Abstract
Background/Objectives: Trefoil factor 1 (TFF1) plays a role in the mucus barrier. Methods: To evaluate the prevalence of TFF1 expression in cancer, a tissue microarray containing 18,878 samples from 149 tumor types and 608 samples of 76 normal tissue types was analyzed through immunohistochemistry (IHC). Results: TFF1 staining was detectable in 65 of 149 tumor categories. The highest rates of TFF1 positivity were found in mucinous ovarian carcinomas (76.2%), colorectal adenomas and adenocarcinomas (47.1–75%), breast neoplasms (up to 72.9%), bilio-pancreatic adenocarcinomas (42.1–62.5%), gastro-esophageal adenocarcinomas (40.4–50.0%), neuroendocrine neoplasms (up to 45.5%), cervical adenocarcinomas (39.1%), and urothelial neoplasms (up to 24.3%). High TFF1 expression was related to a low grade of malignancy in non-invasive urothelial carcinomas of the bladder (p = 0.0225), low grade of malignancy (p = 0.0003), estrogen and progesterone receptor expression (p < 0.0001), non-triple negativity (p = 0.0005) in invasive breast cancer of no special type, and right-sided tumor location (p = 0.0021) in colorectal adenocarcinomas. Conclusions: TFF1 IHC has only limited utility for the discrimination of different tumor entities given its expression in many tumor entities. The link between TFF1 expression and parameters of malignancy argues for a relevant biological role of TFF1 in cancer. TFF1 may represent a suitable therapeutic target due to its expression in only a few normal cell types.
Keywords: TFF1, tissue microarray, immunohistochemistry, human cancers
1. Introduction
Trefoil factor 1 (TFF1/pS2) belongs to one of three highly conserved proteins of the trefoil factor family, which are co-expressed with mucins in the gastrointestinal tract. TFF1 is predominantly expressed in gastric mucosa [1], where it plays a role in protection and healing of the mucus barrier (summarized in [2]). Therefore, TFF1 expression is stimulated in response to acute mucosal injury and chronic inflammation (summarized in [3]). In superficial cells of gastric mucosa, TFF1 is co-expressed and bound to Mucin 5AC (MUC5AC), supporting a role for TFF1 in the packaging and function of the gastric mucosa [4]. Based on the identification of recurrent TFF1 deletions, missense mutations, and hypermethylation in gastric cancer, TFF1 has been considered as a tumor suppressor protein (summarized in [5]). In line with this notion, TFF1-deficient mice serve as a popular model for gastric cancer development [6]. All TFF1-deficient mice develop adenomas in the gastric mucosa, and 30% of them eventually progress to invasive carcinomas [7].
Because the TFF1 expression in normal tissue strongly predominates in stomach mucosa, TFF1 immunohistochemistry (IHC) has been suggested as a marker for gastric cancer [8]. However, studies have shown that not all gastric adenocarcinomas produce TFF1 and that various other tumor entities can express TFF1. In several cancer types, elevated TFF1 expression has been linked to either poor [9,10] or improved patient prognosis [11,12,13,14]. However, the reported frequencies of TFF1 positivity vary considerably for most tumor types. For example, the range of reported TFF1-positive cases ranged from 16% to 90% in gastric cancer [13,15,16,17], from 51% to 98% in cholangiocarcinoma [18,19,20], from 0% to 100% in breast cancer [14,21,22,23], from 0% to 100% in colorectal cancer [24,25,26,27], from 0% to 40% in pulmonary adenocarcinoma [28,29,30,31], from 0% to 14% in serous ovarian carcinoma [28,32,33], from 0% to 25% in clear cell renal cell carcinoma [28,32,34], from 0% to 91% in prostate cancer [28,35,36], and from 55% to 95% in pancreatic cancer [10,37,38]. The use of different antibodies, staining protocols, and criteria to determine TFF1 positivity represents the most likely cause for these controversial data.
To better understand the role of TFF1 protein expression in cancer and to elucidate the potential diagnostic role of TFF1 IHC, an extensive survey of TFF1 immunostaining in a broad range of tumor types and under highly standardized conditions is needed. TFF1 expression was, thus, evaluated in more than 18,000 tumor tissue samples from 149 different tumor types and subtypes as well as 76 different non-neoplastic tissue types by IHC in a tissue microarray (TMA) format in this study.
2. Materials and Methods
2.1. Tissue Microarrays (TMAs)
The normal TMA included 8 samples from 8 different donors from 76 different normal tissue types (608 samples on one slide). The cancer TMAs contained 18,878 primary tumor probes from 149 tumor entities and tumor subtypes. Data on histopathological and molecular parameters were available for cancers of the colorectum (n = 2351), stomach (n = 327), pancreas (n = 598), urinary bladder (n = 1073), and the breast (n = 2139). A detailed description of the composition of both normal and cancer TMAs is available in the Results section. All tumor probes were from the archives of the Institute of Pathology, University Hospital of Hamburg, Germany; the Department of Pathology, Academic Hospital Fuerth, Germany; and the Institute of Pathology, Clinical Center Osnabrueck, Germany. A detailed description of the TMA manufacturing process is available in previous publications [39,40]. Per patient/tumor, one tissue spot (diameter: 0.6 mm) was used. The preparation of TMAs from archived diagnostic tissue remnants and their analysis for research purposes as well as the analysis of patient data were approved by local laws (HmbKHG, §12) and the local ethics committee (Ethics Committee Hamburg, WF-049/09). All work was conducted in accordance with the Declaration of Helsinki. Immunohistochemical data on MUC5AC expression were available from a previous study [41].
2.2. Immunohistochemistry (IHC)
Freshly cut TMA sections were immunostained in one experiment on one day. TMA sections were deparaffinized with xylol, rehydrated using a graded alcohol series, and incubated in an autoclave at 121 °C for 5 min in Dako Target Retrieval Solution, pH9 (Agilent Technologies, Santa Clara, CA, USA; #S2367) for heat-induced antigen retrieval. For blocking of endogenous peroxidase activity, TMA sections were incubated in Dako REAL Peroxidase-Blocking Solution (Agilent Technologies, Santa Clara, CA, USA; #S2023) for 10 min. Primary antibody specific for TFF1 (mouse monoclonal, MSVA-482M, MS Validated Antibodies, Hamburg, Germany; #5784-482M) was applied at 37 °C for 60 min at a dilution of 1:150. For antibody validation, the TMA with normal tissue probes was analyzed by the rabbit recombinant monoclonal TFF1 antibody [EPR3972] (Abcam; Cambridge, UK; #ab92377, dilution 1:900) under identical protocol. Bound antibody was visualized with Dako REAL EnVision Detection System Peroxidase/DAB+, Rabbit/Mouse kit (Agilent Technologies, Santa Clara, CA, USA; #K5007) according to the manufacturer’s directions. The sections were counterstained with hemalaun. For all tumor tissues, the staining intensity was recorded (semi-quantitatively as 0, 1+, 2+, 3+), and the fraction of positive tumor cells was estimated as percentage. For statistical analyses, four groups were determined: negative, tumors without any staining; weak, tumors with 1+ staining intensity in ≤70% of neoplastic cells and 2+ intensity in ≤30% of neoplastic cells; moderate, tumors with 1+ staining intensity in >70% of neoplastic cells, 2+ intensity in 31–70%, or 3+ intensity in ≤30% of neoplastic cells; strong, tumors with 2+ intensity in >70% or 3+ intensity in >30% of neoplastic cells.
2.3. Statistics
Statistical calculations were performed with JMP® software (Version 17) SAS®, Cary, NC, USA). Contingency tables, the chi2-test, and Fisher’s exact test were performed to search for associations between TFF1 immunostaining and tumor phenotype and MUC5AC immunostaining.
3. Results
3.1. Technical Issues
A total of 16,817 (89.1%) of 18,878 tumor samples were interpretable in our TMA analysis. Non-interpretable samples demonstrated the absence of unequivocal tumor cells or a complete lack of individual tissue spots. A sufficient number of samples of each normal tissue type was evaluable (≥4).
3.2. TFF1 in Normal Tissues
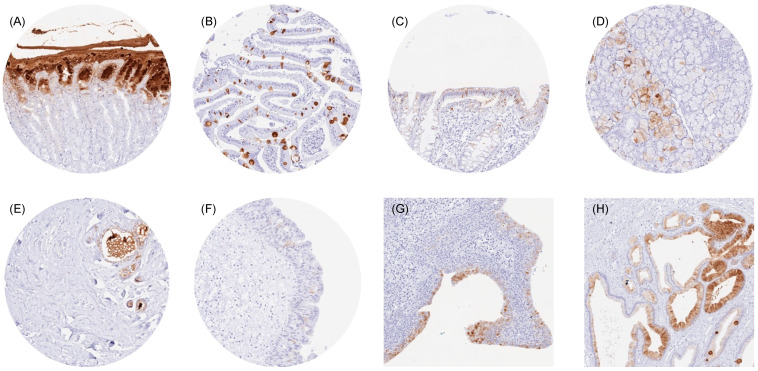
TFF1 immunostaining was always cytoplasmic. It was strongest in superficial epithelial cells of the stomach, while the deeper glandular cells remained TFF1 negative. Scattered goblet cells with a significant TFF1 positivity were also seen in the small intestine and the colorectum. A weak to moderate TFF1 immunostaining was also observed in some goblet cells of the respiratory epithelium; groups of mucinous glandular cells in bronchial, sublingual, and submandibular glands; and in a fraction of luminal breast epithelial cells. Occasionally, a few cells with a distinct TFF1 positivity were also observed in the urothelium (mostly umbrella cells) or in gall bladder epithelial cells. Representative images are shown in Figure 1.
