated A36R gene expressed B5R-GFP in place of the normal B5R IEV membrane protein in order to visualize virion movement. As previously shown (34) and further validated here, fusion of GFP was without discernible effect on B5R function so that the plaque size of vB5R-GFP was the same as that of standard vaccinia virus. Results obtained with vB5R-GFP/A36R-YdF, which contains conservative Phe substitutions for Tyr112 and Tyr132, will be discussed first. Even though the mutated A36R was synthesized and still had four remaining Tyr residues, Tyr phosphorylation was not detected and both actin tail and specialized microvillus formation failed to occur. Thus, the conclusions regarding the regulatory role of Tyr phosphorylation in actin nucleation were corroborated. In addition, we found that the mutant exhibited no discernible defect in wrapping as determined by electron microscopy and that the outer membranes of the abundant IEV reacted with antibody to GFP that was fused to the B5R protein. Therefore, vB5R-GFP/A36R-YdF was ideal for assessing the putative role of actin tails in intracellular IEV movement. The directional, saltatory movement with maximal speeds of >2 μm per s, first demonstrated with vB5R-GFP, was reproduced with vB5R-GFP/A36R-YdF, even though actin tail formation was prevented. A movie demonstrating this movement is available as Supplemental Material no. 2 at http://www.niaid.nih.gov/dir/labs/lvd/moss.htm. These results should eliminate from further consideration any significant role for actin tails in intracellular movement of IEV. The type of movement exhibited by IEV was similar to that of vesicles moving along microtubules arranged parallel to the long axis of the cell (14). Moreover, the movement was inhibited by the microtubule-depolymerizing drug nocodazole. Although nocodazole has also been reported to prevent movement of IMV and the formation of IEV (21, 25), we previously showed that the effect on IEV movement was rapid and reversible (34). Microtubules also may be involved in early postentry stages of vaccinia virus replication (21) and in transport of other viruses (17, 27, 28, 30).
Previous studies had not clearly defined whether virions associated with actin tails are intra- or extracellular. To resolve this issue, a cell line that expressed GFP-actin was infected with vaccinia virus and the unpermeabilized cells were stained with a MAb to the B5R membrane protein. By examination of Z-sections by confocal microscopy, we could find an association of all actin tails with virions on the cell surface. The surface location fits well with evidence, obtained by following the movement of individual GFP-labeled virions, that actin tails form at or near the plasma membrane (34). In addition, van Eijl et al. (32) suggested that Tyr phosphorylation of A36R occurred after fusion of the IEV and plasma membranes. Moreover, this model fits with evidence that Tyr phosphorylation of A36R occurs by a Src family kinase (9), which is presumably plasma membrane associated.
Since actin tails are not involved in the intracellular transport of vaccinia virions, unlike the situation with Listeria and Shigella, what is their role? Actin tails are unnecessary for virions to exit the cell, as the presence of vB5R-GFP/A36R-YdF virions on the cell surface was demonstrated by low-pH-induced cell-to-cell fusion and by scanning electron microscopy. Nevertheless, the mutant plaques were one-third of the size of those formed by viruses with an intact A36R gene, and a lag in virus production occurred following a low-multiplicity spreading infection. These data are consistent with a primary role for actin tails in producing long motile microvilli that promote efficient cell-to-cell virus spread. A movie showing movement of virus-induced actin tails protruding from cells stably expressing GFP-actin is provided as Supplemental Material no. 4 at http://www.niaid.nih.gov/dir/labs/lvd/moss.htm. The extracellular location of virions at the ends of actin tails means that they are unlikely to entirely escape the immune system during cell-to-cell spread.
The mutant vB5R-GFP/ΔA36R, with a deleted A36R ORF, also failed to make actin tails. However, this mutant was much more severely impaired than vB5R-GFP/A36R-YdF. Compared to vB5R-GFP/A36R-YdF, the following were all reduced: plaque size, virus replication and spread, low-pH-induced cell fusion (an indicator of virus on the cell surface), wrapped virus isolated by CsCl centrifugation, and IEV seen by transmission electron microscopy. Although previous studies with an A36R deletion mutant had attributed the severe restriction on virus spread to the failure of actin tail formation (24, 38), our data suggested that the deletion mutant has additional defects. Since the A36R protein has been shown to interact with other IEV membrane proteins (23, 39), its absence may adversely affect the structure of the protein complex forming the wrapping membrane. Although some virion movement was detected in cells infected with vB5R-GFP/ΔA36R, it was extremely infrequent and therefore could not be characterized in detail. Thus, the mutant containing the specific Tyr substitutions was superior to the deletion mutant for studying actin tail-independent IEV movement and virus spread.
