Abstract
Single-cell transcriptome sequencing (scRNA-seq) is a powerful tool for describing the transcriptome dynamics of plant development but has not yet been utilized to analyze the tissue ontology of sweetpotato. This study established a stable method for isolating single protoplast cells for scRNA-seq to reveal the cell heterogeneity of sweetpotato root tip meristems at the single-cell level. The study analyzed 12,172 single cells and 27,355 genes in the root tips of the sweetpotato variety Guangshu 87, which were distributed into 15 cell clusters. Pseudo-time analysis showed that there were transitional cells in the apical development trajectory of mature cell types from stem cell niches. Furthermore, we identified novel development regulators of sweetpotato tubers via trajectory analysis. The transcription factor IbGATA4 was highly expressed in the adventitious roots during the development of sweetpotato root tips, where it may regulate the development of sweetpotato root tips. In addition, significant differences were observed in the transcriptional profiles of cell types between sweetpotato, Arabidopsis thaliana, and maize. This study mapped the single-cell transcriptome of sweetpotato root tips, laying a foundation for studying the types, functions, differentiation, and development of sweetpotato root tip cells.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-024-05574-8.
Keywords: Ipomoea batatas (L.) Lam., scRNA-seq, Root, Transitional cells, Developmental trajectory
Highlights
This is the first single-cell transcriptional atlas of sweetpotato root apex tissue.
Single-cell analysis of stem cell niche initiation showed unique transitional states.
Sweetpotato, Arabidopsis, and maize root tip cell types were correlated.
GATA4 may be involved in regulating sweetpotato root tip development.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-024-05574-8.
Introduction
Ipomoea batatas (L.) Lam. or sweetpotato, belonging to the order Ipomoea of the family Convolvulaceae [1, 2], exhibits homologous hexaploidy (2n = 6x = 90) with a large genome. Sweetpotato tubers are starch rich and rank seventh among starch sources cultivated worldwide [3–5]. Sweetpotato is a short-day crop rich in multiple nutrients and the main crop in regions with barren soil, as the crop has high and stable yields and drought resistance. In addition to it being an nutritious food source, sweetpotatoes are important energy sources for feed and serve as industrial raw materials. Thus, sweetpotatoes are important crops for world food production and highly competitive energy sources [6–8].
Tubers are the edible part of sweetpotatoes, and they have high economic value. Sweetpotato yield is positively correlated with the number of plants, the number of tubers per plant, and the weight of a single tuber [9]. However, sweetpotatoes exhibit self-incompatibility, hindering their sexual reproduction, which can only occur through cross-breeding. Since the vegetative organs, such as the storage root and stem, of sweetpotato have strong regeneration ability and can maintain the traits of improved varieties, seedlings or stem segments with several nodes are commonly used for cultivation and reproduction. Tubers or leaves with petioles can also be used for propagation. The adventitious roots from sweetpotato stem nodes, tubers, leaves, and petioles undergo secondary growth to form a secondary cambium and begin abnormal growth activities, resulting in the formation and expansion of root tubers [10]. The changes in cell types and molecular regulation mechanisms during sweetpotato root development are very important but have been less explored owing to their complexity. The development of single-cell RNA sequencing (scRNA-seq) technology has enabled high-level analysis of sweetpotatoes at the transcriptome level.
scRNA-seq is a high-throughput sequencing technology used to analyze the individual cell level [11]. The technology can be used to explore the expression profile of individual cells at a high-throughput and single-cell resolution while revealing the heterogeneity of cell populations and avoiding the gene expression signals of individual cells being obscured by the averages the cell population. Recent advances in scRNA-seq technology have enabled the analysis of gene expression patterns in heterogeneous tissues and organs at the single-cell level and have been widely applied in plants. However, the plant cell wall inhibits the separation and acquisition of single cells, thus hindering the application of this technology to plant tissues, though scRNA-seq technology has been widely applied to diverse plant tissue [12, 13]. Additionally, scRNA-seq has been used to explain the specific cell types of maize root tips involved in the response to nitrate signal [14] and to reveal the gene expression changes during root tip cell differentiation and fate regulation in Arabidopsis thaliana [15]. This technology has also been used to identify the LBD family core transcription factor responsible for stem root initiation in tomatoes [16].
In this study, 13,966 protoplasts from sweetpotato root tips were used for scRNA-seq analysis, and gene expression profiles of 15 clusters were obtained. Transitional cells were identified in the differentiation and development trajectory through pseudo-time analysis. The differential characteristics of root-cell-type transcriptomes were examined by comparing scRNA-seq data between sweetpotato, A. thaliana, and maize.
Results
Sweetpotato root protoplast isolation and scRNA-seq analysis
Adventitious root tips of sweetpotato grown at 28℃ for 5 days were cut into small pieces and transferred into 10 ml of an enzymatic solution containing cellulase, macerase, and hemicellulase. The suspension was shaken gently for 3 h to isolate protoplasts. More than 100,000 protoplasts were obtained after centrifugation and their cell viability was evaluated by trypan blue staining. Microscopic observation showed that the proportion of living cells was more than 80% (Fig.S1). Total RNA was then extracted from the high-quality protoplasts and used for scRNA-seq on the 10x Genomics platform (Fig. 1a). After removing incompletely divided cells and low-quality cells, we obtained 12,172 single-cell transcriptome information; this comprised 370,228,781 reads, 75.0% of which mapped to the Ipomoea trifida (NSP306) reference genome. Expression of 27,355 genes was detected in the root sample. The median number of unique molecular identifiers (UMIs) per cell was 7142.5, and an average of 2775.5 genes were expressed per cell (Table S1). To assess the robustness of scRNA-seq and the effect of protoplast isolation, conventional bulked RNA-seq was performed on root tips without protoplast isolation, and strong correlations between the scRNA-seq pseudo-bulked expression data and bulked RNA-seq expression data (R = 0.71, P = 0) were observed (Fig. S2 ).
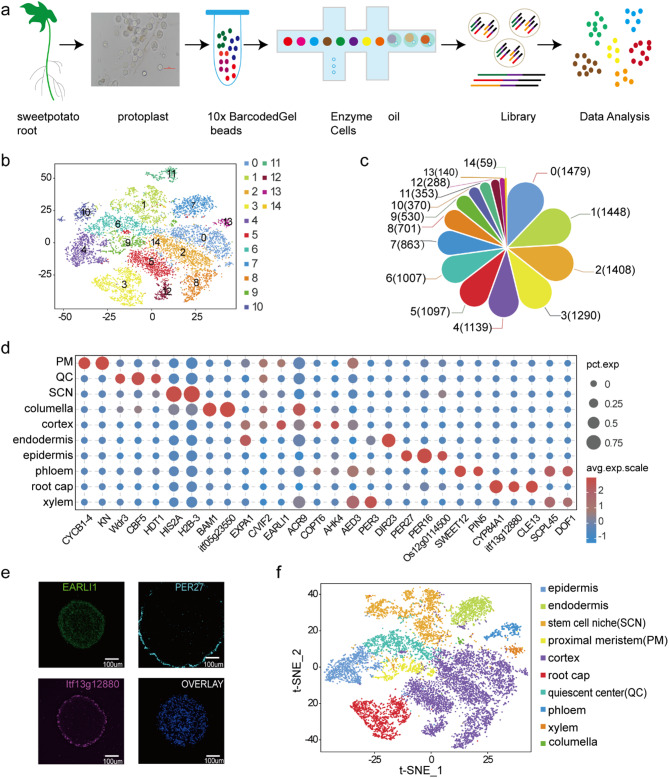
Fig. 1.
