Abstract
The cellular secretory pathway is important during the assembly and envelopment of viruses and also controls the transport of host proteins, such as cytokines and major histocompatibility proteins, that function during the elimination of viruses by the immune system. African swine fever virus (ASFV) encodes at least 26 proteins with stretches of hydrophobic amino acids suggesting entry into the secretory pathway (R. J. Yanez, J. M. Rodriguez, M. L. Nogal, L. Yuste, C. Enriquez, J. F. Rodriguez, and E. Vinuela, Virology 208:249–278, 1995). To predict how and where these potential membrane proteins function, we have studied the integrity of the secretory pathway in cells infected with ASFV. Remarkably, ASFV caused complete loss of immunofluorescence signal for the trans Golgi network (TGN) marker protein TGN46 and dispersed the AP1 TGN adapter complex. Loss of TGN46 signal was not due to degradation of TGN46, suggesting redistribution of TGN46 to other membrane compartments. ASFV markedly slowed transport of cathepsin D to lysosomes, demonstrating that loss of TGN structure correlated with loss of TGN function. ASFV shows a tropism for macrophages, and it is possible that ASFV compromises TGN function to augment the activity of viral membrane proteins or to suppress the function of host immunoregulatory proteins.
The cellular secretory pathway plays an important role during the assembly and envelopment of viruses. Cellular membrane compartments provide the lipids necessary for the production of viral envelopes, and membrane trafficking pathways transport viral envelope proteins to sites of virus budding (reviewed in reference 18). In addition to providing the intracellular pathways necessary for virus envelopment, the secretory pathway controls the transport of host proteins, such as cytokines, adhesion molecules, and major histocompatibility (MHC) proteins, that play key roles during the recognition and elimination of viruses by the immune system. The assembly of MHC class 1 peptide complexes takes place in the endoplasmic reticulum (ER), while endosomes, lysosomes, and late compartments of the Golgi apparatus facilitate the processing of exogenous antigens and the binding of viral peptides to MHC class 2 complexes (20, 44). The importance of control over the secretory pathway for the survival of viruses is underpinned by the observation that many viruses perturb the secretory pathway to subvert recognition by the immune system. The activity of secreted cytokines and the cell surface expression of MHC class 1 and class 2 are inhibited by several viruses, aiding the establishment of persistent infections (1, 42).
African swine fever virus (ASFV) is a large icosahedral enveloped DNA virus that infects the pig genus Suidae. The virus causes a persistent and asymptomatic infection of natural hosts, such as the African warthog and bushpig (43). Infection of domestic pigs, however, causes a fatal hemorrhagic disease for which there are no cures or vaccines. The 170-kDa virus genome encodes at least 150 proteins (45), and as many as 50 are assembled into virions (7). In common with poxviruses, DNA replication and assembly of ASFV take place in the cytoplasm at specialized perinuclear sites called “virus factories.” Virus factories are located close to the microtubule organizing center, require an intact microtubule network for assembly (29), and, in the case of ASFV, are found in aggresome-like structures surrounded by the intermediate filament protein vimentin (19). The envelope of ASFV is derived from the ER. Current models suggest that interactions between capsid subunits recruited from the cytosol, and possibly other viral proteins targeted to the ER, cause constriction of ER cisternae (3, 8–10, 36). The cisternae subsequently bend through a complex series of angular intermediates, eventually producing icosahedral particles with two internal envelopes (3, 36). This process of viral wrapping by membrane cisternae is shared by other large enveloped DNA viruses, such as herpesviruses and poxviruses (18, 37, 38).
Twenty-six proteins encoded by ASFV have stretches of hydrophobic amino acids suggestive of leader sequences or transmembrane domains. Unlike other large enveloped DNA viruses, such as herpesvirus, envelope glycoproteins are present at low levels in ASFV (12), and the functions of most of the ASFV proteins with hydrophobic sequences are unknown. With the exception of a C-type lectin and a CD2 homologue (4, 35), ASFV does not appear to encode homologues of host membrane and secreted proteins with potential to affect the immune system. This differs from large DNA viruses, such as herpesviruses and poxviruses, that manipulate the host through the use of virally encoded chemokine and cytokine receptors, complement control proteins, and secreted growth factors (1, 42). In contrast, many of the proteins encoded by ASFV that have signal or transmembrane sequences are retained within the cell. Members of the multigene 110 family have leader sequences and C-terminal KDEL motifs and are retained in the ER (2, 36). Of the nine other proteins with hydrophobic N-terminal leader peptides, only one (E146l/J16L) is predicted to have a cleaved signal sequence and therefore to be able to be secreted from cells. In common with vaccinia virus (15), many ASFV proteins with hydrophobic sequences lack signal sequences and have central, or, unusually, C-terminal transmembrane domains. The ASFV proteins that have been studied in detail are excluded from the secretory pathway and localize almost exclusively to virus assembly sites (5, 34, 36, 39, 40).
At present, the prediction of how and where membrane proteins encoded by ASFV function in cells is based on the assumption that the secretory pathway is normal in cells infected with ASFV. It is possible, however, that ASFV may compromise protein trafficking through the secretory pathway to augment the activity of viral proteins or suppress the function of host immunoregulatory proteins. In this study, we have investigated the structure and function of the secretory pathway in cells infected with ASFV. Remarkably ASFV caused complete loss of the trans Golgi network (TGN). The TGN is a late compartment of the secretory pathway important for proteolytic processing of bioactive peptides and the sorting of proteins as they leave the Golgi apparatus. Given that ASFV shows a tropism for macrophages, an understanding of the consequences of TGN loss on the processing and sorting of macrophage immunoregulatory proteins may hold the key to understanding the complex cell biology and pathogenesis of ASFV.
MATERIALS AND METHODS
Viruses and cells.
Vero (ECACC 84113001) cells were obtained from the European Collection of Animal Cell Cultures (Porton Down, United Kingdom) and cultured and infected with the tissue culture-adapted BA71 (7). Vaccinia virus strain VTF7.3 was obtained from Bernard Moss (National Institutes of Health, Bethesda, Md.).
Antibodies.
Viral proteins. Antibody 4H3, specific for p73, the major capsid protein of ASFV, is described by Cobbold et al. (8). The monoclonal antibody C18, specific for early phosphoprotein p30 of ASFV, was from Dan Rock (Plumb Island Animal Disease Center, Plumb Island, N.Y.). The rabbit antibodies specific for γ adaptin (AP1) were from Margaret Robinson (Cambridge, United Kingdom). The rabbit and sheep antibodies specific for TGN46 were from S. Ponnambalam (Department of Biochemistry, University of Dundee, Dundee, United Kingdom). H4B4, which recognizes the major lysosomal membrane protein LAMP-2, was obtained from the Developmental Studies Hybridoma Bank (Johns Hopkins University, Baltimore, Md.). The rabbit anti-cathepsin D antibody was provided by Janice Blum (Indiana University, Bloomington).
Immunofluorescence.
