ABSTRACT
Background
Bullous pemphigoid (BP) and atopic dermatitis (AD) are currently thought to be tightly related, yet studies of the mechanisms of co‐morbidities are lacking.
Methods
We obtained GWAS data for BP (N = 376,274) and AD (N = 796,661) from the Finnish Genetic Research Program dataset and the UK Biobank, separately. Then, the following four analyses were performed: (1) cross‐trait linkage disequilibrium score regression (LDSC) to assess the genetic correlation between BP and AD, (2) cross‐phenotype association analysis (CPASSOC) to identify multiple effector loci shared by BP and AD, (3) transcriptome‐wide association study (TWAS) to determine whether their cross‐organizational expression patterns share genes with a common biological mechanism of relevance, and (4) bidirectional Mendelian randomization (MR) analysis to assess bidirectional causal effects of BP and AD.
Results
We found a positive genetic association between BP and AD (rg = 0.5476, p = 0.0495) as well as identified four pleiotropic loci and 59 common genes affecting BP and AD. Bidirectional MR analysis suggested that BP promotes the risk of AD.
Conclusions
We revealed a genetic link between BP and AD, which is associated with biological pleiotropy and causality. Awareness of the association between BP and AD helps dermatologists manage patients with these illnesses.
Keywords: atopic dermatitis, bullous pemphigoid, co‐morbidities, cross‐trait meta‐analysis, genome‐wide genetic correlation analysis, mendelian randomization, transcriptome‐wide association analysis
We revealed a genetic link between atopic dermatitis and bullous pemphigoid, which is associated with biological pleiotropy and causality. Awareness of the association between BP and AD helps dermatologists manage patients with these illnesses.
Abbreviations
- AD
atopic dermatitis
- BP
bullous pemphigoid
- CPASSOC
cross‐phenotype association analysis
- eQTL
expression quantitative trait loci
- GTEx
Genotype‐Tissue Expression Project
- GWAS
genome‐wide association study
- HSV‐1
herpes simplex virus 1
- IVs
instrumental variables
- IVW
inverse variance weighting
- LD
linkage disequilibrium
- LDSC
linkage disequilibrium score regression
- MR
Mendelian randomization
- MR‐PRESSO
MR pleiotropy residual sum and outlier
- RCTs
randomized controlled trials
- SMR
summary‐based Mendelian randomization
- TWAS
transcriptome‐wide association study
1. Introduction
Bullous pemphigoid (BP) is the most common autoimmune bullous skin disease, presenting as pruritic, tense blisters on an erythematous base or normal skin (Bağcı et al. 2017). Commonly found in the elderly population, BP has considerable morbidity and mortality (Persson et al. 2022). The occurrence of BP is associated with autoimmunity, genetic susceptibility, and external triggers, with BP180 and dystonin‐e as target antigens (Bağcı et al. 2017; Moro et al. 2020). Patients with BP may have raised serum total IgE levels and increased peripheral eosinophilia (Yayli et al. 2011).
Atopic dermatitis (AD) is an inflammatory skin disease, with a prevalence of 15 to 20% among children and up to 10% among adults, as well as the highest disease burden among skin diseases (Laughter et al. 2021). The pathogenesis of AD is related to genetics, environmental factors, immune dysregulation, skin barrier function, and skin inflammation and its interactions (Ständer 2021). AD is presented clinically as skin dryness, chronic and recurrent eczema, and severe itching, and one of the main features of AD is elevated serum total IgE levels (Kasperkiewicz et al. 2018).
Currently, it is considered that there is a close relationship between BP and AD, both of which are disorders in which the T‐helper cell (Th2) response predominates, and the patients both have elevated serum IgE levels and Thymic stromal lymphopoietin (Hu and Zhang 2022). Zhang et al. (2018) found that the mice that lacked BP180 function developed spontaneous skin inflammation, similar to AD skin inflammation. Wu et al. (2023) observed that patients with both BP and AD have higher rates of co‐morbid cardiovascular risk factors, neuropsychiatric disorders, and autoimmune connective tissue diseases. This needs to be brought to the attention of clinicians. Yet, there is a lack of research on the co‐morbid relationship between BP and AD.
