Abstract
Background
In the domain of forensic science, the application of kinship identification and mixture deconvolution techniques are of critical importance, providing robust scientific evidence for the resolution of complex cases. Microhaplotypes, as the emerging class of genetic markers, have been widely studied in forensics due to their high polymorphisms and excellent stability.
Results and discussion
In this research, a novel and high-efficient panel integrating 33 microhaplotype loci along with a sex-determining locus was developed by the next generation sequencing technology. In addition, we also assessed its forensic utility and delved into its capacity for kinship analysis and mixture deconvolution. The average effective number of alleles (Ae) of the 33 microhaplotype loci in the Guizhou Han population was 6.06, and the Ae values of 30 loci were greater than 5. The cumulative power of discrimination and cumulative power of exclusion values of the novel panel in the Guizhou Han population were 1-5.6 × 10− 43 and 1-1.6 × 10− 15, respectively. In the simulated kinship analysis, the panel could effectively distinguish between parent-child, full-sibling, half-sibling, grandfather-grandson, aunt-nephew and unrelated individuals, but uncertainty rates clearly increased when distinguishing between first cousins and unrelated individuals. For the mixtures, the novel panel had demonstrated excellent performance in estimating the number of contributors of mixtures with 1 to 5 contributors in combination with the machine learning methods.
Conclusions
In summary, we have developed a small and high-efficient panel for forensic genetics, which could provide novel insights into forensic complex kinships testing and mixture deconvolution.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-024-10880-4.
Keywords: Forensic genetics, Microhaplotype, Guizhou Han, Next generation sequencing, Complex kinships
Background
Short tandem repeat (STR) loci, known for their high polymorphism and widespread distribution, are currently the most commonly used genetic markers in forensic work [1]. However, the presence of stutter during STR typing can pose challenges in the analysis of mixed samples [2]. Moreover, due to high mutation rates of STRs, they have limitations in some kinship analyses [3–7]. Single nucleotide polymorphism (SNP), exhibited great application prospect in forensic genetics since its low mutation rate and widespread distribution in the human genome. Whereas, SNPs usually have lower genetic polymorphism [8], and therefore possess less discriminative power for individual identification. Better forensic performance for SNPs could be achieved with the parallel detection of a larger number of markers [9–13]. Even so, the construction of the multiplex amplification panel for a larger number of SNPs is usually involved with complex chemistry and operation procedures, which is time-consuming and labor-intensive.
In 2013, Kidd and colleagues [14] proposed a new type of genetic marker, microhaplotype, which can solve these problems mentioned above to a certain point. Microhaplotypes are genetic markers that composed of two or more SNPs in the short DNA region (commonly less than 300 bp), combining the advantages of both STRs and SNPs [4, 15]. They exhibit high stability and genetic polymorphism, providing enhanced performance in kinship identification, particularly among individuals with complex kinships. In mixed-sample analysis, it is difficult to reckon the number of contributors (NOCs) due to the loss of alleles and the sharing of alleles among contributors. Previous studies found that microhaplotypes with higher the effective of alleles (Ae) could not only reduce allele sharing among different contributors, but also detect more likely alleles in mixed samples [16–19]. Nowadays, a large number of microhaplotype composite detection systems have been developed for various purposes in forensic genetics [10, 15, 18, 20–29]. For examples, Zhang et al. [23] assessed the potential of three microhaplotype loci for individual identification and ancestry inference in the Hainan Li ethnic group and 26 reference populations from the 1000 Genomes Project; In 2022, Wen et al. [10] developed a panel of 29 microhaplotype loci for paternity testing and sibling testing; Yang et al. [22] employed machine learning algorithms to estimate NOCs in mixtures and they found the feasibility of this panel in inferring the NOCs; Zhao et al. [24] demonstrated that microhaplotypes could be effectively used for personal identification, paternity testing, and ancestry inference even when analyzing highly degraded and unbalanced mixtures, implying that microhaplotypes were also beneficial to assessing degraded samples. However, it should not be overlooked that the uneven performance of selected microhaplotypes in these systems have been observed, which could provide relatively limited information in forensic personal identification, kinship analysis, and mixture deconvolution. Therefore, it is crucial for us to develop a high-efficient panel of microhaplotypes for forensic genetics.
In this study, we firstly screened microhaplotypes with highly genetic diversities in Chinese populations based on previous reported microhaplotypes [18, 30–34]. Next, a multiplex amplification panel composing these microhaplotypes has been constructed by the next generation sequencing (NGS) technology. Furthermore, performance validation of the novel panel like sensitivity, repeatability, species specificity and applicability of mixed samples was conducted. Finally, we also explored the ability of the system for inferring NOC of the mixtures and assessing complex kinship relationships.
Material and method
Locus screening, primer design, and reference populations
Microhaplotypes were selected from previous studies [18, 30–34] according to the following criteria: (1) each region contained at least 3 SNP loci; (2) Ae value was greater than 5 in East Asian populations; (3) microhaplotype loci located on the same chromosome were more than 1 Mb apart. General information of selected microhaplotypes was given in Supplementary Table 1.
For selected microhaplotype loci, we searched flanking sequence information of these microhaplotypes by the Ensembl (http://www.ensembl.org/) database. In addition, we also added a sex-determining locus (Amelogenin) to these microhaplotypes. The primers of these loci were designed by the Oligo tool (version 7) [35].
Based on data of the 1000 Genome Project phase III [36], 26 populations in five continents (African, American, East Asian, South Asian, and European) were used as reference populations for population genetics analysis.
Sample preparation and DNA extraction
A total of 201 bloodstain samples of unrelated healthy Han individuals from Guizhou were collected after obtained their written consent. We performed DNA extraction of these samples by the method of IGT™ Pure Beads (iGeneTech, Beijing, China).
DNA positive sample 9948 was used for dilution of 1ng/µL, 0.5ng/µL, 0.25ng/µL, and 0.125ng/µL for sensitivity analysis. 9948 (1ng/µL) and 9947 (1ng/µL) were mixed at different mix ratio (1:1, 1:2, 2:1, 1:4, 4:1, 1:9, 9:1, 1:19, 19:1, 1:49, and 49:1) for mixture deconvolution evaluation. This study was approved by the Ethics Committee of Guizhou Medical University.
Library preparation, sequencing, and data analysis
The library preparation was carried out according to the specification of the MultipSeq® Custom Panel (iGeneTech, Beijing, China). In brief, the first-round PCR system consisted of 9 µL ddH2O, 3.5 µL Enhancer buffer NB (1 N), 2.5 µL Enhancer buffer M, 5 µL Primer pool, 10 µL IGT-EM808 polymerase mixture and 1ng DNA sample, with a total reaction system of 30 µL. The cycling conditions were listed as below: denaturation for 3 min and 30 s at 95 °C; 22 cycles of 20 s at 98 °C, 60 s at 55 °C, 60 s at 60 °C, and 2 min at 65 °C; extension was performed for 5 min at 72 °C and held at 4 °C. Subsequently, IGT™ Pure Beads were used to purify the first-round PCR amplified product. After that, purified PCR product was used for second-round PCR. The reaction reagents included 13.5 µL purified PCR product, 2.5 µL Enhancer buffer M, 2 µL UDI Index (5 µM), 10 µL IGT-EM808 polymerase mixture, and 2 µL ddH2O. The cycling parameters were set as follows: 3 min and 30 s at 95 °C; 9 cycles of 20 s at 98 °C, 60 s at 58 °C, and 30 s at 72 °C; followed by 5 min at 72 °C and held at 4 °C. Likewise, we used IGT™ Pure Beads to purify the second-round PCR products. The DNA library concentration of each sample was measured by the Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific, USA) on the Qubit® 3.0 Fluorometer. Finally, all samples were mixed into a well and were conducted for sequencing on the DNBSEQ-T7 platform (BGI, Shenzhen, China).
