Abstract
Comparative analysis of the protein sequences encoded in the genomes of three families of large DNA viruses that replicate, completely or partly, in the cytoplasm of eukaryotic cells (poxviruses, asfarviruses, and iridoviruses) and phycodnaviruses that replicate in the nucleus reveals 9 genes that are shared by all of these viruses and 22 more genes that are present in at least three of the four compared viral families. Although orthologous proteins from different viral families typically show weak sequence similarity, because of which some of them have not been identified previously, at least five of the conserved genes appear to be synapomorphies (shared derived characters) that unite these four viral families, to the exclusion of all other known viruses and cellular life forms. Cladistic analysis with the genes shared by at least two viral families as evolutionary characters supports the monophyly of poxviruses, asfarviruses, iridoviruses, and phycodnaviruses. The results of genome comparison allow a tentative reconstruction of the ancestral viral genome and suggest that the common ancestor of all of these viral families was a nucleocytoplasmic virus with an icosahedral capsid, which encoded complex systems for DNA replication and transcription, a redox protein involved in disulfide bond formation in virion membrane proteins, and probably inhibitors of apoptosis. The conservation of the disulfide-oxidoreductase, a major capsid protein, and two virion membrane proteins indicates that the odd-shaped virions of poxviruses have evolved from the more common icosahedral virion seen in asfarviruses, iridoviruses, and phycodnaviruses.
The category of virus is biological, not evolutionary. Viruses are intracellular parasites that depend on the host cell for their protein synthesis, most of the reactions of nucleic acid precursor biosynthesis and, to a variable extent, transcription and replication (15). Clearly, viruses are not a monophyletic group. There is little doubt, for example, that small viruses with single-stranded RNA genomes of only 5 to 10 kb, such as poliovirus or tobacco mosaic virus, on the one hand, and large viruses with double-stranded DNA (dsDNA) genomes of 100 to 500 kb, such as herpesviruses, poxviruses, or iridoviruses, on the other hand, have evolved independently. However, comparative analyses of the genomes of many groups of viruses have suggested common origins for large, heterogeneous assemblages. For example, it appears most likely that all reverse-transcribing viruses and mobile elements, in spite of the extreme diversity of their life cycles and the sets of encoded proteins, have evolved from a common ancestor (17, 56, 70). Even more unexpected evolutionary connections are suggested by the involvement of homologous enzymes, such as superfamily III helicases, in genome replication of both RNA and DNA viruses with small genomes (23), and the central role of the conserved rolling circle replication initiator protein in single-stranded DNA (ssDNA) viruses of eukaryotes and bacteria and in bacterial plasmids (26).
Viruses with large, dsDNA genomes are generally thought to have evolved by capturing multiple genes from the genomes of cellular organisms, their hosts. Indeed, many genes of these viruses, particularly those involved in virus-host interactions, show high levels of protein sequence similarity to their cellular homologs, which is apparently indicative of relatively recent acquisition by the viral genomes (12, 51, 59). However, viruses belonging to a particular large family, such as the herpesvirus family or the poxvirus family, share between themselves a core set of genes encoding proteins involved in DNA replication, transcription, and virion biogenesis, most of which are only moderately similar to cellular homologs, if such are detectable at all (3, 51). The existence of core sets of up to 40 to 50 conserved viral genes (8, 22) establishes beyond reasonable doubt that the extant members of the families Herpesviridae and Poxviridae have diverged from the respective ancestral viruses that already possessed the principal features of genome replication and expression and of virion structure that are typical of these viral families. In contrast, it remains unclear whether there are any evolutionary connections between different viral families. Poxviruses, African swine fever virus (ASFV, the archetypal member of the family Asfarviridae), and iridoviruses are the three families of eukaryotic viruses with large dsDNA genomes that undergo their replication cycle either entirely in the cytoplasm (poxviruses) or start their replication in the nucleus and complete it in the cytoplasm (20, 22, 38, 40, 63, 67), as opposed to herpesviruses and baculoviruses, whose DNA replication and transcription occur exclusively in the nucleus (30, 65). Poxviruses, asfarviruses, and iridoviruses encode their own transcription machinery, which includes, in each case, several RNA polymerase subunits and additional transcription factors, and share several other conserved genes (58, 72). Large DNA viruses isolated from very diverse algae, the Paramecium bursaria chlorella virus (PBCV) and the related Ectocarpus siliculosus virus (ESV), members of the Phycodnaviridae family, also share several genes with nucleocytoplasmic large DNA viruses, although genomes of these viruses are transcribed in the nucleus and, accordingly, they lack genes for RNA polymerase subunits (41, 61). The four families of large eukaryotic DNA viruses, Poxviridae, Asfarviridae, Iridoviridae, and Phycodnaviridae, to which we collectively refer here as nucleocytoplasmic large DNA viruses (NCLDV), have both common and unique features of genomic DNA and virion structure. Poxviruses, ASFV, and PBCV have linear DNA genomes with terminal inverted repeats that form covalently closed hairpins (40, 67, 75), iridoviruses have circularly permuted linear genomes (60), and ESV appears to have a circular genome (41). The virions of ASFV, iridoviruses, and PBCV consist of a DNA-protein core that is surrounded by a lipid bilayer, which in turn is encased in one or more icosahedral capsid shells (58, 63, 66). Poxviruses have a more complex, unique virion structure, with a core surrounded by a “brick-shaped” proteolipid shell (40).
It remains uncertain whether the similarities between the gene repertoires, genome structures, and virion architectures of different families of NCLDV are due to independent recruitment of the same or related host genes driven by the common functional requirements for the viral replication cycles or by origin from a common viral ancestor. This crucial dilemma is not readily amenable to conventional phylogenetic analysis because even homologous proteins of viruses from different families show moderate or weak sequence conservation and may be less similar to each other than to the corresponding cellular homologs (51). At face value, these observations appear to favor the polyphyletic origin of different viral families. However, this aspect of the relationships between viruses needs to be interpreted with caution given the realistic possibility of rapid evolution of viral genes (44). Moreover, such rapid divergence potentially might even preclude the very detection of evolutionary relationships between some viral genes. Given these considerations, we were interested in delineating the complete set of conserved genes among NCLDV by applying the most advanced available methods for sequence similarity detection and assessing the hypothesis of independent recruitment of similar sets of genes from the host as opposed to an origin of several viral families from a single, ancestor virus. We expand the list of conserved genes shared by all or a majority of NCLDV families and show that origin from a common viral ancestor is the most parsimonious scenario for the evolution of all of these viruses.
MATERIALS AND METHODS
Viral genome and protein sequences.
Nucleotide sequences of the complete genomes of large DNA viruses and the corresponding, predicted protein sequences were extracted from the Genomes division of the Entrez system (National Center for Biotechnology Information, National Institutes of Health, Bethesda, Md. [http://www.ncbi.nlm.nih.gov:80/entrez/query.fcgi?db=Genome]). The complete genomes included in this analysis were from the following viruses: poxviruses, including vaccinia virus, strain Copenhagen (VV [21]), variola virus, strain India (VAR [37]), Molluscum contagiosum virus type 1 (MCV [50]), Shope fibroma virus (SFV [66]), Fowlpox virus (FPV [2]), Melanoplus sanguinipes entomopoxvirus (MSV [1]), Amsacta moorei entomopoxvirus (AMV [8]); asfarviruses, including ASFV (72); iridoviruses, including fish lymphocystis disease virus (FLDV [58]), Chilo iridescent virus (CIV [27]); and phycodnaviruses, including PBCV (type 1 [35]) and ESV (type 1; N. Delaroque, G. Bothe, T. Pohl, R. Knippers, D. G. Mueller, and W. Boland [GenBank NC002687]).
Sequence analysis.
Protein sequences were compared to protein sequence databases by using the BLASTP program and to nucleotide sequence databases translated in six frames by using the TBLASTN program (5). Additional searches for detecting subtle similarities were performed by using the PSI-BLAST program with varied cutoffs for including sequences into profiles (4, 5). Multiple alignments of protein sequences were constructed by using the ClustalW (57) and T_coffee programs (43), with subsequent manual refinement on the basis of the PSI-BLAST search results. Protein secondary structure was predicted by using the PHD program, with a multiple alignment submitted as the query (47). Protein sequence-structure threading was performed by using the hybrid fold recognition method (16).
Identification of clusters of orthologous viral proteins.
In order to identify sets of orthologous viral proteins, single-linkage clustering based on BLASTP search results was performed by using the BLASTCLUST program and an empirically determined alignment score cutoff of 0.2 bits/position (I. Dondoshansky, Y. I. Wolf, and E. V. Koonin, unpublished data; ftp://ftp.ncbi.nlm.nih.gov/blast). For resulting clusters that included representatives of two or more viral families, additional PSI-BLAST searches were performed against the NR database, with all sequences from the original cluster used as queries. Position-specific weight matrices obtained through these searches were saved and used for a second round of searching the NCLDV protein sequences. This was done to detect potential members of the given protein cluster encoded in the genomes from other virus families that could have been missed at the first stage due to low sequence conservation.
Cladistic analysis.