For this study, we aim to extend previous findings by a large‐scale genome‐wide cross‐trait analysis. First, we analyzed the overall genetic correlation between BP and AD, then cross‐trait meta‐analysis was adopted to screen potential functional genes shared by both, meanwhile transcriptome association studies (TWAS) were utilized to search for common underlying biological mechanisms. Last, Mendelian randomization (MR) was applied to analyze whether there is a causal relationship between BP and AD.
2. Materials and Methods
2.1. GWAS Data Sources for Bullous Pemphigoid and Atopic Dermatitis
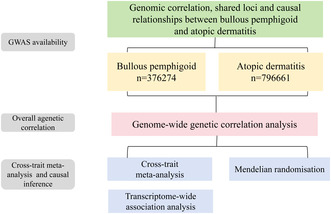
GWAS statistics for BP are summarized from the FinnGen research project, a dataset consisting of 507 patients with BP and 375,767 control participants of European ancestry. We acquired GWAS data for AD from the UK Biobank, which included 22,474 European ancestry cases and 774,187 European ancestry controls. Most importantly, the diagnosis of BP and AD follows the 10th edition of ICD codes (Kurki et al. 2023). We obtained indexed SNPs from GWAS and summarized the statistics. In addition, we used all genetic analyses that were in contrast to the human reference genome build 37 (or hg19). The detailed research steps in this paper are shown in Figure 1.
FIGURE 1.
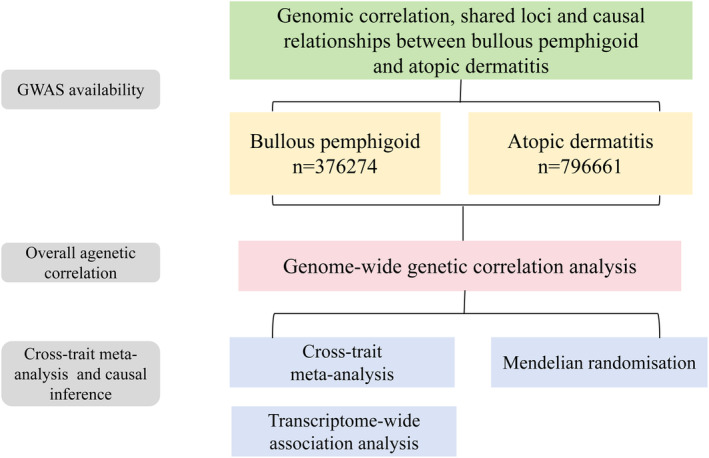
The overall design of the study.
2.2. Genetic Correlation Analysis
In the current study, we analyzed the overall genetic correlation between BP and AD using the linked disequilibrium score regression (LDSC) (Bulik‐Sullivan et al. 2015), which yielded estimates ranging between −1 and 1, such that 1 denotes a complete positive genetic correlation, as opposed to −1, which indicates a complete negative genetic correlation. In this study, we utilized pre‐calculated linkage disequilibrium (LD) scores acquired from the common SNP of European origin represented in the HapMap3 reference panel, which are usually recognized as SNPs with high quality.
2.3. Cross‐Trait Meta‐Analysis
We performed a cross‐trait meta‐analysis on the single SNP level by cross‐phenotype association analysis (CPASSOC), which integrates a summary of GWAS statistics for multiple correlated traits (Zhu et al. 2015). CPASSOC provides two test statistics, S Hom and S Het. Here, we applied paired S Het, which is an expansion of S Hom with greater capability in the presence of multiple genetic effects and which is more commonly used (Liu et al. 2022). We also obtained individual SNPs utilizing the PLINK clumping function that was parameterized to clump‐p 0.99, clump‐r2 0.2, and clump‐kb 500 (Purcell et al. 2007). The major pleiotropic SNPs were necessary to achieve genome‐wide conspicuousness in paired traits (p CPASSOC < 5 × 10−8) and single traits (p single trait < 1 × 10−3). Moreover, we performed specific functional annotations via the dbSNP database.