Figure 1.
TFF1 immunostaining of normal tissues. The panels show a cytoplasmic staining of surface epithelial cells but not of glands in the stomach (A), subsets of goblet cells in the duodenum (B) and the colon (C), a subset of mucinous cells in the submandibulary gland (D), a subset of luminal epithelial cells and intraluminal mucus in the breast (E), a small subset of urothelial cells (mostly umbrella cells) in the renal pelvis (F), a large subset of urothelial cells in an inflamed urinary bladder (G), and (focally) in epithelial cells of the gallbladder (H).
All these staining patterns were seen by both MSVA-482M and EPR3972 (Supplementary Figure S1). TFF1 staining was absent in skeletal muscle, heart muscle, smooth muscle, myometrium of the uterus, corpus spongiosum of the penis, ovarian stroma, fat, skin (including hair follicle and sebaceous glands), surface epithelium of the tonsil, ectocervix, squamous epithelium of the esophagus, decidua, placenta, bone marrow; lymph node, spleen, thymus, tonsil, liver, pancreas, Brunner gland of the duodenum, cortex and medulla of the kidney, prostate, seminal vesicle, testis, epididymis, lung, endocervix, endometrium, fallopian tube, ovary (including corpus luteum and theca and granulosa cells of follicular cysts) adrenal medulla, thyroid, parathyroid gland, cerebellum, cerebrum and the pituitary gland.
3.3. TFF1 in Tumor Tissues
TFF1 immunostaining was detectable in 3347 (19.9%) of the 16,817 analyzable tumors, including 2093 (12.4%) with weak, 740 (4.4%) with moderate, and 514 (3.1%) with strong positivity. Overall, 65 (43.6%) of 149 tumor categories showed detectable TFF1 expression, with 32 (21.5%) tumor categories containing at least one case with strong positivity (Table 1).
Table 1.
TFF1 immunostaining in human tumors: neg. = negative, mod. = moderate, str. = strong, ca. = carcinoma.
| TFF1 Immunostaining | |||||||
|---|---|---|---|---|---|---|---|
| Tumor Category | Tumor Entity | on TMA (n) | Analy-Zable (n) | Neg. (%) | Weak (%) | Mod. (%) | Str. (%) |
| Tumors of the skin | Basal cell carcinoma of the skin | 89 | 81 | 100.0 | 0.0 | 0.0 | 0.0 |
| Benign nevus | 29 | 21 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Squamous cell carcinoma of the skin | 145 | 132 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Malignant melanoma | 65 | 58 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Malignant melanoma lymph node metastasis | 86 | 86 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Merkel cell carcinoma | 2 | 2 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Tumors of the head and neck | Squamous cell carcinoma of the larynx | 109 | 98 | 98.0 | 2.0 | 0.0 | 0.0 |
| Squamous cell ca. of the pharynx | 60 | 60 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Oral squamous cell carcinoma | 130 | 128 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Pleomorphic adenoma of the parotid gland | 50 | 38 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Warthin tumor of the parotid gland | 104 | 91 | 98.9 | 1.1 | 0.0 | 0.0 | |
| Adenocarcinoma, NOS (Papillary Cystadenocarcinoma) | 14 | 10 | 90.0 | 0.0 | 0.0 | 10.0 | |
| Salivary duct carcinoma | 15 | 10 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Acinic cell carcinoma of the salivary gland | 181 | 110 | 99.1 | 0.0 | 0.9 | 0.0 | |
| Adenocarcinoma NOS of the salivary gland | 109 | 54 | 88.9 | 1.9 | 1.9 | 7.4 | |
| Adenoid cystic carcinoma of the salivary gland | 180 | 61 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Basal cell adenocarcinoma of the salivary gland | 25 | 19 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Basal cell adenoma of the salivary gland | 101 | 60 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Epithelial-myoepithelial carcinoma of the salivary gland | 53 | 44 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Mucoepidermoid carcinoma of the salivary gland | 343 | 250 | 86.0 | 8.0 | 4.0 | 2.0 | |
| Myoepithelial carcinoma of the salivary gland | 21 | 15 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Myoepithelioma of the salivary gland | 11 | 9 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Oncocytic carcinoma of the salivary gland | 12 | 6 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Polymorphous adenoca. low grade, of the salivary gland | 41 | 15 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Pleomorphic adenoma of the salivary gland | 53 | 30 | 93.3 | 6.7 | 0.0 | 0.0 | |
| Tumors of the lung, pleura and thymus | Adenocarcinoma of the lung | 196 | 188 | 89.4 | 6.9 | 2.1 | 1.6 |
| Squamous cell carcinoma of the lung | 80 | 74 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Mesothelioma, epithelioid | 40 | 34 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Mesothelioma, biphasic | 29 | 29 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Thymoma | 29 | 24 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Lung, neuroendocrine tumor (NET) | 29 | 27 | 92.6 | 0.0 | 0.0 | 7.4 | |
| Tumors of the female genital tract | Squamous cell carcinoma of the vagina | 78 | 71 | 100.0 | 0.0 | 0.0 | 0.0 |
| Squamous cell carcinoma of the vulva | 157 | 144 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Squamous cell carcinoma of the cervix | 136 | 129 | 98.4 | 1.6 | 0.0 | 0.0 | |
| Adenocarcinoma of the cervix | 23 | 23 | 60.9 | 17.4 | 13.0 | 8.7 | |
| Endometrioid endometrial carcinoma | 338 | 295 | 95.6 | 3.7 | 0.3 | 0.3 | |
| Endometrial serous carcinoma | 86 | 73 | 98.6 | 1.4 | 0.0 | 0.0 | |
| Carcinosarcoma of the uterus | 57 | 53 | 98.1 | 0.0 | 1.9 | 0.0 | |
| Endometrial carcinoma, high grade, G3 | 13 | 8 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Endometrial clear cell carcinoma | 9 | 6 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Endometrioid carcinoma of the ovary | 130 | 115 | 94.8 | 4.3 | 0.0 | 0.9 | |
| Serous carcinoma of the ovary | 580 | 507 | 99.8 | 0.2 | 0.0 | 0.0 | |
| Mucinous carcinoma of the ovary | 101 | 84 | 23.8 | 21.4 | 15.5 | 39.3 | |
| Clear cell carcinoma of the ovary | 51 | 45 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Carcinosarcoma of the ovary | 47 | 47 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Granulosa cell tumor of the ovary | 44 | 42 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Leydig cell tumor of the ovary | 4 | 4 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Sertoli cell tumor of the ovary | 1 | 1 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Sertoli Leydig cell tumor of the ovary | 3 | 3 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Steroid cell tumor of the ovary | 3 | 3 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Brenner tumor | 41 | 41 | 92.7 | 7.3 | 0.0 | 0.0 | |
| Tumors of the breast | Invasive breast carcinoma of no special type | 1764 | 1692 | 55.5 | 26.6 | 10.9 | 7.0 |
| Lobular carcinoma of the breast | 363 | 346 | 47.7 | 27.2 | 17.1 | 8.1 | |
| Medullary carcinoma of the breast | 34 | 34 | 88.2 | 2.9 | 8.8 | 0.0 | |
| Tubular carcinoma of the breast | 29 | 23 | 60.9 | 21.7 | 13.0 | 4.3 | |
| Mucinous carcinoma of the breast | 65 | 59 | 27.1 | 35.6 | 13.6 | 23.7 | |
| Phyllodes tumor of the breast | 50 | 46 | 47.8 | 30.4 | 21.7 | 0.0 | |
| Tumors of the digestive system | Adenomatous polyp, low-grade dysplasia | 50 | 50 | 32.0 | 52.0 | 12.0 | 4.0 |
| Adenomatous polyp, high-grade dysplasia | 50 | 48 | 25.0 | 50.0 | 16.7 | 8.3 | |
| Adenocarcinoma of the colon | 2483 | 2256 | 52.9 | 35.3 | 7.0 | 4.7 | |
| Gastric adenocarcinoma, diffuse type | 215 | 176 | 50.0 | 13.6 | 17.6 | 18.8 | |
| Gastric adenocarcinoma, intestinal type | 215 | 192 | 52.1 | 33.9 | 7.8 | 6.3 | |
| Gastric adenocarcinoma, mixed type | 62 | 57 | 59.6 | 22.8 | 8.8 | 8.8 | |
| Adenocarcinoma of the esophagus | 83 | 62 | 62.9 | 24.2 | 8.1 | 4.8 | |
| Squamous cell carcinoma of the esophagus | 76 | 55 | 98.2 | 0.0 | 1.8 | 0.0 | |
| Squamous cell carcinoma of the anal canal | 91 | 85 | 98.8 | 0.0 | 1.2 | 0.0 | |
| Cholangiocarcinoma | 121 | 114 | 63.2 | 21.1 | 7.9 | 7.9 | |
| Gallbladder adenocarcinoma | 51 | 50 | 48.0 | 22.0 | 20.0 | 10.0 | |
| Gallbladder Klatskin tumor | 42 | 38 | 57.9 | 21.1 | 15.8 | 5.3 | |
