Abstract
The identification and characterization of host cell membranes essential for positive-strand RNA virus replication should provide insight into the mechanisms of viral replication and potentially identify novel targets for broadly effective antiviral agents. The alphanodavirus flock house virus (FHV) is a positive-strand RNA virus with one of the smallest known genomes among animal RNA viruses, and it can replicate in insect, plant, mammalian, and yeast cells. To investigate the localization of FHV RNA replication, we generated polyclonal antisera against protein A, the FHV RNA-dependent RNA polymerase, which is the sole viral protein required for FHV RNA replication. We detected protein A within 4 h after infection of Drosophila DL-1 cells and, by differential and isopycnic gradient centrifugation, found that protein A was tightly membrane associated, similar to integral membrane replicase proteins from other positive-strand RNA viruses. Confocal immunofluorescence microscopy and virus-specific, actinomycin D-resistant bromo-UTP incorporation identified mitochondria as the intracellular site of protein A localization and viral RNA synthesis. Selective membrane permeabilization and immunoelectron microscopy further localized protein A to outer mitochondrial membranes. Electron microscopy revealed 40- to 60-nm membrane-bound spherical structures in the mitochondrial intermembrane space of FHV-infected cells, similar in ultrastructural appearance to tombusvirus- and togavirus-induced membrane structures. We concluded that FHV RNA replication occurs on outer mitochondrial membranes and shares fundamental biochemical and ultrastructural features with RNA replication of positive-strand RNA viruses from other families.
Positive-strand RNA viruses are responsible for a wide range of diseases in humans, animals, and plants. Clinically relevant members of this group cause significant morbidity and mortality and include viruses from the Picornaviridae, Caliciviridae, Togaviridae, and Flaviviridae families. Although these pathogens represent a prominent component of the growing list of emerging and potentially devastating viral diseases (40), current therapies for positive-strand RNA virus infections are limited to a few marginally effective drugs (36). The design and investigation of novel and broadly effective therapies require the identification and characterization of fundamental mechanisms in positive-strand RNA virus replication and pathogenesis, such as replication complex formation.
Flock house virus (FHV) and the closely related black beetle virus (BBV) are the best-studied alphanodaviruses in the Nodaviridae family (2). FHV was originally isolated from the grass grub Costelytra zealandica (12, 57) and contains one of the smallest known genomes of any animal RNA virus. The 4.5-kb FHV genome is bipartite, with two capped but nonpolyadenylated RNAs copackaged into a 29-nm nonenveloped virion with an icosahedral (T=3) capsid (56, 57). The larger 3.1-kb RNA species (RNA1) encodes protein A (2, 11), a 112-kDa protein with several conserved motifs characteristic of RNA-dependent RNA polymerases (44), including the GDD motif of a polymerase catalytic domain (29). Protein A is both necessary and sufficient for FHV RNA replication (1, 28, 45) and belongs to group 2, supergroup I, in the RNA-dependent RNA polymerase classification scheme of Koonin (30). The smaller 1.4-kb RNA species (RNA2) encodes a 43-kDa capsid precursor protein that is dispensable for viral RNA replication but is required for the production of whole virions (15, 56). RNA1 also encodes a subgenomic 0.4-kb RNA species (RNA3) that corresponds to the 3′ terminus of RNA1 (11, 16). RNA3 encodes protein B, a 10-kDa protein whose function is unknown but which is not required for RNA replication (1, 45). FHV replicates in insect (18, 59), plant (58), mammalian (1, 28), and yeast (45, 46) cells, which suggests that any host components required for FHV replication are widely conserved. The small genome and robust growth characteristics of FHV make it a useful model with which to study mechanisms of positive-strand RNA virus replication.
A common, if not universal, feature of positive-strand RNA virus replication is the involvement of host cell membranes (8). The replicase proteins and sites of viral RNA synthesis for numerous animal and plant viruses have been localized to structures derived from diverse intracellular membranes, including the lysosomes, endoplasmic reticulum (ER), and Golgi for poliovirus (55); lysosomes and endosomes for rubella virus (38), Sindbis virus (17), and Semliki Forest virus (32); and ER for equine arterivirus (42), brome mosaic virus (48), and tobacco mosaic virus (39). Previous studies with FHV and BBV suggest that membranes are also involved in alphanodavirus RNA replication. Viral RNA-dependent RNA polymerase activity is associated with a membrane fraction from lysates of Drosophila cells infected with FHV (64) or BBV (22). Moreover, the membrane and phospholipid dependence of FHV RNA positive-strand synthesis in vitro implies that membrane association is crucial for at least some steps of viral RNA replication (65). Morphological studies with two related alphanodaviruses also provide clues to the potential intracellular localization of FHV RNA replication (3, 19). Electron microscopy (EM) studies with wax moth larvae and suckling mice after infection with Nodamura virus (NOV), the prototypic alphanodavirus (2), demonstrated the appearance of vesiculated bodies in the cytoplasm of infected cells (19). The vesiculated bodies contain RNA, as detected by RNase-gold labeling, have morphological characteristics of mitochondria at early stages of infection, and are associated with virus particles at later stages of infection. The authors hypothesized that mitochondria serve as either a support structure or the energy suppliers for NOV replication (19). Bashiruddin and Cross (3) investigated the ultrastructural pathology of cultured Drosophila cells after infection with Boolarra virus (BOV), an alphanodavirus isolated from the grass grub Oncopera intricoides (47). Similar to the NOV-induced ultrastructural changes, there are vesicular bodies whose appearance correlates with the disappearance of mitochondria in BOV-infected cells (3). Thus, these studies suggest that one or more steps of alphanodavirus replication may occur on or near mitochondria.
In this report, we describe the use of antibodies against protein A, the FHV RNA-dependent RNA polymerase, to more definitively identify and characterize the intracellular localization of FHV RNA replication. We used confocal microscopy to localize protein A and viral RNA synthesis in FHV-infected Drosophila melanogaster DL-1 cells. We demonstrate that FHV protein A was tightly associated with intracellular membranes and localized to mitochondria together with viral RNA synthesis. Selective membrane permeabilization and immunogold EM experiments showed that protein A was located on outer mitochondrial membranes. In addition, we describe the ultrastructural appearance of outer mitochondrial membrane spherules in FHV-infected DL-1 cells, which show close similarities to ultrastructural changes induced on other intracellular membranes by positive-strand RNA viruses from several other families. These results imply that FHV RNA replication embodies widely conserved characteristics of positive-strand RNA virus replication and provide a foundation for the further investigation of viral replication mechanisms with FHV.
MATERIALS AND METHODS
Cells, virus stocks, and infection protocol.
Drosophila DL-1 cells were grown at 26°C in Schneider's insect medium supplemented with 10% fetal calf serum unless otherwise indicated. Plaque-purified FHV was amplified in DL-1 cells, virions were purified from cell lysates by sucrose gradient centrifugation, and virus titers were determined by plaque assay as previously described (59). DL-1 cells were infected at a multiplicity of infection of 100 as previously described (18), with minor modifications. Subconfluent DL-1 cells were dislodged by gentle scraping, pelleted, and resuspended at 107/ml. Virus was allowed to attach for 1 h at 26°C on a rotary shaker at 1,000 rpm, cells were diluted to 106/ml, plated onto tissue culture dishes or chamber slides, and incubated at 26°C without rotation until harvested for analysis. Uninfected control cells were prepared in an identical fashion, excluding virus in the attachment period.
Protein A antibody production.
The immunogen used to generate FHV protein A-specific rabbit antiserum was a recombinant C terminally hexahistidine (His6)-tagged protein A expressed in Escherichia coli. The expression plasmid pET-FHVPA was generated by mutually primed extension of overlapping oligonucleotides that contained the 3′-terminal 30 nucleotides from the protein A coding region of FHV RNA1 and 42 nucleotides that encoded an eight-amino-acid spacer (GGSGGSGG), followed by six histidines and a stop codon. The annealed and extended fragment was inserted into the BlpI/HindIII region of pBDL7, a yeast shuttle plasmid designed for galactose-inducible full-length protein A expression in Saccharomyces cerevisiae (B. Lindenbach and P. Ahlquist, unpublished results). The resulting intermediate plasmid pBDL7-C/H6 was digested with PstI and HindIII, and the 3.4-kb fragment that contained full-length FHV RNA1 with the 3′ insertion was cloned into the NotI/HindIII site of pET28a (Novagen, Madison, Wis.) to generate the expression plasmid pET-FHVPA.
E. coli BL21-CodonPlus (RIL) cells (Stratagene, La Jolla, Calif.) were transformed with pET-FHVPA and induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h at 30°C. Cells were harvested, resuspended in a solution containing 50 mM Tris (pH 7.5) and 100 mM NaCl with 8 M urea and a bacterial protease inhibitor cocktail (Sigma, St. Louis, Mo.), lysed by two freeze-thaw cycles, and centrifuged at 10,000 × g to pellet inclusion bodies. The insoluble protein pellet was extracted with 6 M guanidine HCl and 1% (vol/vol) Triton X-100 in a solution containing 10 mM Tris (pH 7.5) and 100 mM NaCl to further remove marginally soluble proteins from the largely insoluble protein A. The final insoluble pellet was dissolved in sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) sample buffer (62.5 mM Tris [pH 6.8], 2% SDS, 5% [wt/vol] glycerol, 14.4 mM 2-mercaptoethanol, 0.02% bromophenol blue), heated to 100°C for 10 min, separated on 10% acrylamide gels, and transferred by electroblotting to polyvinylidene (PVDF) membranes (Millipore, Bedford, Mass.). Membranes were stained with Coomassie blue, and the IPTG-induced full-length protein A band was excised, frozen with liquid nitrogen, and pulverized with a mortar and pestle. The powdered membrane was resuspended in lactated Ringer solution, emulsified with complete Freund's adjuvant, and injected intradermally into New Zealand White rabbits. Rabbits were boosted three times at 4-week intervals with the same antigen emulsified in incomplete Freund's adjuvant, and serum was isolated for 2 to 4 weeks after the final boost and tested for FHV protein A specificity by immunoblotting with FHV-infected DL-1 cell lysates. All animal procedures were done by Harlan Bioproducts for Science, Inc. (Madison, Wis.).
