Abstract
Infection of microglia and other cells of the macrophage/monocyte lineage in the central nervous system (CNS) by human immunodeficiency virus type I (HIV-1) underlies the development of giant cell encephalitis (GCE). It is currently unknown whether GCE depends on the emergence of virus populations specifically adapted to replicate in cells of the monocyte/macrophage lineage and whether this also leads to the specific targeting of macrophages in other nonlymphoid tissues. Autopsy samples from lymph node, brain (frontal region), lung, and full-thickness colon sections were obtained from nine study subjects with GCE and from nine without. The two groups showed no significant differences in CD4 counts, disease progression, or treatment history before death. Genetic relatedness between variants recovered from lymph node and nonlymphoid tissues was assessed by sequence comparison of V3 and p17gag regions using a newly developed method that scores the sample composition at successive nodes in a neighbor-joining tree. The association index enabled objective, numerical comparisons on the degree of tissue compartmentalization to be made. High proviral loads and p24 antigen expression in the brain were confined to the nine individuals with GCE. GCE was also associated with significantly higher proviral loads in colon samples (median of the GCE+ group: 1,010 copies/106 cells; median of GCE− group, 10/106 cells; P = 0.006). In contrast, there were no significant differences in proviral load between the GCE+ and GCE− groups in lymph node or lung samples, where HIV infection was manifested predominantly by infiltrates of lymphoid cells. V3 sequences from brain samples of individuals with GCE showed the greatest compartmentalization from those of lymph node, although samples from other tissues, particularly the colon, frequently contained variants phylogenetically related to those found in brain. The existence of shared, distinct populations of HIV specifically distributed in cells of the monocyte/macrophage lineage was further indicated by immunocytochemical detection of CD68+, multinucleated giant cells expressing p24 antigen in samples of lung and colon in two individuals with GCE. This study provides the basis for future investigation of possible phenotypic similarities that underline the shared distributions of HIV variants infecting microglia and tissue macrophages outside the CNS.
The cellular tropism of human immunodeficiency virus type 1 (HIV-1) is governed at a variety of entry and postentry steps, including the attachment, fusion, and entry of HIV into the cell, reverse transcription, integration, and gene expression (16, 31, 39). Differences between CD4 lymphocytes, the principal targets of HIV-1 in vivo, and other potential cellular targets for HIV-1, such as macrophages and microglia in the brain, exist at many of these levels, particularly in the expression of CD4 and chemokine coreceptors required for virus entry. The ability of HIV-1 to target and productively infect these different cell types in vivo may therefore depend on strain-specific differences of HIV-1 or on the evolution of adaptive differences during the course of infection.
Primary and laboratory isolates show a wide range of cellular tropisms (including ability to grow in transformed T-cell lines, primary cultures of monocyte-derived macrophages), cytopathology (syncytium induction) and coreceptor usage (CXCR4, CCR5, CCR3). These differences have in the past been linked to variability in the rate of disease progression in HIV-infected individuals, in whom the emergence of CXCR4-using, nonmacrophage tropic isolates of HIV-1 is accompanied by a more rapid decline in CD4 lymphocyte numbers and the onset of AIDS-related disease (3, 5, 10, 15, 20, 26, 29, 30, 47, 70, 78). Much less is understood about the existence of differential cellular tropism of HIV variants infecting different anatomical locations and tissue types in vivo, and it is not known whether adaptive changes are responsible for direct virus-mediated outcomes of infection, such as the invasion of the central nervous system (CNS) and the subsequent development of giant cell encephalitis (GCE). It is also unknown whether the ability of HIV-1 to productively infect nonlymphoid tissues, such as the brain, is dependent on the same adaptive changes that underline the CCR5-using, macrophage-tropic phenotype characterized in in vitro studies.
In this study we have used a combination of immunocytochemical detection of p24 antigen (75), PCR for quantitation of proviral DNA sequences (69), and genetic characterization to examine the cell types and virological characteristics of HIV infecting samples of lung and colon collected at autopsy from a large number of HIV-seropositive individuals. We examined the genetic relationships between HIV variants infecting different cell types in these tissues and those recovered from lymph nodes, where lymphocytes are the predominant cell type infected, and also those present in the brains of individuals with GCE, where the principal target cells are cells of the monocyte/macrophage lineage (infiltrating macrophages and microglia). Our findings provide evidence for the existence of genetically distinct populations of HIV targeting cells of the monocyte/macrophage lineage with shared distributions in the CNS and tissue macrophages in the lung and gastrointestinal (GI) tract. Biological characterization of variants recovered from lymphoid and nonlymphoid cells identified in this study will allow the phenotypic differences underlying the in vivo differences in cellular tropism to be identified.
MATERIALS AND METHODS
Study subjects.
All of the tissue samples used in this study were held in the Brain and Tissue Bank of Edinburgh (Western General Hospital, Edinburgh, United Kingdom). Tissue samples, including brain, lung, colon, and lymph node, were obtained from 18 autopsies (NA425, NA021, NA020, NA017, NA272, NA371, NA025, NA446, NA199, NA284, NA038, NA116, NA369, NA270, NA308, NA246, NA420, NA118) from the cohort of HIV-infected individuals in Edinburgh. All study subjects died of complications associated with HIV infection, including opportunistic infections or neoplasms. Their ages ranged from 32 to 49, and they had histories of HIV-1 infections lasting 4 to 13 years before death. Other clinical and background information for the study group is listed in Table 1. Previously determined proviral loads, V3 sequences, and immunocytochemical detection of p24 antigen from three study subjects, NA246, NA118, and NA420 (corresponding to p4, p5, and p6 [ref. 24]), were incorporated into the larger series analyzed in this study.
TABLE 1.
Proviral loads in lymphoid and nonlymphoid autopsy samples from study subjects
| Subject | HIV risk factora | No. of lymphocytes (counts/μl) at time of death
|
GCE diagnosisb | Duration of infection
|
Interval between death and autopsy (h) | V3 predictionc | ||
|---|---|---|---|---|---|---|---|---|
| CD4 | CD8 | HIV (yr) | AIDS (mo) | |||||
| NA371 | M | 0.5 | 56 | N | 4 | 30 | 11 | Y |
| NA425 | I | 1 | 314 | N | 13 | 14 | 29 | Y |
| NA116 | I | 4 | 60 | N | NKd | 18 | 39 | Y |
| NA20 | M | 8 | 388 | N | 13 | 48 | 21 | Y |
| NA17 | I | 17 | 109 | N | 12 | 19 | 59 | N |
| NA308 | I | 23 | 212 | N | 13 | 8 | 59 | N |
| NA199 | I | 28 | 164 | N | 13 | 6 | 54 | N |
| NA272 | I | 105 | 411 | N | 9 | 10 | 21 | N |
| NA270 | I | 246 | 510 | N | 4 | 56 | N | |
| Median GCE− | 17 | 212 | 12 | 16 | 39 | |||
| NA38 | I | 3 | 210 | P | 10 | 7 | 65 | N |
| NA420 | I | 7 | P | NK | NK | 14 | N | |
| NA246 | I | 8 | P | 5 | 24 | 57 | N | |
| NA284 | I | 8 | 110 | P | NK | 21 | 30 | N |
| NA369 | I | 23 | 440 | P | 2 | 22 | 44 | Y |
| NA118 | I | 56 | P | 9 | NK | 70 | N | |
| NA21 | I | 87 | 1,779 | P | 13 | 48 | 63 | N |
| NA446 | I | 137 | 2,611 | P | 14 | 8 | 6 | N |
| NA25 | H | 297 | 232 | P | 4 | 26 | N | |
| Median GCE+ | 23 | 336 | 9 | 22 | 44 | |||
During the autopsies, which were carried out within 3 days of death (Table 1), approximately 1- to 2-cm3 samples of the left frontal region of brain, colon, lung, and lymph node from each individual were dissected and stored at −70°C for PCR and sequence analysis. The remaining tissues were fixed in 10% formalin for histopathological examination. The intervals between death and autopsy were comparable for the GCE+ and GCE− groups (Table 1), indicating that this variable was not a compounding factor in the differences observed between the two groups in proviral loads found in autopsy samples of brain and colon.
DNA extraction and provirus quantitation.
DNA was extracted from frozen brain, lymph node, colon, and lung tissues as previously described (69). Total DNA concentration was estimated by spectrophotometry at 260 nm. Proviral load was determined by previously described limiting-dilution nested PCR (69) using the previously described p17gag primer set (85). Twenty-four replicates at last positive dilution were used to indicate the minimum proviral load in the sample, assuming a Poisson distribution for each sample by the formula −ln (1 − p)/d, where p is the proportion of positive samples and d is the dilution (69). Viral load was expressed as copies per million cells, based on the DNA composition of human diploid cells of 6.6 pg of DNA.
Immunohistochemical examination.
Approximately 2 weeks after formalin fixation, autopsy tissues were processed through a routine 41-h program in the Vacuum Infiltration Processor (Tissue Tek), followed by paraffin wax embedding using a Tissue Tek embedding console. Five-micrometer sections of formalin-fixed paraffin-embedded tissues from all of the study subjects were examined by immunohistological staining with anti-HIV-1 p24 antibody (DuPont) and double labeling with several cell markers, PGM1 (DAKO) for microglial/macrophage cells, CD3 and CD8 (DAKO) for T cells, GFAP (DAKO) for astrocytes, and CD21 (DAKO) for follicular dendritic cells, using the tyramide signal amplification technique, as previously described (75).
Nucleotide sequencing and analysis.
Nucleotide sequences from p17gag and V3 region were amplified using previously described primers (68, 85). For the majority of study subjects, single molecules of HIV-1 provirus were isolated by limiting dilution and sequenced directly using the Thermal-cycle Sequenase kit (Amersham). For seven of the study subjects (NA425, NA021, NA020, NA017, NA272, NA371, and NA025), 1-μg aliquots of extracted DNA were amplified and cloned into pGEM, using T overhangs (pGEM-T easy vector system; Promega). Miniprepped DNA from clones were sequenced using the Sequenase version 2.0 kit (United States Biologicals) following the manufacturers' instructions. To avoid template resampling that may occur when the cloning method is used to obtain sequences from samples with low frequencies of amplifiable sequences, its application was restricted to samples with proviral loads greater than 60 copies per 106 cells. Dideoxynucleotide sequencing of cloned DNA sequences was carried out using USB sequenase 2.0 kit (Amersham Life Science) with 35S-dATP, the thermosequenase-radiolabeled terminator cycle sequencing kit, according to the manufacturer's instructions.