Cellular heterogeneity cluster analysis of the sweetpotato root single-cell transcriptome. (a) Brief summary of the process of isolation of sweetpotato root protoplast cells and their analysis using the 10x Genomics platform. (b) Classification of distinct sweetpotato root cell clusters visualized by t-SNE. Each dot indicates individual cells that are colored based on their cell cluster. (c) The flower plot shows the number of cells in the 15 cell clusters. (d) Bubble plot of representative cluster-specific marker genes in different cell types. (e) RNA in situ hybridization validation of representative cell type-specific marker genes for the cortex, root cap, and epidermis cells; scale bars = 100 μm. (f) Classification of distinct sweetpotato root cell types visualized by t-SNE. Each dot indicates individual cells that are colored according to their cell type
Identification of major cell clusters in sweetpotato root
To identify distinct cell populations of root tips, 12,172 cells were classified into 15 major clusters, with the number of cells ranging from 59 to 1479 (Fig. 1b, c and Table S2). To assign cell identities to the clusters, first, we examined the transcript specificity of homologous known marker genes in A. thaliana roots [17–23], and the results of previous studies helped us to predict the cell types (Fig. 1d and Table S3). Finally, in situ RNA hybridization was performed on some representative marker genes from this study (Fig. 1e and Table S12 ). After annotation, 15 clusters were categorized into ten cluster clouds corresponding to epidermis cells, columella cells, root cap cells, cortex cells, endodermis cells, xylem cells, phloem cells, quiescent center cells, stem cell niche cells, and proximal meristem cells (Fig. 1f and Fig. S3).
The cortex cell population consisted of five clusters (clusters 0, 2, 5, 8, and 12), the representative marker genes EARLI1, C/VIF2, AED3, EXPA1, ACR9, COPT6, and AHK4 were predominantly expressed. For stem cell niche cells, the representative marker genes histone superfamily protein (HIS2A, H2B-3) were specifically expressed in clusters 1 and 10. CBF5 and Wdr3 were identified as marker genes of quiescent center cells in A. thaliana and specifically expressed in cluster 6. CYP84A1, CLE13, and itf13g12880 showed higher expression in cluster 3, as root cap cells marker genes. In proximal meristem cells, two marker genes (KN, CYCB1-4) were specifically expressed in cluster 9, and their homologs have been previously identified as specific marker genes of proximal meristem cells in Arabidopsis roots. DIR23 and PER3 were the representative marker genes in Arabidopsis endodermis cells, and they were specially expressed in cluster 7. PIN5, SWEET12, SCPL45, and DOF1 were expressed in clusters 11 and 13, respectively, as the marker genes in phloem and xylem cells. PER27, PER16, and Os12g0114500 were the representative marker genes of the epidermal cells and expressed in cluster 4. Among the columella cells, itf05g23550 and BAM1 were specifically expressed in cluster 14 (Fig. S4). Altogether, the results indicate that the sweetpotato root was composed of highly heterogeneous cells.
Identification of new marker genes in each cell cluster
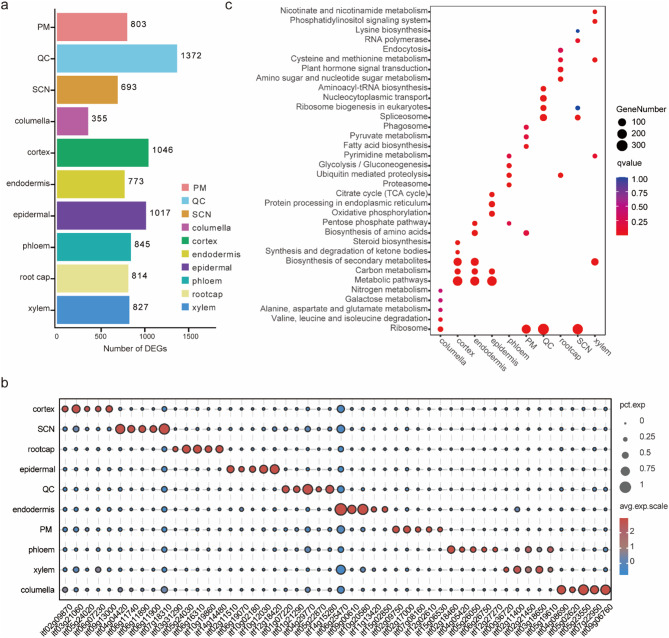
A total of 8,545 significantly up-regulated differentially expressed genes (DEGs) were identified, with 59 to 4,793 of these DEGs among each of the distinct clusters (Fig. 2a). The greatest number of elevated DEGs was observed in the cortex cells (4,973 cells). The top five genes with the highest expression level in each cluster were identified, and the expression profiles are described in a heatmap (Fig. 2b and Table S4). The ten representative genes with the highest expression in each cell type were selected for display as a t-distributed stochastic neighbor embedding (t-SNE) map (Fig. S5). Gene Ontology (GO) enrichment analysis revealed that all ten cell-type clusters were significantly enriched in the “metabolic process” and “cellular process” biological process categories. The molecular function annotation enrichment results showed PM, SCN, and quiescent center cells were significantly enriched for the “structural molecule activity” terms. Root cap and xylem clusters were significantly enriched for “catalytic activity” and “binding” (Fig. S6). Furthermore, Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis revealed that the PM, SCN, and quiescent center clusters were significantly enriched in the “ribosome” pathway. The epidermal, cortex, and endodermis clusters were significantly enriched in the “Metabolic,” “Carbon metabolism,” and “Biosynthesis of secondary metabolites” pathways. The columella cluster was significantly enriched for “Alanine, aspartate and glutamate metabolism,” “Galactose metabolism,” and “Valine, leucine and isoleucine degradation” pathways. The phloem cluster was significantly enriched for the “Glycolysis/Gluconeogenesis” and “Ubiquitin mediated proteolysis” pathways. Root cap and xylem clusters were significantly enriched for the “Cysteine and methionine metabolism” and “ Nicotinate and nicotinamide metabolism” pathways respectively (Fig. 2c).
Fig. 2.
Identification of new marker genes within cell type clusters. (a) The number of differentially expressed genes (DEGs) in each cell type cluster. (b) Bubble plot showing the top five DEGs with the highest expression levels in each cell cluster. (c) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enrichment analysis of all clusters of DEGs
The transition of meristematic cell clusters to mature cell types
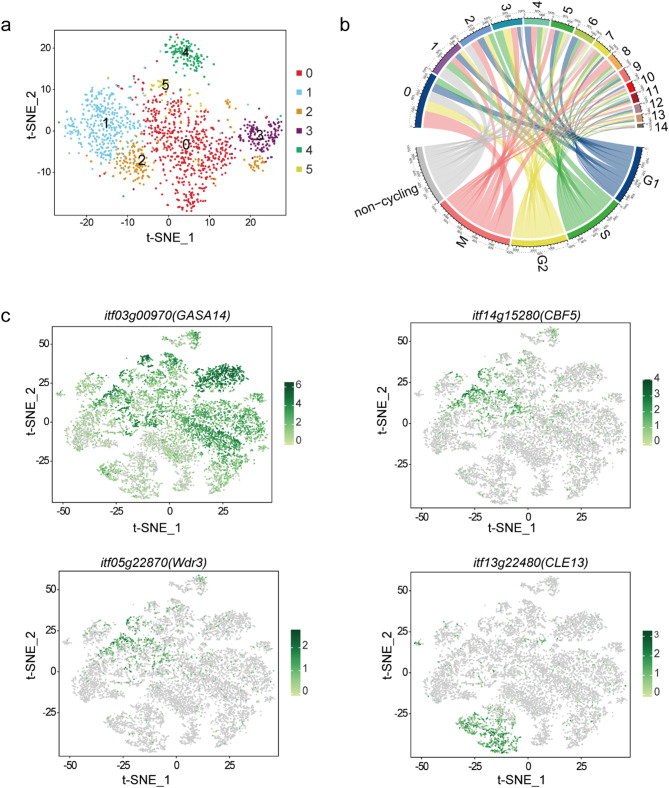
To determine whether there is a transition state in the differentiation of stem cells into mature cell types, we further divided cluster 1 into 6 subclusters and analyzed the DEGs of these subclusters (Fig. 3a and Table S5). We found that UBC20 (itf07g01660) was enriched in subcluster 1 − 0 of the proximal meristem cell, while EARLI1 (itf13g08300, itf08g01110) and AED3(itf08g06010) were highly expressed in subcluster 1–1 of the cortex cell. Moreover, At3g16150, CBF5 (itf14g15280, itf04g29770), and Wdr3(itf05g22870) were significantly enriched in subclusters 1–2 and 1–3 of the quiescent center cell, while GASA14 (itf03g00970), CLE13 (itf13g22480), and CYP84A1 (itf05g24030) were significantly enriched in subcluster 1–4 of the endodermis cell, subcluster 1–5 of the root cap cell, respectively. Clusters exhibiting obvious mature cell characteristics were distinct from the stem cells, indicating an overall cluster arrangement reflecting the developmental time. Subclusters revealed the cell identity of some subclusters, but not as obviously as among clusters corresponding to the cortex, root cap, and endodermis cells, for example. The subclusters showed the gene expression patterns of stem cells and mature cell types. The expression levels of marker genes were mapped to the t-SNE visualization map of the sweetpotato root tip (Fig. 3c), and the genes were distributed in the meristematic cell cluster and the corresponding mature cell type cluster. This indicated the proximity of mature and developmental cells with the same final fate. Thus, this study observed transitional cells during the development of sweetpotato root tip cells, which have the potential to differentiate into mature cell types.
