Abstract
Simple Summary
This first comprehensive genomic analysis of rapidly growing non-tuberculous mycobacteria (RGM) isolates from Betta splendens has led to the re-identification of certain isolates and the discovery of a potential new species, Mycobacterium mucogenicum subsp. phocaicum sp. nov. The in silico analysis is consistent with and confirms previously reported in vitro studies on virulence and antibiotic susceptibility profiles. This study emphasizes the crucial role of whole genome sequencing in the accurate identification and classification of mycobacterial species.
Abstract
This study aims to genomically elucidate six isolates of rapidly growing non-tuberculous mycobacteria (RGM) derived from Siamese fighting fish (Betta splendens). These isolates had previously undergone phenotypic and biochemical characterization, antibiotic susceptibility testing, and in vivo virulence assessment. Initial DNA barcoding using the 16S rRNA sequence assigned these six isolates to five different species, namely Mycobacterium chelonae (BN1983), M. cosmeticum (BN1984 and N041), M. farcinogenes (SNSK5), M. mucogenicum (BN1956), and M. senegalense (BN1985). However, the identification relied solely on the highest percent identity of the 16S rRNA gene, raising concerns about the taxonomic ambiguity of these species. Comprehensive whole genome sequencing (WGS) and extended genomic comparisons using multilocus sequence typing (MLST), average nucleotide identity (ANI), and digital DNA–DNA hybridization (dDDH) led to the reclassification of BN1985 and SNSK5 as M. conceptionense while confirming BN1983 as M. chelonae and BN1984 and N041 as M. cosmeticum. Notably, the analysis of the BN1956 isolate revealed a potential new species that is proposed here as M. mucogenicum subsp. phocaicum sp. nov. Common genes encoding “mycobacterial” virulence proteins, such as PE and PPE family proteins, MCE, and YrbE proteins, were detected in all six isolates. Two species, namely M. chelonae and M. cosmeticum, appear to have horizontally acquired T6SS-II (clpB), catalase (katA), GroEL (groel), and capsule (rmlb) from distantly related environmental bacteria such as Klebsiella sp., Neisseria sp., Clostridium sp., and Streptococcus sp. This study provides the first draft genome sequence of RGM isolates currently circulating in B. splendens and underscores the necessity of WGS for the identification and classification of mycobacterial species.
Keywords: average nucleotide identity, Betta splendens, genomic sequencing, non-tuberculous mycobacteria, whole genome
1. Introduction
The genus Mycobacterium is characterized by the presence of mycolic acids in the cell wall, aerobic growth, and a bacillary form, with Mycobacterium tuberculosis serving as the type strain [1]. Currently, it comprises over 190 species, including significant human pathogens such as M. tuberculosis and Mycobacterium leprae, as well as opportunistic non-tuberculous mycobacteria (NTM) [2]. Traditionally, the genus has been classified by growth rate and further subdivided into complexes or groups [3]. The GenBank database (https://www.ncbi.nlm.nih.gov/, accessed on 19 May 2024) and the partners of the International Nucleotide Sequence Database Collaboration (http://www.insdc.org/, accessed on 19 May 2024) now contain the 16S rRNA sequences of thousands of officially recognized mycobacterial strains. It is, therefore, not surprising that taxonomic and phylogenetic studies of mycobacterial species have been based primarily on 16S rRNA for several decades [4]. The 16S rRNA gene-based taxonomic classification has been largely adequate in the separation of rapidly and slowly growing species and the grouping of most species into well-defined phylogenetic groups (complexes) [5]. However, the utility of 16S rRNA gene-based classification in differentiating closely related mycobacterial species is limited due to a very high sequence similarity among mycobacteria, making precise phylogenetic differentiation difficult [6,7]. In contrast, whole genome sequencing has been shown to be more effective than traditional genotyping in delineating species, discovering new species, and investigating specific traits [8,9,10]. Therefore, the application of whole genome analysis represents a significant step forward in mycobacterial research and provides deeper insights into the genetic diversity and complexity of the genus Mycobacterium.
We recently conducted a comprehensive investigation into the phenotypic and biochemical characteristics, as well as the antibiotic and disinfectant susceptibility of rapidly growing non-tuberculous mycobacteria (RGM) isolated from Siamese fighting fish (Betta splendens) [11]. Specifically, the study included six isolates representing five RGM species, namely Mycobacterium chelonae (BN1983), M. cosmeticum (BN1984 and N041), M. farcinogenes (SNSK5), M. mucogenicum (BN1956), and M. senegalense (BN1985), that were initially identified based on the 16S rRNA gene sequence. Additionally, we evaluated the virulence and pathogenicity of these isolates towards their host species by experimentally infecting fish through intraperitoneal injection. The results demonstrated the pathogenic nature of these rapidly growing NTMs to B. splendens and revealed the presence of multiple antibiotic-resistant patterns using in vitro assays. Notably, the M. chelonae (BN1983) isolate exhibited the highest virulence and resistant to multiple antibiotics. This underscores the significance of this group of RGM and provides the impetus for a comparative genomic analysis of these closely related mycobacterial species.
As a follow-up of our previous studies [11,12,13] we present the draft genomes of all six isolates derived from Siamese fighting fish (B. splendens), marking the first comprehensive genomic analysis of these isolates. The whole genome sequence analysis for each isolate focuses on describing genomic features; clarifying taxonomic identification using MLST, ANI, and dDDH; and profiling virulence factors and antimicrobial resistance genes. The findings identified the BN1956 isolate as a potential novel species, which we propose to name Mycobacterium mucogenicum subsp. phocaicum sp. nov. This study underscores the potential risks of misidentifying Mycobacterium species when relying solely on a single or limited set of target genes, highlighting the complex taxonomic landscape of these pathogens.
2. Materials and Methods
2.1. DNA Extraction and Sequencing
The bacterial samples were isolated from Siamese fighting fish, B. splendens, as described in our previous studies [11,14]. The total of six RGM isolates used in this study were obtained from naturally occurring fish displaying signs of either big belly syndrome (BBS) or skin nodule syndrome (SNS), as previously described by Dong et al. [14].
The isolates were cultured at 30 °C for 2 days in 5 mL sterile Middlebrook 7H9 broth (HiMedia, Mumbai, India) containing 10% oleic albumin dextrose catalase growth supplement. The bacterial cultures were then pelleted by centrifugation at 5000× g for 5 min, and total genomic DNA was extracted using the phenol–chloroform extraction method [15,16]. Briefly, the harvested bacterial cells were lysed using EDTA-saline, RNase A, and lysozyme. Subsequently, SDS and proteinase K were added, followed by 3 rounds of phenol:chloroform:isoamyl alcohol extraction. After purification, DNA was precipitated using sodium acetate and ethanol, washed with 70% ethanol, and resuspended in nuclease-free water. The quality and quantity of the extracted DNA were assessed using Nanodrop and Qubit instruments. Short-insert library preparation and sequencing were performed at BGI-Hong Kong (Hong Kong, China) using the DNBseqTM platform.
2.2. Whole Genome Assembly and Annotation
The raw data were filtered to remove contaminants, adapters, and low-quality sequences using SOAPnuke v2.1.8 [17]. The following filter parameters were applied: “ -n 0.01 -l 20 -q 0.4 –adaMis 3 –outQualSys 1 –minReadLen 150”, which removed reads that matched ≥ 50.0 with the adapter sequence and mismatched, at most, 3 bases. In addition, sequence reads with a quality score < 20 and a proportion ≥40.0% of the total reads, as well as reads shorter than 150 bp and reads with N content ≥ 1.0%, were discarded.
Whole genome de novo assembly of six isolates was performed with Read Assembly and Annotation Pipeline Tool (RAPT) build rapt-45639894 using SKESA version 2.5.1 [18], the taxonomic assignment tool ANI [19], and Prokaryotic Genome Annotation Pipeline (PGAP) 2023-05-17 build6771 [20]. In addition, another round of annotation was performed with Prokka version 1.14.6 [21] for enhanced confirmation. Genome maps for the draft genomes were generated using Proksee (https://proksee.ca, accessed on 27 February 2024) [22]. Mobile genetic elements were discovered and characterized with mobileOG-db [23].