Cladistic analysis was performed by using the PAUP* version 4.0 package (55). A maximum of four states, namely, the primitive state (0) and up to three derived states (1, 2, and 3), were considered. The relationship between the derived states was assumed to be unordered, that is, a primitive character could make the transition to any of the derived states if more than one derived state existed for the given character. Gain of a novel protein, domain, or sequence motif was scored as a derived character with respect to its complete absence, which was defined as the primitive state. The size ranges and domain architectures of proteins were also used as characters scored in the matrix. The shortest trees were determined by using the Branch and Bound and the Exhaustive Search algorithms. The consensus of the shortest trees was obtained by using the Consensus Tree routine of PAUP. The character state transitions for each node of the shortest trees were derived by using the Show Apomorphy routine of PAUP, and this was used to determine the synapomorphies supporting a given clade.
RESULTS AND DISCUSSION
Clusters of orthologous viral proteins.
Viral proteins tend to evolve faster than their cellular counterparts, which makes it difficult to detect homologous relationships for some of them. Therefore, the detection of orthologous sets of viral proteins is not a trivial task and, in some cases, requires application of the most advanced sequence analysis methods. Furthermore, for detecting clusters of viral orthologs, it was important to compare viral proteins among themselves only, to limit the search space and thus increase the sensitivity. Once the clusters were identified, their relationships with non-NCLDV proteins were investigated by additional sequence comparisons; the results of these comparisons were then used for refinement of the NCLDV clusters.
The present study resulted in the identification of 9 clusters of apparent orthologs that are shared by all NCLDV, 8 clusters that are represented in all families (although missing in one or more species), and 14 clusters that are conserved in all but one family (Table 1). To our knowledge, the conservation of five of these proteins in all viral families has not been described previously. These include the predicted helicase D5R (hereinafter we use the systematic nomenclature of proteins from VV Copenhagen, whenever possible), the packaging ATPase A32L, the transcription factor A1L, the capsid protein D13L, and the myristoylated virion membrane protein L1R/F9L (Table 1). The critical aspect of these clusters of conserved viral proteins is that, although they did not necessarily show a high level of sequence conservation, each of them had distinct features that appeared to be synapomorphies (shared derived characters) of the NCLDV class. Despite systematic searches, we were unable to identify direct counterparts (orthologs) of any of these proteins outside this class of viruses, with the possible exception of D5R orthologs from some bacteriophages. Furthermore, for the two virion proteins, no non-NCLDV homologs at all were detected. We briefly describe each of these signature NCLDV protein families below, with an emphasis on the features that support their status as synapomorphies.
TABLE 1.
Distribution of conserved genes in large, cytoplasmic DNA viruses and Phycodnaviridae
| Gene group and protein familya | Distribution of conserved genes inb:
|
Comments | ||||||
|---|---|---|---|---|---|---|---|---|
| Chordopoxvirus | Entomopoxvirus | ASFV | Iridovirus | PBCV | ESV | Other viruses and plasmids | ||
| I | ||||||||
| VV D5 ATPase | D5R | AMV087, MSV089 | C962R | LDV1-ORF6, CIV-184R | A456L | ORF109 | – | A member of the superfamily III helicases within the AAA+ superclass ATPases; involved in poxvirus DNA replication, most likely as the principal helicase |
| DNA polymerase (B family) | E9L | AMV050, MSV036 | G1211R | LDV1-ORF5, CIV-037L | A185R | ORF93 | BV, HV, T4, KP | Members of the B family of DNA polymerases that also includes archaeoeukaryotic replicative DNA polymerases and polymerases of herpes-, adeno- and baculoviruses and many bacteriophages |
| VV A32 ATPase | A32L | MSV171, AMV150 | B354L | LDV1-ORF46, CIV-075L | A392R | ORF26 | – | Distinct family of ATPases required for virion packaging |
| VV A18 helicase | A18R | AMV059, MSV148 | QP509L | CIV-161L | A153R | ORF66 | T4 | Superfamily II helicase required for transcription termination; absent in FLDV |
| Capsid protein | D13L | AMV122, MSV069 | B646L | LDV1-MCP, CIV-274L | A622L | ORF116 | – | PBCV encodes six members of this family |
| Thiol-oxidoreductase | E10R | AMV114, MSV093 | B119L | LDV1-ORF79, CIV-347L | A465R | ORF161 | – | Required for the formation of cytoplasmic disulfide bonds in poxvirus proteins; homologous to the cellular ERV1/2 family but differs from them in having only two conserved cysteines |
| VV D6R/D11L-like helicase | D11, D6 | AMV192, MSV053, AMV174, MSV113 | D1133L, Q706L | LDV1-ORF4, CIV-022L | A363R | ORF23 | KP | Superfamily II helicase required for transcription in poxviruses |
| S/T protein kinase | F10L | MSV154, AMV153 | R298L | LDV1-ORF17, CIV-380R | A617R | ORF156 | BV, HV | Distinct S/T kinases that show no obvious eukaryotic orthologs |
| Transcription factor VLTF2 | A1L | AMV047, MSV187 | B175L | LDV-ORF102, CIV-350L | A482R | ORF96 | – | Small proteins containing an FCS-type Zn-finger; entomopoxviruses have a duplication of the FCS domain |
| II | ||||||||
| TFIIS-like Zn-ribbon-containing transcription factor | E4L | AMV120, MSV082 | I243L | LDV1-ORF105, CIV-349L | A125L | – | – | The viral TFIIS lacks the α-helical TFIIN domain typical of eukaryotic TFIIS |
| Nudix (MutT-like) NTP pyrophosphohydrolase | D9R/D10R | AMV058, MSV150 | D250R | LDV1-ORF78, CIV-414L | A326L | – | – | These nucleotidases are typically involved in repair of oxidative damage to DNA; their functions in NCLDV remain unclear, but they might regulate expression via mRNA cap hydrolysis (53) |
| Myristoylated virion protein A | L1R, F9L | AMV217, AMV243, MSV094, MSV183 | E248R | LDV1-ORF20, CIV-118L, CIV-458R | A565R | – | – | Components of the external lipid membrane, the PBCV form is extremely divergent and lacks the cysteines that are conserved in other members of this family |
| PCNA | G8R | – | E301R | LDV1-ORF45, CIV-436R | A193L, A574L | ORF132 | BV, HV, T4 | DNA sliding clamp, essential for DNA replication; viral forms are extremely divergent from the cellular forms; G8R is a late transcription factor in poxviruses; PBCV A193L is most closely related to the single PCNA ortholog in ESV and these in turn group with other viral |
| PCNAs; PBCV A574L groups weakly but specifically with the divergent poxviral PCNAs | ||||||||
| Ribonucleotide reductase, large subunit | 14L | – | F778R | LDV1-ORF12, CIV-085L | A629R | ORF180 | BV, HV, T4 | Absent in FPV and MCV |
| Ribonucleotide reductase, small subunit | F4L | – | F334L | LDV1-ORF26, CIV-376L | A476R | ORF128 | BV, HV, T4 | Absent in FPV and MCV |
| Thymidylate kinase | A48R | – | A240L | LDV1-ORF60, CIV-143R, CIV-251L | A416R | – | BV, HV | Absent in MCV |
| dUTPase | F2L | AMV002, AMV107 | E165R | CIV-438L | A551L | – | BV, HV | Absent in MCV, FLDV, and MSV |
| III | ||||||||
| Uncharacterized protein | – | – | B385R | LDV1-ORF43, CIV-282R | A494R | ORF101 | – | |