Fig. 3.
Subcluster analysis of stem cell niche cells. (a) The t-distributed stochastic neighbor embedding (t-SNE) map embedding of subcluster assignments following a second round of clustering within stem cell cluster 1. (b) Cell clusters and corresponding cell cycle circos diagram. (c) The expression patterns of mature cell-type marker genes in the stem cell subcluster are distributed in the t-SNE map
Regulatory differences of the root apical meristem cells
Since increased cell division rate increases the probability of spontaneous mutations during genome replication, we analyzed whether there were differences in the cell division rate among sweetpotato root apical meristem (RAM) cells. The cell-cycle stage estimations generated during cell-cycle regression analysis indicated that compared with the stem cell niche, the fraction of meristematic cells in the G2 phase increased significantly, indicative of higher cell division rates (Fig. 3b and Table S6). In addition, we observed that the dividing cells were mostly concentrated closer to the stem cell niches and meristematic cell clusters, and the proportion of G1 phase cells was lower relative to those of other cell types. This indicated that stem cell niches and meristematic [24] cell clusters had higher cell division rates, revealing the cell division characteristics of adventitious RAM cells of sweetpotato.
The feedback inhibition pathway involving WOX5/CLE40, similar to that involving WUS/CLV in the shoot apical meristem, was found in the A. thaliana RAM. In this pathway, the homologous polypeptide CLE40 of CLV3 was perceived by the receptor ACR4 on the membrane, and the expression of WOX5 (WUSCHEL-RELATED HOMEOBOX5) was inhibited, thus regulating the homeostasis of distal root meristem [24]. Cells with IbWUS expression were not detected in the RAM of sweetpotato, while the homologous gene IbWOX8 was differentially expressed in clusters 3, 8, and 10. This indicated that IbWOX8 may regulate meristematic cells and play an important role in root cap development. In addition, the expression of IbWOX5 was detected in clusters 8 and 11, but its expression pattern was not cell type-specific, inconsistent with the well-defined organization center. Therefore, our data contained no candidate WOX gene expressed in the Guangshu 87 RAM as the stem cell organizing center and no homolog of WUS in A. thaliana. To further verify these findings, we performed qRT-PCR analysis (Table S12) and found no expression of IbWUS and IbWOX5 transcripts in the root apical meristem. These results suggest that the canonical CLE40 pathway has been lost in the Guangshu 87 RAM.
Pseudotime trajectory analysis of sweetpotato root tip cells
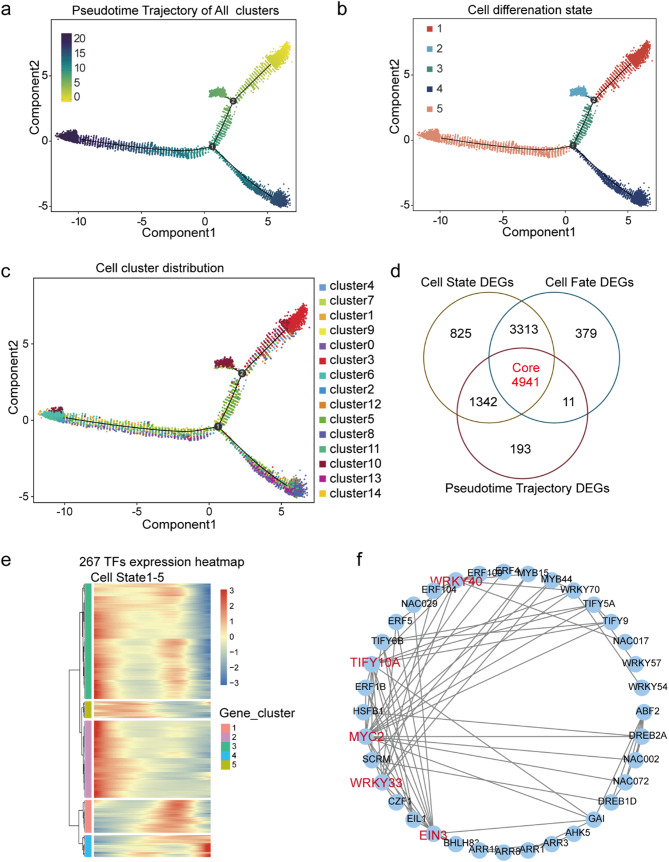
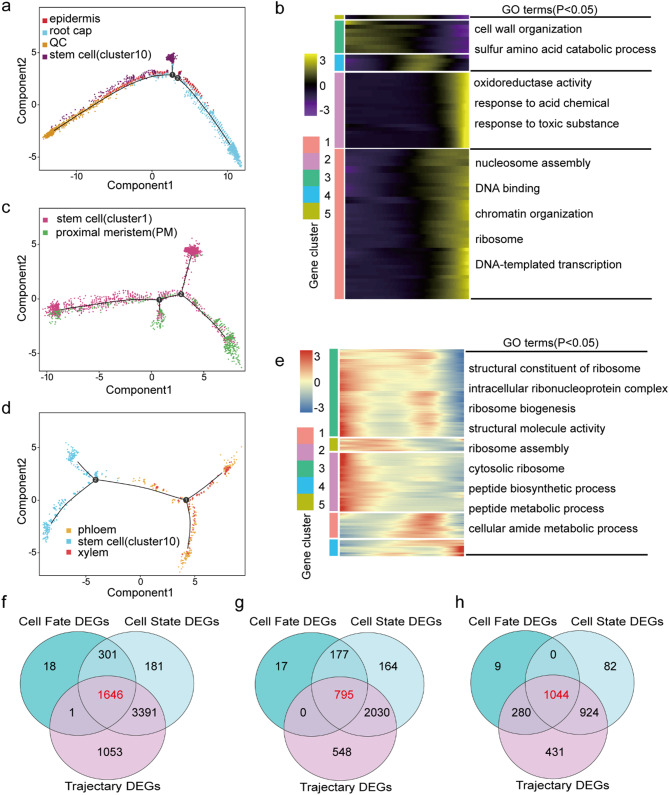
Single-cell RNA-seq allows for simultaneous unbiased analysis of each cell type at each developmental stage [25]. The present study conducted pseudo-time analysis across all cells to determine how central clusters differentiate from the beginning of cell transition into mature cell types outside the cluster cloud (Fig. 4a). There were three main branches observed in the development process, which could be divided into five states (Fig. 4b). Root cap and epidermis cells (clusters 3 and 4) were mainly distributed in state 1, while stem cells (cluster 10) were mainly distributed in state 2, and endodermis cells (cluster 7) were mainly distributed in state 3. Moreover, root cortex (clusters 0, 2, 5, 8, and 12), phloem (cluster11), and xylem (cluster13) cells were mainly distributed in state 4. State 5 mainly contained stem cells (cluster 1), proximal meristem cells (cluster 9), quiescent center cells (cluster 6), and columella cells (cluster 14). Notably, the root tip development trajectory of sweetpotato started from state 5 and gradually differentiated into two branches; one branch included epidermal and root cap cell types, while the other had stele and cortex cell types. In addition, the stem cells distributed in state 2 tended to differentiate into root cap cells (Fig. 4c).