2.4. Transcriptome‐Wide Association Study (TWAS)
CPASSOC does not take into account gene expression or tissue specificity, however, many genetic variants affect complex traits by regulating gene expression levels (Gusev et al. 2016). Thus, we performed a TWAS by utilizing Summary‐based MR software that identifies relevant genes whose cross‐tissue expression patterns indicate the occurrence of a shared biological mechanism. At first, we obtained summary‐level data on blood expression quantitative trait loci (eQTL) provided by the eQTLGen consortium, as well as skin datasets published by the Genotype‐Tissue Expression Project (GTEx) Publishes Final Dataset (V8), which were combined to perform single‐trait TWAS analyses. Then, the above TWAS results were cross‐tabulated to test whether they were commensurable across traits. The Reactome knowledgebase contains signal transduction, transporter, DNA replication, metabolism, etc. Finally, we analyzed the intersecting genes of BP and AD by the Reactome knowledgebase.
2.5. Bidirectional MR
In this study, we assessed the causal relationship between BP and AD via bidirectional MR analysis. The inverse variance weighting (IVW) technique was used, which assumes that all instrumental variables (IVs) are useful since biased results result even from an ineffective instrument (Burgess, Butterworth, and Thompson 2013). Moreover, we conducted sensitivity analyses such as the MR Egger, the weighted median, the simple model, and the weighted models. We calculated Cochran's Q‐value for assessing IV heterogeneity. Besides, we used the MR pleiotropy residual sum and outlier (MR‐PRESSO) framework. Crucially, to account for pleiotropy, the outlier test eliminates outliers in the global test for pleiotropy detection between IVs, if noteworthy (Verbanck et al. 2018). We conducted a leave‐one‐out analysis, which repeatedly eliminated each SNP and used the remaining SNPs for the IVW approach, to investigate if causality estimates were caused by specific SNPs. To ascertain if BP is impacted by AD genetic predisposition, we conducted bidirectional MR in the meantime. Overall, BP and AD were intersected for exposure and outcome for MR analysis. This section involves MR analyses which were run in the “Two‐Sample MR” and “MR‐PRESSO” packages in R software (version 4.3.0).
3. Results
3.1. Overall Genetic Correlation
The LDSC is a tool that uses summary statistics to estimate global correlation (Finucane et al. 2015). We used LDSC to perform preliminary analyses of genetic relationships for the BP and AD data. Significantly, we found a positive correlation between BP and AD (rg = 0.5476, p = 0.0495).
3.2. Cross‐Trait Meta‐Analysis
Table 1 displays the results of our CPASSOC analysis, which identified four distinct pleiotropic SNPs with genome‐wide significance in paired traits (p CPASSOC < 5 × 10−8) and significance in a single trait (p single trait < 1 × 10−3). BP and AD shared four SNPs (rs7746553, rs943451, rs968155, rs28383305), in which rs943451 is present in both. Especially, none of these four SNPs have been previously described with BP or AD. The most important shared locus was rs968155 (p CPASSOC = 6.67099 × 10−7), and the second significant shared locus was rs28383305 (p CPASSOC = 4.48776 × 10−4), yet both are located in non‐coding sequences. Besides, rs7746553, (p CPASSOC = 4.90332 × 10−4), was near C2, CYP21A2, and CFB. As well as rs943451 (p CPASSOC = 8.8038 × 10−4) is located near PRKCQ and PRKCQ‐AS1.
TABLE 1.
Cross‐trait meta‐analysis of BPand AD.
| SNP | CHR | Position | A1 | A2 | EAF | Beta_bullous pemphigoid | Beta_ atopic dermatitis | p bullous pemphigoid | p atopic dermatitis | p CPASSOC | Nearest gene |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs7746553 | 6 | 31928196 | C | G | 0.8284 | 0.275647 | 0.0699 | 4.90332 × 10−4 | 1.457 × 10−7 | 4.90332 × 10−4 | C2, CYP21A2, CFB |
| rs943451 | 10 | 6579811 | T | C | 0.3092 | 0.218655 | 0.0559 | 8.8038 × 10−4 | 1.345 × 10−6 | 8.8038 × 10−4 | PRKCQ, PRKCQ‐AS1 |
| rs968155 | 6 | 32412938 | T | C | 0.5288 | −0.303489 | −0.0388 | 6.67099 × 10−7 | 1.479 × 10−4 | 6.67099 × 10−7 | NA |
| rs28383305 | 6 | 32618944 | T | G | 0.85 | 0.372426 | 0.0934 | 4.48776 × 10−4 | 4.153 × 10−6 | 4.48776 × 10−4 | NA |
Abbrevaitions: A1, effect allele; A2, alternative allele; Beta, effect allele beta coefficient; physical position of SNP (base pairs); CHR, chromosome; EAF, effect allele frequency; NA, not available; p CPASSO, p value for cross‐phenotype association.