| RuvC-like HJR | A22R | AMV162, MSV106 | – | CIV-170L | – | ORF108 | Phage bIL170 | Distantly related to fungal mitochondrial RuvC, a possible degenerate version present in PBCV; absent in FLDV |
| BV BroA-like N-terminal domain | – | MSV194, AMV057 | – | CIV-201R | – | ORF117 | BV phage N15 | A DNA-binding domain (BRO) widely distributed in phages and expanded in baculoviruses, entomopoxviruses, and CIV (73) |
| Capping enzyme (guanylyltransferase) | D1R | AMV135, MSV067 | NP868R | – | A103R | – | KP | ASFV and poxviruses capping enzymes contain RNA triphosphatase, guanylyl transferase, and methyltransferase domains, and the capping enzyme from KP has the same domain architecture; PBCV encodes distinct proteins with RNA triphosphatase and methyltransferase activities |
| ATP-dependent DNA ligase | A50R | – | NP419L | – | A544R | – | T4, BV | Lacks the BRCT domains seen in eukaryotes; absent in MCV |
| RNA polymerase, largest subunit | J6R | AMV221, MSV043 | NP1450L | LDV1-ORF1, CIV-176R | – | – | KP | CIV-343L is only the C-terminal region of this polymerase |
| RNA polymerase, subunit 2 | A24R | AMV066, MSV155 | EP1242L | LDV1-ORF3, CIV-428L | – | – | KP | |
| Thioredoxin/glutaredoxin | G4L | AMV079, MSV087 | – | CIV-196R, CIV-453L | A427L | ORF128 | T4 | |
| Dual-specificity serine/tyrosine phosphatase | H1L | AMV078, AMV246 | – | CIV-123R, CIV-197R | A305L | – | BV | Dual-specificity phosphatases involved in early transcription in poxviruses; absent in FLDV and MSV |
| BIR domains | – | AMV021, MSV242 | A224L | CIV-193R | – | – | BV | Inhibitor of apoptosis in BV and ASFV; the entomopoxviruses, CIV, and BV have a RING finger fused to the C terminus of the BIR domain; the AmEPV and the BV proteins have a duplication of the BIR domain; absent in FLDV |
| Virion-associated membrane proteins | J5L, A16L, G9R | AMV232, MSV142, AMV035, MSV121, AMV118, MSV090 | E199L | LDV1-ORF29, CIV-337L | – | – | – | |
| Topoisomerase II | – | – | P1192R | CIV-045L | A583L | – | Probably involved in the resolution of replication intermediates | |
| SW1/SNF2 family helicase | – | MSV224 | – | CIV-172L | A548L | – | BV | Superfamily II helicase; MSV244 protein is fused to an ariadne-like Parkin domain |
| RNA polymerase, subunit 10 | G5.5R | – | CP80R | CIV-107L | – | – | – | Accessory transcription factor of the helix-turn-helix fold; absent in FLDV |
| IV | ||||||||
| Phage P1-like KilA N-terminal domain | N1R (SFV) | AMV100 | – | CIV-313L | – | – | Phage P1 | DNA-binding protein, widely distributed in phages and expanded in AMV and FPV; absent in MSV, MCV, and FLDV; the chordopoxviral proteins are fused to a RING finger; the N1R protein of SFV has been shown to bind DNA and inhibit apoptosis (10) |
| VV I8-like helicase | I8R | AMV081, MSV086 | B962L | – | – | – | – | Superfamily II helicase required for early transcription in poxviruses |
| RNA polymerase, subunit 5 | – | – | D205R | CIV-455L | – | – | – | |
| Lambda-type exonuclease | – | – | D345L | – | A166R | ORF64 | BV, HV | An exonuclease of the restriction endonuclease fold that, in phage lambda, is involved in recombination |
| RNase III | – | – | – | LDV1-ORF44, CIV-142R | A464R | – | – | Fused to a Staufen-like dsRNA-binding domain |
| 3β-Hydroxysteroid dehydrogenase, steroid isomerase | A44L | – | – | LDV1-ORF31 | – | – | – | |
| Thymidine kinase | J2R | AMV016 | K196R | – | – | – | T4 | Absent in MSV |
| Ankyrin repeats | B17R | – | A238L | – | A672R | ORF157 | – | Multiple paralogs in FPV, PBCV, and ESV; ESV ORF142 is fused to a RING finger; absent in MCV |
| Smt4/adenovirus-like protease | 17L | AMV181, MSV189 | S273R | – | – | – | Adenovirus | Thiol protease related to eukaryotic SUMO-deconjugating enzyme (Smt4) and adenovirus protease, which is involved in virion maturation (64) |
| Cu-Zn superoxide dismutase | A45R | AMV255 | – | – | A245R | – | BV | Absent in MSV |
| RecB-like nuclease | – | AMV240 | – | – | A467L | – | – | A protein with the restriction endonuclease fold, homologous to archaeal proteins containing a stand-alone RecB nuclease domain (7); absent in MSV |
| C-type lectin | A34R | – | EP153R | – | – | – | HV | Essential for infectivity of the extracellular enveloped form of chordopoxviruses; multiple paralogs in FPV |
| Uncharacterized protein | – | AMV193 | DP71L | – | – | – | HV | Uncharacterized proteins that share a domain with GADD34/MyD116; missing in MSV |
| UvrC-like nuclease (URI domain) | – | – | – | CIV-146R | A134L | – | T4 | Related to intron-encoded nucleases (7); CIV-146R is additionally fused to a domain present in CIV-118L (see below); multiple paralogs in PBCV; absent in FLDV |
| Uncharacterized protein | – | – | – | CIV-136R | A521L | – | HV | Predicted metal-dependent hydrolase (unpublished results) |
| Cathepsin B | – | – | – | LDV1-ORF24, CIV-224L, CIV-361L | – | ORF75 | BV | Cysteine protease |
| Thymidylate synthase | – | MSV238 | – | CIV-225R | – | – | T4, HV | Absent in AMV |
| Bcl2/Bax | FPV039 | – | A179L | LDV1-ORF81 | – | – | HV | Apoptosis inhibitor; absent in variola and MCV |
| Lipase | – | AMV133, MSV048 | – | – | – | ORF185 | – | |
| Lysophospholipase | K5L | – | – | – | A271L | – | – | Absent in variola, MCV, and FPV |
| Matrix metalloprotease | – | AMV070, MSV175, MSV176, MSV179 | – | CIV-165R | – | – | BV | Absent in FLDV |
| Uncharacterized protein | – | – | – | LDV1-ORF70, CIV-067R | A324L | ORF103 | – | |
| Ariadne-like Parkin-domain-containing protein | – | MSV224 | – | LDV1-ORF36 | – | – | – | A regulatory domain with a potential role in ubiquitin-mediated signaling; MSV224 is fused to a SW1/SNF2-like superfamily II helicase |
| NAD-dependent DNA ligase | – | AMV199, MSV162 | – | CIV-205R | – | – | – | A distinct DNA ligase family that is distantly related to ATP-dependent DNA ligases and is ubiquitous in bacteria but uncharacteristic of eukaryotes |
| Very short patch repair endonuclease | – | MSV229, MSV196, AMV257 | – | CIV-069L | – | – | – | A nuclease of the restriction enzyme fold (6); CIV-069L and four of its orthologs in MSV are fused to the baculovirus-like BRO DNA-binding domain |
| MACRO domain | – | AMV247, MSV139 | – | CIV-031R, CIV-032R | – | – | T4 | A phosphoesterase domain present in chromatin and splicing associated complexes |
| Methyltransferase | – | AMV004 | – | CIV-235L | – | – | – | A distinct class of non-purine methyltransferase; absent in MSV |
| Uncharacterized protein | – | MSV198, AMV194 | – | CIV-118L | – | – | – | Expanded in CIV and entomopoxviruses; several entomopoxvirus genes are fused to a BRO-like DNA-binding domain; CIV-146R is fused to a URI domain nuclease |
| Predicted esterase | – | – | – | CIV-463L | A173L | – | – | α/β Hydrolase fold protein |
| Uncharacterized domain | – | – | – | CIV-378R, CIV-232R, CIV-380R, LDV1-ORF14, LDV1-ORF16, LDV1-ORF25 | A676R | – | – | The FLDV proteins and CIV 232R and 280R are fused to an S/T protein kinase domain; the domain in PBCV-A676R is fused to a PBCV-specific domain that is also present in several PBCV S/T kinases |
Gene groups: I, genes conserved in all NCLDV; II, genes conserved in all four families of NCDLV but missing in one or more lineages within families; III, genes conserved in three families of NCLDV; IV, genes conserved in two families of NCLDV.
Abbreviations: BV, baculoviruses; HV, herpesviruses; T4, phage T4; KP, yeast killer plasmids; ORF, open reading frame; –, Not found.
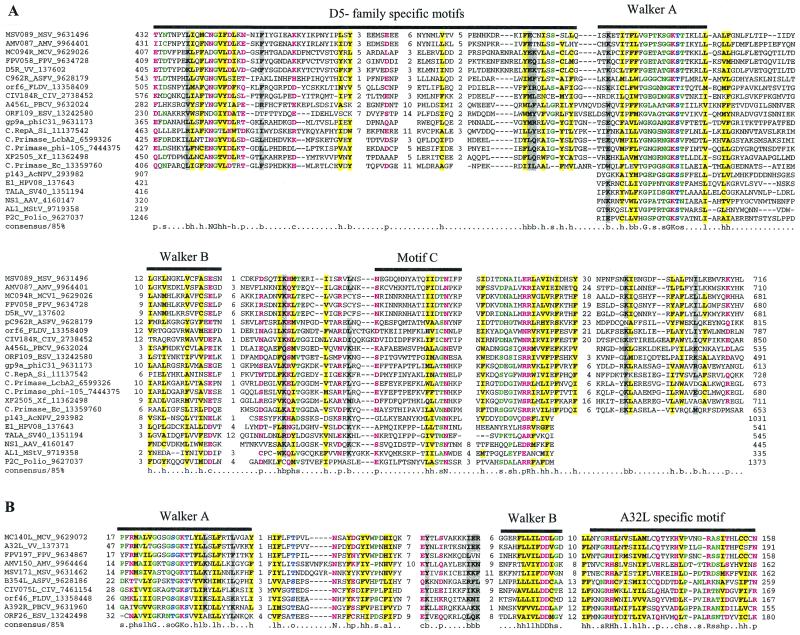
D5 NTPase and helicase.