For obtained raw data, we firstly removed low-quality and index sequences by the Trimmomatic software (version 0.38) [37]. Next, these clean sequences were aligned and annotated onto the human reference genome by the BWA software (version 0.7.12) [38] with the method of mem. The parameters for mem were listed as below: -M, -k 40, and -t 8. Finally, we used perl-based script (https://github.com/moonlightfurion/microhaplotype) to conduct microhaplotype analysis of these samples according to the following parameters: haplotypes frequency ≥ 15% and depth of coverage ≥ 20X. For mixed samples, analytical threshold was set to 5X.
Statistical analysis
Haplotype frequencies and forensic statistical parameters including expected heterozygosity (He), observed heterozygosity (Ho), polymorphic information content (PIC), match probability (PM), power of discrimination (PD), and probability of exclusion (PE) of selected microhaplotype loci in East Asian and Guizhou Han populations were estimated by the STRAF online program (version 2.1.5) [39]. The Ae values of these loci in Guizhou Han and five continental populations were calculated according to the formula: Ae = 1/pi2 (pi is the allele/haplotype frequency of ith allele/haplotype in one population). The allele coverage ratio (ACR) of each locus was calculated by the ratio of the coverage depth of the minor allele to the coverage depth of the major allele.
Population genetic analyses of 26 reference populations from different continents were conducted to evaluate ancestral resolution of selected microhaplotypes. Firstly, pairwise Fst genetic distances of these 26 populations were estimated by the Genepop program (version 4.0.10) [40]. Next, the vegan (2.6-8) and ggplot2 packages (version 3.5.1) in R software (version 4.4) were used to perform the multidimensional scaling analysis (MDS) of these 26 populations based on their pairwise Fst values. Subsequently, the phylogenetic tree of these populations was constructed with the neighbor-joining (NJ) method by the MEGA software (version X) [41] based on their pairwise Fst values. Finally, population structure analysis of these populations was performed using STRUCTURE software (version 2.3.4) [42]. The detailed parameters for STRUCTURE were: K from 2 to 7 and 10 iterations for each K value with 10,000 burn-in and 10,000 Markov Chain Monte Carlo. We used the StructureHarvester software (version cpython-312) [43] to determine the optimal K value and CLUMPP program (version 1.1) [44] to process the data to avoid random effects. Genetic structure of these populations were visually shown by the pophelper online program (version 1.0.10) [45] based on the output results of CLUMPP.
Based on haplotype frequencies of selected microhaplotype loci in Guizhou Han population, the Familias3 software (version 3.3.1) [46] was used to simulate six types of kinship relationships: parent-child, full-sibling, half-sibling, grandfather-grandson, aunt-nephew, and first cousins. Two hypotheses of relationships were set: H1 assumes a certain kinship, and H2 assumes unrelated individuals. Each kinship relationship was simulated 1000 times, and the Log10 (LR) value of the likelihood ratio was calculated. The density plot of Log10 (LR) for different kinship relationships was drawn using the ggplot2 package of the R software. Furthermore, we also set t1 and t2 thresholds based on the Log10 (LR). If the Log10 (LR) was greater than t2, it is judged to have a certain kinship. If the Log10 (LR) was less than t1, it was judged to be unrelated individuals. When t1 < Log10 (LR) < t2, it cannot be determined whether there was a certain kinship. Based on t1 and t2 thresholds, we calculated the sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), error rate, uncertainty, and system effectiveness of the system in identifying different kinship relationships.
Genetic profile of mixed samples with different NOCs was simulated by R software based on the haplotype frequencies of selected microhaplotype loci in the Guizhou Han population, with 2000 samples for each type of mixture. Based on the number of alleles observed on each microhaplotype, we explored the performance of six machine learning algorithms (Naive Bayes, random forest, decision tree, XGBoost, classification and regression trees, and linear discriminant analysis) for inferring NOC of these mixtures by the rtemis package (version 0.97.54) of R software.
Result
Loci information and preliminary evaluation for forensic efficacy of selected microhaplotypes
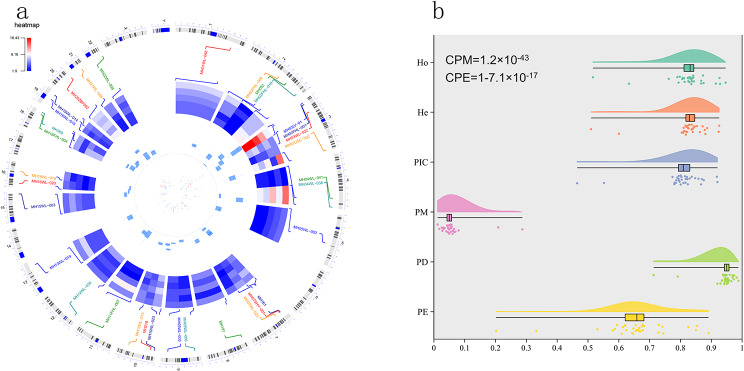
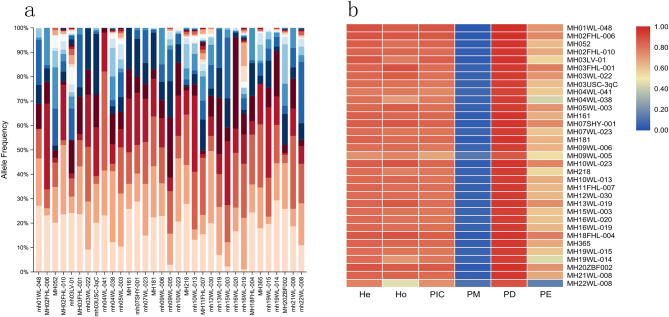
According to the criteria mentioned in Material and Methods, we totally screened 33 microhaplotypes. General information of these loci was shown in Fig. 1 and Supplementary Table (1) As we can see, these 33 microhaplotypes mainly distributed 19 autosomes except for 6, 14, and 17 chromosomes. These 33 microhaplotypes totally included 166 SNPs and the number of SNPs in these loci ranged from 3 to 11. The length of the microhaplotypes ranged from 48 bp (MH19WL-015) to 111 bp (MH365), with an average length of 82 bp, and the length of 29 loci was less than 100 bp. The Ae values of these 33 microhaplotypes in five different continental populations (African, European, East Asian, South Asian, and American) were shown in Fig. 1a and Supplementary Table (2) The average Ae values of these 33 microhaplotypes in these five continental populations were 5.48 (African), 4.78 (European), 6.36 (East Asian), 5.60 (South Asian), and 5.38 (American), respectively. Overall, the majority of loci showed high Ae values (> 3) in these five continental populations, especially in East Asian population. Next, we also estimated forensic parameters of 33 microhaplotype loci in East Asian population, as shown in Fig. 1b and Supplementary Table (3) The average Ho, He, PIC, PM, PD, and PE values of these 33 microhaplotypes in East Asian population were 0.8246, 0.8238, 0.8021, 0.0604, 0.9396, and 0.6533, respectively. Among these 33 microhaplotypes, MH052 locus displayed the lowest Ho (0.5159), He (0.5101), PIC (0.4648), PD (0.713), and PE (0.2018) values; whereas, MH03USC-3qC locus showed the highest Ho (0.9464), He (0.9257), PIC (0.9200), PD (0.9880), and PE (0.8909) values. The cumulative PM (CPM) and PE (CPE) values of these 33 loci in East Asian population were 1.2 × 10− 43 and 1-7.1 × 10− 17, implying that these microhaplotypes showed extremely high forensic application values in personal identification and paternity analysis.