3.3. Transcriptome‐Wide Association Studies
We identified 1173 genes that were associated considerably with AD (Table S1) and 908 genes that were strongly related to BP (Table S2). Subsequent cross‐analysis of the single‐trait TWAS data showed that 59 genes were common to both BP and AD (Table S3). Analyzing the shared genes by Reactome Knowledgebase revealed that the shared genes were mainly enriched in herpes simplex virus 1 (HSV‐1) infection and ubiquitin‐mediated proteolysis.
3.4. Bidirectional MR
3.4.1. The Causal Effect of BP on AD
Table S4 shows that we found 2433 independent SNPs with a median (minimum, maximum) F‐statistic of 34.79(20.85,108.83) that were used as genetic instrumental factors for BP. BP was shown to increase the risk of AD (OR 1.04, 95% CI 1.018–1.068) using the IVW technique, and comparable results were achieved using other approaches including weighted median (Figure S1A). By using MR‐PRESSO, we were unable to identify any possible outliers, and the MR‐Egger intercept (p = 0.7338) did not provide any indication of multiple effects. Cochran's Q test result (Q = 2.56, p = 0.634) did not reveal any heterogeneity in any of the instrument estimations. The scatter plots demonstrate how each MR method's overall impact and each SNP's specific impact on each outcome are displayed (Figure S1B). According to the leave‐one‐out plots, it seems improbable that a few extreme SNPs are the source of the association (Figure S1C).
3.4.2. The Causal Effect of AD on BP
Within our study, 4684 independent genetic variants were associated with AD at genome‐wide significant levels (Table S5), with a median F‐statistic (minimum, maximum) of 28.09 (20.64, 195.17). The IVW analysis showed that the risk of BP was not increased by AD as an exposure factor (OR 1.258, 95% CI 0.954, 1.658). The IVW results and the outcome of the analysis employing MR Egger, weighted median, simple mode, and weighted mode were in agreement (Figure S2A). There was no discernible heterogeneity, according to Cochran's Q statistic (Q = 70.15, p = 0.174). The MR‐Egger intercept test yielded no pleiotropy (p = 0.1862), while the MR‐PRESSO global test revealed no evidence of horizontal multidirectional outliers. The scatterplot displays the composite impacts of each approach as well as the individual SNP effects, as shown in Figure S2B. Furthermore, leave‐one‐out plots indicated that certain extreme SNPs were unlikely to be responsible for the correlation (Figure S2C).
4. Discussion
In this study, the largest specific GWAS statistics to date were applied to conduct a genome‐wide cross‐trait analysis study of BP and AD, including the genetic correlations, shared gene loci, and their potential causal relationships.
Overall, we discovered that BP and AD had a positive genetic association. Using cross‐trait meta‐analysis, we found four independent pleiotropic SNPs in this study. The first novel SNP we identified, rs7746553, is positioned close to C2, CYP21A2, and CFB.C2, namely Complement C2. Ring et al. (1979) examined the sera of patients with AD and discovered that the hemolytic activity of serum C2 was significantly decreased in five patients. The skin has been shown to express CYP21A2 (Slominski, Ermak, and Mihm 1996). CYP21A2 is associated with adrenal steroidogenesis, and the dysregulation of extra‐adrenal glucocorticoid secretion affects a variety of autoimmune diseases, including AD (Slominski et al. 2020). CFB, known as Complement Factor B, is a protein‐coding gene, which has been demonstrated to be markedly lower than normal in children with AD before treatment (Gora et al. 2014). Also, CFB can be detected in the basement membrane zone of the skin in patients with BP (Nelson et al. 2006). The second novel SNP, rs943451, is located near PRKCQ and PRKCQ‐AS1. PRKCQ, called Protein Kinase C Theta, is a protein‐coding gene, which is associated with Crohn's disease (Zhang et al. 2019). PRKCQ‐AS1 is an RNA gene belonging to the lncRNA class. There is no literature on PRKCQ or PRKCQ‐AS1 in association with AD or BP. Both the third and fourth SNPs, rs968155 and rs28383305, are located in the non‐coding zone, which is likely the result of LD.