VV D5R protein is an NTPase that is essential for viral DNA replication (14). The D5R protein and its orthologs in other NCLDV are peripheral members of the AAA+ class of NTPases (42), as demonstrated by the detection of these sequences in iterative database searches started with many AAA+ NTPase sequences. Within the AAA+ class, the D5R family belongs to the so-called helicase superfamily III (SFIII), which consists entirely of viral and plasmid proteins (Fig. 1A). Originally, SFIII has been identified as an assemblage of (predicted) helicases encoded by small RNA and DNA viruses (23, 31). We found that, in PSI-BLAST searches seeded with the sequence of the predicted ATPase domains of poxvirus D5R proteins, statistically significant similarity to E1 proteins of papillomaviruses (bona fide members of SFIII) was detected in the fifth iteration. The closest homologs of the predicted NCLDV helicases are encoded by certain bacteriophages, in some cases integrated into bacterial chromosomes (Fig. 1A). The predicted helicases of NCLDV and this subset of bacteriophage helicases share a distinct, conserved region upstream of the ATPase domain that is not found in any other proteins (Fig. 1A). The NCLDV group also has several unique motifs within the predicted ATPase domain (Fig. 1A).
FIG. 1.
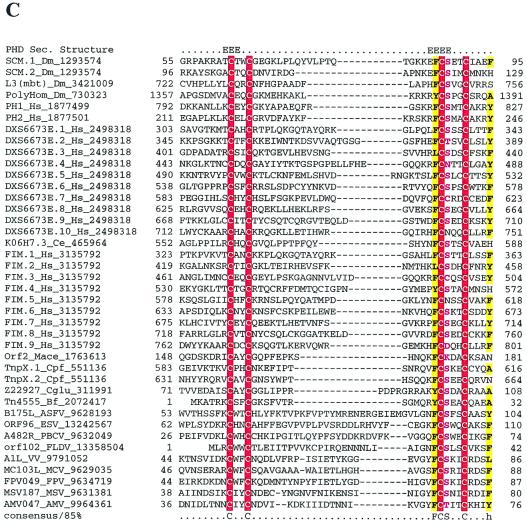
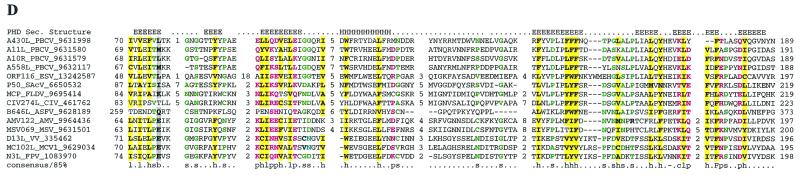
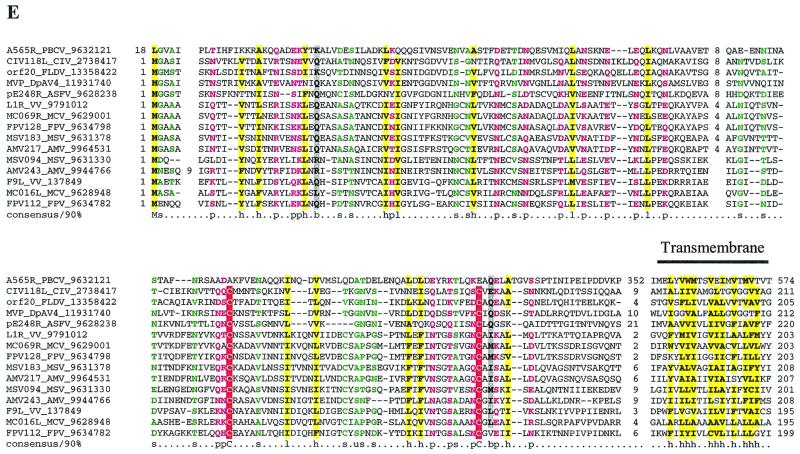
Multiple alignments of conserved proteins that define the cytoplasmic DNA virus clade. (A) D5R-like helicases. With the PBCV ATPase as the seed, the ESV ortholog and many phage primases were recovered with highly significant Expectation (E) values in the first iteration. Proteins from the other NCLDV and the distantly related papillomavirus, parvovirus, and positive-strand RNA viruses were recovered in the second and third iterations with E-values of <10−3. For example, ASFV C962R was recovered with an E-value of 10−8 in the third iteration. Further transitive searches identified all of the members of superfamily III helicase. (B) A32L-like ATPases. With the PBCV ATPase as the seed, iridoviral orthologs were recovered in the first iteration with an E-value of <10−5. Orthologs from all other NCLDV were recovered by the third iteration with significant E-values such as 3 × 10−19 for MCV and 2 × 10−04 for ASFV orthologs. (C) A1L-like transcription factors. A profile made with previously detected FCS domains from the polyhomeotic and FIM families of proteins, when run against the NCLDV protein sets, with an inclusion cutoff of 0.01, recovered all members of this family; VV A1L, for example, was recovered with an E-value of 10−4. (D) D13L-like capsid proteins. With p50 of the Spodoptera exigua ascovirus as the seed, the PBCV and other iridoviral capsid proteins were recovered with E-values of <2 × 10−8. The ASFV ortholog was detected in the third iteration with an E-value of 3 × 10−3, and the poxviral D13L-like proteins were recovered at borderline E-values (0.14) in the fourth iteration. When a profile made from the alignment of the PBCV, iridovirus, and ASFV sequences was run against a database of all NCLDV proteins, the poxviral orthologs were detected as top hits, with E-values of <10−5. The probability of the conserved motifs shown here to occur in these proteins by chance was <10−15, as computed by using the MACAW program (49). (E) L1R/F9L-like virion membrane proteins. With CIV 048L as the seed, the ASFV and PBCV orthologs were recovered in the second iteration, with E-values of 8 × 10−4 and 10−3, respectively. The entomopoxviral orthologs were detected in the third iteration with an E-value of 2 × 10−4. A transitive search with the entomopoxviral proteins recovered the other poxviral proteins with E-values of <10−3. Each protein is denoted by the corresponding gene name followed by species abbreviation and the GenBank Identifier (GI) number. The numbers preceding and following the alignments indicate the positions of the first and last residues of the aligned regions in the corresponding protein sequences. The numbers between aligned blocks indicate the number of inserted residues that were omitted from the figure. The coloring reflects the conservation of amino acid residues at 85% consensus. The coloring scheme and the consensus abbreviations are as follows: hydrophobic residues (LIYFMWACV) are designated “h” in the consensus line, aliphatic (LIAV) residues are also shaded yellow and designated “l,” alcohol (S,T) is blue and designated “o,” charged (KERDH) residues are purple and designated “c,” polar (STEDRKHNQ) residues are purple and designated “p,” small (SACGDNPVT) residues are green and designated “s,” big (LIFMWYERKQ) residues are shaded gray and designated “b.” Conserved cysteines predicted to form a Zn-finger structure (C) or a disulfide bond (E) are indicated by white letters against a red background. Secondary structure elements predicted by using the PHD program are indicated in panels C and D; where “E” indicates extended conformation (b-strand) and “H” indicates the α-helix. The abbreviations for the NCLDV are defined in Materials and Methods. Additional abbreviations: AAV, adeno-associated virus 5; AcNPV, Autographa californica nucleopolyhedrovirus; Bf, Bacteroides fragilis, Ce, Caenorhabditis elegans; Cglu, Corynebacterium glutamicum; Cpf, Clostridium perfringens; Dm, Drosophila melanogaster; DpAV4, Diadromus pulchellus ascovirus; Ec, Escherichia coli; HPV08, human papillomavirus type 8; Hs, Homo sapiens; LcbA2, Lactobacillus casei bacteriophage A2; Mace, Methanosarcina acetivorans; MStV, maize streak virus; phi-105, Bacteriophage phi-105; phiC31, Bacteriophage phiC31; Polio, human poliovirus 1; SacV, Spodoptera exigua ascovirus; Si, Sulfolobus islandicus; SV40, Simian virus 40; Xf, Xylella fastidiosa.
Packaging ATPase A32L.
The A32L gene product has been predicted to possess ATPase activity, primarily on the basis of the conservation of the P-loop and Mg2+-binding motifs (33), and subsequently has been shown to be involved in DNA packaging into virions (13). Comparisons of the NCLDV protein sets and iterative database searches detected apparent orthologs of A32L in all NCLDV (Fig. 1B). Although these predicted ATPases may be distantly related to the AAA+ superclass, they showed no specific relationship with any other ATPase family. In particular, other ATPases do not contain readily detectable counterparts of the C-terminal motifs of A32L, which should be considered a synapomorphy of NCLDV (Fig. 1B).
Transcription factor A1L.
A1L is a small protein that contains a Zn-finger-domain that we designated the FCS-finger (so named after a characteristic amino acid signature) and functions as a transcriptional transactivator of late VV genes (28); A1L orthologs were found in all NCLDV. The FCS-finger is a previously undetected Zn-binding domain that we identified in several eukaryotic chromatin proteins such as the Drosophila Sex Combs on Middle Leg, Polyhomeotic, Lethal 3 of Malignant Brain Tumor, and vertebrate FIM. This domain is also found fused to the C termini of recombinases from certain prokaryotic transposons. However, A1L orthologs from NCLDV are a distinct stand-alone form of the FCS domain and thus should be considered an NCLDV synapomorphy (Fig. 1C).