2.3. Analysis of the Assembled Draft Genomes
2.3.1. Genome-Based Taxonomic Analysis
The assembled genomes were analyzed using rMLST [24] and the Type (Strain) Genome Server (TYGS) (https://tygs.dsmz.de, accessed on 1 January 2024), employing a 70% digital DNA–DNA hybridization (dDDH) cutoff for a whole genome-based taxonomic analysis [25,26]. Briefly, the determination of the genomes of the closest type strains was performed in two complementary ways. First, all genomes in our study were compared to genomes of type strains available in the TYGS database using the MASH algorithm and a fast approximation of intergenomic relatedness [27]. Then, ten type strains with the smallest MASH distances per genome were selected. Secondly, an additional group of ten closely related type strains was determined based on the 16S rDNA gene sequences. These were extracted from our study genomes using RNAmmer [28], and each sequence was then compared to the 16S rDNA gene sequence of each of the 20,173 type strains currently available in the TYGS database using BLASTed [29]. This was used as a proxy to find the best 50 matching type strains (according to the bitscore) for each of our study genomes; then, exact distances were calculated using the Genome BLAST Distance Phylogeny (GBDP) approach under the “coverage” algorithm and distance formula d5 [30]. These distances were finally used to determine the ten closest type-strain genomes for each of the six genomes we sequenced. For phylogenomic inference, all pairwise comparisons between genomes were performed using GBDP, and exact intergenomic distances were inferred using the “trimming” algorithm and distance formula d5 [30]. For each comparison, 100 distance replicates were calculated. The dDDH values and confidence intervals were calculated using the recommended settings of GGDC version 4.0 [26,30]. The average nucleotide identity (ANI) values of our study genomes compared to the respective reference genomes determined in the previous step were calculated using OrthoANIu [31] and FastANI version 1.34 [32].
2.3.2. Clustering and Phylogenetic Inference
Species clustering was performed using a 70% dDDH threshold, grouping around each of the 60 type strains to differentiate species [25]. For subspecies, a stricter 79% dDDH threshold was applied to ensure more precise clustering [33]. The resulting intergenomic distances were used to construct a balanced minimum-evolution tree that was generated using FASTME version 2.1.6.1, with subtree pruning and regrafting (SPR) post processing to optimize the tree topology [34]. Branch support was calculated from 100 pseudo-bootstrap replicates to enhance the reliability of the tree. The final tree was centrally rooted to provide a clearer visualization of evolutionary relationships [35] and was displayed using the interactive PhyD3 tool for better interpretability [36].
2.3.3. Plasmid Identification and Antimicrobial Resistance Profiling
Multilocus sequence typing analysis (MLST) was performed for housekeeping genes S142Z, L35, S19, L19, S12, S8, L16, and S7, together with their corresponding sequence types (STs). Plasmid sequences were identified within the draft genomes using Plasmer version 0.1 20220816 [37], which employs machine learning to analyze shared k-mers and genomic features. The identified plasmid sequences were then taxonomically classified by comparing them to all plasmids in the NCBI RefSeq database using Kraken 2 version 2.1.2 [38]. Antimicrobial resistance genes (ARGs) were detected using several complementary tools to ensure comprehensive identification across various databases and detection methods. ResFinder version 4.4.2 [39] was chosen for its accuracy in identifying acquired antimicrobial resistance genes based on curated reference sequences. abriTAMR version 3.10.42 [40], supported by AMRFinderPlus [41], was employed to enhance sensitivity in detecting ARGs by comparison against multiple databases. Additionally, ABRicate v1.0.1 (https://github.com/tseemann/abricate, accessed on 28 February 2024) was used to scan contig sequences against various ARG databases, including NCBI AMRFinderPlus [41], CARD [42], Resfinder [43], ARG-ANNOT [44], VFDB [45], PlasmidFinder [46], EcOH [47], and MEGARes 2.0 [48]. The Resistance Gene Identifier (RGI) online portal version 6.0.3 (https://card.mcmaster.ca/analyze/rgi, accessed on 28 February 2024) was also used to predict resistomes by integrating homology and SNP models, leveraging the Comprehensive Antibiotic Resistance Database (CARD; version 3.2.8) [49]. The use of multiple tools and databases allows for cross-validation of the results, enhancing both accuracy and coverage in detecting known and novel ARGs across the genome.
2.3.4. Pangenome Analysis
Pangenome analysis of the six isolates from this study was conducted using Roary version 3.13.0 [50] to determine the gene clusters and delineate the core and accessory genomes across the isolates. The analysis was executed using the command “roary -f roary_out -e -n -v -p 30 *.gff”, which enabled parallel processing, excluded splitting paralogs, and allowed for refined gene clustering. This approach facilitated the identification of shared and unique genetic elements among the isolates, providing insights into their genomic diversity and potential functional differences. Following the pangenome analysis, phylogenetic inference for both core and accessory genes was conducted using the maximum likelihood method implemented in IQ-TREE version 2.2.0 [51]. IQ-TREE analysis was performed with the command “iqtree2 -s core_gene_alignment.aln -m MFP -bb 1000 -alrt 1000 -nt 30”. This command employed the ModelFinder Plus algorithm for best-fit model selection, ensuring the most appropriate evolutionary model was used for the data. Additionally, 1000 ultrafast bootstrap replicates and SH-aLRT tests were conducted to assess branch support, providing robust statistical validation for the resulting phylogenetic trees.
3. Results
3.1. Data Summary
A comprehensive summary of the key metrics for each of the six mycobacterium isolates sequenced in this study is provided in Table 1. These metrics include the total number of clean reads, the sum of clean bases, the average read length, quality scores represented by Q20 and Q30, and GC content. The data reflect acceptable quality standards, and subsequent analyses were then conducted as described below.
Table 1.
Quality statistics of sequence data generated in this study.
| Sample ID | Clean Reads | Clean Bases (bp) | Read Length | Q20 (%) | Q30 (%) | GC (%) |
|---|---|---|---|---|---|---|
| BN1956 | 4,050,677 | 1,215,203,100 | PE150 | 95.00 | 87.02 | 66.64 |
| BN1983 | 4,043,521 | 1,213,056,300 | PE150 | 95.42 | 87.77 | 64.06 |
| BN1984 | 4,037,827 | 1,211,348,100 | PE150 | 95.09 | 87.31 | 64.05 |
| BN1985 | 4,038,265 | 1,211,479,500 | PE150 | 95.16 | 87.36 | 65.94 |
| SNSK5 | 4,037,574 | 1,211,272,200 | PE150 | 95.22 | 87.73 | 66.22 |
| N041 | 4,048,072 | 1,214,421,621 | PE150 | 95.22 | 87.69 | 67.27 |
3.2. Genome Assembly and Annotation
The results of de novo assembly and annotation are shown in Table 2, highlighting key genomic features, including genome size, the quality of assembled genomes, and the number of predicted genes. The analysis revealed high-quality draft genomes for all six isolates with genomic features that matched the respective reference genomes in terms of size and predicted gene count. However, the higher number of contigs observed in the annotated genomes is likely due to the limitations of short-read sequencing, which struggles to resolve repetitive regions and complex genomic structures, resulting in fragmented assemblies.
Table 2.