Cells were grown on 13- or 19-mm-diameter round sterile glass coverslips to approximately 70% confluency. Following the appropriate drug treatments and infection or transfection protocols, the cells were fixed in −20°C methanol, −20°C methanol followed by −20°C acetone, or 4% paraformaldehyde. Cells were permeabilized in Tris-buffered saline containing 0.2% gelatin and 0.5% Nonidet P-40 and then blocked with the same buffer containing 30% goat serum (blocking buffer). Primary antibodies were added to samples diluted in blocking buffer and visualized by second antibodies conjugated to Alexa 488 (green) or Alexa 594 (red) purchased from Molecular Probes (Leiden, The Netherlands). Viral and cellular DNA was stained with DAPI (4′-6-diamidino-2-phenylindole) purchased from Sigma, St. Louis, Mo. Cells were mounted in Fluoromount-G (Southern Biotechnology Associates, Birmingham, Ala.), and in most studies, the cells were viewed at ×60/1.4 NA with a Nikon E800 microscope. The images from 0.2-μm-thick optical sections were captured with a Hamamatsu C-4746A DCC camera and deconvolved with Improvision Openlab software (Warwick, United Kingdom).
Metabolic labeling and immunoprecipitation.
Cells infected with ASFV were labeled metabolically with [35S]methionine and [35S]cysteine by using 35S-Pro-mix (Amersham Pharmacia, United Kingdom) as described previously (8–10). At the appropriate time intervals, cells were washed once in phosphate-buffered saline (PBS) and lysed in immunoprecipitation buffer (10 mM Tris [pH 7.5]; 150 mM NaCl; 10 mM iodoacetamide; 1 mM EDTA; 1 mM phenymethylsulfonyl fluoride; 1 μg each of pepstatin, chymostatin, and antipain per ml) containing 1% Brij35 or Nonidet P-40. Antigens were immunoprecipitated by overnight incubation with antibodies immobilized on protein A or G coupled to Sepharose beads. For digestion with endo-β-N-acetylglucosamine (endo H), immunoprecipitates were boiled for 3 min after addition of 10 μl of 1% sodium dodecyl sulfate (SDS), and enzyme digestions were performed at 37°C overnight after the addition of 50 μl of endo H buffers and 1 μU of enzyme (Boehringer Mannheim). Immunoprecipitates were resolved by SDS-polyacrylamide gel electrophoresis (PAGE), and proteins were detected by autoradiography.
Western blotting.
Proteins were resolved by SDS-PAGE and transferred to cellulose nitrate membranes (Schleicher and Schuell, Dassel, Germany). The membrane was blocked overnight at 4°C in PBS containing 5% Marvel and Tween 20. Blocked and washed membranes were incubated with primary antibodies and then incubated with goat anti-mouse or anti-rabbit antibody coupled to horseradish peroxidase. Blots were visualized by using the ECL enhanced chemiluminescence system (Amersham Life Sciences, United Kingdom) and fluorography.
RESULTS
ASFV infection causes loss of the TGN.
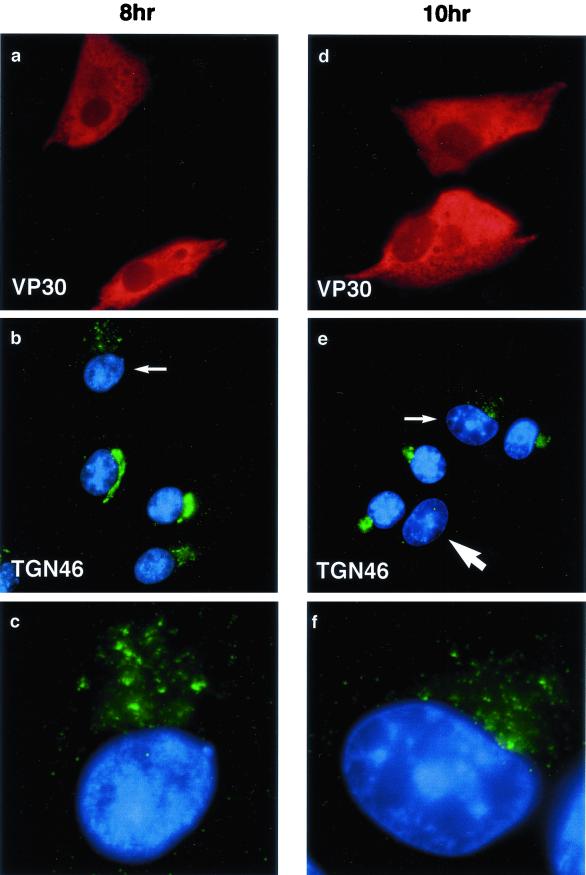
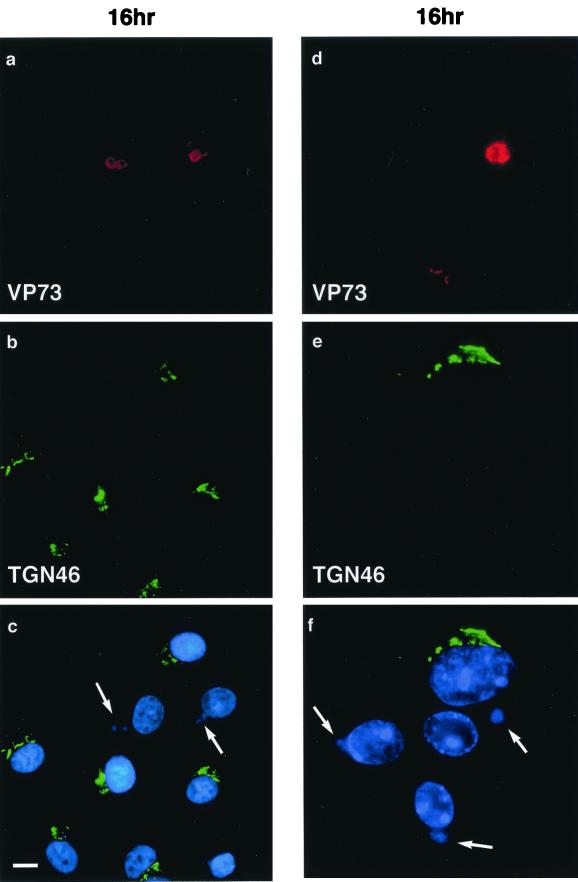
The TGN is important for sorting proteins as they leave the Golgi apparatus for transport to the cell surface, lysosomes, or endosomal systems. TGN46 is a type 1 membrane protein that recycles between the cell surface and the TGN (13, 32), and at steady-state distribution, most of the protein is located to the TGN, making TGN46 an accepted marker for the organelle. Figure 1 shows the effect of ASFV on the distribution of TGN46 at 8 and 10 h after infection. Infected cells were identified by positive staining for the early ASFV protein, vp30 (Fig. 1a and d). In the cells negative for vp30, the TGN46 signal was concentrated in a characteristic compact perinuclear crescent (Fig. 1b and e). At both time points, however, a fragmented TGN was observed in infected cells. The fragmentation of the TGN is shown at higher magnification in panels c and f. In some cells observed at 10 h, the signal for TGN46 was difficult to detect (large arrow, Fig. 1e). The effects of the virus 16 h after infection are shown in Fig. 2. Infected cells were identified by positive staining for the major capsid protein p73 (Fig. 2a and d) and extranuclear DAPI staining of viral DNA in viral factories (Fig. 2c and f). Strikingly, the TGN46 marker was absent from all infected cells. This effect is particularly evident in panel f, where three of the four cells shown have viral factories indicated by arrows. The green signal indicating TGN46 forms a compact crescent to one side of the nucleus of the cell lacking viral markers, but is completely absent from the three infected cells.