| Hepatocellular carcinoma | 312 | 304 | 89.5 | 5.3 | 2.0 | 3.3 | |
| Ductal adenocarcinoma of the pancreas | 659 | 613 | 42.6 | 28.1 | 15.0 | 14.4 | |
| Pancreatic/Ampullary adenocarcinoma | 98 | 96 | 37.5 | 32.3 | 18.8 | 11.5 | |
| Acinar cell carcinoma of the pancreas | 18 | 18 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Gastrointestinal stromal tumor (GIST) | 62 | 59 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Appendix, neuroendocrine tumor (NET) | 25 | 16 | 81.3 | 6.3 | 6.3 | 6.3 | |
| Colorectal, neuroendocrine tumor (NET) | 12 | 11 | 54.5 | 18.2 | 27.3 | 0.0 | |
| Ileum, neuroendocrine tumor (NET) | 53 | 51 | 92.2 | 5.9 | 2.0 | 0.0 | |
| Pancreas, neuroendocrine tumor (NET) | 101 | 92 | 83.7 | 7.6 | 4.3 | 4.3 | |
| Colorectal, neuroendocrine carcinoma (NEC) | 14 | 12 | 75.0 | 16.7 | 8.3 | 0.0 | |
| Ileum, neuroendocrine carcinoma (NEC) | 8 | 7 | 85.7 | 0.0 | 14.3 | 0.0 | |
| Gallbladder, neuroendocrine carcinoma (NEC) | 4 | 4 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Pancreas, neuroendocrine carcinoma (NEC) | 14 | 14 | 92.9 | 0.0 | 7.1 | 0.0 | |
| Tumors of the urinary system | Non-invasive papillary urothelial ca. pTa G2 low grade | 177 | 152 | 75.7 | 19.7 | 3.3 | 1.3 |
| Non-invasive papillary urothelial ca., pTa G2 high grade | 141 | 116 | 82.8 | 13.8 | 2.6 | 0.9 | |
| Non-invasive papillary urothelial carcinoma, pTa G3 | 219 | 131 | 90.1 | 7.6 | 2.3 | 0.0 | |
| Urothelial carcinoma, pT2-4 G3 | 735 | 600 | 94.5 | 4.0 | 1.5 | 0.0 | |
| Squamous cell carcinoma of the bladder | 22 | 22 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Small cell neuroendocrine carcinoma of the bladder | 5 | 5 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Sarcomatoid urothelial carcinoma | 25 | 23 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Urothelial carcinoma of the kidney pelvis | 62 | 61 | 88.5 | 11.5 | 0.0 | 0.0 | |
| Clear cell renal cell carcinoma | 1286 | 1230 | 99.8 | 0.2 | 0.0 | 0.0 | |
| Papillary renal cell carcinoma | 368 | 327 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Clear cell (tubulo) papillary renal cell carcinoma | 26 | 23 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Chromophobe renal cell carcinoma | 170 | 151 | 98.7 | 1.3 | 0.0 | 0.0 | |
| Oncocytoma | 257 | 230 | 80.9 | 9.6 | 9.1 | 0.4 | |
| Tumors of the male genital organs | Adenocarcinoma of the prostate, Gleason 3 + 3 | 83 | 79 | 92.4 | 6.3 | 1.3 | 0.0 |
| Adenocarcinoma of the prostate, Gleason 4 + 4 | 80 | 69 | 81.2 | 14.5 | 4.3 | 0.0 | |
| Adenocarcinoma of the prostate, Gleason 5 + 5 | 85 | 79 | 89.9 | 8.9 | 1.3 | 0.0 | |
| Adenocarcinoma of the prostate (recurrence) | 258 | 216 | 92.6 | 6.0 | 1.4 | 0.0 | |
| Small cell neuroendocrine carcinoma of the prostate | 2 | 2 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Seminoma | 682 | 586 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Embryonal carcinoma of the testis | 54 | 41 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Leydig cell tumor of the testis | 31 | 31 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Sertoli cell tumor of the testis | 2 | 2 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Sex cord stromal tumor of the testis | 1 | 1 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Spermatocytic tumor of the testis | 1 | 1 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Yolk sac tumor | 53 | 42 | 97.6 | 2.4 | 0.0 | 0.0 | |
| Teratoma | 53 | 39 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Squamous cell carcinoma of the penis | 92 | 90 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Tumors of endocrine organs | Adenoma of the thyroid gland | 113 | 108 | 99.1 | 0.9 | 0.0 | 0.0 |
| Papillary thyroid carcinoma | 391 | 374 | 97.3 | 2.7 | 0.0 | 0.0 | |
| Follicular thyroid carcinoma | 154 | 146 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Medullary thyroid carcinoma | 111 | 101 | 77.2 | 19.8 | 2.0 | 1.0 | |
| Parathyroid gland adenoma | 43 | 42 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Anaplastic thyroid carcinoma | 45 | 42 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Adrenal cortical adenoma | 50 | 37 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Adrenal cortical carcinoma | 28 | 28 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Pheochromocytoma | 50 | 50 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Tumors of haemotopoetic and lymphoid tissues | Hodgkin’s lymphoma | 103 | 90 | 100.0 | 0.0 | 0.0 | 0.0 |
| Small lymphocytic lymphoma, B-cell type (B-SLL/B-CLL) | 50 | 50 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Diffuse large B cell lymphoma (DLBCL) | 113 | 113 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Follicular lymphoma | 88 | 88 | 100.0 | 0.0 | 0.0 | 0.0 | |
| T-cell non-Hodgkin’s lymphoma | 25 | 25 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Mantle cell lymphoma | 18 | 18 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Marginal zone lymphoma | 16 | 16 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Diffuse large B-cell lymphoma (DLBCL) in the testis | 16 | 16 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Burkitt lymphoma | 5 | 5 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Tumors of soft tissue and bone | Granular cell tumor | 23 | 21 | 76.2 | 9.5 | 14.3 | 0.0 |
| Leiomyoma | 50 | 45 | 97.8 | 2.2 | 0.0 | 0.0 | |
| Leiomyosarcoma | 94 | 88 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Liposarcoma | 96 | 93 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Malignant peripheral nerve sheath tumor (MPNST) | 15 | 13 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Myofibrosarcoma | 26 | 26 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Angiosarcoma | 42 | 40 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Angiomyolipoma | 91 | 89 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Dermatofibrosarcoma protuberans | 21 | 17 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Ganglioneuroma | 14 | 12 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Kaposi sarcoma | 8 | 6 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Neurofibroma | 117 | 116 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Sarcoma, not otherwise specified (NOS) | 74 | 72 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Paraganglioma | 41 | 40 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Ewing sarcoma | 23 | 18 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Rhabdomyosarcoma | 7 | 7 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Schwannoma | 122 | 119 | 99.2 | 0.8 | 0.0 | 0.0 | |
| Synovial sarcoma | 12 | 11 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Osteosarcoma | 19 | 18 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Chondrosarcoma | 15 | 11 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Rhabdoid tumor | 5 | 5 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Solitary fibrous tumor | 17 | 14 | 100.0 | 0.0 | 0.0 | 0.0 | |
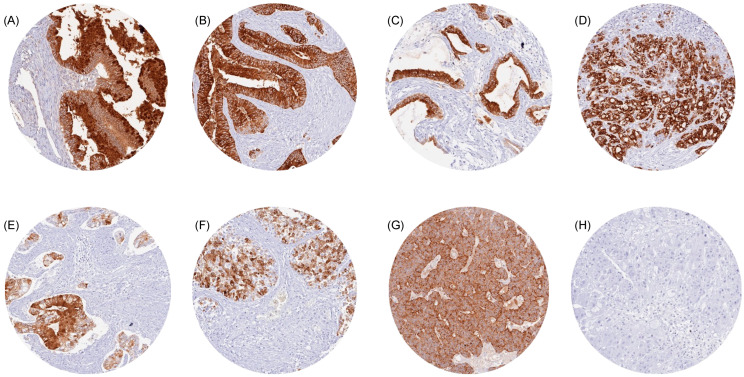
Representative images of TFF1-positive tumors are shown in Figure 2. The highest rate of TFF1 positivity was found in mucinous carcinomas of the ovary (76.2%), colorectal adenomas and adenocarcinomas (47.1–75.0%), neoplasms of the breast (11.8–72.9%), bilio-pancreatic adenocarcinomas (42.1–62.5%), gastro-esophageal adenocarcinomas (40.4–50.0%), neuroendocrine neoplasms of various sites of origin (7.1–45.5%), adenocarcinomas of the cervix (39.1%), and in urothelial neoplasms (5.5–24.3%).
Figure 2.
TFF1 immunostaining in cancer. The panels show TFF1 positivity in a mucinous carcinoma of the ovary (A), a colorectal adenocarcinoma (B), a pancreatic adenocarcinoma (C), a gastric adenocarcinoma (D), an adenocarcinoma of the cervix (E), urothelial carcinoma of the bladder (F), and a neuroendocrine tumor of the lung (G). TFF1 staining is absent in a hepatocellular carcinoma (H).
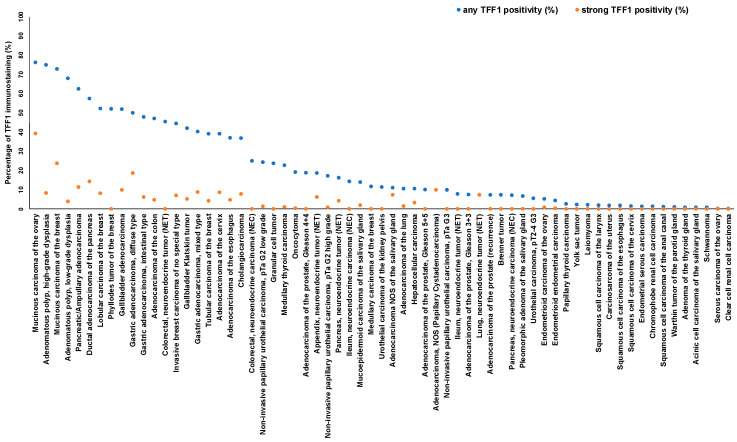
The ranking order of all TFF1-positive entities is shown in Figure 3. The relationship between TFF1 expression and clinically important histopathological and molecular tumor features in colorectal, gastric, pancreatic, urinary bladder, and breast cancer is shown in Table 2.
