Abstract
Epstein-Barr virus nuclear antigen 2 (EBNA2) is essential for viral transformation of B cells and transactivates cellular and viral target genes by binding RBPJκ tethered to cognate promoter elements. EBNA2 interacts with the DEAD-box protein DP103 (DDX20/Gemin3), which in turn is complexed to the survival motor neuron (SMN) protein. SMN is implicated in RNA processing, but a role in transcriptional regulation has also been suggested. Here, we show that DP103 and SMN are complexed in B cells and that SMN coactivates the viral LMP promoter in the presence of EBNA2 in reporter gene assays and in vivo. Subcellular localization studies revealed that nuclear gems and/or coiled bodies containing DP103 and SMN are targeted by EBNA2. Protein-protein interaction experiments demonstrated that DP103 binds to SMN exon 6 and that both EBNA2 and SMN interact with the C terminus of DP103. Furthermore, a DP103 binding-deficient SMN mutant was released from nuclear gems and/or coiled bodies and further enhanced coactivation. In addition, impaired transactivation of a DP103 binding-deficient EBNA2 mutant was rescued by overexpression of SMN. Testing different promoter constructs in luciferase assays showed that RBPJκ is required but not sufficient for coactivation by EBNA2 and SMN. Overall, our data suggest that EBNA2 might target spliceosomal complexes by binding to DP103, thereby releasing SMN which subsequently exerts a coactivational function within the RNA-polymerase II transcription complex on the LMP1 promoter.
The Epstein-Barr virus (EBV) causes infectious mononucleosis and is linked to the genesis of several human lymphoproliferative diseases (for a review, see reference 33). The EBV-encoded nuclear antigen 2 (EBNA2) is a viral transactivator essential for EBV-induced transformation of resting human B lymphocytes, by promoting the expression of the transforming latent membrane proteins LMP1 and 2, the nuclear EBV Cp promoter-driven EBNA proteins, and the cellular genes CD23 and c-fgr (for review, see reference 15). EBNA2 does not bind directly to DNA but exerts its function by interacting with the cellular proteins RBPJκ (CBF1) and, on the more complex LMP1 promoter, also Spi1 (PU.1), tethered to cognate response elements (12, 17, 20, 45, 46). Transcriptional activation is induced by binding of the C-terminal acidic domain (5) to components of the basal RNA polymerase II transcription machinery, such as RPA70, TAF40, TFIIB, and TFIIH (38, 39), and recruitment of the coactivators p300, CBP, and PCAF histone deacetylase (14, 41). In addition, by attracting the hSWI/SNF complex (42, 43) and targeting histone H1 (9, 34), EBNA2 likely promotes relief of nucleosome-mediated gene repression.
We have recently shown that EBNA2 binds to DP103, a novel member of the DEAD-box family of putative RNA helicases (10). DP103 is a ubiquitously expressed 103-kDa phosphoprotein with an RNA-dependent ATPase activity; its other functions, in particular with regard to its interaction with EBNA2, remained unknown. While the work presented here was in progress, an interaction of DP103 (alternatively called Gemin3 [2]) and the survival motor neuron (SMN) protein, and their respective murine homologues, were described in two independent studies (1, 2).
SMN is part of a multiprotein complex containing SIP1, DP103 (Gemin3), GIP1 (Gemin4), and several Sm proteins that is involved in the assembly and nuclear regeneration of snRNPs and spliceosomes (2, 7, 25, 31). Both SMN and DP103 are localized in the cytoplasm and distinct nuclear structures, described as coiled bodies and gems (gemini of coiled bodies) (2, 21, 31). Mutations in the SMN gene result in spinal muscular atrophy (SMA), a recessive genetic disease with loss of α-motor neurons in the spinal cord, leading to muscle weakness and subsequent death. The SMN gene exists in two inverted copies within the same chromosomal region on chromosome 5q13 (19). In most SMA patients, a mutated telomeric form of the SMN gene results in a nonfunctional exon 7-deleted SMN, unable to self-associate (22), which cannot be compensated by the low amounts of full-length SMN protein expressed from the centromeric allele (24). In a few cases of SMA, point mutations were described which exchange amino acid (aa) 272 (Y272C) (19) or aa 134 (E134K) (4), affecting a putative RNA binding tudor domain (26). Furthermore, knockout of the murine SMN gene or its yeast homologue Yab8p resulted in a lethal phenotype (11, 28, 35).
Interestingly, a role for SMN in transcriptional regulation has been implicated, since SMN was shown to interact with the bovine papillomavirus E2 transactivator and to coactivate an E2-responsive viral promoter (1, 36). Furthermore, Ou et al. demonstrated that murine dp103 is also involved in transcriptional regulation by negatively modulating the expression of steroidogenic factor-1 (27). Finally, the SMN complex has recently been shown to associate with the C-terminal domain (CTD) of RNA polymerase II (30), although the functional consequences of this interaction have not yet been elucidated.