Fig. 1.
General information of selected 33 microhaplotype loci. Physical positions of selected 33 microhaplotypes in different chromosomes (a) and their forensic parameters in East Asian population (b). The heatmap in the circle diagram was the Ae values of selected 33 microhaplotypes in different continental populations
Population genetic analyses of different continental populations based on 33 microhaplotypes
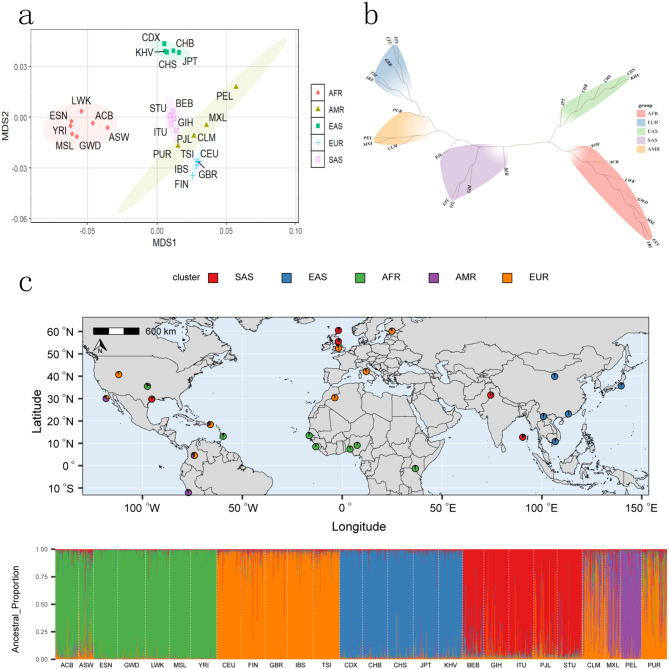
Firstly, we estimated pairwise Fst genetic distances of 26 reference populations based on selected 33 microhaplotypes, as presented in Supplementary Table 4. Results indicated that populations from the same continent possessed low Fst genetic distances; whereas, populations from different continents displayed high Fst genetic distances. Next, the MDS of these 26 populations was plotted based on their Fst genetic distances, as shown in Fig. 2a. It could be seen from Fig. 2a that the MDS1 could distinguish seven African populations from other continental populations, and MDS2 could distinguish five East Asian populations from other continental populations. In addition, five South Asian populations were mainly located on the right part and five European populations were mainly situated on the bottom right part. However, four American populations were mainly scattered among South Asian and European populations. Subsequently, the phylogenetic tree of these 26 populations was constructed, as shown in Fig. 2b. Five different branches could be observed from the phylogenetic tree, which exactly corresponded to their continental origins. Last, we assessed genetic structure of these 26 reference populations by the STRUCTURE software, as shown in Supplementary Fig. 1. At K = 2, African populations mainly showed yellow genetic components; whereas, other populations mainly displayed pink genetic components. At K = 3, African, East Asian, and European populations mainly exhibited pinked, yellow, and blue genetic components. At K = 4, we found that African, East Asian, South Asian and European populations displayed different genetic components, which could be differentiated from each other. At K = 5, we found that the PEL population exhibited distinct genetic components compared to other continental populations. Nonetheless, no further genetic components among these populations were discerned at higher K values. According to the mean LnP(K) values of each K (Supplementary Fig. 2), we found that the plateau could be observed at K = 5, indicating that K = 5 is the best K value. Therefore, we further provided genetic structure analyses of these 26 populations at K = 5, as shown in Fig. 2c.
Fig. 2.
Population genetic analyses of 26 reference populations from different continents based on selected 33 microhaplotypes. a, MDS analysis of 26 populations; b, the phylogenetic tree of 26 populations; c, population genetics structure analyses of 26 reference populations
Constructing the panel of 33 microhaplotypes and amelogenin locus by the NGS platform
Based on the NGS platform, we successfully developed a multiplex amplification panel of one sex-determining locus (the Amelogenin locus) and 33 microhaplotypes. The primer sequences of these loci were presented in Table 1. Results revealed that the amplicon size of these loci ranged from 106 to 271, with an average size of 221.
Table 1.
Primer sequences and amplicon sizes of the amelogenin locus and 33 microhaplotypes
| Loci | Forward primer (5’-3’) | Reverse primer (5’-3’) | Amplicon length |
|---|---|---|---|
| MH01WL-048 | TGAGCGATCTAGGCCTTAGTGA | GGAACTCGGTCAAGGTGAAGAA | 255 |
| MH02FHL-006 | TGCACTAAGGCAGAGTTTTCATG | AAGCTTTACCTCCCGGGTTTG | 258 |
| MH052 | AAAGACGTACTTGAGACTGGGTAAT | ACCTGGTGGGAGACACATGA | 255 |
| MH02FHL-010 | AGGGTACTTTCCGTAGGACCAA | TGGTCTGGGAGGTCCTCTTTAA | 210 |
| MH03LV-01 | GCTAGACTCCAAGTCGAAACTGAG | GATCACTTGAGCCCAGGAGGA | 232 |
| MH03FHL-001 | CTCCCCCAAGCATACTCATATCTC | CGTAAAATGTTCTGGAAGGGACTG | 221 |
| MH03WL-022 | ACAAAAGCTTCACTCAGGAAGGA | CCATTGTTCACAGACATGAGCG | 240 |
| MH03USC-3qC | CAGTAAAATTCAGGATGCCCAGTG | ACAGGAGTCATTGCCTTGTGG | 259 |
| MH04WL-041 | GGGAGACTAGTGCATGAGAAGAAG | CAGAGGATACTCATTAGCCTGCTC | 211 |