FIG. 1.
The early effects of ASFV on the TGN. BSC40 cells were fixed 8 h (a to c) or 10 h (d to f) after infection with the Ba71v strain of ASFV. Samples were incubated with a monoclonal antibody specific for early viral protein vp30 (a and d) and a rabbit antibody specific for TGN46 (b and e). Cellular and viral DNA was visualized by DAPI (b and e). Antigens were visualized by second antibodies coupled to Alexa 488 or Alexa 594. Samples were viewed at ×60, and 0.2-μm digital sections were digitally deconvolved with Openlab software from Improvision. Panels b and e show a digital merge of the DAPI and TGN46 distributions. The small arrows indicate cells with disrupted TGN, which are shown at higher magnification in panels c and f. The large arrow indicates a cell lacking TGN signal.
FIG. 2.
Effects of ASFV on TGN at 16 h postinfection. BSC40 cells were fixed 16 h after infection with the Ba71v strain of ASFV. Samples were incubated with a monoclonal antibody specific for the major capsid protein p73 (a and d) and a rabbit antibody specific for TGN46 (b and e). Cellular and viral DNA was visualized by DAPI (c and f). Antigens were visualized by second antibodies coupled to Alexa 488 or Alexa 594. Samples were viewed at ×60, and 0.2-μm digital sections were digitally deconvolved with Openlab software from Improvision. The bar in panel c represents 10 μm. Note that the cells in panels d, e, and f are shown at higher magnification. Panels c and f show a digital merge of the DAPI and TGN46 distributions. The arrows indicate cells with extranuclear DAPI staining of virus factories and a lack of TGN46 signal.
ASFV infection causes scattering of TGN clathrin adapter protein AP1.
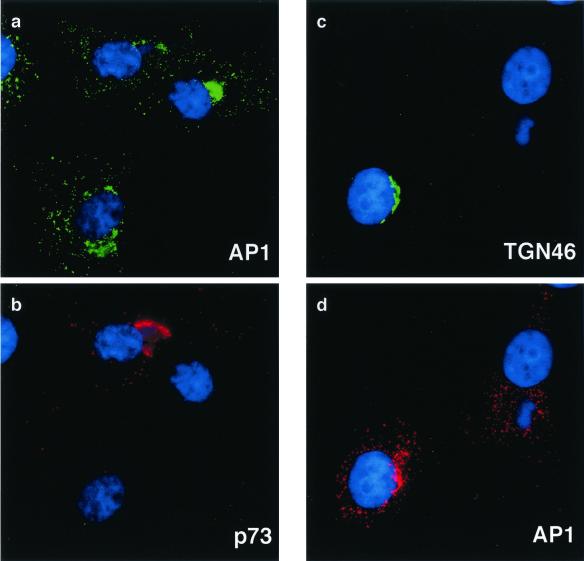
The AP1 adapter protein complex assembles with clathrin coats formed on the TGN (26, 28). AP1 recognizes sorting motifs in the cytoplasmic domains of membrane proteins entering the TGN and facilitates their transport to endosomes and lysosomes. The experiments described above have shown that ASFV infection induced loss of immunofluorescence signal for the integral membrane protein TGN46. Antibodies specific for the γ chain of AP1 were therefore used to follow the effects of ASFV on the TGN adapter protein complex. Figure 3 shows the effect of the virus 16 h postinfection, a time when the virus causes the loss of the TGN46 signal from cells. Each image compares infected and uninfected cells. Infected cells were indicated by the presence of the viral capsid protein p73 (Fig. 3b) or extranuclear DAPI staining in virus factories (Fig. 3c and d). At steady state, AP1 is distributed between the TGN and endosomes. This distribution is seen in the cells lacking viral markers. The AP1 complex localized at the TGN is identified as a dense perinuclear staining that colocalizes with TGN46 (Fig. 3c and d), while the endosomal signal is revealed as a punctate stain seen throughout the cytoplasm. In the cells infected with virus, the perinuclear concentration of AP1 stain was lost. In Fig. 3a and b, a compact perinuclear signal for AP1 was seen in two cells lacking viral markers, but was dispersed in the infected cell positive for p73. In Fig. 3c and d, the infected cell is identified by a perinuclear viral factory and loss of the TGN 46 signal. In this cell, the AP1 stain was dispersed. In the cell lacking a virus factory, approximately half of the AP1 signal was compact and perinuclear and colocalized with TGN46. ASFV therefore causes loss of both integral (TGN46) and peripheral (AP1) membrane proteins from the TGN.
FIG. 3.
Effects of ASFV on TGN adapter protein AP1. BSC40 cells were fixed 16 h after infection with the Ba71v strain of ASFV. Samples were incubated with a monoclonal antibody specific for the major capsid protein p73 and a rabbit antibody specific for AP1 (a and b) or a goat antibody specific for TGN46 and a rabbit antibody specific for AP1 (c and d). In each image, cellular and viral DNA was visualized by DAPI. Antigens were visualized by second antibodies coupled to Alexa 488 or Alexa 594. Samples were viewed at ×60, and 0.2-μm digital sections were digitally deconvolved with Openlab software from Improvision. Panels a and b compare the AP1 and p73 distributions, while panels c and d compare AP1 and TGN46.
ASFV slows delivery of cathepsin D to lysosomes.
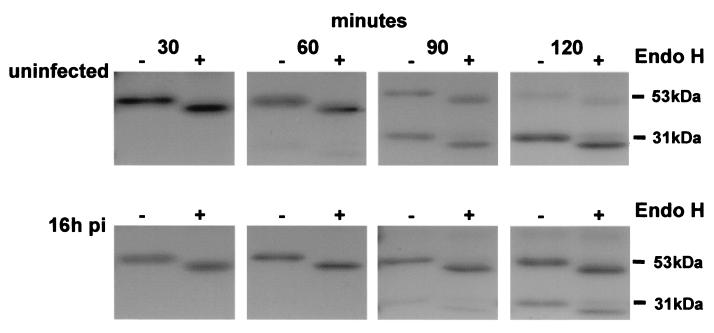
The lysosomal aspartinyl protease cathepsin D is synthesized as a preproenzyme of approximately 53 kDa. A short-lived 47-kDa intermediate proenzyme is formed on delivery to the TGN and endosomes, and this is further cleaved in lysosomes to the mature enzyme characterized by noncovalently associated 31- and 14-kDa polypeptides (11, 16, 25, 33). Delivery of cathepsin D from the TGN to lysosomes is mediated by the mannose-6-phosphate receptor. This sorting step is controlled by AP1 adapter complexes present in clathrin-coated pits on the TGN. An analysis of cathepsin D transport in infected cells therefore allowed us to test the effects of ASFV on the sorting function of the TGN.
