Abstract
Phylogenetic analyses of retroviral elements, including endogenous retroviruses, have relied essentially on the retroviral pol gene expressing the highly conserved reverse transcriptase. This enzyme is essential for the life cycle of all retroid elements, but other genes are also endowed with conserved essential functions. Among them, the transmembrane (TM) subunit of the envelope gene is involved in virus entry through membrane fusion. It has also been reported to contain a domain, named the immunosuppressive domain, that has immunosuppressive properties most probably essential for virus spread within the host. This domain is conserved among a large series of retroviral elements, and we have therefore attempted to generate phylogenetic links between retroviral elements identified from databases following tentative alignments of the immunosuppressive domain and adjacent sequences. This allowed us to unravel a conserved organization among TM domains, also found in the Ebola and Marburg filoviruses, and to identify a large number of human endogenous retroviruses (HERVs) from sequence databases. The latter elements are part of previously identified families of HERVs, and some of them define new families. A general phylogenetic analysis based on the TM proteins of retroelements, and including those with no clearly identified immunosuppressive domain, could then be derived and compared with pol-based phylogenetic trees, providing a comprehensive survey of retroelements and definitive evidence for recombination events in the generation of both the endogenous and the present-day infectious retroviruses.
Among the gag, pol, and env retroviral genes, the pol gene, encoding reverse transcriptase (RT), is by far the most conserved among the retroid elements (33). RT is actually the key enzyme in the retroviral replicative cycle, being involved in the synthesis of the proviral DNA from the viral RNA genome. Due to most probably very stringent constraints for enzymatic activity, this gene is highly conserved not only among retroviral elements but also among a large series of elements requiring a reverse transcription step, including endogenous retroviruses (ERVs) and retrotransposons, group II introns, and the cellular telomerase genes, as well as some plasmidic elements from procaryotes (51). Consequently, sequence alignments including the RT domains from these diverse elements have led to the unraveling of phylogenetic links between them (51). Furthermore, RT contains signature motifs allowing an easy search for RT-containing elements within genomes, especially in the case of humans, where systematic sequencing should now enable rapid and extensive identification of retroelements. Accordingly, it has been shown that the human genome contains numerous ERVs (HERVs) distributed in several multigenic families comprising a few to several hundreds elements (26, 45, 48). These elements are hallmarks of ancient infections of the germ line by retroviruses which have thereafter been “endogenized” and can be used as molecular markers of evolution (4, 21).
In contrast to the pol gene, the env gene, encoding the protein involved in virus entry, has long been considered a highly diverging sequence in relation to the highly diverse sequences of the receptor molecules with which the env proteins interact for virus-cell interaction and entry. The env gene encodes a polypeptide which is cleaved into two proteins (Fig. 1), the surface protein (SU), which is involved in receptor recognition, and the transmembrane (TM) subunit, which anchors the whole env complex to the membrane and is directly responsible for cell membrane fusion and virus entry. TM structures have been elucidated in the case of Moloney murine leukemia virus (Mo-MuLV) (15), human immunodeficiency virus type 1 (HIV-1) (10, 47), and human T-cell leukemia virus type 1 (HTLV-1) (23) and show a highly conserved organization also found in proteins of nonretroviral elements such as influenza virus (50) and Ebola virus (28). This structural conservation is most probably relevant to a common mechanism for the triggering of the fusion process and viral entry (9, 17). Finally, there is a region with significant homology among retroviruses, namely, the immunosuppressive domain, so called because 17-mer peptides derived from this relatively conserved sequence have immunosuppressive properties as assayed in vitro by their effects on the proliferation and/or differentiation of lymphocytes (12, 41). We have recently shown that the env protein of the murine Mo-MuLV and the primate Mason-Pfizer monkey virus (MPMV) are actually immunosuppressive in vivo, based on an assay involving rejection of tumor cells engrafted into immunocompetent mice (6, 30). Moreover, we have shown that an HERV envelope is also immunosuppressive in this assay, thus strengthening the importance of this domain (30a). Taking into account this conservation, we have therefore attempted to identify from databases all of the sequences showing an immunosuppressive domain. Doing so, we have been able to align the TMs of most retroviral elements and generate phylogenetic trees including both endogenous and exogenous retroviruses. Comparison with pol-based phylogenetic trees provides hints of definite recombination events for both endogenous and infectious retroviruses in the course of their natural history.
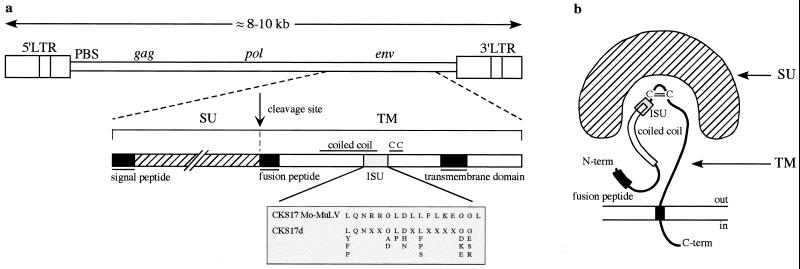
FIG. 1.
Schematic representation of the proviral form of a retrovirus and its env gene products. (a) Genomic proviral structure and delineation of the TM subunit encoded by the env gene. The Mo-MuLV immunosuppressive domain (ISU) is shown, as is the degenerate CKS17d motif used for the initial screening of the databases (see text and Table 1; an extended optimized universal CKS17u motif [see Materials and Methods] which recognizes at least one member from each retroelement family of the CKS17-positive group in Fig. 3 and 5 was also devised). (b) Schematic structure of the env products, adapted from reference 17. CC, disulfide bond.
MATERIALS AND METHODS
Screening for sequences encoding a CKS17-like domain.
The BioMotif program (G. Mennessier, http://www.lpm.univ-montp2.fr/software.html) searches for protein motifs along the six frames of nucleotide sequences. The designed motifs can be degenerate. The program allows frameshifts but not mismatches. Motifs used for the screenings are (using the BioMotif syntax, with | for degenerate positions, X for any amino acid excluding stop codons, and a triplet of n for any amino acid, including stop codons) as follows: the degenerate immunosuppressive CKS17d consensus motif (L|Y|F|P) QN [6,6]n (G|A|D) (L|P) (D|H|N) [3,3]n (L|F|P|S) [12,12]n (G|D|K|E) (G|E|S|R), an optimized universal CKS17u motif (L|Y|F|P|W|A) (Q|N|E)N [6,6]n (G|A|D|M) (L|P|I) (D|H|N) [3,3]n (L|F|P|S|V|T|I) [12,12]n (G|D|K|E|S) (G|E|S|R|H) designed from the general TM alignment, and the reverse transcriptase RT7 consensus motif LPQ [57,162]n YXDD, which allows frameshifts between the LPQ and YXDD residues. This search combines amino acid residues by codon translation and nucleotide gaps.
LTR test.