Genomic feature distribution of the six bacterial isolates in this study.
| Features | Isolates | ||||||
|---|---|---|---|---|---|---|---|
| BN1956 | BN1983 | BN1984 | BN1985 | SNSK5 | N041 | ||
| GenBank accession | JAYXBQ000000000 | JAYXBR000000000 | JAYXBS000000000 | JAYXBT000000000 | JAYXBU000000000 | JAYXBV000000000 | |
| Size (bp) | 6,369,566 | 5,056,078 | 7,557,134 | 6,818,352 | 6,342,755 | 7,556,912 | |
| Minimum sequence length | 350 | 374 | 361 | 378 | 378 | 361 | |
| Maximum sequence length | 446,856 | 1,063,941 | 1,115,510 | 1,179,774 | 903,710 | 861,703 | |
| Contigs | 84 | 19 | 57 | 39 | 30 | 59 | |
| N50 (bp) | 205,143 | 800,484 | 398,421 | 540,107 | 520,742 | 385,935 | |
| Completeness (CheckM) (%) | 99.32 | 99.48 | 99.62 | 100 | 99.66 | 99.62 | |
| Contamination (%) | 1.06 | 0.38 | 5.29 | 1.15 | 0.98 | 5.29 | |
| L50 | 10 | 2 | 7 | 5 | 5 | 8 | |
| Genes | 6184 | 4937 | 7512 | 6644 | 6118 | 7511 | |
| CDSs | 6088 | 4885 | 7425 | 6558 | 6063 | 7424 | |
| Genes (coding) | 6010 | 4854 | 7324 | 6506 | 6013 | 7322 | |
| Genes (RNA) | 96 | 52 | 87 | 86 | 55 | 87 | |
| rRNA | 5S | 2 | 1 | 2 | 2 | 2 | 2 |
| 16S | 2 | 1 | 2 | 1 | 1 | 2 | |
| 23S | 6 | 1 | 3 | 2 | 2 | 3 | |
| tRNAs | 83 | 46 | 77 | 77 | 47 | 77 | |
| ncRNAs | 3 | 3 | 3 | 4 | 3 | 3 | |
| Pseudo Genes | 78 | 31 | 101 | 52 | 50 | 102 | |
3.3. Species Identification
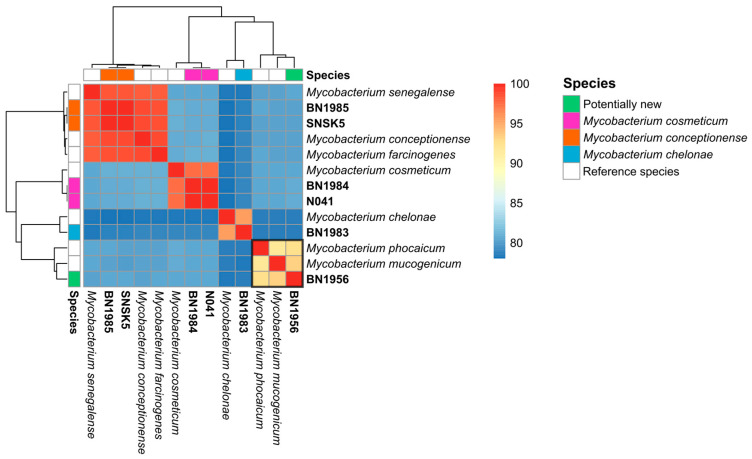
A comparison of species identification results based on the previous 16S rRNA sequence-based homology [11] and the current analysis using the assembled whole genome is summarized in Table 3. Initially, the following six isolates belonging to five different species were identified by DNA barcoding using the 16S rRNA amplicon sequence: Mycobacterium chelonae (BN1983), M. cosmeticum (BN1984 and N041), M. farcinogenes (SNSK5), M. senegalense (BN1985), and M. mucogenicum (BN1956). WGS and extended genomic comparisons with MLST, ANI, and dDDH led to the reclassification of these isolates (see Table 3). Isolates BN1985 and SNSK5 were identified as M. conceptionense, while BN1983 was confirmed as M. chelonae and BN1984 and N041 were confirmed as M. cosmeticum. There were discrepancies in species identification for some isolates when comparing the 16S rRNA sequences with the assembled draft genomes, particularly for isolates BN1985 and SNSK5. Analysis of isolate BN1956 revealed a previously unrecognized species that is proposed here as M. mucogenicum subsp. phocaicum sp. nov. The MLST analysis not only confirmed the identity of these isolates but also revealed similarities in the allele frequencies of housekeeping genes, particularly between isolates BN1984 and N041 and between isolates BN1985 and SNSK5. The similarity of the isolates was further supported by ANI (Figure 1) and dDDH scores (Supplementary Material Table S1). The MLST analysis also suggested that isolate BN1956 might represent a potential new species, as it had an ANI score < 95% compared to its closest relatives, implying that it is not the same species (Table 4).
Table 3.
Bacterial species identification based on 16S rRNA sequence homology compared to the assembled whole genome analyzed using multiple pipelines and databases.
| Sample ID | 16s rRNA * | MLST | PubMLST | TYGS | RAPT | DFAST | Conclusion |
|---|---|---|---|---|---|---|---|
| BN1956 | M. mucogenicum | Mycobacteria | M. phocaicum | Potential new species | Inconclusive | Inconclusive | M. mucogenicum subsp. phocaicum sp. nov. |
| BN1983 | M. chelonae | Mycobacteria | M. chelonae | M. chelonae | M. chelonae | M. chelonae | M. chelonae |
| BN1984 | M. cosmeticum | Mycobacteria | M. cosmeticum | M. cosmeticum | M. cosmeticum | M. cosmeticum | M. cosmeticum |
| BN1985 | M. senegalense | Mycobacteria | M. conceptionense | M. senegalense | M. conceptionense | M. conceptionense | M. conceptionense |
| SNSK5 | M. farcinogenes | Mycobacteria | M. conceptionense | M. senegalense | M. conceptionense | M. conceptionense | M. conceptionense |
| N041 | M. cosmeticum | Mycobacteria | M. cosmeticum | M. cosmeticum | M. cosmeticum | M. cosmeticum | M. cosmeticum |
* based on the work of Dinh-Hung et al. [11].
Figure 1.
Genomic relatedness heat map. This figure displays the relationships between the study isolates and their tentative reference genomes based on 16S rRNA and MLST proposed taxa. The genomes were compared using ANI. A darker red color indicates higher similarity, while shades of blue suggest greater differences.
Table 4.
ANI scores of isolate BN1956 against two closely related reference strains.