Sequences were aligned using the Simmonic 2000 Sequence Editor package. Phylogenetic trees were constructed by the neighbor-joining method using Jukes-Cantor corrected sequence distances in the MEGA package (48). The nucleotide sequences from p17gag and V3 regions from each of the study subjects were compared with each other and with a range of standard HIV-1 variants. Each set of sequences from the 12 study subjects compared was monophyletic in both genomic regions and distinct from those of the published sequences of subtype B: HIV-1SF2 (K02007), HIV-1RF (M17451), HIV-1OYI (M26727), HIV-1LAI (K02013), HIV-1JRFL (M74978), HIV-1YU2 (M93258), HIV-1CAMI (D10112), HIV-1NY5CG (M38431), HIV-1HAN (U43131), HIV-1WMJ22 (M12507), and HIV-1SFAAA (M65024). This comparison provided no evidence for coinfection with epidemiologically unrelated HIV strains, nor for intersample or exogenous laboratory contamination.
Analysis of phylogenetic groupings.
The degree of genetic segregation between variants recovered from different samples was scored using a novel method for scoring phylogenetic trees. For each sequence comparison, a phylogeny was calculated with the programs DNADIST and NEIGHBOR in the PHYLIP package (28) using an epidemiologically unlinked sequence (HIV-1SF2) as an outgroup. Starting from the root of the tree, the composition of sequences in each successive bifurcating node was calculated. An association value, d, for the tree was calculated by summation of values individually calculated from each node, according to the formula d = (1 − f)/2n−1, where n is the number of sequences below the node and f is the frequency of most common sample type. Values of d expected from the null hypothesis (i.e., for samples showing no phylogenetic grouping) were calculated by random reassignment of the sequences to different samples. Finally, the influence of tree robustness on the association value was indicated by bootstrap resampling using the program SEQBOOT in the PHYLIP package. The association index (AI) represents the mean ratio of 100 bootstrap replicates of the association value calculated from the test sequences to those of 10 sample-reassigned controls.
Confidence intervals for AI values were difficult to calculate from first principles, as the variance depended on the number of sequences compared, the degree of sequence divergence, and the phylogeny of the sequences. For the purpose of this study, in which each comparison of tissue samples contained similar numbers of sequence with similar divergence, we estimated confidence intervals empirically for AI values using (sample-reassigned) control values from independent lymph node and brain comparisons from 10 of the study subjects. The distribution of AI values corresponded closely to a normal distribution around 1, and confidence intervals for 2 standard deviations ranged from 0.74 to 1.36.
Nucleotide sequence accession numbers.
The nucleotide sequences obtained in the present study have been submitted to GenBank and have been assigned accession no. AF353734 through AF35394 and AF409200 through AF409685.
RESULTS
Detection of HIV-1 proviral sequences in nonlymphoid tissue.
DNA was extracted from the left frontal region of the cerebral cortex in the brain, from lung, and from full-thickness autopsy samples of colon from 18 HIV-infected individuals. Proviral loads were measured by limiting-dilution PCR using highly conserved primers from the p17gag region (Table 1). High frequencies of infected cells in the lymph node samples were detected in most individuals (range, 15 to 110,000 proviral copies/106 cells).
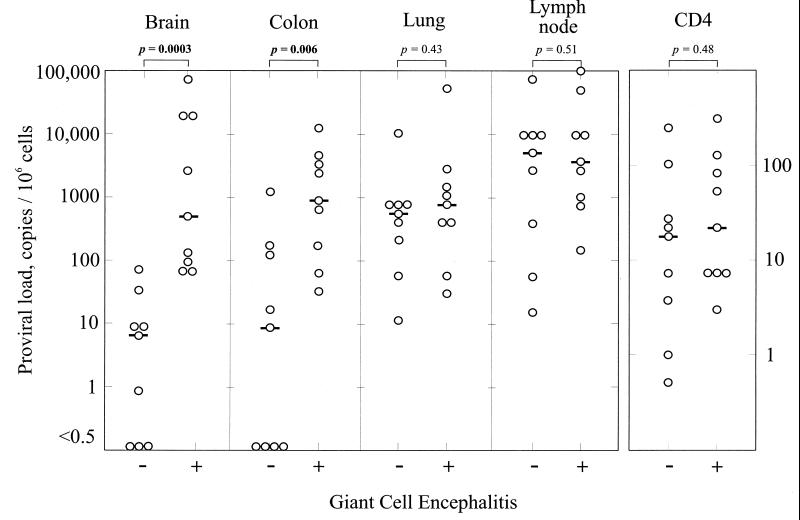
Proviral loads in nonlymphoid tissue were more variable. In brain, high proviral loads were observed only in study subjects with a postmortem diagnosis of GCE (median proviral load, 540/106 cells compared with 6/106 cells in those without encephalitis; P = 0.0003) (Fig. 1). Detection of proviral sequences in the colon samples was also more frequent in those with GCE (Table 1), and there was a marked difference in median proviral load between the GCE and nonGCE groups (1,010/106 cells compared with 10/106 cells; P = 0.006) (Fig. 1). In contrast, there was no significant difference between the GCE and non-GCE groups in the degree of infection of either lung (median values, 600 [non-GCE] and 881 [GCE]; P = 0.43) or lymph node samples (median values 6,000 [non-GCE] and 4,100 [GCE], P = 0.51) (Fig. 1). On individual comparison, proviral loads from only brain and colon correlated with each other (R = 0.629 by Spearman's rank correlation test; P < 0.01); all other pairwise comparisons of tissues produced low R values (0.27553, 0.074, 0.20, 0.23, 0.35; P > 0.1).
FIG. 1.
Comparison of proviral loads (copies per 106 cells) in samples from brain, colon, lung, and lymph node, and CD4 lymphocyte counts between study subjects with and without GCE. Median proviral loads or CD4 counts are indicated by horizontal bars. Differences in distributions of values between GCE and non-GCE groups were estimated using Mann-Whitney U test; P values are indicated above each graph.
The detection of higher proviral loads in brain and colon tissues of individuals with GCE could not be attributed to a greater degree of disease progression. CD4 lymphocyte counts of the two categories were similar at time of death (median values of 17 [non-GCE] and 23 [GCE]; Fig. 1). As described above, there was also no evidence for higher proviral loads in lymph node autopsy specimens from the GCE group.
Localization and cytopathology of HIV-1 in vivo.
Evidence of productive infection by HIV-1 in each tissue was demonstrated by immunocytochemical staining for p24 antigen using the tyramide signal amplification method (Table 2). HIV-expressing cells were identified by double labeling or immunostaining sequential serial sections with anti-p24 antibody and monoclonal antibodies (MAbs) to the cell surface markers CD3 and CD8 (T lymphocytes), PG-M1 (macrophages and brain microglia cells), L26 (B cells), and CD21 (follicular dendritic cells). Any p24-positive cells that were not recognized by the cell markers listed above were stained using other cell markers, such as glial fibrillary protein GFAP (for astrocytes), depending on the observed morphology.
TABLE 2.
Detection of p24 antigen in lymphoid and nonlymphoid autopsy samples from study subjects
| Subject | No. of CD4 (counts/μl) | GCE diagnosisa | LNb | Nonlymphoid samplesc
|
||
|---|---|---|---|---|---|---|
| LF | CO | LU | ||||
| NA371 | 0.5 | N | + | –d | – | – |
| NA425 | 1 | N | + | – | – | + |
| NA116 | 4 | N | ++ | – | – | – |
| NA20 | 8 | N | – | – | – | – |
| NA17 | 17 | N | – | – | – | – |
| NA308 | 23 | N | + | – | – | – |
| NA199 | 28 | N | +++ | – | – | + |
| NA272 | 105 | N | + | – | – | – |
| NA270 | 246 | N | +++ | – | – | – |
| NA38 | 3 | P | +++ | ⊕⊕⊕ | – | – |
| NA420 | 7 | P | ++ | ⊕ | – | – |
| NA246 | 8 | P | ++ | ⊕ | – | ⊕⊕⊕ |
| NA284 | 8 | P | ++ | ⊕⊕⊕ | ++, ⊕⊕ | – |
| NA369 | 23 | P | ++ | ⊕⊕ | – | – |
| NA118 | 56 | P | +++ | ⊕⊕ | – | ++ |
| NA21 | 87 | P | +++ | ⊕⊕⊕ | – | ++ |
| NA446 | 137 | P | +++ | ⊕⊕ | – | – |
| NA25 | 297 | P | +++ | ⊕ | + | + |
LN, lymph node; N, negative; P, positive.
Detection of p24 antigen in autopsy samples by immunocytochemistry; positive samples were graded on a scale from + (sparse) to +++ (frequent), based on numbers of infected cells.
p24 antigen expression detected in cells of monocyte/macrophage lineages is indicated on a scale of ⊕ (sparse), to ⊕⊕⊕ (frequent). LF, left frontal region of the brain; CO, colon; LU, lung.
–, not detected.