Searching for DP103-associated cellular proteins by using the yeast two-hybrid system, we also identified SMN as an interaction partner of DP103. Here, we show that this interaction is also relevant in B cells and that SMN is able to coactivate the viral LMP1 promoter in the presence of EBNA2 in vitro and in vivo. Data obtained from analyzing different EBNA2, DP103, and SMN mutants regarding their binding domains, subcellular distribution, and influence on EBNA2-mediated transactivation suggest that SMN is a novel factor involved in EBNA2-mediated transactivation of the viral LMP1 promoter: by targeting of DP103 within spliceosomal complexes, EBNA2 subsequently releases transcriptionally active SMN, which functions as a coactivator, likely within the RNA polymerase II transcription complex.
MATERIALS AND METHODS
Cell lines and antibodies.
Raji cells, derived from an EBV-positive Burkitt's lymphoma, EBV-negative BJAB B-lymphoma cells, and EBV-positive P3HR1 cells, harboring an EBNA2-deleted virus strain, were maintained in RPMI 1640 supplemented with 10% fetal calf serum, antibiotics and 1 mM sodium pyruvate as described previously (10). Human HeLa and 293GP cells were maintained in Dulbecco modified Eagle medium supplemented as described above. Monoclonal antibodies (MAbs) 9A3 and 8H4 directed against DP103, MAb R3 directed against EBNA2, and MAb S12 directed against LMP1 have been described (10, 16, 23). A polyclonal goat antiserum directed against the N terminus of human SMN protein was purchased from Santa Cruz Biochemicals; anti-β-actin MAb was obtained from Sigma. Anti-SMN MAb 7B10 (25) was kindly provided by U. Fischer (MPI für Biochemie, Martinsried, Germany). MAb 3F10 directed against the hemagglutinin (HA)-epitope sequence YPYDVPDYA and MAb 9E10 against the c-myc-epitope sequence EQKLISEEDL were from Roche Molecular Biochemicals.
Plasmids.
DNA manipulations were carried out according to standard procedures. pACT2 SMN wild type (WT) was derived from a positive scoring yeast clone, subcloned into pGEM-T (pGEM-T SMN WT), and sequenced. A WT SMN fragment was PCR amplified from from pACT2 SMN WT by using primers EcoHASMN (5′-GCGGAATTCCACCATGTACCCTTACGATGTACCGGATTACGCAGCGATGAGCAGCGGCGGCAGTGGT-3′) and SMN3′XhoBam (5′-CGCGGATCCTCGAGCTGCTCTATGCCAGCA-3′), EcoRI/BamHI digested, and ligated into pSG5 (Stratagene) to generate pSG5-HA SMN WT. Exon mutants of SMN (ΔEx7, ΔEx6/7, and ΔEx5-7) were PCR amplified from pACT2 SMN WT by using 5′-primer SMN5′Bam1 (5′-CGCGGATCCATGGCGATGAGCAGCGG-3′) and the respective 3′-primers 3′SMNd7stop (5′-CGCCTCGAGTTACATATAATAGCCAGT-3′), 3′SMNd6,7stop (5′-CGCCTCGAGTTATGGTGGTCCAGAAGG-3′), and 3′SMNd5,6,7stop (5′-CGCCTCGAGTTACTTTCCTGGTCCCAG-3′). PCR products were BamHI/XhoI digested and ligated into PACT2 for yeast two-hybrid analysis. BglII fragments, including the sequences encoding the HA tag from these pACT2 constructs, were ligated into pSG5 to generate the corresponding pSG5-HA constructs. pSG5-myc SMN and pSG5-HA SMN ΔN27 were synthesized by PCR using the 5′-primer Myc5′EcoSMN (5′-GCGGAATTCCATATGGAGCAAAAGCTAATATCGGAAGAAGATCTCGCGATGAGCAGCGGCGGCAGTGG-3′) or Eco-HA-SMN27 (5′-GCGGAATTCCACCATGTACCCTTACGATGTACCGGATTACGCAGCGAGCGATGATTCTGACATTTGG-3′) and 3′-primer SMN3′XhoBam (5′-CGCGGATCCTCGAGCTGCTCTATGCCAGCA-3′) and ligation of the EcoRI/BamHI-digested fragments into pSG5. Ligation of an NcoI/SalI fragment from pGEM-T DP103 (10) into pACT2 resulted in pACT2 DP103. pSG5-HA DP103 WT, Δ456-547, and Δ341-461 were generated by ligation of a DP103 BglII fragment from the respective pACT2 DP103 mutants into pSG5. Point mutations (pSG5-HA SMN E134K and Y272C; pSG55-HA DP103 and K112N) were introduced by site-directed mutagenesis of the WT pSG5-HA constructs by using Pfu Turbo DNA-Polymerase (Stratagene) and primers introducing the respective mutation and a new, unique restriction site. For expression as enhanced green fluorescence fusion protein (EGFP), an EBNA2 EcoRI/BglII fragment was cloned from pSG5 into pEGFP-C1 (Clontech). A WT SMN fragment was synthesized by PCR from pGEM-T SMN WT by using primers C1EGFP-SMN5′ (5′-GCGAATTCCATGGCGATGAGCAGCGGC-3′) and SMN3′XhoBam (see above), EcoR/BamHI digested, and ligated into pEGFP-C1 (Clontech) to yield pEGFP-C1 SMN WT. pSG5-luc was generated by ligating a SalI/Bg II fragment to a SalI/BglII-digested LL0 luciferase plasmid (18), thereby replacing the LMP1 promoter by the simian virus 40 promoter and a β-globin intron. The EBNA2 mutants pSG5 EBNA2 Δ121-216 and pSG5 EBNA2 322 were generated by replacing an internal M-ABA strain BamHI fragment of pSG5-EBNA2 WT by BamHI fragments which were PCR amplified from pSG5 EBNA2 WT or pAC EBNA2-HaeII1 (34) with the primers BamHI213E25′ (5′-GCGGATCCGCCACCAAGGCCTACCCGTC-3′) and E2wtBglII3′ (5′-AGATCTTACTGGATGGAGGGGCGA-3′) or the primers EcoXho5E2 (5′-GCCGAATTCTCGAGGCCATCATGCCTACATTCTATCTTGCGTTA-3′) and BglXho-3E2 (5′-CGAAGATCTCGAGTTACTGGATGGAGGGGCGAGGTCT-3′). All constructs were sequenced by using the Biozym sequencing kit. pSG5 EBNA2 WT was a generous gift from M. Rowe (University of Wales, Cardiff, United Kingdom). Luciferase constructs LL0 to LL9 (18) and pGa981-21 and pGa50-7 (37) were kindly provided by G. Laux and U. Zimber-Strobl (GSF).