| Isolates | BN1956 | Mycobacterium mucogenicum | Mycobacterium phocaicum |
|---|---|---|---|
| GenBank accession | JAYXBQ000000000 | GCF_005670685.2 | GCF_010731115.1 |
| Size (bp) | 6,326,040 | 6,098,580 | 5,853,197 |
| GC (%) | 67.11 | 67.23 | 67.05 |
| OrthoANIu (%) | 93.26 | 92.51 | |
| dDDH (d4, in %) | 51.0 | 70.3 |
3.4. Type-Based Species and Subspecies Clustering
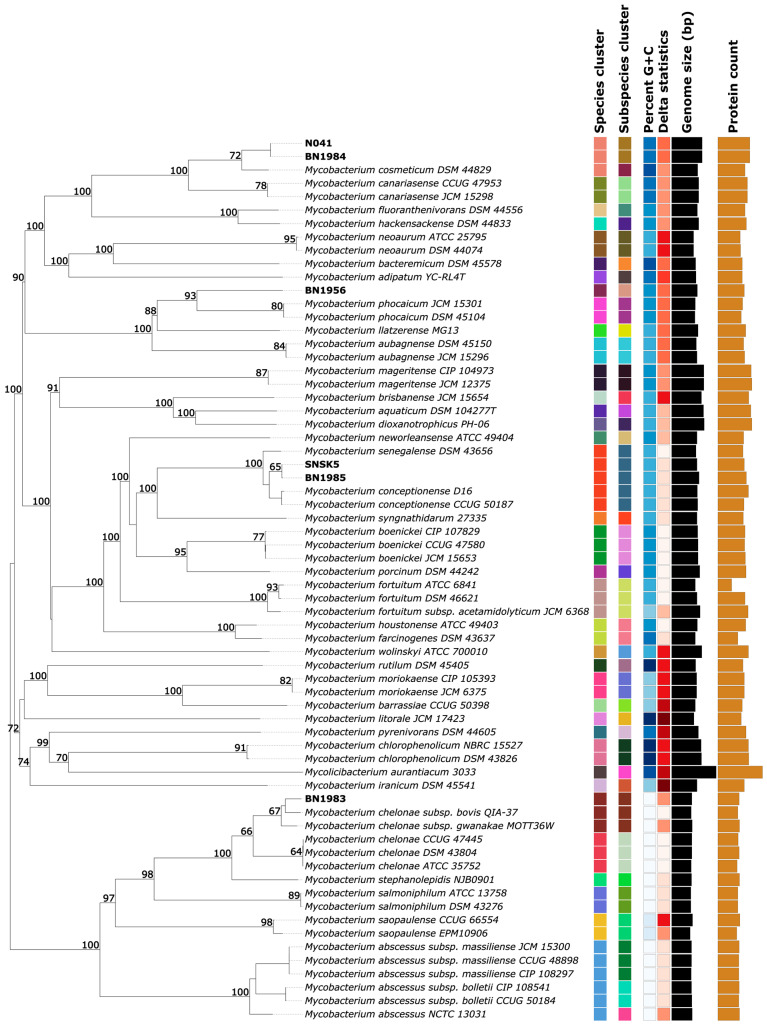
The taxonomic identification of the query strains, together with the respective results of dDDH, can be found in Supplementary Material Table S1. Isolates N041 and BN1984 formed a common cluster, indicating a close relationship, while isolates SNSK5 and BN1985 formed another distinct cluster (see Figure 2). Isolate BN1983 was clustered with its reference species, indicating a close evolutionary relationship. Notably, isolate BN1956 formed a separate cluster with a longer branch, suggesting a more distant relationship compared to the other isolates. This shows that our study species are included in 4 of the 40 subspecies clusters. Detailed information on the strains used for the phylogenetic analysis can be found in Supplementary Material Table S2.
Figure 2.
Tree derived with FastME 2.1.6.1 [34] from GBDP distances calculated from genome sequences. Branch lengths are scaled with respect to GBDP distance formula d5. The numbers above branches are GBDP pseudo-bootstrap support values > 60% from 100 replications, with an average branch support of 81.5%. The tree was rooted at the midpoint [35].
3.5. Comparative Genomic Analysis
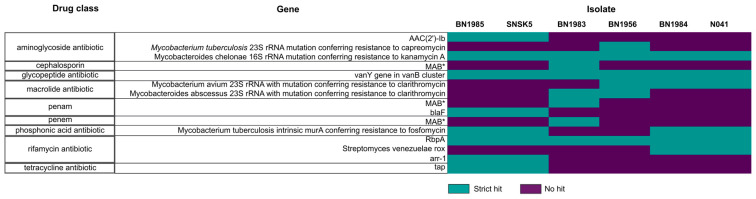
The analysis revealed plasmid sequences in the genomes of five isolates, except for isolate BN1983, and a BLAST search showed that these sequences matched significantly with those of other Mycobacterium species, suggesting a potential role for plasmid recombination within their natural context (Supplementary Material Table S3). Additionally, various ARGs were found in all six isolate genomes, conferring resistance to nine major drug classes (Figure 3). Furthermore, five major types of mobile genetic elements (MGEs) were detected in the genomes of the study isolates, including integration/excision elements, phages, replication/recombination/repair elements, transfer elements, and stability/transfer/defense elements. The MGEs were distributed across both putative plasmid and chromosome contigs. This highlights the importance of MGEs in horizontal gene transfer and the evolution of bacterial pathogens.
Figure 3.
Distribution of antimicrobial resistance genes among the 6 Mycobacterium isolates in this study. Genes with an asterisk (*) appear more than once because they belong to more than one drug class in the Antibiotic Resistance Ontology (ARO). Strict hit represents genes with a bitscore ≥500 when matched to the reference database.
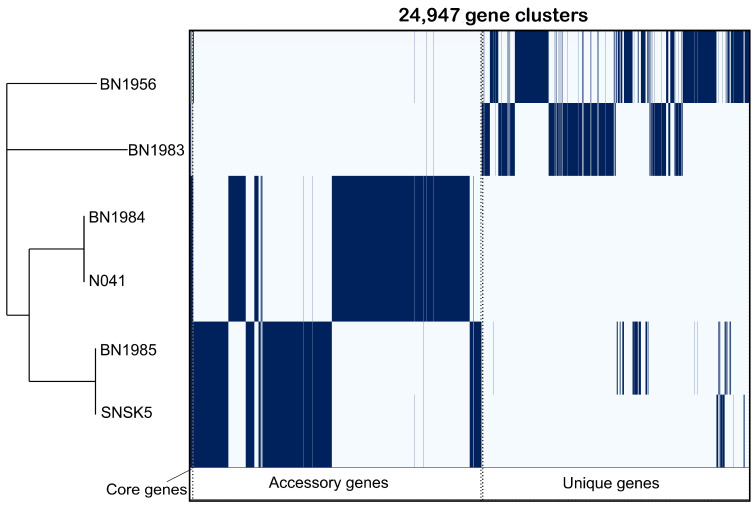
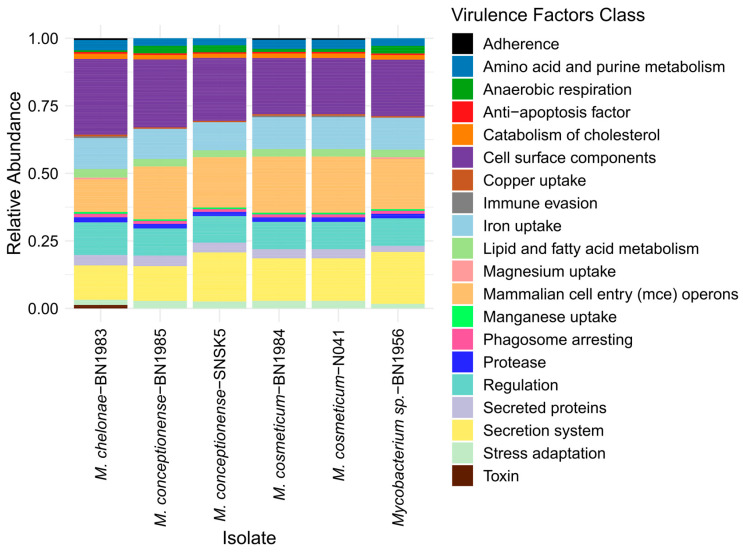
The pangenome analysis revealed a total of 24,947 gene clusters in the six isolates examined in this study. Among these, 25 were identified as core genes shared by all strains, while the remaining 24,922 were classified as shell genes (Figure 4). Remarkably, a considerable number of unique genes was detected in these isolates, underscoring the presence of isolate-specific genomic features. Functional annotation of the accessory genome revealed a variety of gene functions, including potential virulence factors, antibiotic resistance genes, and metabolic pathways. All six mycobacterial isolates in this study carried multiple virulence factors, as described in Supplementary Material Table S4. The distribution of these virulence factors varied between isolates, indicating differences in pathogenicity and adaptive mechanisms, as shown in Figure 5. The results indicated the presence of important virulence factors in Mycobacterium, such as the antigen 85 complex, DevR, and DevS, among many others. The findings were particularly intriguing, especially because M. chelonae-BN1983 exhibited virulence-associated toxin mycolactone, a feature that was absent in the other isolates. Additionally, M. chelonae BN1983 and M. cosmeticum (BN1984 and N041) were found to possess an additional class of adherence virulence factors encoded by the groEL gene, which distinguishes them from the other isolates analyzed in this study.
Figure 4.