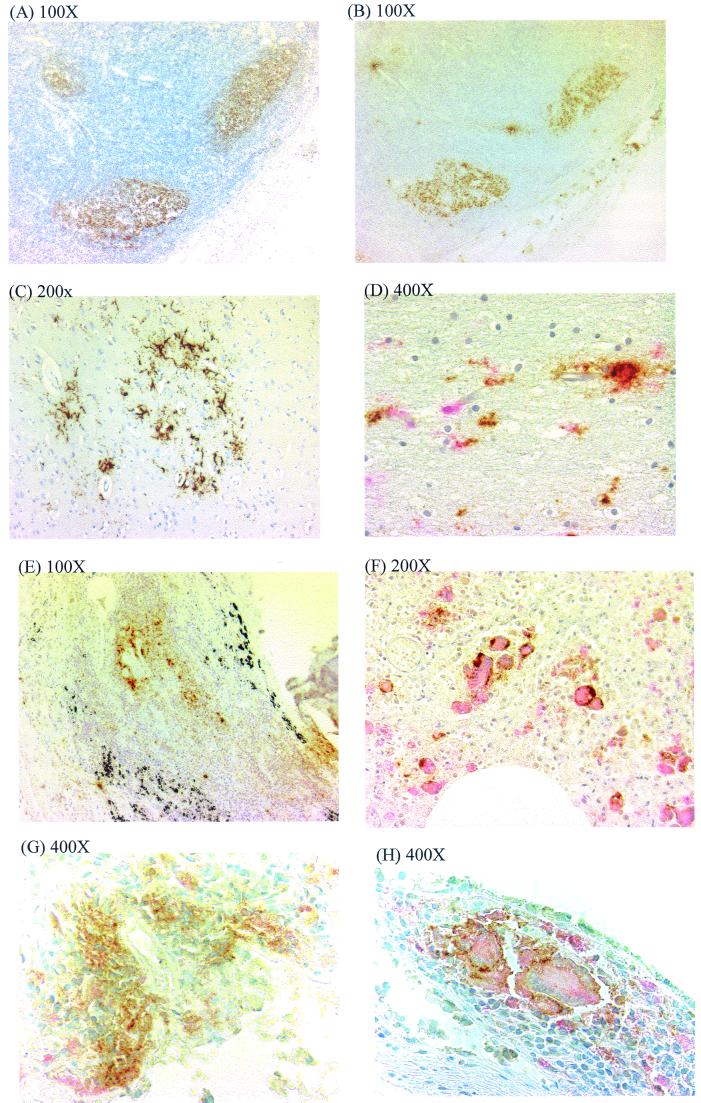
The majority of p24 in lymph node tissue was detected in germinal centers of lymphoid follicles, colocalizing with CD21 (Fig. 2A and B). In only a few instances were p24-positive cells detected in the paracortical area, indicating infrequent productive infection of T lymphocytes. The detection of HIV-1 p24 immunopositivity in the brain was strongly correlated with the stage of disease. p24 detection was restricted to study subjects diagnosed with GCE (Table 2). p24 detection was strongly associated with proviral load measured by limiting-dilution PCR (Table 1; P = 0.0003). The main cell types infected with HIV-1 in the brain were those of the monocyte/macrophage lineage; p24 was detected primarily in multinucleated giant cells (MGCs), perivascular mononuclear macrophages, and microglial cells, all of which could be double stained with PGM-1 (anti-CD68 MAb). The p24 staining in these cells was granular and cytoplasmic and was present not only in cell bodies but also in the processes of microglial cells (Fig. 2C and D).
FIG. 2.
Identification of HIV-expressing cells from different tissues in vivo. (A and B) Serial sections of lymph node from NA25 immunostained with diaminobenzidine, using monoclonal antibodies to CD21 (A) and p24 antigen (B), localizing HIV to follicle center. (C) Low-power magnification of section of brain tissue from NA21 immunostained for p24 antigen, showing cluster of p24 antigen-expressing cells. (D) Higher magnification of same section as for panel C, costained for p24 antigen with DAB and for CD68 with new fuchsin red. (E) Lung tissue from NA21 showing p24 antigen expression in lymphoid follicle. (F) Same section as for panel E, identifying p24 expression (DAB) on MGCs expressing CD68 (new fuchsin red). (G) Colon tissue from NA284 showing p24 antigen expression in lymphoid follicle. (H) Same section as for panel G, identifying p24 expression (DAB) on MGCs expressing CD68 (new fuchsin red).
HIV-1 p24 antigen was detected in lung samples of 6 of the 18 study subjects examined. p24 detection was more frequent in samples with high proviral loads (Table 1; P = 0.005; Spearman's rank correlation test). p24 detection in lung was generally restricted to germinal centers in peribronchial lymphoid aggregates, and most of the p24-positive cells were morphologically mononucleated (Fig. 2E). However, a large number of infected MGCs which double stained with anti-p24 MAb and the PGM-1 marker, were observed in subject NA246 (Fig. 2F).
p24 immunopositivity was detected in colon samples from only two study subjects, in differing cell types. In the sample from NA25, p24 was detected in lymphoid infiltrate in the colon wall (Table 2). In contrast, p24 antigen expression was detected in both the lymphoid infiltrates (Fig. 2G) and CD68-positive cells with a tissue macrophage morphology in the colon sample from NA284 (Fig. 2H). In this sample, infected cells frequently formed pronounced multinucleated syncytia, reminiscent of the appearance of infected macrophages in the lung and microglia in the brain (Fig. 2D and F).
Nucleotide sequence analysis.
The association between GCE and high proviral loads in nonlymphoid tissue and the histological evidence for infection of cells of the monocyte/macrophage lineage in lung and colon in NA284 and NA246 suggested that HIV populations in nonlymphoid tissue may be related to those infecting the brain.
To investigate genetic relationships between variants in different tissues, we sequenced the V3 hypervariable and flanking regions from proviral sequences recovered from lymph node tissue of each study subject and, where appropriate, from brain, lung, and colon tissues. The proviral loads determined by limiting dilution were essential in interpreting the sequencing results obtained, in view of the possibility that residual blood contamination (particularly in highly vascular tissue such as the lung) may contribute partly or entirely to the sequences amplified by PCR.
Problems associated with analysis of sequences to define distributions of sequences from different samples are the arbitrariness of the phylogenetic groups assigned and the absence of any defensible statistical test to detect differences in the composition of the clades. We have developed a method to analyze the large amount of sequence information generated in the genetic comparison of V3 sequences from different tissues from the 18 study subjects. The method is based on the detection of deviation from randomness in the position of sequences from different tissues in phylogenetic trees. A number of methods can be used to score the grouping of sequences in trees; in this study, we calculated a numerical index (grouping value) that is derived from the composition of descendants from each bifurcating node in a standard neighbor-joining tree. Trees showing a high degree of compartmentalization of sequences would contain relatively few nodes with descendants from different samples (in this case, from lymph node, brain, lung, or colon), and would be assigned a lower grouping score than for one where each descendant node contained mixed variants. The grouping value for a sequence dataset without compartmentalization (the null hypothesis) is also influenced by the number of sequences analyzed and the shape of phylogenetic tree. Therefore, compartmentalization has to be demonstrated by comparison of the grouping value with that calculated for a control dataset that retains the same phylogeny. The most convenient way to achieve this computationally is to randomly reassign the sequence labels, while retaining the relative numbers of sequences from each compartment and the tree structure. In the analysis presented here, a mean value from 10 label reassignments was used for each control value.
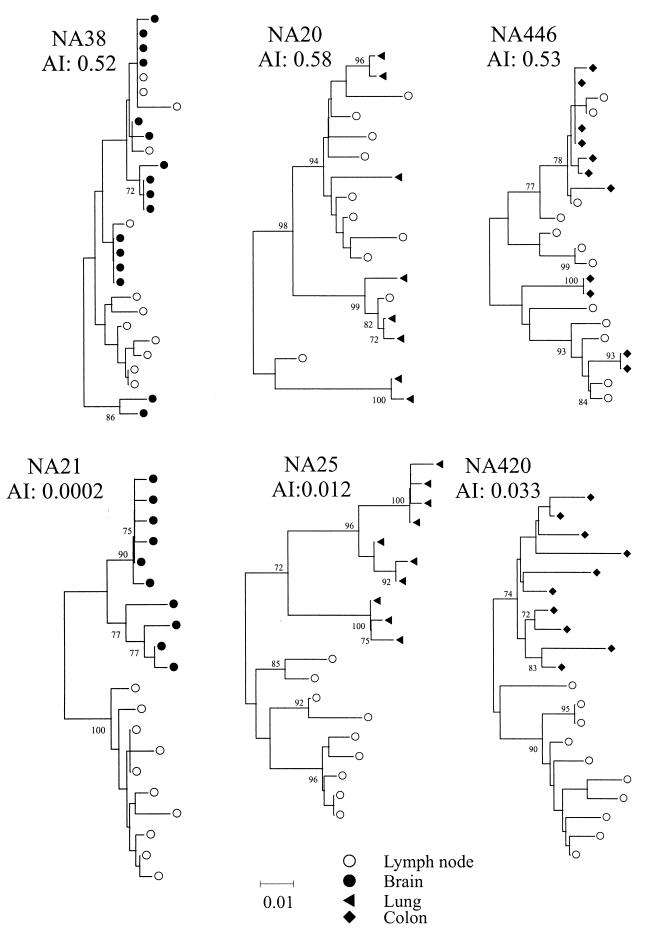
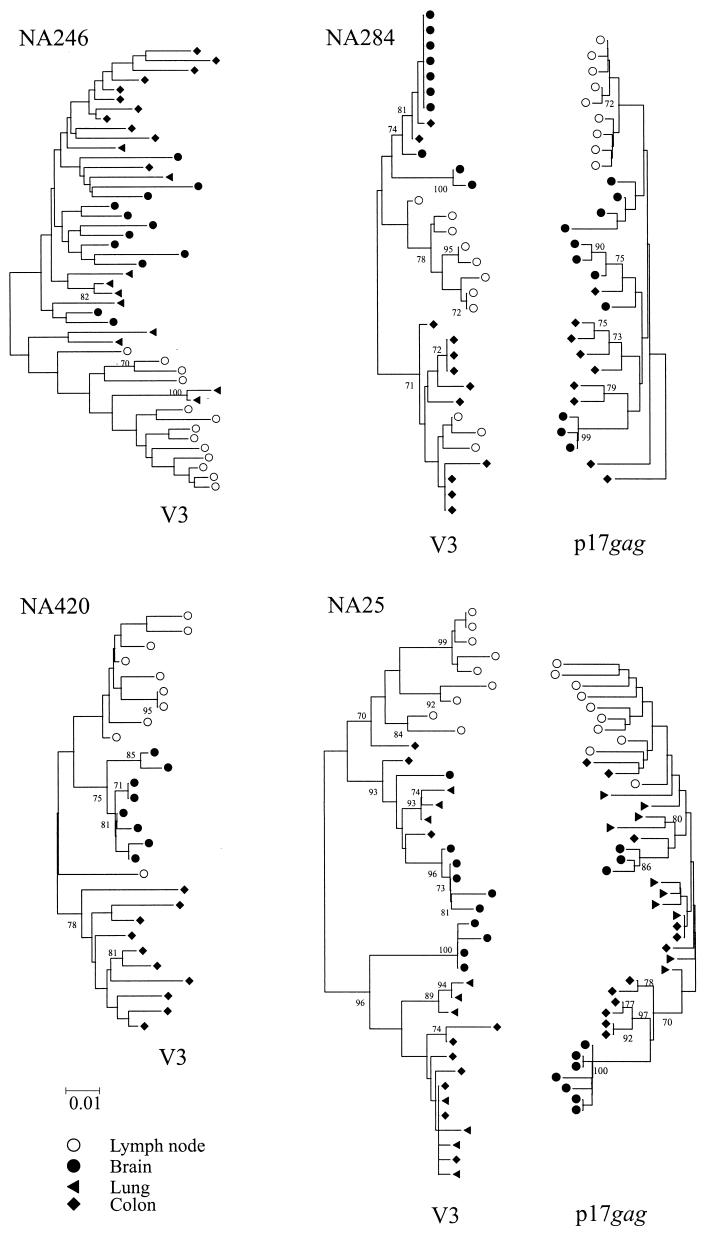
Finally, the grouping value is also dependent on the robustness of the tree analyzed. For this reason, grouping values were calculated on 100 sets of bootstrap-resampled replicates of the sequences, each with 10 sample label reassignments. Sequence datasets with grouping values consistently lower than label-reassigned controls therefore showed evidence for tissue-specific compartmentalization. Although the method can detect groupings in datasets containing sequences from any number of tissue compartments, grouping values are most relevant for comparing pairs of samples (such as brain and lymph node). To allow for different values from control sequences, results from the association test are expressed as an association index, AI, which is the ratio of the association value of sequences over that of controls). Examples of phylogenetic trees and their derived AI values for sequence comparisons between lymphoid and nonlymphoid tissues are shown in Fig. 3.