Metabolic labeling and immunoprecipitation experiments were initially conducted with uninfected Vero cells in order to ensure that precursor and mature forms of cathepsin D could be detected. Cells were pulse-labeled for 15 min and chased for increasing periods of time. Lysates were immunoprecipitated, and the extent of N-linked glycosylation and oligosaccharide processing was tested by digesting half of each sample with endo H. The first two lanes of the upper panel of Fig. 4 show that cathepsin D migrated at 53 kDa after a 30-min chase and at approximately 44 kDa following endo H digestion. These sizes were consistent with documented properties of this protein. After 1 h of chase in uninfected cells, a small quantity of the 31-kDa lysosomal form was detected, and this increased at 90 min, indicating processing of cathepsin D to the mature enzyme in lysosomes had occurred. After 2 h, the almost complete loss of the 53-kDa preproenzyme was mirrored by increased levels of the mature 31-kDa protein. The intermediate 47-kDa form was present at very low levels, and detectable only after long exposure of gels (data not shown). The effects of ASFV infection on the transport of cathepsin D are shown in the lower panel of Fig. 4. In parallel experiments, the multiplicity of infection was checked by immunofluorescence staining of cells and was shown to be greater than 70%. Infection with ASFV caused an obvious delay in the processing of cathepsin D. At 60 min, the lysosomal form of the enzyme was absent, and at 90 min, only a very small fraction of cathepsin D had reached lysosomes and cleaved to the mature 31-kDa form. After 2 h, less than half of the cathepsin D in ASFV-infected cells was present as the mature enzyme, suggesting that the majority of the protein had not yet reached lysosomes. The results show that ASFV compromised the ability of the TGN to sort lysosomal enzymes to lysosomes.
FIG. 4.
Effect of ASFV infection on transport of cathepsin D to lysosomes. Vero cells were pulse-labeled for 15 min with [35S]methionine and [35S]cysteine and then chased in complete media. At the indicated time points, cells were lysed and immunoprecipitated with an antibody specific for cathepsin D. One half of each immunoprecipitate was digested with endo H (+). Samples were analyzed by SDS-PAGE followed by autoradiography. The preproenzyme (53 kDa) and mature enzyme (31 kDa) are indicated. (Top panel) Control cells. (Bottom panel) Cells analyzed 16 h postinfection (pi) with ASFV.
TGN46 is not degraded in cells infected with ASFV.
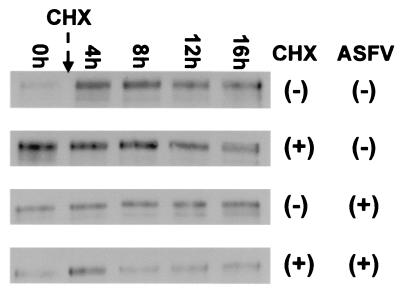
The loss of the TGN46 immunofluorescence signal seen in cells infected with ASFV could have resulted from degradation of TGN46 or from extensive redistribution of the protein within cells. To distinguish between these possibilities, the effects of ASFV infection on the turnover of TGN46 were investigated. In initial experiments (not shown), cells infected with ASFV were incubated with concanamycin A to neutralize lysosomal pH and inhibit proteolysis in the organelle. Cells were then probed for the presence of TGN46 by immunofluorescence microscopy. Concanamycin A did not rescue the TGN46 signal, suggesting it was unlikely that TGN46 was degraded in lysosomes. The mature form of TGN38 lacks methionine or cysteine residues and cannot be labeled by incorporation of [35S]methionine or cysteine. The stability of TGN46 was therefore tested by adding cycloheximide to cells and assaying TGN46 levels at increasing times by Western blotting. The top two panels of Fig. 5 compare the TGN46 signals in the presence or absence of cycloheximide. The similarity of the signals recovered over 16 h shows that TGN46 is stable in cells. The lower panels of Fig. 5 show the same experiment carried out on cells infected with ASFV. As for the experiments with cathepsin D, the multiplicity of infection judged by immunofluorescence staining of cells was approximately 70%. There was no noticeable loss of TGN46 in cells incubated with cyclohexmide, showing again that ASFV does not induce degradation of the protein.
FIG. 5.
Loss of the TGN46 signal does not involve degradation of TGN46. The stability of TGN46 in Vero cells was determined by adding cycloheximide (10 μg/ml) to cell cultures. The cells were then lysed at the indicated times, and the levels of TGN46 were determined by Western blotting. For infected cells, cycloheximide was added 4 h after addition of virus to allow expression of early genes prior to the block in protein synthesis with cycloheximide. The addition of virus (ASFV ±) or cycloheximide (CHX ±) is indicated.
TGN loss does not require DNA-dependent late protein expression.
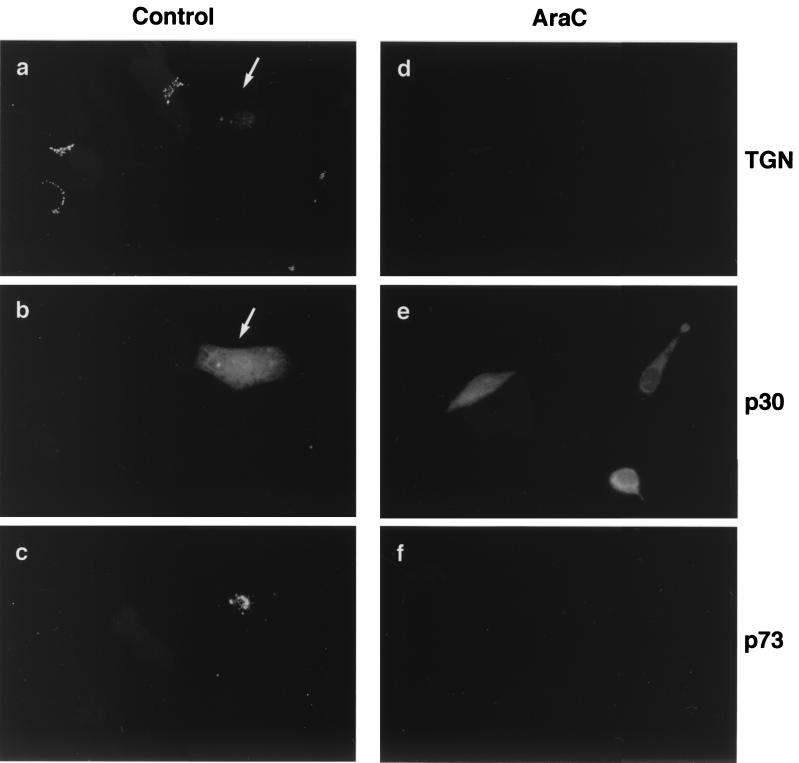
Cytosine α-d-arabinofuranoside (Ara-C) is a selective inhibitor of DNA replication. The drug has no inhibitory effect on the synthesis of RNA and can be used to prevent expression of late DNA-dependent proteins. To test whether late viral proteins caused TGN loss, Vero cells were incubated with 50 μg of Ara-C per ml at 3 h postinfection, a time calculated to allow for the attachment and entry of ASFV into cells, but before the onset of viral DNA replication. Cells were incubated for a further 13 h and then processed for immunofluorescence analysis. Figure 6 compares the expression of early and late ASFV gene products in the presence or absence of Ara-C at 16 h postinfection. In control cells (Fig. 6a to c), incubated in the absence of Ara-C, both the early viral protein p30 (Fig. 6b) and the late viral protein p73 (Fig. 6c) were detected. Note that the characteristic morphology of the TGN was retained in all uninfected cells (Fig. 6a), yet the fluorescence signal for TGN46 was lost from the cells infected with ASFV (arrow). The cells observed in Fig. 6d to f were incubated in the presence of Ara-C. All three cells shown were infected with ASFV, indicated by a positive signal for p30 (Fig. 6e). Of importance to this study, there was no expression of the late major capsid protein p73 (Fig. 6f) in any of the infected cells present. Ara-C had therefore inhibited ASFV DNA replication effectively. Significantly, there was a loss of TGN46 signal in all of the infected cells incubated with Ara-C. Since the loss of immunostaining for TGN46 was observed in the absence of viral DNA replication, and further control experiments showed that Ara-C itself did not cause loss of TGN46 signal (not shown), the results implied that one or more early ASFV proteins were responsible for the loss of the TGN46 signal. Evidence for this also comes from close inspection of Fig. 1e. The cell indicated by the large arrow has lost TGN46 signal, but lacks extranuclear DNA. In this cell, the TGN was lost before it was possible to detect replication of viral DNA in viral factories.
