Abstract
Maximal gene expression in retroviruses requires that polyadenylation in the 5′ long terminal repeat (LTR) is suppressed. In human immunodeficiency virus type 1 (HIV-1) the promoter-proximal poly(A) site is blocked by interaction of U1 snRNP with the closely positioned major splice donor site (MSD) 200 nucleotides downstream. Here we investigated whether the same mechanism applies to down-regulate 5′ LTR polyadenylation in Moloney murine leukemia virus (MoMLV). Although the same molecular architecture is present in both viruses, the MoMLV poly(A) signal in the 5′ LTR is active whether or not the MSD is mutated. This surprising difference between the two retroviruses is not due to their actual poly(A) signals or MSD sequences, since exchange of either element between the two viral sequences does not alter their ability to regulate 5′ LTR poly(A) site use. Instead we demonstrate that sequence between the cap and AAUAAA is required for MSD-dependent poly(A) regulation in HIV-1, indicating a key role for this part of the LTR in poly(A) site suppression. We also show that the MoMLV poly(A) signal is an intrinsically weak RNA-processing signal. This suggests that in the absence of a poly(A) site suppression mechanism, MoMLV is forced to use a weak poly(A) signal.
The life cycle of retroviruses includes a transition stage where the RNA genome is reverse transcribed into DNA (11, 31). Reverse transcription results in the duplication of U3-R-U5 sequences termed long terminal repeats (LTRs) at either end of the proviral DNA. Consequently, transcriptional regulatory elements and poly(A) signals are present at both termini of the provirus. In particular, the presence of poly(A) signals in both LTRs produces a regulatory obstacle to viral expression. An active 5′ poly(A) site would dramatically impair viral gene expression by premature polyadenylation (5, 12, 28). The exact location of the poly(A) signals in the LTR effectively divides retroviruses into two classes. In the cases of human T-cell leukemia virus type 1 (HTLV-1), HTLV-2, bovine leukemia virus, Rous sarcoma virus, and murine mammary tumor virus, ingenious use is made of the bipartite nature of poly(A) signals. This comprises an AAUAAA sequence (binding site for the cleavage and polyadenylation specificity factor, CPSF) positioned 15 to 30 nucleotides (nt) upstream of the cleavage site and a more variant GU/U-rich sequence (binding site for the cleavage stimulation factor, CstF) normally positioned immediately downstream. In each of these retroviruses the AAUAAA sequence is located in U3 just upstream of the transcription start site (which defines the beginning of R) and the GU/U-rich sequence at the beginning of U5, thus placing the cleavage site of the poly(A) signal at the end of the R sequence. Consequently, the full poly(A) signal is transcribed only in the 3′ LTR. For Rous sarcoma virus and murine mammary tumor virus, R is kept to a short length (30 to 50 nt) to allow a functional spacing of the poly(A) signals. In contrast, the R sequence in HTLV-1 is 275 nt. Correct spacing between AAUAAA and the GU/U-rich sequences is achieved by RNA secondary structure. This also provides the binding site for the viral Rex protein, which promotes nuclear export of unspliced genomic transcripts (1, 6).
In the second group of retroviruses, including human immunodeficiency virus type 1 (HIV-1), HIV-2, equine infectious anemia virus, and Moloney murine leukemia virus (MoMLV), both AAUAAA and the GU/U-rich sequence are located within R. Therefore, a functional poly(A) site is present at either end of the retroviral transcript. To maximize gene expression, the promoter-proximal (5′ LTR) poly(A) site must be suppressed (occluded), while the same RNA-processing signal in the 3′ LTR has to be efficiently used to polyadenylate all resulting viral RNAs. How this mechanistic dilemma is resolved in HIV-1 has been extensively investigated. First, U3 sequences that are uniquely transcribed at the 3′ end of the provirus have been shown to enhance polyadenylation in vivo (2, 34), as well as in vitro (17, 35) by increasing binding of CPSF to the AAUAAA sequence (18, 19). This enhancer activity, which is associated with a U-rich upstream sequence element, ensures efficient use of the poly(A) site at the 3′ end of viral transcripts. Second, structural predictions for the HIV-1 poly(A) site suggest that it is part of a stem-loop structure (8) that may play a role in poly(A) site suppression in the 5′ LTR (15, 22).
Neither of the above-described mechanisms fully accounts for the suppression of the promoter-proximal poly(A) site, since it is known to function efficiently in the absence of the U3 upstream sequence element (36). We have recently shown that neither the close proximity of the 5′ LTR poly(A) site to the cap/promoter nor the HIV-1 promoter itself is essential for the occlusion process (4). Instead, we demonstrated that the suppression of the 5′ poly(A) site is dependent on the presence of the downstream major splice donor site (MSD). Inactivation of the MSD results in efficient promoter-proximal polyadenylation (2, 3). Detailed analysis of this mechanism revealed that the interaction of the U1 snRNP rather than splicing accounts for the inactivation of the poly(A) site, since tethering of a modified U1 snRNP close to a mutant MSD rescued suppression. Furthermore, we have also shown that the U1 snRNP stem-loop I and possibly the associated 70-kDa protein play a crucial role in this suppression mechanism (4).
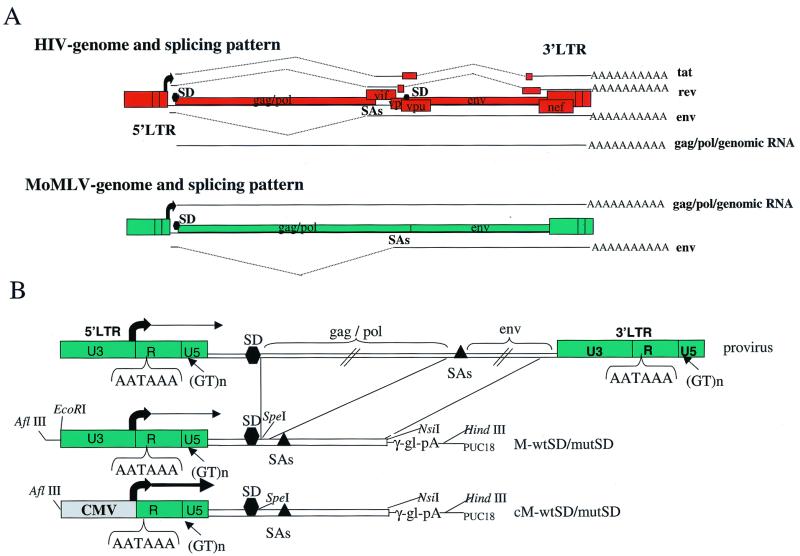
We wished to establish if the above-described mechanisms for 5′ LTR poly(A) site suppression in HIV-1 represent a general strategy for other retroviruses with poly(A) signals in R-U5. We have therefore analyzed 5′ LTR polyadenylation in MoMLV, since this retrovirus is frequently used in the construction of retroviral gene delivery systems for human gene therapy. As such gene delivery systems require maximal gene expression, it is important to understand the nature of 5′ LTR poly(A) site regulation in this virus. In particular, disruption of elements involved in poly(A) site suppression could dramatically decrease gene expression by premature polyadenylation in the 5′ LTR. Figure 1A demonstrates that the MoMLV gene structure is much simpler than that of HIV-1. In contrast to HIV-1, which produces multiply spliced RNAs, MoMLV expresses only unspliced and singly spliced RNA species. As in HIV-1, the MoMLV poly(A) signals are situated within the R-U5 sequences and in the 5′ LTR are followed by a downstream MSD (30). Given these close similarities, we have investigated whether the promoter-proximal poly(A) site in an MoMLV minigene context is repressed by the MSD, as is the case for HIV-1. In sharp contrast to that in HIV-1, we show that 5′ LTR polyadenylation in MoMLV is unaffected by mutational inactivation of the major splice donor. Comparison of the two systems provides evidence for the mechanistic importance of sequences between the cap and the poly(A) site in HIV-1 for the occlusion process. Finally, the weakness of the MoMLV poly(A) site is instrumental in preventing premature polyadenylation of the majority of viral transcripts in the 5′ LTR.