From the TWAS analysis, we identified a total of 61 shared genes between BP and AD. Subjecting the above genes to Reactome Knowledgebase, they were mainly enriched in HSV‐1 infection and ubiquitin‐mediated protein hydrolysis. The infection with HSV‐1 may trigger severe skin infections in some patients with AD and may even spread throughout the body. The proteins of HSV‐1, especially gD, could induce significant T cell‐ and mast cell‐driven skin inflammation in a mouse model of AD (Novak et al. 2021). Möckel et al. (2022) demonstrate that HSV‐1 can indeed penetrate and initiate infection in AD skin. Viral coinfections are a potentially serious complication in patients with autoimmune bullous disease (Lehman and el‐Azhary RA. 2016). Nakamura et al. (2016) have reported an elderly female patient diagnosed with BP who had a positive cerebrospinal fluid sample for HSV1‐DNA by PCR. Ubiquitin protein hydrolysis is involved in basic physiological functions such as immunological and inflammatory response control and cell cycle regulation (Ciechanover, Orian, and Schwartz 2000). The current research related to ubiquitin hydrolysis involves neurodegenerative diseases (Layfield, Cavey, and Lowe 2003), whereas studies related to immune skin diseases are lacking.
Recently, a population‐based retrospective cohort study in Israel found that patients with a pre‐existing diagnosis of AD were at increased odds of suffering from BP, as well as an increased risk of subsequent AD in patients with BP (Kridin et al. 2022). Besides, in a population‐based case–control study in Taiwan, the relationship between AD and BP was explored by logistic regression analysis, it was observed that AD increased the risk of BP by 76% (Wu et al. 2023). However, our findings are not completely consistent with these conclusions. With bidirectional MR analysis, we found that BP, as an exposure factor, increases the risk of developing AD. Surprisingly, AD, as an exposure factor, is not causally related to the development of BP. This may be related to population differences in inclusion.
This is the first MR investigation to examine the cause‐and‐effect connection between BP and AD. MR bears similarities to RCTs or randomized controlled trials, but RCTs may be influenced by other uncontrolled residual confounders, such as lifestyle, socioeconomic status, and other factors that affect the results. Compared to RCTs, MR is less impacted by a variety of confounding factors. The two‐sample MR approach can partially or totally avoid the aforementioned issues when compared to RCTs, and the statistical reliability of the study can be increased because the analysis in this study was based on the GWAS study's summary statistics.
There are undoubtedly some limitations to this study. Since our GWAS data sources are primarily limited to European populations, the findings may not always hold for other populations. Following, other types of bullous skin diseases exist, such as mucous membrane pemphigoid, linear IgA bullous dermatosis, and epidermolysis bullosa acquisita (Baum et al. 2014). The BP statistics used do not yet differentiate between the various types. Besides, AD can be categorized as mild, moderate, and severe (Ständer 2021), and at present we have not studied the effect of AD severity on BP. Lastly, although both AD and BP incidence peaks are found in the elderly population (Bağcı et al. 2017; Laughter et al. 2021), there are variations in the incidence of the disease in the different age groups. This research did not stratify the age of the patients.
In conclusion, our research revealed a strong genetic correlation between BP and AD, indicating a shared genetic foundation. Then, we identified four pleiotropic SNPs. Meanwhile, through single‐trait TWAS results across traits, we found that BP and AD share 61 genes. Ultimately, using two‐sample MR to examine the causative link between BP and AD, we found that BP increases the risk of AD. The above reveals the existence of a common genetic background for BP and AD. The newly identified SNPs in this study may provide new entry points for the treatment of BP and AD. Moreover, the co‐morbid mechanisms of BP and AD deserve further in‐depth exploration, and clinical dermatologists need to focus on the disease management of both.