Yeast two-hybrid analysis.
For a review of the yeast two-hybrid system, see Phizicky and Fields (32). The complete open reading frame of DP103 was PCR amplified by using the original isolate pGEM DP103 (10) and the primers BglII5′DP103 (5′-GGAAGATCTGCCATGGCGGCGGCATTTGAAGC-3′) and Sp6 (5′-TATTTAGGTGACACTATAG-3′) and then BglII/PstI digested and inserted into the BamHI/PstI-digested vector pAS2-1 (Matchmaker Two-Hybrid System 2; Clontech) to generate vector pAS DP103 expressing a Gal4 DNA-binding domain fused to DP103. The expression of a fusion protein with the appropriate size in the yeast strain Y190 was verified by using the DP103-specific MAb 8H4. This construct was used to screen a lymphocyte-derived cDNA library (Clontech Matchmaker Library; Clontech). Resulting clones were segregated by using cycloheximide and appropriate media and retested for specific binding by mating with yeast containing either DP103 or an unspecific bait. Clones scoring positive were rescued in Escherichia coli and sequenced with the Biozym sequencing kit. Quantification of β-galactosidase activity was carried out by liquid culture assay by using ONPG (o-nitrophenyl-β-d-galactopyranoside) according to the Clontech Matchmaker 2 protocol.
Immunoprecipitations.
For immunoprecipitation of endogenous proteins, Raji cells were washed in phosphate-buffered saline (PBS), extracted in lysis buffer (100 mM Tris-HCl [pH 8.0], 100 mM NaCl, 5 mM KCl, 0.5 mM MgCl2, 1 mM CaCl2, 0.5% [vol/vol] Triton X-100, and protease inhibitors), and incubated for 1 h at 4°C with anti-DP103 MAb 9A3 or an irrelevant control MAb (anti-trpE 3A6) absorbed to protein G-Sepharose (Pharmacia). Bound immune complexes were washed 10 times (5 times with 1 M NaCl and five times without NaCl) with radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris-HCl [pH 7.5], 0.5% [vol/vol] deoxycholate, 0.5% [vol/vol] NP-40, 0.1% sodium dodecyl sulfate [SDS], 0.1 mM EDTA, and protease inhibitors), released by boiling in gel loading buffer, and then subjected to SDS-polyacrylamide gel electrophoresis (PAGE) and Western blotting. Proteins were detected by using MAb 8H4 or anti-SMN-N serum (Santa Cruz Biotechnology). For mapping of the SMN and DP103 interaction domains, 293GP cells were grown in 10-cm dishes and transfected with HA-tagged SMN mutants or cotransfected with myc-tagged WT SMN and HA-tagged DP103 mutants by the calcium phosphate method. After growth for 36 to 48 h, cells were extracted in lysis buffer. Supernatants were incubated with anti-HA MAb 3F10 or unspecific control MAbs (anti-trpE 3A6 and anti-BDV p40 9C2) absorbed to protein G-Sepharose for 1 h at 4°C. Bound complexes of endogenous DP103 and transfected HA-tagged SMN mutants were washed 10 times (5 times with 1 M NaCl and five times without NaCl) with RIPA buffer; complexes of transfected myc-tagged WT SMN and HA-tagged DP103 mutants were washed two times in RIPA buffer (1 M NaCl) and four times in lysis buffer. Proteins were separated by boiling with gel loading buffer and then subjected to SDS-PAGE and Western blot analysis.
Transfection of B lymphocytes and luciferase assays.