Capsid protein D13L.
The virions of different NCLDV have dramatically different structures. The major capsid proteins of iridoviruses and phycodnaviruses, both of which have icosahedral capsids surrounding an inner lipid membrane, showed a high level of sequence conservation. A more limited, but statistically significant sequence similarity was observed between these proteins and the major capsid protein (p72) of ASFV, which also has an icosahedral capsid. It was surprising, however, to find that all of these proteins shared a conserved domain with the poxvirus protein D13L, which is an integral virion component thought to form a scaffold for the formation of viral crescents and immature virions (54). In spite of low sequence similarity, D13L sequences share a common domain with conserved predicted structural elements with the major capsid proteins of the other NCLDV (Fig. 1D). The capsid proteins of iridoviruses, phycodnaviruses, and ASFV have an additional C-terminal domain that is predicted to adopt the jelly roll fold typical of capsid proteins of numerous DNA and RNA viruses (46). In poxvirus D13L proteins, the jelly roll domain is replaced by a distinct β-strand-rich domain that showed no detectable relationship with any known domains. This difference in the C-terminal domains of poxvirus D13L proteins compared to the major capsid proteins of other NCLDV probably reflects the new function of D13L as a scaffold for viral crescents.
Virion membrane protein L1R/F9L.
Paralogous poxvirus genes L1R and F9L encode membrane proteins that have a conserved domain architecture, with a single, C-terminal transmembrane helix, and an N-terminal, multiple-disulfide-bonded domain (51). The L1R protein is myristoylated and has been implicated in virion assembly (45, 68). Homologs of the L1R/F9L family proteins so far have not been detected outside poxviruses. However, our comparisons revealed apparent representatives of this family in all NCLDV, with the single exception of ESV (Fig. 1E). With the exception of PBCV, all NCLDV share two of the disulfide-bond-forming cysteine residues and have a transmembrane helix C-terminal to the core domain. The PBCV protein is highly divergent and seems to have lost the disulfide-bonding cysteines; however, it has an additional cysteine-rich, EGF-like domain that is also found in other PBCV proteins (data not shown). This domain is inserted between the core L1R-like domain and the C-terminal transmembrane helix.
A conserved structural role for this protein is compatible with the existence of a lipid membrane in all NCLDV, in spite of the major differences in virion structure. Furthermore, the conservation of the myristoylated, disulfide-bonded protein in most of the NCLDV correlates with the conservation of the thiol-disulfide oxidoreductase E10R which, in VV, is required for the formation of disulfide bonds in L1R and F9L (52).
Other apparent synapomorphies of NCLDV.
Even when apparent orthologs of a viral protein are present in cellular life forms, the viral version may have unique features. An example is the thiol-disulfide oxidoreductase E10R. The proteins of this family encoded by different NCLDV show limited sequence similarity to each other, and some are more similar to apparent orthologs from eukaryotes, such as the yeast ERV1/2 proteins (52). However, all nonviral members of this family share two pairs of conserved cysteines, whereas only one pair is conserved in the proteins from NCLDV.
Another notable ancestral protein family of NCLDV consists of homologs of proliferating cell nuclear antigen (PCNA), a protein that is ubiquitous in cellular life forms and functions as the sliding clamp during DNA replication (11). The members of the PCNA superfamily identified in NCLDV show limited sequence similarity to the cellular homologs; in fact, the poxvirus PCNA homologs (G8R) were identified in this study only through the use of the sequence-structure threading technique. Phylogenetic analyses on the PCNA superfamily indicated that the NCLDV PCNA homologs tend to cluster together, to the exclusion of eukaryotic homologs, but typically form longer branches than any cellular PCNAs, suggesting rapid divergence during NCLDV evolution (unpublished data). Poxvirus G8R is the most divergent member of the PCNA superfamily. The available experimental evidence points to a principal role of this protein in vaccinia virus late gene transcription, rather than replication (69, 74), suggesting a causal connection between rapid sequence divergence and the change of function.
Among the proteins that are conserved in three of the four NCLDV families, the most notable one is the membrane protein that, in poxviruses, is represented by three paralogs, J5L, G9R, and A16L, which are predicted to form multiple disulfide bonds (51). These proteins resemble the virion membrane proteins of the L1R/F9L group in domain architecture, but appear not to be homologous to them or to any other proteins.
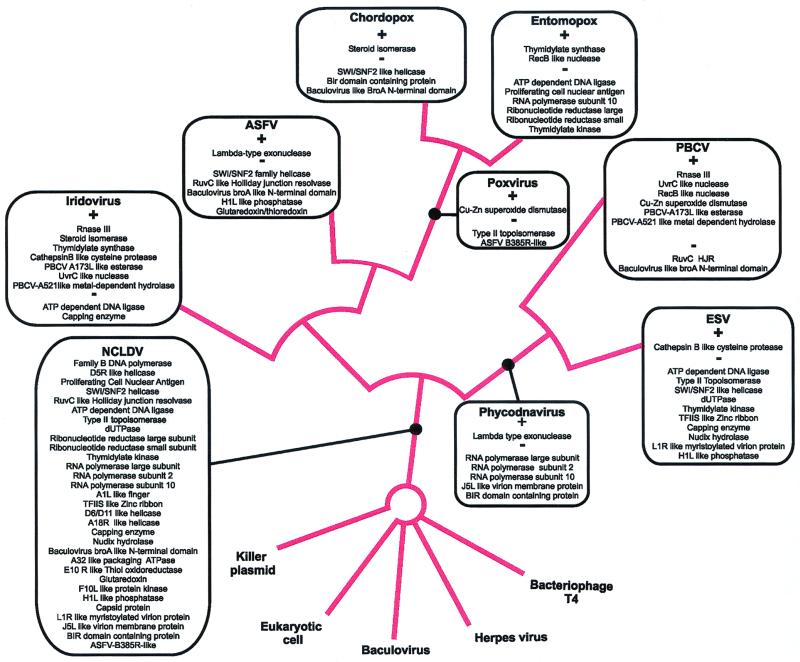
Cladistic analysis suggests monophyly of NCLDV.
Phylogenetic tree analysis of those NCLDV proteins that have homologs in other viruses and in cellular life forms, such as DNA polymerase, helicases and others (Table 1), fails to support monophyly of NCLDV (26; unpublished observations). However, this cannot be considered strong evidence against monophyly because viral genomes tend to evolve rapidly, resulting in distortions of phylogenetic tree topologies. Indeed, as discussed above, even those groups of orthologous NCLDV proteins that comprise clear synapomorphies show only limited sequence conservation. Therefore, as an alternative approach for assessing the evolutionary relationships among the NCLDV, we undertook formal cladistic analysis (25) of viral gene sets after identifying probable orthologs in other viruses and cellular organisms (Table 1). All genes that occur in at least two families of NCLDV were scored as described in Materials and Methods to obtain character states for the terminal taxa under examination. The 11 terminal taxa considered in this analysis were chordopox viruses, entomopox viruses, asfarviruses (ASFV), iridoviruses (CIV and FLDV), PBCV, ESV, herpesviruses, baculoviruses, bacteriophage T4, and the eukaryotic cell (host cell). A total of 59 characters were scored over these 11 taxa to construct the data matrix used in the cladistic analysis (data not shown [available as supplementary material from the authors]).
Trees that provided the shortest path of character state changes to result in the character configuration observed in the terminal taxa were identified by using the Branch and Bound method and the Exhaustive Search algorithm that evaluates all possible tree topologies for the given terminal taxa. One most parsimonious tree was found that supported the monophyly of the NCLDV by 16 synapomorphies. As expected, the monophyly of the so-called phycodnavirus clade (PBCV plus ESV) and the poxvirus clade (entomopox viruses plus chordopoxviruses) was strongly supported (Fig. 2). In addition, there was a weaker support for the monophyly of the animal viruses (poxviruses plus ASFV plus iridoviruses), to the exclusion of the phycodnaviruses, by six synapomorphies. Furthermore, the tree contained a clade consisting of poxviruses and asfarviruses, to the exclusion of the iridoviruses, which was supported by eight synapomorphies. This tree was used to extract a list of derived shared characters for the NCLDV clade that were used in reconstructing the repertoire of genes present in the hypothetical NCLDV (see below). The monophyly of the three animal viral families, namely, asfarviruses, iridoviruses, and poxviruses, emerged consistently with different sets of characters, but the relationships among these families were highly sensitive to minor changes in characters used in the analysis (data not shown). Thus, the actual branching pattern within the animal NCLDV clade requires additional data for confident resolution.
FIG. 2.
Consensus cladogram of cytoplasmic DNA viruses. The cladistic analysis was performed as described in the text. The proteins that were probably present in the common ancestor of the universally supported NCLDV clade are superimposed on the consensus tree. Also shown on the consensus tree are the state changes in each of the terminal lineages and the strictly supported clades. The plus sign indicates a character that is most parsimoniously explained as an independent gain that was most likely acquired through horizontal transfer between the viral genome or through transfer from the host genome. The minus sign denotes the loss of an ancestral character in a particular lineage.