Fig. 4.
Pseudotime trajectory analysis of cell types in the sweetpotato root. (a) The color represents the pseudotime score. (b) The color represents the different states. (c) The color represents the different clusters. (d) A Venn diagram of the three differentially expressed gene (DEG) sets in root cells. (e) Heatmap of 267 core transcription factors (TFs) with cell state expression trend. (f) The interaction network of 36 differentially expressed TFs
Monocle2 was used to screen DEGs in all cells based on the pseudo-time value, differentiation status, and differentiation fate of each cell. A total of 6,487, 10,421, and 11,043 DEGs were obtained with respect to the pseudo-time axis, different differentiation statuses, and different differentiation fates, respectively (Table S7). Furthermore, 4,941 core DEGs were specifically involved in multiple biological pathways in the cell development trajectory (Fig. 4d and Table S7). A total of 267 important transcription factors (TFs) were identified from the 4,941 core DEGs, which provided potential transcriptional dynamics for cell differentiation by participating in the Plant hormone signal transduction, MAPK signaling pathway - plant, Plant-pathogen interaction and Circadian rhythm – plant pathways (Fig. 4e and Table S7). Additionally, interactive network analysis of these 267 TFs showed that among them 36 TFs were in protein–protein interaction networks, including MYC2, WRKY33, WRKY40, TIFY10A, and EIN3, which play important roles in cell development of sweetpotato root tissue (Fig. 4f).
The developmental trajectories of different cell types in sweetpotato root tips
The distal stem cells produce the root cap, and the lateral root cap cells lateral to the QC generate the epidermis; accordingly, the lateral root cap and epidermis share a common initial cell in dicots [26, 27]. Therefore, pseudotime analysis was performed on the quiescent center cells, stem cells (cluster 10), and root cap/epidermal cells (clusters 3 and 4). The development trajectory began from the quiescent center and stem cells (cluster 10), which gradually differentiated into epidermis cells and root cap cells (Fig. 5a). In addition, the branch-dependent genes were subjected to heatmap analysis, which identified five different modules (Fig. 5b and Table S8). The epidermal cell-related genes were found mainly in module 5, which was enriched for roles in cell wall organization and sulfur amino acid catabolic process. The root cap cell-related genes were found mainly in module 2, which was significantly enriched for involvement in oxidoreductase activity, response to acid chemicals, and response to toxic substances (Fig. 5b). Furthermore, 79 TFs were screened by analyzing 1,646 core DEGs, which showed expression trends that drove the differences in stem cells to root cap cell differentiation (Fig. 5f and Table S8).
Fig. 5.
Pseudotime trajectory analysis identified the trajectory map of different cell types. (a) Colored pseudotime trajectory of epidermis, root cap, quiescent center, and stem cells (cluster 10). (b) Pseudotime heatmap of Gene Ontology (GO) analysis of DEGs (P < 0.05). The bar color indicates the relative gene expression level. (c) Colored pseudotime trajectory of stem cell and proximal meristem cells. (d) Colored pseudotime trajectory of xylem, phloem, and stem cells (cluster 10). (e) Pseudotime heatmap of the GO analysis of DEGs (P < 0.05). The bar color indicates the relative gene expression level. f–h. A Venn diagram showing the core DEGs with respect to cell differentiation trajectory, cell differentiation states, and cell fate. (f) root cap/epidermis cell DEGs. (g) Stem cell DEGs. (h) Stele cell DEGs
Pseudotime analysis was also performed on the proximal meristem cells and stem cell niche cells to illustrate their differentiation trajectory, which has previously been analyzed by scRNA-seq of the Arabidopsis root and cotton roots [18, 28]. Proximal meristem cells and stem cell niche cells were separately clustered at the two ends of the pseudotime backbone (Fig. 5c). The developmental trajectory began from the stem cell niche cells, which gradually differentiated into proximal meristem cells. Similarly, five gene modules were identified from the expression heat map. The proximal meristem/stem cell-related genes were found mainly in modules 3 and 1. The genes were significantly enriched for roles in ribosome biogenesis, cellular amide metabolic processes, and ribosome assembly (Fig. 5e). Furthermore, 15 candidate key regulatory factors were identified by analyzing 795 DEGs (Fig. 5g and Table S9). The expression of the transcription factors ARF18, WRKY69, ERF3, and NAC081 increased gradually with differentiation time, which may be the key regulatory factors leading to the differentiation of stem cells into proximal meristem cells (Fig. S7).
The development of sweetpotato rootlets into tuberous roots underwent two growth periods: the primary and the secondary cambium activity periods. During the primary cambium activity period, the primary cambium was formed between the primary xylem and phloem of the rootlets, which divided, thus forming secondary xylem and phloem. The secondary phloem is formed outwards, while the secondary xylem is formed inwards. In the secondary cambium activity period, secondary cambium cells are generated inside the primary xylem vessels, and the secondary cambium is formed inside the secondary xylem vessels and around the metaxylem vessels, where it divides into several parenchyma cells. Since these two cambiums are located in the middle column of the root tip, we reconstructed the developmental trajectory of the middle column cells. The developmental trajectory of stele cells showed that their differentiation began from stem cells (cluster 10) and differentiated into two branches. One of the branches differentiated into phloem cells, and the other differentiated into xylem cells (Fig. 5d). Subsequently, 48 candidate key regulatory factors (Fig. 5h) were screened by analyzing the 1,044 core DEGs across the pseudo-timeline, cell states, and cell fates (Table S10). Among these transcription factors, 13 were unique to the fate of stele cell differentiation (Fig. S8).
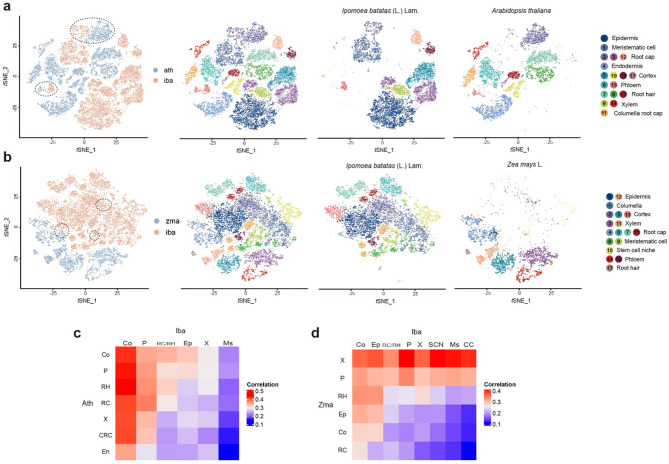
Similarity among sweetpotato, A. thaliana, and maize cell types
Some tissues and organs exhibit similar functions among different angiosperms but with different characteristics. To assess the similarities and differences among angiosperms, we chose the root tip cell types of A. thaliana [29] and maize [14], representing dicots and monocots, respectively. We integrated the sweetpotato scRNA-seq data with the published data sets of A. thaliana and maize root tips, respectively, and re-clustered the cells into 18 cell clusters (Fig. 6a, c). Additionally, t-SNE visualization and Pearson correlation coefficient analysis showed that A. thaliana matched a specific subset of phloem cells and root cap cells of sweetpotato, indicating conservation of some functions of the phloem and root cap in these two species (Fig. 6a). Significant differences in the epidermis, xylem, and meristematic cell populations were observed between sweetpotato and A. thaliana. In addition, sweetpotato cortical cells were highly correlated with all cell populations of A. thaliana, possibly because most genes in A. thaliana had the same expression patterns as those in the sweetpotato cortex (Fig. 6c). Stem cell niches, meristematic cells, and columella cells of sweetpotato had high correlations with stele cells but low correlations with epidermis and cortex cells of maize (Fig. 6b and d). These cell types also partially overlapped in the t-SNE space (Fig. 6b).
Fig. 6.