Author Contributions
The concept was conceptualized and proposed by Qing Wang. Xuehua Wang, Qizhen Zhuang, Yuan Wu, and Qing Wang handled data processing and manuscript writing. Yue Lu, Junhong Zhang, and Jingjing Wu all contributed to the gathering and analysis of the data. The manuscript was critically evaluated and discussed by Juanjuan Liu, Xiangyu Hu, and Ling Han. The essay was written by all writers, who also reviewed and approved the submitted version.
Ethics Statement
Since the study's data came from open‐source sources, there were no conflicts of interest or ethical concerns. The patients who are listed in the database have ethical clearance. Users may obtain the data for free to do research and write relevant articles.
Conflicts of Interest
The authors declare no conflicts of interest.
Supporting information
Figure S1. (A) Scatter plots of estimates of causal effect for bullous pemphigoid on atopic dermatitis. (B) Estimates of causal effect for bullous pemphigoid on atopic dermatitis. (C) Leave‐one‐out plots of causal effect bullous pemphigoid on atopic dermatitis. MR, Mendelian randomization; SNP, single nucleotide polymorphisms.
Figure S2. (A) Scatter plots of estimates of causal effect for atopic dermatitis on bullous pemphigoid. (B) Estimates of causal effect for atopic dermatitis on bullous pemphigoid. (C) Leave‐one‐out plots of causal effect for atopic dermatitis on bullous pemphigoid. MR, Mendelian randomization; SNP, single nucleotide polymorphisms.
Figure S3.
Table S1.
Table S2.
Table S3.
Table S4.
Table S5.
Acknowledgments
We are grateful for the GWAS data that were kindly supplied by the FinnGen genomes research project and the UK Biobank.
Funding: This work was supported by National Natural Science Foundation of China (82374313), State Key Laboratory of Dampness Syndrome of Chinese Medicine Special Fund (SZ2021ZZ29), and Research Fund for Bajian Talents of Guangdong Provincial Hospital of Chinese Medicine (BJ2022KY05), the Guangzhou Science and Technology Plan Project‐Municipal School (College) Joint Funding Project (202201020332, 202201020320, and 202201020317).
Contributor Information
Juanjuan Liu, Email: 705816727@qq.com.
Xiangyu Hu, Email: woshihuxiangyu@163.com.
Ling Han, Email: linghan99@gzucm.edu.cn.
Data Availability Statement
The GWAS data used in this investigation are openly accessible, and upon reasonable request, the corresponding author can provide the code covered.
References
- Bağcı, I. S. , Horváth O. N., Ruzicka T., and Sárdy M.. 2017. “Bullous Pemphigoid.” Autoimmunity Reviews 16, no. 5: 445–455. [DOI] [PubMed] [Google Scholar]
- Baum, S. , Sakka N., Artsi O., Trau H., and Barzilai A.. 2014. “Diagnosis and Classification of Autoimmune Blistering Diseases.” Autoimmunity Reviews 13, no. 4–5: 482–489. [DOI] [PubMed] [Google Scholar]
- Bulik‐Sullivan, B. , Finucane H. K., Anttila V., et al. 2015. “An Atlas of Genetic Correlations Across Human Diseases and Traits.” Nature Genetics 47, no. 11: 1236–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess, S. , Butterworth A., and Thompson S. G.. 2013. “Mendelian Randomization Analysis With Multiple Genetic Variants Using Summarized Data.” Genetic Epidemiology 37, no. 7: 658–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover, A. , Orian A., and Schwartz A. L.. 2000. “Ubiquitin‐Mediated Proteolysis: Biological Regulation via Destruction.” BioEssays 22, no. 5: 442–451. [DOI] [PubMed] [Google Scholar]