| MH04WL-038 | CTTCTGGAGACAGGACATGTCAAG | GGAAGTATGATGGTTATGGGTGGG | 160 |
| MH05WL-003 | ACTCTGCTGTGATGCCCTCT | AGCAACTTTTCAGGATGCCCA | 259 |
| MH161* | TAGGAGCATAGGCAGAGCCTT | CATCCAGTCAGTGTCTGCAGAT | 229 |
| MH07SHY-001 | GCGCTATGCTTGTGGTAGTGA | AGACATACCTGAGACTGGGAAGA | 204 |
| MH07WL-023 | GAGATATGTGTGAGGTGCTTAGCA | GACAGAAGCTTTTACTGCTCCGA | 251 |
| MH181 | GATGGGAGAAAAACCTCCCTATGG | GTAGGAGGGCAGGGTGATAATAAG | 213 |
| MH09WL-006 | TCAGAGAACTGGTGAAGACAAAGG | GGATCTCACTACCTGACAATTCCC | 251 |
| MH09WL-005 | ATGAAGTCAGGAGTCCTGCTGT | TGCTGGGCACACTCATTTATTTC | 222 |
| MH10WL-023 | TCAACCAAAGTCCCATTCTGACA | CAGAAAATACCATATGGCCACTGC | 249 |
| MH218 | CACTAAAGGCTGTCAGCAAAGG | CATGTGCCACTACACCCAGATA | 205 |
| MH10WL-013 | AGGGCTCTGCACGTATTAAGTG | TGTCTCAGCAGAGAGGTGGTTA | 187 |
| MH11FHL-007 | AATCCCCATGAAGGACAGTGT | GAGACTCCGTCTCGAGGAAA | 199 |
| MH12WL-030 | AAGTCACACTGGCTGAAGTCAG | AAGCCATCAGTAGTGTGAGGATC | 205 |
| MH13WL-019 | CGGATAATGGCGATTTCAATGCT | GACTTACAGCCCTGTGGTTGTT | 248 |
| MH15WL-003 | GCAACCATCATCTCTTGCCTCA | AGTGAGGTGGTCAGGGTGATAG | 181 |
| MH16WL-020 | CTGTGTGCTTTCTGTAGGCAGA | AAGGCTTTGACACGCATTATCTG | 258 |
| MH16WL-019 | TTCAGCAAGTCCAGCAGATGTG | TGCTCAGCCAACATTTAGCTGT | 218 |
| MH18FHL-004 | GTATTTTTAGTAGAGACAGGGCTTCAC | ACAGTAAATGGAGAGAGAGTTGTCTG | 194 |
| MH365 | GGAAGTCTAGCCAGAGCAATTAGG | GTTGTCATGCAGGGAGTGTCTAC | 271 |
| MH19WL-015 | GGAGGTGATCAGGGATGACGA | ACCAGAGACAGACACTCGTTTC | 194 |
| MH19WL-014 | GTGCATGACCAGGAGATAAATGC | ACACCTGAACAGCAAATCGTTTG | 260 |
| MH20ZBF002 | TGTACCAACCTCCTTGCTCTCT | GAACGCCCTTGGGATTCACTTA | 203 |
| MH21WL-008 | GACAAATTATTCCGCCTCCCTGA | CATGTTTCATGATATTGCCTATAACCTT | 249 |
| MH22WL-008 | AGACTGGACAGAGCCTTGGA | CACAGGCTGGTCTAAGTCTCC | 169 |
| Amelogenin | CCCTGGGCTCTGTAAAGAATAGTG | ATCAGAGCTTAAACTGGGAAGCTG | 106 |
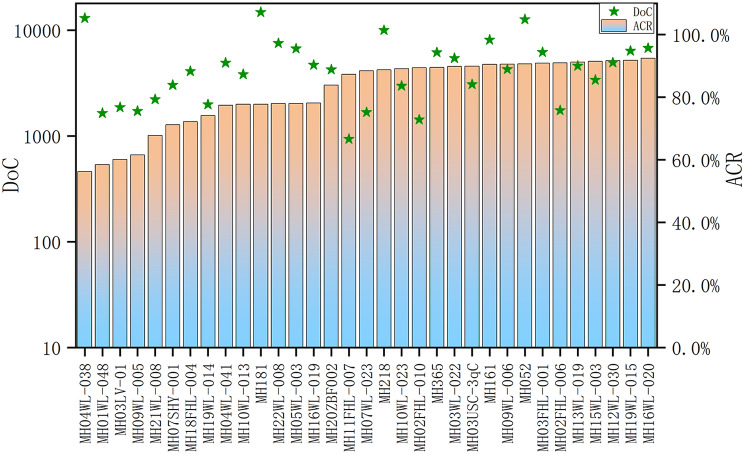
Next, we used the panel to detect 201 Guizhou Han individuals. Sequencing results of these 201 samples for these 33 microhaplotypes were shown in Fig. 3. Results showed that the average depth of coverage (DoC) values of 33 microhaplotypes in these samples ranged from 937 to 14,795, with the median of 4,270. The ACRs of these 33 loci ranged from 56.33% to 92.47%, with the mean value of 81.75%, indicating the developed panel showed relatively good allele balances.
Fig. 3.
Sequencing results (depth of coverage and allele coverage ratio) of the 33 microhaplotypes in the Guizhou Han population
Performance validation of the developed panel
To verify the sensitivity of the panel, we performed gradient dilution on 9948 DNA positive sample. Next, different concentrations of 9948 samples (1-0.125ng) were detected by the developed panel. Results revealed that all alleles could be detected in these samples regardless of their concentrations. Even when the amount of DNA template was as low as 0.125ng, the mean ACR of these microhaplotypes were still more than 80%. Furthermore, the typing results of each locus were consistent for different concentrations of 9948 samples.
We used 9947 DNA sample to construct three DNA libraries, respectively. Each DNA library was sequenced at 1µL loading volume under the same reaction conditions and repeated three times. The typing results for each locus were consistent, indicating that the panel had good repeatability.
Next, we evaluated the species specificity of the developed panel by the online tool BLAST. Obtained results exhibited that these loci were only blasted to human DNA sequences and some primate species. And no cross-reaction among common species (like dog, cat, chicken, horse, rat, and so on) was observed for the developed panel, indicating that the panel had good species specificity.
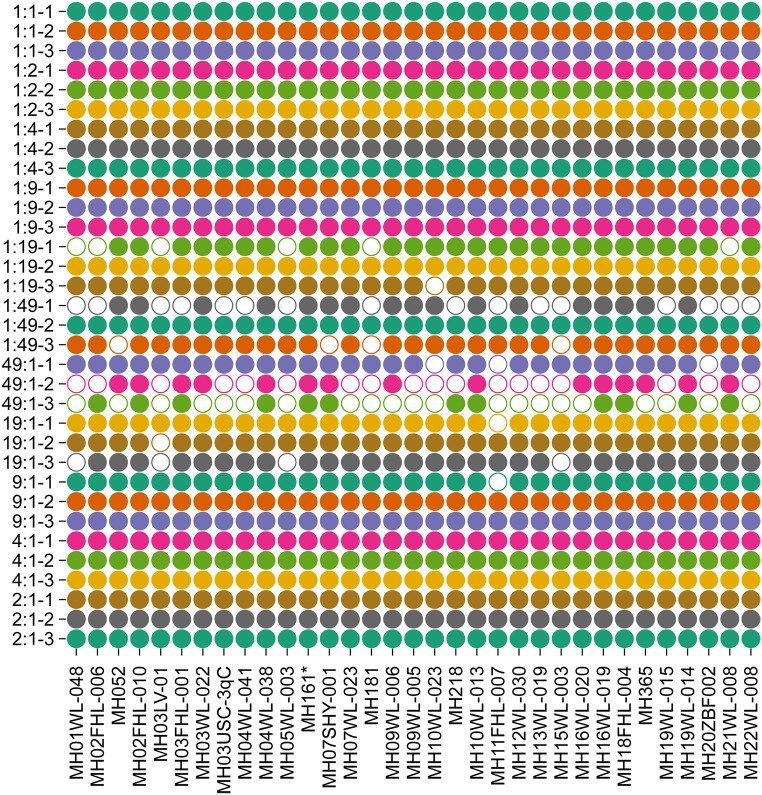
Subsequently, we also assessed the power of the developed panel to dissect mixtures. Different ratios of 9948 and 9947 DNA samples were detected by the developed panel. Loci detection ratio of 33 microhaplotypes was shown in Fig. 4. It could be seen that all alleles could be observed for these 33 microhaplotypes when mixture ratio ranged from 1:1 to 1:4. When mixing ratios reached 1:9, loci detection ratio was more than 96%. In addition, more than one third of loci could be still detected when mixing ratios were 1:19 and 1:49.