Comparison of sweetpotato with Arabidopsis thaliana and Zea mays root gene expression patterns. (a) The t-distributed stochastic neighbor embedding (t-SNE) visualization of sweetpotato with Arabidopsis thaliana (each color represents different species and different clusters, respectively). (b) The t-SNE visualization of sweetpotato with Zea mays (each color represents different species and different clusters, respectively). (c) Pearson’s correlation coefficients of gene expression levels between sweetpotato and Arabidopsis thaliana. (d) Pearson’s correlation coefficients of gene expression levels between sweetpotato and Zea mays
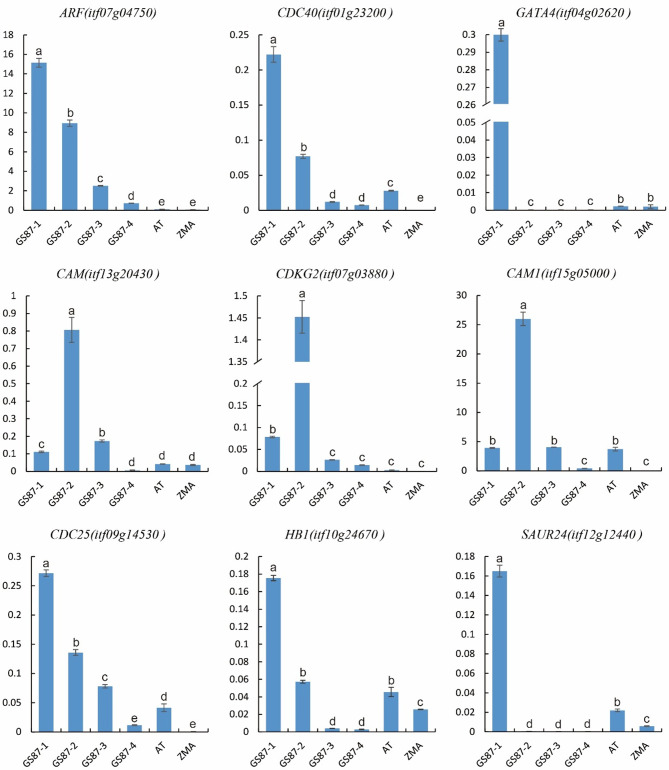
Moreover, we analyzed the DEGs of the three species and found that seven DEGs were highly expressed in sweetpotato (Table S11). These included SAUR24 (auxin-responsive protein SAUR21-like, itf12g12440), CDC40 (Transducin/WD40 repeat-like superfamily protein, itf01g23200), CDKG2 (cyclin-dependent kinase G-2 isoform X1, itf07g03880), HB1 (non-symbiotic hemoglobin 1, itf10g24670), CAM (calmodulin-like protein 8, itf13g20430), GATA4 (GATA transcription factor 4, itf04g02620), ARF (ADP-ribosylation factor A1F, itf07g04750), CAM1 (calmodulin-7-like, itf15g05000), and CDC25 (dual specificity phosphatase Cdc25, itf09g14530). The differential expression of these genes might have caused the differences in cell function and root morphology between sweetpotato, A. thaliana, and maize. The expression of root development-related genes in the adventitious roots of sweetpotato and root tips of A. thaliana and maize was analyzed using RT-qPCR to further verify their expression patterns (Table S12). These genes were highly expressed in sweetpotato roots compared to A. thaliana and maize roots. In addition, expression levels of these genes were analyzed at the different developmental stages of the adventitious roots of sweetpotato. Genes such as GATA4, ARF, CDC40, CDC25, HB1, and SAUR24 were highly expressed in the early stage of adventitious root development, namely the GS87-1 stage, but gradually decreased with root differentiation and development (Fig. 7). Similarly, CAM, CAM1, and CDKG2 were highly expressed in the GS87-2 stage of adventitious roots of sweetpotato, but their expression levels gradually decreased with the continued differentiation and development of roots.
Fig. 7.
Expression of candidate sweetpotato root differentiation and development genes (expression levels of candidate genes in roots of sweetpotato in different developmental states compared with Arabidopsis thaliana and Zea mays roots; GS87-1, < 2 mm; GS87-2, 2–4 mm; GS87-3, 3–5 cm, GS87-4, 9–12 cm, AT, Arabidopsis thaliana; ZMA, Zea mays
Discussion
Sweetpotato tubers are nutritious plant organs with high economic value, and they are critical for understanding the development process of the sweetpotato root system. The root apical meristem (RAM) is located at the root apex, and the stem cell niche [30], including the tissue center and surrounding stem cells, is the core of the RAM [31]. Root stem cells can produce various root cell types through cell division and differentiation. Thus, it is necessary to define the main cell types in specific tissues at the anatomical and molecular levels to understand how stem cells produce different cell types. Therefore, we used scRNA-seq to sequence the transcriptome of the adventitious root tips of sweetpotato and obtained high-level cell heterogeneity information based on the transcriptome data. This study preliminarily constructed the transcriptome map of adventitious root tips of sweetpotato at the single cell level, revealed the cell heterogeneity of sweetpotato root tip, and described the development trajectory of root tip cells. The study also identified the transitional cells involved in the differentiation and development processes of the stem cell niche into differentiated cell types. There was a lower correlation between the root tip cell types of the dicotyledonous plants sweetpotato and Arabidopsis thaliana and the monocotyledonous plant maize. Thus, the correlation between sweetpotato and A. thaliana was higher than that between either of them and maize. In addition, some root differentiation and development-related genes were screened, highlighting that GATA4 may regulate the early development of sweetpotato root tips.
The technical difficulty for high-throughput scRNA-seq of sweetpotato root tip tissues is preparing qualified single-cell suspension, which provides crucial insights into cell development at specific locations. This study established a high-quality protoplast preparation system for sweetpotato root tips, providing the potential for genetic transformation, gene editing, and transient gene expression in protoplasts. Self-incompatibility and cross-incompatibility are common in sweetpotatoes, but can be overcome by protoplast fusion cell hybridization, which provides important genetic resources for sweetpotato breeding [32]. High-throughput scRNA-seq was used to analyze the complex adventitious roots of sweetpotato, and the transcriptome map of the main root tip cell types was generated, providing the temporal and spatial expression dynamics of cell differentiation. A high-resolution map of more than 10,000 root cells captured all major cell types, including rare cell types, such as the resting center and meristem cells of the differentiation stage. Using this map, we were able to accurately predict the spatio-temporal patterns of gene expression in the roots and provide valuable insights for gene expression and functional studies at the single-cell level. Several studies [18, 33–35] have shown that scRNA-seq can reveal the heterogeneity of cell populations and help to discover the intermediate states during biological differentiation and development. The present study found that the transcriptome of stem cells (cluster 1) expressed different cell markers, which could be grouped into six subclusters. The DEGs in the six subclusters corresponded to their related differentiated cells, but each subcluster expressed more than one cell type marker gene, indicating that early cells have different cell fates and that cell fate exhibits some instability. When performing the pseudo-time analysis on different cell types, these subclusters were merged into each group of cell types. The results showed that these stem cell subclusters were located at the initial position of the development trajectory of each cell type, further indicating that there were transitional cell types involved in the differentiation of root tip stem cells of sweetpotato into mature cell types.
A previous association analysis showed that the cell-type transcription profiles of A. thaliana and rice were very different [36]. Only the root hair cell type had a high correlation between rice and A. thaliana, while the correlation between cortex cells was more moderate, and those between other cell types were quite different. These species-specific characteristics emphasize the importance of analyzing tissues of different species. Similarly, this study compared the cell types of sweetpotato, A. thaliana, and maize and found that the cell populations of sweetpotato were significantly different from those of A. thaliana and maize. These cell populations had less overlap in t-SNE space and exhibited several genes with different expression patterns. GATA transcription factors are a group of DNA binding proteins widely distributed among eukaryotes, and they contain class IV zinc finger DNA binding motifs. In plants, the DNA motif of GATA is involved in light-dependent and nitrate-dependent transcriptional regulation. For example, GATA4 is involved in biological processes such as light stimulation responses, cell differentiation, and positive regulation of RNA polymerase II promoter transcription in A. thaliana [37]. In sweetpotatoes, the GATA family genes may be regulatory factors associated with the development of tuberous root enlargement. Thus, this study analyzed the expression of GATA4, which may be involved in regulating the early development of sweetpotato root tips. Consistent with the traditional transcriptome data corresponding to sweetpotato tuberous root enlargement, the GATA4 gene was highly expressed in the early stage of root development but gradually decreased with the development and enlargement of roots [38]. In addition, the expression of GATA4 in the stem cell niche, meristematic cell, and stele was higher than that of other cell types, probably because these cells are involved in root cambium activity which drives sweetpotato root development.