BJAB and P3HR1 cells were transfected by electroporation by using a Bio-Rad Gene Pulser at 250 V and 950 μF, with slight modifications as described previously (45). Briefly, 107 cells were washed once and resuspended in 0.25 ml of ice-cold RPMI 1640 without supplements and placed on ice. Then, 4 μg of reporter plasmid, 10 μg of each respective effector plasmid, and 2 μg of pEGFP-C1 (Clontech) were added. Parental pSG5 vector (Stratagene) was used to adjust DNA amounts. After electroporation, cells were kept on ice for 10 min, suspended in 10 ml of RPMI with 20% fetal calf serum, and grown for 48 h. To determine the transfection efficiency, 100 μl of the cells was fixed and analyzed in a Becton Dickinson FACScan analyzer for EGFP-positive cells, gated on the living population. The remainder of cells were washed in PBS and lysed by three cycles of freeze-thawing in 250 mM Tris-HCl (pH 7.8). The luciferase activity of the supernatants was determined in a Lumat LB9501 (Berthold) by using the Promega luciferase assay system (Promega) as recommended by the manufacturer.
Immunofluorescence.
Immunofluorescence analysis with HeLa or BJAB cells was performed with slight modifications as described previously (6). Briefly, HeLa cells grown on cover slides in six-well plates were lipofected with 5 μg of DNA by SuperFect (Qiagen) according to the manufacturer's protocol. BJAB cells were electroporated with 10 μg of DNA as described above. After 24 h, the cells were washed with PBS, fixed with 4% paraformaldehyde–PBS at room temperature for 15 min, permeabilized with 0.2%Triton X-100–PBS (2 min, 4°C), and blocked with 2% bovine serum albumin (BSA)–PBS (15 min, 37°C). HA-tagged proteins were detected by using anti-HA MAb 3F10, and endogenous SMN protein was detected by using MAb 7B10, diluted in 3 μg of BSA-PBS/ml (45 min, 37°C), and TRITC (tetramethyl rhodamine isocyanate)-labeled anti-rat (Dianova) or anti-mouse (Sigma) MAb in 3 μg of BSA-PBS/ml as secondary antibodies (30 min, 37°C). Cover slides were mounted in Elvanol and subjected to immunofluorescence microscopy by using a Zeiss Axiovert 100 TV microscope and a Sony 3CCD camera (see Fig. 3D). Confocal images were taken by using a Nikon Eclipse E600 micoscope equipped with a PCM 2000 confocal laser-scanning system and analyzed by using Confocal Assistant 4.02 and use of Corel Photo Paint and Draw version 8.0 software.
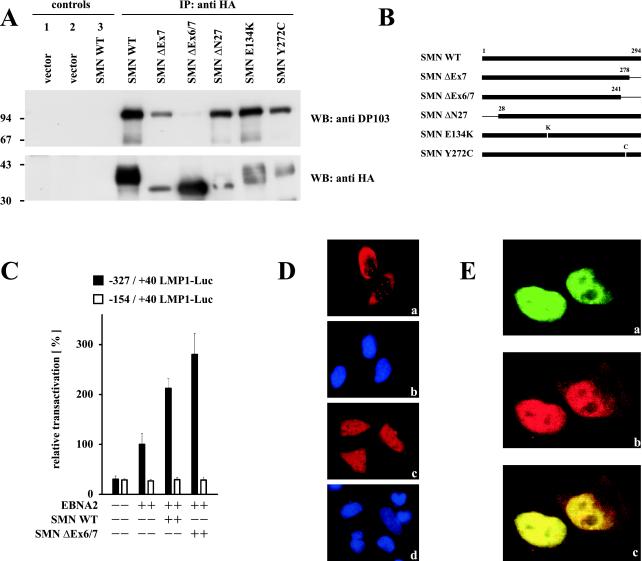
FIG. 3.
Enhanced coactivation of the LMP1 promoter by DP103 binding-deficient SMN ΔEx6/7. (A) Mapping of the DP103 binding site on SMN. 293GP cells were transfected with pSG5 constructs encoding HA-tagged SMN mutants (10 μg) as indicated. After 36 h native cell extracts were immunoprecipitated with anti-HA MAb 3F10 (IP: anti HA and control 2) or unspecific MAb 3A6 (controls 1 and 3) and analyzed by SDS–10% PAGE and Western blotting. Precipitated transfected SMN mutants were detected by using anti-HA MAb 3F10 (WB: anti HA), and coprecipitated endogenous DP103 was detected by using anti-DP103 MAb 8H4 (WB: anti DP103). The positions of the molecular mass markers (in kilodaltons) are indicated on the left side of each panel. Deletion of SMN exon 6 abolished coprecipitation of endogenous DP103. (B) Schematic representation of the SMN mutants tested. (C) Coexpression of EBNA2 and the HA-tagged DP103 binding-deficient SMN mutant SMN ΔEx6/7 further increased coactivation of the −327/+40 LMP1 promoter luciferase construct. Assays were performed as described for Fig. 1B. Graphs represent the mean values of three independent experiments performed in duplicate (±SEM). (D) Immunofluorescence of HA-tagged WT SMN (a) and SMN ΔEx6/7 (c) expressed in HeLa cells and stained with 3F10 anti-HA/anti-rat TRITC MAbs. (b and d) Nuclei were visualized by using DAPI (4′,6′-diamidino-2-phenylindole). Loss of binding to DP103 released SMN ΔEx6/7 from nuclear gems/coiled bodies. (E) Enhanced colocalization of EBNA2 and DP103 binding-deficient SMN ΔEx6/7. HeLa cells coexpressing EGFP-EBNA2 (a) and HA-tagged SMN ΔEx6/7 (b) were stained by using anti-HA 3F10/anti-rat TRITC MAbs and subjected to confocal laser scanning microscopy. In the merged image (subpanel c), colocalization results in a yellow signal.