Antibodies and fluorescent reagents.
MitoTracker Red CM-H2Xros and Oregon Green 488-labeled concanavalin A were from Molecular Probes (Eugene, Oreg.). Polyclonal rabbit anti-voltage-dependent anion channel (VDAC), an outer mitochondrial membrane porin, was from Affinity Bioreagents (Golden, Colo.). Monoclonal mouse antibromodeoxyuridine was from Sigma, and monoclonal mouse anti-His6 was from Clontech (Palo Alto, Calif.). Monoclonal mouse antibiotin and all secondary immunoblot, immunofluorescence, and immunogold antibodies were from Jackson Immunoresearch (West Grove, Pa.).
Immunoblot analysis.
Protein samples were solubilized in SDS-PAGE sample buffer, separated on 10% acrylamide gels, and transferred to Immobilon-P PVDF membranes by electroblotting in a solution containing 25 mM Tris (pH 8.3), 192 mM glycine, and 10% methanol. Blotted membranes were blocked with Tris-buffered saline (50 mM Tris [pH 7.4], 100 mM NaCl) that contained 0.1% sodium azide, 2% nonfat milk, and 0.2% (vol/vol) Tween 20, incubated with primary antibodies and alkaline phosphatase-conjugated secondary antibodies, and developed with Immunostar chemiluminescence substrate in accordance with the manufacturer's (Bio-Rad, Hercules, Calif.) instructions. Chemiluminescence was detected with a Boehringer Mannheim Lumi-Imager.
Northern blot analysis.
Total RNA was isolated with an RNeasy kit (Qiagen, Valencia, Calif.). RNA samples were prepared in 60% formamide sample buffer, heated to 65°C for 10 min, separated on formaldehyde–1.4% agarose gels, and transferred to Nytran nylon membranes (Schleicher & Schuell, Inc., Keene, N.H.) via horizontal capillary blotting in a solution containing 1.5 M NaCl and 150 mM sodium citrate. Membranes were UV cross-linked, prehybridized with a solution containing 50 mM phosphate (pH 6.8), 0.75 M NaCl, 75 mM sodium citrate, 1% SDS, 50% formamide, 5× Denhardt's solution, and 0.1 mg of salmon sperm DNA per ml, and hybridized at 60°C with a strand-specific, 32P-labeled riboprobe that corresponded to nucleotides 2718 to 3064 from FHV RNA1 (45). Radioactive signals were detected with a Molecular Dynamics model 425 PhosphorImager imaging system.
Differential centrifugation and membrane association assays.
Cells were recovered by scraping and centrifugation and resuspended in lysis buffer that contained 50 mM piperazine-N,N′-bis[2-ethanesulfonic acid] (PIPES; pH 7.5), 50 mM KCl, 5 mM EDTA, 2 mM MgCl2, and a mammalian protease inhibitor cocktail (Sigma). Cell plasma membranes were disrupted with 10 strokes of a glass pestle Dounce homogenizer, and unbroken cells, nuclei, and large debris were removed by centrifugation at 500 × g for 5 min to obtain the initial total lysate. The total lysate was centrifuged at 20,000 × g for 10 min to pellet membranes and associated proteins, and the supernatant were carefully removed and used as the soluble fraction. The pellet was washed once with lysis buffer and resuspended in SDS-PAGE sample buffer as the pellet fraction. For membrane association assays, total lysates were incubated with 0.1 M NaCO3 at pH 11.5, 4 M urea, 1 M NaCl, or 1 M NaCl with 1.5% (vol/vol) Triton X-100 for 30 min on ice prior to differential centrifugation. Equilibrium density gradient centrifugation was performed with Nycodenz gradients and cell lysates prepared as described above. Nycodenz was added to total lysates to a final concentration of 37.5% (wt/vol), and samples were loaded under a 5 to 25% discontinuous Nycodenz gradient prepared in lysis buffer. Samples were centrifuged at 100,000 × g for 20 h at 4°C, and equal-volume gradient fractions were recovered manually, separated by SDS-PAGE, and immunoblotted as described above.
Immunofluorescence staining and confocal microscopy.
Cells were grown on eight-well glass chamber slides (Nalge Nunc, Naperville, Ill.) coated with 1% (wt/vol) polyethyleneimine. Medium was removed, cells were washed once with phosphate-buffered saline (PBS; 50 mM phosphate [pH 7.4], 100 mM NaCl, 10 mM KCl), fixed overnight with 4% paraformaldehyde in PBS at 4°C, permeabilized with 0.2% Triton X-100 in PBS for 10 min, and blocked with PBS that contained 0.1% azide, 1% nonfat milk, 1% bovine serum albumin, and 0.1% Tween 20. For immunofluorescence assays with MitoTracker Red CM-H2Xros, live cells were labeled with the mitochondrion-specific dye for 1 h in accordance with the manufacturer's instructions, fixed with 4% paraformaldehyde, and permeabilized with ice-cold acetone-methanol (1:1 ratio) for 1 min. For selective membrane permeabilization experiments, cells were permeabilized with 0.002% (wt/vol) saponin for 10 min and the blocking and wash buffers contained no Tween 20. Blocked cells were incubated with primary antibodies and fluorescein isothiocyanate (FITC)- or Texas Red (TR)-labeled secondary antibodies, and slides were coverslipped with 10% MOWIOL (Hoechst) in a solution containing 100 mM Tris (pH 8.5), 25% glycerol, and 25 μg of 1,4-diazobicyclo-[2.2.2]-octane per ml to prevent fading. Differential interference contrast (DIC) and immunofluorescence images were obtained with a Bio-Rad MRC 1024 confocal microscope equipped with an Ar/Kr laser (excitation at 488 and 568 nm) and 506- to 538-nm/664- to 696-nm emission filters for simultaneous FITC/TR visualization.
Viral RNA labeling and localization.
Newly synthesized FHV RNA was detected by bromouridine (BrU) incorporation after lipofection-mediated bromo-UTP (BrUTP) uptake as previously described (62), with minor modifications. Liposomes were formed by following the Lipofectin manufacturer's (Gibco BRL, Rockville, Md.) directions for DNA transfections. Serum-free medium was incubated with 10 mM BrUTP and 200 μg of Lipofectin per ml at room temperature for 30 min, diluted 10-fold with Schneider's insect medium that contained 10% fetal calf serum and 20 μg of actinomycin D per ml, and added to DL-1 cells. Unless otherwise indicated, cells were incubated with 20 μg of actinomycin D per ml for 30 min prior to transfection to inhibit endogenous DL-1 RNA polymerase activity. Cells were transfected for 1 h at room temperature, fixed with 4% paraformaldehyde, and processed for indirect immunofluorescence as described above with primary antibodies that detect incorporated BrU. For specificity control, some samples were treated with RNase A at 100 μg/ml and RNase T1 at 2 mg/ml in PBS for 30 min after cell fixation and permeabilization but before immunostaining.
EM.
Cells were fixed in 4% paraformaldehyde and 2% glutaraldehyde, postfixed in 1% osmium tetroxide with 1% potassium ferricyanide, stained with 1% uranyl acetate, dehydrated in a graded series of ethanols, and embedded in Spurr's resin (Electron Microscopy Sciences, Ft. Washington, Pa.). Seventy-nanometer sections were cut and placed on copper grids, counterstained with 8% uranyl acetate in 50% methanol and Reynold's lead citrate, and analyzed with a Philips CM120 transmission electron microscope. For immunogold EM, cells were similarly fixed except that the pre-embedding osmium tetroxide and uranyl acetate steps were omitted and samples were embedded in LR White resin (Polysciences, Inc., Warrington, Pa.). Grids were blocked with 0.5% gelatin, immunostained with rabbit protein A antiserum and 12-nm gold-labeled secondary antibodies, counterstained, and analyzed by transmission EM as described above.
RESULTS
Production of FHV protein A-specific antisera.
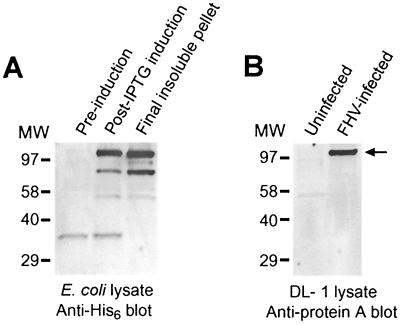
To investigate FHV RNA replication, we generated polyclonal rabbit antisera specific for protein A, the FHV RNA-dependent RNA polymerase. We produced recombinant C terminally His6-tagged protein A in E. coli with a pET28-based expression vector and obtained IPTG-induced expression of a 115-kDa His6-tagged protein (Fig. 1A) with the approximate predicted molecular weight of full-length FHV protein A (2). There were less prominent induced bands between 50 and 80 kDa that likely were protein A degradation products. Initial attempts to purify His6-tagged protein A by metal affinity chromatography were unsuccessful, as protein A overexpressed in E. coli was insoluble even in 8 M urea or 6 M guanidine. However, we used this insolubility to design a purification strategy that produced a final pellet enriched for protein A (Fig. 1A). We isolated the 115-kDa band from the final pellet by SDS-PAGE and injected it into rabbits to generate antisera that reacted specifically with a 110- to 115-kDa protein in immunoblots of lysates from FHV-infected Drosophila DL-1 cells but not uninfected cells (Fig. 1B). Preimmune rabbit sera showed no significant reactivity in immunoblots of lysates from uninfected or FHV-infected DL-1 cells (data not shown). In addition, protein A antisera showed prominent immunofluorescence in yeast transformed with an FHV RNA1 expression plasmid but not in control plasmid-transformed yeast (data not shown). We concluded from these results that the rabbit antisera reacted specifically with FHV protein A.