FIG. 3.
Examples of phylogenetic comparisons between sequences from lymph node with brain (left), lung (center), and colon (right) from samples showing high (upper panel) and low (lower panel) AI values. Trees were constructed by the neighbor-joining method using the MEGA package; frequency of bootstrap replicates supporting individual clades is indicated on branches (only values ≥70% are shown). All trees were plotted to the scale indicated by the bar at the bottom of the figure.
Comparisons of HIV proviral sequences from different tissues.
Approximately 10 V3 and flanking region sequences were obtained from the frontal region tissues of the nine study subjects with GCE. These sequences were compared with those amplified from lymph node (Table 3). With one exception (NA38; AI value, 0.52), brain V3 sequences were phylogenetically distinct from the lymphoid HIV population (AIs from 0.03 to 0.0002).
TABLE 3.
Association indices for pairwise comparison of proviral sequences from different tissuesa
| Subject | Association index for sequences from:
|
|||
|---|---|---|---|---|
| LN and LF | LN and CO | LN and LU | CO and LF | |
| NA371 | –b | – | 0.25 | – |
| NA425 | – | 0.41 | – | – |
| NA116 | – | 0.26c | NDd | – |
| NA20 | – | – | 0.56 | – |
| NA199 | – | 0.44 | – | – |
| NA38 | 0.52 | 0.56 | – | 0.50 |
| NA420 | 0.019 | 0.033 | – | 0.0087 |
| NA246 | 0.029 | 0.030 | 0.18 | 0.26 |
| NA284 | 0.0020 | 0.095 | – | 0.067 |
| NA369 | 0.0086 | 0.71 | – | 0.036 |
| NA118 | 0.059 | – | 0.40 | – |
| NA21 | 0.00024 | 0.28 | 0.85 | 0.0052 |
| NA446 | 0.044 | 0.52 | – | 0.0066 |
| NA25 | 0.0027 | 0.12 | 0.012 | 0.089 |
LN, lymph node; LF, left frontal region of the brain; CO, colon; LU, lung.
Sequences unavailable for comparison.
Association index values indicating separate virus populations are set in bold.
ND, not done.
For other tissues, phylogenetic relationships with lymphoid populations were more variable. Most samples from lung autopsy tissue contained proviral populations that were interspersed with lymph node sequences (Table 3), with only two study subjects showing clearly distinct lung populations. All lung-derived sequences from NA25 segregated separately from lymph node sequences (Table 3), while lung sequences from NA246 comprised two viral populations, one interspersed with lymph node sequences and another grouping separately (see below). There was no association between proviral loads in lung and their degree of sequence relatedness to lymphoid-cell populations (Tables 1 and 3).
Sequences from colon also showed variable relationships with the lymphoid populations; approximately half of the colon samples (from NA420, NA246, NA284, NA21, and NA25, all with evidence of GCE) contained sequences grouping separately from those in lymph node samples (Table 3). There was a tendency for colon samples with high viral loads to show greater genetic segregation, although this difference did not reach statistical significance.
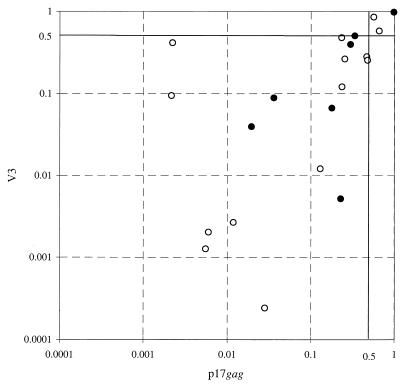
To provide independent evidence for genetic segregation of proviral populations in lymphoid and nonlymphoid cells, samples from seven study subjects were sequenced in the p17gag region and association indices were calculated (Fig. 4). There was a close correlation between sequence relationships from the two genomic regions (R = 0.71; P = 0.0002 by Spearman's rank correlation test), demonstrating substantial concordance about which populations were genetically distinct and which were interspersed. An outlier from this association was NA425, for which samples from colon were genetically distinct from lymph node samples in the p17gag region (AI, 0.0022) but interspersed in V3 (AI, 0.41).
FIG. 4.
Comparison of association indices calculated from p17gag sequences (x axis) and V3 sequences (y axis). Symbols: ○, comparison between lymph node- and nonlymphoid tissue-derived sequences; ●, comparison between nonlymphoid sequences.
Genetic relationships between nonlymphoid populations.
The combination of the previously described findings suggests that virus populations in the brain (in which the principal target cells are microglia; Fig. 2C and D) may be genetically linked to those in nonlymphoid populations, particularly in samples with infection of tissue macrophages (NA284 and NA246). In all study subjects with GCE, except for NA38, variants infecting the brain were distinct from those in lymphoid tissue, potentially allowing the identification of macrophage-associated populations elsewhere. Analysis was concentrated on the four individuals with evidence for infection of the colon and/or lung with variants of HIV genetically distinct from those in lymph node (NA420, NA246, NA284, and NA25; Fig. 5). All four individuals showed evidence for GCE, and comparison was therefore also made with variants amplified from the left frontal region.
FIG. 5.
Phylogenetic comparison of lymph node, lung, colon, and brain sequences from the four study subjects with evidence for separate proviral populations in colon and/or lung samples. p17gag region sequences from NA284 and NA25 are placed in reversed orientation on the right. Trees were constructed by the neighbor-joining method using the MEGA package; the frequencies of bootstrap replicates supporting individual clades are indicated on branches (only values ≥70% are shown). All trees were plotted to the scale indicated by the bar at the bottom of the figure.
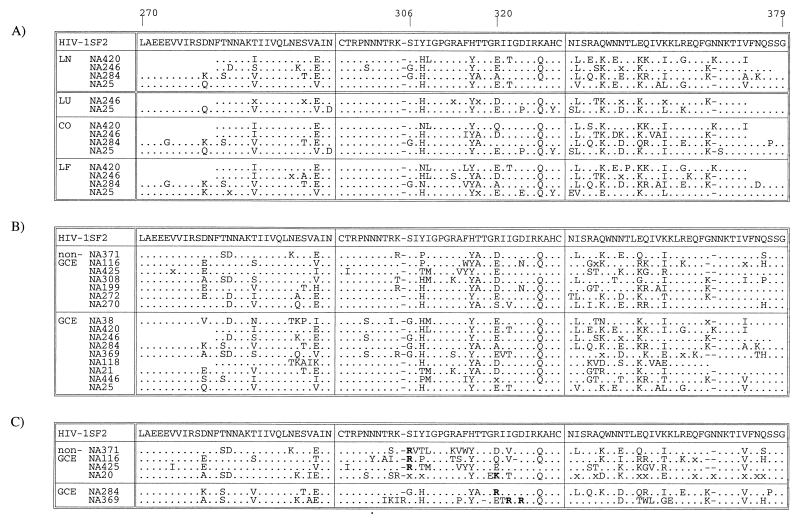
V3 sequences from lymph node, brain, lung, and colon of NA246 grouped into two main phylogenetic groups. One contained exclusively variants from nonlymphoid tissue, with sequences from brain, colon, and lung interspersed with each other. The other lineage contained predominantly lymph node-derived variant, with some from lung. The genetic division between lymphoid- and nonlymphoid-derived variants was also observed for NA25; this subject's sequences from brain, colon, and lung were mixed in the two clades which contained exclusively nonlymphoid tissue variants. Sequence relationships for the remaining two individuals were more complex. Sequences from brain, colon, and lymph node from NA246 each grouped separately from each other, with a colon-specific population distinct from both lymphoid variants and those from brain. Finally, most colon-derived variants from NA284 formed a separate clade which also contained lymph node sequences. These sequences shared a basic amino acid at position 320 in the V3 loop, associated with a CXCR4-dependent phenotype (Fig. 6). The remaining colon sequences grouped in a separate clade which contained the sequences from brain; these sequences had a predicted CCR5-dependent phenotype.
FIG. 6.
(A) Comparison of consensus V3 region amino acid sequences from lymphoid and nonlymphoid samples from the four study subjects with distinct colon and/or lung proviral populations. (B and C) Comparison of lymph node-derived sequences from study subjects with and without GCE; separate consensus sequences are shown for V3 sequences with predicted CCR5-dependent (B) and CXCR4-dependent (C) phenotypes. Symbols: x, no overall consensus; ., sequence identity with HIV-1SF2.
Sequences in the p17gag regions were obtained from NA284 and NA25 (Fig. 5). Sequence relationships in the p17gag region were generally congruent with those in V3, with genetic compartmentalization of lymph node sequences similar to those derived from nonlymphoid tissue. A noticeable difference was the grouping of colon-derived sequences from NA284. While the majority of V3 sequences grouped in a separate clade with lymph node sequences, there was no evidence for shared grouping of colon and lymph node sequences in the p17gag region. Instead, all sequences from the colon were interspersed with those from the brain.