FIG. 1.
(A) Schematic comparison of the proviral organizations of HIV-1 and MoMLV (32), highlighting the complexity of the HIV-1 genome and gene expression. The Tat and Rev splicing patterns are indicated by the dotted line and represent examples of multiply spliced HIV-1 RNA species. The splicing of the singly spliced env gene is indicated, as well as the unspliced genomic RNA. In contrast, with the much simpler MoMLV only two species, spliced RNA (env) and the unspliced genomic RNA, are shown. (B) Diagram of the MoMLV proviral DNA and construction of the minigene. In both LTRs the U3, R, and U5 elements are shown. The locations of the bipartite poly(A) signals are indicated. The bent arrow at the end of U3 represents transcription initiation. The filled diamond and triangle highlight the splice donor and acceptor sites, respectively. The minigene was constructed by excision of the gag-pol genes followed by fusion with sequences immediately downstream of the splice donor to the splice acceptors and the env gene. The 3′ LTR was replaced with the γ-globin poly(A) signal. The U3 sequences were then precisely replaced by the CMV promoter elements without affecting the site of transcription initiation. M, MoMLV-based minigene; wtSD, wild-type MSD (G/GU); mutSD, inactivated MSD (GCA); c, constructs in which the CMV promoter is replacing the original MoMLV U3 promoter.
The radically different approaches to poly(A) site suppression in MoMLV and HIV-1 may reflect the extreme differences in the levels of gene expression observed for these two viruses. Thus, HIV-1 achieves high levels of gene expression in the infected T cell, while MoMLV expression is maintained at much lower levels.
MATERIALS AND METHODS
Plasmid construction. (i) MoMLV minigene.
The MoMLV minigene was constructed from proviral DNA containing plasmid pMLV-K (25). First, the 5′ LTR and downstream sequences to nt 1580 (30) were amplified by PCR with oligonucleotides containing EcoRI and BamHI overhangs and subcloned into the EcoRI and BamHI sites of pUC18. This initial plasmid was then cut with SpeI (site located 75 nt downstream of the MSD) and HindIII within the polylinker of pUC18, resulting in the M vector. Next, MoMLV sequences between nt 5367 and 7063 containing the splice acceptor sites and most of the env gene were amplified by PCR using oligonucleotides containing a SpeI or NsiI overhang, respectively. The γ-globin poly(A) site was amplified by PCR with oligonucleotides containing an NsiI or HindIII overhang. Both PCR products were then cut with NsiI, gel purified, and ligated. The ligated product was amplified by PCR, digested with SpeI and HindIII, and ligated into the M vector, resulting in the construct M-wtSD (Fig. 1B). The splice donor mutation (nt 206 and 207) G/GT to GCA was introduced by using the megaprimer PCR technique (27).
(ii) CMV promoter-driven constructs cM-wt and cM-mutSD.
The cytomegalovirus (CMV) promoter-driven constructs cM-wt and cM-mutSD were constructed in two PCR steps. First, the CMV promoter was amplified from the CMV HIV-1 mg construct (4). The second PCR amplified MoMLV nt 1 to 290. The two PCR products were then blunt-end ligated, reamplified, cut with AflIII and SpeI, and cloned into M-wtSD and M-mutSD digested with the same enzymes. A two-step PCR was used to create the construct cM-xho-mutSD. First, the CMV promoter plus downstream sequences immediately 5′ of the AATAAA were amplified from cM-mutSD and ligated to the PCR product obtained using primers complementary to the sequence immediately downstream of the AATAAA and downstream of the SpeI site, respectively. The forward primer contains an XhoI (CTCGAG) overhang replacing the AATAAA hexamer. The PCR products were ligated and reamplified, cut with AflIII and SpeI, and ligated into the backbone of the cM-mutSD construct, which was digested with the same enzymes.
(iii) Constructs cM-HpA-wtSD and cM-HpA-mutSD.
The constructs cM-HpA-wtSD and cM-HpA-mutSD were obtained by performing round-the-plasmid PCR using the cM-wtSD and cM-mutSD plasmids, respectively, and primers complementary to nt 20 to 42 and 157 to 179, respectively. The HIV poly(A) site was obtained by digesting the cM constructs with AflII and NarI. This fragment was blunt ended with T4 DNA polymerase and ligated into the round-the-plasmid PCR products.
(iv) cH-MpA-wtSD and cH-MpA-mutSD plasmids.
The cH-MpA-wtSD and cH-MpA-mutSD plasmids were constructed by digesting the cH-wtSD and cH-mutSD plasmids (CMVHIV-1 mg plasmids [4]) with AflII and NarI. The MoMLV poly(A) site (nt 43 to 156) was then amplified and cloned into the latter plasmids.
(v) Substitution of MoMLV MSD.
Substitution of the MoMLV MSD (GAG/GTAAGCTG) with the HIV-1 MSD (CTG/GTGAGTAC), resulting in cM-HwtSD, was achieved by round-the-plasmid PCR using corresponding primers and the cM-wtSD plasmid as a template.
(vi) Constructs cH-5′M-wtSD and cH-5′M-mutSD.
Constructs cH-5′M-wtSD and cH-5′M-mutSD were obtained by digesting cH-wtSD and cH-mutSD, respectively, with NcoI (located within the CMV promoter sequence) and AflII (located 10 nt upstream of the AATAAA hexamer). PCR was then performed using primers that amplify MoMLV sequences from immediately upstream of the AATAAA (reverse primer containing an AflII overhang) and the entire CMV promoter. This PCR product was then digested with AflII and NcoI and cloned into the above-described plasmids.
(vii) Constructs cM-5′H-wtSD and cM-5′H-mutSD.
Constructs cM-5′H-wtSD and cM-5′H-mutSD were created by three PCRs. First, sequences immediately upstream of the AATAAA and the CMV promoter were amplified using the cH-wtSD construct as a template. Next, MoMLV nt 47 to 280 were amplified from either the cM-wtSD or the cM-mutSD construct. The two PCR products were then ligated and reamplified, cut with AflIII and SpeI, and cloned into cM-wtSD digested with the same enzymes.
(viii) cH-5′RAT-wtSD and cH-5′RAT-mutSD.
cH-5′RAT-wtSD and cH-5′RAT-mutSD were constructed in two steps. First, a BamHI site was introduced by replacing the nucleotides CTCTCTGG (+5 to +12) with the sequence TGGATCCT in cH-wtSD and cH-mutSD. Insertion of the BamHI site showed no effect on poly(A) occlusion in an RNase protection assay (data not shown). Next the double-stranded oligonucleotide (RAT sequence) GATCCTAATCTGGTCTAGACTCGGACCCTCGAGAGACCGATTGATCCCTTGGGTGACC was inserted into the BamHI-AflII-digested plasmid as described above.
(ix) Construction of poly(A) competition plasmids.