The long terminal repeat (LTR) test is based on the LFASTA program. Two-LTR structures were searched for on extracted sequences 9 kb upstream and 3 kb downstream of the CKS17d motif with the following parameters: greater than 80% identity between the two LTRs, which could be 350 to 1,000 nucleotides long and separated by 3 to 10 kb.
PBS search.
Potential primer binding sites (PBS) were searched for with the BLAST program (1) using a database of tRNAs (http://www.uni-bayreuth.de/departments/biochemie/sprinzl/trna/index.html) in the region downstream of the 5′ LTR of sequences positive for a two-LTR structure.
Clusterization.
The 544 sequences were extracted on a 120-nucleotide segment upstream and downstream of the CKS17d motif and translated, since the program determines clusters on peptide sequences to avoid degeneracy of the genetic code. The sequences were clustered by the means of pair comparison with the LFASTA program and subsequent classification in groups with a minimum of 90% identity.
Alignments.
The Clustalw program (44) was used to perform multiple alignments, which were manually refined with the Seaview program (18; http://pbil.univ-lyon1.fr/software/seaview.html).
Frame search.
We used the Framesearch program in the Wisconsin package, version 10.0 (Genetics Computer Group, Madison, Wis.). It searches for the correct frame in a nucleotide sequence compared to a protein sequence. It may change the frame if needed, which is an important feature for defective HERV sequences which can encompass frameshift mutations.
Prediction of coiled coils.
The LearnCoil-VMF program developed by Singh et al. (38; http://nightingale.lcs.mit.edu/cgi-bin/vmf) for identifying coiled-coil-like regions in viral membrane fusion protein envelopes was applied to TM sequences.
Hydrophobicity plots.
Hydropathy was calculated by the Kyte-Doolittle method implemented with the DNA Strider program (31).
Phylogenetic methods.
The phylogenetic methods used were from the PHYLIP (Phylogeny Inference Package) version 3.5c developed by Joseph Felsenstein (16) and the University of Washington (http://evolution.genetics.washington.edu/phylip.html). For the distance method, the Dnadist program with the Kimura two-parameter correction (CKS17 nucleotide tree) or the Protdist program with the PAM matrix correction of M. Dayhoff (TM and RT trees), both followed by the Neighbor program (neighbor-joining method), were run on 100 bootstrap replicates, and then the Fitch program was used to obtain proportional branch lengths in the calculated trees. For the parsimony method, the Dnapars and Protpars programs were run on 100 bootstrap replicates.
RESULTS
Initial screening for env sequences with an immunosuppressive motif and rationale of the search procedure.
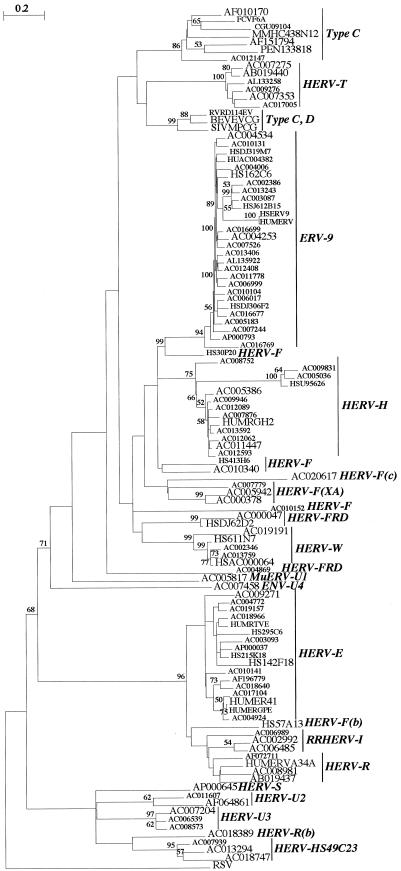
To screen the databases for envelopes, we first designed a common motif based on the Mo-MuLV CKS17 immunosuppressive domain. To this end, TMs of known retroviral envelopes of exogenous and endogenous origin were aligned, and a consensus degenerate CKS17d motif was designed (Fig. 1) (see Materials and Methods). This motif was positive for the majority of known TMs, with a few exceptions, including HIV, mouse mammary tumor virus (MMTV), and the HERVs of the HERV-K family. Sequences from all divisions of GenBank were screened for the CKS17d motif using the BioMotif program, which is a highly sensitive approach that permits the detection of conserved, but not necessarily contiguous, amino acids. The positive hits obtained, essentially among mammals (457 out of 544) and with the majority of human origin as expected from the relative abundance of human sequences in the databases, were then analyzed for additional criteria: (i) an RT motif (RT7 motif, see below) upstream of the CKS17d motif (still using the BioMotif program) and (ii) a two-LTR structure, i.e., a typical proviral organization (using a two-LTR test program), combined with a search for a potential PBS downstream of the 5′LTR (using the BLAST program) (see Materials and Methods). As 544 sequences cannot be aligned, we reduced their number by clustering their translated sequences (see Materials and Methods). Finally, a set of 110 sequences (Table 1) was extracted based on a region of approximately 300 nucleotides centered on the CKS17d motif, which was aligned with the Clustalw program (44). Phylogenetic trees were determined by the neighbor-joining (Fig. 2) and parsimony (data not shown) methods, which allowed the assignment of each sequence within a definite retroelement family (Table 1; Fig. 2). The validity of the search procedure could be evaluated based on the well-characterized group of the HERVs, since 18 families were sorted out simply based on the CKS17 search, compared to the 22 families, including CKS17-negative ones, previously identified by Tristem (45) using an RT domain screen. Interestingly, new groups could be identified (Table 1; Fig. 2), namely, four new HERV families [HERV-T, HERV-F(c) (also comprising a murine sequence), HERV-U2, and HERV-U3] and a new murine ERV (MuERV) family (MuERV-U1). Conversely, several infectious retroviruses (e.g., HIV and MMTV) as well as some HERV families (e.g., HERV-K) were not identified by the CKS17 screen, for reasons mentioned above, but all of these sequences could finally be included in a larger (200-amino-acid) alignment (see below) which exceeded the sole CKS domain and comprised almost all of the TM sequence.
TABLE 1.