FIG. 6.
Loss of the TGN46 signal does not require late ASFV gene expression. Vero cells were fixed 16 h after infection with the Ba71v strain of ASFV. (a to c) Control cells incubated without Ara-C. (d to f) Cells incubated with 50 μg of Ara-C per ml added 4 h postinfection. Cells were stained with antibodies specific for TGN46 (a and d), biotinylated antibody specific for the early ASFV protein p30 (b and e), and a monoclonal antibody binding the late structural protein, p73 (c and f). Antigens were visualized with secondary antibodies coupled to Alexa 488 (a and d), Alexa 594 (c and f), and avidin coupled to Cascade Blue (b and e). Arrows identify an infected cell.
ASFV does not affect the distribution of lysosomes.
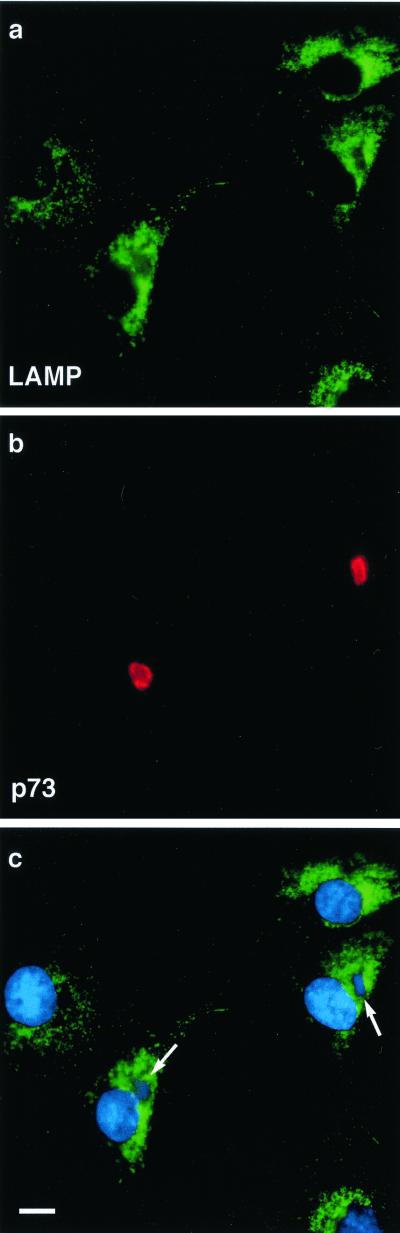
Since the TGN was lost following infection by ASFV, and delivery of cathepsin D to lysosomes was slowed, the effect of ASFV infection on the integrity and distribution of lysosomes in cells was studied. Figure 7a shows that the majority of the staining obtained with antibodies specific for lysosome-associated membrane protein (LAMP2) was found associated with vesicles in the juxtanuclear region near the Golgi. Vesicles were also found in smaller numbers throughout the cell cytoplasm. Infected cells were identified by the presence of p73 (Fig. 7b) and extranuclear DAPI staining of viral factories (Fig. 7c). Infection of cells by ASFV did not noticeably affect the distribution of lysosomes (Fig. 7a and c); there was, however, a diminished LAMP2 signal in the area of the virus factory.
FIG. 7.
Effect of ASFV on lysosomes. BSC40 cells were fixed 16 h after infection with the Ba71v strain of ASFV and incubated with a monoclonal antibody (H4B4) specific for LAMP2 (a and c) and a rabbit antibody specific for viral capsid protein p73 (b). Cellular and viral DNA was visualized by DAPI (c). Antigens were visualized by second antibodies coupled to Alexa 488 or Alexa 594. Samples were viewed at ×60, and 0.2-μm digital sections were digitally deconvolved with Openlab software from Improvision. Panels c shows a digital merge of the DAPI and LAMP2 distributions. Bar, 10 μm. The arrows indicate extranuclear DAPI staining of viral DNA in virus factories.
Vaccinia virus does not cause loss of TGN46.
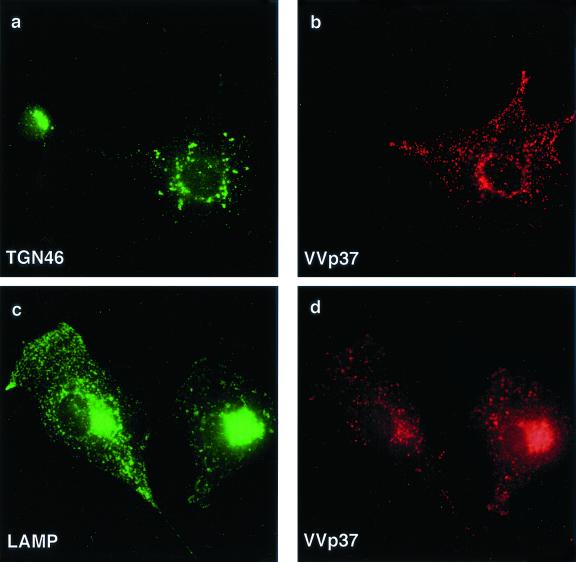
The next experiment determined whether loss of TGN was unique to ASFV or whether it was a general consequence of infecting cells with large cytoplasmic DNA viruses. The morphogenesis of vaccinia virus shares many features with ASFV. Assembly of both viruses takes place at sites of DNA replication in perinuclear viral factories (19, 29). In addition, ASFV and vaccinia virus gain membrane envelopes by being wrapped by membrane cisternae originating from the membrane compartments of the secretory pathway (36–38). The distribution of TGN and lysosomal markers in cells infected with vaccinia virus was therefore studied (Fig. 8). Cells infected with vaccinia virus were identified by using an antibody to the viral envelope protein p37, which produced a punctate stain at the cell surface and within the cytoplasm of cells (Fig. 8b and d). TGN46 staining (Fig. 8a) was scattered in the cells infected with virus, however, unlike ASFV, vaccinia virus did not cause loss of the TGN46 signal. Figure 8d shows that, in contrast to ASFV, which excluded lysosomes from assembly sites, vaccinia virus replication areas appeared to recruit lysosomes.