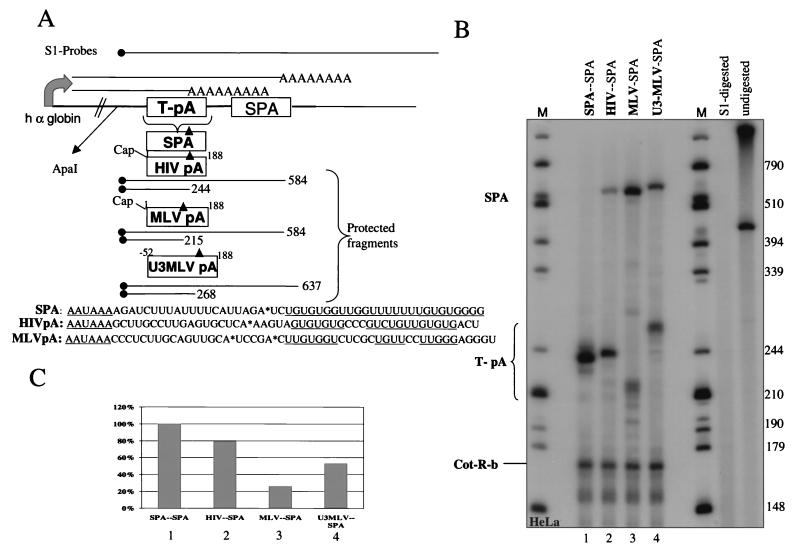
The construction of the poly(A) competition plasmids is illustrated in Fig. 7A. The T-pA inserts were obtained by PCR and cloned into the PvuII site of the poly(A) competition construct (26).
FIG. 7.
Measurement of MoMLV poly(A) strength versus HIV-1 using a poly(A) site competition assay. (A) Basic outline of the poly(A) site competition gene construct. The strong SPA follows a test poly(A) site box (T-pA). The sequences between the cap and nt 188 of both HIV-1 (HIV pA) and MoMLV (MLV pA) were inserted into the test position as indicated. In the case of U3MLV pA, the region between −52 (from the site of transcription initiation) to nt 188 was inserted. The lengths of the S1-protected fragments are indicated below each construct. The sequences of all minimal poly(A) sites are outlined. Sites of cleavage are indicated (asterisks), and the AAUAAA and GU-rich sequences are underlined. (B) S1 protection of cytoplasmic RNA from HeLa cells transfected with the various poly(A) site competition gene constructs. Lanes M, size markers (in nucleotides); boldface letters at top of gel, origin of the poly(A) site inserted into the T-pA box; S1 digested, fully digested S1 probe; undigested, S1 probe for MLV-SPA and rabbit β-globin incubated without S1 nuclease; SPA, transcripts that read through the test poly(A) site and were processed at the downstream SPA site; T-pA, transcripts polyadenylated at the inserted poly(A) site; Cot-R-b, protected fragments from the cotransfected control plasmid containing the rabbit β-globin gene. (C) Percent use of the test poly(A) site.
(x) Riboprobe plasmids.
The riboprobe plasmids for the HIV-1 minigenes were constructed by PCR amplification of the sequence from −54 (within the CMV promoter) to +362 (71 nt downstream of the major splice donor) using oligonucleotides with EcoRI-XbaI overhangs. The fragments were then subcloned into pGEM-4 digested with EcoRI-XbaI. The MoMLV riboprobes were obtained by PCR amplification of the fragment between −54 (within the CMV promoter) and the SpeI site, which was then ligated into pGEM-4 digested with EcoRI-XbaI.
Cell culture, transfection, and RNA isolation.
Subconfluent NIH 3T3 and HeLa cells were transfected using the Qiagen Superfect transfection reagent. Transfections were carried out as follows. Three micrograms of retroviral minigene plasmid and 0.5 μg of VA plasmid (pUC plasmid containing the adenovirus VAI gene) in 150 μl of serum-free minimal essential medium were mixed with 25 μl of Superfect reagent and incubated at room temperature for 20 min. The transfection mix was then added to subconfluent HeLa or NIH 3T3 cells in 90-mm-diameter plates in a total volume of 4 ml of minimal essential medium or Dulbecco modified Eagle medium, respectively (medium supplemented with serum), and incubated at 37°C with 5% CO2 for 5 to 8 h. Subsequently, the medium was replaced. RNA isolations were performed at 24 h posttransfection. Cytoplasmic RNA was isolated as previously described (16). Total RNA was isolated using the hot-phenol method. Volumes of 450 μl of NTE buffer (0.1 M NaCl, 10 mM Tris [pH 8], 1 mM EDTA) and 50 μl of 10% sodium dodecyl sulfate were added to 500 μl of phenol and heated to 90°C. Subsequently, the cell pellets were added to the hot phenol mix, phenol-chloroform extracted twice, and precipitated. Total and cytoplasmic RNA pellets were resuspended in 65 to 90 μl of R loop buffer (2).
RNA analysis.
Initially, 3 to 5 μl of RNA sample was used for a quantitative RNase protection analysis of the cotransfected VA gene (13). The RNA was annealed to 500 cps of VA riboprobe in a total volume of 30 μl of R loop buffer at 56°C (15 to 18 h) and then diluted with 300 μl of RNase protection buffer (10 mM Tris [pH 7.5], 20 mM EDTA, 30 U of RNase I [Roche Boehringer] per ml). Digestion was carried out at 20°C for 1 h, followed by proteinase K digestion, phenol-chloroform extraction, and ethanol precipitation. The protected fragments were fractionated on a 6% polyacrylamide gel and subjected to PhosphorImager quantitation. Subsequently, retroviral minigene transcription was normalized according to the VA quantitation and RNase protection was carried out as described above, with the exception that the hybridization temperature for RNAs obtained from cells transfected with MoMLV constructs was set at 50°C.
In Fig. 3B, lanes 1 and 2, and Fig. 4A, lanes 1 to 4, equal volumes of RNA were analyzed without prior VA normalization.
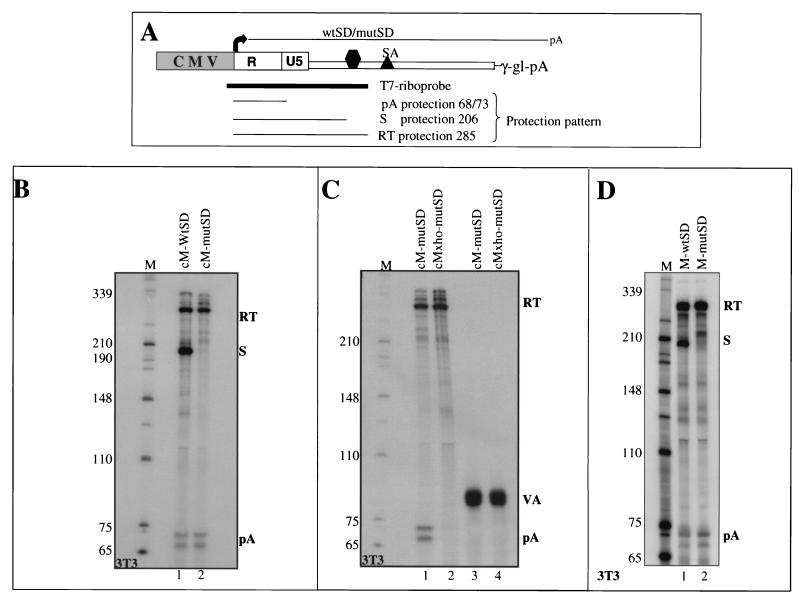
FIG. 3.
(A) Diagram of minigene constructs and the expected lengths of protected fragments from the T7 riboprobes (thick line) after RNase digestion. (B through D) RNase protection analysis of total RNA from NIH 3T3 cells transfected with MoMLV minigene constructs as indicated. (C) Lanes 1 and 2, effect of mutation of the MoMLV AATAAA hexamer with an XhoI site on RNase protection products. Lanes 3 and 4, cotransfection controls probing for the cotransfected VA gene, confirming equal transfection efficiency and loading of the RNA. (D) RNase protection analysis with constructs containing the original MoMLV promoter. The use of the MoMLV promoter results in a substantially lower yield of viral transcripts, and therefore longer exposures were necessary to detect the 5′ LTR transcripts. Lanes M, size markers (in nucleotides).