Pangenome analysis of Mycobacterium isolates. The analysis was based on six isolates, categorizing the genes into core genes (shared by all strains), accessory genes (present in some strains), and unique genes (specific to individual strains). Each blue bar represents a gene.
Figure 5.
Illustration of the distribution of virulence factor classes among the six Mycobacterium isolates analyzed in this study. Each bar represents the relative abundance of the virulence factors and gives insight into the different virulence profiles of the isolates.
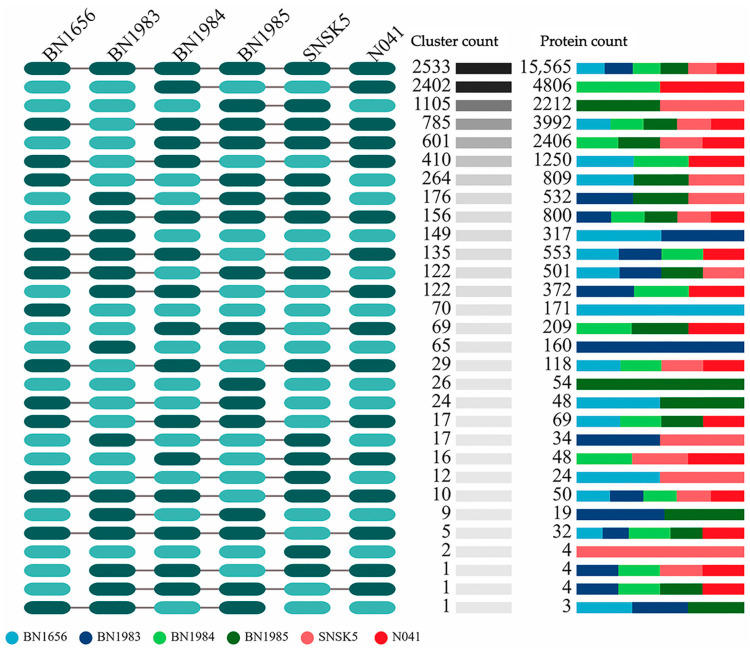
A comprehensive analysis of gene family clusters in six Mycobacterium isolates identified a total of 9334 clusters, of which 2533 occur together and 3215 occur individually (Figure 6). This analysis revealed different patterns in the number of unique gene clusters in the different species. Isolate BN1985, representing M. conceptionense, had 26 unique gene clusters, while another M. conceptionense isolate, SNSK5, had only 2 unique clusters. In contrast, M. chelonae isolate BN1983 had a significantly higher number of 65 unique gene clusters. Remarkably, M. cosmeticum isolates BN1984 and N041 had no unique gene clusters, indicating a greater similarity in gene content compared to other studied Mycobacterium species. The most significant discovery was a potential new Mycobacterium species represented by isolate BN1956, which had the highest number of unique gene clusters, at 70. These variations in the profiles of unique gene clusters illustrate the great genetic diversity within the Mycobacterium genus and provide insights into its evolutionary pathways and potential functional specializations.
Figure 6.
Illustration of the gene family clusters within the six mycobacterial isolates from this study. The diagram shows both the common and unique gene clusters with the respective number of proteins. The oval shapes in the inverted box on the left represent overlaps between the isolates. The dark-green shading represents overlapping gene families, while the light-green shading represents no overlap.
4. Discussion
In this study, we present the complete genomic sequences of six mycobacterial isolates that were previously analyzed for their phenotypic properties and pathogenicity [11,14]. Our research highlights the limitations of the 16S rRNA gene in accurately distinguishing between closely related species. Whole genome sequencing overcomes this limitation by enabling a more detailed analysis. For instance, the use of whole genome ANI and phylogenetic analysis of core genes provided convincing evidence for the reclassification of isolate BN1956 as a potential new species. Although we have provisionally assigned the designation Mycobacterium mucogenicum subsp. phocaicum sp. nov. to isolate BN1956 based on comprehensive genomic analyses, formal recognition of this new species is contingent upon compliance with the International Code of Nomenclature of Prokaryotes (ICNP) [52]. This process involves the description of the species in the International Journal of Systematic and Evolutionary Microbiology (IJSEM), deposition of the species in international culture collections, and validation by the International Committee on Systematics of Prokaryotes (ICSP). The formal description of the species will be the focus of a subsequent investigation, requiring the completion of all prescribed taxonomic procedures. On the other hand, isolates BN1985 and SNSK5, which were initially identified by 16S rRNA analysis as M. senegalense and M. farcinogenes, respectively, were reclassified as M. conceptionense following whole genome sequencing. These results highlight the challenges and potential inaccuracies in identifying Mycobacterium species based solely on a single or limited number of target genes, emphasizing the importance of comprehensive genomic analysis for accurate taxonomic classification.
The accuracy of taxonomic classifications is fundamentally linked to the scope and quality of the reference databases used. This relationship is emphasized by the variability of taxonomic conclusions that can result from the use of different reference databases (see Table 3). To maintain consistency and ensure the reliability of our classifications, this study adhered to established standards for species delimitation by adopting ANI (average nucleotide identity) and dDDH (digital DNA–DNA hybridization) thresholds. Specifically, we followed a criterion of 95%+ coverage for ANI [19] and a threshold of 70% for dDDH [25], both of which are recognized as critical benchmarks for accurate species delimitation.
The identification of matching plasmid sequences in different Mycobacterium isolates in our study emphasizes the dynamic and adaptive nature of Mycobacterium genomes. It also underscores the crucial role of horizontal gene transfer in their evolutionary processes. This observation is supported by the findings of Chen et al. [53] and Redondo-Salvo et al. [54], which, together, emphasize the importance of plasmid-mediated genetic exchange for the rapid adaptation and diversification of bacterial species. All these results reflect the inherent genomic adaptability and shared evolutionary pathways between closely related bacterial species. Consequently, they prompt a re-evaluation of species delimitation criteria, taxonomic classification, and the influence of conserved plasmid sequences on functional traits. Furthermore, understanding the distribution of plasmids and their dynamic interactions is crucial for the assessment of potential risks associated with antibiotic resistance, virulence, and environmental adaptability [55]. Further research is essential to decipher the importance and functional impact of these plasmids within bacterial communities and their impact on ecological systems and public health. In addition, all six Mycobacterium isolates examined in this study exhibited a variety of ARGs that confer resistance to multiple classes of antibiotics. The results of in silico analysis are consistent with previously reported antibiotic susceptibility profiles for these isolates [11], highlighting the diversity of resistance mechanisms, including the alteration of antibiotic targets, the protection of these targets, inactivation of antibiotics, and the enhancement of antibiotic efflux. This multi-layered resistance underlines the complexity of dealing with these infections, requiring a deeper understanding of the underlying genetic mechanisms to develop effective treatment strategies.
The identification of virulence factors plays a central role in the assessment of bacterial pathogenicity, as these components are crucial for the ability of the bacterium to infect a host and cause clinical manifestation [56]. The genome sequence analysis of six isolates revealed the presence of significant number of virulence factors, including multiple copies of the antigen 85 complex (Ag85), which plays a key role in adherence. Ag85, which consists of mycolyltransferases found in mycobacteria, is instrumental in the pathogenesis of several species of this genus, particularly Mycobacterium tuberculosis, by contributing to the synthesis of the mycobacterial cell wall [57]. In addition, we identified the presence of DevR (also known as DosR) and DevS, regulatory proteins in mycobacteria that play a crucial role in the adaptation of the bacterium to environmental stress, especially under hypoxic conditions. The genes encoding proteins associated with “mycobacterial” virulence, such as the PE and PPE family proteins, MCE and YrbE proteins, were also detected in all six isolates, highlighting their widespread role in the virulence mechanism of these bacteria. These proteins play a crucial role in the adaptation strategy of the bacterium to survive under low-oxygen conditions [58]. All isolates possess catalase–peroxidase (katG), an enzyme of great importance for various mycobacterial species, including M. tuberculosis. katG is essential for the detoxification of reactive oxygen species and, thus, increases the bacterium’s resistance to oxidative stress [59]. The observed virulence factor profile is characteristic of the Mycobacterium genus and enables these bacteria to resist environmental stressors and effectively modulate or evade host immune defenses. Mycobacterium chelonae and M. cosmeticum appear to have acquired novel virulence genes from distantly related environmental bacteria, including Klebsiella sp., Neisseria sp., Clostridium sp., and Streptococcus sp. Horizontally transferred genes include the T6SS-II (clpB) system, as well as catalase (katA), GroEL (groel), and capsule (rmlb) genes. We hypothesize that these genes arose by horizontal gene transfer (HGT) and confer significant functional advantages to the recipient mycobacterial species. The T6SS-II system, for instance, is involved in bacterial pathogenesis and host–microbe interactions, potentially increasing the virulence of M. chelonae and M. cosmeticum [60]. Similarly, the catalase and GroEL genes may have enhanced the oxidative stress response and protein-folding ability of these bacteria, contributing to their environmental fitness [61].