Summarizing the results of this comparison, most variants from colon and lung showed common genetic lineages with proviral sequences recovered from brain, while sequences resembling those from lymphoid tissue were generally in the minority in both V3 and p17gag regions. However, sequences from nonlymphoid tissue were not always monophyletic, and in the case of NA246 there was a tripartite division between brain-, colon-, and lymph node-derived variants. Taken together, however, the comparisons reveal a closer genetic relationship between sequences within nonlymphoid tissue than with those derived from infected lymphocytes.
Comparison of V3 hypervariable region amino acid sequences.
To investigate whether variants from nonlymphoid tissue shared sequence motifs in the V3 region which distinguished them from lymphoid populations, a consensus amino acid sequence for variants from each tissue from the four study subjects was calculated (Fig. 6A). Each consensus sequence in this comparison showed acidic or neutral residues at positions 306 and 320 in the V3 loop. By χ2 analysis, there were no residues differentiating lymphoid and nonlymphoid sample types or individual nonlymphoid tissues that were shared between different study subjects in the V3 loop or flanking regions.
To investigate whether the occurrence of GCE was associated with particular sequences in the V3 region, consensus sequences from lymph node were calculated from each study subject. This analysis was complicated by the occurrence of various proportions of V3 sequences associated with CXCR4 usage in samples from six of the study subjects (Table 1). As these are genetically distinct from viruses with predicted CCR5 usage, separate V3 consensus sequences were calculated for each (Fig. 6B and C). For either predicted phenotype, there was no evidence for any systematic difference between sequences from individuals with and without GCE in the distribution of amino acids at a particular site or combination of sites.
DISCUSSION
Limiting-dilution PCR was used to quantify proviral sequences in a range of lymphoid and nonlymphoid autopsy tissues from HIV-infected individuals. Consistent with previous studies (1, 7–9, 25, 57), there was a strong association between the development of HIV-related GCE and proviral load measured in a representative sample of the brain. For the subjects analyzed in this study, ranges of proviral loads between GCE+ and GCE− groups were almost nonoverlapping (Fig. 1). Identification of infected cells in the brain and in other tissues was carried out by immunocytochemical staining for p24 antigen expression and was therefore restricted to cells containing actively replicating virus. p24 antigen staining was localized to CD68+ cells of the macrophage/monocyte lineage in the CNS. Morphologically, CD68+ cells corresponded both to perivascular microglia and macrophages and to microglia in the brain parenchyma, although more precise identification awaits the development of markers to distinguish cells of this lineage. None of the findings presented in this paper rule out the possibility for additional restricted or latent infection by HIV of other cell types in the CNS. For example, there may be nonproductive infection of astrocytes (reviewed in reference 11), associated with the expression of nonstructural gene products such as Nef (21, 44, 65), although whether they contribute significantly to the proviral population detected by PCR awaits more precise quantitative studies based on methods such as in situ PCR (4, 76) which do not rely on gene expression for detection of infected cells.
CNS viral populations in brain autopsy samples from the study subjects were relatively uniform in the V3 region of env and genetically distinct from variants infecting lymph node tissue in eight of the nine study subjects (Fig. 3 and 6), consistent with a large number of previous studies (6, 22, 24, 27, 33, 34, 36, 43, 45, 52, 53, 56, 58, 62, 63, 74, 81). While this overwhelming body of evidence of compartmentalization suggests the existence of adaptive differences involved in infection of the CNS, it has been alternatively hypothesized that differences in the rates of virus turnover in different cell types may lead to the population differences observed between brain and lymph node, given the rapid temporal change in HIV populations over time in peripheral blood mononuclear cells and other lymphoid cell types (45). However, the strict association between proviral load and the detection of replicating virus in the CNS by immunocytochemistry and the likelihood of continuous trafficking of HIV-infected cells through the blood brain barrier and intermingling of HIV variants at this interface (59, 83) are inconsistent with the concept that CNS-derived sequences represent an inactive or slower-replicating archival HIV population. Further indirect evidence for a functional difference with lymphoid- and CNS-derived virus variants is provided by the comparison of sequence relationships in the env gene with regions elsewhere in the genome not involved in cellular tropism, such as gag (41, 56). To summarize a large amount of comparative sequence information, only sequences derived from the V1/V2 and V3 regions demonstrated consistent tissue specificity; brain-derived sequences in the p17gag region showed much greater sequence diversity and in some individuals or from certain brain regions may be genetically indistinct from those recovered from lymphoid tissue. Greater diversity and weaker partitioning are also evident from the comparison of V3 and p17gag sequences in the present study (Fig. 5). Given the propensity of HIV to frequently recombine in vivo, the restriction of tissue specificity to V1/V2 and V3 provides evidence for phenotypic selection in this region not exerted elsewhere in the genome.
Direct evidence for differences in cellular tropism between HIV variants recovered from CNS and lymphoid tissues has been obtained by comparison of the biological properties of virus isolated from the CNS and virus isolates passaged through microglia or through the characterization of infectious clones or pseudotypes constructed from env gene sequences amplified from brain and lymphoid tissue (13, 23, 35, 55, 67, 72, 84). All brain-derived or microglia-passaged variants analyzed to date require CD4 for virus attachment and use predominantly or exclusively the chemokine receptor CCR5 for entry (2, 13, 18, 23, 37, 38, 51, 67, 72, 84). This clearly is not the defining feature of CNS-derived viruses, as the majority of HIV variants derived from lymphoid cell types also use CCR5. Indeed, while there is a consensus that CCR5 is the principal coreceptor by which HIV enters cells of the macrophage/monocyte lineage, additional phenotypic differences are likely to underlie the specific macrophage tropism demonstrated by certain HIV-1 isolates.
Evidence supporting the hypothesis for additional factor(s) required for productive infection of macrophages includes the observations that many CCR5-using isolates of HIV fail to infect macrophages in vitro (14, 18, 23, 42, 71). Macrophages can be infected in vitro through the CXCR4 receptor by dual-tropic isolates of HIV (17, 38), while still retaining an apparent preferential tropism for macrophages (18). In vitro passaging of a blood-derived CCR5-using isolate of HIV-1 in microglia led to the emergence of a variant with an enhanced ability to replicate in macrophages, a change in phenotype dependent on amino acid changes in the V1/V2 hypervariable region (55, 67). These changes had no apparent effect on its usage of CCR5 or other coreceptors but enhanced its ability to use low cell surface concentration of CD4 for attachment. This may have direct adaptive advantages for infection of cells of the macrophage/monocyte lineage, where levels of CD4 expression are lower than on the surfaces of CD4 lymphocytes (55, 60, 67).
Together, these observations indicate the complexity of the HIV-macrophage interaction and support the original hypothesis for multiple entry or postentry restrictions to their productive infection (31), additional to requirements for CD4 and CCR5 cell surface expression. Adaptive changes required for replication in macrophages, and not coreceptor usage, may therefore underlie the consistent sequence differences between brain and lymph node populations in this study and previous genetic analyses of autopsy-derived tissues. This is particularly relevant for published comparisons of brain- and lymph node-derived variants of HIV-1 in which there are no differences in net V3 charge that might suggest use of different coreceptors.
A close association between macrophage tropism and the ability of isolates to infect microglia has been long recognized (35, 46, 66, 77, 82), although whether infection of microglia or neurovirulence requires additional adaptive changes remains unclear. In the simian immunodeficiency virus macaque model of AIDS, it has been shown that the gp120 gene determines macrophage tropism, while sequences in gp41 and/or nef are required for neurotropism (54). The lack of an animal model has precluded comparable investigations of neurovirulence of HIV-1, although there is evidence for similar or identical replication kinetics of various CCR5-using isolates in macrophages and microglia in vitro (35, 38, 77), indicating similarity in the phenotypic determinants underlying tropism for the two cell types. The development of GCE may therefore reflect the evolution of HIV variants, capable of infecting not only cells of the CNS but also macrophage populations in other nonlymphoid tissues. The relationship between the development of GCE and infection of macrophage populations elsewhere in the body was the focus of our quantitative and genetic comparison of HIV proviral sequences in different autopsy tissues.
Relationship between infections of the CNS and of other nonlymphoid tissues.
The most striking link between the occurrence of GCE and infection of other nonlymphoid tissues was the observation of significantly higher proviral loads in colon samples of the study subjects with GCE (Fig. 1). This phenomenon was not the result of differences in disease progression, treatment history, or extent of destruction of lymphoid tissue between the GCE+ and GCE− groups on autopsy examination (data not shown). Similarly, there was no significant difference in CD4 count before death, nor were there differences in proviral loads in lymph node samples between the two groups (Fig. 2). These observations led us to the hypothesis that the development of GCE reflects a broader tendency of some virus infections to spread beyond lymphocytes to other cell types in nonlymphoid tissues.
The finding of increased proviral loads in colon samples in the GCE+ group was associated with the frequent detection of V3 sequences closely related to those found in the CNS and distinct from those in lymphoid tissues (NA246, NA284, NA25; Fig. 5). The finding of virus populations distinct from lymphoid tissue in some colon autopsy specimens is consistent with previous observations for partial genetic partitioning of HIV between GI-and blood-derived samples. For example, different frequencies of antiviral resistant variants in gut mucosal biopsy samples with circulating virus in plasma were obtained from a minority of individuals (61). Similarly, V3 sequence differences were frequently found between variants of HIV recovered from feces and those found in peripheral blood (79, 80).
In one case, it was possible to detect p24 antigen-positive CD68+ MGCs in the mucosal layer of the colon, representing productive infection of tissue macrophages. The associated syncytial cytopathology was strikingly reminiscent of giant cells associated with GCE in the brain. V3 sequences from the colon of this individual comprised two populations; one group contained positively charged residues at positions 322 and 324 and grouped with lymph node-derived variants (Fig. 5, 6). The others were closely similar to those recovered from the CNS (Fig. 5). As this sample showed both macrophage infection and lymphoid cell infiltration (Table 2), it is possible that the former cell types harbored the HIV variants similar to those in the CNS. We are currently using microdissection to allow the separate genetic analysis of macrophage- and lymphoid-associated infected cells in this autopsy sample to confirm this hypothesis. Despite this example, detection of productively infected macrophages in the GI tract by p24 antigen immunocytochemistry was infrequent in this series and has been previously described in only one case report (49). It is possible that restricted replication of HIV in macrophages prevented the detection of infected macrophages in other colon samples where there was evidence for the presence of CNS-related variants. Restricted or latent infection may result from the reduced expression of CCR5 found in gut-derived macrophages compared with those derived from blood monocytes or in microglia (50, 73). More extensive analysis of sections by immunocytochemistry or the application of in situ PCR to identify latently infected cells may help resolve this issue.