Figure 3.
Ranking order of TFF1 immunostaining in tumors. Both the percentage of positive cases (blue dots) and the percentage of strongly positive cases (orange dots) are shown.
Table 2.
TFF1 immunostaining and tumor phenotype.
| TFF1 Immunostaining | ||||||||
|---|---|---|---|---|---|---|---|---|
| n | Negative (%) | Weak (%) | Moderate (%) | Strong (%) | p | Remarks | ||
| Invasive breast carcinoma of no special type | pT1 | 802 | 448 (55.9) | 203 (25.3) | 94 (11.7) | 57 (7.1) | 0.3766 | |
| pT2 | 635 | 331 (52.1) | 185 (29.1) | 67 (10.6) | 52 (8.2) | |||
| pT3-4 | 126 | 71 (56.3) | 35 (27.8) | 15 (11.9) | 5 (4) | |||
| G1 | 202 | 106 (52.5) | 52 (25.7) | 32 (15.8) | 12 (5.9) | <0.0001 | ||
| G2 | 845 | 423 (50.1) | 238 (28.2) | 110 (13) | 74 (8.8) | |||
| G3 | 564 | 354 (62.8) | 146 (25.9) | 36 (6.4) | 28 (5) | |||
| pN0 | 692 | 361 (52.2) | 174 (25.1) | 102 (14.7) | 55 (8) | 0.3307 | ||
| pN+ | 946 | 521 (55.1) | 241 (25.5) | 111 (11.7) | 73 (7.7) | |||
| pM0 | 217 | 121 (55.8) | 52 (24) | 28 (12.9) | 16 (7.4) | 0.7665 | ||
| pM1 | 112 | 60 (53.6) | 32 (28.6) | 14 (12.5) | 6 (5.4) | |||
| HER2 negative | 890 | 479 (53.8) | 258 (29) | 97 (10.9) | 56 (6.3) | 0.9505 | ||
| HER2 positive | 122 | 69 (56.6) | 33 (27.1) | 13 (10.7) | 7 (5.7) | |||
| ER negative | 213 | 184 (86.4) | 21 (9.9) | 3 (1.4) | 5 (2.4) | <0.0001 | ||
| ER positive | 745 | 334 (44.8) | 251 (33.7) | 105 (14.1) | 55 (7.4) | |||
| PR negative | 408 | 277 (67.9) | 90 (22.1) | 27 (6.6) | 14 (3.4) | <0.0001 | ||
| PR positive | 598 | 270 (45.2) | 197 (32.9) | 82 (13.7) | 49 (8.2) | |||
| non-triple negative | 783 | 366 (46.7) | 258 (33) | 102 (13) | 57 (7.3) | <0.0001 | ||
| triple negative | 144 | 133 (92.4) | 7 (4.9) | 1 (0.7) | 3 (2.1) | |||
| Urothelial bladder carcinoma | pTa G2 low | 152 | 115 (75.7) | 30 (19.7) | 5 (3.3) | 2 (1.3) | <0.0001 | |
| pTa G2 high | 116 | 96 (82.8) | 16 (13.8) | 3 (2.6) | 1 (0.9) | 0.0072 | 1 | |
| pTa G3 | 99 | 91 (91.9) | 5 (5.1) | 3 (3) | 0 (0) | |||
| pT2-4 | 453 | 429 (94.7) | 19 (4.2) | 5 (1.1) | 0 (0) | |||
| pT2 | 125 | 122 (97.6) | 3 (2.4) | 0 (0) | 0 (0) | 0.3037 | 2,3,4 | |
| pT3 | 219 | 206 (94.1) | 9 (4.1) | 4 (1.8) | 0 (0) | |||
| pT4 | 98 | 91 (92.9) | 6 (6.1) | 1 (1) | 0 (0) | |||
| G2 | 23 | 20 (87) | 3 (13) | 0 (0) | 0 (0) | 0.0995 | 2,4 | |
| G3 | 429 | 408 (95.1) | 16 (3.7) | 5 (1.2) | 0 (0) | |||
| pN0 | 271 | 261 (96.3) | 8 (3) | 2 (0.7) | 0 (0) | 0.0414 | 2,3 | |
| pN+ | 161 | 148 (91.9) | 11 (6.8) | 2 (1.2) | 0 (0) | |||
| Adenocarcinoma of the pancreas | pT1 | 14 | 3 (21.4) | 6 (42.9) | 2 (14.3) | 3 (21.4) | 0.3769 | 5 |
| pT2 | 66 | 18 (27.3) | 22 (33.3) | 15 (22.7) | 11 (16.7) | |||
| pT3 | 378 | 133 (35.2) | 125 (33.1) | 58 (15.3) | 62 (16.4) | |||
| pT4 | 27 | 12 (44.4) | 5 (18.5) | 8 (29.6) | 2 (7.4) | |||
| G1 | 17 | 5 (29.4) | 7 (41.2) | 2 (11.8) | 3 (17.6) | 0.8595 | 6 | |
| G2 | 343 | 115 (33.5) | 111 (32.4) | 61 (17.8) | 56 (16.3) | |||
| G3 | 103 | 39 (37.9) | 32 (31.1) | 17 (16.5) | 15 (14.6) | |||
| pN0 | 101 | 28 (27.7) | 34 (33.7) | 18 (17.8) | 21 (20.8) | 0.3281 | ||
| pN+ | 383 | 137 (35.8) | 125 (32.6) | 65 (17) | 56 (14.6) | |||
| R0 | 244 | 80 (32.8) | 90 (36.9) | 39 (16) | 35 (14.3) | 0.4142 | ||
| R1 | 201 | 72 (35.8) | 59 (29.4) | 37 (18.4) | 33 (16.4) | |||
| Adenocarcinoma of the stomach | pT1-2 | 62 | 31 (50) | 21 (33.9) | 7 (11.3) | 3 (4.8) | 0.8258 | 3 |
| pT3 | 128 | 70 (54.7) | 28 (21.9) | 13 (10.2) | 17 (13.3) | |||
| pT4 | 122 | 64 (52.5) | 26 (21.3) | 14 (11.5) | 18 (14.8) | |||
| pN0 | 85 | 42 (49.4) | 23 (27.1) | 10 (11.8) | 10 (11.8) | |||
| pN+ | 226 | 123 (54.4) | 50 (22.1) | 24 (10.6) | 29 (12.8) | |||
| MMR proficient | 257 | 136 (52.9) | 60 (23.3) | 34 (13.2) | 27 (10.5) | 0.6225 | 3 | |
| MMR deficient | 41 | 20 (48.8) | 14 (34.1) | 1 (2.4) | 6 (14.6) | |||
| Adenocarcinoma of the colon | pT1 | 75 | 30 (40) | 39 (52) | 4 (5.3) | 2 (2.7) | 0.6503 | 5 |
| pT2 | 421 | 220 (52.3) | 141 (33.5) | 39 (9.3) | 21 (5) | |||
| pT3 | 1200 | 604 (50.3) | 455 (37.9) | 86 (7.2) | 55 (4.6) | |||
| pT4 | 435 | 227 (52.2) | 153 (35.2) | 28 (6.4) | 27 (6.2) | |||
| pN0 | 1125 | 575 (51.1) | 413 (36.7) | 82 (7.3) | 55 (4.9) | 0.9973 | ||
| pN+ | 1003 | 516 (51.4) | 364 (36.3) | 73 (7.3) | 50 (5) | |||
| V0 | 1533 | 783 (51.1) | 555 (36.2) | 112 (7.3) | 81 (5.3) | 0.6924 | ||
| V1 | 563 | 291 (51.7) | 208 (36.9) | 41 (7.3) | 23 (4.1) | |||
| L0 | 701 | 369 (52.6) | 240 (34.2) | 54 (7.7) | 38 (5.4) | 0.4669 | ||
| L1 | 1404 | 706 (50.3) | 529 (37.7) | 102 (7.3) | 67 (4.8) | |||
| right side | 427 | 188 (44) | 168 (39.3) | 47 (11) | 24 (5.6) | 0.0021 | ||
| left side | 1173 | 622 (53) | 416 (35.5) | 75 (6.4) | 59 (5) | |||
| MMR proficient | 1110 | 586 (52.8) | 402 (36.2) | 75 (6.8) | 47 (4.2) | 0.0292 | 3 | |
| MMR deficient | 84 | 34 (40.5) | 35 (41.7) | 11 (13.1) | 4 (4.8) | |||
| RAS wildtype | 450 | 274 (60.9) | 131 (29.1) | 23 (5.1) | 22 (4.9) | <0.0001 | ||
| RAS mutation | 340 | 148 (43.5) | 138 (40.6) | 38 (11.2) | 16 (4.7) | |||
| BRAF wildtype | 111 | 70 (63.1) | 30 (27) | 8 (7.2) | 3 (2.7) | 0.0331 | 3 | |
| BRAF V600E mutation | 19 | 7 (36.8) | 6 (31.6) | 3 (15.8) | 3 (15.8) | |||
Abbreviations: pT: pathological tumor stage, G: Grade, pN: pathological lymph node status, pM: pathological status of distant metastasis, L: lymphatic infiltration, V: venous infiltration, R: resection margin status, MMR: mismatch repair. Remarks: p-values corespond to 1: pTa only, 2: pT2-4 only, 3: TFF1 negative vs. TFF1 positive (weak, mod. str.), 4: Fisher’s excact test, 5: pT1-2 vs. pT3-4, 6: G1-2 vs. G3.