These discoveries affirm the crucial role of HGT as an evolutionary mechanism that enables bacteria to rapidly acquire new genetic traits from diverse sources, influencing their phenotypic characteristics and ecological adaptations [62]. The identification of these genes in M. chelonae and M. cosmeticum underscores the importance of comparative genomics in unraveling the complex evolutionary pathways of non-tuberculous mycobacterial species. Notably, M. chelonae-BN1983 was the only isolate in our study that produced mycolactones, which are complex macrolide toxins typically associated with Mycobacterium ulcerans and linked to Buruli ulcer, a necrotizing skin disease. These toxins have immunosuppressive and cytotoxic effects [63]. The elevated mortality rate observed in B. splendens following experimental exposure to M. chelonae-BN1983 [11,12] could be attributed to the strain’s unique genetic profile, which includes mycolactone-encoding genes responsible for producing potent toxins usually found in highly virulent mycobacterial species. Furthermore, its genome contains numerous genes encoding cell surface components, including members of the Mycobacterial membrane protein Large (MmpL) family, such as MmpL4a. MmpL proteins, particularly MmpL4a, which are known to play key roles in mycobacterial virulence and drug resistance [64], suggesting that this strain possesses a unique potential for drug resistance and high virulence.
5. Conclusions
In summary, this study provides valuable insights into the genomic characterization of six rapidly growing non-tuberculous mycobacteria (RGM) from Siamese fighting fish (Betta splendens). Initial identification via 16S rRNA sequences revealed taxonomic ambiguities that led to comprehensive whole genome sequencing (WGS) and advanced genomic analyses, such as multilocus sequence typing (MLST), average nucleotide identity (ANI), and digital DNA–DNA hybridization (dDDH). These methods led to the reclassification of certain species and the identification of a potential new species, namely Mycobacterium mucogenicum subsp. phocaicum sp. nov. The detection of shared virulence genes and horizontally acquired virulence factors in M. chelonae and M. cosmeticum emphasizes the pathogenicity and adaptability of these bacteria. This study provides the first draft genome sequences of RGM isolates in B. splendens and highlights the crucial role of WGS in understanding the genetic diversity and pathogenic potential of RGMs, contributing to improved diagnostic, therapeutic, and health management strategies for these infections.
Acknowledgments
N.D.-H. would like to thank Miho Furukawa for guidance on initial genomic analysis techniques.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ani14192833/s1, Table S1: Pairwise comparison for dDDH; Table S2: Strains for phylogeny; Table S3: Plasmids; Table S4: Virulence factors.
Author Contributions
Conceptualization, N.D.-H., S.M.M., H.T.D. and S.S.; methodology, W.M.; software, S.M.M.; validation, H.T.D. and S.S.; formal analysis, N.D.-H. and S.M.M.; writing—original draft preparation, N.D.-H. and S.M.M.; writing—review and editing, H.T.D., C.R., N.V.L., H.N.M., A.K.D., I.H., S.S. and S.C.; funding acquisition, S.S. and S.C. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
This study is in silico work. We obtained bacterial isolates from a previous study and conducted sequence and analysis using software only, with no related work involving animals or humans.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data that support the findings of this study are available upon request. The draft genomes of the six isolates were submitted to NCBI under BioProject PRJNA1063977.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This project was funded by the National Research Council of Thailand (NRCT) under grant no. N24A640089 and the National Science, Research, and Innovation Fund of Thailand Science Research and Innovation (TSRI) under grant no. FFB670076/0337.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Wolinsky E. Mycobacterial Diseases Other than Tuberculosis. Clin. Infect. Dis. 1992;15:1–12. doi: 10.1093/clinids/15.1.1. [DOI] [PubMed] [Google Scholar]
- 2.Tortoli E. Nontuberculous Mycobacteria (NTM): Microbiological, Clinical and Geographical Distribution. Academic Press; London, UK: Elsevier; London, UK: 2019. The Taxonomy of the Genus Mycobacterium; pp. 1–10. [DOI] [Google Scholar]
- 3.Shinnick T.M., Good R.C. Mycobacterial Taxonomy. Eur. J. Clin. Microbiol. Infect. Dis. 1994;13:884–901. doi: 10.1007/BF02111489. [DOI] [PubMed] [Google Scholar]
- 4.Tortoli E. Phylogeny of the Genus Mycobacterium: Many Doubts, Few Certainties. Infect. Genet. Evol. 2012;12:827–831. doi: 10.1016/j.meegid.2011.05.025. [DOI] [PubMed] [Google Scholar]
- 5.Gupta R.S., Lo B., Son J. Phylogenomics and Comparative Genomic Studies Robustly Support Division of the Genus Mycobacterium into an Emended Genus Mycobacterium and Four Novel Genera. Front. Microbiol. 2018;9:67. doi: 10.3389/fmicb.2018.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gauthier D.T., Rhodes M.W. Mycobacteriosis in Fishes: A Review. Vet. J. 2009;180:33–47. doi: 10.1016/j.tvjl.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 7.Rogall T., Wolters J., Flohr T., Bottger E.C. Towards a Phylogeny and Definition of Species at the Molecular Level within the Genus Mycobacterium. Int. J. Syst. Bacteriol. 1990;40:323–330. doi: 10.1099/00207713-40-4-323. [DOI] [PubMed] [Google Scholar]
- 8.Roetzer A., Diel R., Kohl T.A., Rückert C., Nübel U., Blom J., Wirth T., Jaenicke S., Schuback S., Rüsch-Gerdes S., et al. Whole Genome Sequencing versus Traditional Genotyping for Investigation of a Mycobacterium tuberculosis Outbreak: A Longitudinal Molecular Epidemiological Study. PLoS Med. 2013;10:e1001387. doi: 10.1371/journal.pmed.1001387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shelenkov A. Whole-Genome Sequencing of Pathogenic Bacteria—New Insights into Antibiotic Resistance Spreading. Microorganisms. 2021;9:2624. doi: 10.3390/microorganisms9122624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraser C.M., Eisen J.A., Salzberg S.L. Microbial Genome Sequencing. Nature. 2000;406:799–803. doi: 10.1038/35021244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dinh-Hung N., Dong H.T., Senapin S., Pimsannil K., Thompson K.D., Shinn A.P., Soontara C., Sirimanapong W., Chatchaiphan S., Rodkhum C. Insight into Characteristics and Pathogenicity of Five Rapidly Growing Non-Tuberculous Mycobacterium Species Isolated from the Siamese Fighting Fish, Betta Splendens. Aquaculture. 2023;575:739822. doi: 10.1016/j.aquaculture.2023.739822. [DOI] [Google Scholar]
- 12.Dinh-Hung N., Dong H.T., Senapin S., Linh N.V., Shinn A.P., Pirarat N., Hirono I., Chatchaiphan S., Rodkhum C. Infection and Histopathological Consequences in Siamese Fighting Fish (Betta Splendens) Due to Exposure to a Pathogenic Mycobacterium Chelonae via Different Routes. Aquaculture. 2024;579:740191. doi: 10.1016/j.aquaculture.2023.740191. [DOI] [Google Scholar]