GenBank accession number.
The nomenclature committee authorized by the human genome project (HUGO) proposed to rename the DP103 gene to DDX20 in keeping with the guidelines for nomenclature of DEAD/H-box proteins of putative RNA and DNA helicases (GenBank accession no. NM_007204).
RESULTS
DP103 (DDX20/Gemin3) and the SMN protein interact in B cells.
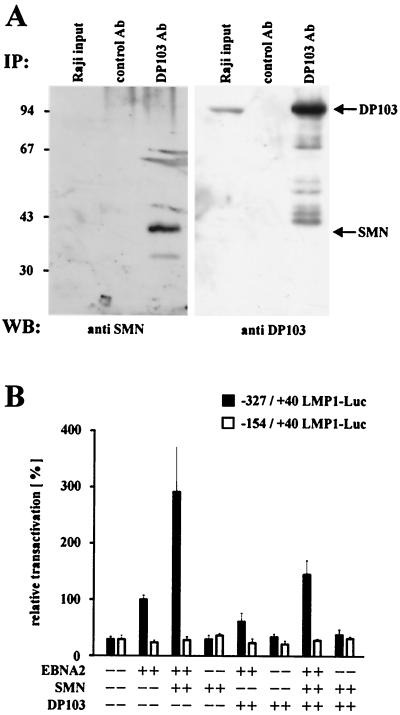
Recently, we have shown that EBNA2 associates with the cellular DEAD-box protein DP103 (10). Since the functional consequences of this interaction remained unknown, we sought to identify cellular proteins associating with DP103 in the yeast two-hybrid system. The screening of a B-lymphocyte cDNA library with full-length DP103 yielded nine individual clones: four containing the complete SMN gene and the remaining five containing 5′-truncated SMN genes (data not shown). This result was confirmed by an alternative experimental approach with data published while the present manuscript was in preparation (1, 2). Since EBV is a lymphotrophic virus, we sought to determine whether this interaction of DP103 and SMN could also be detected in B lymphocytes. Therefore, both endogenous proteins were coimmunoprecipitated from EBV-positive Raji lymphocytes by using the DP103-specific MAb 9A3 (Fig. 1A). An irrelevant control MAb neither precipitated DP103 nor coprecipitated SMN. Note that, due to the small amount of total cell lysate used as input and the low affinity of the goat anti SMN antibody, the amount of SMN in the input lane was below the level of detection.
FIG. 1.
(A) Interaction of DP103 and SMN in B lymphocytes. Coimmunoprecipitations (IP) from Raji cell extracts were performed with DP103-specific MAb 9A3 (IP: DP103 Ab) or an irrelevant control antibody (anti-TrypE 3A6, IP: control Ab), followed by SDS–10% PAGE and Western blotting. Precipitated proteins were detected with anti-SMN-N serum (Santa Cruz Biochemicals) (left panel, WB: anti SMN) or anti-DP103 MAb 8H4 (right panel, WB: anti DP103). The positions of SMN and DP103 are indicated by arrows. Lanes designated Raji input represent ca. 1% of unprecipitated Raji cell extract. The positions of the molecular mass marker proteins are indicated on the left side (in kilodaltons). (B) SMN coactivates the viral LMP1 promoter in the presence of EBNA2. BJAB cells were transfected with luciferase reporter constructs encoding positions −327/+40 (EBNA2 responsive) or −154/+40 (nonresponsive) of the LMP1 promoter (4 μg) and the indicated combinations of pSG5 constructs encoding EBNA2 or HA-tagged SMN and DP103 (10 μg). After 48 h, the cells were lysed by freeze-thawing, and the luciferase activity was measured. The transfection efficiency was determined by scanning the expression of cotransfected pEGFP-C1 vector (2 μg) by FACS analysis prior to lysis of the cells. For each experiment, luciferase values standardized for transfection efficiency were calculated relative to the values obtained by EBNA2 and the respective full-length promoter construct (set to 100%). Graphs represent the mean values of five independent experiments (± the standard error of the mean [SEM]).
SMN coactivates the viral LMP1 promoter in the presence of EBNA2.