FIG. 8.
Effect of vaccinia virus on the TGN and lysosomes. BSC40 cells were fixed 16 h after infection with the VTF7 strain of vaccinia virus and incubated with a rabbit antibody specific for TGN46 (a) or monoclonal antibody H4B4 specific for LAMP2 (c). Vaccinia virus infection was detected with a rat antibody (15B6) that recognizes viral protein VV-p37 (b and d). Antigens were visualized by second antibodies coupled to Alexa 488 or Alexa 594.
DISCUSSION
This study has shown that ASFV infection causes a complete loss of the TGN, as judged by immunofluorescence staining for TGN46 and the TGN adapter complex AP1. The TGN is normally a compact crescent-shaped organelle located to one side of the nucleus. Between 8 and 10 h of infection, the immunofluorescence signal for TGN46 fragmented and scattered throughout the cytosol, and at 16 h, the signal disappeared. Similar results were recorded for sialyltransferase, an enzyme that also localizes predominantly to the TGN (data not shown). The protein levels of TGN46 remained constant throughout infection, suggesting TGN46 was not degraded, but was redistributed throughout the cell. Since the TGN46 signal was undetectable in infected cells, the site of redistribution of the protein was not obvious. Large organelles such as the plasma membrane or the ER are candidates. The surface areas of these membrane compartments are relatively much larger than the TGN, and it is possible that the TGN46 signal was lost through redistribution and dilution.
A study of the trafficking of the lysosomal enzyme cathepsin D enabled us to test the effects of ASFV on the function of the TGN. The recognition of mannose-6-phosphate residues in the TGN by the mannose-6-phosphate receptor is crucial for recruitment of lysosomal enzymes into clathrin-coated vesicles for transport to endosomal compartments and from there to lysosomes (6, 14, 16, 17, 25, 33). In uninfected cells, newly synthesized cathepsin D reached lysosomes within 2 h of synthesis; however, in cells infected with virus, only a small proportion of the enzyme was delivered to lysosomes within this period. The precise site of the block in the transport of cathepsin D to lysosomes caused by ASFV is not known. Lysosomal targeting of cathepsin D is dependent on phosphorylation of mannose residues on N-linked oligosaccharides added to the protein in the ER. Addition of mannose-6-phosphate to N-linked oligosaccharides in the ER and/or cis Golgi compartments prevents acquisition of resistance to endo H. The ability of endo H to remove sugars from cathepsin D was unaffected by ASFV, showing that addition of mannose-6-phosphate had occurred correctly in cells infected with the virus. Inhibition of mannose phosphorylation did not, therefore, explain the slow delivery of cathepsin D to lysosomes. An alternative explanation is that the slowed delivery of cathepsin D to lysosomes results from compromised binding of cathepsin D to the mannose-6-phosphate receptor in the TGN. This would be expected in the absence of an organized TGN to concentrate both receptor and ligand. An alternative possibility is that ASFV inhibited the proteases responsible for cleaving cathepsin D to the mature form in lysosomes. This could be mediated by a virally encoded protease inhibitor, neutralization of endosomal or lysosomal acid pH, or by preventing delivery of the processing proteases to lysosomes.
It is interesting to speculate about the mechanism of virus-induced loss of the TGN. Resident TGN proteins, such as TGN46 and furin, recycle between the TGN, endosomes, and the plasma membrane. It has been calculated that TGN46 molecules traffic between the TGN and the plasma membrane, with half-times of 45 to 60 min (13). Rapid internalization and delivery of TGN38 or -46 back to the TGN, coupled with a slower exit of the protein from the TGN, results in the bulk of the protein being localized to the TGN. It is likely that the supply of TGN46 to late Golgi compartments and the TGN is normal in cells infected with ASFV. ASFV did not, for example, cause accumulation of TGN46 in pre-TGN compartments that could be detected by immunofluorescence. The loss of TGN signal was, therefore, most likely caused by disruption of the TGN46 recycling pathway. The rate of budding and transport of TGN46-positive vesicles from the TGN to the plasma membrane could have been increased, or the virus may slow return of TGN46 to the TGN by inhibition of endocytosis pathways. Both would result in dilution of TGN46 within the endosomal system and plasma membrane, which eventually results in the loss of immunofluorescence signal of the protein. As TGN46, and possibly other integral membrane proteins, continually leave the TGN and fail to return, this causes an eventual breakdown of the structure of the TGN. At early stages, this would produce a scattered vesicle population, as we observed between 8 and 10 h of infection.
At present, it is not clear exactly how TGN38 or -46 reaches the cell surface, although it is suspected that transport does not involve recruitment into clathrin-coated vesicles at the TGN (26). Several proteins are recruited to areas of the TGN where TGN46 is concentrated: these include the cytoplasmic phosphoprotein p62, a phosphatidylinositol-specific 3-kinase (21–23, 30), and cytoplasmic factors, such as Rab6 and dynamin-2 (24). These molecules may be involved in a signaling pathway that leads to the recruitment of factors involved in the biogenesis of vesicles containing TGN46. The activity of these kinases and GTPases could potentially be altered by ASFV, and enhanced exit of TGN46 from the TGN would, for example, lead to a greater quantity of TGN46 at the cell surface, resulting in a dilution of the protein and an eventual loss of signal. TGN38 or -46 is rapidly internalized from the cell surface via clathrin-coated vesicles. ASFV may modulate the factors necessary for either nucleating clathrin coat formation or later steps in the biogenesis of clathrin-coated vesicles at the plasma membrane. Either effect would slow internalization of the protein and result in a greater amount of TGN46 at the cell surface of infected cells, leading to a dilution and subsequent loss of immunofluorescence signal.
Vaccinia virus and ASFV are both large double-stranded DNA viruses that replicate in the cytosol of infected cells. In common with ASFV, vaccinia virus acquires envelopes through enwrapment (36–38). The effects of vaccinia virus on the TGN were therefore studied. There was no loss of the TGN46 signal following infection of cells with vaccinia virus. Instead, the TGN46 signal was scattered throughout the cytosol. The data suggested that the loss of the TGN was unique to infection by ASFV and was not a common feature of infection of cells with pox-like viruses.