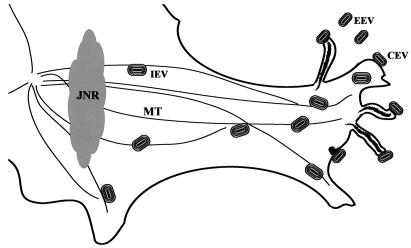
A scheme depicting directional IEV movement along microtubules and actin tail formation at the plasma membrane is shown in Fig. 11. Because microtubules tend to be arranged parallel to the long axis of the cell, the IEV accumulate at the plus ends near the plasma membrane. The direction of IEV movement could be explained by interaction with a kinesin family microtubule motor, either directly or through additional cell protein adapters.
FIG. 11.
Diagram representing the movement of enveloped virions. After wrapping of IMV in the juxtanuclear region (JNR), IEV move out to the cell periphery along microtubules (MT). Actin polymerization occurs at the plasma membrane, forming motile CEV-tipped microvilli.
ACKNOWLEDGMENTS
We thank members of the Laboratory of Viral Diseases, especially Andrea Weisberg for electron microscopy and Norman Cooper for tissue culture cells, and Tim Maugel at the Laboratory for Biological Ultrastructure at the University of Maryland for help with the scanning electron microscopy. Some of the work was carried out in the NIAID imaging facility with the guidance of Owen Schwartz.
REFERENCES
- 1.Blasco R, Moss B. Extracellular vaccinia virus formation and cell-to-cell virus transmission are prevented by deletion of the gene encoding the 37,000-dalton outer envelope protein. J Virol. 1991;65:5910–5920. doi: 10.1128/jvi.65.11.5910-5920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cossart P. Actin-based motility of pathogens: the Arp2/3 complex is a central player. Cell Microbiol. 2000;2:195–205. doi: 10.1046/j.1462-5822.2000.00053.x. [DOI] [PubMed] [Google Scholar]
- 3.Cudmore S, Cossart P, Griffiths G, Way M. Actin-based motility of vaccinia virus. Nature. 1995;378:636–638. doi: 10.1038/378636a0. [DOI] [PubMed] [Google Scholar]
- 4.da Fonseca F G, Wolffe E J, Weisberg A, Moss B. Characterization of the vaccinia virus H3L envelope protein: topology and posttranslational membrane insertion via the C-terminal hydrophobic tail. J Virol. 2000;74:7508–7517. doi: 10.1128/jvi.74.16.7508-7517.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dales S, Siminovitch L. The development of vaccinia virus in Earle's L strain cells as examined by electron microscopy. J Biophys Biochem Cytol. 1961;10:475–503. doi: 10.1083/jcb.10.4.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doms R W, Blumenthal R, Moss B. Fusion of intra- and extracellular forms of vaccinia virus with the cell membrane. J Virol. 1990;64:4884–4892. doi: 10.1128/jvi.64.10.4884-4892.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engelstad M, Smith G L. The vaccinia virus 42-kDa envelope protein is required for the envelopment and egress of extracellular virus and for virus virulence. Virology. 1993;194:627–637. doi: 10.1006/viro.1993.1302. [DOI] [PubMed] [Google Scholar]
- 8.Falkner F G, Moss B. Transient dominant selection of recombinant vaccinia viruses. J Virol. 1990;64:3108–3111. doi: 10.1128/jvi.64.6.3108-3111.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frischknecht F, Moreau V, Rottger S, Gonfloni S, Reckmann I, Superti-Furga G, Way M. Actin-based motility of vaccinia virus mimics receptor tyrosine kinase signalling. Nature. 1999;401:926–929. doi: 10.1038/44860. [DOI] [PubMed] [Google Scholar]
- 10.Gong S C, Lai C F, Esteban M. Vaccinia virus induces cell fusion at acid pH and this activity is mediated by the N-terminus of the 14-kDa virus envelope protein. Virology. 1990;178:81–91. doi: 10.1016/0042-6822(90)90381-z. [DOI] [PubMed] [Google Scholar]