Hypothetical ancestral NCLDV.
Given the support for a monophyletic NCLDV clade, the possibility emerges for an approximate reconstruction of the hypothetical ancestral virus. The genes that are shared by all viruses within this clade are obvious candidates for ancestral origin but, additionally, other genes identified as synapomorphies of the NCLDV clade are also, according to the parsimony principle, likely to have been present in their last common ancestor. These typically are genes present in the majority of the NCLDV taxa considered in this analysis. Under this reasoning, the absence of otherwise conserved genes in one lineage is attributed to gene loss, in case of essential genes accompanied by nonorthologous gene displacement (32). Lineage-specific gene loss obviously occurred also within individual NCLDV families, particularly in ESV, which does not have many genes conserved in all or most NCLDV, including PBCV, and, among poxviruses, in MCV that has lost all genes involved in nucleotide metabolism (51). A probable example of displacement is the topoisomerase function that is represented by the predicted ancestral form, type II topoisomerase, in asfarviruses, iridoviruses, and phycodnaviruses (except for ESV, which apparently has lost this gene), whereas poxviruses have an unrelated type IB topoisomerase. Some of the genes that are conserved in only two of the NCLDV families also might be part of the legacy of the ancestral virus, but in these cases, it is difficult to rule out alternative scenarios, such as independent acquisition from the host or horizontal gene transfer.
Under these assumptions, we arrive at a conservative list of 31 ancestral viral genes (Table 1); for comparison, all poxviruses share ca. 50 genes (8). Considering that the ancestral virus might have been a simpler entity than its extant descendants, even this conservative reconstruction may be a reasonable approximation of the ancestral set of essential viral genes. Examination of this list suggests that the ancestral NCLDV already had fairly elaborate systems for genome replication and expression, some enzymes of nucleotide metabolism, a packaging mechanism, capsid and membrane virion proteins, an electron-transfer system for disulfide-bond formation in the latter, a mechanism of protein phosphorylation-dephosphorylation probably involved in the regulation of virion morphogenesis, and possibly an apoptosis inhibitor (Table 2).
TABLE 2.
Predicted functional systems of the ancestral nucleocytoplasmic DNA virus
| Function and/or pathway | Proteins |
|---|---|
| DNA replication | DNA polymerase, D5R-like helicase, RuvC-like Holliday junction resolvase, PCNA (DNA clamp), ATP-dependent DNA ligase, type II topoisomerase, dUTPase |
| DNA precursor synthesis | Ribonucleotide reductase (two subunits), thymidylate kinase |
| Transcription and RNA processing | RNA polymerase (two large subunits and subunit 10), A1L-like and TFIIS-like transcription factors, D6R-like, A18R-like, SWI/SNF2-like helicases, capping enzyme, BRO-like DNA-binding protein, Nudix hydrolase |
| Virion morphogenesis | A32-like packaging ATPase, E10R-like thiol-oxidoreductase, glutaredoxin-thioredoxin |
| Regulation of morphogenesis | F10L-like protein kinase, H1L-like phosphatase |
| Virion structure | D13L-like capsid protein, L1R-family and J5L-family virion membrane proteins |
| Inhibition of apoptosis | BIR-domain-containing protein |
Given the presence of nucleocytoplasmic, purely cytoplasmic, and nuclear life cycles in the monophyletic assemblage of NCLDV, it appears most likely that their last common ancestor had both nuclear and cytoplasmic phases in its life cycle. From this ancestral state, some of the descendant lineages, such as phycodnaviruses, appear to have moved to an entirely nuclear replication. The wholly nuclear replication of vertebrate iridoviruses (22, 36) also appears to be a secondary adaptation because FLDV has lost several essential enzymes that are essential for viruses that replicate in the cytoplasm, such as DNA ligase, capping enzyme, and topoisomerase.
The ancestral virus can be inferred to have had an icosahedral capsid with an inner membrane layer, a structure most similar to those of iridoviruses and PBCV. This notion is supported by the presence of icosahedral capsids in three of the four NCLDV families, which correlates with the presence of the jelly roll domain in the major capsid protein, and the general consideration of the icosahedron being one of the basic virion structures in numerous, diverse viruses. The more complex organization of poxvirus virions appears to be a derived state. With the previously described conservation of the ERV-family thiol-oxidoreductase and glutaredoxin (with the apparent exception of ASFV) that contribute to the formation of disulfide bonds in virion membrane proteins (51, 52) and the present demonstration of the conservation of three structural proteins of the virion, the evolutionary connection between the poxvirus virions and those of other NCLDV appears certain.
The genes of the ancestral NCLDV that were responsible for virus-host interaction cannot be inferred from the comparison of extant viral genomes because the repertoires of such genes in different NCLDV families are largely different and, based on the existence of highly similar cellular homologs for most of them, must have been acquired independently. The BIR domain-containing apoptosis inhibitor could be an exception to this general pattern (Table 1). We are unlikely to get any insight into this aspect of the ancestral NCLDV until clear indications are obtained as to what kind of host it infected. If the fungal connections mentioned below point to the original host, a relatively simple genome with a small number of host-interaction genes seems a plausible possibility.
Relationships between NCLDV and other genetic elements and origin of NCLDV.
Many NCLDV genes have homologs or even apparent orthologs in other viruses and plasmids (Table 1). In particular, multiple relationships have been previously noticed to exist between NCLDV genes (specifically, those of poxviruses) and genes of T-even bacteriophages (34, 62). However, neither T-even phages nor herpesviruses or baculoviruses possess a significant subset of the core gene set of the NCLDV (Table 1). Furthermore, the genes that are shared do not show appreciable synapomorphic features. Therefore, direct evolutionary relationships between these classes of viruses apparently cannot be positively established. The observed overlaps between gene sets can be explained largely by independent acquisition of genes that are generically required for DNA virus replication (for example, DNA polymerase, ribonucleotide reductase, or thymidylate kinase) and, possibly, some cases of horizontal gene exchange.
A more coherent relationship appears to exist between the NCLDV and linear DNA plasmids from fungal mitochondria, with five shared genes (of the 10 to 12 genes that are typically present on these plasmids [18, 39]) (Table 1). Importantly, these seem to be the principal genes that are required for DNA virus genome expression in the cytoplasm, including two RNA polymerase subunits, a helicase involved in transcription, and a capping enzyme with a conserved domain architecture (Table 1). In at least one case, that of the D6R-type helicase, the NCLDV proteins show high sequence similarity to the plasmid homolog, to the exclusion of other homologous helicases (data not shown). It seems plausible that the fungal plasmids indeed contain a part of the core gene set of the hypothetical ancestral NCLDV. However, the fungal plasmid genomes have a terminal protein that functions in replication priming and, in this respect, resemble adenoviruses and protein-priming DNA phages (48), rather than NCLDV; the monophyly of DNA polymerases from protein-priming viruses and plasmids is supported by phylogenetic tree analysis (29). Thus, the data suggest complex evolutionary relationships, with components of the replication and expression systems drawn from different types of genetic elements, rather than a direct link between the NCLDV and fungal plasmids.
A complex evolutionary scenario for the origin of the NCLDV, including multiple gene exchanges between different types of genomes, is suggested by the phyletic provenance of several other genes shared by all or a subset of NCLDV families. These include the replicative helicase D5R, the Holliday junction resolvase (HJR) A22R, and the predicted protease I7L (Table 1). The distribution of the D5R homologs is particularly unusual. As shown above (Fig. 1), true orthologs of the NCLDV replicative helicase were detected only in certain bacteriophages. More distant members of the helicase III superfamily are encoded by diverse small genetic elements, including ssDNA viruses (geminiviruses and parvoviruses), small dsDNA viruses (papovaviruses), positive-strand RNA viruses (for example, picornaviruses), some phages, and plasmids. So far, no members of this superfamily encoded in genomes of cellular life forms (some prophages notwithstanding) have been detected. This distribution pattern of an essential viral gene suggests a long history of dissemination between (relatively) small genomes, perhaps tracing back to the ancient RNA world.
A different evolutionary history appears plausible for the RuvC-like HJR A22R, which is present in poxviruses, at least some iridoviruses, and phycodnaviruses, suggesting that it might have been inherited from the common ancestor of the NCLDV. This enzyme belongs to a family of resolvases that are common in bacteria but not detectable in eukaryotes, except for a nuclease that functions in fungal mitochondria; the latter shows the strongest (albeit limited) sequence similarity to the resolvases of NCLDV (19). This suggests at least two horizontal transfers, from protomitochondria to fungi and from fungi to the ancestral NCLDV (assuming that this resolvase indeed is inherited by NCLDV from their common ancestor). In the lineages which lack the RuvC-like HJR, such as PBCV and ASFV, it might have been displaced by an alternative enzyme, namely, the Lambda-type exonuclease that is present in these viruses (6) (Table 1) or the RecB-like nuclease in PBCV.