FIG. 1.
Production of rabbit polyclonal antisera specific for FHV protein A. (A) Total cell lysates from uninduced and IPTG-induced BL21 CodonPlus (RIL) E. coli cells transformed with a plasmid expressing C terminally His6-tagged protein A were separated by SDS-PAGE, transferred to a PVDF membrane, and immunoblotted with an anti-His6 monoclonal antibody. The induced lysate was subjected to sequential extractions with 8 M urea and 6 M guanidine as described in Materials and Methods, and the final protein A-enriched insoluble pellet was used as the starting material for immunogen preparation. The prominent band at 115 kDa was excised from PVDF membranes, powdered, and injected into rabbits. (B) Total cell lysates from uninfected and FHV-infected Drosophila DL-1 cells were separated by SDS-PAGE, transferred to a PVDF membrane, and immunoblotted with rabbit antisera generated with the immunogen from panel A. There is prominent reactivity with a 110- to 115-kDa band only in the infected-cell lysate (arrow), consistent with the predicted molecular weight of full-length FHV protein A (2). Molecular size (MW) markers, in kilodaltons, are shown on the left.
Temporal pattern of FHV protein A expression and viral RNA replication in Drosophila DL-1 cells.
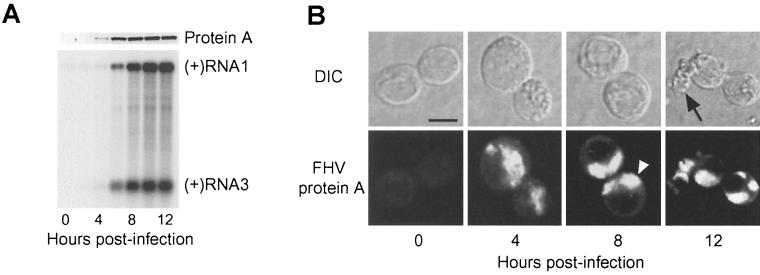
To identify a time point with maximal protein A expression and viral RNA synthesis early after infection but prior to the development of gross cytopathology, we examined FHV protein A and RNA levels by immunoblot and Northern blot analyses at 2-h intervals up to 12 h postinfection (hpi) (Fig. 2A). Protein A was first detectable at 4 hpi, increased to a maximal level by 10 hpi, and declined slightly thereafter (Fig. 2A, upper blot). Positive-strand RNA1 was detectable at all time points, consistent with its presence in whole virions, but amplification of positive-strand RNA1 only initiated at around 4 to 6 hpi and continued thereafter for the remainder of the time course (Fig. 2A, lower blot). Similar to protein A, positive-strand RNA3 was detectable at 4 hpi and increased to a maximal level by 10 hpi (Fig. 2A, lower blot). The levels of negative-strand RNA1 and RNA3 paralleled the protein A and positive-strand RNA3 temporal expression patterns (data not shown).
FIG. 2.
FHV protein A expression correlates with genomic RNA1 replication and subgenomic RNA3 synthesis, and protein A localizes to discrete intracellular structures in infected Drosophila DL-1 cells. (A) Temporal pattern of protein A expression and FHV RNA replication. Total protein from an equivalent number of cells per sample was separated by SDS-PAGE, transferred to a PVDF membrane, and immunoblotted with rabbit anti-protein A (upper blot). Total-protein staining showed equivalent protein loading in all lanes (data not shown). One microgram of total RNA per sample was separated on denaturing formaldehyde-agarose gels, transferred to a nylon membrane, and blotted with a 32P-labeled complementary riboprobe that detected positive-strand RNA1 and RNA3 (lower blot). The positions of RNA1 and RNA3 are indicated on the right. (B) Temporal pattern of protein A localization. FHV-infected cells were incubated on polyethyleneimine-coated chamber slides, fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100, and immunostained with rabbit anti-protein A, followed by TR-labeled secondary antibodies. DIC and confocal immunofluorescence images of the same cells are shown for each indicated time postinfection. The open arrowhead in the 8-h time point immunofluorescence image indicates the clustering and polarization of protein A in infected cells, and the closed arrow in the 12-h time point DIC image indicates a degenerating cell, characteristic of FHV-induced cytopathic effects (57). Bar = 5 μm.
We also examined the temporal pattern of FHV protein A localization in infected DL-1 cells by confocal immunofluorescence microscopy (Fig. 2B). We detected protein A by 4 hpi and at all time points thereafter, consistent with the immunoblot results (Fig. 2A). The pattern of protein A localization was suggestive of an intracellular organelle, with a discrete intracytoplasmic distribution distinct from the nucleus. At 4 hpi, protein A was present in a clustered cytoplasmic distribution, which became more prominent by 8 and 12 hpi and tended to polarize to discrete regions of infected cells (open arrowhead, Fig. 2B, bottom). The pattern of discrete intracytoplasmic protein A distribution was indistinguishable in cells fixed with paraformaldehyde, methanol, or acetone (Fig. 2B and data not shown). By 12 hpi, there was an initial clustering of infected cells, followed by eventual cell degeneration (closed arrow, Fig. 2B, top), consistent with previously observed FHV-induced cytopathic effects in Drosophila cells (57). Extensive cell destruction was present by 24 hpi, and by 48 hpi, few intact cells remained (data not shown). From these immunofluorescence results and the immunoblot and Northern blot results described above, we chose 8 hpi as the time point at which to evaluate FHV RNA replication in all subsequent experiments, unless otherwise indicated.
Membrane association of FHV protein A.
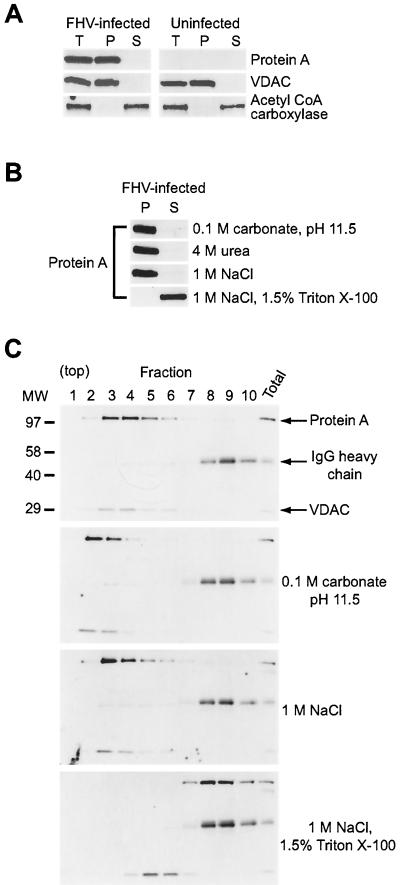
The intracytoplasmic distribution of protein A in FHV-infected DL-1 cells suggested that protein A was associated with an intracellular membrane-bound organelle. To investigate the potential intracellular membrane association of protein A, we performed differential centrifugation experiments with uninfected and FHV-infected DL-1 cell lysates (Fig. 3). Protein A partitioned into the 20,000 × g pellet fraction (Fig. 3A), similar to the distribution of VDAC, an outer mitochondrial integral membrane protein (52). In contrast, the cytoplasmic protein acetyl coenzyme A carboxylase (20) was recovered in the soluble fraction. We also investigated the membrane association of protein A under buffer conditions designed to remove peripheral proteins loosely associated with intracellular membranes (Fig. 3B). Most peripheral membrane proteins are dissociated from membranes by high pH, chaotropic agents, or high ionic strength, whereas more tightly associated proteins, such as integral membrane proteins, require the presence of a detergent for solubilization (7). FHV protein A remained associated with the pellet fraction, even at pH 11.5 or in the presence of 4 M urea or 1 M NaCl (Fig. 3B). However, protein A was recovered in the soluble fraction when the nonionic detergent Triton X-100 was combined with 1 M NaCl. The integral membrane protein VDAC behaved similarly under these differential centrifugation conditions (data not shown). We concluded from these results that FHV protein A was membrane associated through a mechanism that imparted significant stability to the protein-membrane interaction.
FIG. 3.
FHV protein A is tightly membrane associated in infected Drosophila DL-1 cells. (A) Differential centrifugation of FHV-infected and uninfected cell lysates. At 8 hpi, cells were dislodged, washed, and lysed by Dounce homogenization. Nuclei, debris, and unlysed cells were removed by centrifugation at 500 × g for 5 min, and the resultant total lysate (T) was centrifuged at 20,000 × g for 10 min to obtain pellet (P) and soluble (S) fractions. Samples were separated by SDS-PAGE, transferred to a PVDF membrane, and immunoblotted. We analyzed partitioning of the 30-kDa outer mitochondrial membrane protein VDAC (52) and the 265-kDa cytoplasmic protein acetyl coenzyme A carboxylase (20) as controls for the pellet and soluble fractions, respectively. (B) Differential centrifugation of protein A from FHV-infected cell lysates after treatment with buffer conditions designed to dislodge peripherally associated membrane proteins or a nonionic detergent. Total cell lysates were prepared as described above, incubated with 0.1 M carbonate at pH 11.5, 4 M urea, 1 M NaCl, or 1 M NaCl with 1.5% (vol/vol) Triton X-100 for 30 min on ice, and subjected to differential centrifugation and immunoblot analysis with antibodies to protein A. (C) Equilibrium density Nycodenz gradient fractionation of lysates from FHV-infected DL-1 cells. Total cell lysates were prepared as described above, except that 1 μg of rabbit immunoglobulin per ml was added to the total lysate and Nycodenz was added to a final concentration of 37.5% (wt/vol). Samples were loaded under a 5 to 25% discontinuous Nycodenz gradient and centrifuged at 100,000 × g for 20 h. Equal-volume fractions were recovered from the top of the gradient, and samples were separated by SDS-PAGE, transferred to PVDF membrane, and immunoblotted with rabbit anti-protein A and rabbit anti-VDAC simultaneously. The positions of protein A (112 kDa) and VDAC (30 kDa) are indicated on the right. Molecular size (MW) markers in kilodaltons are shown on the left of the upper blot. The 50-kDa rabbit immunoglobulin G (IgG) heavy chain was detected by the secondary reagent, and its position is also indicated on the right. Lysates for the lower three blots were incubated with the indicated buffers prior to Nycodenz gradient fractionation.