- Finucane, H. K. , Bulik‐Sullivan B., Gusev A., et al. 2015. “Partitioning Heritability by Functional Annotation Using Genome‐Wide Association Summary Statistics.” Nature Genetics 47, no. 11: 1228–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gora, N. V. , Kozlov L. V., Logunov O. V., Bashkina O. A., Rubalskiĭ O. V., and Aleshkin V. A.. 2014. “Activity of B and D Factors of Complement Alternative Pathway in Children With Atopic Dermatitis.” Zhurnal Mikrobiologii, Epidemiologii, i Immunobiologii 3: 15–20. [PubMed] [Google Scholar]
- Gusev, A. , Ko A., Shi H., et al. 2016. “Integrative Approaches for Large‐Scale Transcriptome‐Wide Association Studies.” Nature Genetics 48, no. 3: 245–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Y. Q. , and Zhang J. Z.. 2022. “A Comparison for Type 2 Cytokines and Lesional Inflammatory Infiltrations in Bullous Pemphigoid and Atopic Dermatitis.” Clinical, Cosmetic and Investigational Dermatology 15: 2313–2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasperkiewicz, M. , Schmidt E., Ludwig R. J., and Zillikens D.. 2018. “Targeting IgE Antibodies by Immunoadsorption in Atopic Dermatitis.” Frontiers in Immunology 9: 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kridin, K. , Hammers C. M., Ludwig R. J., et al. 2022. “The Association of Bullous Pemphigoid With Atopic Dermatitis and Allergic Rhinitis‐A Population‐Based Study.” Dermatitis 33, no. 4: 268–276. [DOI] [PubMed] [Google Scholar]
- Kurki, M. I. , Karjalainen J., Palta P., et al. 2023. “FinnGen Provides Genetic Insights From a Well‐Phenotyped Isolated Population.” Nature 613, no. 7944: 508–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughter, M. R. , Maymone M. B. C., Mashayekhi S., et al. 2021. “The Global Burden of Atopic Dermatitis: Lessons From the Global Burden of Disease Study 1990‐2017.” British Journal of Dermatology 184, no. 2: 304–309. [DOI] [PubMed] [Google Scholar]
- Layfield, R. , Cavey J. R., and Lowe J.. 2003. “Role of Ubiquitin‐Mediated Proteolysis in the Pathogenesis of Neurodegenerative Disorders.” Ageing Research Reviews 2, no. 4: 343–356. [DOI] [PubMed] [Google Scholar]
- Lehman, J. S. , and el‐Azhary R. A.. 2016. “Kaposi Varicelliform Eruption in Patients With Autoimmune Bullous Dermatoses.” International Journal of Dermatology 55, no. 3: e136–e140. [DOI] [PubMed] [Google Scholar]
- Liu, Q. , Zhu Z., Kraft P., Deng Q., Stener‐Victorin E., and Jiang X.. 2022. “Genomic Correlation, Shared Loci, and Causal Relationship Between Obesity and Polycystic Ovary Syndrome: A Large‐Scale Genome‐Wide Cross‐Trait Analysis.” BMC Medicine 20, no. 1: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möckel, M. , De La Cruz N. C., Rübsam M., et al. 2022. “Herpes Simplex Virus 1 Can Bypass Impaired Epidermal Barriers Upon Ex Vivo Infection of Skin From Atopic Dermatitis Patients.” Journal of Virology 96, no. 17: e0086422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moro, F. , Fania L., Sinagra J. L. M., Salemme A., and Di Zenzo G.. 2020. “Bullous Pemphigoid: Trigger and Predisposing Factors.” Biomolecules 10, no. 10: 1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, Y. , Kakiuti K., Tani H., Nakajima H., Kimura F., and Hanafusa T.. 2016. “Herpes Simplex Encephalitis Without Cerebrospinal Fluid Pleocytosis in a Patient With Bullous Pemphigoid: A Case Report.” Rinshō Shinkeigaku 56, no. 6: 435–438. [DOI] [PubMed] [Google Scholar]
- Nelson, K. C. , Zhao M., Schroeder P. R., et al. 2006. “Role of Different Pathways of the Complement Cascade in Experimental Bullous Pemphigoid.” Journal of Clinical Investigation 116, no. 11: 2892–2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak, N. , Weighardt H., Valdelvira R., Izquierdo E., Förster I., and Cabanillas B.. 2021. “Herpes Simplex Virus 1 Proteins Can Induce Skin Inflammation in an Atopic Dermatitis‐Like Mouse Model.” Experimental Dermatology 30, no. 11: 1699–1704. [DOI] [PubMed] [Google Scholar]