To elucidate the role of DP103 and SMN regarding the function of EBNA2, the transactivation of its main target, the viral LMP1 promoter, was examined in EBV-negative BJAB lymphocytes. Coexpression of EBNA2 and SMN reproducibly increased EBNA2-mediated transactivation of a cotransfected full-length (−327/+40) LMP1 promoter luciferase construct (LL0) (18) by almost 300% (Fig. 1B), relative to the values obtained by EBNA2 alone (set to 100%). This effect could further be titrated by coexpressing EBNA2 and increasing amounts of SMN (Fig. 2A), indicating that SMN coactivates the viral LMP1 promoter in the presence of EBNA2. However, coexpression of both SMN and DP103 in the presence of EBNA2 resulted only in an increase of 150%. Furthermore, coexpression of DP103 and EBNA2 slightly decreased EBNA2-mediated transactivation to 75%. Note that in the absence of EBNA2 neither SMN, DP103, nor both showed any effect. A truncated, EBNA2 nonresponsive (−154/+40) reporter construct (LL4) (18) served as a negative control. Thus, SMN is capable of coactivating the LMP1 promoter in the presence of the viral transactivator EBNA2, resulting in increased amounts of luciferase transcripts and protein.
FIG. 2.
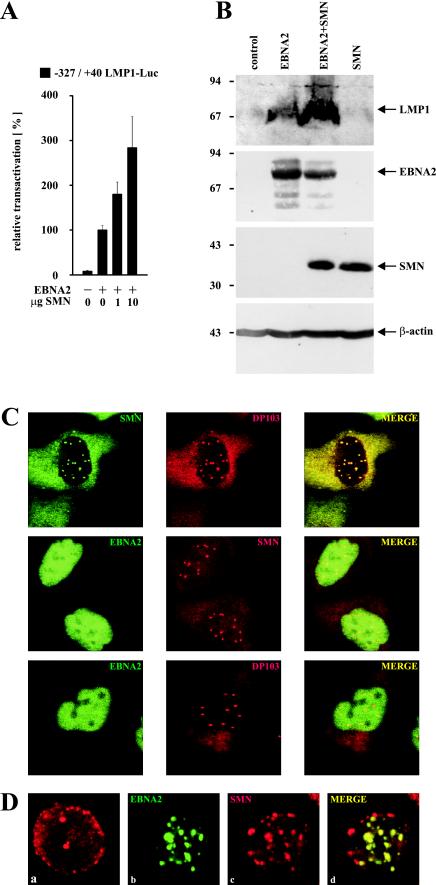
(A) Dose-dependent coactivation of the −327/+40 LMP1 promoter by coexpression of EBNA2 and increasing amounts of HA-tagged SMN (indicated in micrograms). Assays were performed as described for Fig. 1B. Graphs represent the mean values of three independent experiments performed in duplicate (±SEM). (B) SMN increases EBNA2-mediated induction of endogenous LMP1 protein. EBV-positive P3HR1 cells were transfected with pSG5 constructs (15 μg) encoding EBNA2 or HA-tagged SMN, as indicated. Cells were harvested and subjected to SDS–10% PAGE and Western blotting by using MAbs S12 (anti-LMP1), R3 (anti-EBNA2), 3F10 (anti-HA), and anti-β-actin MAb (Sigma). The positions of the respective proteins are indicated by arrows. The positions of the molecular mass markers (in kilodaltons) are indicated on the left side. (C) Subcellular distribution of EBNA2, DP103, and SMN. HeLa cells transfected with pSG5-HA (red signals) or pEGFP-C1 (green signals) constructs (5 μg) encoding the corresponding fusion proteins of EBNA2, DP103, or SMN were immunostained and analyzed by confocal laser scanning microscopy. HA-tagged proteins were visualized by using anti-HA 3F10/anti-rat TRITC MAbs. The localizations of coexpressed SMN and DP103 (upper panel), EBNA2 and SMN (middle panel), or EBNA2 and DP103 (lower panel) are shown. In the merged images, colocalization results in a yellow signal. (D) Subcellular localization of endogenous SMN and EGFP-EBNA2 in BJAB cells. BJAB cells mock transfected (a) or transfected with 10 μg of pEGFP-C1 EBNA2 (b, c, and d) were immunostained by using anti-SMN 7B10/anti-mouse TRITC MAbs and subjected to confocal laser scanning microscopy. In the merged image (subpanel d), colocalization results in a yellow signal.
To test whether these findings were also reflected in elevated levels of endogenous viral LMP1 protein, Western blot analysis of cotransfected P3HR1 cells was performed (Fig. 2B). In P3HR1 cells transfected with empty vector no expression of LMP1 could be detected, since these cells harbor an EBNA2-deleted, nontransforming EBV genome. Expression of EBNA2 induced LMP1 expression; coexpression of EBNA2 and SMN further increased the strength of the LMP1 signal, suggesting that coactivation of the LMP1 promoter by EBNA2 and SMN is also relevant in the context of latently EBV-infected B cells. Since SMN has been implicated in RNA processing, it is interesting to note that in this assay the level of EBNA2 expression was not elevated by coexpressed SMN. Furthermore, neither expression of EBNA2 nor coexpression of EBNA2 and SMN increased the activity of a pSG5 luciferase construct (pSG5-luc) tested in three independent experiments in BJAB cells (data not shown). Since all expression vectors used were pSG5 constructs containg the simian virus 40 promoter and a β-globin intron to facilitate protein synthesis, these results argue against a possible increase in luciferase activity or LMP1 expression due to a more efficient splicing by the overexpression of SMN. A second control experiment showed that the coactivation demonstrated above was also not due to an increased viability of cells overexpressing SMN, since a synergistic antiapoptotic activity of SMN and Bcl-2 had been described (13). Fluorescence-activated cell sorting (FACS) analysis of transfected P3HR1 cells from three independent experiments revealed that the relative quantity of gated, living cells did not significantly differ, irrespective whether they overexpressed EBNA2 and/or SMN (data not shown).