The sedimentation behavior of FHV protein A in differential centrifugation assays might have represented self-aggregation rather than membrane association, as protein A overexpressed in E. coli was insoluble (Fig. 1A), presumably due to aggregation and the formation of inclusion bodies. To address this possibility, we used equilibrium density Nycodenz gradient centrifugation of FHV-infected DL-1 lysates to examine the flotation behavior of protein A (Fig. 3C). Cell lysates in 37.5% Nycodenz were loaded under a 5 to 25% Nycodenz gradient and centrifuged to equilibrium. We analyzed equal-volume gradient fractions by immunoblot assay for protein A, VDAC, and rabbit immunoglobulin heavy chain, which we added to cell lysates prior to centrifugation in order to easily follow soluble protein distribution on a single blot. Consistent with membrane association, protein A floated up into and was recovered in lower density gradient fractions that also contained the integral membrane protein VDAC (Fig. 3C, upper blot). Protein A remained membrane associated in Nycodenz gradients after incubation with 0.1 M carbonate, pH 11.5, or with 1 M NaCl (Fig. 3C, middle blots) but was removed from membranes by 1 M NaCl with 1.5% Triton X-100 (Fig. 3C, lower blot). Surprisingly, the latter buffer condition was insufficient to completely disrupt the membrane association of VDAC, potentially due to its predicted 15 or 16 transmembrane domains (52). A similar distribution of protein A and anomalous behavior of VDAC were observed when the nonionic detergent n-octyl β-glucoside was used in isotonic buffer (data not shown). These results support the conclusion from differential centrifugation that FHV protein A is tightly membrane associated in infected DL-1 cells.
Subcellular localization of FHV protein A in infected Drosophila DL-1 cells.
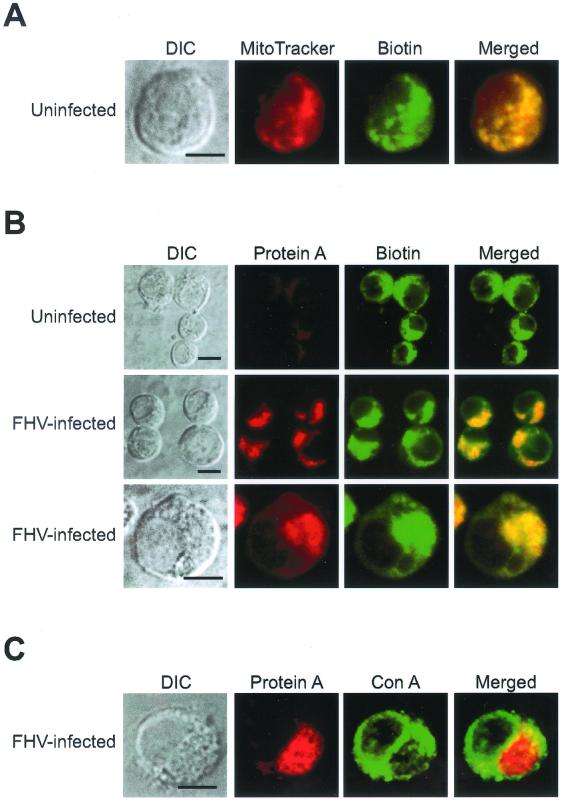
Previous studies have suggested an association of one or more steps in alphanodavirus replication with mitochondria (3, 19). Therefore, we used confocal immunofluorescence microscopy to compare the intracellular localization of FHV protein A to that of mitochondria in infected Drosophila DL-1 cells (Fig. 4). Hollinshead et al. have shown that antibodies against biotin can be used to identify mitochondria by immunofluorescence microscopy in normal rat kidney cells (25), and we obtained similar results with Drosophila DL-1 cells (Fig. 4A). Uninfected DL-1 cells were labeled with the mitochondrion-specific dye MitoTracker Red CM-H2Xros, fixed, permeabilized, and immunostained with a monoclonal antibody against biotin. Biotin immunoreactivity was distributed in a punctate intracellular pattern that colocalized with the MitoTracker Red dye signal. On the basis of these results, we used biotin as a marker for mitochondria in all subsequent confocal immunofluorescence experiments.
FIG. 4.
FHV protein A localizes to mitochondria in infected Drosophila DL-1 cells. (A) Biotin immunofluorescence identifies mitochondria in DL-1 cells. Uninfected DL-1 cells were labeled with the mitochondrion-specific dye MitoTracker Red for 1 h, fixed with 4% paraformaldehyde, permeabilized with methanol-acetone, and immunostained with mouse antibiotin, followed by FITC-labeled goat anti-mouse secondary antibodies. A DIC image (left image) and confocal immunofluorescence images for MitoTracker Red (left middle image, red), biotin (right middle image, green), and merged signals (right image) are shown. The merged image represents a digital superimposition of red and green signals, where areas of fluorescence colocalization are yellow. (B) Protein A and biotin immunofluorescence colocalize in infected DL-1 cells. At 8 hpi, cells were fixed, permeabilized with 0.2% Triton X-100, and immunostained with rabbit anti-protein A and mouse antibiotin, followed by TR-labeled goat anti-rabbit and FITC-labeled goat anti-mouse secondary antibodies. DIC images (left column) and confocal immunofluorescence images for protein A (left middle column, red), biotin (right middle column, green), and merged signals (right column) from uninfected (top) and FHV-infected (lower two panels) DL-1 cells are shown. (C) Protein A does not colocalize with concanavalin A (Con A) reactivity in infected DL-1 cells. At 8 hpi, cells were fixed, permeabilized with 0.2% Triton X-100, and immunostained with rabbit anti-protein A, followed by TR-labeled goat anti-rabbit secondary antibodies and Oregon Green 488-labeled concanavalin A. A DIC image (left image) and confocal images for protein A immunofluorescence (left middle image, red), concanavalin A reactivity (right middle image, green), and merged signals (right image) are shown. Bars = 5 μm.
Uninfected cells labeled with antibodies to protein A and biotin showed a punctate and dispersed mitochondrial distribution and no protein A immunofluorescence (Fig. 4B, top). In contrast, FHV-infected DL-1 cells showed a clustered cytoplasmic protein A immunofluorescence pattern that localized to mitochondria, as demonstrated by colocalization with biotin immunofluorescence (Fig. 4B, middle and bottom). The clustering of protein A and biotin immunofluorescence represented a polarization of mitochondria in FHV-infected cells, which we demonstrated further by the EM ultrastructural analysis described later. Although the colocalizing patterns of protein A and biotin immunofluorescence showed some occasional variations in intensity, we found no readily identifiable cells with distinct protein A and biotin immunofluorescence patterns, which suggested that protein A localized to the majority of mitochondria. In contrast, protein A localization in FHV-infected cells was clearly distinct from that of concanavalin A (Fig. 4C), a lectin that labels the ER and Golgi. We also analyzed protein A localization by confocal immunofluorescence microscopy at earlier (4 h) and later (12 h) times postinfection and found that protein A uniformly localized to mitochondria (data not shown). We concluded from these results that FHV protein A localized to mitochondria in infected DL-1 cells and that by 8 hpi protein A was associated with most mitochondria in infected cells.
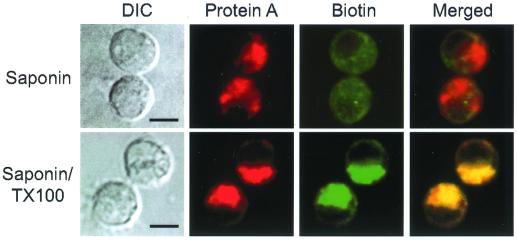
To further characterize the mitochondrial association of protein A in FHV-infected DL-1 cells, we used a selective membrane permeabilization strategy coupled with confocal immunofluorescence microscopy (Fig. 5). Mitochondrial membranes and cell plasma membrane have different lipid compositions, and therefore, low concentrations of detergents such as saponin and digitonin can selectively permeabilize the plasma membrane while leaving mitochondrial membranes intact (53). We permeabilized paraformaldehyde-fixed DL-1 cells with 0.002% saponin and found the same intensity and distribution of protein A immunofluorescence (Fig. 5, top) that was produced by Triton X-100 permeabilization (Fig. 4B). In contrast, biotin immunofluorescence was significantly reduced, consistent with the matrix and inner membrane localization of biotinylated mitochondrial proteins (25). When we permeabilized cells with 0.002% saponin and 0.2% Triton X-100, both protein A and biotin immunofluorescence patterns were unchanged (Fig. 5, bottom) compared to those produced by permeabilization with Triton X-100 alone (Fig. 4B). These results implied that the protein A epitopes recognized by the polyclonal antisera were exposed to the cytoplasm on the outer mitochondrial membrane.
FIG. 5.
FHV protein A is located on the outer mitochondrial membrane and exposed to the cytoplasm in infected Drosophila DL-1 cells. At 8 hpi, cells were fixed with 4% paraformaldehyde, permeabilized with either 0.002% saponin (top) or 0.002% saponin and 0.2% Triton X-100 (bottom) for 10 min, and immunostained with antibodies to protein A and biotin as described in the Fig. 4 legend. DIC images (left column) and confocal immunofluorescence images for protein A (left middle column, red), biotin (right middle column, green), and merged signals (right column) are shown. The merged images represent a digital superimposition of red and green signals, where areas of immunofluorescence colocalization are yellow. Bars = 5 μm.
Colocalization of FHV protein A with sites of viral RNA synthesis in infected Drosophila DL-1 cells.