The available data are insufficient to reconstruct a complete evolutionary scenario for the origin of the ancestral NCLDV. Genome sequencing of representatives of additional viral families has the potential to shed light on the evolutionary source(s) of NCLDV as suggested, for example, by the recent preliminary analysis of the genome of the archaeal virus SIRV1 (9). This virus has a relatively small genome of 32 kB with covalently closed hairpins at the ends, which resembles the genome structure of poxviruses, asfaviruses, and phycodnaviruses. However, the HJR and dUTPase of SIRV1 show clear archaeal affinities, emphasizing a difference from NCLDV (unpublished data). Taken together, the above observations show that the ancestral viral genome probably assembled via gradual accretion of genes from different genetic sources, including host genomes, plasmids, and other viruses. It appears that a complex history of multiple horizontal genes transfers and gene losses both preceded and succeeded the emergence of the ancestral NCLDV. Thus, it is all the more notable that this evolutionary focal point can be identified and some basic aspects of the replication of the ancestral virus can be reconstructed with reasonable confidence on the basis of a detailed comparison of extant viral genomes.
ACKNOWLEDGMENTS
We thank Bernard Moss for critical reading of the manuscript and useful suggestions and Stewart Shuman for a helpful discussion.
ADDENDUM IN PROOF
While this article was being processed for production, a paper describing the sequence of the ESV1 genome was published (N. Delaroque, D. G. Muller, G. Bothe, T. Pohl, R. Knippers, and W. Boland, Virology 287:112–132, 2001).
REFERENCES
- 1.Afonso C L, Tulman E R, Lu Z, Oma E, Kutish G F, Rock D L. The genome of Melanoplus sanguinipes entomopoxvirus. J Virol. 1999;73:533–552. doi: 10.1128/jvi.73.1.533-552.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Afonso C L, Tulman E R, Lu Z, Zsak L, Kutish G F, Rock D L. The genome of fowlpox virus. J Virol. 2000;74:3815–3831. doi: 10.1128/jvi.74.8.3815-3831.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alba M M, Das R, Orengo C A, Kellam P. Genomewide function conservation and phylogeny in the Herpesviridae. Genome Res. 2001;11:43–54. doi: 10.1101/gr.149801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altschul S F, Koonin E V. PSI-BLAST—a tool for making discoveries in sequence databases. Trends Biochem Sci. 1998;23:444–447. doi: 10.1016/s0968-0004(98)01298-5. [DOI] [PubMed] [Google Scholar]
- 5.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aravind L, Makarova K S, Koonin E V. Holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res. 2000;28:3417–3432. doi: 10.1093/nar/28.18.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aravind L, Walker D R, Koonin E V. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 1999;27:1223–1242. doi: 10.1093/nar/27.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bawden A L, Glassberg K J, Diggans J, Shaw R, Farmerie W, Moyer R W. Complete genomic sequence of the Amsacta moorei entomopoxvirus: analysis and comparison with other poxviruses. Virology. 2000;274:120–139. doi: 10.1006/viro.2000.0449. [DOI] [PubMed] [Google Scholar]
- 9.Blum H, Zillig W, Mallok S, Domdey H, Prangishvili D. The genome of the archaeal virus SIRV1 has features in common with genomes of eukaryal viruses. Virology. 2001;281:6–9. doi: 10.1006/viro.2000.0776. [DOI] [PubMed] [Google Scholar]
- 10.Brick D J, Burke R D, Schiff L, Upton C. Shope fibroma virus RING finger protein N1R binds DNA and inhibits apoptosis. Virology. 1998;249:42–51. doi: 10.1006/viro.1998.9304. [DOI] [PubMed] [Google Scholar]
- 11.Bruck I, O'Donnell M. The ring-type polymerase sliding clamp family. Genome Biol. 2001;2:3001.1–3001.3. doi: 10.1186/gb-2001-2-1-reviews3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bugert J J, Darai G. Poxvirus homologues of cellular genes. Virus Genes. 2000;21:111–133. [PubMed] [Google Scholar]
- 13.Cassetti M C, Merchlinsky M, Wolffe E J, Weisberg A S, Moss B. DNA packaging mutant: repression of the vaccinia virus A32 gene results in noninfectious, DNA-deficient, spherical, enveloped particles. J Virol. 1998;72:5769–5780. doi: 10.1128/jvi.72.7.5769-5780.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans E, Klemperer N, Ghosh R, Traktman P. The vaccinia virus D5 protein, which is required for DNA replication, is a nucleic acid-independent nucleoside triphosphatase. J Virol. 1995;69:5353–5361. doi: 10.1128/jvi.69.9.5353-5361.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fields B N. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. [Google Scholar]
- 16.Fischer D. Hybrid fold recognition: combining sequence derived properties with evolutionary information. Pac Symp Biocomput. 2000;2000:119–130. [PubMed] [Google Scholar]
- 17.Flavell A J. Retroelements, reverse transcriptase and evolution. Comp Biochem Physiol B Biochem Mol Biol. 1995;110:3–15. doi: 10.1016/0305-0491(94)00122-b. [DOI] [PubMed] [Google Scholar]
- 18.Fukuhara H. Linear DNA plasmids of yeasts. FEMS Microbiol Lett. 1995;131:1–9. doi: 10.1016/0378-1097(95)00201-f. [DOI] [PubMed] [Google Scholar]
- 19.Garcia A D, Aravind L, Koonin E V, Moss B. Bacterial-type DNA Holliday junction resolvases in eukaryotic viruses. Proc Natl Acad Sci USA. 2000;97:8926–8931. doi: 10.1073/pnas.150238697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia-Beato R, Salas M L, Vinuela E, Salas J. Role of the host cell nucleus in the replication of African swine fever virus. DNA Virol. 1992;188:637–649. doi: 10.1016/0042-6822(92)90518-t. [DOI] [PubMed] [Google Scholar]
- 21.Goebel S J, Johnson G P, Perkus M E, Davis S W, Winslow J P, Paoletti E. The complete DNA sequence of vaccinia virus. Virology. 1990;179:247–266. doi: 10.1016/0042-6822(90)90294-2. ; 517–563. [DOI] [PubMed] [Google Scholar]
- 22.Goorha R. Frog virus 3 DNA replication occurs in two stages. J Virol. 1982;43:519–528. doi: 10.1128/jvi.43.2.519-528.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorbalenya A E, Koonin E V, Wolf Y I. A new superfamily of putative NTP-binding domains encoded by genomes of small DNA and RNA viruses. FEBS Lett. 1990;262:145–148. doi: 10.1016/0014-5793(90)80175-i. [DOI] [PubMed] [Google Scholar]
- 24.Hannenhalli S, Chappey C, Koonin E V, Pevzner P A. Genome sequence comparison and scenarios for gene rearrangements: a test case. Genomics. 1995;30:299–311. doi: 10.1006/geno.1995.9873. [DOI] [PubMed] [Google Scholar]
- 25.Harvey P H, Pagel M D. The comparative method in evolutionary biology. Oxford, England: Oxford University Press; 1991. [Google Scholar]
- 26.Ilyina T V, Koonin E V. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 1992;20:3279–3285. doi: 10.1093/nar/20.13.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jakob N J, Muller K, Bahr U, Darai G. Analysis of the first complete DNA sequence of an invertebrate iridovirus: coding strategy of the genome of Chilo iridescent virus. Virology. 2001;286:182–196. doi: 10.1006/viro.2001.0963. [DOI] [PubMed] [Google Scholar]
- 28.Keck J G, Kovacs G R, Moss B. Overexpression, purification, and late transcription factor activity of the 17-kilodalton protein encoded by the vaccinia virus A1L gene. J Virol. 1993;67:5740–5748. doi: 10.1128/jvi.67.10.5740-5748.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knopf C W. Evolution of viral DNA-dependent DNA polymerases. Virus Genes. 1998;16:47–58. doi: 10.1023/a:1007997609122. [DOI] [PubMed] [Google Scholar]
- 30.Kool M, Ahrens C H, Vlak J M, Rohrmann G F. Replication of baculovirus DNA. J Gen Virol. 1995;76:2103–2118. doi: 10.1099/0022-1317-76-9-2103. [DOI] [PubMed] [Google Scholar]
- 31.Koonin E V. A common set of conserved motifs in a vast variety of putative nucleic acid-dependent ATPases including MCM proteins involved in the initiation of eukaryotic DNA replication. Nucleic Acids Res. 1993;21:2541–2547. doi: 10.1093/nar/21.11.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koonin E V, Mushegian A R, Bork P. Non-orthologous gene displacement. Trends Genet. 1996;12:334–336. [PubMed] [Google Scholar]
- 33.Koonin E V, Senkevich T G, Chernos V I. Gene A32 product of vaccinia virus may be an ATPase involved in viral DNA packaging as indicated by sequence comparisons with other putative viral ATPases. Virus Genes. 1993;7:89–94. doi: 10.1007/BF01702351. [DOI] [PubMed] [Google Scholar]