The direct interaction of EBNA2 and DP103 (10) and the functional cooperation of EBNA2 and SMN described here should be reflected in the subcellular distribution of these proteins. Therefore, EBNA2, DP103, and SMN were transiently coexpressed in HeLa cells as EGFP- or HA-tagged fusion proteins, immunostained, and subjected to confocal laser scanning microscopy (Fig. 2C). DP103 and SMN were detected dispersed in cytoplasm and distinct punctate subnuclear structures (gems or coiled bodies) (Fig. 2C, upper panel), as previously reported (2). Interestingly, these structures were also included within the strictly nuclear distribution pattern of EBNA2, when SMN (Fig. 2C, middle panel) or DP103 (Fig. 2C, lower panel) were coexpressed with EBNA2. In contrast, unlike its dispersed nuclear distribution in HeLa cells, expression of EGFP-EBNA2 in BJAB cells resulted in a speckled nuclear pattern. Endogenous SMN protein could be detected by immunostaining in cytoplasm and nuclear gems and/or coiled bodies (Fig. 2D, subpanel a) of mock-transfected BJAB cells. A portion of the cells, however, lacked the nuclear gems and/or coiled bodies or showed a diffusely dispersed nuclear staining (data not shown). Furthermore, the number of gems or coiled bodies detectable in BJAB cells (2 to 4 per cell) was lower than in HeLa cells (6 to 16 per cell). Strikingly, a relocation of endogenous SMN to a speckled nuclear pattern, partially colocalizing with the EGFP-EBNA2 signals, could be detected in cells transfected with EGFP-EBNA2 (Fig. 2D, b to d). Thus, if we take into account the limits of resolution provided by the system used, EBNA2 colocalizes with SMN and DP103 in HeLa cells within the same punctate subnuclear structures, most likely nuclear gems and/or coiled bodies. Furthermore, EGFP-EBNA2 expression in BJAB cells results in a relocation of endogenous SMN to nuclear speckles, partially colocalizing with EBNA2. These data suggest that EBNA2 can target nuclear gems or coiled bodies and, depending on the cell type, release SMN from these structures.
Enhanced coactivation of the LMP1 promoter by EBNA2 and a DP103 binding-deficient SMN mutant.
The results outlined in Fig. 1 showed that coactivation by EBNA2 and SMN was slightly decreased by the coexpression of DP103. This raised the question of whether binding to DP103 influences the ability of SMN to functionally cooperate with EBNA2. Therefore, several SMN mutants (depicted in Fig. 3B) were expressed in 293GP cells and tested for their ability to coimmunoprecipitate endogenous DP103 (Fig. 3A). The deletion of both exons 6 and 7 resulted in a loss of binding, whereas the SMA-associated deletion of exon 7 alone (SMN ΔEx7) or the two patient-derived point mutations (E134K and Y272C) did not abolish the interaction with DP103. Deletion of the first exon (SMN ΔN27), shown to be negative in spliceosomal assembly (7), also did not affect binding to DP103. This mapping of the DP103 binding site to exon 6 of SMN was confirmed by testing several deletion mutants, including those deleting exon 7 or exons 6 and 7 in the yeast two-hybrid system (data not shown).
Indirect immunofluorescence of transfected HeLa cells further revealed that, in contrast to WT SMN which was localized in the cytoplasm and nuclear gems or coiled bodies (Fig. 3D, a and b), the DP103 binding-deficient SMN ΔEx6/7 mutant was now also detectable within the whole nucleus. (Fig. 3D, c and d). Furthermore, confocal laser scanning microscopy of cotransfected HeLa cells showed colocalization of EBNA2 and SMN ΔEx6/7 within the same nuclear staining pattern (Fig. 3E), indicating that the loss of binding to DP103 releases significant amounts of this SMN mutant from nuclear gems or coiled bodies. Interestingly, compared to WT SMN, SMN ΔEx6/7 had an increased potential to coactivate the viral LMP1 promoter in the presence of EBNA2, as demonstrated by luciferase assays performed in BJAB cells (Fig. 3C). In contrast, neither the SMN mutant negative in spliceosomal assembly (SMN ΔN27) nor the SMA-associated SMN mutants (ΔEx7, E134K, and Y272C) were able to coactivate the LMP1 promoter above the level of WT SMN in the presence of EBNA2 (data not shown), suggesting that the shown coactivation involves properties of SMN distinct from its function in spliceosomal assembly.
EBNA2-mediated transactivation of the LMP1 promoter involves binding of EBNA2 to DP103.