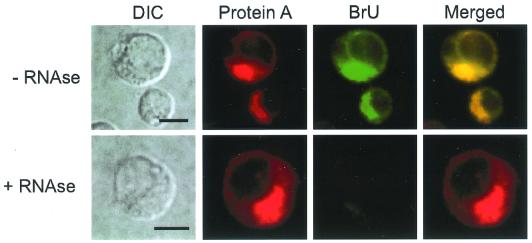
We used actinomycin D treatment and BrUTP incorporation to colocalize FHV protein A with sites of viral RNA synthesis in infected DL-1 cells (Fig. 6). Preliminary in vitro RNA-dependent RNA polymerase assays demonstrated that a crude membrane fraction from FHV-infected DL-1 cells (64) efficiently incorporated BrUTP into newly synthesized viral RNA (data not shown). In FHV-infected cells pretreated with actinomycin D, protein A and BrU immunofluorescence colocalized in a pattern that recapitulated the previously identified protein A localization (Fig. 6, top). BrU reactivity was eliminated by RNase treatment after transfection (Fig. 6, bottom), which indicated that the BrU signal was due to ribonucleotide incorporation into newly synthesized viral RNA and not to localized pools of unincorporated BrUTP. In the absence of actinomycin D, which inhibits cellular but not FHV RNA polymerase activity (2), nuclei showed prominent BrU labeling, which was eliminated with actinomycin D at 20 μg/ml (data not shown). We concluded from these results that the intracellular sites of FHV protein A localization were also the sites of viral RNA synthesis in infected DL-1 cells.
FIG. 6.
FHV protein A colocalizes with sites of viral RNA synthesis in infected Drosophila DL-1 cells. At 8 hpi, cells were incubated with actinomycin D at 20 μg/ml for 30 min to inhibit endogenous DL-1 RNA polymerase activity and then incubated for 1 h with preformed liposomes that contained 1 mM BrUTP, actinomycin D at 20 μg/ml, and Lipofectin at 20 μg/ml. Cells were fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100, and immunostained with rabbit anti-protein A and mouse anti-bromodeoxyuridine, followed by TR-labeled goat anti-rabbit and FITC-labeled goat anti-mouse secondary antibodies (top). As a specificity control, BrUTP-labeled cells were treated with RNase A/T1 after fixation and permeabilization but before immunostaining (bottom). DIC images (left column) and confocal immunofluorescence images for protein A (left middle column, red), BrU (right middle column, green), and merged signals (right column) are shown. The merged images represent a digital superimposition of red and green signals, where areas of immunofluorescence colocalization are yellow. Bars = 5 μm.
EM ultrastructural analysis of FHV-infected Drosophila DL-1 cells.
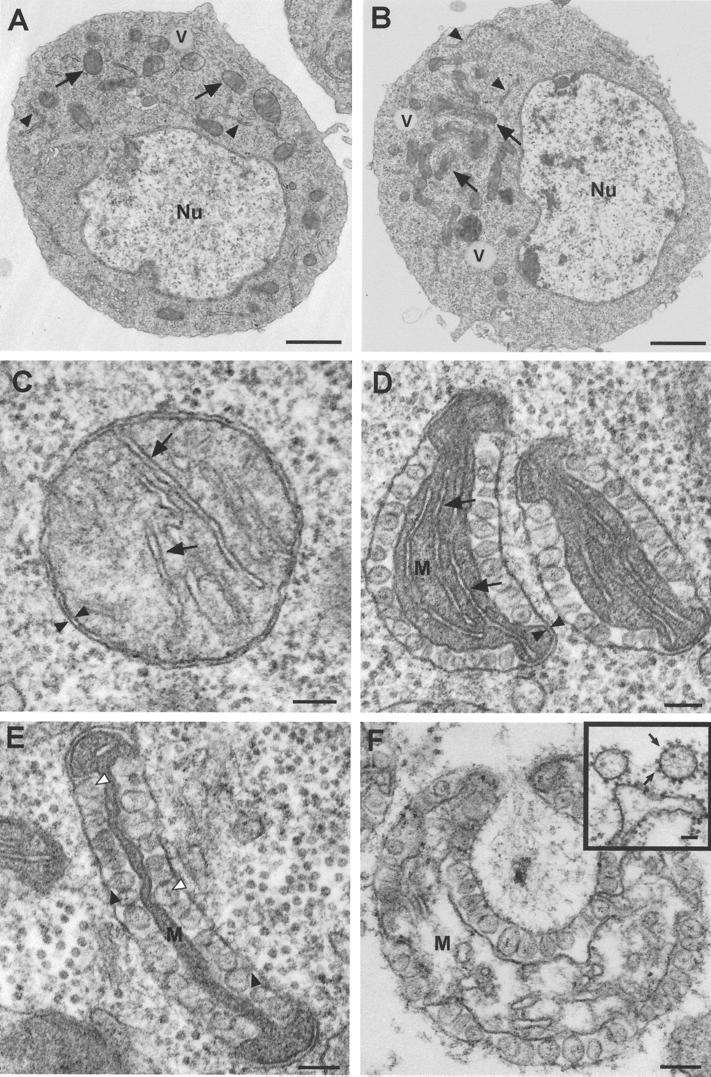
To further verify and extend the confocal microscopy results that showed the involvement of mitochondria in FHV RNA replication, we used transmission EM to analyze the ultrastructural pathology in FHV-infected DL-1 cells (Fig. 7). Even at low magnification, FHV-infected DL-1 cells showed subtle but distinctive intracellular changes (Fig. 7B) compared to uninfected cells (Fig. 7A). In FHV-infected cells, we observed clustering of unique intracellular membrane-bound organelles (arrows in Fig. 7B) and the absence of normal-appearing mitochondria. Otherwise, there was little or no disruption of intracellular architecture early after infection. The plasma membrane was unaltered, nuclei appeared intact with a distinct nuclear envelope and dispersed chromatin, and organelles such as the ER and vacuoles also appeared intact (Fig. 7B), as in uninfected cells (Fig. 7A).
FIG. 7.
FHV infection induces ultrastructural changes in the mitochondria of infected Drosophila DL-1 cells. Cells were fixed in 4% paraformaldehyde and 2% glutaraldehyde, postfixed in osmium, embedded, sectioned, counterstained with uranyl acetate and Reynold's lead citrate, and analyzed by transmission EM. (A) An uninfected DL-1 cell, showing a normal nucleus (Nu) and distribution of intracellular organelles, including the ER (closed arrowheads), vacuoles (V), and mitochondria (arrows). (B) An FHV-infected DL-1 cell at 8 hpi also has a normal-appearing nucleus (Nu), vacuoles (V), and ER (closed arrowheads), but there is clustering and polarization of membrane-bound organelles (arrows) and an absence of normal-appearing mitochondria. (C) Higher magnification of a normal mitochondrion from an uninfected DL-1 cell. Note the characteristic double-membrane structure (closed arrowheads), sparse intermembrane space, and cristae (arrows) characteristic of normal mitochondria. (D) Early ultrastructural changes in mitochondria of FHV-infected DL-1 cells at 8 hpi are restricted primarily to the outer mitochondrial membrane. There is still an identifiable matrix (M), cristae (arrows), and double membrane (closed arrowheads), but there are numerous 40- to 60-nm spherules in the intermembrane space. (E) Later ultrastructural changes in the mitochondria of FHV-infected DL-1 cells include a collapse of the matrix (M) and loss of identifiable cristae. The membrane-bound spherules have visible necks that connect with the outer mitochondrial membrane (closed arrowheads) and an electron-dense substance in the center (open arrowheads). (F) Marked disruption of mitochondria 24 h after FHV infection. Although the outer and inner membranes are still visible, there is swelling and disruption of the matrix (M) with the appearance of membrane vesicles. (Inset) Electron-dense virus-like particles (arrows) are associated with membrane stacks and vesicles late after FHV infection. The two vesicles at the top of the image are adjacent to the residual plasma membrane. Bars = 1 μm in panels A and B, 100 nm in panels C to F, and 50 nm in panel F (inset).
We further examined the clustered membrane-bound organelles in FHV-infected DL-1 cells, as these structures were the most prominent difference between uninfected and infected cells early after infection. Like mitochondria from uninfected cells (Fig. 7C), the clustered membrane-bound organelles in FHV-infected cells had a double external membrane and crista-like internal membrane structures within an electron-dense matrix (Fig. 7D). However, a characteristic feature of the membrane-bound organelles from FHV-infected cells was the presence of numerous round vesicles in an expanded intermembrane space between the double external membranes. These vesicles frequently encompassed almost the entire external membrane with compression of the matrix (Fig. 7E). The predominantly round appearance of these vesicle structures and their various diameters suggested that the structures were spheric or ellipsoid. These FHV-induced spherules were membrane bound and 40 to 60 nm in diameter, contained an electron-dense amorphous material (open arrowheads in Fig. 7E), and appeared to be formed from an invagination of the outer organellar membrane, as necks that connected spherules to the outer membrane were frequently visible (closed arrowheads in Fig. 7E). We did not observe spherules associated with other intracellular membranes or free in the cytoplasm of FHV-infected cells. Based on the clustered distribution; the presence of a double external membrane, a matrix, and internal crista-like structures; and the absence of normal-appearing mitochondria in infected cells, we concluded that the unique membrane-bound organelles with intermembrane spherules in FHV-infected cells were altered mitochondria.
We made several observations regarding the likely temporal pattern of FHV-induced mitochondrial alterations by examining cells at earlier (4 h) and later (24 h) times postinfection. At 4 hpi, we found mitochondria with few spherules and minimal overall ultrastructural changes more frequently than in cells at 8 hpi, when most mitochondria showed ultrastructural changes, including many mitochondria with numerous spherules, the disappearance of cristae, and compression of the mitochondrial matrix (Fig. 7E). The mitochondrial alterations were even more pronounced at 24 hpi (Fig. 7F). Although spherules maintained their ultrastructural morphology, the mitochondrial matrix was swollen and disrupted. These results suggest that the pattern of mitochondrial changes progressed from spherule formation to initial matrix compaction, followed by matrix swelling, disruption, and dissolution of the overall mitochondrial structure. We concluded from these EM results that FHV infection induced significant mitochondrial changes that included the formation of membrane-bound spherules in the intermembrane space in DL-1 cells.