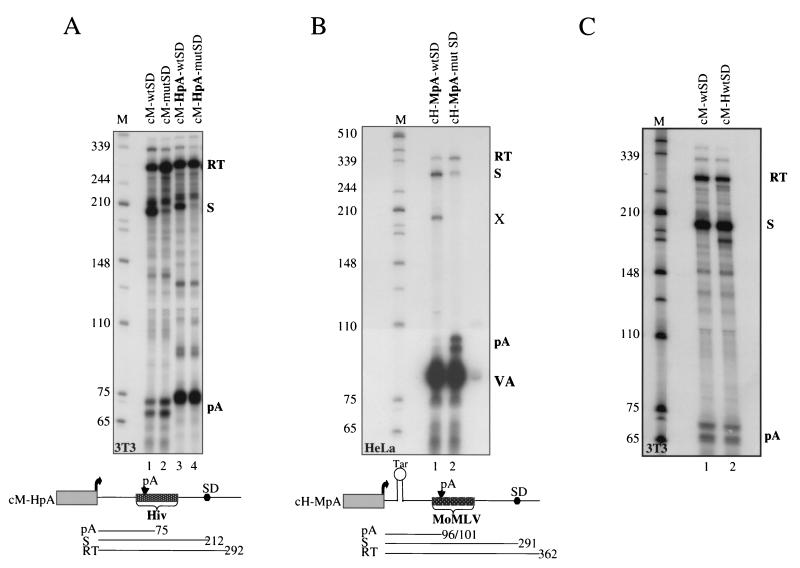
FIG. 4.
RNase protection analysis using HIV-1 and MoMLV minigenes with interchanged poly(A) and MSD sites. Labeling is as indicated in Fig. 3A. The cM-HpA [M, MoMLV minigene backbone; HpA, HIV-1 poly(A) signal] and cH-MpA [H, HIV-1 minigene backbone; MpA, MoMLV poly(A) signal] constructs were used for this series of experiments, and the resulting protected fragments are indicated below the panels. The cell lines used for each transfection are indicated in the bottom left corners of the panels. (A) RNase protection analysis of the indicated minigenes using total RNA from transfected NIH 3T3 cells. (B) RNase protection analysis of cytoplasmic RNA from HeLa cells transfected with the indicated HIV-1 minigene constructs. To ensure equal efficient transfection and RNA loading, a riboprobe for the cotransfected VA gene was included (VA). An additional band, X, appears in lane 1. The intensity of this band varied between experiments and is most likely an RNase protection artifact. (C) RNase protection analysis of cM-wtSD and the same MoMLV minigene, but containing the HIV-1 splice donor sequence (cM-HwtSD), transfected into NIH 3T3 cells. Lanes M, size markers (in nucleotides).
S1 nuclease analysis was performed as described previously (3).
RESULTS
Previous studies on the regulation of polyadenylation in HIV-1 revealed that results obtained in the physiological context of the whole provirus (2) could be reproduced in a more experimentally tractable HIV-1 minigene construct (3). It was demonstrated that in both the HIV-1 provirus and minigene, the MSD acts to suppress the 5′ LTR poly(A) signal. Furthermore, replacement of the HIV-1 U3 promoter by the heterologous CMV promoter has no effect on poly(A) site suppression by the MSD (4). In the present studies we have aimed to compare the 5′ LTR poly(A) site regulation of MoMLV with that of HIV-1. We therefore generated MoMLV minigene constructs in an fashion analogous to that used for the constructs previously constructed for HIV-1 (3). As indicated in Fig. 1B, MoMLV 5′ LTR sequences up to the MSD were fused to the splice acceptor sites of the env gene (23, 30), followed by the poly(A) signal of the human γ-globin gene to generate the MoMLV minigene construct (M). The weak U3 promoter of MoMLV was replaced by the highly active CMV promoter to allow easier transcription analysis (construct cM). In addition, RNA polymerase III transcripts initiating from the MoMLV U3 element have been reported (10) and could interfere with the RNase protection analysis of RNA polymerase II transcripts. HIV-1 and MoMLV 5′ LTRs have quite similar arrangements of their mRNA-processing signals. Thus, the distance between the cap and poly(A) signal differs by only 23 nt, while the spacing between the AAUAAA and the MSD in HIV-1 is 216 nt, compared to 159 nt in MoMLV.
5′ LTR polyadenylation in MoMLV is not repressed by the MSD.
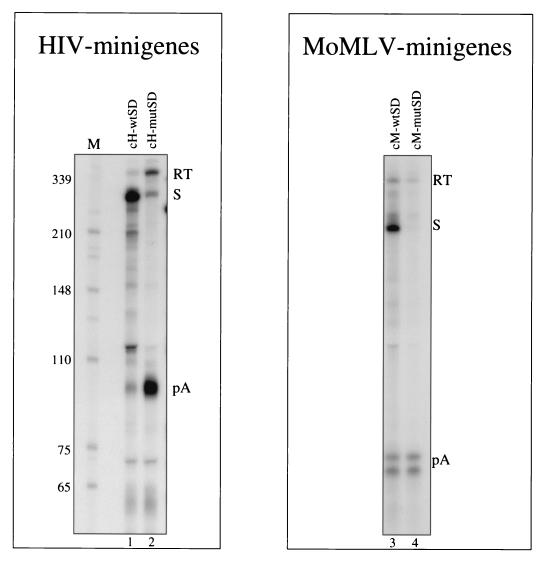
We initially carried out a direct comparison of the ability of the MSD to suppress 5′ LTR polyadenylation in the HIV-1 and MoMLV minigene constructs (Fig. 2). RNase protection analysis of cytoplasmic RNA isolated from HeLa cells transiently transfected with these minigene plasmids revealed a striking difference between the two retroviruses. We have previously demonstrated the near-complete dependence of 5′ LTR polyadenylation on the inactivation of the downstream MSD (3). This result is reproduced in Fig. 2 (lanes 1 and 2), where the HIV-1 pA band is dramatically increased following MSD inactivation. Instead, the presence or absence of the MSD in the MoMLV minigene showed little effect on the absolute levels of 5′ LTR polyadenylation (lanes 3 and 4). As described below, this poly(A) site produces a characteristic doublet band. The expected sizes of the RNase protection products are presented in Fig. 3A.
FIG. 2.
RNase protection analysis of cytoplasmic RNAs from HIV-1 and MoMLV minigenes containing a wild-type or mutant splice donor transfected into HeLa cells. The sizes and identities of bands designated RT, S, and pA are indicated in Fig. 3A. Lanes are labeled with plasmids used in transfections. Lane M, size markers (in nucleotides).
The RNase protection analysis was controlled for transfection efficiency and RNA loading (see Materials and Methods). Although mutation of the MoMLV MSD results in the disappearance of the splicing-derived band (S band), the level of poly(A) bands (pA bands) is unaffected by the MSD mutation. Surprisingly, there was no commensurate increase in the readthrough band (RT band). Presumably, transcripts that cannot be spliced at the MSD are unstable, so that no increase in readthrough signal is detected.
Figure 3 provides additional controls for the lack of regulation of the MoMLV poly(A) site by the downstream MSD, mutated by a GU→CA point mutation. The murine NIH 3T3 cell line was employed in these experiments since it is permissive to MoMLV infection (as HeLa cells are to HIV-1 infection). Identical results were obtained with both cell lines. Figure 3B reveals that for both constructs, readthrough products (lanes 1 and 2) of the expected size are obtained. The larger amount of these RT bands in NIH 3T3 transfections may reflect differences in splicing efficiency between this cell line and HeLa cells (Fig. 2). As before, a band of 206 nt coinciding with the expected splice product (S) is present for cM-wtSD (Fig. 3B, lane 1) but is clearly absent when the splice donor is inactivated in cM-mutSD (lane 2), and doublet pA bands about 70 nt in length are present at equal intensities for both wild-type and mutant SD transcripts.