- 11.Goode B L, Drubin D G, Barnes G. Functional cooperation between the microtubule and actin cytoskeletons. Curr Opin Cell Biol. 2000;12:63–71. doi: 10.1016/s0955-0674(99)00058-7. [DOI] [PubMed] [Google Scholar]
- 12.Hiller G, Weber K. Golgi-derived membranes that contain an acylated viral polypeptide are used for vaccinia virus envelopment. J Virol. 1985;55:651–659. doi: 10.1128/jvi.55.3.651-659.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiller G, Weber K, Schneider L, Parajsz C, Jungwirth C. Interaction of assembled progeny pox viruses with the cellular cytoskeleton. Virology. 1979;98:142–153. doi: 10.1016/0042-6822(79)90533-6. [DOI] [PubMed] [Google Scholar]
- 14.Hirschberg K, Miller C M, Ellenberg J, Presley J F, Siggia E D, Phair R D, Lippincott-Schwartz J. Kinetic analysis of secretory protein traffic and characterization of golgi to plasma membrane transport intermediates in living cells. J Cell Biol. 1998;143:1485–1503. doi: 10.1083/jcb.143.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ichihashi Y, Matsumoto S, Dales S. Biogenesis of poxviruses: role of A-type inclusions and host cell membranes in virus dissemination. Virology. 1971;46:507–532. doi: 10.1016/0042-6822(71)90056-0. [DOI] [PubMed] [Google Scholar]
- 16.Ishii K, Moss B. Role of vaccinia virus A20R protein in DNA replication: construction and characterization of temperature-sensitive mutants. J Virol. 2001;75:1656–1663. doi: 10.1128/JVI.75.4.1656-1663.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu W J, Qi Y M, Zhao K N, Liu Y H, Liu X S, Frazer I H. Association of bovine papillomavirus type 1 with microtubules. Virology. 2001;282:237–244. doi: 10.1006/viro.2000.0728. [DOI] [PubMed] [Google Scholar]
- 18.Moreau V, Frischknecht F, Reckmann I, Vincentelli R, Rabut G, Stewart D, Way M. A complex of N-WASP and WIP integrates signalling cascades that lead to actin polymerization. Nat Cell Biol. 2000;2:441–448. doi: 10.1038/35017080. [DOI] [PubMed] [Google Scholar]
- 19.Morgan C. Vaccinia virus reexamined: development and release. Virology. 1976;73:43–58. doi: 10.1016/0042-6822(76)90059-3. [DOI] [PubMed] [Google Scholar]
- 20.Parkinson J E, Smith G L. Vaccinia virus gene A36R encodes a M(r) 43–50 K protein on the surface of extracellular enveloped virus. Virology. 1994;204:376–390. doi: 10.1006/viro.1994.1542. [DOI] [PubMed] [Google Scholar]
- 21.Ploubidou A, Moreau V, Ashman K, Reckmann I, Gonzalez C, Way M. Vaccinia virus infection disrupts microtubule organization and centrosome function. EMBO J. 2000;19:3932–3944. doi: 10.1093/emboj/19.15.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roper R L, Wolffe E J, Weisberg A, Moss B. The envelope protein encoded by the A33R gene is required for formation of actin-containing microvilli and efficient cell-to-cell spread of vaccinia virus. J Virol. 1998;72:4192–4204. doi: 10.1128/jvi.72.5.4192-4204.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rottger S, Frischknecht F, Reckmann I, Smith G L, Way M. Interactions between vaccinia virus IEV membrane proteins and their roles in IEV assembly and actin tail formation. J Virol. 1999;73:2863–2875. doi: 10.1128/jvi.73.4.2863-2875.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanderson C M, Frischknecht F, Way M, Hollinshead M, Smith G L. Roles of vaccinia virus EEV-specific proteins in intracellular actin tail formation and low pH-induced cell-cell fusion. J Gen Virol. 1998;79:1415–1425. doi: 10.1099/0022-1317-79-6-1415. [DOI] [PubMed] [Google Scholar]
- 25.Sanderson C M, Hollinshead M, Smith G L. The vaccinia virus A27L protein is needed for the microtubule-dependent transport of intracellular mature virus particles. J Gen Virol. 2000;81:47–58. doi: 10.1099/0022-1317-81-1-47. [DOI] [PubMed] [Google Scholar]