Set of 110 CKS17d-positive sequences from GenBank release 115
| IDa | CKS17db | RTc | LTRd | Family or viruse |
|---|---|---|---|---|
| AB019437 | 44076 (+) | + | + | HERV-R |
| AB019440 | 34816 (+) | + | + | HERV-T∗ |
| AC000047 | 30757 (+) | + | HERV-FRD | |
| AC000378 | 94638 (+) | + | + | HERV-F(XA) |
| AC002346 | 84258 (+) | + | + | HERV-W |
| AC002386 | 73583 (−) | + | ERV9 | |
| AC002992 | 17206 (+) | + | RRHERV-I | |
| AC003087 | 46235 (+) | + | + | ERV9 |
| AC003093 | 25376 (−) | + | HERV-E | |
| AC004006 | 9034 (+) | + | ERV9 | |
| AC004253 | 9911 (+) | + | + | ERV9 |
| AC004534 | 81583 (−) | ERV9 | ||
| AC004772 | 83200 (+) | + | HERV-E | |
| AC004869 | 99245 (+) | + | HERV-FRD | |
| AC004924 | 89383 (+) | + | HERV-E | |
| AC005036 | 171165 (+) | HERV-H | ||
| AC005183 | 27600 (+) | + | ERV9 | |
| AC005386 | 178754 (+) | + | + | HERV-H |
| AC005817 | 97156 (−) | + | MuERV-UI∗ | |
| AC005942 | 22750 (+) | + | + | HERV-F(XA) |
| AC006017 | 142708 (−) | + | ERV9 | |
| AC006485 | 14188 (+) | + | RRHERV-I | |
| AC006539 | 12323 (−) | + | HERV-U3∗ | |
| AC006989 | 4728 (−) | RRHERV-I | ||
| AC006999 | 71755 (−) | + | ERV9 | |
| AC007204 | 10189 (−) | + | HERV-U3∗ | |
| AC007244 | 137080 (+) | ERV9 | ||
| AC007275 | 129167 (−) | + | HERV-T∗ | |
| AC007353 | 129702 (+) | + | + | HERV-T∗ |
| AC007458 | 8944 (−) | ENV-U4∗ | ||
| AC007526 | 97589 (+) | + | + | ERV9 |
| AC007779 | 89968 (+) | + | HERV-F(XA) | |
| AC007876 | 114202 (−) | + | + | HERV-H |
| AC007939 | 26365 (−) | + | HERV-HS49C23 | |
| AC008573 | 51728 (−) | HERV-U3∗ | ||
| AC008752 | 33833 (−) | + | HERV-H | |
| AC008981 | 26664 (−) | HERV-R | ||
| AC009271 | 92913 (+) | + | + | HERV-E |
| AC009276 | 33461 (−) | HERV-T∗ | ||
| AC009831 | 67534 (+) | HERV-H | ||
| AC009946 | 93859 (−) | + | HERV-W | |
| AC010104 | 60731 (−) | + | ERV9 | |
| AC010131 | 67248 (−) | + | ERV9 | |
| AC010141 | 120496 (−) | HERV-E | ||
| AC010152 | 57664 (−) | + | HERV-F | |
| AC010340 | 69706 (−) | + | HERV-F | |
| AC011447 | 133957 (−) | + | + | HERV-H |
| AC011607 | 170698 (+) | HERV-U2∗ | ||
| AC011778 | 104300 (−) | + | + | ERV9 |
| AC012062 | 13750 (+) | + | HERV-H | |
| AC012089 | 136656 (−) | + | HERV-H | |
| AC012147 | 174249 (+) | + | + | Type C |
| AC012408 | 156301 (+) | + | + | ERV9 |
| AC012593 | 152461 (+) | + | HERV-H | |
| AC013243 | 33932 (−) | + | + | ERV9 |
| AC013294 | 33838 (+) | + | HERV-HS49C23 | |
| AC013406 | 20757 (−) | + | + | ERV9 |
| AC013592 | 101976 (−) | + | HERV-H | |
| AC013759 | 144260 (+) | + | + | HERV-W |
| AC016677 | 152001 (−) | + | + | ERV9 |
| AC016699 | 53279 (−) | ERV9 | ||
| AC016769 | 8915 (+) | ERV9 | ||
| AC017005 | 45085 (+) | + | HERV-T∗ | |
| AC017104 | 72857 (−) | + | HERV-E | |
| AC018389 | 141038 (+) | + | HERV-R(b) | |
| AC018640 | 113549 (−) | + | HERV-E | |
| AC018747 | 40719 (−) | HERV-HS49C23 | ||
| AC018966 | 21146 (+) | + | HERV-E | |
| AC019157 | 137529 (+) | + | HERV-E | |
| AC019191 | 9682 (−) | HERV-W | ||
| AC020617 | 76159 (+) | HERV-F(c)∗ | ||
| AF010170 | 7946 (+) | + | + | Type C |
| AF064861 | 85519 (+) | + | HERV-U2∗ | |
| AF072711 | 11331 (−) | + | HERV-R | |
| AF151794 | 7502 (+) | + | + | Type C |
| AF196779 | 74784 (−) | + | HERV-E | |
| AL133258 | 47537 (+) | + | HERV-T∗ | |
| AL135922 | 2076 (−) | + | ERV9 | |
| AP000037 | 85657 (+) | + | HERV-E | |
| AP000645 | 20257 (+) | + | HERV-S | |
| AP000793 | 24743 (−) | ERV9 | ||
| BEVEVCG | 7552 (+) | + | + | BaEV |
| CGU09104 | 7879 (+) | + | + | Type C |
| FCVF6A | 7520 (+) | + | mRNA | Type C |
| HS142F18 | 82679 (+) | + | + | HERV-E |
| HS162C6 | 63858 (−) | + | + | ERV9 |
| HS215K18 | 30913 (−) | + | HERV-E | |
| HS295C6 | 22922 (+) | HERV-E | ||
| HS30P20 | 50221 (+) | + | HERV-F | |
| HS413H6 | 111259 (−) | HERV-F | ||
| HS57A13 | 50357 (−) | + | HERV-F(b) | |
| HS611N7 | 102072 (+) | + | HERV-W | |
| HSAC000064 | 37016 (+) | + | HERV-W | |
| HSDJ306F2 | 20288 (−) | ERV9 | ||
| HSDJ319M7 | 93125 (−) | + | + | HERV-H |
| HSDJ62D2 | 147474 (−) | HERV-FRD | ||
| HSERV9 | 2938 (+) | + | mRNA | ERV9 |
| HSJ612B15 | 7532 (+) | + | ERV9 | |
| HSU95626 | 65824 (−) | + | + | HERV-H |
| HUAC004382 | 171855 (−) | + | + | ERV9 |
| HUMER41 | 7853 (+) | + | + | HERV-E |
| HUMERGPE | 7476 (+) | + | HERV-E | |
| HUMERV | 2932 (+) | + | mRNA | ERV9 |
| HUMERVA34A | 2329 (+) | HERV-R | ||
| HUMRGH2 | 7659 (+) | + | HERV-H | |
| HUMRTVE | 3986 (+) | HERV-E | ||
| MMHC438N12 | 122487 (−) | + | Type C | |
| PEN133818 | 7683 (+) | + | + | Type C |
| RVRD114EV | 1927 (+) | RD114 | ||
| SIVMPCG | 7613 (+) | + | + | MPMV |
ID, Identifier.
Position and orientation (sense, +; antisense, −) of the CKS17d motif in the sequence, according to GenBank release 115 for the high-throughput-genomic sequences (and still unfinished in release 120) or to the definitive positions.
+, positive for the RT7 consensus motif.
+, positive for a proviral structure (two-LTR screen); mRNA, sequence corresponds to the RNA retroviral genome and not to the provirus.
Infectious retrovirus or endogenous retrovirus family (45), including families newly described in this paper (∗). ERVs are grouped in families designated by a letter corresponding to the amino acid whose tRNA is used as a primer for reverse transcription by annealing to the PBS. Families devoid of a PBS (in the CKS17d-positive sequences or in homologous ones found by BLAST searches) were thus designated U for unknown, followed by a number. In the ENV-U4 human family, only env sequences were found.