Fig. 4.
Loci detection rates of 33 microhaplotypes in different mixed ratios of 9948 and 9947 A samples. The hollow circle indicated the missing locus
Genetic distributions and forensic efficiency evaluation of the novel panel in the of Guizhou Han population
Genetic distributions and forensic statistical parameters of 33 microhaplotypes in the Guizhou Han population were assessed. Firstly, we assessed Hardy-Weinberg equilibrium (HWE) and linkage disequilibrium (LD) of these microhaplotypes in the Guizhou Han population, as presented in Supplementary Tables 5–6. For these 33 loci, we found that three loci (MH19WL-014, MH22WL-008, and MH03LV-01) did not conform to HWE in the Guizhou Han population after applying to Bonferroni correction (p < 0.05/33). Even so, no LD among these 33 loci was observed in the Guizhou Han population, implying that these loci could be employed as independent loci from each other.
Next, we evaluated allele frequency distributions of these 33 microhaplotypes in the Guizhou Han population, as shown in Fig. 5a. A total of 439 alleles were detected at these microhaplotypes, with the number of alleles ranging from 6 to 27. We also estimated Ae values of these 33 loci in the Guizhou Han population, as shown in Supplementary Table 5. The Ae values of 33 microhaplotype loci ranged from 3.60 to 8.72, and the average values were 6.06. Except for MH04WL-041 (4.59), MH09WL-005 (3.6), and MH22WL-008 (4.19), the Ae values of the other loci were greater than 5. The forensic parameters of these loci were shown in Fig. 5b and Supplementary Table 5. The average Ho, He, PIC, PM, PD and PE values of these 33 loci in the Guizhou Han population were 0.8112, 0.8316, 0.8084, 0.0554, 0.9446 and 0.6286, respectively. In addition, we found that the MH01WL-048 locus showed the highest He, PIC, and PD values; whereas, the MH09WL-005 locus displayed the lowest He, PIC, and PD values. The cumulative PD and PE values of these 33 microhaplotypes in the Guizhou Han population were 1-5.6 × 10− 43 and 1-1.6 × 10− 15, respectively, implying that the novel panel could be viewed as the high-efficient tool for forensic individual identification and paternity testing in the Guizhou Han population.
Fig. 5.
Allele frequency distributions (a) and forensic parameters (b) of the 33 microhaplotypes in the Guizhou Han population
Performance evaluation of the novel panel for assessing different degree of kinships
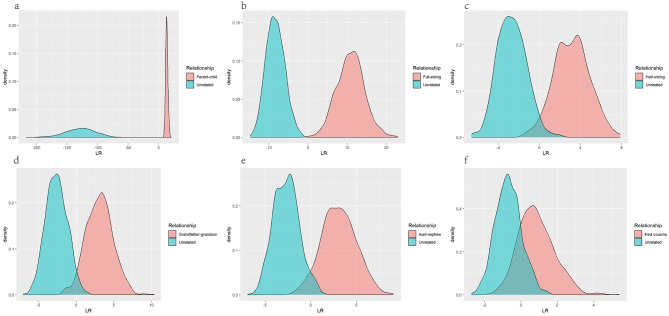
By simulating six different kinships (parent-child, full-sibling, half-sibling, grandparent-grandson, aunt-nephew, and first cousins), we further evaluated the performance of the developed panel for differentiating these kinships from unrelated individuals. The density plot of Log10(LR) of each kinship and unrelated individuals was shown in Fig. 6. Furthermore, we also set different thresholds to evaluate the system efficacy of the novel panel for assessing these kinships, as listed in Table 2. For parent-child, no overlapped between simulated parent-child and unrelated-individual cases was observed (Fig. 6a). In addition, all parent-child pairs could be correctly predicted as parent-child regardless of the thresholds. For full-sibling, there were no overlapping between the density plots of full-sibling and unrelated-individual cases (Fig. 6b). The PPV and NPV of the panel for full-sibling cases were 100% and 100% when the threshold was set to -1/1 or -2/2. Less than 1% full-sibling or unrelated cases were identified as uncertainty cases. For half-sibling, grandfather-grandson, and aunt-nephew kinships, a slight overlapped area between these kinships and unrelated individuals were observed for the density plot of Log10(LR) (Fig. 6c-e). The system effectiveness of the panel for half-sibling, grandfather-grandson, and aunt-nephew kinships was basically over 70% regardless of the threshold set to -1/1 or -2/2. For first cousins, almost one-third overlapped area between first cousins and unrelated-individual cases was seen from the density plot (Fig. 6f). Besides, the sensitivity and specificity of the system in identifying first cousins and unrelated individuals were low, and the uncertainty rate for first cousin cases increased significantly. Under the threshold value of -1/1, the efficiency of the system for first cousin kinships was only 37.25%.
Fig. 6.
The density plot of Log10(LR) for different relationships. Log10(LR) were drawn for true relationships (blue curve) and true unrelated pairs (pink curves)
Table 2.
System powers of the 33 microhaplotypes for different pairwise kinships at different thresholds
| Relationship | Threshold (t1\t2) | Sensitivity | Specificity | PPV | NPV | Error rate | Uncertainty rate | Effectiveness |
|---|---|---|---|---|---|---|---|---|
| Parent-child | -1\1 | 100.00% | 100.00% | 100.00% | 100.00% | 0.00% | 0.00% | 100.00% |
| -2\2 | 100.00% | 100.00% | 100.00% | 100.00% | 0.00% | 0.00% | 100.00% | |
| Full-sibling | -1\1 | 99.70% | 100.00% | 100.00% | 100.00% | 0.00% | 0.30% | 99.85% |
| -2\2 | 99.40% | 99.50% | 100.00% | 100.00% | 0.00% | 0.60% | 99.45% | |
| Half-sibling | -1\1 | 89.90% | 88.10% | 99.01% | 99.32% | 0.75% | 4.75% | 89.00% |
| -2\2 | 73.60% | 68.30% | 99.86% | 100.00% | 0.05% | 13.20% | 70.95% | |
| Grandfather-grandson | -1\1 | 88.50% | 86.40% | 99.55% | 98.18% | 1.00% | 4.95% | 87.45% |
| -2\2 | 73.10% | 67.10% | 100.00% | 100.00% | 0.00% | 13.45% | 70.10% | |
| Aunt-nephew | -1\1 | 86.70% | 88.90% | 99.66% | 98.45% | 0.85% | 5.95% | 87.80% |
| -2\2 | 69.80% | 69.50% | 100.00% | 99.86% | 0.05% | 15.05% | 69.65% | |
| First cousins | -1\1 | 40.60% | 33.90% | 97.60% | 94.69% | 1.45% | 28.75% | 37.25% |
| -2\2 | 13.00% | 3.10% | 100.00% | 100.00% | 0.00% | 43.50% | 8.05% |
The efficiency of the novel panel for inferring number of contributors of mixtures
The NOC for mixtures is the pivotal index for mixture deconvolution, which usually need to be firstly determined before dissecting alleles of each contributor. In this study, we also estimated the power of the novel panel to infer NOCs of mixtures consisting of 1–5 contributors.