High TFF1 expression was significantly associated with a low grade of malignancy in non-invasive (pTa) urothelial carcinomas of the urinary bladder (p = 0.0072), low grade of malignancy (p < 0.0001), estrogen (ER) and progesterone receptor (PR) expression and non-triple negativity (p < 0.0001 each) in invasive breast cancer of no special type, as well as with right-sided tumor location (p = 0.0021), MMR deficiency (p = 0.0292), RAS mutations (p < 0.0001), and BRAF mutation (p = 0.0331) in colorectal adenocarcinomas. The extent of TFF1 staining was unrelated to histopathological parameters of malignancy in 312 gastric and 485 pancreatic adenocarcinomas.
3.4. Comparison with MUC5AC
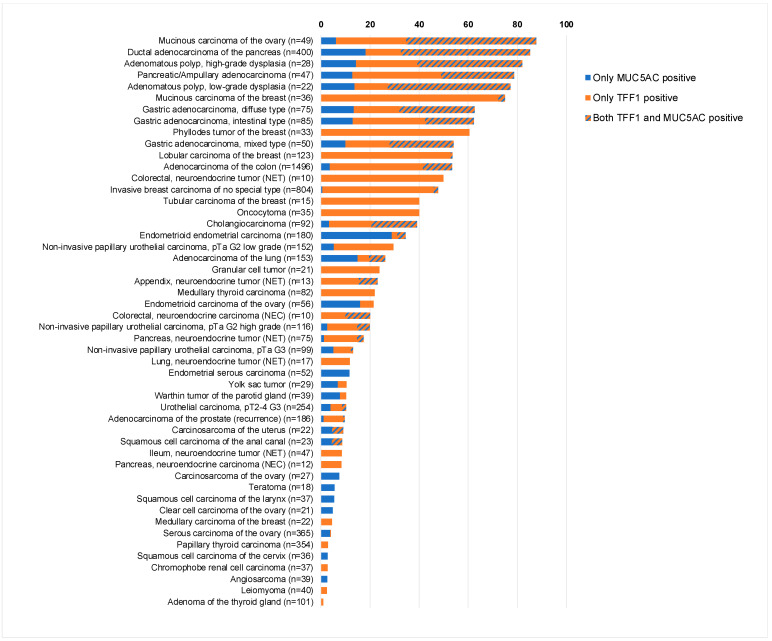
Data on MUC5AC expression were available for 7870 of our tumors, which were evaluated for TFF1. A total of 50 entities showed expression of either TFF1 or MUC5AC. There was a tendency towards a co-expression of TFF1 and MUC5AC, but this relationship was rather weak. Among 7870 tumor samples with available TFF1 and MUC5AC data, 573 (7.3%) showed expression of both TFF1 and MUC5AC, 1391 (17.7%) showed expression of only TFF1, 325 (4.1%) showed expression of only MUC5AC, and 5581 (70.9%) showed expression of none of the two proteins (p < 0.0001). MUC5AC/TFF1 co-expression was particularly common in mucinous carcinoma of the ovary, gastro-intestinal, and bilio-pancreatic neoplasms but also occurred in other entities. A graphical representation of the expression of MUC5AC and TFF1 in 50 tumor entities is given in Figure 4.
Figure 4.
Comparison of TFF1 and MUC5AC immunostaining.
4. Discussion
The successful analysis of 16,817 tumors from 149 tumor types and subtypes resulted in a comprehensive overview on the expression of TFF1 in human tumors. The ranking order of 65 tumor entities according to their rate of TFF1 positivity is a key result of our study (Figure 3). The comparative representation of our data and previously published data (Figure 5) demonstrates that this information could not be retrieved from literature databases as many tumor entities have, so far, not been analyzed, and data were often highly discordant for entities that have been analyzed in multiple studies. Our data show that TFF1 positivity occurs most commonly in mucinous carcinomas of the ovary, colorectal adenocarcinomas, breast cancer, bilio-pancreatic, and gastro-esophageal adenocarcinomas, neuroendocrine neoplasms, adenocarcinomas of the cervix uteri, and in urothelial neoplasms. Considering that 65 different tumor entities contained TFF1-positive cases and that the rate of positivity did not exceed 80% in any of the most frequently positive cancer entities, we do not believe that TFF1 IHC has a relevant role for the distinction of cancer entities. However, TFF1 IHC may be useful to select patients for potential future therapies using, for example, antibody drug conjugates. Only recently, the de novo expression of TFF1 has been associated with gemcitabine resistance in pancreatic adenocarcinomas, and TFF1 silencing increased sensitivity to gemcitabine in vitro and in vivo [42].
Figure 5.
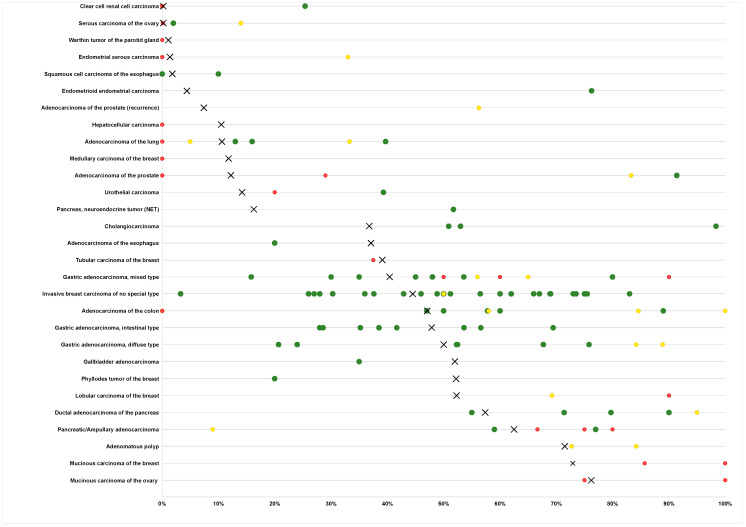
Comparison with previous TFF1 literature. An “X” indicates the fraction of TFF1 positive cancers in the present study, dots indicate the reported frequencies from the literature for comparison: red dots mark studies with ≤10 analyzed tumors, yellow dots mark studies with 11 to 25 analyzed tumors, and green dots mark studies with >25 analyzed tumors.
The striking discrepancy between TFF1 expression in only a few normal tissues and TFF1 expression in so many different cancer types demonstrates that TFF1 upregulation is a common phenomenon in cancer, which may have a significant functional role. The increase in TFF1 staining that was seen in colon adenomas as compared to normal colon mucosa further supports this notion. Earlier studies confirmed TFF1 expression in colon adenomas [8,43,44] and also reported TFF1 staining from other preneoplastic lesions, including, for example, intestinal metaplasia [45]. An oncogenic role would be most conceivable for a protein that is often overexpressed in cancer as compared to its cell of origin. In line with this notion, several authors have indeed described evidence for the oncogenic effects of TFF1. For example, TFF1 promoted anchorage-independent growth in colon carcinoma cells [43], phosphoinositide 3-kinases (PI3K) dependent migration and invasion in gastric carcinoma cells [46], and cyclooxygenase-2 (COX-2) and EGF receptor (EGFR)-dependent angiogenesis in kidney and colonic cancer cells [47]. The molecular mechanism underlying a putative oncogenic function is largely unknown. Rodrigues et al. [43] reported that TFF1 upregulates the phosphatase CDC25A, one of the most crucial cell cycle regulators and activator of CDKs [48], suggesting that both factors cooperate with other oncogenic pathways to drive a adenoma–carcinoma transition in colon carcinoma. However, the significant associations found between TFF1 expression and several favorable tumor features in bladder and breast cancer in combination with the absence of significant links with unfavorable prognostic parameters in colorectal, gastric, and pancreatic cancers at least argues against a significant role of TFF1 for driving cancer aggressiveness. TFF1 RNA expression data derived from The Cancer Genome Atlas (TCGA) database also pinpoint a favorable prognosis for breast and endometrium cancer with high TFF1 expression (https://www.proteinatlas.org/ENSG00000160182-TFF1/pathology, accessed on 9 January 2024). In line with these findings, several functional studies have rather supported a tumor-suppressive than oncogenic role for TFF1. For example, the overexpression of TFF1 suppressed nuclear factor kappa-light-chain-enhancer of activated B-cells (NF-kB) signaling and, therefore, pro-tumorigenic and metastatic properties in colon and gastric cancer cells [49,50] as well as ß-catenin signaling in hepatocellular carcinoma cells [51]. TFF1 knockdown enhanced anchorage-independent growth in breast cancer cells in vivo and in vitro [52], reduced apoptosis and induced proliferation in gastric cancer cells [53], and accelerated the development of esophageal adenocarcinoma [54] and hepatocellular carcinoma in vivo [51].
Previous studies analyzing the prognostic role of TFF1 have provided rather controversial results. For breast cancer, gastric adenocarcinoma and pancreatic ampullary adenocarcinoma studies have found associations between high TFF1 expression levels and both favorable [12,13,14,17,55,56] and unfavorable tumor features [10,57,58]. Significant associations of high TFF1 expression with favorable tumor features or favorable prognosis were also found in gallbladder adenocarcinoma [11], while high TFF1 expression was linked to unfavorable tumor parameters or poor patient outcomes in adenocarcinoma of the lung [30,31], colorectal adenocarcinoma [9,59], and in urothelial carcinoma [60]. Another study in renal cell carcinomas did not suggest any relationship between TFF1 expression and parameters of cancer aggressiveness [34].