FIG. 2.
CKS17 nucleotide tree. A phylogenetic tree of a set of 110 nucleotide sequences positive for the CKS17d motif is shown, with the RSV sequence as an outgroup. This tree was determined by the neighbor-joining method with branch lengths proportional to the degrees of divergence between the sequences. Percent bootstrap values obtained from 100 replicates are indicated on the branches only when they are >50%. The names of the sequences correspond to the GenBank identifiers. The sequences retained for the TM alignment are indicated by larger characters. The HERV families or retrovirus types are indicated.
TM amino acid sequence alignment and phylogeny.
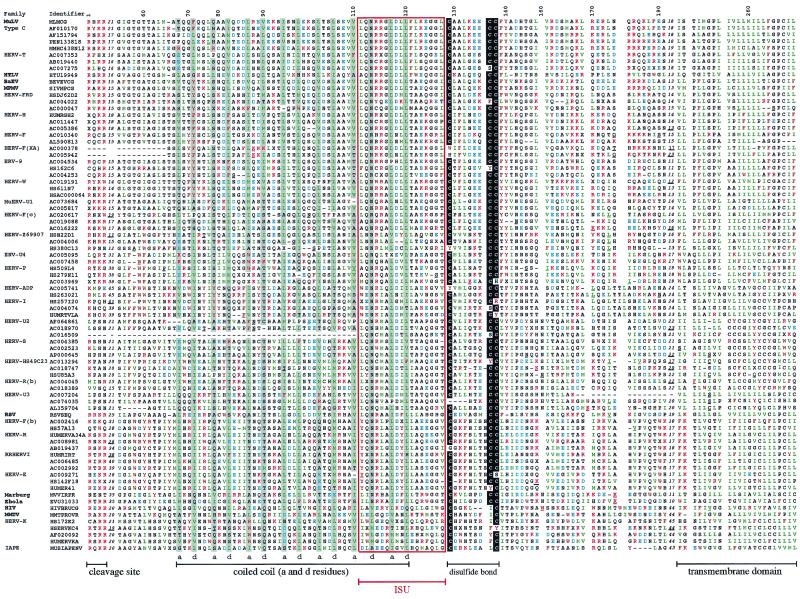
Although the TM primary sequences seem not to be conserved, biochemical analyses and X-ray crystallographic data for some retroviral TMs (HTLV-I gp21 [23], Mo-MuLV p15E [15], and HIV-1 gp41 [10, 47]) disclose a well-conserved general organization (Fig. 1), which includes, from the N to the C terminus, the following: (i) an extended hydrophobic region, generally A and G rich, at or near the amino terminus, corresponding to the fusion peptide (13 to 24 amino acids long) adjacent to the cleavage site (R-X-R/K-R) between the SU and the TM subunits; (ii) a coiled-coil-forming sequence which overlaps the immunosuppressive domain; (iii) an adjacent short disulfide-bonded loop; (iv) a variable C-terminal extracellular segment containing alpha-helical elements and numerous aromatic residues; (v) a hydrophobic region corresponding to the membrane-spanning domain, 19 to 27 amino acids long; and (vi) a cytoplasmic domain highly variable in both sequence and length. Based on these characteristic features, we attempted to align the extracellular and transmembrane domains of the TMs identified by the CKS17 screen (retaining, in Table 1, all members from small families and at least three members from large ones), as well as those of the other retroelements previously defined in the literature (45). The alignment (Fig. 3) was anchored on the central conserved cysteines of the internal disulfide-bonded loop together with, when present, the CKS17 domain and was then extended progressively to the rest of the sequences, taking advantage of the conserved residues or domains that had been identified by structural or biochemical approaches and making use of hydrophobic plots as well as of coiled-coil structure predictions that we made for each sequence (see Materials and Methods). As illustrated in Fig. 3, the resulting alignment shows a highly conserved organization. First, the overall lengths of the TMs, after exclusion of the cytoplasmic domain, are closely related. They can be bordered at the N terminus by the SU-TM cleavage site (R,X,R/K,R) and at the C terminus by the hydrophobic transmembrane domain (L/I/V/M amino acids in green). In the central part, the highly conserved cysteine residues can be found in almost all TMs, together with a large domain (approximately 50 amino acids) showing alignments of the a and d positions in the heptad repeats and corresponding to the predicted coiled-coil domains. The immunosuppressive domain, encompassing the C-terminal end of the coiled-coil region, can also be easily positioned, even among retroelements which do not possess a canonical CKS17-like sequence (e.g., those of HIV-1 and HERV-K). Some regions show only reduced conservation, among which is the domain corresponding to the fusion peptide located downstream of the SU-TM cleavage site, as well as the variable region just upstream of the transmembrane anchor. The latter domains also differ slightly in length (by approximately 20 amino acids) between retroelements possessing and those not possessing a canonical CKS17 domain (i.e., the last seven sequences in Fig. 3). Finally, it should be noted that the TM of human foamy virus (HFV), which is actually more than twice the length of the other TMs and includes an internal specific beta-sheet and loop region (46), could not be included in the alignment. Conversely, and rather interestingly, the TMs (GP2) of the Ebola and Marburg filoviruses, which had been shown to share structure and sequence homologies with the Mo-MuLV TM (8, 28, 49), could actually be aligned with the retroviral sequences (but we could not align the hemagglutinin of influenza virus, despite reported structural similarities with retroviral TMs [17]).
FIG. 3.
TM protein sequence alignment of ERVs and exogenous retroviruses (extracellular and TM domains). Sequences not selected from the CKS17 search and added in the alignment (see text) were found by BLAST searches or were derived from the literature (45). Sequence names correspond to the GenBank identifiers. The HERV or MuERV families are indicated on the left, as are the virus names. The order is the same as that of the TM tree (see Fig. 5, left) from top to bottom. Variable regions are indicated with the number of omitted residues, underlined positions correspond to insertions or frameshifts, and dashes correspond to deletions. The numbers above the alignment are relative to the full-length alignment (including insertions), which is 269 positions long. The region aligned to establish the initial CKS17 nucleotide phylogeny (Fig. 2) corresponds to positions 54 to 181. The immunosuppressive domain is boxed in red. The cysteine residues potentially involved in a short internal disulfide bond are highlighted in black, and the a and d positions in the heptad repeats within the coiled coil are in grey. Basic residues are in red, acidic residues are in blue, aromatic residues are in brown, and hydrophobic (aliphatic) residues are in green.