- 26.Schmelz M, Sodeik B, Ericsson M, Wolffe E J, Shida H, Hiller G, Griffiths G. Assembly of vaccinia virus: the second wrapping cisterna is derived from the trans Golgi network. J Virol. 1994;68:130–147. doi: 10.1128/jvi.68.1.130-147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith G A, Gross S P, Enquist L W. Herpesviruses use bidirectional fast-axonal transport to spread in sensory neurons. Proc Natl Acad Sci USA. 2001;98:3466–3470. doi: 10.1073/pnas.061029798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sodeik B. Mechanisms of viral transport in the cytoplasm. Trends Microbiol. 2000;8:465–472. doi: 10.1016/s0966-842x(00)01824-2. [DOI] [PubMed] [Google Scholar]
- 29.Stokes G V. High-voltage electron microscope study of the release of vaccinia virus from whole cells. J Virol. 1976;18:636–643. doi: 10.1128/jvi.18.2.636-643.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suomalainen M, Nakano M Y, Keller S, Boucke K, Stidwill R P, Greber U F. Microtubule-dependent plus- and minus end-directed motilities are competing processes for nuclear targeting of adenovirus. J Cell Biol. 1999;144:657–672. doi: 10.1083/jcb.144.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tooze J, Hollinshead M, Reis B, Radsak K, Kern H. Progeny vaccinia and human cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur J Cell Biol. 1993;60:163–178. [PubMed] [Google Scholar]
- 32.van Eijl H, Hollinshead M, Smith G L. The vaccinia virus A36R protein is a type Ib membrane protein present on intracellular but not extracellular enveloped virus particles. Virology. 2000;271:26–36. doi: 10.1006/viro.2000.0260. [DOI] [PubMed] [Google Scholar]
- 33.Ward B M, Moss B. Golgi network targeting and plasma membrane internalization signals in vaccinia virus B5R envelope protein. J Virol. 2000;74:3771–3780. doi: 10.1128/jvi.74.8.3771-3780.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ward B M, Moss B. Visualization of intracellular movement of vaccinia virus virions containing a green fluorescent protein-B5R membrane protein chimera. J Virol. 2001;75:4802–4813. doi: 10.1128/JVI.75.10.4802-4813.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolffe E J, Isaacs S N, Moss B. Deletion of the vaccinia virus B5R gene encoding a 42-kilodalton membrane glycoprotein inhibits extracellular virus envelope formation and dissemination. J Virol. 1993;67:4732–4741. doi: 10.1128/jvi.67.8.4732-4741.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolffe E J, Katz E, Weisberg A, Moss B. The A34R glycoprotein gene is required for induction of specialized actin-containing microvilli and efficient cell-to-cell transmission of vaccinia virus. J Virol. 1997;71:3904–3915. doi: 10.1128/jvi.71.5.3904-3915.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolffe E J, Vijaya S, Moss B. A myristylated membrane protein encoded by the vaccinia virus L1R open reading frame is the target of potent neutralizing monoclonal antibodies. Virology. 1995;211:53–63. doi: 10.1006/viro.1995.1378. [DOI] [PubMed] [Google Scholar]
- 38.Wolffe E J, Weisberg A S, Moss B. Role for the vaccinia virus A36R outer envelope protein in the formation of virus-tipped actin-containing microvilli and cell-to-cell virus spread. Virology. 1998;244:20–26. doi: 10.1006/viro.1998.9103. [DOI] [PubMed] [Google Scholar]
- 39.Wolffe E J, Weisberg A S, Moss B. The vaccinia virusA33R protein provides a chaperone function for viral membrane localization and tyrosine phosphorylation of the A36R protein. J Virol. 2001;75:303–310. doi: 10.1128/JVI.75.1.303-310.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]