The availability of data from an earlier study enabled us to compare our TFF1 results with MUC5AC data [41]. Although these proteins are frequently co-expressed and act cooperatively in normal tissues, the relationship between the expression of TFF1 and MUC5AC was rather weak in cancer, since only 7% of 2289 cancer samples with TFF1 and/or MUC5AC expression expressed both proteins. We, therefore, assume that MUC5AC and TFF1 are functionally less dependent on each other in neoplastic than in normal tissues. That mucinous carcinoma of the ovary, gastro-intestinal, and bilio-pancreatic neoplasms showed co-expression of TFF1 and MUC5AC (20% to 53%) more commonly than other tumor entities is in line with some previous studies describing high rates of MUC5AC/TFF1 co-expression in neoplasms of the colon, liver, stomach, and gallbladder [8,18,61,62]. However, other authors found lower co-expression rates in gastric [63] and colorectal adenocarcinomas [25], and markedly higher frequencies of co-expression were reported for cancers from the esophagus, lung, and bladder [31,64,65].
To validate our assay, our IHC results in normal tissue were compared with RNA results from three different publicly available databases [66,67,68,69] and with immunohistochemical data from an independent anti-TFF1 antibody. The antibody comparison study included 76 different categories of normal tissues to ensure the broadest possible range of expressed proteins. The assay validity was supported by the detection of TFF1 strong immunostaining in the stomach, which was the only normal organ for which RNA expression had previously been reported. The additional positive TFF1 stainings in scattered goblet cells in the small intestine, colorectum, and respiratory epithelium, some mucinous glandular cells in salivary and in bronchial glands, a fraction of luminal breast epithelial cells as well as in some cells of the urothelium and gallbladder epithelium were all validated by identical stainings seen by the independent antibody EPR3972. Given that these TFF1-positive cells constituted very small subpopulations of the respective organs, we assume that TFF1 RNA had not been detected due to a massive dilution if RNAs from total organs were analyzed.
5. Conclusions
Our data provide a comprehensive overview on the expression of TFF1 in human cancer. Given that TFF1 is expressed in a broad range of tumor entities, TFF1 IHC may have only limited utility for the discrimination of different tumor entities. Upregulation in multiple tumor entities and the significant link between TFF1 expression and parameters of malignancy in several tumors argue for a relevant biological role of TFF1 in cancer, however. TFF1 may also represent a suitable therapeutic target.
Acknowledgments
We are grateful to Melanie Steurer, Laura Behm, Inge Brandt, and Sünje Seekamp for excellent technical assistance.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/diagnostics14192157/s1. Supplementary Figure S1. Immunohistochemistry validation by comparison of two antibodies. The panels show immunostaining results obtained by two independent TFF1 antibodies. Using MSVA-482M, a distinct cytoplasmic staining was seen in surface epithelial cells (but not of glands) in the stomach (A), subsets of goblet cells in the duodenum (B) and the colon (C), a subset of mucinous cells in the submandibulary gland (D), a subset of luminal epithelial cells and intraluminal mucus in the breast (E), a small subset of urothelial cells (mostly umbrella cells) in the renal pelvis (F), a large subset of urothelial cells in an inflamed urinary bladder (G), and in epithelial cells of the gallbladder (H). Using clone EPR3972, a comparable staining was seen in the stomach (a), the duodenum (b), the colon (c), the submandibulary gland (d), the breast (e), the renal pelvis (f), the bladder (g), and the gallbladder (h). The images A-H and a-h are from consecutive tissue sections. Supplementary Table S1. Comparison of TFF1 staining in normal tissues and corresponding tumor types.
Author Contributions
F.L., S.-Y.H., M.K., R.S. (Ronald Simon) and F.J.: contributed to conception, design, data collection, data analysis and manuscript writing. F.L., S.B., K.M., F.V., R.S. (Ria Schlichter), A.M., A.M.L., A.A.B., V.R., A.H., S.W., M.L., D.D., C.B., P.L., A.H.M., T.K., C.F., N.G., E.B., S.M., S.S., F.J. and T.S.C.: participated in pathology data analysis, data interpretation, and collection of samples. F.L., R.S. (Ria Schlichter), M.K. and C.H.-M.: data analysis. F.J., R.S. (Ronald Simon) and G.S.: study supervision. All authors agree to be accountable for the content of the work. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The usage of archived diagnostic leftover tissues for manufacturing of TMAs and their analysis for research purposes as well as patient data analysis has been approved by local laws (HmbKHG, §12,1) and by the local ethics committee (Ethics commission Hamburg, WF-049/09). All work has been carried out in compliance with the Helsinki Declaration.
Informed Consent Statement
Patient consent was waived due to local laws (HmbKHG, §12,1) that permit research with anonymized diagnostic leftover tissue samples.
Data Availability Statement
Dataset available on request from the authors.
Conflicts of Interest
The mouse monoclonal antibody, clone MSVA-482M, was provided from MS Validated Antibodies GmbH, Hamburg, Germany (owned by a family member of GS).
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Madsen J., Nielsen O., Tornoe I., Thim L., Holmskov U. Tissue localization of human trefoil factors 1, 2, and 3. J. Histochem. Cytochem. 2007;55:505–513. doi: 10.1369/jhc.6A7100.2007. [DOI] [PubMed] [Google Scholar]
- 2.Taupin D., Podolsky D.K. Trefoil factors: Initiators of mucosal healing. Nat. Rev. Mol. Cell Biol. 2003;4:721–732. doi: 10.1038/nrm1203. [DOI] [PubMed] [Google Scholar]
- 3.Aihara E., Engevik K.A., Montrose M.H. Trefoil Factor Peptides and Gastrointestinal Function. Annu. Rev. Physiol. 2017;79:357–380. doi: 10.1146/annurev-physiol-021115-105447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ruchaud-Sparagano M.H., Westley B.R., May F.E. The trefoil protein TFF1 is bound to MUC5AC in human gastric mucosa. Cell. Mol. Life Sci. 2004;61:1946–1954. doi: 10.1007/s00018-004-4124-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Katoh M. Trefoil factors and human gastric cancer (review) Int. J. Mol. Med. 2003;12:3–9. doi: 10.3892/ijmm.12.1.3. [DOI] [PubMed] [Google Scholar]
- 6.Salm F., Znalesniak E.B., Laskou A., Harder S., Schluter H., Hoffmann W. Expression Profiling along the Murine Intestine: Different Mucosal Protection Systems and Alterations in Tff1-Deficient Animals. Int. J. Mol. Sci. 2023;24:12684. doi: 10.3390/ijms241612684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lefebvre O., Chenard M.P., Masson R., Linares J., Dierich A., LeMeur M., Wendling C., Tomasetto C., Chambon P., Rio M.C. Gastric mucosa abnormalities and tumorigenesis in mice lacking the pS2 trefoil protein. Science. 1996;274:259–262. doi: 10.1126/science.274.5285.259. [DOI] [PubMed] [Google Scholar]
- 8.Khaidakov M., Lai K.K., Roudachevski D., Sargsyan J., Goyne H.E., Pai R.K., Lamps L.W., Hagedorn C.H. Gastric Proteins MUC5AC and TFF1 as Potential Diagnostic Markers of Colonic Sessile Serrated Adenomas/Polyps. Am. J. Clin. Pathol. 2016;146:530–537. doi: 10.1093/ajcp/aqw142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tuna B., Sokmen S., Sarioglu S., Fuzun M., Kupelioglu A., Ellidokuz H. PS2 and HSP70 expression in rectal adenocarcinomas: An immunohistochemical investigation of 45 cases. Appl. Immunohistochem. Mol. Morphol. 2006;14:31–36. doi: 10.1097/01.pai.0000141544.28862.35. [DOI] [PubMed] [Google Scholar]
- 10.Sunagawa M., Yamaguchi J., Kokuryo T., Ebata T., Yokoyama Y., Sugawara G., Nagino M. Trefoil factor family 1 expression in the invasion front is a poor prognostic factor associated with lymph node metastasis in pancreatic cancer. Pancreatology. 2017;17:782–787. doi: 10.1016/j.pan.2017.07.188. [DOI] [PubMed] [Google Scholar]
- 11.Kornprat P., Rehak P., Lemmerer M., Gogg-Kamerer M., Langner C. Analysis of trefoil factor family protein 1 (TFF1, pS2) expression in chronic cholecystitis and gallbladder carcinoma. Virchows Arch. 2005;446:505–510. doi: 10.1007/s00428-005-1240-4. [DOI] [PubMed] [Google Scholar]