From the TM protein alignment, phylogenetic TM trees could be derived by the neighbor-joining (see Fig. 5, left) and the parsimony (data not shown) methods, with very similar results. Two major branches are observed. One of them corresponds to the CKS17-negative sequences (among which are the HERV-K elements and the MMTV and HIV-1 retroviruses), and the other corresponds to the CKS17-positive sequences. Each branch defines rather well-identified and unambiguously distinct groups of sequences. Importantly, a tree calculated from an alignment omitting the CKS17 motif (not shown) showed the same general pattern, thus demonstrating that the TM sequences of the two major branches differ over the full length of the protein and not only in the CKS17 motif. Moreover, a tree calculated from the CKS17-positive sequences only (not shown) gives a topology similar to that of the CKS17-positive branch of the complete TM tree, showing that the large distances from the CKS17-negative sequences do not artifactually modify the internal topology of the CKS17-positive branch. Accordingly, the two major branches most probably correspond to distinct “master” or progenitor sequences, from which most envelope proteins have derived. At a more refined level, the CKS17-positive sequences are themselves distributed into major subgroups (highlighted by different colors in Fig. 5). Retroelements in red include sequences closely related to the type C retroviruses exemplified by MuLV, koala retrovirus (AF151794), or porcine ERV (PEN133818), while retroelements in light blue and green, as well as the Ebola and Marburg viruses, are more distantly related, as illustrated by the longer branches.
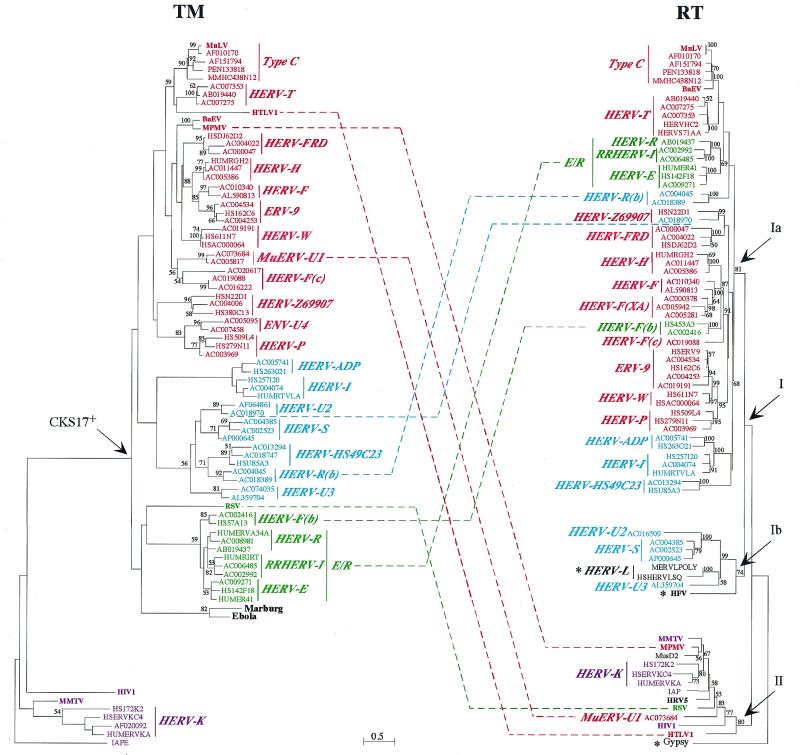
FIG. 5.
TM and RT protein phylogenetic trees. Both trees were determined by the neighbor-joining method with horizontal branch lengths proportional to the degree of divergence between the sequences (common scale for both trees). Vertical bars are only for presentation, with a few of them lengthened to highlight the major groups of sequences. The TM tree (left) is presented with the IAPE sequence (a murine retrotransposon envelope) as an outgroup, and the RT tree (right) is presented with Gypsy (a Drosophila melanogaster retrotransposon) as an outgroup. Percent bootstrap values obtained from 100 replicates are indicated on the branches only when they are >50%. Asterisks in the RT tree are for families devoid of the TM region (HERV-L family) or for which the TM could not be aligned (HFV and Gypsy). Major chimerisms are indicated by dotted lines between sequences from both trees. All identifiers and ERV families names are the same as in the TM alignment (Fig. 3). The few RT-only sequences are as follow: MERVLPOLY and HSHERVLSQ, the murine and human prototypic ERV-L sequences, respectively; HFV (HSPGAGPOL); MusD2 (AF246633) (27), a new mouse ERV devoid of the env region; HRV5 (HRU46939); IAP (AC006584); and Gypsy (AF033821). Within the HERV-T family, HERVHC2 and HUMS71AA (in red) are two known HERV sequences, but they carry internal deletions in the PBS (among others) which had precluded the definite naming of the family. For the sequences exhibiting large deletions in the TM (Fig. 3) and thus not included in the TM tree, a phylogeny calculated on reduced TM alignments led to the expected conclusions that the HERV-F(XA) sequences (AC000378 and AC005942) branch with the HERV-F family, the AC016509 sequence branches with the HERV-U2 family, and the AC007204 sequence branches with the HERV-U3 family.
RT tree and comparison with TM tree.
To compare the present TM phylogenetic tree with the RT-based trees (13, 24, 45), we performed an alignment of the RT domains of the sequences shown in the TM alignment in Fig. 3 with, in addition, those of the HFV and ERV-L sequences (the TM of the former could not be aligned, and the latter is devoid of env gene). The RT alignment (Fig. 4) included approximately 180 amino acids corresponding to the region selected by Tristem (45) and comprising domains 1 to 5 as defined by Xiong and Eickbush (51). This alignment was unambiguously and rather easily determined because of the high conservation of the RT protein between retroviral elements (33). An RT tree (Fig. 5, right) was calculated by the neighbor-joining method as for the TM tree to allow a comparison of branch lengths. RT phylogeny determined by the parsimony method (not shown) was congruent with the neighbor-joining tree, as well as with previously published RT trees (see, e.g., reference 45 [but that tree did not contain some of the present sequences and had one group {HERV-I/HERV-ADP} branching differently]). The RT tree is composed of two major branches corresponding to the two major HERV classes, i.e., sequences related to the mammalian type C infectious retroviruses, with the HFV-related sequences being the most distant ones (they are often considered a third class), and the HERV-K sequences clustering with the majority of the infectious retroviruses, including HIV-1, MMTV, MPMV, human retrovirus 5 (19), Rous sarcoma virus (RSV), and HTLV-1.
FIG. 4.
Partial RT protein sequence alignment. Sequence names correspond to the GenBank identifiers for ERVs, while common names are used for infectious retroviruses (with their GenBank identifiers given in Fig. 3 and 5 and their legends). The order of the sequences is the same as that for the RT tree (Fig. 4, right). Underlined positions correspond to insertions, and dashes correspond to deletions. The first five common peptide domains among the seven domains described by Xiong and Eickbush (51) are delineated below the sequences (the fifth is partial).