To establish the identity of the pA doublet bands, an XhoI site replacing the AATAAA hexamer was introduced into the cM-mutSD construct. RNase protection analysis of this construct reveals that the doublet pA bands disappear, confirming that they represent mRNAs that are polyadenylated within the 5′ LTR (Fig. 3C, lanes 1 and 2). It appears that two nearly equally efficient cleavage sites are defined by the MoMLV poly(A) signal, a feature quite often observed in other poly(A) signals. As before, these transfections were controlled by cotransfection with a second plasmid containing the adenovirus VA gene (transcribed by RNA polymerase III). Figure 3C (lanes 3 and 4) shows the RNase protection product for this VA transcript, confirming the equal transfection efficiency, RNA recovery, and gel loading for this experiment. As pointed out above, a heterologous CMV promoter drives transcription of the MoMLV minigene. To ensure that the lack of splice donor-dependent regulation in MoMLV is not an indirect consequence of the efficient CMV promoter substitution, constructs containing the original MoMLV promoter (M-wtSD and M-mutSD [Fig. 1B]) were also analyzed. As shown in Fig. 3D (lanes 1 and 2), substitution of the CMV promoter for the original MoMLV U3 promoter has no significant effect on the regulation of 5′ LTR polyadenylation when normalized to the VA cotransfection control (see Materials and Methods). However, we note that a larger fraction of transcripts fail to be processed at the poly(A) or MSD signals, resulting in a stronger readthrough band. Also, the longer exposure necessary to detect the poly(A) and MSD signals resulted in higher levels of background bands. These results clearly demonstrate a distinct difference between HIV-1 and MoMLV. Whereas MSD inactivation in HIV-1 triggers efficient 5′ LTR poly(A) site use (Fig. 2), surprisingly no effect is observed for MoMLV. Indeed, 5′ LTR polyadenylation appears to occur at similar levels whether or not the MSD is present.
Exchanging of poly(A) and MSD signals between HIV-1 and MoMLV.
The fact that the simple positioning of a poly(A) site upstream of an active donor site is not sufficient to induce poly(A) site suppression is clearly revealed by our analysis of MoMLV. To try to understand the specificity of the HIV-1 poly(A) site suppression mechanism and why this is absent in MoMLV, we exchanged poly(A) signals between the two retroviral minigenes. The HIV-1 poly(A) site (including downstream sequences; see Material and Methods) was inserted in place of the MoMLV poly(A) site into the cM constructs to generate cM-HpA-wtSD and cM-HpA-mutSD. Figure 4A shows the RNA analysis of these two constructs transfected into murine NIH 3T3 cells. As clearly indicated by these results, the HIV-1 poly(A) site loses its ability to be suppressed by a splice donor when placed within the MoMLV sequence background. Thus, lanes 3 and 4 reveal equivalent amounts of HIV-1 pA site use with or without a functional downstream MSD. A similar result was obtained when these constructs were transfected into HeLa cells (data not shown). Figure 4B shows the reciprocal experiment, in which the MoMLV poly(A) signal is used to replace the HIV-1 poly(A) signal in the HIV-1 minigene. In this case, full MSD suppression of the MoMLV poly(A) signal occurs. The VA cotransfectional control is shown in the same lane for this particular experiment. Taken together, these results clearly suggest that splice donor-induced suppression in HIV-1 is not dependent on the origin of the poly(A) site (plus the immediate 3′ flanking region). Furthermore, the lack of poly(A) site regulation in the MoMLV system cannot be attributed to a specific sequence of the MoMLV polyadenylation signal.
We also investigated the effect of replacing the MoMLV MSD with the HIV-1 MSD. Even though the HIV-1 MSD is a closer match to the optimal donor site consensus than the MoMLV MSD, the unregulated use of the MoMLV poly(A) signal was still observed. As shown in Fig. 4C, the HIV-1 MSD failed to induce suppression of the MoMLV poly(A) signal when these cM constructs were transfected into NIH 3T3 cells. Similarly, when the MoMLV MSD was used to replace the HIV-1 MSD, poly(A) suppression was still observed in the HIV-1 minigene (data not shown). These results indicate that the difference in poly(A) site regulation between these two retroviruses is not attributable to their MSD sequences. Either additional elements in HIV-1 are required to allow MSD-dependent poly(A) site occlusion or regulatory elements in MoMLV are able to counteract the occlusion effect.
Role of sequence between the cap and poly(A) in HIV-1.
We have tested whether sequences between the cap and poly(A) signal in the HIV-1 5′ LTR might also contribute to the poly(A) site suppression phenomenon. In HIV-1 this sequence encompasses the TAR element, which forms a well-characterized stem-loop structure of 59 nt (20). Furthermore, TAR acts as a transcriptional regulatory element through its interaction with the viral Tat protein (7) and other cellular factors. TAR has also been implicated in posttranscriptional events such as translation (9) and viral packaging (24). Part of the sequence between the cap and AAUAAA in MoMLV is also thought to form a hairpin structure and plays a role in promoting the accumulation of unspliced MoMLV transcripts in the cytoplasm. However, it is unknown whether this is a direct effect on nuclear export or an indirect effect through the inhibition of splicing (32).
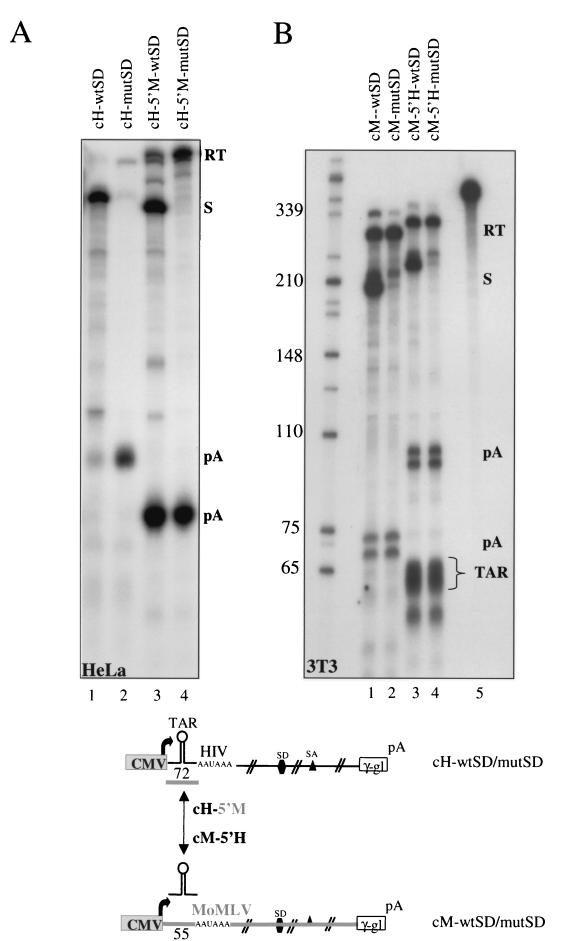
To test the possible roles of the cap-AAUAAA sequence in 5′ LTR poly(A) site suppression, we exchanged these sequences between HIV-1 (nt +1 to +72) and MoMLV (nt +1 to +55) (Fig. 5). The two plasmids cH-5′M-wtSD and cH-5′M-mutSD plus the parental cH constructs were then transfected into HeLa cells, and cytoplasmic RNA was analyzed. As before, the cH constructs resulted in a suppression pattern where the 5′ poly(A) site is activated by the splice donor mutation (Fig. 5A, lanes 1 and 2). However, surprisingly a complete loss of regulation was evident if the cap-AAUAAA sequence of HIV-1 was replaced by the equivalent MoMLV sequence (Fig. 5A, lanes 3 and 4). Clearly, insertion of the MoMLV sequence abolishes the ability of the HIV-1 MSD to suppress polyadenylation. The pattern obtained with chimeric cH-5′M constructs now resembles that obtained with MoMLV (cM). Since we were able to transform regulated poly(A) site suppression of the HIV-1 minigene into the unregulated MoMLV arrangement, we also tested whether the insertion of the HIV-1 cap-AAUAAA sequence into the MoMLV minigene can induce poly(A) site suppression. As shown in Fig. 5B, the constructs cM-5′H-wtSD and cM-5′H-mutSD were transfected into NIH 3T3 cells, and the resulting transcripts were analyzed by RNase protection. Replacement of the MoMLV 5′ element with the HIV-1 TAR-containing region did not induce the suppression of the promoter-proximal poly(A) site (identical results were obtained using HeLa cells). Furthermore, the presence of RNA transcripts corresponding to the TAR sequence is clearly visible in lanes 3 and 4, as is often observed when mapping HIV-1 5′ LTR transcripts (29). It is clear from these results that the HIV-1 TAR sequence is not sufficient to induce poly(A) site regulation in an MoMLV background. Presumably other sequence elements in MoMLV still prevent poly(A) site regulation.