- 12.Ioachim E., Tsanou E., Briasoulis E., Batsis C., Karavasilis V., Charchanti A., Pavlidis N., Agnantis N.J. Clinicopathological study of the expression of hsp27, pS2, cathepsin D and metallothionein in primary invasive breast cancer. Breast. 2003;12:111–119. doi: 10.1016/S0960-9776(02)00290-4. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka T., Nakamura J., Kitajima Y., Kai K., Miyake S., Hiraki M., Ide T., Koga Y., Noshiro H. Loss of trefoil factor 1 is regulated by DNA methylation and is an independent predictive factor for poor survival in advanced gastric cancer. Int. J. Oncol. 2013;42:894–902. doi: 10.3892/ijo.2013.1759. [DOI] [PubMed] [Google Scholar]
- 14.Soubeyran I., Wafflart J., Bonichon F., de Mascarel I., Trojani M., Durand M., Avril A., Coindre J.M. Immunohistochemical determination of pS2 in invasive breast carcinomas: A study on 942 cases. Breast Cancer Res. Treat. 1995;34:119–128. doi: 10.1007/BF00665784. [DOI] [PubMed] [Google Scholar]
- 15.Machado J.C., Nogueira A.M., Carneiro F., Reis C.A., Sobrinho-Simoes M. Gastric carcinoma exhibits distinct types of cell differentiation: An immunohistochemical study of trefoil peptides (TFF1 and TFF2) and mucins (MUC1, MUC2, MUC5AC, and MUC6) J. Pathol. 2000;190:437–443. doi: 10.1002/(SICI)1096-9896(200003)190:4<437::AID-PATH547>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 16.Fox C.A., Sapinoso L.M., Zhang H., Zhang W., McLeod H.L., Petroni G.R., Mullick T., Moskaluk C.A., Frierson H.F., Hampton G.M., et al. Altered expression of TFF-1 and CES-2 in Barrett’s Esophagus and associated adenocarcinomas. Neoplasia. 2005;7:407–416. doi: 10.1593/neo.04715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Im S., Yoo C., Jung J.H., Choi H.J., Yoo J., Kang C.S. Reduced expression of TFF1 and increased expression of TFF3 in gastric cancer: Correlation with clinicopathological parameters and prognosis. Int. J. Med. Sci. 2013;10:133–140. doi: 10.7150/ijms.5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thuwajit P., Chawengrattanachot W., Thuwajit C., Sripa B., May F.E., Westley B.R., Tepsiri N.N., Paupairoj A., Chau-In S. Increased TFF1 trefoil protein expression in Opisthorchis viverrini-associated cholangiocarcinoma is important for invasive promotion. Hepatol. Res. 2007;37:295–304. doi: 10.1111/j.1872-034X.2007.00045.x. [DOI] [PubMed] [Google Scholar]
- 19.Kosriwong K., Menheniott T.R., Giraud A.S., Jearanaikoon P., Sripa B., Limpaiboon T. Trefoil factors: Tumor progression markers and mitogens via EGFR/MAPK activation in cholangiocarcinoma. World J. Gastroenterol. 2011;17:1631–1641. doi: 10.3748/wjg.v17.i12.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaewlert W., Sakonsinsiri C., Namwat N., Sawanyawisuth K., Ungarreevittaya P., Khuntikeo N., Armartmuntree N., Thanan R. The Importance of CYP19A1 in Estrogen Receptor-Positive Cholangiocarcinoma. Horm. Cancer. 2018;9:408–419. doi: 10.1007/s12672-018-0349-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henry J.A., Piggott N.H., Mallick U.K., Nicholson S., Farndon J.R., Westley B.R., May F.E. pNR-2/pS2 immunohistochemical staining in breast cancer: Correlation with prognostic factors and endocrine response. Br. J. Cancer. 1991;63:615–622. doi: 10.1038/bjc.1991.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hurlimann J., Gebhard S., Gomez F. Oestrogen receptor, progesterone receptor, pS2, ERD5, HSP27 and cathepsin D in invasive ductal breast carcinomas. Histopathology. 1993;23:239–248. doi: 10.1111/j.1365-2559.1993.tb01196.x. [DOI] [PubMed] [Google Scholar]
- 23.Gohring U.J., Scharl A., Ahr A. [Value of immunohistochemical determination of receptors, tissue proteases, tumor suppressor proteins and proliferation markers as prognostic indicators in primary breast carcinoma] Geburtshilfe Frauenheilkd. 1996;56:177–183. doi: 10.1055/s-2007-1022256. [DOI] [PubMed] [Google Scholar]
- 24.Hirota Y., Tanaka S., Haruma K., Yoshihara M., Sumii K., Kajiyama G., Shimamoto F., Kohno N. pS2 expression as a possible diagnostic marker of colorectal carcinoma in ulcerative colitis. Oncol. Rep. 2000;7:233–239. doi: 10.3892/or.7.2.233. [DOI] [PubMed] [Google Scholar]
- 25.Gurbuz Y., Kloppel G. Differentiation pathways in duodenal and ampullary carcinomas: A comparative study on mucin and trefoil peptide expression, including gastric and colon carcinomas. Virchows Arch. 2004;444:536–541. doi: 10.1007/s00428-004-1008-2. [DOI] [PubMed] [Google Scholar]
- 26.Welter C., Theisinger B., Rio M.C., Seitz G., Schuder G., Blin N. Expression pattern of breast-cancer-associated protein pS2/BCEI in colorectal tumors. Int. J. Cancer. 1994;56:52–55. doi: 10.1002/ijc.2910560110. [DOI] [PubMed] [Google Scholar]
- 27.Kim D.H., Kim J.W., Cho J.H., Baek S.H., Kakar S., Kim G.E., Sleisenger M.H., Kim Y.S. Expression of mucin core proteins, trefoil factors, APC and p21 in subsets of colorectal polyps and cancers suggests a distinct pathway of pathogenesis of mucinous carcinoma of the colorectum. Int. J. Oncol. 2005;27:957–964. doi: 10.3892/ijo.27.4.957. [DOI] [PubMed] [Google Scholar]
- 28.Luqmani Y.A., Ryall G., Shousha S., Coombes R.C. An immunohistochemical survey of pS2 expression in human epithelial cancers. Int. J. Cancer. 1992;50:302–304. doi: 10.1002/ijc.2910500222. [DOI] [PubMed] [Google Scholar]
- 29.Minegishi K., Dobashi Y., Koyama T., Ishibashi Y., Furuya M., Tsubochi H., Ohmoto Y., Yasuda T., Nomura S. Diagnostic utility of trefoil factor families for the early detection of lung cancer and their correlation with tissue expression. Oncol. Lett. 2023;25:139. doi: 10.3892/ol.2023.13725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Higashiyama M., Doi O., Kodama K., Yokouchi H., Inaji H., Nakamori S., Tateishi R. Prognostic significance of pS2 protein expression in pulmonary adenocarcinoma. Eur. J. Cancer. 1994;30A:792–797. doi: 10.1016/0959-8049(94)90294-1. [DOI] [PubMed] [Google Scholar]
- 31.Matsubara D., Yoshimoto T., Soda M., Amano Y., Kihara A., Funaki T., Ito T., Sakuma Y., Shibano T., Endo S., et al. Reciprocal expression of trefoil factor-1 and thyroid transcription factor-1 in lung adenocarcinomas. Cancer Sci. 2020;111:2183–2195. doi: 10.1111/cas.14403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henry J.A., Bennett M.K., Piggott N.H., Levett D.L., May F.E., Westley B.R. Expression of the pNR-2/pS2 protein in diverse human epithelial tumours. Br. J. Cancer. 1991;64:677–682. doi: 10.1038/bjc.1991.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davidson B., Stavnes H.T., Holth A., Chen X., Yang Y., Shih Ie M., Wang T.L. Gene expression signatures differentiate ovarian/peritoneal serous carcinoma from breast carcinoma in effusions. J. Cell. Mol. Med. 2011;15:535–544. doi: 10.1111/j.1582-4934.2010.01019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kraus S., Abel P.D., Nachtmann C., Linsenmann H.J., Weidner W., Stamp G.W., Chaudhary K.S., Mitchell S.E., Franke F.E., Lalani E.N. MUC1 mucin and trefoil factor 1 protein expression in renal cell carcinoma: Correlation with prognosis. Hum. Pathol. 2002;33:60–67. doi: 10.1053/hupa.2002.29682. [DOI] [PubMed] [Google Scholar]
- 35.Vestergaard E.M., Borre M., Poulsen S.S., Nexo E., Torring N. Plasma levels of trefoil factors are increased in patients with advanced prostate cancer. Clin. Cancer Res. 2006;12:807–812. doi: 10.1158/1078-0432.CCR-05-1545. [DOI] [PubMed] [Google Scholar]
- 36.Abdou A.G., Aiad H.A., Sultan S.M. pS2 (TFF1) expression in prostate carcinoma: Correlation with steroid receptor status. APMIS. 2008;116:961–971. doi: 10.1111/j.1600-0463.2008.01009.x. [DOI] [PubMed] [Google Scholar]
- 37.Collier J.D., Bennett M.K., Bassendine M.F., Lendrum R. Immunolocalization of pS2, a putative growth factor, in pancreatic carcinoma. J. Gastroenterol. Hepatol. 1995;10:396–400. doi: 10.1111/j.1440-1746.1995.tb01590.x. [DOI] [PubMed] [Google Scholar]
- 38.Terris B., Blaveri E., Crnogorac-Jurcevic T., Jones M., Missiaglia E., Ruszniewski P., Sauvanet A., Lemoine N.R. Characterization of gene expression profiles in intraductal papillary-mucinous tumors of the pancreas. Am. J. Pathol. 2002;160:1745–1754. doi: 10.1016/S0002-9440(10)61121-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kononen J., Bubendorf L., Kallioniemi A., Barlund M., Schraml P., Leighton S., Torhorst J., Mihatsch M.J., Sauter G., Kallioniemi O.P. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat. Med. 1998;4:844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 40.Dancau A.M., Simon R., Mirlacher M., Sauter G. Tissue Microarrays. Methods Mol. Biol. 2016;1381:53–65. doi: 10.1007/978-1-4939-3204-7_3. [DOI] [PubMed] [Google Scholar]