- 34.Kutter E, Gachechiladze K, Poglazov A, Marusich E, Shneider M, Aronsson P, Napuli A, Porter D, Mesyanzhinov V. Evolution of T4-related phages. Virus Genes. 1995;11:285–297. doi: 10.1007/BF01728666. [DOI] [PubMed] [Google Scholar]
- 35.Li Y, Lu Z, Sun L, Ropp S, Kutish G F, Rock D L, Van Etten J L. Analysis of 74 kb of DNA located at the right end of the 330-kb chlorella virus PBCV-1 genome. Virology. 1997;237:360–377. doi: 10.1006/viro.1997.8805. [DOI] [PubMed] [Google Scholar]
- 36.Martin J P, Aubertin A M, Tondre L, Kirn A. Fate of frog virus 3 DNA replicated in the nucleus of arginine-deprived CHO cells. J Gen Virol. 1984;65:721–732. doi: 10.1099/0022-1317-65-4-721. [DOI] [PubMed] [Google Scholar]
- 37.Massung R F, Esposito J J, Liu L I, Qi J, Utterback T R, Knight J C, Aubin L, Yuran T E, Parsons J M, Loparev V N, et al. Potential virulence determinants in terminal regions of variola smallpox virus genome. Nature. 1993;366:748–751. doi: 10.1038/366748a0. [DOI] [PubMed] [Google Scholar]
- 38.McAuslan B R, Armentrout R W. The biochemistry of icosahedral cytoplasmic deoxyviruses. Curr Top Microbiol Immunol. 1974;68:77–105. doi: 10.1007/978-3-642-66044-3_4. [DOI] [PubMed] [Google Scholar]
- 39.Meinhardt F, Schaffrath R, Larsen M. Microbial linear plasmids. Appl Microbiol Biotechnol. 1997;47:329–336. doi: 10.1007/s002530050936. [DOI] [PubMed] [Google Scholar]
- 40.Moss B. Poxviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2637–2671. [Google Scholar]
- 41.Muller D G, Kapp M, Knippers R. Viruses in marine brown algae. Adv Virus Res. 1998;50:49–67. doi: 10.1016/s0065-3527(08)60805-2. [DOI] [PubMed] [Google Scholar]
- 42.Neuwald A F, Aravind L, Spouge J L, Koonin E V. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 43.Notredame C, Higgins D G, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 44.Pagel M. Inferring the historical patterns of biological evolution. Nature. 1999;401:877–884. doi: 10.1038/44766. [DOI] [PubMed] [Google Scholar]
- 45.Ravanello M P, Hruby D E. Characterization of the vaccinia virus L1R myristylprotein as a component of the intracellular virion envelope. J Gen Virol. 1994;75:1479–1483. doi: 10.1099/0022-1317-75-6-1479. [DOI] [PubMed] [Google Scholar]
- 46.Rossmann M G, Johnson J E. Icosahedral RNA virus structure. Annu Rev Biochem. 1989;58:533–573. doi: 10.1146/annurev.bi.58.070189.002533. [DOI] [PubMed] [Google Scholar]
- 47.Rost B, Sander C. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins. 1994;19:55–72. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- 48.Salas M. Protein-priming of DNA replication. Annu Rev Biochem. 1991;60:39–71. doi: 10.1146/annurev.bi.60.070191.000351. [DOI] [PubMed] [Google Scholar]
- 49.Schuler G D, Altschul S F, Lipman D J. A workbench for multiple alignment construction and analysis. Proteins. 1991;9:180–190. doi: 10.1002/prot.340090304. [DOI] [PubMed] [Google Scholar]
- 50.Senkevich T G, Bugert J J, Sisler J R, Koonin E V, Darai G, Moss B. Genome sequence of a human tumorigenic poxvirus: prediction of specific host response-evasion genes. Science. 1996;273:813–816. doi: 10.1126/science.273.5276.813. [DOI] [PubMed] [Google Scholar]
- 51.Senkevich T G, Koonin E V, Bugert J J, Darai G, Moss B. The genome of molluscum contagiosum virus: analysis and comparison with other poxviruses. Virology. 1997;233:19–42. doi: 10.1006/viro.1997.8607. [DOI] [PubMed] [Google Scholar]
- 52.Senkevich T G, White C L, Koonin E V, Moss B. A viral member of the ERV1/ALR protein family participates in a cytoplasmic pathway of disulfide bond formation. Proc Natl Acad Sci USA. 2000;97:12068–12073. doi: 10.1073/pnas.210397997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shors T, Keck J G, Moss B. Down regulation of gene expression by the vaccinia virus D10 protein. J Virol. 1999;73:791–796. doi: 10.1128/jvi.73.1.791-796.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sodeik B, Griffiths G, Ericsson M, Moss B, Doms R W. Assembly of vaccinia virus: effects of rifampin on the intracellular distribution of viral protein p65. J Virol. 1994;68:1103–1114. doi: 10.1128/jvi.68.2.1103-1114.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Swofford D L. PAUP*: phylogenetic analysis using parsimony (and other methods). Sunderland, Mass: Sinauer Associates; 2000. [Google Scholar]
- 56.Temin H M. Reverse transcription in the eukaryotic genome: retroviruses, pararetroviruses, retrotransposons, and retrotranscripts. Mol Biol Evol. 1985;2:455–468. doi: 10.1093/oxfordjournals.molbev.a040365. [DOI] [PubMed] [Google Scholar]
- 57.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tidona C A, Darai G. The complete DNA sequence of lymphocystis disease virus. Virology. 1997;230:207–216. doi: 10.1006/viro.1997.8456. [DOI] [PubMed] [Google Scholar]
- 59.Tidona C A, Darai G. Iridovirus homologues of cellular genes—implications for the molecular evolution of large DNA viruses. Virus Genes. 2000;21:77–81. [PubMed] [Google Scholar]
- 60.Tidona C A, Darai G. Molecular anatomy of lymphocystis disease virus. Arch Virol Suppl. 1997;13:49–56. doi: 10.1007/978-3-7091-6534-8_5. [DOI] [PubMed] [Google Scholar]
- 61.Van Etten J L, Meints R H. Giant viruses infecting algae. Annu Rev Microbiol. 1999;53:447–494. doi: 10.1146/annurev.micro.53.1.447. [DOI] [PubMed] [Google Scholar]
- 62.Villarreal L P, DeFilippis V R. A hypothesis for DNA viruses as the origin of eukaryotic replication proteins. J Virol. 2000;74:7079–7084. doi: 10.1128/jvi.74.15.7079-7084.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vinuela E. African swine fever virus. Curr Top Microbiol Immunol. 1985;116:151–170. doi: 10.1007/978-3-642-70280-8_8. [DOI] [PubMed] [Google Scholar]
- 64.Webster A, Hay R T, Kemp G. The adenovirus protease is activated by a virus-coded disulphide-linked peptide. Cell. 1993;72:97–104. doi: 10.1016/0092-8674(93)90053-s. [DOI] [PubMed] [Google Scholar]
- 65.Whitley R J. Herpes simplex viruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2297–2342. [Google Scholar]
- 66.Willer D O, McFadden G, Evans D H. The complete genome sequence of shope (rabbit) fibroma virus. Virology. 1999;264:319–343. doi: 10.1006/viro.1999.0002. [DOI] [PubMed] [Google Scholar]
- 67.Williams T. The iridoviruses. Adv Virus Res. 1996;46:345–412. doi: 10.1016/s0065-3527(08)60076-7. [DOI] [PubMed] [Google Scholar]
- 68.Wolffe E J, Vijaya S, Moss B. A myristylated membrane protein encoded by the vaccinia virus L1R open reading frame is the target of potent neutralizing monoclonal antibodies. Virology. 1995;211:53–63. doi: 10.1006/viro.1995.1378. [DOI] [PubMed] [Google Scholar]
- 69.Wright C F, Coroneos A M. Purification of the late transcription system of vaccinia virus: identification of a novel transcription factor. J Virol. 1993;67:7264–7270. doi: 10.1128/jvi.67.12.7264-7270.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xiong Y, Eickbush T H. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yan X, Olson N H, Van Etten J L, Bergoin M, Rossmann M G, Baker T S. Structure and assembly of large lipid-containing dsDNA viruses. Nat Struct Biol. 2000;7:101–103. doi: 10.1038/72360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yanez R J, Rodriguez J M, Nogal M L, Yuste L, Enriquez C, Rodriguez J F, Vinuela E. Analysis of the complete nucleotide sequence of African swine fever virus. Virology. 1995;208:249–278. doi: 10.1006/viro.1995.1149. [DOI] [PubMed] [Google Scholar]
- 73.Zemskov E A, Kang W, Maeda S. Evidence for nucleic acid binding ability and nucleosome association of Bombyx mori nucleopolyhedrovirus BRO proteins. J Virol. 2000;74:6784–6789. doi: 10.1128/jvi.74.15.6784-6789.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Y, Keck J G, Moss B. Transcription of viral late genes is dependent on expression of the viral intermediate gene G8R in cells infected with an inducible conditional-lethal mutant vaccinia virus. J Virol. 1992;66:6470–6479. doi: 10.1128/jvi.66.11.6470-6479.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang Y, Strasser P, Grabherr R, Van Etten J L. Hairpin loop structure at the termini of the chlorella virus PBCV-1 genome. Virology. 1994;202:1079–1082. doi: 10.1006/viro.1994.1444. [DOI] [PubMed] [Google Scholar]