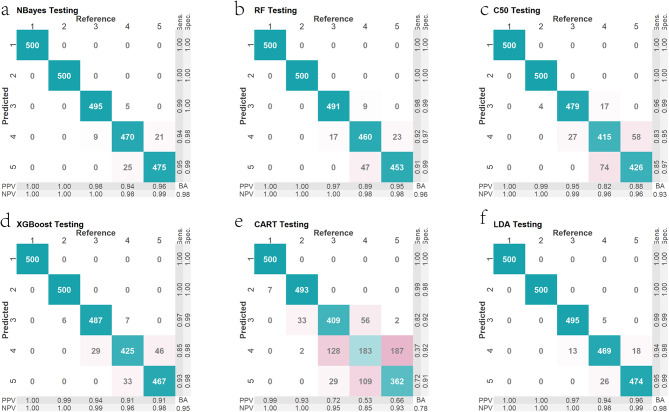
Firstly, we conducted simulations of mixture samples ranging from 1 to 5 contributors based on allelic frequencies of 33 microhaplotypes in the Guizhou Han population by the R software. Next, a set of features were defined according to the number of alleles observed in each locus. These features included the number of alleles per locus, the maximum allele count, the minimum allele count, total allele count, mean allele count, and median allele count. Thirdly, based on these features observed in simulated mixtures, we constructed six different machine learning models to evaluate the efficiency of the novel panel to infer NOCs of these mixtures. The predicted and actual NOCs in training samples were shown in Supplementary Fig. 3. Results revealed that the NOC of most samples could be correctly estimated, especially for models built by Naive Bayes, random forest, decision tree, XGBoost, and linear discriminant analysis algorithms. Confusion matrix of predicted and actual NOCs in testing samples was shown in Fig. 7. We found that these models displayed good performance for inferring NOCs of mixed samples (1–5 contributors) except for classification and regression trees.
Fig. 7.
Confusion matrices of predicted and actual results for the number of contributors in testing samples by the Naive Bayes (a), random forest (b), decision tree (c), XGBoost (d), classification and regression trees (e) and linear discriminant analysis methods (f)
Discussion
Microhaplotype, as the novel genetic markers, showed great potential in forensic genetics given that it had the advantages of high genetic diversities and extremely low mutation rate. In the current study, we assembled a set of microhaplotypes with highly polymorphisms in East Asian population and developed a small panel of 33 microhaplotype loci and a sex-determination locus for forensic genetics based on the NGS technology. For the novel panel, He and PIC values of 33 microhaplotypes were more than 0.7000 and 0.6500 in the Guizhou Han population, indicating that these loci showed highly genetic diversities in the Guizhou Han population. In addition, the average Ae value of these loci in the Guizhou Han population was more than 6, which were superior to the results reported by Wu et al. [18] (the average Ae value was 5.44) and Yang et al. [22] (the average Ae value was 4.41), implying that the panel had better efficiency for dissecting mixtures. As listed in Supplementary Table 7, CPD and CPE values of different panels were also compared. We found that the panel in this study showed higher CPD and CPE values than the a previously developed 40-plex panel [22]. Even CPD and CPE values in this study were less than other studies [15, 47, 48], which might be related to more microhaplotypes included in these studies. Thus, the panel in this study is expected to reach comparable or greater performance in comparison to these studies if the panel had the same number of microhaplotypes. More critically, the developed panel displayed high sensitivity, good resolution for two mixtures and species specificity. From the above results, the developed panel could be viewed as high-efficient tool for forensic personal identification and paternity testing in the Guizhou Han population.
For population genetics of different continental populations, we found that selected 33 microhaplotypes could achieve ancestral resolution for different continental populations, especially for African, European, East Asian and South Asian populations. Accordingly, these 33 microhaplotypes were not only highly genetic diversities in these continental populations, they also could be viewed as ancestry informative markers to infer biogeographical origins of these populations.
In the simulation of kinship analysis, the discriminatory efficiency of the novel panel for parent-child relationships and unrelated individuals was 100% at 1/-1 and 2/-2 thresholds. However, the system effectiveness of the panel gradually decreased with the degree of kinships increased, especially for first cousins and unrelated individual cases. In a previous study, Du et al. developed a 188-plex panel for 2nd-degree kinship testing [49]. They found that 188 microhaplotypes could obtain clear opinions for 83.36% of 2nd-degree identifications; whereas, these loci were not enough to assess 3rd-degree kinship (like first cousin), and more loci (more than 800) were needed to identify the 3rd-degree kinship. In order to improve the identification efficiency of distant relatives, the panel in this study need to be further optimized by the following methods: (1) combined with different types of genetic markers, like autosomal STR, SNP, InDels, and allosomal genetic markers; (2) added more microhaplotypes with highly genetic diversities in the existing panel.
The machine learning algorithm has been proved to be feasible in inferring the NOC to the mixtures [22]. In the typing results of the mixed samples, the number of unique alleles is the key to inferring NOC of the mixture [16, 50]. In this study, we also employed six machine learning algorithms to evaluate the efficiency of the novel panel in deducing NOC based on the number of unique alleles per locus. The results showed a slight increase in the frequency of false estimates as NOCs increased, especially for the classification and regression trees model. Even so, high accuracy could be achieved by the remaining five machine learning methods. Accordingly, Naive Bayes, random forest, decision tree, XGBoost, and linear discriminant analysis methods could be used to construct models for predicting NOCs of mixed samples based on the novel panel, which could provide more valuable information for mixture deconvolution. However, NOC inferences were affected by mixed ratio, mixture concentration, mixture status, and other factors in forensic practice. Therefore, the performance of the novel panel for predicting NOCs should be further evaluated in more forensic-related biological samples.
Conclusion
In this study, we have developed a novel panel of 33 microhaplotypes and a sex-determining locus for forensic genetics based on the NGS platform. The novel panel not only showed good sensitivity and species specificity, it was also suitable for mixture deconvolution and complex kinships testing. More importantly, microhaplotypes included in the panel were also used as ancestry informative markers for biogeographical origin inferences of different continental populations. To conclusion, the novel panel could be viewed as an independent and high-efficient tool for forensic genetics.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Material 1: Figure S1. Population genetic structure analyses of 26 reference populations with K values ranging from 2 to 7 based on selected 33 microhaplotypes
Supplementary Material 2: Figure S2. Mean LnP(K) values of each K value.
Supplementary Material 3: Figure S3. Confusion matrices of predicted and actual results for the number of contributors in training samples by the Naive Bayes (a), random forest (b), decision tree (c), XGBoost (d), classification and regression trees (e) and linear discriminant analysis methods (f).
Acknowledgements
Not applicable.