- 41.Rico S.D., Mahnken M., Buscheck F., Dum D., Luebke A.M., Kluth M., Hube-Magg C., Hinsch A., Hoflmayer D., Moller-Koop C., et al. MUC5AC Expression in Various Tumor Types and Nonneoplastic Tissue: A Tissue Microarray Study on 10 399 Tissue Samples. Technol. Cancer Res. Treat. 2021;20:15330338211043328. doi: 10.1177/15330338211043328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shah A., Jahan R., Kisling S.G., Atri P., Natarajan G., Nallasamy P., Cox J.L., Macha M.A., Sheikh I.A., Ponnusamy M.P., et al. Secretory Trefoil Factor 1 (TFF1) promotes gemcitabine resistance through chemokine receptor CXCR4 in Pancreatic Ductal Adenocarcinoma. Cancer Lett. 2024;598:217097. doi: 10.1016/j.canlet.2024.217097. [DOI] [PubMed] [Google Scholar]
- 43.Rodrigues S., Rodrigue C.M., Attoub S., Flejou J.F., Bruyneel E., Bracke M., Emami S., Gespach C. Induction of the adenoma-carcinoma progression and Cdc25A-B phosphatases by the trefoil factor TFF1 in human colon epithelial cells. Oncogene. 2006;25:6628–6636. doi: 10.1038/sj.onc.1209665. [DOI] [PubMed] [Google Scholar]
- 44.Sugai T., Osakabe M., Eizuka M., Tanaka Y., Yamada S., Yanagawa N., Matsumoto T., Suzuki H. Genome-wide analysis of mRNA expression identified the involvement of trefoil factor 1 in the development of sessile serrated lesions. Pathol. Res. Pract. 2022;236:153987. doi: 10.1016/j.prp.2022.153987. [DOI] [PubMed] [Google Scholar]
- 45.Song J.Y., Kim B.W., Lee A.W., Lee K.Y., Chung I.S., Lee B.I., Choi H., Ji J.S., Chae H.S., Choi K.Y. Expression of MUC5AC and Trefoil Peptide 1 (TFF1) in the Subtypes of Intestinal Metaplasia. Clin. Endosc. 2012;45:151–154. doi: 10.5946/ce.2012.45.2.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yio X., Diamond M., Zhang J.Y., Weinstein H., Wang L.H., Werther L., Itzkowitz S. Trefoil factor family-1 mutations enhance gastric cancer cell invasion through distinct signaling pathways. Gastroenterology. 2006;130:1696–1706. doi: 10.1053/j.gastro.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 47.Rodrigues S., Van Aken E., Van Bocxlaer S., Attoub S., Nguyen Q.D., Bruyneel E., Westley B.R., May F.E., Thim L., Mareel M., et al. Trefoil peptides as proangiogenic factors in vivo and in vitro: Implication of cyclooxygenase-2 and EGF receptor signaling. FASEB J. 2003;17:7–16. doi: 10.1096/fj.02-0201com. [DOI] [PubMed] [Google Scholar]
- 48.Shen T., Huang S. The role of Cdc25A in the regulation of cell proliferation and apoptosis. Anticancer. Agents Med. Chem. 2012;12:631–639. doi: 10.2174/187152012800617678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saha A., Gavert N., Brabletz T., Ben-Ze’ev A. Downregulation of the Tumor Suppressor TFF1 Is Required during Induction of Colon Cancer Progression by L1. Cancers. 2022;14:4478. doi: 10.3390/cancers14184478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soutto M., Belkhiri A., Piazuelo M.B., Schneider B.G., Peng D., Jiang A., Washington M.K., Kokoye Y., Crowe S.E., Zaika A., et al. Loss of TFF1 is associated with activation of NF-kappaB-mediated inflammation and gastric neoplasia in mice and humans. J. Clin. Investig. 2011;121:1753–1767. doi: 10.1172/JCI43922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ochiai Y., Yamaguchi J., Kokuryo T., Yokoyama Y., Ebata T., Nagino M. Trefoil Factor Family 1 Inhibits the Development of Hepatocellular Carcinoma by Regulating beta-Catenin Activation. Hepatology. 2020;72:503–517. doi: 10.1002/hep.31039. [DOI] [PubMed] [Google Scholar]
- 52.Buache E., Etique N., Alpy F., Stoll I., Muckensturm M., Reina-San-Martin B., Chenard M.P., Tomasetto C., Rio M.C. Deficiency in trefoil factor 1 (TFF1) increases tumorigenicity of human breast cancer cells and mammary tumor development in TFF1-knockout mice. Oncogene. 2011;30:3261–3273. doi: 10.1038/onc.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ge Y., Zhang J., Cao J., Wu Q., Sun L., Guo L., Wang Z. TFF1 inhibits proliferation and induces apoptosis of gastric cancer cells in vitro. Bosn. J. Basic. Med. Sci. 2012;12:74–81. doi: 10.17305/bjbms.2012.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hasebe K., Yamazaki K., Yamaguchi J., Kokuryo T., Yokoyama Y., Miyata K., Fukaya M., Nagino M., Ebata T. Trefoil factor 1 inhibits the development of esophageal adenocarcinoma from Barrett’s epithelium. Lab. Investig. 2022;102:885–895. doi: 10.1038/s41374-022-00771-1. [DOI] [PubMed] [Google Scholar]
- 55.Stonelake P.S., Baker P.G., Gillespie W.M., Dunn J.A., Spooner D., Morrison J.M., Bundred N.J., Oates G.D., Lee M.J., Neoptolemos J.P., et al. Steroid receptors, pS2 and cathepsin D in early clinically node-negative breast cancer. Eur. J. Cancer. 1994;30A:5–11. doi: 10.1016/S0959-8049(05)80008-5. [DOI] [PubMed] [Google Scholar]
- 56.Sagol O., Tuna B., Coker A., Karademir S., Obuz F., Astarcioglu H., Kupelioglu A., Astarcioglu I., Topalak O. Immunohistochemical detection of pS2 protein and heat shock protein-70 in pancreatic adenocarcinomas. Relationship with disease extent and patient survival. Pathol. Res. Pract. 2002;198:77–84. doi: 10.1078/0344-0338-00190. [DOI] [PubMed] [Google Scholar]
- 57.Bruce D.M., Heys S.D., Payne S., Miller I.D., Eremin O. Male breast cancer: Clinico-pathological features, immunocytochemical characteristics and prognosis. Eur. J. Surg. Oncol. 1996;22:42–46. doi: 10.1016/S0748-7983(96)91418-3. [DOI] [PubMed] [Google Scholar]
- 58.Muller W, Borchard F: pS2 protein in gastric carcinoma and normal gastric mucosa: Association with clincopathological parameters and patient survival. J. Pathol. 1993;171:263–269. doi: 10.1002/path.1711710406. [DOI] [PubMed] [Google Scholar]
- 59.Hackel C., Falkenberg B., Gunther T., Lippert H., Roessner A. The pS2 protein in colorectal carcinomas and metastases. Pathol. Res. Pract. 1998;194:171–176. doi: 10.1016/S0344-0338(98)80018-6. [DOI] [PubMed] [Google Scholar]
- 60.Lipponen P.K., Eskelinen M.J. Expression of pS2 protein in transitional cell bladder tumours. J. Pathol. 1994;173:327–332. doi: 10.1002/path.1711730407. [DOI] [PubMed] [Google Scholar]
- 61.Fujimoto A., Ishikawa Y., Ishii T., Yamada A., Igarashi Y., Ohmoto Y., Kaise M. Differences between gastric signet-ring cell carcinoma and poorly differentiated adenocarcinoma: A comparison of histopathologic features determined by mucin core protein and trefoil factor family peptide immunohistochemistry. Pathol. Int. 2017;67:398–403. doi: 10.1111/pin.12559. [DOI] [PubMed] [Google Scholar]
- 62.Sasaki M., Tsuneyama K., Nakanuma Y. Aberrant expression of trefoil factor family 1 in biliary epithelium in hepatolithiasis and cholangiocarcinoma. Lab. Investig. 2003;83:1403–1413. doi: 10.1097/01.LAB.0000092230.59485.9E. [DOI] [PubMed] [Google Scholar]
- 63.Kouznetsova I., Laubinger W., Kalbacher H., Kalinski T., Meyer F., Roessner A., Hoffmann W. Biosynthesis of gastrokine-2 in the human gastric mucosa: Restricted spatial expression along the antral gland axis and differential interaction with TFF1, TFF2 and mucins. Cell Physiol. Biochem. 2007;20:899–908. doi: 10.1159/000110450. [DOI] [PubMed] [Google Scholar]
- 64.Kunze E., Krassenkova I., Fayyazi A. Tumor-associated neoexpression of the pS2 peptide and MUC5AC mucin in primary adenocarcinomas and signet ring cell carcinomas of the urinary bladder. Histol. Histopathol. 2008;23:539–548. doi: 10.14670/HH-23.539. [DOI] [PubMed] [Google Scholar]
- 65.Van De Bovenkamp J.H., Korteland-Van Male A.M., Warson C., Buller H.A., Einerhand A.W., Ectors N.L., Dekker J. Gastric-type mucin and TFF-peptide expression in Barrett’s oesophagus is disturbed during increased expression of MUC2. Histopathology. 2003;42:555–565. doi: 10.1046/j.1365-2559.2003.01619.x. [DOI] [PubMed] [Google Scholar]
- 66.Consortium G.T. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lizio M., Abugessaisa I., Noguchi S., Kondo A., Hasegawa A., Hon C.C., de Hoon M., Severin J., Oki S., Hayashizaki Y., et al. Update of the FANTOM web resource: Expansion to provide additional transcriptome atlases. Nucleic Acids Res. 2019;47:D752–D758. doi: 10.1093/nar/gky1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lizio M., Harshbarger J., Shimoji H., Severin J., Kasukawa T., Sahin S., Abugessaisa I., Fukuda S., Hori F., Ishikawa-Kato S., et al. Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol. 2015;16:22. doi: 10.1186/s13059-014-0560-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thul P.J., Akesson L., Wiking M., Mahdessian D., Geladaki A., Ait Blal H., Alm T., Asplund A., Bjork L., Breckels L.M., et al. A subcellular map of the human proteome. Science. 2017;356:eaal3321. doi: 10.1126/science.aal3321. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Dataset available on request from the authors.