- 13.Dinh-Hung N., Dong H.T., Senapin S., Shinn A.P., Linh N.V., Dien L.T., Soontara C., Hirono I., Chatchaiphan S., Rodkhum C. Using Ozone Nanobubbles to Mitigate the Risk of Mycobacteriosis in Siamese Fighting Fish (Betta Splendens) Aquaculture. 2024;581:740390. doi: 10.1016/j.aquaculture.2023.740390. [DOI] [Google Scholar]
- 14.Dong H.T., Senapin S., Phiwsaiya K., Techatanakitarnan C., Dokladda K., Ruenwongsa P., Panijpan B. Histopathology and Culturable Bacteria Associated with “Big Belly” and “Skin Nodule” Syndromes in Ornamental Siamese Fighting Fish, Betta Splendens. Microb. Pathog. 2018;122:46–52. doi: 10.1016/j.micpath.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 15.Gautam A. Phenol-Chloroform DNA Isolation Method. Springer; Cham, Switzerland: 2022. pp. 33–39. [DOI] [Google Scholar]
- 16.Salvà Serra F., Salvà-Serra F., Svensson-Stadler L., Busquets A., Jaén-Luchoro D., Karlsson R., Moore E.R.B., Gomila M. A Protocol for Extraction and Purification of High-Quality and Quantity Bacterial DNA Applicable for Genome Sequencing: A Modified Version of the Marmur Procedure. Protoc. Exch. 2018 doi: 10.1038/protex.2018.084. [DOI] [Google Scholar]
- 17.Chen Y., Chen Y., Shi C., Huang Z., Zhang Y., Li S., Li Y., Ye J., Yu C., Li Z., et al. SOAPnuke: A MapReduce Acceleration-Supported Software for Integrated Quality Control and Preprocessing of High-Throughput Sequencing Data. Gigascience. 2018;7:gix120. doi: 10.1093/gigascience/gix120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Souvorov A., Agarwala R., Lipman D.J. SKESA: Strategic k-Mer Extension for Scrupulous Assemblies. Genome Biol. 2018;19:153. doi: 10.1186/s13059-018-1540-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ciufo S., Kannan S., Sharma S., Badretdin A., Clark K., Turner S., Brover S., Schoch C.L., Kimchi A., DiCuccio M. Using Average Nucleotide Identity to Improve Taxonomic Assignments in Prokaryotic Genomes at the NCBI. Int. J. Syst. Evol. Microbiol. 2018;68:2386–2392. doi: 10.1099/ijsem.0.002809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tatusova T., Dicuccio M., Badretdin A., Chetvernin V., Nawrocki E.P., Zaslavsky L., Lomsadze A., Pruitt K.D., Borodovsky M., Ostell J. NCBI Prokaryotic Genome Annotation Pipeline. Nucleic Acids Res. 2016;44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seemann T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 22.Grant J.R., Enns E., Marinier E., Mandal A., Herman E.K., Chen C.Y., Graham M., Van Domselaar G., Stothard P. Proksee: In-Depth Characterization and Visualization of Bacterial Genomes. Nucleic Acids Res. 2023;51:W484–W492. doi: 10.1093/nar/gkad326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown C.L., Mullet J., Hindi F., Stoll J.E., Gupta S., Choi M., Keenum I., Vikesland P., Pruden A., Zhang L. MobileOG-Db: A Manually Curated Database of Protein Families Mediating the Life Cycle of Bacterial Mobile Genetic Elements. Appl. Environ. Microbiol. 2022;88:e00991-22. doi: 10.1128/aem.00991-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jolley K.A., Bliss C.M., Bennett J.S., Bratcher H.B., Brehony C., Colles F.M., Wimalarathna H., Harrison O.B., Sheppard S.K., Cody A.J., et al. Ribosomal Multilocus Sequence Typing: Universal Characterization of Bacteria from Domain to Strain. Microbiology. 2012;158:1005–1015. doi: 10.1099/mic.0.055459-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meier-Kolthoff J.P., Göker M. TYGS Is an Automated High-Throughput Platform for State-of-the-Art Genome-Based Taxonomy. Nat. Commun. 2019;10:2182. doi: 10.1038/s41467-019-10210-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meier-Kolthoff J.P., Carbasse J.S., Peinado-Olarte R.L., Göker M. TYGS and LPSN: A Database Tandem for Fast and Reliable Genome-Based Classification and Nomenclature of Prokaryotes. Nucleic Acids Res. 2022;50:D801–D807. doi: 10.1093/nar/gkab902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ondov B.D., Treangen T.J., Melsted P., Mallonee A.B., Bergman N.H., Koren S., Phillippy A.M. Mash: Fast Genome and Metagenome Distance Estimation Using MinHash. Genome Biol. 2016;17:132. doi: 10.1186/s13059-016-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lagesen K., Hallin P., Rødland E.A., Stærfeldt H.H., Rognes T., Ussery D.W. RNAmmer: Consistent and Rapid Annotation of Ribosomal RNA Genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meier-Kolthoff J.P., Auch A.F., Klenk H.P., Göker M. Genome Sequence-Based Species Delimitation with Confidence Intervals and Improved Distance Functions. BMC Bioinform. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoon S.H., Ha S.M., Lim J., Kwon S., Chun J. A Large-Scale Evaluation of Algorithms to Calculate Average Nucleotide Identity. Antonie Leeuwenhoek Int. J. General. Mol. Microbiol. 2017;110:1281–1286. doi: 10.1007/s10482-017-0844-4. [DOI] [PubMed] [Google Scholar]
- 32.Jain C., Rodriguez-R L.M., Phillippy A.M., Konstantinidis K.T., Aluru S. High Throughput ANI Analysis of 90K Prokaryotic Genomes Reveals Clear Species Boundaries. Nat. Commun. 2018;9:5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meier-Kolthoff J.P., Hahnke R.L., Petersen J., Scheuner C., Michael V., Fiebig A., Rohde C., Rohde M., Fartmann B., Goodwin L.A., et al. Complete Genome Sequence of DSM 30083T, the Type Strain (U5/41T) of Escherichia coli, and a Proposal for Delineating Subspecies in Microbial Taxonomy. Stand. Genom. Sci. 2014;9:2. doi: 10.1186/1944-3277-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lefort V., Desper R., Gascuel O. FastME 2.0: A Comprehensive, Accurate, and Fast Distance-Based Phylogeny Inference Program. Mol. Biol. Evol. 2015;32:2798–2800. doi: 10.1093/molbev/msv150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farris J.S. Estimating Phylogenetic Trees from Distance Matrices. Am. Nat. 1972;106:645–668. doi: 10.1086/282802. [DOI] [Google Scholar]
- 36.Kreft L., Botzki A., Coppens F., Vandepoele K., Van Bel M. PhyD3: A Phylogenetic Tree Viewer with Extended PhyloXML Support for Functional Genomics Data Visualization. Bioinformatics. 2017;33:2946–2947. doi: 10.1093/bioinformatics/btx324. [DOI] [PubMed] [Google Scholar]