In contrast to infection of the colon, that of the lung was almost invariably the result of lymphoid infiltration. There was no association between proviral load in lung and the development of GCE (Fig. 1), and V3 and p17gag sequences from variants infecting lung tissue were generally interspersed with those from lymph node (Table 3). The evidence for lack of genetic partitioning between lymph node- and lung-derived HIV variants is consistent with previously observed similarities between lymph node and lung env sequences (71) and suggests extensive trafficking and infection of similar cell types between the two tissue types. Supporting this, p24 antigen detection was, with one exception, confined to lymphoid follicle centers (Table 2).
However, the sample from NA246 showed a high frequency of p24 antigen-positive CD68+ giant cells, representing the productive infection of alveolar macrophages. This histological picture was remarkably similar to the observed infection of infected macrophages in the colon samples from NA284 (see above). Genetic analysis of V3 sequences from the lung sample of NA246 revealed two distinct virus populations; the majority form (6 of 10) grouped with brain-derived variants, while two were interspersed with lymph node-derived sequences. As with the colon samples, it is possible that these different populations of lung-derived variants have specific associations with the observed giant cells in the alveoli and the lymphoid cells infiltrating the lung.
Mechanism of infection of nonlymphoid tissue.
The combined genetic and immunohistochemical findings in this study provide evidence of at least two distinct mechanisms for infection of nonlymphoid tissue. Infiltration of lymphoid cells, perhaps in response to inflammatory processes associated with opportunistic infection, was most evident in the lung and was observed to a lesser extent in the colon, but it was not a feature of the lymphoid infiltrates in the CNS. Infection of nonlymphoid tissue was alternatively or additionally manifested by the appearance of productively infected CD68+ cells of the macrophage/monocyte lineage, in which giant cell formation was a marked histological feature. This pattern of infection typified that of the CNS but was also found in colon and lung samples of two study subjects, both of which harbored virus populations closely related to those found in the brain.
This study forms the basis for future functional analysis of macrophage-associated populations in vivo. In particular, it will be important to determine whether the colon and lung-associated variants from NA284 and NA21 show evidence for greater phenotypic similarities to CNS-derived variants than the lymphoid-related variants recovered from corresponding samples of other study subjects. Secondly, it will be relevant to establish whether the particular ability of certain variants of HIV to infect cells of the macrophage/monocyte lineage reflects larger-scale epidemiological differences between HIV variants infecting injecting drug users (IDUs) in Edinburgh and other risk groups. HIV infection in the majority of Edinburgh IDUs, including those described in the present study, originated from a common source introduced into a specific suburb of Edinburgh around 1982-1983 (12, 40, 64). Despite careful analysis of compounding factors, such as treatment compliance and risk-taking behavior, it has remained difficult to account for the much higher incidence of GCE in this group than in other risk groups in Edinburgh or in IDU cohorts in other cities (8, 9). One possibility is that the virus introduced into this group has a greater propensity for infection of macrophages than other subtype B variants of HIV. Phenotypic analysis of the variants genetically characterized in the present study, with particular attention to the existence of specific adaptive changes, such as efficient use of lower cell surface CD4 concentration for attachment previously associated with macrophage tropism (55, 60, 67), will provide an important test of this theory and an insight into the disease manifestations outside the CNS in this group.
ACKNOWLEDGMENTS
We are grateful to Francis Brennan for preparation of the sections for immunocytochemistry and for the provision of frozen samples from the MRC Edinburgh HIV Brain and Tissue Bank (SG9708080).
The virological studies were supported by a Medical Research Council strategic project grant (G9632414).
REFERENCES
- 1.Achim C L, Wang R, Miners D K, Wiley C A. Brain viral burden in HIV infection. J Neuropathol Exp Neurol. 1994;53:284–294. doi: 10.1097/00005072-199405000-00010. [DOI] [PubMed] [Google Scholar]
- 2.Albright A V, Shieh J T C, Itoh T, Lee B, Pleasure D, O'Connor M J, Doms R W, Gonzalez-Scarano F. Microglia express CCR5, CXCR4, and CCR3, but of these, CCR5 is the principal coreceptor for human immunodeficiency virus type 1 dementia isolates. J Virol. 1999;73:205–213. doi: 10.1128/jvi.73.1.205-213.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: A RANTES, MIP-1 alpha, MIP-1 beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 4.An S F, Groves M, Gray F, Scaravilli F. Early entry and widespread cellular involvement of HIV-1 DNA in brains of HIV-1 positive asymptomatic individuals. J Neuropathol Exp Neurol. 1999;58:1156–1162. doi: 10.1097/00005072-199911000-00005. [DOI] [PubMed] [Google Scholar]
- 5.Asjo B, Albert J, Karlsson A, Morfeldt Manson L, Biberfeld G, Lidman K, Fenyo E M. Replicative capacity of human immunodeficiency virus from patients with varying severity of HIV infection. Lancet. 1986;ii:660–662. [PubMed] [Google Scholar]
- 6.Ball J K, Holmes E C, Whitwell H, Desselberger U. Genomic variation of human immunodeficiency virus type 1 (HIV-1)—molecular analyses of HIV-1 in sequential blood samples and various organs obtained at autopsy. J Gen Virol. 1994;75:867–879. doi: 10.1099/0022-1317-75-4-867. [DOI] [PubMed] [Google Scholar]
- 7.Bell J E, Brettle R P, Chiswick A, Simmonds P. HIV encephalitis, proviral load and dementia in drug users and homosexuals with AIDS. Effect of neocortical involvement. Brain. 1998;121:2043–2052. doi: 10.1093/brain/121.11.2043. [DOI] [PubMed] [Google Scholar]
- 8.Bell J E, Busuttil A, Ironside J W, Rebus S, Donaldson Y K, Simmonds P, Peutherer J F. Human immunodeficiency virus and the brain—investigation of virus load and neuropathologic changes in pre-AIDS subjects. J Infect Dis. 1993;168:818–824. doi: 10.1093/infdis/168.4.818. [DOI] [PubMed] [Google Scholar]
- 9.Bell J E, Donaldson Y K, Lowrie S, McKenzie C A, Elton R A, Chiswick A, Brettle R P, Ironside J W, Simmonds P. Influence of risk group and zidovudine therapy on the development of HIV encephalitis and cognitive impairment in AIDS patients. AIDS. 1996;10:493–499. doi: 10.1097/00002030-199605000-00007. [DOI] [PubMed] [Google Scholar]
- 10.Berson J F, Long D, Doranz B J, Rucker J, Jirik F R, Doms R W. A seven-transmembrane domain receptor involved in fusion and entry of T-cell-tropic human immunodeficiency virus type 1 strains. J Virol. 1996;70:6288–6295. doi: 10.1128/jvi.70.9.6288-6295.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brack-Werner R. Astrocytes: HIV cellular reservoirs and important participants in neuropathogenesis. AIDS. 1999;13:1–22. doi: 10.1097/00002030-199901140-00003. [DOI] [PubMed] [Google Scholar]
- 12.Brown A J L, Lobidel D, Wade C M, Rebus S, Phillips A N, Brettle R P, France A J, Leen C S, McMenamin J, McMillan A, Maw R D, Mulcahy F, Robertson J R, Sankar K N, Scott G, Wyld R, Peutherer J F. The molecular epidemiology of human immunodeficiency virus type 1 in six cities in Britain and Ireland. Virology. 1997;235:166–177. doi: 10.1006/viro.1997.8656. [DOI] [PubMed] [Google Scholar]
- 13.Chan S Y, Speck R F, Power C, Gaffen S L, Chesebro B, Goldsmith M A. V3 recombinants indicate a central role for CCR5 as a coreceptor in tissue infection by human immunodeficiency virus type 1. J Virol. 1999;73:2350–2358. doi: 10.1128/jvi.73.3.2350-2358.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng-Mayer C, Liu R, Landau N R, Stamatatos L. Macrophage tropism of human immunodeficiency virus type 1 and utilization of the CC-CKR5 coreceptor. J Virol. 1997;71:1657–1661. doi: 10.1128/jvi.71.2.1657-1661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng-Mayer C, Seto D, Tateno M, Levy J A. Biological features of HIV-1 that correlate with virulence in the host. Science. 1988;240:80–82. doi: 10.1126/science.2832945. [DOI] [PubMed] [Google Scholar]
- 16.Clapham P R, Reeves J D, Simmons G, Dejucq N, Hibbitts S, McKnight A. HIV coreceptors, cell tropism and inhibition by chemokine receptor ligands. Mol Membr Biol. 1999;16:49–55. doi: 10.1080/096876899294751. [DOI] [PubMed] [Google Scholar]
- 17.Collman R G, Yi Y. Cofactors for human immunodeficiency virus entry into primary macrophages. J Infect Dis. 1999;179(Suppl 3):S422–S426. doi: 10.1086/314797. [DOI] [PubMed] [Google Scholar]
- 18.Cunningham A L, Li S, Juarez J, Lynch G, Alali M, Naif H. The level of HIV infection of macrophages is determined by interaction of viral and host cell genotypes. J Leukoc Biol. 2000;68:311–317. [PubMed] [Google Scholar]