Comparison of the TM and RT trees discloses the following characteristic features. First, the evolution rates appear much lower for the RT tree than for the TM tree, as exemplified by the overall branch lengths as well as by the higher bootstrap values; this most probably corresponds to the greater constraint imposed by conservation of the RT enzymatic function. A second important feature is that the previously described CKS17-negative sequences, which are quite distinct in the TM tree (Fig. 5, retroelements in violet), again branch together on the separate class II branch of the RT tree, with the RT data being congruent with and thus strengthening the TM approach. Third, among the TM major branch, branching is also on the whole congruent with that obtained for the RT tree (with some minor differences most probably due to phylogenetic uncertainties), but clearly major chimerisms between the RT and TM domains can be observed (dotted lines in Fig. 5), which involve both endogenous retroelements and infectious retroviruses. For instance, among ERVs, at least five families or isolated sequences exhibit such a chimeric structure when the TM and RT trees are compared. The HERV-E/HERV-R/RRHERV-I (E/R) group (in green) appears to be closely related to the type C group in the RT tree, while these two groups are distant in the TM tree. A similar observation can be made for the HERV-R(b) group. The HERV-F(b) group, which is very closely related to the E/R group in the TM tree, is closely related to the HERV-F and HERV-F(XA) families at the RT level and not to the E/R group. It is also noteworthy that the HERV-F family [F, F(XA), F(b), and F(c)], which is rather homogenous at the RT level, is finally chimeric, with three divergent TMs. Conversely, the HERV-U2 sequences, which are grouped in the TM tree, are divergent at the RT level. At least one member of the MuERV-U1 family, whose TM belongs to the type C group (in red) in the CKS17-positive branch of the TM tree, is also chimeric, with its RT sequence in the class II group. Interestingly, such chimerisms between sequences of the CKS17-positive branch on the one hand and the class II RT on the other hand are also observed for three infectious retroviruses, namely MPMV, HTLV-1, and RSV. The MPMV TM is highly related to that of baboon endogenous virus (both viruses belong to the same interference group [40]), but these viruses are highly divergent in their RTs, thus strongly suggesting the occurrence of specific recombination events for these viruses (see also references 29 and 41). HTLV-1 and RSV, both of which are related to the class II group in the RT tree, also exhibit TM proteins which belong to the CKS17-positive group of sequences. Interestingly, the HTLV-1 TM appears to be closely related to that of the type C retroviruses, with which it could therefore share a common ancestor. This would be consistent with the recently reported similarities between the HTLV-1 and MuLV envelope SU moieties at the functional level (22). It is also noteworthy that the chimeric origin of the bovine HTLV-1 homologue, i.e., bovine leukemia virus, has been documented (37).
In conclusion, comparison of the TM and RT phylogenies strongly suggests that recombination has been a common and important event for the generation of both endogenous and exogenous retroviral sequences.
DISCUSSION
One important issue in the present investigation is the alignment of the TM moieties of retroviral envelopes based upon the so-called immunosuppressive domain. Although this motif is not systematically present in a canonical form among all elements, it allowed the identification of conserved residues within TMs and the inclusion of almost all retroviral envelopes within phylogenetic trees. Most interestingly, it also allowed the inclusion of the envelopes of the two nonretroviral filoviruses Ebola virus and Marburg virus, which then appear to have “borrowed” a retroviral structure to their own benefit. Overall, the achieved alignment made possible the identification of several new endogenous retroviral elements from databases, leading to the identification of a total of 26 HERV families, the identification of major phylogenetic branches containing both endogenous and exogenous elements, and the proposal that generation of retroviral diversity involved exchange of pol and env genes among elements from distinct branches, resulting in chimeric retroviruses. The screening procedure should be of great help to identify within genomes env or env-like genes which could be involved either in protective effects against infection through interference or in pathological processes through immunosuppressive effects (see below). The method could also lead to the identification of putative ancestral envelopes of cellular origin, from which viral envelopes would have emerged.
Phylogeny of retroviral elements.
Comparison of TM and RT phylogenies provided a specific tool for studying ERVs or exogenous retroviruses. The RT tree discloses two major branches: one containing most of the infectious retroviruses (e.g., MMTV, HIV-1, and HTLV-1) and another containing the majority of the ERVs (22 of 26 families for the human ERVs). Two important exceptions to this scheme concern (i) the type C infectious retroviruses (which cluster with the ERVs) as well as the foamy retroviruses and (ii) the endogenous HERV-K retroviruses clustering with the infectious retrovirus group. This dichotomy is consistent with that mentioned by Chiu et al. (11), who proposed that infectious retroviruses have evolved from two divergent pol genes leading to the type C virus lineage on the one hand and the type A, B, and D lineages, as well as RSV, on the other hand, to which we can now add HIV and HTLV. McClure et al. (33) have also reported that the type C RT sequences are the most distantly related among those of the infectious retroviruses. The abundance of type C-related ERVs could attest to a more successful expansion of type C retroviruses during evolution or could indicate that retroelements of the second branch are more recent ones for which “endogenization” has not yet widely occurred. The second alternative is most probably true for HIV, as well as for the HERV-K family which has invaded the primate branch recently, after the divergence of Old World and New World monkeys (32). Alternatively, one could hypothesize that germ line cells are more prone to infection by class I retroviruses (although one would expect that this property should be determined primarily by the env gene rather than by the RT gene) or even more simply that class I retroelements have a higher replicative capacity, possibly amplified by intracellular retrotransposition (a property not requiring the env gene [42]).
The TM tree also discloses two groups, not strictly overlapping those of the RT tree: one group, corresponding to the CKS17-negative sequences, is associated, as observed for the RT tree, with infectious retroviruses of group II, whereas the other group contains the majority of the ERVs. Again, the TM tree shows that the majority of ERVs are related to type C retroviruses. Interestingly, the HERV-K group, which is excluded from the branch containing all of the other ERVs in the RT tree, is also excluded in the TM tree. Overall, as well as at the more refined level of major branchings, the RT and TM trees show congruent clustering. However, important deviations from this scheme are observed, with evidence for chimeric structures: within the RT class II retroelements for the infectious MPMV, RSV, and HTLV-1 viruses; and similarly among the RT class I retroelements for several ERVs.
Several mechanisms could account for recombination between retroviral sequences (43). Among them, recombination occurring between copackaged genomic retroviral RNA in the course of reverse transcription is a common retroviral process for retroviral RNAs with identical packaging sequences but can also take place for heterologous sequences (52). Such events might not be rare, as chimeric retroelements have been documented to reproducibly emerge in the mouse, leading to the generation of recombinant and highly pathogenic retroelements (14). Recombination events between lentiviruses have also been identified by the comparison of the phylogeny of the gag or pol gene with that of the env gene (35).
Identification of ERVs and of potentially functional env genes.
A compilation of our data based on TM and RT sequences and previous data based on RT sequences (45) discloses that the most extensively sequenced mammalian genome, i.e., the human genome, contains 26 (and possibly not significantly more) families of ERVs, still comprising altogether approximately 8% of the human genome when the numerous solo LTRs are included (25). Actually, our complementary approaches with the RT and TM protein sequences from approximately 25% of the human genome can be considered almost complete, if not complete, taking into account that with HERV being a multigenic family, only very small families might have been missed. Accordingly, the present study already provides a catalog of human sequences and a method for updating and extending the search to other genomes when they are entirely sequenced. In the case of the mouse genome, for instance, we have already detected two new mouse ERV sequences. One of them, MuERV-U1, is likely to be mouse specific, while the second (AC020617) is homologous to the human HERV-F(c) family. The latter case is reminiscent of the HERV-L family, which is shared by all mammalian species and most probably corresponds to an ancestral retroelement already present in living species before the mammalian radiation and which therefore constitutes an evolution marker among mammals (4).