FIG. 5.
Role of sequences between the cap and AAUAAA in HIV-1 poly(A) site occlusion. As indicated below the panels, sequence between the cap and the AAUAAA in the HIV-1-minigene was replaced by the equivalent MoMLV element. The distance between the cap and the AAUAAA hexamer differs by 17 nt between the two viruses, explaining the change in mobility of the protected fragments. (A) Lanes 1 and 2, RNase protection analysis confirming the splice donor-dependent occlusion of the 5′ LTR poly(A) site in HIV-1. Lanes 3 and 4, loss of 5′ LTR poly(A) site inhibition by insertion of the MoMLV sequence. (B) Insertion of the HIV-1 cap-AAUAAA sequence element into the MoMLV minigene fails to trigger 5′ LTR occlusion in MoMLV minigenes when transfected into NIH 3T3 cells. Compare the RNase protection analysis shown in lanes 1 and 2 with that in lanes 3 and 4. Lane 5 shows a fifth of the undigested riboprobe used for the cM-5′H-wtSD RNase protection. Numbers on the left in panel B are size markers (in nucleotides).
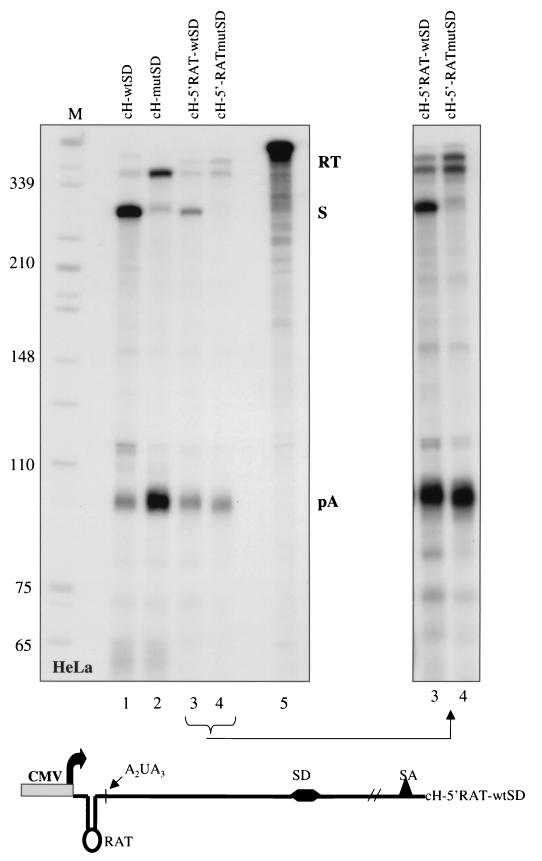
To further define the requirement of cap-proximal sequences in HIV-1 poly(A) site suppression, we constructed HIV-1 minigenes in which nt +12 to +63 were replaced by their antisense sequence (Fig. 6, bottom). The resulting transcripts have the same distance between the cap and the poly(A) site and also have the potential to form an antisense TAR hairpin structure (RAT). Figure 6 shows an RNase protection analysis of the cH-5′RAT-wtSD and cH-5′RAT-mutSD constructs transfected into HeLa cells. Consistent with the results obtained with the constructs cH-5′M-wtSD and cH-5′M-mutSD, regulation of 5′ promoter-proximal poly(A) site use is virtually abolished (Fig. 6, compare lanes 3 and 4). Since the U contents of the riboprobes for the cH-5′RAT and cH constructs are identical, we can directly compare the ratios of the splice products and the 5′-polyadenylated RNAs. It is clear that the RAT mutation substantially reduces the level of 5′ LTR transcripts obtained and favors the use of the poly(A) site over the MSD site (by a sixfold level compared to cH-wtSD). Even so, no suppression of the poly(A) site is observed in the presence of the downstream MSD.
FIG. 6.
RNase protection analysis establishing the importance of the sequences between the cap and AAUAAA in 5′ LTR poly(A) suppression in HIV-1. A diagram of the HIV-1 minigene used is shown below panels. Lanes 1 and 2, splice donor-dependent poly(A) inhibition in the HIV-1 minigene. Lanes 3 and 4, loss of poly(A) site regulation when the region between the cap and AAUAAA is replaced by its antisense sequence (RAT) (cH-5′RAT-wtSD/mutSD). A longer exposure of lanes 3 and 4 can be seen on the right. Lane 5 shows a fifth of the undigested riboprobe used for the cH5′RAT-wtSD RNase protection analysis. Constructs containing the RAT sequence consistently produced lower levels of steady-state transcripts. Lane M, size markers (in nucleotides).
We conclude from these experiments that the sequences between the cap and poly(A) site in HIV-1 play a critical role in the suppression of polyadenylation. Replacement or inversion of these sequences abolishes the inhibitory effect of the MSD on the 5′ poly(A) site use. However, insertion of the HIV-1 cap-AAUAAA sequences alone into an MoMLV background is insufficient to trigger splice donor-dependent inhibition of 5′ LTR polyadenylation.
Strength of the MoMLV poly(A) site.
Our results indicate that MoMLV lacks the ability to suppress the promoter-proximal poly(A) site by the presence of an active downstream MSD. We therefore wondered if the inability to suppress 5′ polyadenylation as observed in MoMLV requires the use of a weak polyadenylation signal. A weak poly(A) signal would allow a substantial number of transcripts to escape the 5′ LTR poly(A) site. In addition, as is the case in HIV-1, U3 sequences could ensure more efficient polyadenylation at the 3′ ends of all viral RNAs.
To test this hypothesis, we compared the relative strengths of the MoMLV and HIV-1 poly(A) sites. A poly(A) competition assay was employed as previously described (26). In this system, test poly(A) sites are inserted upstream of a strong synthetic poly(A) site (SPA) placed in an α-globin gene construct. The strength of each poly(A) site is represented by the ratio of cleavage at the test poly(A) signal to that at the SPA. A 189-nt fragment extending from the cap to downstream of the poly(A) signal in both HIV-1 and MoMLV was inserted into the test box as indicated in Fig. 7A. The plasmids were transfected into HeLa cells, and cytoplasmic RNA was subjected to S1 analysis. The lengths of the resulting protected fragments are indicated (Fig. 7A). Insertion of a strong SPA into the test position resulted in an almost 100% use of the test poly(A) site versus the second SPA (Fig. 7B and C, lane 1 and bar 1), and 70% of transcripts are polyadenylated at the HIV-1 poly(A) site (Fig. 7B and C, lane 2 and bar 2). However, the use of the test poly(A) site was significantly reduced when the MoMLV poly(A) site was inserted. Only 30% of the transcripts were processed at this site (Fig. 7B and C, lane 3 and bar 3). Insertion of an additional 52 nt from the U3 element increased the use of the MoMLV poly(A) site twofold (Fig. 7B and C, lane and bar 4). This observation is in agreement with the HIV-1 system, where polyadenylation at the 3′ LTR is increased by U3 sequences (2, 17).