The potential consequences of TGN loss are numerous. The TGN is important for sorting proteins to their correct destination as they leave the Golgi apparatus. A loss of the TGN could result in compromised secretion of proteins and slowed movement of proteins through the endocytic pathway to lysosomes, as demonstrated for cathepsin D in this study. ASFV replicates in macrophages, and an ability to block cytokine secretion following infection could exert an anti-inflammatory effect. NF-κB-dependent cytokine transcription is suppressed in infected macrophages by a virally encoded homolog of IκB (31, 41). A block in the secretory pathway at the TGN would offer a second means of preventing secretion of these potent modulators of the proinflammatory immune response. Macrophages are professional antigen-presenting cells, and perturbation of MHC class 2 processing and peptide loading in the TGN and endosome compartments offers a second site for inhibition of the immune response by ASFV. Infection of macrophages by ASFV causes bystander apoptosis of T and B cells “in vivo” (27); a block in the surface expression of macrophage proteins that rescue lymphocytes from apoptosis may provide a mechanism. It is clear that a thorough characterization of the effects of virus-induced TGN loss on the processing of proteins by macrophages is now worthy of study and may provide answers to the complex pathology of African swine fever.
ACKNOWLEDGMENTS
We thank Dan Rock, Geoff Smith, Jenny Hirst, Margaret Robinson, and Janice Blum for gifts of antibodies. The figures would not have been possible without help with graphics from Steve Archibald.
This project was supported by the BBSRC, the BBSRC Bioimaging Initiative, and DEFRA.
REFERENCES
- 1.Alcami A, Koszinowski U H. Viral mechanisms of immune evasion. Trends Microbiol. 2000;8:410–418. doi: 10.1016/S0966-842X(00)01830-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almendral J M, Almazán F, Blasco R, Viñuela E. Multigene families in African swine fever virus: family 110. J Virol. 1990;64:2064–2072. doi: 10.1128/jvi.64.5.2064-2072.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrés G, Garcia-Escudero R, Simón-Mateo C, Viñuela E. African swine fever virus is enveloped by a two-membraned collapsed cisterna derived from the endoplasmic reticulum. J Virol. 1998;72:8988–9001. doi: 10.1128/jvi.72.11.8988-9001.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borca M V, Carillo C, Zsak L, Laegreid W W, Kutish G F, Neilan J G, Burrage T G, Rock D L. Deletion of a CD2-like gene, 8DR, from African swine fever virus affects viral infection in domestic swine. J Virol. 1998;72:2881–2889. doi: 10.1128/jvi.72.4.2881-2889.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brookes S M, Sun H, Dixon L K, Parkhouse R M E. Characterisation of African swine fever virion proteins j5R and j13L: immunolocalization in virus particles and assembly sites. J Gen Virol. 1998;79:1179–1188. doi: 10.1099/0022-1317-79-5-1179. [DOI] [PubMed] [Google Scholar]
- 6.Brown W J, Goodhouse J, Farquhar M G. Mannose 6-phosphate receptors for lysosomal enzymes cycle between the Golgi complex and endosomes. J Cell Biol. 1986;103:1235–1247. doi: 10.1083/jcb.103.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carrascosa A L, del Val M, Santarén J F, Viñuela E. Purification and properties of African swine fever virus. J Virol. 1985;54:337–344. doi: 10.1128/jvi.54.2.337-344.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cobbold C, Whittle J T, Wileman T. Involvement of the endoplasmic reticulum in the assembly and envelopment of African swine fever virus. J Virol. 1996;70:8382–8390. doi: 10.1128/jvi.70.12.8382-8390.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cobbold C, Wileman T. The major structural protein of African swine fever virus, p73, is packaged into large structures, indicative of viral capsid or matrix precursors, on the endoplasmic reticulum. J Virol. 1998;72:5215–5223. doi: 10.1128/jvi.72.6.5215-5223.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cobbold C, Brookes S M, Wileman T. Biochemical requirements of virus wrapping by the endoplasmic reticulum: involvement of ATP and endoplasmic reticulum calcium store during envelopment of African swine fever virus. J Virol. 2000;74:2151–2160. doi: 10.1128/jvi.74.5.2151-2160.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delbrück R, Desel C, von Figura K, Hille-Rehfeld A. Proteolytic processing of cathepsin D in prelysosomal organelles. Eur J Cell Biol. 1994;64:7–14. [PubMed] [Google Scholar]
- 12.Del Val M, Vinuela E. Glycosylated components induced in African swine fever virus-infected Vero cells. Virus Res. 1987;7:297–308. doi: 10.1016/0168-1702(87)90044-x. [DOI] [PubMed] [Google Scholar]
- 13.Ghosh R N, Mallet W G, Soe T T, McGraw T E, Maxfield F R. An endocytosed TGN38 chimeric protein is delivered to the TGN after trafficking through the endocytic recycling compartment in CHO cells. J Cell Biol. 1998;142:923–936. doi: 10.1083/jcb.142.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goda Y, Pfeffer S R. Selective recycling of the mannose 6-phosphate/IGF-II receptor to the trans Golgi network in vitro. Cell. 1988;55:309–320. doi: 10.1016/0092-8674(88)90054-2. [DOI] [PubMed] [Google Scholar]
- 15.Goebel S J, Johnson G P, Perkus M E, Davis S W, Winslow J P, Paoletti E. The complete DNA sequence of vaccinia virus. Virology. 1990;179:247–266. doi: 10.1016/0042-6822(90)90294-2. [DOI] [PubMed] [Google Scholar]
- 16.Goldberg D E, Kornfeld S. Evidence for extensive subcellular organization of asparagine-linked oligosaccharide processing and lysosomal enzyme phosphorylation. J Biol Chem. 1983;258:3159–3165. [PubMed] [Google Scholar]
- 17.Griffiths G, Hoflack B, Simons K, Mellman I, Kornfeld S. The mannose 6-phosphate receptor and the biogenesis of lysosomes. Cell. 1988;52:329–341. doi: 10.1016/s0092-8674(88)80026-6. [DOI] [PubMed] [Google Scholar]
- 18.Griffiths G, Rottier P. Cell biology of viruses that assemble along the biosynthetic pathway. Semin Cell Biol. 1992;3:367–381. doi: 10.1016/1043-4682(92)90022-N. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heath C M, Windsor M, Wileman T. Aggresomes resemble sites specialized for virus assembly. J Cell Biol. 2001;153:449–455. doi: 10.1083/jcb.153.3.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heemels M T, Ploegh H. Generation, translocation and presentation of MHC class I-restricted peptides. Annu Rev Biochem. 1995;64:463–491. doi: 10.1146/annurev.bi.64.070195.002335. [DOI] [PubMed] [Google Scholar]