We could not definitively identify mature virions at 4 and 8 hpi, as FHV particles are 29 nm in diameter (12) and were easily confused with similar-sized ribosomes present in the cytoplasm of both uninfected (Fig. 7C) and FHV-infected (Fig. 7D and E) cells. However, by 24 hpi, we observed aggregates of dense particles that were morphologically similar to previously described alphanodavirus arrays (3, 19). We also observed dense particles associated with stacked membrane structures and membrane vesicles, particularly near the plasma membrane (Fig. 7F, inset). These ultrastructural features of virion dispersal have been previously described for both NOV (19) and BOV (3).
Ultrastructural localization of protein A in FHV-infected Drosophila DL-1 cells.
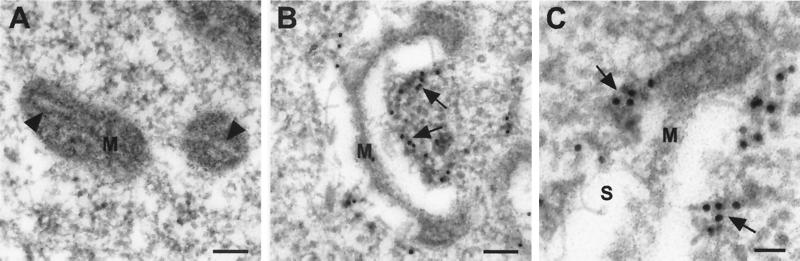
To further investigate the subcellular localization of protein A and connect the mitochondrial ultrastructure changes with viral RNA replication, we performed immunogold EM with protein A antisera (Fig. 8). Since preliminary experiments showed that osmium abolished protein A antigenicity, we omitted osmium fixation during sample preparation for immunogold labeling, which decreased the ultrastructural detail of cellular membranes. However, mitochondria could still be identified based on their intracellular distribution, the presence of cristae and an electron-dense matrix in uninfected cells (Fig. 8A), and their clustered distribution and characteristic compacted matrix and large intermembrane space in FHV-infected cells (compare Fig. 8B to Fig. 7E). By immunogold EM, preimmune sera showed no reactivity with either uninfected or FHV-infected DL-1 cells (data not shown). When immunolabeled with protein A antisera, sections from both infected and uninfected cells showed scattered rare gold particles in nuclei and the cytoplasm but no gold particles were present around mitochondria from uninfected cells (Fig. 8A). In contrast, gold particles were clustered around the altered mitochondria in FHV-infected cells, predominantly at the outer membrane (Fig. 8B), consistent with the selective membrane permeabilization experiments that implied an outer mitochondrial membrane localization of protein A (Fig. 5). At higher magnification, the clustering of gold particles at the outer mitochondrial membrane was particularly evident (Fig. 8C). We did not see gold particles over spherule remnants in the mitochondrial intermembrane space. However, this observation was complicated by poor preservation of spherule ultrastructure in the absence of osmium. Nevertheless, we concluded from these results that protein A was localized to the outer mitochondrial membranes in infected DL-1 cells.
FIG. 8.
Immunogold EM localizes FHV protein A to the outer mitochondrial membrane in infected Drosophila DL-1 cells. Cells were fixed in 4% paraformaldehyde and 2% glutaraldehyde, embedded, sectioned, immunostained with protein A antisera and 12-nm gold-labeled secondary antibodies, counterstained with uranyl acetate and Reynold's lead citrate, and analyzed by transmission EM. (A) Mitochondria from uninfected DL-1 cells show no reactivity with protein A antisera. Cristae (closed arrowheads) are visible in the matrix (M). (B) Mitochondria from FHV-infected DL-1 cells labeled with gold particles along the outer mitochondrial membrane (arrows). Although FHV-induced spherules are not well preserved, their location can be inferred from the position of the compressed matrix (M) and comparison to Fig. 7E. (C) Higher magnification of mitochondria from FHV-infected DL-1 cells. Localization of protein A along the outer mitochondrial membrane is demonstrated by the clustering of gold particles (arrows) in areas distinct from the residual compressed matrix (M) and remnants of FHV-induced spherules (S). Bars = 100 nm in panels A and B and 50 nm in panel C.
DISCUSSION
In this report, we have provided a detailed description of the cellular and ultrastructural changes induced by FHV infection in Drosophila DL-1 cells and identified mitochondria as the key cellular organelle involved in FHV RNA replication. We have demonstrated that protein A, the FHV RNA-dependent RNA polymerase, was tightly membrane associated, was localized to outer mitochondrial membranes, and colocalized with sites of viral RNA synthesis. We have also shown that FHV infection induced the formation of membrane-bound spherules in the mitochondrial intermembrane space. These observations implicate the outer mitochondrial membrane and the associated intermembrane spherules as important structures in FHV RNA replication.
The membrane association of FHV protein A is consistent with previously identified associations between host intracellular membranes and positive-strand RNA virus replication (8). FHV provides the opportunity to investigate the mechanisms of positive-strand RNA virus replicase membrane association, localization, and function based on a single viral protein. In contrast, many positive-strand RNA viruses encode accessory proteins that determine the intracellular localization or membrane association of the viral RNA polymerase. For example, the poliovirus 3AB protein (26), the Semliki Forest virus nsP1 protein (43), and the brome mosaic virus 1a protein (10) all target or anchor the nonoverlapping viral RNA polymerase to intracellular membranes. In contrast, protein A is the sole FHV protein required for RNA replication (1, 45) and preliminary results indicate that when FHV protein A is expressed in yeast in the absence of other viral factors, it becomes membrane associated and localizes to mitochondria (D. Miller and P. Ahlquist, unpublished data). The differential and gradient centrifugation results presented in this report demonstrated that FHV protein A remained membrane associated even in the presence of a high pH, a chaotropic agent, or high ionic strength (Fig. 3), which suggests that protein A might be an integral membrane protein, similar to replicase proteins from some other animal (27, 61, 62) and plant (49, 54) positive-strand RNA viruses. Mutational studies intended to determine the precise mechanisms of the FHV protein A-membrane association are currently in progress.
The membrane association of protein A, the colocalization of protein A and viral RNA synthesis to mitochondria, and the ultrastructural pathology in FHV-infected DL-1 cells provide evidence for a direct connection between nodavirus replication and mitochondrial membranes, as suggested by previous ultrastructural studies with other alphanodaviruses. Infection with NOV (19) or BOV (3) induces the formation of membrane-bound spherules in the mitochondrial intermembrane space. Although the intracellular localization of NOV and BOV proteins and viral RNA has not been established, Garzon et al. used RNase-gold labeling to identify RNA in and around NOV-altered mitochondria (19), consistent with our virus-specific BrUTP-labeling results (Fig. 6). In addition, double-stranded RNA-specific antibodies labeled mitochondrial spherules formed in Drosophila DL-1 cells after infection with a virus-like particle isolated in Australia from the sheep blowfly Lucilia cuprina (4, 5). Interestingly, FHV (12, 57), BBV (37), and BOV (47) were all isolated from insects in Australia or New Zealand, which suggests that the L. cuprina isolate might be an uncharacterized alphanodavirus. These observations, together with the data presented in this report, suggest that mitochondrial membrane involvement and mitochondrial spherule formation may be general features of alphanodavirus replication.
Mitochondrial spherules have also been reported after infection with several plant viruses (14, 23, 24), particularly members of the Tombusviridae family (51). Tombusviruses have several genomic and structural similarities to alphanodaviruses, which include a 4.7-kb genome with capped but nonpolyadenylated RNA, a 41-kDa capsid protein and two replicase proteins with a combined molecular mass of 125 to 131 kDa, and a 30-nm nonenveloped virion with an icosahedral (T=3) capsid (50). The best-studied tombusvirus whose infection of plants induces the ultrastructural feature of mitochondrial spherule formation is carnation Italian ringspot virus (CIRV). CIRV infection induces the formation of multivesicular bodies, which represent transformed mitochondria with 80- to 150-nm spherules in the intermembrane space that are connected to the outer membrane via thin, neck-like structures (14). Apart from the slightly larger size, the CIRV-induced spherules are similar to the mitochondrial spherules we observed after FHV infection in DL-1 cells (Fig. 7). CIRV-induced spherules contain RNA, as detected by RNase susceptibility (14), and their formation and localization are dependent on determinants encoded by the 5′-terminal region of the CIRV genome (9), a region that encodes the essential 36-kDa viral replicase integral membrane protein responsible for intracellular membrane targeting (49). In addition, immunogold EM studies of a closely related tombusvirus, cymbidium ringspot virus, have shown that viral replicase proteins are localized to multivesicular bodies (6), similar to our immunogold EM results with FHV protein A (Fig. 8).
Membrane-bound spherule formation is not limited to infection with alphanodaviruses and plant viruses. Another well-studied group of viruses that induce spherules with ultrastructural similarities to FHV-induced structures are members of the Togaviridae family. Mammalian cells infected with the alphaviruses Semliki Forest virus or Sindbis virus develop two types of intracellular cytopathic vacuoles (CPV), termed CPV-I and CPV-II (21). CPV-I are derived from cellular endosomes and lysosomes, and they contain 50-nm spherules attached to the inner surface of the endosomal/lysosomal membrane via narrow, neck-like structures (17, 21). Newly synthesized viral RNA and all four nonstructural alphavirus proteins, including the catalytic subunit of the RNA polymerase, localize to membrane-bound CPV-I (17, 21, 32). Apart from the different organellar membrane association, the ultrastructural appearance of alphavirus spherules is similar to that of outer mitochondrial membrane spherules in FHV-infected Drosophila cells (Fig. 7).