This study utilized scRNA-seq to quantify gene expression in various cell types, including single cells, providing important insights into cell development at specific stages and elucidating the complex gene regulatory networks controlling sweetpotato root development. Since the root development-related gene GATA4 screened in this study had not been functionally verified before, its regulatory mechanism could not be understood at a deeper level. Fluorescence-activated cell sorting technology can be used to enrich specific cell types and verify gene function, thus enabling the detection of function and regulatory networks involved in different cell types. At present, methods such as glass microcapillary [39], microdissection [16], or fluorescence-activated cell sorting (FACS) [40] have been successfully used to obtain target plant cell types and a small amount of single-cell transcriptome data. However, advances in plant cell isolation and scRNA-seq technology have enabled the application of single-cell transcriptomics to more plant cell tissues and species. Furthermore, combining various technologies with scRNA-seq technology could further deepen the understanding of plants at the molecular, subcellular, cellular, and tissue levels, enabling the generation of high-resolution plant cell molecular spatiotemporal maps, thus promoting the study of basic plant science problems. Such technologies include assay for targeting accessible-chromatin with high-throughput sequencing (ATAC-seq) [41], high-through chromosome conformation capture (Hi-C) [42], spatial transcriptomics-seq (ST-seq) [43], single-cell genome analysis [44, 45], epigenome analysis [46], and protein analysis [47] techniques. When coupled with scRNA-seq technology, these techniques can be used to investigate plants at the molecular, subcellular, cellular and tissue levels, making it possible to infer high-resolution plant cell molecular spatiotemporal maps, which can promote the study of basic problems in plant science.
Method details
Plant materials and growth conditions
Sweetpotato (Ipomoea batatas [L.] Lam., cv. ‘Guangshu 87’) plants were grown in the facilities of the Zhongluotan Academy of Agricultural Sciences, Guangdong Province, China (113.440470°N, 23.388464°E). ‘Guangshu 87’ is a high-quality and high-yield sweetpotato cultivar bred by the Guangdong Academy of Agricultural Sciences, and it ranks among the top five cultivated sweetpotato varieties in terms of planting area in China [48]. Branch cuttings (20 cm) of sweetpotato seedlings were soaked in sterile water for 5 days (28℃ with 12-h light/12-h dark cycles, with a light intensity of 13,000 lx) .
Isolation of protoplasts
Thirty tips (5 mm) of adventitious roots were cut into small pieces of about 1 mm and incubated in 10 ml of an enzyme solution (1.5% cellulase RS, 1.5% macerozyme R10, 1.5% hemicellulase, 0.4 M mannitol, 20 mM MES, and 0.1% bovine serum albumin [BSA]) in the dark. The cell solution was sequentially filtered through 70-µm (FALCON, Cat # 352350) and 40-µm (FALCON, Cat # 352340) cell strainers after digestion at 28 °C for 3 h at 80 rpm on an orbital shaker. Protoplasts were collected after being softly washed twice (centrifuged for 10 min at 300 × g) and were re-suspended in 0.4 M mannitol solution. Cell activity was detected via trypan blue staining, and the number of protoplasts was measured with a hemocytometer.
scRNA-Seq library construction and sequencing
The root single-cell suspensions were loaded onto a Chromium Single Cell Instrument (10× Genomics, Pleasanton, CA) to generate single-cell gel-bead emulsions (GEMs). scRNA-seq libraries were constructed using the Chromium Single Cell 3′ GEM Gel Bead and Library Kit v3 (10x Genomics, Cat No./ID: PN-1000076 and 1000078), as per the user guide (Chromium Single Cell 3ʹ Reagent Kits v3, CG000183 Rev A). The qualitative analyses of DNA libraries were performed with an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA), and the libraries were sequenced with an Illumina Nova-Seq sequencer 6000 (Illumina, San Diego, CA, USA) by Genergy Biotechnology (Shanghai, China) using two 150-bp paired-end kits. The raw scRNA-seq datasets comprised 28 bp Read1, 150 bp Read2, and 8 bp i7 index reads.
RT-qPCR
Arabidopsis thaliana(Col-0 wild-type) and maize inbred line B73 seeds were planted in Murashige and Skoog medium (22℃ with 16-h light/8-h dark cycles and a light intensity of 11,000 lx). Whole roots were separated from shoots and collected in bulk for RNA extraction using the RNAprep pure plant kit (Tiangen, Beijing). Complementary DNA (cDNA) synthesis and genomic DNA (gDNA) removal steps were performed using FastKing gDNA dispelling RT superMix (Tiangen, Beijing). Each obtained cDNA library served as a template for gene expression level analysis via real-time quantitative PCR on the ABI step one plus system. The relative expression level was calculated by the delta-delta Ct method (2−ΔΔCt). The primers for the target genes were designed using primer 5 software (Table S12), and the β-actin gene was used as the internal control.
Pre-processing of raw scRNA-seq data
The raw files were analyzed by Cell Ranger 3.1.0 (10× Genomics), which pre-processed the raw scRNA-seq data by aligning reads and generating gene-cell matrices. The sweetpotato genome (I. trifida v3 JBrowse) was downloaded from the sweetpotato genomics resource website (http://sweetpotato.plantbiology.msu.edu). The reference genome was generated by implementing ‘cellranger mkref’ with ‘‘–genome, –fasta and –genes’’ arguments in Cell Ranger. However, ‘cellranger count’ with ‘‘–id, –fastqs, –sample and –transcriptome’’ arguments were used to generate single-cell gene counts. More than 89.3% of reads in all the samples were successfully aligned to the I. trifida reference genome (v3 JBrowse) by the aligner STAR (Spliced Transcripts Alignment to a Reference, v. 2.6.0a). The gene-cell matrices were used as raw data for further analyses, where each row was a feature (gene expression level), and each column was a valid cell barcode.
Identification of highly variable genes
The gene-cell matrices of 13,966 cells were loaded into the Seurat package (v. 3.0.0.9) for analysis [49]. To remove the low-quality cells and likely multiple captures, we removed the cells with unique gene counts of > 5000 or < 200 and those with more than 5% mitochondrial sequence. Doublet GEMs were also removed using the tool DoubletFinder (v2.0.3) [50], which generates artificial doublets using the principal component (PC) distance matrix to find each cell’s proportion of artificial k-nearest neighbors (pANN) and ranks them according to the expected number of doublets. After filtering, 32,301 genes from 12,172 cells were used for further analysis. The scaled data were first normalized by ‘LogNormalize’ and used to identify the highly variable genes using ‘FindVariableFeatures’ with “vst” parameters in Seurat.
Dimension reduction and cell clustering
Principal component analysis (PCA) [51] was performed to reduce the dimensionality of the log-transformed gene-barcode matrices of the most variable genes. Cells were clustered via a graph-based approach and visualized in two dimensions. The scRNA-seq datasets were explored using the t-SNE tool [52], which enabled the assignment of cells with similar local neighborhoods in high-dimensional and low-dimensional spaces. We also used t-SNE to identify differentially expressed genes (DEGs) between clusters. A likelihood-ratio test was used to identify differential expression when comparing a single cluster to all other cells. We also performed uniform manifold approximation and projection (UMAP) analysis to confirm the cell clusters identified by t-SNE. Specifically, the total number of PCs (npcs), dimensions (dims), the number of neighboring points (n.neighbors), and resolution and minimum distance (min.dist) parameters were tuned to represent the best approximation of the underlying topology of each subcluster.