Abbreviations
- Ae
Average effective number of alleles
- STR
Short tandem repeat
- SNP
Single nucleotide polymorphism
- NOC
Number of contributor
- NGS
Next-Generation Sequencing
- He
Expected heterozygosity
- Ho
Observed heterozygosity
- PIC
Polymorphic information content
- PM
Match probability
- PD
Power of discrimination
- PE
Probability of exclusion
- CPM
Cumulative match probability
- CPE
Cumulative probability of exclusion
- MDS
Multidimensional scaling analysis
- LR
Likelihood ratio
- PPV
Positive predictive value
- NPV
Negative predictive value
- DoC
Depth of coverage
- ACR
Allele coverage ratio
Author contributions
GCY wrote the main text; HXL, CL, TSY, RQC and ZM collected samples; HWP, WK and SDL performed experiment; GCY conducted statistical analysis; RZ, WQY, YMQ, JJY and LYB commented on the manuscript and provided valuable feedback. JXY and ZHL revised the manuscript; HJ, ZHL and JXY designed the work and provided the conception. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Guizhou Provincial Science and Technology Projects (ZK [2024] General 162, ZK [2022] General 355, and Qian Foundation [2024] Youth 240), Guizhou Education Department Young Scientific and Technical Talents Project, Qian Education KY NO. [2022] 215, Guizhou Scientific Support Project, Qian Science Support [2021] General 448, Guizhou “Hundred” High-level Innovative Talent Project, Qian Science Platform Talents [2020] 6012, National Natural Science Foundation [No. 82160324] and the Guizhou Innovation training program for college students (S202310660094, S202210660028).
Data availability
The original contributions presented in the study are publicly available. The datasets generated and/or analysed during the current study are available in the CNGB Sequence Archive (CNSA) of the China National Genebank Database, accession number CNP0006055. [https://db.cngb.org/].
Declarations
Ethics approval and consent to participate
The research adhered to the guidelines outlined in the Declaration of Helsinki. Informed consent was obtained from all individual participants included in the study. Additionally, this study was approved by the Ethics Committee of Guizhou Medical University (No. 2021 − 224).
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jiang Huang, Email: mmm_hj@126.com.
Hongling Zhang, Email: 229598103@qq.com.
Xiaoye Jin, Email: 1115259825@qq.com.
References
- 1.Tao RY, Dong XY, Chen AQ, Lü YH, Zhang SH, Li CT. Application progress of massively parallel sequencing technology in STR genetic marker detection. Fa Yi Xue Za Zhi. 2022;38(2):267–79. [DOI] [PubMed] [Google Scholar]
- 2.Warshauer DH, Churchill JD, Novroski N, King JL, Budowle B. Novel Y-chromosome short Tandem repeat variants detected through the Use of massively parallel sequencing. Genomics Proteom Bioinf. 2015;13(4):250–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun S, Liu Y, Li J, Yang Z, Wen D, Liang W, Yan Y, Yu H, Cai J, Zha L. Development and application of a nonbinary SNP-based microhaplotype panel for paternity testing involving close relatives. Forensic Sci Int Genet. 2020;46:102255. [DOI] [PubMed] [Google Scholar]
- 4.Zhou J, Wang Y, Xu E. Research progress on application of microhaplotype in forensic genetics. Zhejiang Da Xue Xue Bao Yi Xue Ban. 2021;50(6):777–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chandra D, Mishra VC, Raina A, Raina V. Mutation rate evaluation at 21 autosomal STR loci: paternity testing experience. Leg Med (Tokyo). 2022;58:102080. [DOI] [PubMed] [Google Scholar]
- 6.Martinez J, Braganholi DF, Ambrósio IB, Polverari FS, Cicarelli RMB. Mutation rates for 20 STR loci in a population from São Paulo state, Southeast, Brazil. Ann Hum Biol. 2017;44(7):659–62. [DOI] [PubMed] [Google Scholar]
- 7.Puch-Solis R, Pope S, Tully G. Considerations on the application of a mutation model for Y-STR interpretation. Sci Justice. 2024;64(2):180–92. [DOI] [PubMed] [Google Scholar]
- 8.Ge J, Budowle B, Planz JV, Chakraborty R. Haplotype block: a new type of forensic DNA markers. Int J Legal Med. 2010;124(5):353–61. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y, Wang S, He H, Wang X, Zhu D, Wen X, Zhang S. Evaluation of three microhaplotypes in individual identification and ancestry inference. Forensic Sci Int. 2021;320:110681. [DOI] [PubMed] [Google Scholar]
- 10.Wen D, Xing H, Liu Y, Li J, Qu W, He W, Wang C, Xu R, Liu Y, Jia H, Zha L. The application of short and highly polymorphic microhaplotype loci in paternity testing and sibling testing of temperature-dependent degraded samples. Front Genet. 2022;13:983811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pontes L, Sousa JC, Medeiros R. SNPs and STRs in forensic medicine. A strategy for kinship evaluation. Arch Med Sadowej Kryminol. 2017;67(3):226–40. [DOI] [PubMed] [Google Scholar]
- 12.Lee HJ, Lee JW, Jeong SJ, Park M. How many single nucleotide polymorphisms (SNPs) are needed to replace short tandem repeats (STRs) in forensic applications? Int J Legal Med. 2017;131(5):1203–10. [DOI] [PubMed] [Google Scholar]
- 13.Phillips C, Amigo J, Tillmar AO, Peck MA, de la Puente M, Ruiz-Ramírez J, Bittner F, Idrizbegović Š, Wang Y, Parsons TJ, Lareu MV. A compilation of tri-allelic SNPs from 1000 genomes and use of the most polymorphic loci for a large-scale human identification panel. Forensic Sci Int Genet. 2020;46:102232. [DOI] [PubMed] [Google Scholar]
- 14.Kidd KK, Pakstis AJ, Speed WC, Lagace R, Chang J, Wootton S, Ihuegbu N. Microhaplotype loci are a powerful new type of forensic marker. Forensic Sci International: Genet Supplement Ser. 2013;4(1):e123–4. [Google Scholar]
- 15.Zhang R, Xue J, Tan M, Chen D, Xiao Y, Liu G, Zheng Y, Wu Q, Liao M, Lv M et al. An MPS-Based 50plex microhaplotype assay for forensic DNA analysis. Genes (Basel). 2023;14(4):865. [DOI] [PMC free article] [PubMed]
- 16.Wang H, Zhu Q, Huang Y, Cao Y, Hu Y, Wei Y, Wang Y, Hou T, Shan T, Dai X et al. Using simulated microhaplotype genotyping data to evaluate the value of machine learning algorithms for inferring DNA mixture contributor numbers. Forensic Sci Int Genet. 2024;69:103008. [DOI] [PubMed]
- 17.Kidd KK, Speed WC. Criteria for selecting microhaplotypes: mixture detection and deconvolution. Investig Genet. 2015;6:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu R, Li H, Li R, Peng D, Wang N, Shen X, Sun H. Identification and sequencing of 59 highly polymorphic microhaplotypes for analysis of DNA mixtures. Int J Legal Med. 2021;135(4):1137–49. [DOI] [PubMed] [Google Scholar]
- 19.Kidd KK, Pakstis AJ. State of the art for microhaplotypes. Genes (Basel). 2022;13(8):1322. [DOI] [PMC free article] [PubMed]