The present env-based approach should also be especially interesting for the detection of genes not necessarily associated with a complete RT-containing proviral structure but endowed with important physiological functions. Endogenous retroviral genes without a surrounding proviral structure have already been described, such as the ERV-L gag-related Fv1 gene (5) or the env-related Fv4 gene (20) (positive in our CKS17d screen), both of which are involved in resistance of the mouse to infection by leukemia viruses. In this respect, it is noteworthy that in the present search we have identified several envelope sequences which are also not in a proviral structure (no LTRs or gag or pol genes were detected), such as the ENV-U4 sequences, which, together with other sequences with large open reading frames (e.g., AB019440, AC018389, HSDJ62D2, and AC016222), clearly constitute interesting candidate genes for further investigations. Some of them could even constitute progenitor envelopes that ancestral, env-negative retroelements (such as the ERV-L elements) would have acquired in the course of evolution, for instance, by capture mechanisms similar to those described for the present-day oncogene-containing retroviruses (43). Envelope proteins displaying a fusogenic function (e.g., the HERV-W env product [7, 34]), displaying immunosuppression (6, 30, 30a, 36, 39), acting as cofactors for infection (e.g., the FELIX gene product [3]), or even conferring infectivity in pseudotypes (2) have also been described and could now be searched for systematically.
ACKNOWLEDGMENTS
We acknowledge the INFOBIOGEN Bioinformatics Resource Centre (http://www.infobiogen.fr/), where most of the computing was carried out, and C. Lavialle for critical reading of the manuscript.
This work was supported by a grant from the Ligue Nationale contre le Cancer (Equipe Labellisée).
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.An D S, Xie Y M, Chen I S Y. Envelope gene of the human endogenous retrovirus HERV-W encodes a functional retrovirus envelope. J Virol. 2001;75:3488–3489. doi: 10.1128/JVI.75.7.3488-3489.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson M M, Lauring A S, Burns C C, Overbaugh J. Identification of a cellular cofactor required for infection by feline leukemia virus. Science. 2000;287:1828–1830. doi: 10.1126/science.287.5459.1828. [DOI] [PubMed] [Google Scholar]
- 4.Bénit L, Lallemand J B, Casella J F, Philippe H, Heidmann T. ERV-L elements: a family of endogenous retrovirus-like elements active throughout the evolution of mammals. J Virol. 1999;73:3301–3308. doi: 10.1128/jvi.73.4.3301-3308.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Best S, Le Tissier P, Towers G, Stoye J P. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature. 1996;382:826–829. doi: 10.1038/382826a0. [DOI] [PubMed] [Google Scholar]
- 6.Blaise S, Mangeney M, Heidmann T. The envelope of Mason-Pfizer monkey virus has immunosuppressive properties. J Gen Virol. 2001;82:1597–1600. doi: 10.1099/0022-1317-82-7-1597. [DOI] [PubMed] [Google Scholar]
- 7.Blond J L, Lavillette D, Cheynet V, Bouton O, Oriol G, Chapel-Fernandes S, Mandrand B, Mallet F, Cosset F L. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J Virol. 2000;74:3321–3329. doi: 10.1128/jvi.74.7.3321-3329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bukreyev A, Volchkov V E, Blinov V M, Netesov S V. The GP-protein of Marburg virus contains the region similar to the ‘immunosuppressive domain’ of oncogenic retrovirus P15E proteins. FEBS Lett. 1993;323:183–187. doi: 10.1016/0014-5793(93)81476-g. [DOI] [PubMed] [Google Scholar]
- 9.Chambers P, Pringle C R, Easton A J. Heptad repeat sequences are located adjacent to hydrophobic regions in several types of virus fusion glycoproteins. J Gen Virol. 1990;71:3075–3080. doi: 10.1099/0022-1317-71-12-3075. [DOI] [PubMed] [Google Scholar]
- 10.Chan D C, Fass D, Berger J M, Kim P S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 11.Chiu I M, Callahan R, Tronick S R, Schlom J, Aaronson S A. Major pol gene progenitors in the evolution of oncoviruses. Science. 1984;223:364–370. doi: 10.1126/science.6197754. [DOI] [PubMed] [Google Scholar]
- 12.Cianciolo G, Copeland T D, Orozlan S, Snyderman R. Inhibition of lymphocyte proliferation by a synthetic peptide homologous to retroviral envelope proteins. Science. 1985;230:453–455. doi: 10.1126/science.2996136. [DOI] [PubMed] [Google Scholar]
- 13.Doolittle R F, Feng D F, McClure M A, Johnson M S. Retrovirus phylogeny and evolution. Curr Top Microbiol Immunol. 1990;157:1–18. doi: 10.1007/978-3-642-75218-6_1. [DOI] [PubMed] [Google Scholar]
- 14.Elder J H, Gautsch J W, Jensen F C, Lerner R A, Hartley J W, Rowe W P. Biochemical evidence that MCF murine leukemia viruses are envelope (env) gene recombinants. Proc Natl Acad Sci USA. 1977;74:4676–4680. doi: 10.1073/pnas.74.10.4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fass D, Harrison S C, Kim P S. Retrovirus envelope domain at 1.7 angstrom resolution. Nat Struct Biol. 1996;3:465–469. doi: 10.1038/nsb0596-465. [DOI] [PubMed] [Google Scholar]
- 16.Felsenstein J. PHYLIP—phylogeny inference package. Cladistics. 1989;5:164–166. [Google Scholar]
- 17.Gallaher W R, Ball J M, Garry R F, Griffin M C, Montelaro R C. A general model for the transmembrane proteins of HIV and other retroviruses. AIDS Res Hum Retroviruses. 1989;5:431–440. doi: 10.1089/aid.1989.5.431. [DOI] [PubMed] [Google Scholar]
- 18.Galtier N, Gouy M, Gautier C. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- 19.Griffiths D J, Venables P J, Weiss R A, Boyd M T. A novel exogenous retrovirus sequence identified in humans. J Virol. 1997;71:2866–2872. doi: 10.1128/jvi.71.4.2866-2872.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ikeda H, Sugimura H. Fv-4 resistance gene: a truncated endogenous murine leukemia virus with ecotropic interference properties. J Virol. 1989;63:5405–5412. doi: 10.1128/jvi.63.12.5405-5412.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson W E, Coffin J M. Constructing primate phylogenies from ancient retrovirus sequences. Proc Natl Acad Sci USA. 1999;96:10254–10260. doi: 10.1073/pnas.96.18.10254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim F J, Seiliez I, Denesvre C, Lavillette D, Cosset F L, Sitbon M. Definition of an amino-terminal domain of the human T-cell leukemia virus type 1 envelope surface unit that extends the fusogenic range of an ecotropic murine leukemia virus. J Biol Chem. 2000;275:23417–23420. doi: 10.1074/jbc.C901002199. [DOI] [PubMed] [Google Scholar]