- 19.de Jong J J, de Ronde A, Keulen W, Tersmette M, Goudsmit J. Minimal requirements for the human immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: analysis by single amino acid substitution. J Virol. 1992;66:6777–6780. doi: 10.1128/jvi.66.11.6777-6780.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng H K, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Dimarzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 21.Di Rienzo A M, Aloisi F, Santarcangelo A C, Palladino C, Olivetta E, Genovese D, Verani P, Levi G. Virological and molecular parameters of HIV-1 infection of human embryonic astrocytes. Arch Virol. 1998;143:1599–1615. doi: 10.1007/s007050050401. [DOI] [PubMed] [Google Scholar]
- 22.Di Stefano M, Wilt S, Gray F, Dubois-Dalcq M, Chiodi F. HIV type 1 V3 sequences and the development of dementia during AIDS. AIDS Res Hum Retrovir. 1996;12:471–476. doi: 10.1089/aid.1996.12.471. [DOI] [PubMed] [Google Scholar]
- 23.Dittmar M T, Simmons G, Donaldson Y, Simmonds P, Clapham P R, Schulz T F, Weiss R A. Biological characterization of human immunodeficiency virus type 1 clones derived from different organs of an AIDS patient by long-range PCR. J Virol. 1997;71:5140–5147. doi: 10.1128/jvi.71.7.5140-5147.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Donaldson Y K, Bell J E, Holmes E C, Hughes E S, Brown H K, Simmonds P. In vivo distribution and cytopathology of variants of human immunodeficiency virus type 1 showing restricted sequence variability in the V3 loop. J Virol. 1994;68:5991–6005. doi: 10.1128/jvi.68.9.5991-6005.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Donaldson Y K, Bell J E, Ironside J W, Brettle R P, Robertson J R, Busuttil A, Simmonds P. Redistribution of HIV outside the lymphoid system with onset of AIDS. Lancet. 1994;343:382–385. doi: 10.1016/s0140-6736(94)91222-x. [DOI] [PubMed] [Google Scholar]
- 26.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y X, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4(+) cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 27.Epstein L G, Kuiken C, Blumberg B M, Hartman S, Sharer L R, Clement M, Goudsmit J. HIV-1 V3 domain variation in brain and spleen of children with AIDS: tissue-specific evolution within host-determined quasispecies. Virology. 1991;180:583–590. doi: 10.1016/0042-6822(91)90072-j. [DOI] [PubMed] [Google Scholar]
- 28.Felsenstein J. PHYLIP—phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 29.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 30.Fenyo E M, Morfeldt Manson L, Chiodi F, Lind B, von Gegerfelt A, Albert J, Olausson E, Asjo B. Distinct replicative and cytopathic characteristics of human immunodeficiency virus isolates. J Virol. 1988;62:4414–4419. doi: 10.1128/jvi.62.11.4414-4419.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fouchier R A M, Brouwer M, Kootstra N A, Huisman H G, Schuitemaker H. HIV-1 macrophage tropism is determined at multiple levels of the viral replication cycle. J Clin Investig. 1994;94:1806–1814. doi: 10.1172/JCI117529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fouchier R A M, Groenink M, Kootstra N A, Tersmette M, Huisman H G, Miedema F, Schuitemaker H. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J Virol. 1992;66:3138–3187. doi: 10.1128/jvi.66.5.3183-3187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gartner S, McDonald R A, Hunter E A, Bouwman F, Liu Y, Popovic M. gp120 sequence variation in brain and in T-lymphocyte human immunodeficiency virus type 1 primary isolates. J Hum Virol. 1997;1:3–18. [PubMed] [Google Scholar]
- 34.Gatanaga H, Oka S, Ida S, Wakabayashi T, Shioda T, Iwamoto A. Active HIV-1 redistribution and replication in the brain with HIV encephalitis. Arch Virol. 1999;144:29–43. doi: 10.1007/s007050050483. [DOI] [PubMed] [Google Scholar]
- 35.Ghorpade A, Nukuna A, Che M, Haggerty S, Persidsky Y, Carter E, Carhart L, Shafer L, Gendelman H E. Human immunodeficiency virus neurotropism: an analysis of viral replication and cytopathicity for divergent strains in monocytes and microglia. J Virol. 1998;72:3340–3350. doi: 10.1128/jvi.72.4.3340-3350.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haggerty S, Stevenson M. Predominance of distinct viral genotypes in brain and lymph node compartments of HIV-infected individuals. Viral Immunol. 1991;4:123–131. doi: 10.1089/vim.1991.4.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He J L, Chen Y Z, Farzan M, Choe H Y, Ohagen A, Gartner S, Busciglio J, Yang X Y, Hofmann W, Newman W, Mackay C R, Sodroski J, Gabuzda D. CCR3 and CCR5 are co-receptors for HIV-1 infection of microglia. Nature. 1997;385:645–649. doi: 10.1038/385645a0. [DOI] [PubMed] [Google Scholar]
- 38.Hibbitts S, Reeves J D, Simmons G, Gray P W, Epstein L G, Schols D, De Clercq E, Wells T N, Proudfoot A E, Clapham P R. Coreceptor ligand inhibition of fetal brain cell infection by HIV type 1. AIDS Res Hum Retrovir. 1999;15:989–1000. doi: 10.1089/088922299310502. [DOI] [PubMed] [Google Scholar]
- 39.Hoffman T L, Doms R W. HIV-1 envelope determinants for cell tropism and chemokine receptor use. Mol Membr Biol. 1999;16:57–65. doi: 10.1080/096876899294760. [DOI] [PubMed] [Google Scholar]
- 40.Holmes E C, Zhang L Q, Robertson P, Cleland A, Harvey E, Simmonds P, Brown A J L. The molecular epidemiology of human immunodeficiency virus type 1 in Edinburgh. J Infect Dis. 1995;171:45–53. doi: 10.1093/infdis/171.1.45. [DOI] [PubMed] [Google Scholar]
- 41.Hughes E S, Bell J E, Simmonds P. Investigation of the dynamics of the spread of human immunodeficiency virus to brain and other tissues by evolutionary analysis of sequences from the p17gag and env genes. J Virol. 1997;71:1272–1280. doi: 10.1128/jvi.71.2.1272-1280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hung C S, Pontow S, Ratner L. Relationship between productive HIV-1 infection of macrophages and CCR5 utilization. Virology. 1999;264:278–288. doi: 10.1006/viro.1999.0013. [DOI] [PubMed] [Google Scholar]
- 43.Keys B, Karis J, Fadeel B, Valentin A, Norkrans G, Hagberg L, Chiodi F. V3 sequences of paired HIV-1 isolates from blood and cerebrospinal fluid cluster according to host and show variation related to the clinical stage of disease. Virology. 1993;196:475–483. doi: 10.1006/viro.1993.1503. [DOI] [PubMed] [Google Scholar]
- 44.Kleinschmidt A, Neumann M, Moller C, Erfle V, Brack-Werner R. Restricted expression of HIV1 in human astrocytes: molecular basis for viral persistence in the CNS. Res Virol. 1994;145:147–53. doi: 10.1016/s0923-2516(07)80016-1. [DOI] [PubMed] [Google Scholar]
- 45.Korber B T M, Kunstman K J, Patterson B K, Furtado M, Mcevilly M M, Levy R, Wolinsky S M. Genetic differences between blood- and brain-derived viral sequences from human immunodeficiency virus type 1-infected patients: evidence of conserved elements in the V3 region of the envelope protein of brain-derived sequences. J Virol. 1994;68:7467–7481. doi: 10.1128/jvi.68.11.7467-7481.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koyanagi Y, Miles S, Mitsuyasu R T, Merrill J E, Vinters H V, Chen I S. Dual infection of the central nervous system by AIDS viruses with distinct cellular tropisms. Science. 1987;236:819–822. doi: 10.1126/science.3646751. [DOI] [PubMed] [Google Scholar]
- 47.Kozak S L, Platt E J, Madani N, Ferro F E, Peden K, Kabat D. CD4, CXCR-4, and CCR-5 dependencies for infections by primary and laboratory-adapted isolates of human immunodeficiency virus type 1. J Virol. 1997;71:873–882. doi: 10.1128/jvi.71.2.873-882.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar S, Tamura K, Nei M. MEGA: molecular evolutionary genetics analysis, version 1.0. University Park, Pa: The Pennsylvania State University; 1993. [Google Scholar]
- 49.Lewin-Smith M, Wahl S M, Orenstein J M. Human immunodeficiency virus-rich multinucleated giant cells in the colon: a case report with transmission electron microscopy, immunohistochemistry, and in situ hybridization. Mod Pathol. 1999;12:75–81. [PubMed] [Google Scholar]
- 50.Li L, Meng G, Graham M F, Shaw G M, Smith P D. Intestinal macrophages display reduced permissiveness to human immunodeficiency virus 1 and decreased surface CCR5. Gastroenterology. 1999;116:1043–1053. doi: 10.1016/s0016-5085(99)70007-7. [DOI] [PubMed] [Google Scholar]
- 51.Li S, Juarez J, Alali M, Dwyer D, Collman R, Cunningham A, Naif H M. Persistent CCR5 utilization and enhanced macrophage tropism by primary blood human immunodeficiency virus type 1 isolates from advanced stages of disease and comparison to tissue-derived isolates. J Virol. 1999;73:9741–9755. doi: 10.1128/jvi.73.12.9741-9755.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Y X, Kappes J C, Conway J A, Price R W, Shaw G M, Hahn B H. Molecular characterization of human immunodeficiency virus type 1 cloned directly from uncultured human brain tissue: identification of replication-competent and -defective viral genomes. J Virol. 1991;65:3973–3985. doi: 10.1128/jvi.65.8.3973-3985.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu Z Q, Wood C, Levy J A, Cheng Mayer C. The viral envelope gene is involved in macrophage tropism of a human immunodeficiency virus type 1 strain isolated from brain tissue. J Virol. 1990;64:6148–6153. doi: 10.1128/jvi.64.12.6148-6153.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mankowski J L, Flaherty M T, Spelman J P, Hauer D A, Didier P J, Amedee A M, Murphey-Corb H, Kirstein L M, Munoz A, Clements J E, Zink M C. Pathogenesis of simian immunodeficiency virus encephalitis: viral determinants of neurovirulence. J Virol. 1997;71:6055–6060. doi: 10.1128/jvi.71.8.6055-6060.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martin J, Labranche C C, Gonzalez-Scarano F. Differential CD4/CCR5 utilization, gp120 conformation, and neutralization sensitivity between envelopes from a microglia-adapted human immunodeficiency virus type 1 and its parental isolate. J Virol. 2001;75:3568–3580. doi: 10.1128/JVI.75.8.3568-3580.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morris A, Marsden M, Halcrow K, Hughes E S, Brettle R P, Bell J E, Simmonds P. Mosaic structure of the human immunodeficiency virus type 1 genome infecting lymphoid cells and the brain: evidence for frequent in vivo recombination events in the evolution of regional populations. J Virol. 1999;73:8720–8731. doi: 10.1128/jvi.73.10.8720-8731.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pang S, Koyanagi Y, Miles S, Wiley C, Vinters H V, Chen I S. High levels of unintegrated HIV-1 DNA in brain tissue of AIDS dementia patients. Nature. 1990;343:85–89. doi: 10.1038/343085a0. [DOI] [PubMed] [Google Scholar]