Screening and identification of cluster marker genes
Cluster identification depends on marker genes. Genotyping multiple marker genes is necessary to determine their cell types [53]. DEGs or marker genes can be identified by differential expression analysis of clusters, which can also reflect the specificity of clusters. The FindAllMarkers function of Seurat was used to identify marker genes for each cluster. The known marker genes of the cell types in sample tissues were screened based on the published literature and databases, and the upregulated/down-regulated genes in each cluster were matched to identify the cell types of each cell subgroup.
Pseudotime analysis
The pseudo-time analysis [54] of cell differentiation and cell fate was performed using the Monocle R package (Version 3.0). For pseudo-time trajectory analysis, the raw count in the Seurat object was first converted into a CellDataSet object with the importCDS (object, import_all = F) function in Monocle. We then normalized the differences in mRNA recovered across cells using the ‘estimateSizeFactors’ function and conducted the differential expression analysis using ‘estimateDispersions’ function. First, the variance in the expression of each gene across cells was calculated by the ‘dispersionTable’ function. Variable genes were then chosen based on average expression level to define developmental progress. Second, we used the ‘differentialGeneTest’ function of the Monocle 2 package to order genes (qval < 0.01) that were likely to be informative for ordering cells along the pseudo-time trajectory. The ordered genes were then marked with the ‘setOrderingFilter’ function, and we reduced the dimensionality of the data to two (set myoblast, method = ‘DDRTree’). With the expression data in a lower dimensional space, the state transition of single cells was described by the ‘orderCells’ function. The cell trajectory was plotted by ‘plot_cell_trajectory’ in Monocle. The ‘orderCells’ function was implemented again, setting the ‘‘root_state’’ argument to specify an a priori beginning of the trajectory. The branch point was selected to analyze branches in differentiation trajectories. The pseudo-time-dependent or branch-dependent genes were analyzed using branched expression analysis modeling (BEAM). The genes that were significantly branch-dependent were visualized by the ‘plot_genes_branched_heatmap’ function.
Association analysis of multispecies scRNA-seq data
The data were obtained from A. thaliana single-cell transcriptome sequences (GSE123013) and maize single-cell transcriptome sequences (GSE183171) in the Gene Expression Omnibus (GEO) database. To compare the expression of genes between each group of species, we used OrthoMCL to define one-to-one homologous genes between each group (Supplementary Table 11). We used the homologous genes to arrange and cluster the cells by dimensionality reduction.
Differentially expressed gene functional analysis
Differentially expressed genes (DEGs) were identified using the Seurat package [22]. Thresholds of P < 0.01 and |log2(fold change in expression)| > 0.36 were set as the thresholds for significantly differential expression. Gene Ontology (GO) annotation enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses of DEGs were performed using R based on the hypergeometric distribution.
RNA in situ hybridization
RNA in situ hybridization was performed as previously described [55]. In brief, the specific targeting probes were designed by spatial FISH Ltd. (Table S12). Sweetpotato root tips were fixed with 4% paraformaldehyde and then covered with a reaction chamber to perform the following reactions. After dehydration and denaturation of root tips with methanol, the hybridization buffer with specific targeting probes was added to the chamber for incubation at 37℃ overnight. Then, root tips were washed three times with PBST, followed by ligation of targeting probes in ligation solution at 25℃ for 3 h. Next, root tips were washed three times with PBST and subjected to rolling circle amplification by Phi29 DNA polymerase at 30℃ overnight. Subsequently, the fluorescent detection probes in hybridization buffer were applied to root tips. Finally, root tips were dehydrated with an ethanol series and mounted with mounting medium. After capturing images with a Leica THUNDER Imaging System (Leica, Wetzlar, Germany), 20× (NA = 0.80) signal dots were decoded to interpret RNA spatial position information.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Not applicable.
Author contributions
Lifei Huang, Shuyan Xie, Xiawei Ding, Nan Zhao conceived and designed the experiments. Nan Zhao, Hongda Zou, Shuyan Xie, Xiawei Ding performed the experiments. Nan Zhao, Xiawei Ding, Hao Liu analyzed the data. Nan Zhao, Xiawei Ding wrote the paper. Lifei Huang, Xuelian Liang eviewed of the manuscript.
Funding
This work was supported by grants from the earmarked fund for CARS-10-Sweet Potato, the Key R&D Program of the Key-Area Research and Development Program of Guangdong Province (no. 2020B020219001), and the Sweetpotato Potato Innovation Team of Modern Agricultural Industry Technology System in Guangdong Province (2023KJ111).
Data availability
All relevant data are provided in the article and its supplementary file. Raw sequencing data have been uploaded to China National Center for Bioinformation (CNCB) under Bio-project accession PRJCA020029(https://ngdc.cncb.ac.cn/).
Declarations
Ethics approval and consent to participate
Sweetpotato (Ipomoea batatas [L.] Lam., cv. ‘Guangshu 87’) plants were grown at the Zhongluotan Academy of Agricultural Sciences, Guangdong Province, China (113.440470°N, 23.388464°E). The research site is a public institution. Data collection was approved by the management of the facility.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Nan Zhao and Xiawei Ding co-first author.
Contributor Information
Xue lian Liang, Email: liangxuelian2005@sina.com.
Lifei Huang, Email: hlf157@163.com.
References
- 1.Kyndt T, Quispe D, Zhai H, et al., et al. The genome of cultivated sweet potato contains Agrobacterium T-DNAs with expressed genes: an example of a naturally transgenic food crop. Proc Natl Acad Sci. 2015;112(18):5844–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Isobe S, Shirasawa K, Hirakawa H. Challenges to genome sequence dissection in sweetpotato. Breed Sci. 2017;67(1):35–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zirui Z, Guiming L, Yuhong L. Development Situation of Sweet Potato Harvester at Home and Abroad.Agricultural Engineering, 2015.
- 4.Ren Z, He S, Zhao N, et al. A sucrose non-fermenting‐1‐related protein kinase‐1 gene, IbSnRK1, improves starch content, composition, granule size, degree of crystallinity and gelatinization in transgenic sweet potato. Plant Biotechnol J. 2019;17(1):21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fan W, Wang H, Wu Y, et al. H+-pyrophosphatase ib VP 1 promotes efficient iron use in sweet potato [Ipomoea batatas (L.) Lam]. Plant Biotechnol J. 2017;15(6):698–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ishida H, Suzuno H, Sugiyama N, Innami S, Tadokoro T, Maekawa A, et al. Nutritive evaluation on chemical components of leaves, stalks and stems of sweet potatoes (Ipomoea batatas poir). Food Chem. 2000;68(3):359–67. [Google Scholar]
- 7.Park SC, Kim SH, Park S, Lee HU, Lee JS, Park WS, et al. Enhanced accumulation of carotenoids in sweetpotato plants overexpressing IbOr-Ins gene in purple-fleshed sweetpotato cultivar. Plant Physiol Biochem. 2015;86:82–90. [DOI] [PubMed] [Google Scholar]
- 8.Luo CL, Zhou Q, Yang ZW, Wang RD, Zhang JL. Evaluation of structure and bioprotective activity of key high molecular weight acylated anthocyanin compounds isolated from the purple sweet potato (Ipomoea batatas L. Cultivar Eshu 8). Food Chem. 2018;241:23–31. [DOI] [PubMed] [Google Scholar]
- 9.Villordon AQ, Ginzberg I, Firon N. Root architecture and root and tuber crop productivity. Trends Plant Sci. 2014;19(7):419–25. [DOI] [PubMed] [Google Scholar]
- 10.Firon N, LaBonte D, Villordon A, et al. Transcriptional profiling of sweetpotato (Ipomoea batatas) roots indicates down-regulation of lignin biosynthesis and up-regulation of starch biosynthesis at an early stage of storage root formation. BMC Genomics. 2013;14:1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kolodziejczyk AA, Kim JK, Svensson V, et al. The technology and biology of single-cell RNA sequencing. Mol Cell. 2015;58(4):610–20. [DOI] [PubMed] [Google Scholar]