The ultrastructural similarities between alphavirus and FHV infections are not limited to spherule formation. In contrast to CPV-I, alphavirus CPV-II do not have spherules on the inner membrane surface but rather are studded along the outer membrane surface with electron-dense particles, which are thought to be mature nucleocapsids awaiting envelope attachment and budding (21). We observed similar membrane structures studded with electron-dense particles in FHV-infected Drosophila cells late in infection (Fig. 7F, inset). Although mature FHV particles do not acquire a lipid envelope (56, 57), we suspect that the electron-dense particles in FHV-infected cells were virions based on closely parallel results of ultrastructural studies with NOV (19) and BOV (3) and the observation that the use of nonionic detergents to disrupt membrane association can double virion recovery from FHV-infected Drosophila cells (57).
Membrane-bound spherules are also seen after infection with rubella virus, another member of the Togaviridae family. Rubella virus-induced CPV are also derived from endosomes and lysosomes and contain 60-nm membrane-bound spherules along the inner surface (38), similar to alphavirus CPV-I (17, 21). However, rubella virus induces some unique mitochondrial abnormalities not seen with alphaviruses, which include mitochondrial clustering around virus-induced CPV, the formation of an electron-dense zone between the outer membrane of juxtaposed mitochondria, and the close association of core particles with mitochondria (34). We also observed mitochondrial clustering in FHV-infected Drosophila cells (Fig. 4B and 7B), and although we did not see a clear association of virus-like particles with mitochondrial membranes, both NOV (19) and BOV (3) particles have been closely associated with mitochondria.
The similar intracellular pathologies induced by togaviruses and alphanodaviruses are remarkable in light of their genomic and structural differences. In contrast to alphanodaviruses, togaviruses have a 10- to 12-kb polyadenylated genome, multiple replicase-associated nonstructural proteins, and a 34- to 38-nm enveloped virion with an icosahedral (T=4) capsid (60). Despite these differences, the similar ultrastructural changes induced by alphanodaviruses, tombusviruses, and togaviruses suggest a conservation of fundamental mechanisms in RNA replication, which include the formation of membrane-bound spherules. Intracellular membranes have been postulated to provide either a structural framework for replication complex component assembly (32, 42, 63) or a protective structure that shields nascent viral RNA or viral replication complexes from degradation (33). However, detailed structure-function relationships for spherules and other unique virus-induced membrane structures identified by detailed EM studies have not been definitively established. Although the association of viral RNA replication with togavirus-induced spherules is firmly established (17, 21, 31–33, 38) and the data presented in this report and previous studies (3, 5, 19) strongly implicate mitochondrial spherules in nodavirus RNA replication, the precise function of virus-induced spherules is unclear. Studies are currently in progress to elucidate FHV spherule formation and function.
The utility of FHV as a model with which to investigate viral replication and identify novel therapeutics derives, in part, from its genomic simplicity, the defined localization of replication as demonstrated in this report, the presence of ultrastructural features common to many positive-strand RNA viruses, and the ability of FHV to complete its replication cycle in S. cerevisiae (45, 46), a host that can facilitate the identification and study of virus-host interactions and host functions required for viral replication (13, 35, 41). Further analysis of FHV replication should provide insights into basic mechanisms of positive-strand RNA virus replication and potentially identify targets for broadly effective antiviral agents that inhibit fundamental steps in viral replication.
ACKNOWLEDGMENTS
We thank Kathleen Wessels for assistance and Brett Lindenbach for helpful comments on the manuscript. We performed the confocal immunofluorescence microscopy and transmission EM at the Keck Neural Imaging Laboratory and the Medical School Electron Microscope Facility at the University of Wisconsin—Madison, respectively.
This work was supported by National Institutes of Health grants K08 AI01770-01 and GM35072. P.A. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Ball L A. Requirements for the self-directed replication of flock house virus RNA 1. J Virol. 1995;69:720–727. doi: 10.1128/jvi.69.2.720-727.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ball L A, Johnson K L. Nodaviruses of insects. In: Miller L K, Ball L A, editors. The insect viruses. New York, N.Y: Plenum Publishing Corporation; 1998. pp. 225–267. [Google Scholar]
- 3.Bashiruddin J B, Cross G F. Boolarra virus: ultrastructure of intracytoplasmic virus formation in cultured Drosophila cells. J Invertebr Pathol. 1987;49:303–315. [Google Scholar]
- 4.Binnington K C. Structural transformation of blowfly mitochondria by a putative virus: similarities with virus-induced changes in plant mitochondria. J Gen Virol. 1987;68:201–206. [Google Scholar]
- 5.Binnington K C, Brooks L. Gold-labelling of RNA in virus-induced mitochondrial vesicles in the sheep blowfly Lucilia cuprina. Tissue Cell. 1992;24:411–416. doi: 10.1016/0040-8166(92)90057-e. [DOI] [PubMed] [Google Scholar]
- 6.Bleve-Zacheo T, Rubino L, Melillo M T, Russo M. The 33K protein encoded by cymbidium ringspot tombusvirus localizes to modified peroxisomes of infected cells and of uninfected transgenic plant. J Plant Pathol. 1997;79:179–202. [Google Scholar]
- 7.Bonifacino J S. Characterization of cellular proteins. In: Morgan K S, editor. Current protocols in cell biology. New York. N.Y: John Wiley & Sons, Inc.; 2000. pp. 5.0.1–5.5.11. .. [Google Scholar]
- 8.Buck K W. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv Virus Res. 1996;47:159–251. doi: 10.1016/S0065-3527(08)60736-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burgyan J, Rubino L, Russo M. The 5′-terminal region of a tombusvirus genome determines the origin of multivesicular bodies. J Gen Virol. 1996;7:1967–1974. doi: 10.1099/0022-1317-77-8-1967. [DOI] [PubMed] [Google Scholar]
- 10.Chen J, Ahlquist P. Brome mosaic virus polymerase-like protein 2a is directed to the endoplasmic reticulum by helicase-like viral protein 1a. J Virol. 2000;74:4310–4318. doi: 10.1128/jvi.74.9.4310-4318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dasmahapatra B, Dasgupta R, Ghosh A, Kaesberg P. Structure of the black beetle virus genome and its functional implications. J Mol Biol. 1985;182:183–189. doi: 10.1016/0022-2836(85)90337-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dearing S C, Scotti P D, Wigley P J, Dhana S D. A small RNA virus isolated from the grass grub. Costelytra zealandica (Coleoptera: Scarabaeidae) N Z J Zool. 1980;7:267–269. [Google Scholar]
- 13.Diez J, Ishikawa M, Kaido M, Ahlquist P. Identification and characterization of a host protein required for efficient template selection in viral RNA replication. Proc Natl Acad Sci USA. 2000;97:3913–3918. doi: 10.1073/pnas.080072997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Franco A, Russo M, Martelli G P. Ultrastructure and origin of cytoplasmic multivesicular bodies induced by carnation Italian ringspot virus. J Gen Virol. 1984;65:1233–1237. [Google Scholar]
- 15.Friesen P D, Rueckert R R. Synthesis of black beetle virus proteins in cultured Drosophila cells: differential expression of RNAs 1 and 2. J Virol. 1981;37:876–886. doi: 10.1128/jvi.37.3.876-886.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friesen P D, Rueckert R R. Black beetle virus: messenger for protein B is a subgenomic viral RNA. J Virol. 1982;42:986–995. doi: 10.1128/jvi.42.3.986-995.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Froshauer S, Kartenbeck J, Helenius A. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J Cell Biol. 1988;107:2075–2086. doi: 10.1083/jcb.107.6.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallagher T M, Rueckert R R. Assembly-dependent maturation cleavage in provirions of a small icosahedral insect ribovirus. J Virol. 1988;62:3399–3406. doi: 10.1128/jvi.62.9.3399-3406.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garzon S, Strykowski H, Charpentier G. Implication of mitochondria in the replication of Nodamura virus in larvae of the Lepidoptera. Galleria mellanolla (L.) and in suckling mice. Arch Virol. 1990;113:165–176. doi: 10.1007/BF01316670. [DOI] [PubMed] [Google Scholar]
- 20.Geelen M J, Bijleveld C, Velasco G, Wanders R J, Guzman M. Studies on the intracellular localization of acetyl-CoA carboxylase. Biochem Biophys Res Commun. 1997;233:253–257. doi: 10.1006/bbrc.1997.6437. [DOI] [PubMed] [Google Scholar]
- 21.Grimley P M, Berezesky I K, Friedman R M. Cytoplasmic structures associated with an arbovirus infection: loci of viral ribonucleic acid synthesis. J Virol. 1968;2:1326–1338. doi: 10.1128/jvi.2.11.1326-1338.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guarino L A, Kaesberg P. Isolation and characterization of an RNA-dependent RNA polymerase from black beetle virus-infected Drosophila melanogaster cells. J Virol. 1981;40:379–386. doi: 10.1128/jvi.40.2.379-386.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harrison B D, Stefanac Z, Roberts I M. Role of mitochondria in the formation of X-bodies in cells of Nicotiana clevelandii infected by tobacco rattle virus. J Gen Virol. 1970;6:127–140. [Google Scholar]