- 58.Pang S, Vinters H V, Akashi T, O'Brien W A, Chen I S. HIV-1 env sequence variation in brain tissue of patients with AIDS-related neurologic disease. J Acquir Immune Defic Syndr. 1991;4:1082–1092. [PubMed] [Google Scholar]
- 59.Petito C K, Chen H, Mastri A R, Torres-Munoz J, Roberts B, Wood C. HIV infection of choroid plexus in AIDS and asymptomatic HIV-infected patients suggests that the choroid plexus may be a reservoir of productive infection. J Neurovirol. 1999;5:670–677. doi: 10.3109/13550289909021295. [DOI] [PubMed] [Google Scholar]
- 60.Platt E J, Wehrly K, Kuhmann S E, Chesebro B, Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Poles M A, Elliott J, Vingerhoets J, Michiels L, Scholliers A, Bloor S, Larder B, Hertogs K, Anton P A. Despite high concordance, distinct mutational and phenotypic drug resistance profiles in human immunodeficiency virus type 1 RNA are observed in gastrointestinal mucosal biopsy specimens and peripheral blood mononuclear cells compared with plasma. J Infect Dis. 2001;183:143–148. doi: 10.1086/317640. [DOI] [PubMed] [Google Scholar]
- 62.Power C, Mcarthur J C, Johnson R T, Griffin D E, Glass J D, Perryman S, Chesebro B. Demented and nondemented patients with AIDS differ in brain-derived human immunodeficiency virus type 1 envelope sequences. J Virol. 1994;68:4643–4649. doi: 10.1128/jvi.68.7.4643-4649.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reddy R T, Achim C L, Sirko D A, Tehranchi S, Kraus F G, Wongstaal F, Wiley C A, Grant I, Atkinson J H, Kelly M, Chandler J L, Wallace M R, McCutchan J A, Spector S A, Thal L, Heaton R K, Hesselink J, Jernigan T, Masliah E, Abramson I, Butters N, Patterson T, Zisook S, Jeste D. Sequence analysis of the V3 loop in brain and spleen of patients with HIV encephalitis. AIDS Res Hum Retrovir. 1996;12:477–482. doi: 10.1089/aid.1996.12.477. [DOI] [PubMed] [Google Scholar]
- 64.Robertson J R, Bucknall A B V, Welsby P D, Roberts J J K, Inglis J M, Peutherer J F, Brettle R P. Epidemic of AIDS related virus (HTLV-III/LAV) among intravenous drug abusers. BMJ. 1986;292:527–529. doi: 10.1136/bmj.292.6519.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sabri F, Tresoldi E, Distefano M, Polo S, Monaco M C, Verani A, Fiore J R, Lusso P, Major E, Chiodi F, Scarlatti G. Nonproductive human immunodeficiency virus type 1 infection of human fetal astrocytes: independence from CD4 and major chemokine receptors. Virology. 1999;264:370–384. doi: 10.1006/viro.1999.9998. [DOI] [PubMed] [Google Scholar]
- 66.Sharpless N E, O'Brien W A, Verdin E, Kufta C V, Chen I S, Dubois-Dalcq M. Human immunodeficiency virus type 1 tropism for brain microglial cells is determined by a region of the env glycoprotein that also controls macrophage tropism. J Virol. 1992;66:2588–2593. doi: 10.1128/jvi.66.4.2588-2593.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shieh J T, Martin J, Baltuch G, Malim M H, Gonzalez-Scarano F. Determinants of syncytium formation in microglia by human immunodeficiency virus type 1: role of the V1/V2 domains. J Virol. 2000;74:693–701. doi: 10.1128/jvi.74.2.693-701.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Simmonds P, Balfe P, Ludlam C A, Bishop J O, Leigh Brown A J. Analysis of sequence diversity in hypervariable regions of the external glycoprotein of human immunodeficiency virus type 1. J Virol. 1990;64:5840–5850. doi: 10.1128/jvi.64.12.5840-5850.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Simmonds P, Balfe P, Peutherer J F, Ludlam C A, Bishop J O, Leigh Brown A J. Human immunodeficiency virus-infected individuals contain provirus in small numbers of peripheral mononuclear cells and at low copy numbers. J Virol. 1990;64:864–872. doi: 10.1128/jvi.64.2.864-872.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Simmons G, Wilkinson D, Reeves J D, Dittmar M, Beddows S, Weber J, Carnegie G, Desselberger U, Gray P W, Weiss R A, Clapham P R. Primary, syncytium-inducing human immunodeficiency virus type 1 isolates are dual-tropic and most can use either Lestr or CCR5 as coreceptors for virus entry. J Virol. 1996;70:8355–8360. doi: 10.1128/jvi.70.12.8355-8360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Singh A, Besson G, Mobasher A, Collman R G. Patterns of chemokine receptor fusion cofactor utilization by human immunodeficiency virus type 1 variants from the lungs and blood. J Virol. 1999;73:6680–6690. doi: 10.1128/jvi.73.8.6680-6690.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smit T K, Wang B, Ng T, Osborne R, Brew B, Saksena N K. Varied tropism of HIV-1 isolates derived from different regions of adult brain cortex discriminate between patients with and without AIDS dementia complex (ADC): evidence for neurotropic HIV variants. Virology. 2001;279:509–526. doi: 10.1006/viro.2000.0681. [DOI] [PubMed] [Google Scholar]
- 73.Smith P D, Meng G, Shaw G M, Li L. Infection of gastrointestinal tract macrophages by HIV-1. J Leukoc Biol. 1997;62:72–77. doi: 10.1002/jlb.62.1.72. [DOI] [PubMed] [Google Scholar]
- 74.Steuler H, Storch Hagenlocher B, Wildemann B. Distinct populations of human immunodeficiency virus type 1 in blood and cerebrospinal fluid. AIDS Res Hum Retrovir. 1992;8:53–59. doi: 10.1089/aid.1992.8.53. [DOI] [PubMed] [Google Scholar]
- 75.Strappe P M, Wang T H, McKenzie C A, Lowrie S, Simmonds P, Bell J E. Enhancement of immunohistochemical detection of HIV-1 p24 antigen in brain by tyramide signal amplification. J Virol Methods. 1997;67:103–112. doi: 10.1016/s0166-0934(97)00083-9. [DOI] [PubMed] [Google Scholar]
- 76.Strappe P M, Wang T H, McKenzie C A, Lowrie S, Simmonds P, Bell J E. In situ polymerase chain reaction amplification of HIV-1 DNA in brain tissue. J Virol Methods. 1998;70:119–127. doi: 10.1016/s0166-0934(97)00156-0. [DOI] [PubMed] [Google Scholar]
- 77.Strizki J M, Albright A V, Sheng H, OConnor M, Perrin L, Gonzalez-Scarano F. Infection of primary human microglia and monocyte-derived macrophages with human immunodeficiency virus type 1 isolates: evidence of differential tropism. J Virol. 1996;70:7654–7662. doi: 10.1128/jvi.70.11.7654-7662.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tersmette M, Lange J M, de Goede R E, de Wolf F, Eeftink Schattenkerk J K, Schellekens P T, Coutinho R A, Huisman J G, Goudsmit J, Miedema F. Association between biological properties of human immunodeficiency virus variants and risk for AIDS and AIDS mortality. Lancet. 1989;i:983–985. doi: 10.1016/s0140-6736(89)92628-7. [DOI] [PubMed] [Google Scholar]
- 79.Vanderhoek L, Sol C J A, Maas J, Lukashov V V, Kuiken C L, Goudsmit J. Genetic differences between human immunodeficiency virus type 1 subpopulations in faeces and serum. J Gen Virol. 1998;79:259–267. doi: 10.1099/0022-1317-79-2-259. [DOI] [PubMed] [Google Scholar]
- 80.Vanderhoek L, Sol C J A, Snijders F, Bartelsman J F W, Boom R, Goudsmit J. Human immunodeficiency virus type 1 RNA populations in faeces with higher homology to intestinal populations than to blood populations. J Gen Virol. 1996;77:2415–2425. doi: 10.1099/0022-1317-77-10-2415. [DOI] [PubMed] [Google Scholar]
- 81.Vantwout A B, Ran L J, Kuiken C L, Kootstra N A, Pals S T, Schuitemaker H. Analysis of the temporal relationship between human immunodeficiency virus type 1 quasispecies in sequential blood samples and various organs obtained at autopsy. J Virol. 1998;72:488–496. doi: 10.1128/jvi.72.1.488-496.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Watkins B A, Dorn H H, Kelly W B, Armstrong R C, Potts B J, Michaels F, Kufta C V, Dubois-Dalcq M. Specific tropism of HIV-1 for microglial cells in primary human brain cultures. Science. 1990;249:549–553. doi: 10.1126/science.2200125. [DOI] [PubMed] [Google Scholar]
- 83.Wu D T, Woodman S E, Weiss J M, McManus C M, D'Aversa T G, Hesselgesser J, Major E O, Nath A, Berman J W. Mechanisms of leukocyte trafficking into the CNS. J Neurovirol. 2000;6(Suppl 1):S82–S85. [PubMed] [Google Scholar]
- 84.Zhang K, Hawken M, Rana F, Welte F J, Gartner S, Goldsmith M A, Power C. Human immunodeficiency virus type 1 clade A and D neurotropism: molecular evolution, recombination, and coreceptor use. Virology. 2001;283:19–30. doi: 10.1006/viro.2001.0876. [DOI] [PubMed] [Google Scholar]
- 85.Zhang L Q, MacKenzie P, Cleland A, Holmes E C, Leigh Brown A J, Simmonds P. Selection for specific sequences in the external envelope protein of human immunodeficiency virus type 1 upon primary infection. J Virol. 1993;67:3345–3356. doi: 10.1128/jvi.67.6.3345-3356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]