- 20.Jin X, Zhang X, Shen C, Liu Y, Cui W, Chen C, Guo Y, Zhu B. A highly polymorphic panel consisting of microhaplotypes and compound markers with the NGS and its forensic efficiency evaluations in Chinese two groups. Genes(Basel). 2020;11(9):1027. [DOI] [PMC free article] [PubMed]
- 21.Tomas C, Rodrigues P, Jønck CG, Barekzay Z, Simayijiang H, Pereira V, Børsting C. Performance of a 74-Microhaplotype assay in kinship analyses. Genes(Basel). 2024;15(2):224. [DOI] [PMC free article] [PubMed]
- 22.Yang J, Chen J, Ji Q, Yu Y, Li K, Kong X, Xie S, Zhan W, Mao Z, Yu Y et al. A highly polymorphic panel of 40-plex microhaplotypes for the Chinese Han population and its application in estimating the number of contributors in DNA mixtures. Forensic Sci Int Genet. 2022;56:102600. [DOI] [PubMed]
- 23.Zhang Y, Wang S, He H, Wang X, Zhu D, Wen X, Zhang S. Evaluation of three microhaplotypes in individual identification and ancestry inference. Forensic Sci Int. 2021;320:110681. [DOI] [PubMed]
- 24.Zhao X, Fan Y, Zeye MMJ, He W, Wen D, Wang C, Li J, Hua Z. A novel set of short microhaplotypes based on non-binary SNPs for forensic challenging samples. Int J Legal Med. 2022;136(1):43–53. [DOI] [PubMed] [Google Scholar]
- 25.Kureshi A, Li J, Wen D, Sun S, Yang Z, Zha L. Construction and forensic application of 20 highly polymorphic microhaplotypes. R Soc Open Sci. 2020;7(5):191937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qu N, Lin S, Gao Y, Liang H, Zhao H, Ou X. A microhap panel for kinship analysis through massively parallel sequencing technology. Electrophoresis. 2020;41(3–4):246–53. [DOI] [PubMed] [Google Scholar]
- 27.Staadig A, Tillmar A. Evaluation of microhaplotypes in forensic kinship analysis from a Swedish population perspective. Int J Legal Med. 2021;135(4):1151–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oldoni F, Yoon L, Wootton SC, Lagacé R, Kidd KK, Podini D. Population genetic data of 74 microhaplotypes in four major U.S. population groups. Forensic Sci Int Genet. 2020;49:102398. [DOI] [PubMed] [Google Scholar]
- 29.Bulbul O, Pakstis AJ, Soundararajan U, Gurkan C, Brissenden JE, Roscoe JM, Evsanaa B, Togtokh A, Paschou P, Grigorenko EL, et al. Ancestry inference of 96 population samples using microhaplotypes. Int J Legal Med. 2018;132(3):703–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bai Z, Zhang N, Liu J, Ding H, Zhang Y, Wang T, Gao J, Ou X. Identification of missing persons through kinship analysis by microhaplotype sequencing of single-source DNA and two-person DNA mixtures. Forensic Sci Int Genet. 2022;58:102689. [DOI] [PubMed] [Google Scholar]
- 31.Jin XY, Liu YF, Cui W, Chen C, Zhang XR, Huang J, Zhu BF. Development a multiplex panel of AISNPs, multi-allelic InDels, microhaplotypes, and Y-SNP/InDel loci for multiple forensic purposes via the NGS. Electrophoresis. 2022;43(4):632–44. [DOI] [PubMed] [Google Scholar]
- 32.Yu WS, Feng YS, Kang KL, Zhang C, Ji AQ, Ye J, Wang L. Screening of highly discriminative microhaplotype markers for individual identification and mixture deconvolution in east Asian populations. Forensic Sci Int Genet. 2022;59:102720. [DOI] [PubMed] [Google Scholar]
- 33.Standage DS, Mitchell RN. MicroHapDB: a portable and extensible database of all published microhaplotype marker and frequency data. Front Genet. 2020;11:781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fan H, Xie Q, Wang L, Ru K, Tan X, Ding J, Wang X, Huang J, Wang Z, Li Y, et al. Microhaplotype and Y-SNP/STR (MY): a novel MPS-based system for genotype pattern recognition in two-person DNA mixtures. Forensic Sci Int Genet. 2022;59:102705. [DOI] [PubMed] [Google Scholar]
- 35.Rychlik W. OLIGO 7 primer analysis software. Methods Mol Biol. 2007;402:35–60. [DOI] [PubMed] [Google Scholar]
- 36.Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gouy A, Zieger M. STRAF—A convenient online tool for STR data evaluation in forensic genetics. Forensic Sci Int Genet. 2017;30:148–51. [DOI] [PubMed] [Google Scholar]
- 40.Rousset F. Genepop’007: a complete re-implementation of the genepop software for Windows and Linux. Mol Ecol Resour. 2008;8(1):103–6. [DOI] [PubMed] [Google Scholar]
- 41.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing platforms. Mol Biol Evol. 2018;35(6):1547–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164(4):1567–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Earl DA, vonHoldt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour. 2012;4(2):359–61. [Google Scholar]
- 44.Mattias J. A RN: CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23(14):1801–6. [DOI] [PubMed] [Google Scholar]
- 45.Francis RM. Pophelper: an R package and web app to analyse and visualize population structure. Mol Ecol Resour. 2017;17(1):27–32. [DOI] [PubMed] [Google Scholar]
- 46.Kling D, Tillmar AO, Egeland T. Familias 3 - extensions and new functionality. Forensic Sci Int Genet. 2014;13:121–7. [DOI] [PubMed] [Google Scholar]
- 47.Tao R, Yang Q, Xia R, Zhang X, Chen A, Li C, Zhang S. A sequence-based 163plex microhaplotype assay for forensic DNA analysis. Front Genet. 2022;13:988223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pang JB, Rao M, Chen QF, Ji AQ, Zhang C, Kang KL, Wu H, Ye J, Nie SJ, Wang L. A 124-plex Microhaplotype Panel based on next-generation sequencing developed for forensic applications. Sci Rep. 2020;10(1):1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Du Q, Ma G, Lu C, Wang Q, Fu L, Cong B, Li S. Development and evaluation of a novel panel containing 188 microhaplotypes for 2nd-degree kinship testing in the hebei han population. Forensic Sci Int Genet 2023;65:102855. [DOI] [PubMed]
- 50.Marciano MA, Adelman JD. PACE: Probabilistic Assessment for Contributor Estimation— A machine learning-based assessment of the number of contributors in DNA mixtures. Forensic Sci Int Genet. 2017;27:82–91. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1: Figure S1. Population genetic structure analyses of 26 reference populations with K values ranging from 2 to 7 based on selected 33 microhaplotypes
Supplementary Material 2: Figure S2. Mean LnP(K) values of each K value.
Supplementary Material 3: Figure S3. Confusion matrices of predicted and actual results for the number of contributors in training samples by the Naive Bayes (a), random forest (b), decision tree (c), XGBoost (d), classification and regression trees (e) and linear discriminant analysis methods (f).
Data Availability Statement
The original contributions presented in the study are publicly available. The datasets generated and/or analysed during the current study are available in the CNGB Sequence Archive (CNSA) of the China National Genebank Database, accession number CNP0006055. [https://db.cngb.org/].