- 24.Hatta T, Nakamoto T, Takagi Y, Ushiyama R. Cytological abnormalities of mitochondria induced by infection with cucumber green mottle mosaic virus. Virology. 1971;45:292–297. doi: 10.1016/0042-6822(71)90136-x. [DOI] [PubMed] [Google Scholar]
- 25.Hollinshead M, Sanderson J, Vaux D J. Anti-biotin antibodies offer superior organelle-specific labeling of mitochondria over avidin or streptavidin. J Histochem Cytochem. 1997;45:1053–1057. doi: 10.1177/002215549704500803. [DOI] [PubMed] [Google Scholar]
- 26.Hope D A, Diamond S E, Kirkegaard K. Genetic dissection of interaction between poliovirus RNA-dependent RNA polymerase and viral protein 3AB. J Virol. 1997;71:9490–9498. doi: 10.1128/jvi.71.12.9490-9498.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hügle T, Fehrmann F, Bieck E, Kohara M, Kräusslich H, Rice C M, Blum H E, Moradpour D. The hepatitis C virus nonstructural protein 4B is an integral endoplasmic reticulum membrane protein. Virology. 2001;284:70–81. doi: 10.1006/viro.2001.0873. [DOI] [PubMed] [Google Scholar]
- 28.Johnson K L, Ball L A. Replication of flock house virus RNAs from primary transcripts made in cells by RNA polymerase II. J Virol. 1997;71:3323–3327. doi: 10.1128/jvi.71.4.3323-3327.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamar G, Argos P. Primary structural comparisons of RNA-dependent polymerases from plant, animal and bacterial viruses. Nucleic Acids Res. 1984;12:7269–7282. doi: 10.1093/nar/12.18.7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koonin E V. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J Gen Virol. 1991;72:2197–2206. doi: 10.1099/0022-1317-72-9-2197. [DOI] [PubMed] [Google Scholar]
- 31.Kujala P, Ahola T, Ehsani N, Auvinen P, Vihinen H, Kääriäinen L. Intracellular distribution of rubella virus nonstructural protein P150. J Virol. 1999;73:7805–7811. doi: 10.1128/jvi.73.9.7805-7811.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kujala P, Ikäheimonen A, Ehsani N, Vihinen H, Auvinen P, Kääriäinen L. Biogenesis of the Semliki Forest virus RNA replication complex. J Virol. 2001;75:3873–3884. doi: 10.1128/JVI.75.8.3873-3884.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee J, Marshall J A, Bowden D S. Characterization of rubella virus replication complexes using antibodies to double-stranded RNA. Virology. 1994;200:307–312. doi: 10.1006/viro.1994.1192. [DOI] [PubMed] [Google Scholar]
- 34.Lee J, Marshall J A, Bowden D S. Localization of rubella virus core particles in Vero cells. Virology. 1999;265:110–119. doi: 10.1006/viro.1999.0016. [DOI] [PubMed] [Google Scholar]
- 35.Lee W-M, Ishikawa M, Ahlquist P. Mutation of host Δ9 fatty acid desaturase inhibits brome mosaic virus RNA replication between template recognition and RNA synthesis. J Virol. 2001;75:2097–2106. doi: 10.1128/JVI.75.5.2097-2106.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leyssen P, De Clercq E, Neyts J. Perspectives for the treatment of infections with Flaviviridae. Clin Microbiol Rev. 2000;13:67–82. doi: 10.1128/cmr.13.1.67-82.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Longworth J F, Carey G P. A small RNA virus with a divided genome from Heteronychus arator (F.) [Coleoptera:Scarabaeidae] J Gen Virol. 1976;33:31–40. doi: 10.1099/0022-1317-33-1-31. [DOI] [PubMed] [Google Scholar]
- 38.Magliano D, Marshall J A, Bowden D S, Vardaxis N, Meagner J, Lee J. Rubella virus replication complexes are virus-modified lysosomes. Virology. 1998;240:57–63. doi: 10.1006/viro.1997.8906. [DOI] [PubMed] [Google Scholar]
- 39.Mas P, Beachy R N. Replication of tobacco mosaic virus on endoplasmic reticulum and role of the cytoskeleton and virus movement protein on intracellular distribution of viral RNA. J Cell Biol. 1999;147:945–958. doi: 10.1083/jcb.147.5.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morse S S. The public health threat of emerging viral diseases. J Nutr. 1997;127(Suppl. 5):951S–957S. doi: 10.1093/jn/127.5.951S. [DOI] [PubMed] [Google Scholar]
- 41.Noueriy A O, Chen J, Ahlquist P. A mutant allele of essential, general translation initiation factor DED1 selectively inhibits translation of a viral mRNA. Proc Natl Acad Sci USA. 2000;97:12985–12990. doi: 10.1073/pnas.240460897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pedersen K W, van der Meer Y, Roos N, Snijder E J. Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J Virol. 1999;73:2016–2026. doi: 10.1128/jvi.73.3.2016-2026.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peränen J, Laakkonen P, Hynönen M, Kääriäinen L. The alphavirus replicase protein nsP1 is membrane-associated and has affinity to endocytic organelles. Virology. 1995;208:610–620. doi: 10.1006/viro.1995.1192. [DOI] [PubMed] [Google Scholar]
- 44.Poch O, Sauvaget I, Delarue M, Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Price D B, Roerder M, Ahlquist P. DNA-directed expression of functional flock house virus RNA1 derivatives in Saccharomyces cerevisiae, heterologous gene expression, and selective effects on subgenomic mRNA synthesis. Virology. 2000;74:11724–11733. doi: 10.1128/jvi.74.24.11724-11733.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Price D B, Rueckert R R, Ahlquist P. Complete replication of an animal virus and maintenance of expression vectors derived from it in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:9465–9470. doi: 10.1073/pnas.93.18.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reinganum C, Bashiruddin J B, Cross G F. Boolarra virus: a member of the Nodaviridae isolated from Oncopera intricoides (Lepidoptera:Hepialidae) Intervirology. 1985;20:10–17. doi: 10.1159/000149613. [DOI] [PubMed] [Google Scholar]
- 48.Restropo-Hartwig M, Ahlquist P. Brome mosaic virus helicase- and polymerase-like proteins colocalize in the endoplasmic reticulum at sites of viral RNA synthesis. J Virol. 1996;70:8908–8916. doi: 10.1128/jvi.70.12.8908-8916.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rubino L, Russo M. Membrane targeting sequences in tombusvirus infections. Virology. 1998;252:431–437. doi: 10.1006/viro.1998.9490. [DOI] [PubMed] [Google Scholar]
- 50.Russo M, Burgyan J, Martelli G P. Molecular biology of Tombusviridae. Adv Virus Res. 1994;44:381–428. doi: 10.1016/s0065-3527(08)60334-6. [DOI] [PubMed] [Google Scholar]
- 51.Russo M, Di Franco A, Martelli G P. Cytopathology in the identification and classification of tombusviruses. Intervirology. 1987;28:134–143. doi: 10.1159/000150009. [DOI] [PubMed] [Google Scholar]
- 52.Ryerse J, Blachly-Dyson E, Forte M, Nagel B. Cloning and molecular characterization of a voltage-dependent anion-selective channel (VDAC) from Drosophila melanogaster. Biochim Biophys Acta. 1997;1327:204–212. doi: 10.1016/s0005-2736(97)00059-x. [DOI] [PubMed] [Google Scholar]
- 53.Saks V A, Veksler V I, Kuznetsov A V, Kay L, Sikk P, Tiivel T, Tranqui L, Olivares J, Winkler K, Wiedemann F, Kunz W S. Permeabilized cell and skinned fiber techniques in studies of mitochondrial function in vivo. Mol Cell Biochem. 1998;184:81–100. [PubMed] [Google Scholar]
- 54.Schaad M C, Jensen P E, Carrington J C. Formation of plant RNA virus replication complexes on membranes: role of an endoplasmic reticulum-targeted viral protein. EMBO J. 1997;16:4049–4059. doi: 10.1093/emboj/16.13.4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schlegel A, Giddings T H, Jr, Ladinsky M S, Kirkegaard K. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J Virol. 1996;70:6576–6588. doi: 10.1128/jvi.70.10.6576-6588.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schneemann A, Reddy V, Johnson J E. The structure and function of nodavirus particles: a paradigm for understanding chemical biology. Adv Virus Res. 1998;50:381–446. doi: 10.1016/s0065-3527(08)60812-x. [DOI] [PubMed] [Google Scholar]
- 57.Scotti P D, Dearing S, Mossop D W. Flock house virus: a nodavirus isolated from Costelytra zealandica (White) (Coleoptera: Scarabaeidae) Arch Virol. 1983;75:181–189. doi: 10.1007/BF01315272. [DOI] [PubMed] [Google Scholar]
- 58.Selling B H, Allison R F, Kaesberg P. Genomic RNA of an insect virus directs synthesis of infectious virions in plant. Proc Natl Acad Sci USA. 1990;87:434–438. doi: 10.1073/pnas.87.1.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Selling B H, Rueckert R R. Plaque assay for black beetle virus. J Virol. 1984;51:251–253. doi: 10.1128/jvi.51.1.251-253.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Strauss J H, Strauss E G. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Towner J S, Ho T V, Semler B L. Determinants of membrane association for poliovirus protein 3AB. J Biol Chem. 1996;271:26810–26818. doi: 10.1074/jbc.271.43.26810. [DOI] [PubMed] [Google Scholar]
- 62.van der Meer Y, van Tol H, Locker J K, Snijder E J. ORF1a-encoded replicase subunits are involved in the membrane association of the arterivirus replication complex. J Virol. 1998;72:6689–6698. doi: 10.1128/jvi.72.8.6689-6698.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Westaway E G, Mackenzie J M, Kenney M T, Jones M K, Khromykh A A. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA and of NS2B with NS3, in virus-induced membrane structures. J Virol. 1997;71:6650–6661. doi: 10.1128/jvi.71.9.6650-6661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu S, Kaesberg P. Synthesis of template-sense, single-strand flockhouse virus RNA in a cell-free replication system. Virology. 1991;183:392–396. doi: 10.1016/0042-6822(91)90153-3. [DOI] [PubMed] [Google Scholar]
- 65.Wu S, Ahlquist P, Kaesberg P. Active complete in vitro replication of nodavirus RNA requires glycerophospholipid. Proc Natl Acad Sci USA. 1992;89:11136–11140. doi: 10.1073/pnas.89.23.11136. [DOI] [PMC free article] [PubMed] [Google Scholar]