The above-described results indicate that the MoMLV poly(A) site is significantly weaker than the equivalent HIV-1 processing site. We suggest that the lack of a mechanism to allow the inhibition of the promoter-proximal poly(A) site in MoMLV requires the use of a weak poly(A) signal.
DISCUSSION
As HIV-1 and MoMLV both contain the complete poly(A) signals within the 5′ untranslated region of the viral transcript, suppression of promoter-proximal polyadenylation must be critical for maximal gene expression. In this study, we have compared the abilities of these two retroviral minigene systems to suppress their promoter-proximal poly(A) sites. We have previously demonstrated that in HIV-1, the MSD and its interaction with U1 snRNP play a predominant role in the inactivation of 5′ LTR polyadenylation (2, 3, 4). Instead, these studies show that mutational inactivation of the MSD in an MoMLV-based minigene system has little if any effect on promoter-proximal poly(A) site use. This difference in poly(A) site regulation between HIV-1 and MoMLV is partly explicable by our demonstration that sequences between the cap and the AAUAAA poly(A) signal in HIV-1 are essential for the poly(A) suppression mechanism. However, other sequence features of the MoMLV retrovirus, as yet uncharacterized, appear to prevent poly(A) suppression even if the HIV-1 cap and the AAUAAA sequence are used to replace equivalent MoMLV sequence. It is also possible that further sequence features of HIV-1 are required for MSD suppression in addition to the cap-poly(A) signal region. The fact that the MoMLV poly(A) signal is significantly weaker than that of HIV-1 may also be relevant to the marked differences in poly(A) site regulation between these two retroviruses.
HIV-1 5′ LTR poly(A) site regulation requires sequence between the cap and poly(A) site.
The direct inversion of sequences between the cap and the AAUAAA poly(A) signal in HIV-1 or their replacement with MoMLV sequences clearly abolishes promoter-proximal poly(A) suppression. This points to a further role for this sequence element in HIV-1 beyond its well-established involvement in Tat-dependent promoter activation. The necessity of additional sequences required for poly(A) site suppression in the HIV-1 system was unexpected. For instance, splice donor-dependent suppression of an upstream poly(A) site was demonstrated using an in vitro system with the adenovirus L3 polyadenylation signal (33). However, in contrast to the situation in HIV-1 and MoMLV, where the distance between the processing signals exceeds 100 nt, these studies used short spacers in the range of 13 to 68 nt. This close proximity of the two processing signals might enhance their interaction, independent of additional sequence elements.
The 5′ region of MoMLV used to replace the HIV-1 counterpart contains a sequence element that has been suggested to influence both transcription initiation (14) and accumulation of unspliced RNA in the cytoplasm (32). One suggested mechanism (32) was that accumulation of unspliced RNA might occur through inhibition of the splicing process. If this inhibition reduced the interaction of U1 snRNP at the donor site, then the insertion of this element in the HIV-1 context would lead to the activation of polyadenylation. However, results obtained with the reciprocal construct, in which the HIV-1 5′ region is placed in the MoMLV minigene, argue against this possibility. In this case, no splice donor-dependent regulation is observed.
It could also be argued that the MoMLV 5′ LTR sequences simply abolish poly(A) site suppression by an intrinsic poly(A) enhancer effect. This would lead to a substantially stronger HIV-1 poly(A) site, which might overrule the suppression effect (3). Again, this is unlikely since replacement of the same sequence with the HIV-1 element had no detectable effect on the efficiency of the MoMLV poly(A) site (Fig. 5B). Indeed, the fact that inhibition of 5′ LTR polyadenylation is abolished upon inversion of sequences between the cap and AAUAAA (cH-5′RAT-wt and cH-5′RAT-mutSD) in HIV-1 demonstrates the significance of this specific region in the occlusion process. It has been suggested that the poly(A) site of HIV-1 lies within a hairpin motif (8). Disruption of this hairpin appears to influence polyadenylation (15, 21) by altering its accessibility for poly(A) factors (22). Either substitution or inversion of TAR and flanking regions could result in efficient 5′ LTR polyadenylation by disrupting this poly(A) hairpin and so allow more efficient recognition by poly(A) factors. This increased accessibility for polyadenylation factors could then strengthen the HIV-1 5′ LTR poly(A) site and might partly overrule inhibition by the splice donor site.
The weak poly(A) site might be instrumental for MoMLV gene expression.
In contrast to HIV-1, MoMLV is unable to suppress the promoter-proximal poly(A) site via the U1 snRNP-splice donor interaction. It is clear that MoMLV has to follow a different strategy to overcome the 5′ LTR poly(A) site dilemma. We believe that the basis for the MoMLV strategy can be found in the simplicity of its genome. The HIV-1 genome is much more complex than that of MoMLV (Fig. 1A). In MoMLV only singly spliced and unspliced transcripts appear, whereas in HIV-1 additional multiply spliced RNA species are generated. A strong poly(A) site in HIV-1 would ensure efficient 3′-end processing of all emerging transcripts and therefore facilitate high levels and precise balances between the different RNA species. Thus, the complexity of HIV-1 might have coincided with the evolution of a stronger poly(A) site.
In parallel to the introduction of a stronger poly(A) signal, a mechanism had to evolve to suppress the promoter-proximal site. As described in the introduction, for HTLV the poly(A) signal is divided by placing the AATAAA into the U3 element so that it is transcribed only at the 3′ LTR. However, in HIV-1 and MoMLV the poly(A) sites are in RU5, and therefore different mechanisms must apply. Since HIV-1 possesses a strong poly(A) site, this must be inactivated in the 5′ LTR via a splice donor-dependent mechanism. For MoMLV we suggest that the balance of unspliced and singly spliced RNAs can be achieved by simply using a weak polyadenylation signal. The weak poly(A) signal allows some transcripts to escape premature polyadenylation at the 5′ LTR, and in conjunction with the U3 enhancer element, most of these viral RNAs will then subsequently be polyadenylated in the 3′ LTR. This arrangement does not rely on additional suppression mechanisms even though it must reduce the overall levels of viral transcripts.
ACKNOWLEDGMENTS
We thank Shona Murphy and Mick Dye for many helpful discussions as well as for critical reading of the manuscript. We also acknowledge the contribution of Ana Gonzalez Santis in the initial analysis of MoMLV.
This work was supported by a Medical Research Council project grant (no. G9707608) to N.J.P.