- 37.Zhu Q., Gao S., Xiao B., He Z., Hu S. Plasmer: An Accurate and Sensitive Bacterial Plasmid Prediction Tool Based on Machine Learning of Shared k-Mers and Genomic Features. Microbiol. Spectr. 2023;11:e04645-22. doi: 10.1128/spectrum.04645-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wood D.E., Lu J., Langmead B. Improved Metagenomic Analysis with Kraken 2. Genome Biol. 2019;20:257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Florensa A.F., Kaas R.S., Clausen P.T.L.C., Aytan-Aktug D., Aarestrup F.M. ResFinder—An Open Online Resource for Identification of Antimicrobial Resistance Genes in next-Generation Sequencing Data and Prediction of Phenotypes from Genotypes. Microb. Genom. 2022;8:000748. doi: 10.1099/mgen.0.000748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sherry N.L., Horan K.A., Ballard S.A., Gonçalves da Silva A., Gorrie C.L., Schultz M.B., Stevens K., Valcanis M., Sait M.L., Stinear T.P., et al. An ISO-Certified Genomics Workflow for Identification and Surveillance of Antimicrobial Resistance. Nat. Commun. 2023;14:60. doi: 10.1038/s41467-022-35713-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feldgarden M., Brover V., Haft D.H., Prasad A.B., Slotta D.J., Tolstoy I., Tyson G.H., Zhao S., Hsu C.H., McDermott P.F., et al. Validating the AMRFinder Tool and Resistance Gene Database by Using Antimicrobial Resistance Genotype-Phenotype Correlations in a Collection of Isolates. Antimicrob. Agents Chemother. 2019;63:10–1128. doi: 10.1128/AAC.00483-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jia B., Raphenya A.R., Alcock B., Waglechner N., Guo P., Tsang K.K., Lago B.A., Dave B.M., Pereira S., Sharma A.N., et al. CARD 2017: Expansion and Model-Centric Curation of the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2017;45:D566–D573. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., Aarestrup F.M., Larsen M.V. Identification of Acquired Antimicrobial Resistance Genes. J. Antimicrob. Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gupta S.K., Padmanabhan B.R., Diene S.M., Lopez-Rojas R., Kempf M., Landraud L., Rolain J.M. ARG-ANNOT, a New Bioinformatic Tool to Discover Antibiotic Resistance Genes in Bacterial Genomes. Antimicrob. Agents Chemother. 2014;58:212–220. doi: 10.1128/AAC.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen L., Zheng D., Liu B., Yang J., Jin Q. VFDB 2016: Hierarchical and Refined Dataset for Big Data Analysis—10 Years On. Nucleic Acids Res. 2016;44:D694–D697. doi: 10.1093/nar/gkv1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carattoli A., Zankari E., Garciá-Fernández A., Larsen M.V., Lund O., Villa L., Aarestrup F.M., Hasman H. In Silico Detection and Typing of Plasmids Using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ingle D.J., Valcanis M., Kuzevski A., Tauschek M., Inouye M., Stinear T., Levine M.M., Robins-Browne R.M., Holt K.E. In Silico Serotyping of E. coli from Short Read Data Identifies Limited Novel O-Loci but Extensive Diversity of O:H Serotype Combinations within and between Pathogenic Lineages. Microb. Genom. 2016;2:e000064. doi: 10.1099/mgen.0.000064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Doster E., Lakin S.M., Dean C.J., Wolfe C., Young J.G., Boucher C., Belk K.E., Noyes N.R., Morley P.S. MEGARes 2.0: A Database for Classification of Antimicrobial Drug, Biocide and Metal Resistance Determinants in Metagenomic Sequence Data. Nucleic Acids Res. 2020;48:D561–D569. doi: 10.1093/nar/gkz1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alcock B.P., Huynh W., Chalil R., Smith K.W., Raphenya A.R., Wlodarski M.A., Edalatmand A., Petkau A., Syed S.A., Tsang K.K., et al. CARD 2023: Expanded Curation, Support for Machine Learning, and Resistome Prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2023;51:D690–D699. doi: 10.1093/nar/gkac920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Page A.J., Cummins C.A., Hunt M., Wong V.K., Reuter S., Holden M.T.G., Fookes M., Falush D., Keane J.A., Parkhill J. Roary: Rapid Large-Scale Prokaryote Pan Genome Analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Minh B.Q., Schmidt H.A., Chernomor O., Schrempf D., Woodhams M.D., Von Haeseler A., Lanfear R., Teeling E. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oren A., Arahal D.R., Göker M., Moore E.R.B., Rossello-Mora R., Sutcliffe I.C. International Code of Nomenclature of Prokaryotes. Prokaryotic Code (2022 Revision) Int. J. Syst. Evol. Microbiol. 2023;73:005585. doi: 10.1099/ijsem.0.005585. [DOI] [PubMed] [Google Scholar]
- 53.Chen C.Y., Fuqua C., Jackson C.R., Kadlec K., Top E.M. Editorial: Plasmid Transfer-Mechanisms, Ecology, Evolution and Applications. Front. Microbiol. 2022;13:993628. doi: 10.3389/fmicb.2022.993628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Redondo-Salvo S., Fernández-López R., Ruiz R., Vielva L., de Toro M., Rocha E.P.C., Garcillán-Barcia M.P., de la Cruz F. Pathways for Horizontal Gene Transfer in Bacteria Revealed by a Global Map of Their Plasmids. Nat. Commun. 2020;11:3602. doi: 10.1038/s41467-020-17278-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Movahedzadeh F., Bitter W. Ins and Outs of Mycobacterial Plasmids. Methods Mol. Biol. 2008;465:217–228. doi: 10.1007/978-1-59745-207-6_14. [DOI] [PubMed] [Google Scholar]
- 56.Deng Y., Xu L., Chen H., Liu S., Guo Z., Cheng C., Ma H., Feng J. Prevalence, Virulence Genes, and Antimicrobial Resistance of Vibrio Species Isolated from Diseased Marine Fish in South China. Sci. Rep. 2020;10:14329. doi: 10.1038/s41598-020-71288-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takayama K., Wang C., Besra G.S. Pathway to Synthesis and Processing of Mycolic Acids in Mycobacterium tuberculosis. Clin. Microbiol. Rev. 2005;18:81–101. doi: 10.1128/CMR.18.1.81-101.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saini D.K., Malhotra V., Dey D., Pant N., Das T.K., Tyagi J.S. DevR-DevS Is a Bona Fide Two-Component System of Mycobacterium tuberculosis That Is Hypoxia-Responsive in the Absence of the DNA-Binding Domain of DevR. Microbiology. 2004;150:865–875. doi: 10.1099/mic.0.26218-0. [DOI] [PubMed] [Google Scholar]
- 59.Ng V.H., Cox J.S., Sousa A.O., MacMicking J.D., McKinney J.D. Role of KatG Catalase-Peroxidase in Mycobacterial Pathogenesis: Countering the Phagocyte Oxidative Burst. Mol. Microbiol. 2004;52:1291–1302. doi: 10.1111/j.1365-2958.2004.04078.x. [DOI] [PubMed] [Google Scholar]
- 60.Behra P.R.K., Pettersson B.M.F., Ramesh M., Dasgupta S., Kirsebom L.A. Insight into the Biology of Mycobacterium mucogenicum and Mycobacterium neoaurum Clade Members. Sci. Rep. 2019;9:19259. doi: 10.1038/s41598-019-55464-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Choo S.W., Ang M.Y., Dutta A., Tan S.Y., Siow C.C., Heydari H., Mutha N.V.R., Wee W.Y., Wong G.J. MycoCAP—Mycobacterium Comparative Analysis Platform. Sci. Rep. 2015;5:18227. doi: 10.1038/srep18227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xia X. Horizontal Gene Transfer and Drug Resistance Involving Mycobacterium tuberculosis. Antibiotics. 2023;12:1367. doi: 10.3390/antibiotics12091367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hong H., Demangel C., Pidot S.J., Leadlay P.F., Stinear T. Mycolactones: Immunosuppressive and Cytotoxic Polyketides Produced by Aquatic Mycobacteria. Nat. Prod. Rep. 2008;25:447–454. doi: 10.1039/b803101k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chalut C. MmpL Transporter-Mediated Export of Cell-Wall Associated Lipids and Siderophores in Mycobacteria. Tuberculosis. 2016;100:32–45. doi: 10.1016/j.tube.2016.06.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available upon request. The draft genomes of the six isolates were submitted to NCBI under BioProject PRJNA1063977.