- 21.Hickinson D M, Lucocq J M, Towler M C, Clough S, James J, James S R, Downes C P, Ponnambalam S. Association of a phosphatidylinositol-specific 3-kinase with a human trans-Golgi network resident protein. Curr Biol. 1997;7:987–990. doi: 10.1016/s0960-9822(06)00418-0. [DOI] [PubMed] [Google Scholar]
- 22.Jones S M, Howell K E. Phosphatidylinositol 3-kinase is required for the formation of constitutive transport vesicles from the TGN. J Cell Biol. 1997;139:339–349. doi: 10.1083/jcb.139.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones S M, Alb J G, Phillips S E, Bankaitis V A, Howell K E. A phosphatidylinositol 3-kinase and phosphatidylinositol transfer protein act synergistically in formation of constitutive transport vesicles from the trans-Golgi network. J Biol Chem. 1998;273:10349–10354. doi: 10.1074/jbc.273.17.10349. [DOI] [PubMed] [Google Scholar]
- 24.Jones S M, Howell K E, Henley J R, Cao H, McNiven M A. Role of dynamin in the formation of transport vesicles from the trans-Golgi network. Science. 1998;279:573–577. doi: 10.1126/science.279.5350.573. [DOI] [PubMed] [Google Scholar]
- 25.Kornfeld S, Mellman I. The biogenesis of lysosomes. Annu Rev Cell Biol. 1989;5:483–525. doi: 10.1146/annurev.cb.05.110189.002411. [DOI] [PubMed] [Google Scholar]
- 26.Ohno H, Stewart J, Fournier M C, Bosshart H, Rhee I, Miyatake S, Saito T, Gallusser A, Kirchhausen T, Bonifacino J S. Interaction of tyrosine-based sorting signals with clathrin-associated proteins. Science. 1995;269:1872–1875. doi: 10.1126/science.7569928. [DOI] [PubMed] [Google Scholar]
- 27.Oura C A L, Powell P P, Parkhouse R M E. African swine fever: a disease characterized by apoptosis. J Gen Virol. 1998;79:1427–1438. doi: 10.1099/0022-1317-79-6-1427. [DOI] [PubMed] [Google Scholar]
- 28.Pearse B M F, Robinson M S. Clathrin, adaptors, and sorting. Annu Rev Cell Biol. 1990;6:151–171. doi: 10.1146/annurev.cb.06.110190.001055. [DOI] [PubMed] [Google Scholar]
- 29.Ploubidou A, Moreau V, Ashman K, Reckmann I, Gonzalez C, Way M. Vaccinia virus infection disrupts microtubule organization and centrosome function. EMBO J. 2000;19:3932–3944. doi: 10.1093/emboj/19.15.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ponnambalam S, Clough S, Downes C P, Lucocq J M, McLauchlan H J, Towler M C. Lipid kinases and trans-Golgi network membrane dynamics. Biochem Soc Trans. 1999;27:670–673. doi: 10.1042/bst0270670. [DOI] [PubMed] [Google Scholar]
- 31.Powell P P, Dixon L K, Parkhouse R M E. An IκB homolog encoded by African swine fever virus provides a novel mechanism for downregulation of proinflammatory cytokine responses in host macrophages. J Virol. 1996;70:8527–8533. doi: 10.1128/jvi.70.12.8527-8533.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prescott A R, Lucocq J M, James J, Lister J M, Ponnambalam S. Distinct compartmentalization of TGN46 and beta 1,4-galactosyltransferase in HeLa cells. Eur J Cell Biol. 1997;72:238–246. [PubMed] [Google Scholar]
- 33.Rijnboutt S, Stoorvogel W, Geuze H J, Strous G J. Identification of subcellular compartments involved in biosynthetic processing of cathepsin D. J Biol Chem. 1992;267:15665–15672. [PubMed] [Google Scholar]
- 34.Rodriguez F, Alcarez C, Eiras A, Yáñez R J, Rodriguez J M, Alonso C, Rodriguez J F, Escribano J M. Characterization and molecular basis of heterogeneity of the African swine fever virus envelope protein p54. J Virol. 1994;68:7244–7252. doi: 10.1128/jvi.68.11.7244-7252.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodriguez J M, Yañez R J, Almazán F, Viñuela E, Rodriguez J F. African swine fever virus encodes a CD2 homolog responsible for the adhesion of erythrocytes to infected cells. J Virol. 1993;67:5312–5320. doi: 10.1128/jvi.67.9.5312-5320.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rouiller I, Brookes S M, Hyatt A D, Windsor M, Wileman T. African swine fever virus is wrapped by the endoplasmic reticulum. J Virol. 1998;72:2373–2387. doi: 10.1128/jvi.72.3.2373-2387.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmelz M, Sodiek B, Ericsson M, Wolffe E J, Shida H, Hiller G, Griffiths G. Assembly of vaccinia virus: the second wrapping cisternae is derived from the trans Golgi network. J Virol. 1994;68:130–147. doi: 10.1128/jvi.68.1.130-147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sodeik B, Doms R W, Ericsson M, Hiller G, Machamer C E, Vanthof W, Vanmeer G, Moss B, Griffiths G. Assembly of vaccinia virus—role of the intermediate compartment between the endoplasmic reticulum and the Golgi stacks. J Cell Biol. 1993;121:521–541. doi: 10.1083/jcb.121.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun H, Jacobs S, Smith G L, Dixon L K, Parkhouse R M E. African swine fever virus gene j13L encodes a 25–27 kDa virion protein with variable numbers of amino acid repeats. J Gen Virol. 1995;76:1117–1127. doi: 10.1099/0022-1317-76-5-1117. [DOI] [PubMed] [Google Scholar]
- 40.Sun H, Jenson J S, Dixon L K, Parkhouse R M E. Characterisation of African swine fever virion protein j18L. J Gen Virol. 1996;77:941–946. doi: 10.1099/0022-1317-77-5-941. [DOI] [PubMed] [Google Scholar]
- 41.Tait S W G, Reid E B, Greaves D R, Wileman T, Powell P P. Mechanism of inactivation of NF-kB by a viral homologue of ASFV. J Biol Chem. 2000;275:34656–34664. doi: 10.1074/jbc.M000320200. [DOI] [PubMed] [Google Scholar]
- 42.Tortorella D, Geurz B E, Furman M H, Schurt D J, Ploegh H L. Viral subversion of the immune system. Annu Rev Immunol. 2000;18:861–926. doi: 10.1146/annurev.immunol.18.1.861. [DOI] [PubMed] [Google Scholar]
- 43.Wilkinson P J. African swine fever virus. In: Pensaert M B, editor. Virus infections of porcines. Amsterdam, The Netherlands: Elsevier; 1989. pp. 17–35. [Google Scholar]
- 44.Wolf P R, Ploegh H. How MHC class II molecules acquire peptide cargo: biosynthesis and trafficking through the endocytic pathway. Annu Rev Cell Dev Biol. 1995;II:267–306. doi: 10.1146/annurev.cb.11.110195.001411. [DOI] [PubMed] [Google Scholar]
- 45.Yanez R J, Rodriguez J M, Nogal M L, Yuste L, Enriquez C, Rodriguez J F, Vinuela E. Analysis of the complete nucleotide sequence of African swine fever virus. Virology. 1995;208:249–278. doi: 10.1006/viro.1995.1149. [DOI] [PubMed] [Google Scholar]