- 23.Kobe B, Center R J, Kemp B E, Poumbourios P. Crystal structure of human T cell leukemia virus type 1 gp21 ectodomain crystallized as a maltose-binding protein chimera reveals structural evolution of retroviral transmembrane proteins. Proc Natl Acad Sci USA. 1999;96:4319–4324. doi: 10.1073/pnas.96.8.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li M D, Bronson D L, Lemke T D, Faras A J. Phylogenetic analyses of 55 retroelements on the basis of the nucleotide and product amino acid sequences of the pol gene. Mol Biol Evol. 1995;12:657–670. doi: 10.1093/oxfordjournals.molbev.a040231. [DOI] [PubMed] [Google Scholar]
- 25.Li W H, Gu Z, Wang H, Nekrutenko A. Evolutionary analyses of the human genome. Nature. 2001;409:847–849. doi: 10.1038/35057039. [DOI] [PubMed] [Google Scholar]
- 26.Löwer R, Löwer J, Kurth R. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc Natl Acad Sci USA. 1996;93:5177–5184. doi: 10.1073/pnas.93.11.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mager D L, Freeman J D. Novel mouse type D endogenous proviruses and ETn elements share long terminal repeat and internal sequences. J Virol. 2000;74:7221–7229. doi: 10.1128/jvi.74.16.7221-7229.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malashkevich V N, Schneider B J, McNally M L, Milhollen M A, Pang J X, Kim P S. Core structure of the envelope glycoprotein GP2 from Ebola virus at 1.9-A resolution. Proc Natl Acad Sci USA. 1999;96:2662–2667. doi: 10.1073/pnas.96.6.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mang R, Maas J, van Der Kuyl A C, Goudsmit J. Papio cynocephalus endogenous retrovirus among Old World monkeys: evidence for coevolution and ancient cross-species transmissions. J Virol. 2000;74:1578–1586. doi: 10.1128/jvi.74.3.1578-1586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mangeney M, Heidmann T. Tumor cells expressing a retroviral envelope escape immune rejection in vivo. Proc Natl Acad Sci USA. 1998;95:14920–14925. doi: 10.1073/pnas.95.25.14920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30a.Mangeney M, de Parseval N, Thomas G, Heidmann T. The full-length envelope of an HERV-H human endogenous retrovirus has immunosuppressive properties. J Gen Virol. 2001;82:2515–2518. doi: 10.1099/0022-1317-82-10-2515. [DOI] [PubMed] [Google Scholar]
- 31.Marck C. ‘DNA Strider’: a ‘C’ program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acids Res. 1988;16:1829–1836. doi: 10.1093/nar/16.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mariani-Costantini R, Horn T M, Callahan R. Ancestry of a human endogenous retrovirus family. J Virol. 1989;63:4982–4985. doi: 10.1128/jvi.63.11.4982-4985.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McClure M A, Johnson M S, Feng D F, Doolittle R F. Sequence comparisons of retroviral proteins: relative rates of change and general phylogeny. Proc Natl Acad Sci USA. 1988;85:2469–2473. doi: 10.1073/pnas.85.8.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mi S, Lee X, Li X, Veldman G M, Finnerty H, Racie L, LaVallie E, Tang X Y, Edouard P, Howes S, Keith J C, McCoy J M. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403:785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 35.Robertson D L, Sharp P M, McCutchan F E, Hahn B H. Recombination in HIV-1. Nature. 1995;374:124–126. doi: 10.1038/374124b0. [DOI] [PubMed] [Google Scholar]
- 36.Ruegg C L, Strand M. Inhibition of protein kinase C and anti-CD3-induced Ca2+ influx in Jurkat T cells by a synthetic peptide with sequence identity to HIV-1 gp41. J Immunol. 1990;144:3928–3935. [PubMed] [Google Scholar]
- 37.Sagata N, Yasunaga T, Tsuzuku-Kawamura J, Ohishi K, Ogawa Y, Ikawa Y. Complete nucleotide sequence of the genome of bovine leukemia virus: its evolutionary relationship to other retroviruses. Proc Natl Acad Sci USA. 1985;82:677–681. doi: 10.1073/pnas.82.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh M, Berger B, Kim P S. LearnCoil-VMF: computational evidence for coiled-coil-like motifs in many viral membrane-fusion proteins. J Mol Biol. 1999;290:1031–1041. doi: 10.1006/jmbi.1999.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Snyderman R, Cianciolo G. Immunosuppressive activity of the retroviral envelope protein p15E and its possible relationship to neoplasia. Immunol Today. 1984;5:240–244. doi: 10.1016/0167-5699(84)90097-5. [DOI] [PubMed] [Google Scholar]
- 40.Sommerfelt M A, Weiss R A. Receptor interference groups of 20 retroviruses plating on human cells. Virology. 1990;176:58–69. doi: 10.1016/0042-6822(90)90230-o. [DOI] [PubMed] [Google Scholar]
- 41.Sonigo P, Barker C, Hunter E, Wain-Hobson S. Nucleotide sequence of Mason-Pfizer monkey virus: an immunosuppressive D-type retrovirus. Cell. 1986;45:375–385. doi: 10.1016/0092-8674(86)90323-5. [DOI] [PubMed] [Google Scholar]
- 42.Tchénio T, Heidmann T. Defective retroviruses can disperse in the human genome by intracellular transposition. J Virol. 1991;65:2113–2118. doi: 10.1128/jvi.65.4.2113-2118.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Telesnitsky A, Goff S P. Reverse transcriptase and the generation of retroviral DNA. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 121–160. [PubMed] [Google Scholar]
- 44.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tristem M. Identification and characterization of novel human endogenous retrovirus families by phylogenetic screening of the human genome mapping project database. J Virol. 2000;74:3715–3730. doi: 10.1128/jvi.74.8.3715-3730.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang G, Mulligan M J. Comparative sequence analysis and predictions for the envelope glycoproteins of foamy viruses. J Gen Virol. 1999;80:245–254. doi: 10.1099/0022-1317-80-1-245. [DOI] [PubMed] [Google Scholar]
- 47.Weissenhorn W, Dessen A, Harrison S C, Skehel J J, Wiley D C. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 48.Wilkinson D A, Mager D L, Leong J A C. Endogenous human retroviruses. In: Levy J A, editor. The Retroviridae. Vol. 3. New York, N.Y: Plenum Press; 1994. pp. 465–535. [Google Scholar]
- 49.Will C, Muhlberger E, Linder D, Slenczka W, Klenk H D, Feldmann H. Marburg virus gene 4 encodes the virion membrane protein, a type I transmembrane glycoprotein. J Virol. 1993;67:1203–1210. doi: 10.1128/jvi.67.3.1203-1210.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilson I A, Skehel J J, Wiley D C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 51.Xiong Y, Eickbush T H. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang J, Temin H M. Rate and mechanism of nonhomologous recombination during a single cycle of retroviral replication. Science. 1993;259:234–238. doi: 10.1126/science.8421784. [DOI] [PubMed] [Google Scholar]