REFERENCES
- 1.Ahmed Y F, Gilmartin G M, Hanly S M, Nevins J R, Greene W C. The HTLV-I Rex responsive element mediates a novel form of mRNA polyadenylation. Cell. 1991;64:727–737. doi: 10.1016/0092-8674(91)90502-p. [DOI] [PubMed] [Google Scholar]
- 2.Ashe M P, Griffin P, James W, Proudfoot N J. Poly(A) site selection in HIV-1 provirus: inhibition of promoter-proximal polyadenylation by the downstream major splice donor site. Genes Dev. 1995;9:3008–3025. doi: 10.1101/gad.9.23.3008. [DOI] [PubMed] [Google Scholar]
- 3.Ashe M P, Pearson L H, Proudfoot N J. The HIV-1 5′ LTR poly(A) site is inactivated by U1 snRNP interaction with the downstream major splice donor site. EMBO J. 1997;16:5752–5763. doi: 10.1093/emboj/16.18.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashe M P, Furger A, Proudfoot N J. Stem-loop 1 of the U1 snRNP plays a critical role in the suppression of HIV-1 polyadenylation. RNA. 2000;6:170–177. doi: 10.1017/s1355838200991957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barabino S M L, Keller W. Last but not least: regulated poly(A) tail formation. Cell. 1999;99:9–11. doi: 10.1016/s0092-8674(00)80057-4. [DOI] [PubMed] [Google Scholar]
- 6.Bar-Shira A, Panet A, Honigman A. An RNA secondary structure juxtaposes two remote genetic signals for human T-cell leukemia virus type 1 RNA 3′-end processing. J Virol. 1991;65:5165–5173. doi: 10.1128/jvi.65.10.5165-5173.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berkhout B, Silverman H, Jeang K T. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell. 1989;59:273–282. doi: 10.1016/0092-8674(89)90289-4. [DOI] [PubMed] [Google Scholar]
- 8.Berkhout B, Klaver B, Das A T. A conserved hairpin structure predicted for the poly(A) signal of human and simian immunodeficiency viruses. Virology. 1995;207:276–281. doi: 10.1006/viro.1995.1077. [DOI] [PubMed] [Google Scholar]
- 9.Braddock M, Thorburn A M, Chambers A, Elliot G D, Anderson G J, Kingsman A J, Kingsman S M. A nuclear translational block imposed by the HIV-1 U3 region is relieved by the Tat-TAR interaction. Cell. 1990;62:1123–1133. doi: 10.1016/0092-8674(90)90389-v. [DOI] [PubMed] [Google Scholar]
- 10.Choi S Y, Faller D V. A transcript from the long terminal repeats of a murine retrovirus associated with trans activation of cellular genes. J Virol. 1995;69:7054–7060. doi: 10.1128/jvi.69.11.7054-7060.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coffin J, Hughes S, Varmus H. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 12.Colgan D F, Manley J L. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 13.Cuello P, Boyd D C, Dye M J, Proudfoot N J, Murphy S. Transcription of the human U2 snRNA genes continues beyond the 3′ box in vivo. EMBO J. 1999;10:2867–2877. doi: 10.1093/emboj/18.10.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cupelli L, Okenquist S A, Trubetskoy A, Lenz J. The secondary structure of the R region of a murine leukemia virus is important for stimulation of long terminal repeat-driven gene expression. J Virol. 1998;72:7807–7814. doi: 10.1128/jvi.72.10.7807-7814.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Das A T, Klever B, Berkhout B. A hairpin structure in the R region of human immunodeficiency virus type 1 RNA genome is instrumental in polyadenylation site selection. J Virol. 1999;73:81–91. doi: 10.1128/jvi.73.1.81-91.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eggermont J, Proudfoot N J. Poly(A) signals and transcriptional pause sites combine to prevent interference between RNA polymerase II promoters. EMBO J. 1993;12:2539–2548. doi: 10.1002/j.1460-2075.1993.tb05909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilmartin G M, Fleming E S, Oetjen J. Activation of HIV-I pre-mRNA processing in vitro requires both an upstream element and TAR. EMBO J. 1992;11:4419–4428. doi: 10.1002/j.1460-2075.1992.tb05542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilmartin G M, Fleming E S, Oetjen J, Graveley B R. CPSF recognition of an HIV-I mRNA 3′ processing enhancer: multiple sequence contacts involved in poly(A) site definition. Genes Dev. 1995;9:72–83. doi: 10.1101/gad.9.1.72. [DOI] [PubMed] [Google Scholar]
- 19.Gravely B, Gilmartin G M. A common mechanism for the enhancement of mRNA 3′ processing by U3 sequences in two distantly related lentiviruses. J Virol. 1996;70:1612–1617. doi: 10.1128/jvi.70.3.1612-1617.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaeger J A, Tinoco I. An NMR study of the HIV TAR element hairpin. Biochemistry. 1993;32:12522–12530. doi: 10.1021/bi00097a032. [DOI] [PubMed] [Google Scholar]
- 21.Klasens B I F, Das A T, Berkhout B. Inhibition of polyadenylation by stable RNA secondary structure. Nucleic Acids Res. 1998;26:1870–1876. doi: 10.1093/nar/26.8.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klasens B I F, Thiesen M, Virtanen A, Berkhout B. The ability of the HIV-1 AAUAAA signal to bind polyadenylation factors is controlled by local RNA structure. Nucleic Acids Res. 1999;27:446–454. doi: 10.1093/nar/27.2.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lazo P A, Prasad V, Tsichlis P N. Splice acceptor site for the env message of Moloney murine leukemia virus. J Virol. 1987;61:2038–2041. doi: 10.1128/jvi.61.6.2038-2041.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McBride M S, Schwartz M D, Panganiban A T. Efficient encapsidation of human immunodeficiency virus type 1 vectors and further characterization of cis elements required for encapsidation. J Virol. 1997;71:4544–4554. doi: 10.1128/jvi.71.6.4544-4554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller A D, Verma I M. Two base changes restore infectivity to a noninfectious molecular clone of Moloney murine leukemia virus (pMLV-1) J Virol. 1984;49:214–222. doi: 10.1128/jvi.49.1.214-222.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moreira A, Wollerton M, Monks J, Proudfoot N J. Upstream sequence elements enhance poly(A) site efficiency of the C2 complement gene and are phylogenetically conserved. EMBO J. 1995;14:3809–3819. doi: 10.1002/j.1460-2075.1995.tb00050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Picard V, Ersdal-Badju E, Lu A, Bock S C. A rapid and efficient one-tube PCR based mutagenesis technique using Pfu DNA polymerase. Nucleic Acids Res. 1994;22:2587–2591. doi: 10.1093/nar/22.13.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Proudfoot N J. Poly(A) signals. Cell. 1991;64:671–674. doi: 10.1016/0092-8674(91)90495-k. [DOI] [PubMed] [Google Scholar]
- 29.Ratnasabapathy R, Sheldon M, Johal L, Hernandez N. The HIV-1 long terminal repeat contains an unusual element that induces the synthesis of short RNAs from various mRNA and snRNA promoters. Genes Dev. 1990;4:2061–2074. doi: 10.1101/gad.4.12a.2061. [DOI] [PubMed] [Google Scholar]
- 30.Shinnick T M, Lerner R A, Sutcliffe G. Nucleotide sequence of Moloney murine leukaemia virus. Nature. 1981;293:543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- 31.Tang H, Kuhen K L, Wong-Staal F. Lentivirus replication and regulation. Annu Rev Genet. 1999;33:133–170. doi: 10.1146/annurev.genet.33.1.133. [DOI] [PubMed] [Google Scholar]
- 32.Trubetskoy A M, Okenquist S A, Lenz J. R region sequences in the long terminal repeat of a murine retrovirus specifically increase expression of unspliced RNAs. J Virol. 1999;73:3477–3483. doi: 10.1128/jvi.73.4.3477-3483.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vagner S, Ruegsegger U, Gunderson S I, Keller W, Mattaj I W. Position-dependent inhibition of the cleavage step of pre-mRNA 3′-end processing by U1 snRNP. RNA. 2000;6:178–188. doi: 10.1017/s1355838200991854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valsamakis A, Zeichner S, Carswell S, Alwine J C. The human immunodeficiency virus type 1 polyadenylation signal: a 3′ long terminal repeat element upstream of the AAUAAA necessary for efficient polyadenylation. Proc Natl Acad Sci USA. 1991;88:2108–2112. doi: 10.1073/pnas.88.6.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valsamakis A, Schek N, Alwine J C. Elements upstream of the AAUAAA within the human immunodeficiency virus polyadenylation signal are required for efficient polyadenylation in vitro. Mol Cell Biol. 1992;12:3699–3705. doi: 10.1128/mcb.12.9.3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weichs an der Glon C, Monks J, Proudfoot N J. Occlusion of the HIV poly(A) site. Genes Dev. 1991;5:244–253. doi: 10.1101/gad.5.2.244. [DOI] [PubMed] [Google Scholar]