- 12.Zhang TQ, Chen Y, Liu Y, et al. Single-cell transcriptome atlas and chromatin accessibility landscape reveal differentiation trajectories in the rice root. Nat Commun. 2021;12(1):2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelms B, Walbot V. Defining the developmental program leading to meiosis in maize. Science. 2019;364(6435):52–6. [DOI] [PubMed] [Google Scholar]
- 14.Li X, Zhang X, Gao S, et al. Single-cell RNA sequencing reveals the landscape of maize root tips and assists in identification of cell type-specific nitrate-response genes. Crop J. 2022;10(6):1589–600. [Google Scholar]
- 15.Shahan R, Hsu CW, Nolan TM, et al. A single-cell Arabidopsis root atlas reveals developmental trajectories in wild-type and cell identity mutants. Dev Cell. 2022;57(4):543–60. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Omary M, Gil-Yarom N, Yahav C, et al. A conserved superlocus regulates above-and belowground root initiation. Science. 2022;375(6584):eabf4368. [DOI] [PubMed] [Google Scholar]
- 17.Dorrity MW, Alexandre CM, Hamm MO, et al. The regulatory landscape of Arabidopsis thaliana roots at single-cell resolution. Nat Commun. 2021;12(1):3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang TQ, Xu ZG, Shang GD, et al. A single-cell RNA sequencing profiles the developmental landscape of Arabidopsis root. Mol Plant. 2019;12(5):648–60. [DOI] [PubMed] [Google Scholar]
- 19.Gala HP, Lanctot A, Jean-Baptiste K, et al. A single-cell view of the transcriptome during lateral root initiation in Arabidopsis thaliana. Plant Cell. 2021;33(7):2197–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jean-Baptiste K, McFaline-Figueroa JL, Alexandre CM, et al. Dynamics of gene expression in single root cells of Arabidopsis thaliana. Plant Cell. 2019;31(5):993–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farmer A, Thibivilliers S, Ryu KH, et al. Single-nucleus RNA and ATAC sequencing reveals the impact of chromatin accessibility on gene expression in Arabidopsis roots at the single-cell level. Mol Plant. 2021;14(3):372–83. [DOI] [PubMed] [Google Scholar]
- 22.Ryu KH, Huang L, Kang HM, et al. Single-cell RNA sequencing resolves molecular relationships among individual plant cells. Plant Physiol. 2019;179(4):1444–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song J, Fan B, Shao X, et al. Single-cell transcriptome sequencing atlas of cassava tuberous root. Front Plant Sci. 2023;13:1053669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stahl Y, Wink RH, Ingram GC, et al. A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr Biol. 2009;19(11):909–14. [DOI] [PubMed] [Google Scholar]
- 25.Satija R, Farrell JA, Gennert D, et al. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33(5):495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirschner GK, Stahl Y, Von Korff M, et al. Unique and conserved features of the barley root meristem. Front Plant Sci. 2017;8:276652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L, Chu H, Li Z, et al. Origin and development of the root cap in rice. Plant Physiol. 2014;166(2):603–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li P, Liu Q, Wei Y et al. Transcriptional landscape of cotton roots in response to salt stress at single-cell resolution. Plant Commun, 2024, 5(2).
- 29.Denyer T, Ma X, Klesen S, et al. Spatiotemporal developmental trajectories in the Arabidopsis root revealed using high-throughput single-cell RNA sequencing. Dev Cell. 2019;48(6):840–52. e5. [DOI] [PubMed] [Google Scholar]
- 30.Benfey PN. Defining the path from stem cells to differentiated tissue. Curr Top Dev Biol. 2016;116:35–43. [DOI] [PubMed] [Google Scholar]
- 31.Aichinger E, Kornet N, Friedrich T, et al. Plant stem cell niches. Annu Rev Plant Biol. 2012;63:615–36. [DOI] [PubMed] [Google Scholar]
- 32.Zhang BY, Liu QC, Zhai H et al. Production of fertile interspecific somatic hybrid plants between sweetpotato and its wild relative, Ipomoea lacunosa//I International Conference on Sweetpotato. Food and Health for the Future 583. 2001: 81–85.
- 33.Li H, Dai X, Huang X, et al. Single-cell RNA sequencing reveals a high‐resolution cell atlas of xylem in Populus. J Integr Plant Biol. 2021;63(11):1906–21. [DOI] [PubMed] [Google Scholar]
- 34.Serrano-Ron L, Perez-Garcia P, Sanchez-Corrionero A, et al. Reconstruction of lateral root formation through single-cell RNA sequencing reveals order of tissue initiation. Mol Plant. 2021;14(8):1362–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim JY, Symeonidi E, Pang TY, et al. Distinct identities of leaf phloem cells revealed by single cell transcriptomics. Plant Cell. 2021;33(3):511–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Q, Liang Z, Feng D, et al. Transcriptional landscape of rice roots at the single-cell resolution. Mol Plant. 2021;14(3):384–94. [DOI] [PubMed] [Google Scholar]
- 37.Teakle GR, Manfield IW, Graham JF, et al. Arabidopsis thaliana GATA factors: organisation, expression and DNA-binding characteristics. Plant Mol Biol. 2002;50:43–56. [DOI] [PubMed] [Google Scholar]
- 38.Cai Z, Cai Z, Huang J, et al. Transcriptomic analysis of tuberous root in two sweet potato varieties reveals the important genes and regulatory pathways in tuberous root development. BMC Genomics. 2022;23(1):473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu H, Hu D, Du P, et al. Single-cell RNA‐seq describes the transcriptome landscape and identifies critical transcription factors in the leaf blade of the allotetraploid peanut (Arachis hypogaea L). Plant Biotechnol J. 2021;19(11):2261–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun G, **a M, Li J, et al. The maize single-nucleus transcriptome comprehensively describes signaling networks governing movement and development of grass stomata. Plant Cell. 2022;34(5):1890–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buenrostro JD, Giresi PG, Zaba LC, et al. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10(12):1213–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dong Q, Li N, Li X, et al. Genome-wide Hi‐C analysis reveals extensive hierarchical chromatin interactions in rice. Plant J. 2018;94(6):1141–56. [DOI] [PubMed] [Google Scholar]
- 43.Liao J, Lu X, Shao X, et al. Uncovering an organ’s molecular architecture at single-cell resolution by spatially resolved transcriptomics. Trends Biotechnol. 2021;39(1):43–58. [DOI] [PubMed] [Google Scholar]
- 44.Lu S, Zong C, Fan W, et al. Probing meiotic recombination and aneuploidy of single sperm cells by whole-genome sequencing. Science. 2012;338(6114):1627–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gawad C, Koh W, Quake SR. Single-cell genome sequencing: current state of the science. Nat Rev Genet. 2016;17(3):175–88. [DOI] [PubMed] [Google Scholar]
- 46.Kim TH, Ren B. Genome-wide analysis of protein-DNA interactions. Annu Rev Genomics Hum Genet. 2006;7:81–102. [DOI] [PubMed] [Google Scholar]
- 47.**e H, Ding X. The intriguing landscape of single-cell protein analysis. Adv Sci. 2022;9(12):2105932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu J, Wang X, Qin J, et al. Investigation Report on the development of Sweet Potato planting industry in my country (2017)——Based on the analysis of fixed Observation Point Data of Industrial Economy in National Sweet Potato Industrial Technology System. Jiangsu Agricultural Sci. 2018;46(23):6. [Google Scholar]
- 49.Butler A, Hoffman P, Smibert P, et al. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McGinnis CS, Murrow LM, Gartner ZJ. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 2019;8:329–37. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chung NC, Storey JD. Statistical significance of variables driving systematic variation in high-dimensional data. Bioinformatics. 2014;31(4):545–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van der Maaten L, Hinton G. Visualizing data using t-SNE. J Mach Learn Res, 2008, 9(11).
- 53.Aran D, Looney AP, Liu L, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol. 2019;20(2):163–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qiu X, Mao Q, Tang Y, et al. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods. 2017;14(10):979–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shulse CN, Cole BJ, Ciobanu D, et al. High-throughput single-cell transcriptome profiling of plant cell types. Cell Rep. 2019;27(7):2241–7. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are provided in the article and its supplementary file. Raw sequencing data have been uploaded to China National Center for Bioinformation (CNCB) under Bio-project accession PRJCA020029(https://ngdc.cncb.ac.cn/).