- Persson, M. S. M. , Begum N., Grainge M. J., Harman K. E., Grindlay D., and Gran S.. 2022. “The Global Incidence of Bullous Pemphigoid: A Systematic Review and Meta‐Analysis.” British Journal of Dermatology 186, no. 3: 414–425. [DOI] [PubMed] [Google Scholar]
- Purcell, S. , Neale B., Todd‐Brown K., et al. 2007. “PLINK: A Tool Set for Whole‐Genome Association and Population‐Based Linkage Analyses.” American Journal of Human Genetics 81, no. 3: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring, J. , Senter T., Cornell R. C., Arroyave C. M., and Tan E. M.. 1979. “Plasma Complement and Histamine Changes in Atopic Dermatitis.” British Journal of Dermatology 100, no. 5: 521–530. [DOI] [PubMed] [Google Scholar]
- Slominski, A. , Ermak G., and Mihm M.. 1996. “ACTH Receptor, CYP11A1, CYP17 and CYP21A2 Genes Are Expressed in Skin.” Journal of Clinical Endocrinology and Metabolism 81, no. 7: 2746–2749. [DOI] [PubMed] [Google Scholar]
- Slominski, R. M. , Tuckey R. C., Manna P. R., et al. 2020. “Extra‐Adrenal Glucocorticoid Biosynthesis: Implications for Autoimmune and Inflammatory Disorders.” Genes and Immunity 21, no. 3: 150–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ständer, S. 2021. “Atopic Dermatitis.” New England Journal of Medicine 384, no. 12: 1136–1143. [DOI] [PubMed] [Google Scholar]
- Verbanck, M. , Chen C. Y., Neale B., and Do R.. 2018. “Detection of Widespread Horizontal Pleiotropy in Causal Relationships Inferred From Mendelian Randomization Between Complex Traits and Diseases.” Nature Genetics 50, no. 5: 693–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, P. C. , Wu C. Y., Lyu Y. S., Chang Y. T., and Wu C. Y.. 2023. “Association Between Bullous Pemphigoid and Atopic Dermatitis: A Population‐Based Case‐Control Study in Taiwan.” Archives of Dermatological Research 315, no. 3: 419–427. [DOI] [PubMed] [Google Scholar]
- Yayli, S. , Pelivani N., Beltraminelli H., et al. 2011. “Detection of Linear IgE Deposits in Bullous Pemphigoid and Mucous Membrane Pemphigoid: A Useful Clue for Diagnosis.” British Journal of Dermatology 165, no. 5: 1133–1137. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Hwang B. J., Liu Z., et al. 2018. “BP180 Dysfunction Triggers Spontaneous Skin Inflammation in Mice.” Proceedings of the National Academy of Sciences of the United States of America 115, no. 25: 6434–6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. W. , Xu X. Y., Zhang J., et al. 2019. “Missense Mutation in PRKCQ Is Associated With Crohn's Disease.” Journal of Digestive Diseases 20, no. 5: 243–247. [DOI] [PubMed] [Google Scholar]
- Zhu, X. , Feng T., Tayo B. O., et al. 2015. “Meta‐Analysis of Correlated Traits via Summary Statistics From GWASs With an Application in Hypertension.” American Journal of Human Genetics 96, no. 1: 21–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. (A) Scatter plots of estimates of causal effect for bullous pemphigoid on atopic dermatitis. (B) Estimates of causal effect for bullous pemphigoid on atopic dermatitis. (C) Leave‐one‐out plots of causal effect bullous pemphigoid on atopic dermatitis. MR, Mendelian randomization; SNP, single nucleotide polymorphisms.
Figure S2. (A) Scatter plots of estimates of causal effect for atopic dermatitis on bullous pemphigoid. (B) Estimates of causal effect for atopic dermatitis on bullous pemphigoid. (C) Leave‐one‐out plots of causal effect for atopic dermatitis on bullous pemphigoid. MR, Mendelian randomization; SNP, single nucleotide polymorphisms.
Figure S3.
Table S1.
Table S2.
Table S3.
Table S4.
Table S5.
Data Availability Statement
The GWAS data used in this investigation are openly accessible, and upon reasonable request, the corresponding author can provide the code covered.