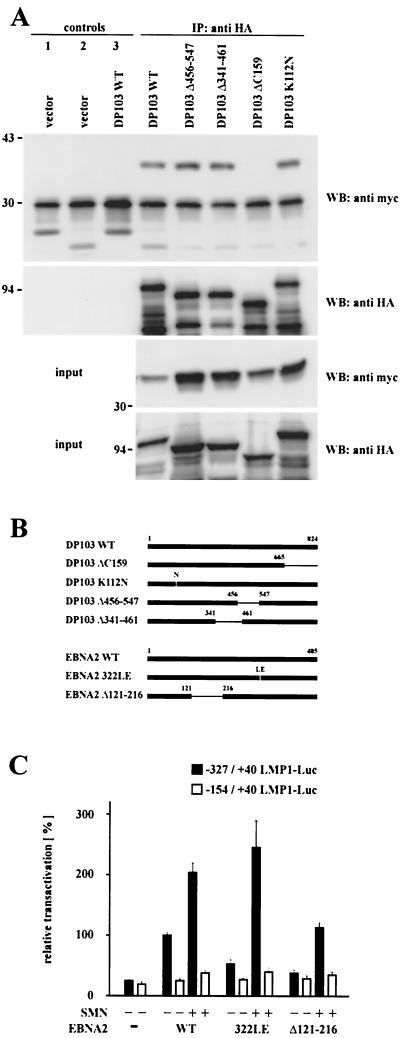
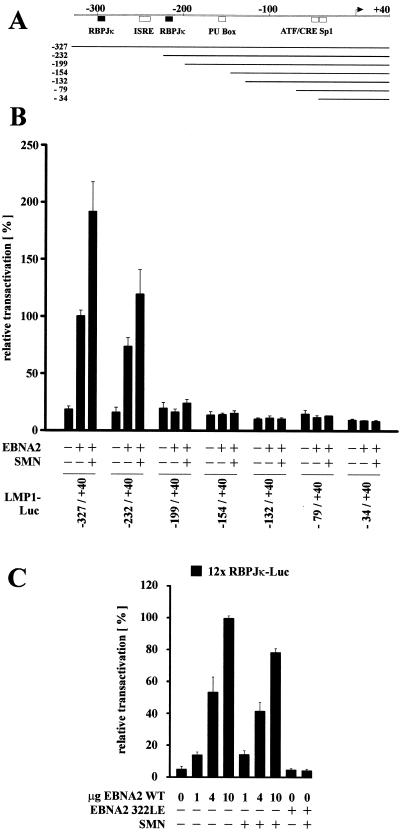
Our previously published data (10) demonstrated that EBNA2 interacts with the C terminus (aa 665 to 824) of DP103. Since it was shown here that coexpression of EBNA2 and a DP103 binding-deficient SMN mutant resulted in an enhanced coactivation of the LMP1 promoter, we asked where the binding site of SMN on DP103 is located. Therefore, several HA-tagged DP103 mutants (depicted in Fig. 4B) were expressed in 293GP cells and tested for their ability to coimmunoprecipitate coexpressed myc-tagged SMN (Fig. 4A). Deletion of aa 665 to 824 (ΔC159) abolished binding to SMN, whereas mutants deleting more central parts of DP103 (Δ341-461 and Δ456-547) still bound as efficiently as WT DP103. Note that the two mutants affecting consensus DEAD-Box motives, DP103 Δ341-461 deleting a putative ATP binding site and DP103 K112N mutating a putative ATPase motive (GK112T to GN112T), described as essential for eIF4A activity (29), were still able to interact with SMN. Thus, the binding sites for both SMN and EBNA2 are located within the C terminus (aa 665 to 824) of DP103, leading to the hypothesis that the binding of EBNA2 to the C terminus of DP103 releases DP103-complexed SMN. To test this, luciferase assays were performed in BJAB cells by using the −327/+40 and −154/+40 LMP1 promoter constructs (Fig. 4C). Compared to WT EBNA2, the DP103 binding-deficient mutant EBNA2 Δ121-216 was severely impaired in transactivating the LMP1 promoter, whereas coexpression of SMN rescued transactivation to WT levels. This result suggests that DP103 binding-deficient EBNA2 Δ121-216 is not able to release sufficient amounts of endogenous SMN from its DP103-bound state and, further, that the mechanism of EBNA targeting DP103 to release SMN is relevant for transactivation. Surprisingly, a similar result could be obtained by testing EBNA2 322LE (Fig. 4C), an insertion mutant shown to be unable to bind to RBPJκ (34) and therefore to be impaired in both transactivation and transformation (5, 12). Coexpression of SMN rescued transactivation of EBNA2 322 LE above WT levels, indicating that targeting DP103 and binding to RBPJκ are separate mechanisms involving distinct properties of EBNA2. To further dissect the role of RBPJκ in this context, a set of mutated LMP1 promoter luciferase constructs (LL0 to LL9) (18), schematically represented in Fig. 5A, were examined in BJAB cells (Fig. 5B). The full-length −327/+40 promoter construct, containing two RBPJκ sites and, to a lesser extent, a truncated −232/+40 construct, containing only one RBPJκ-site, were EBNA2 responsive and coactivated by EBNA2 and SMN. All further truncated constructs tested (−199/+40 to −34/+40) were not responsive to either EBNA2 or EBNA2 and SMN, suggesting that the presence of RBPJκ bound to its cognate promoter sequences is essential for coactivation by EBNA2 and SMN. To further investigate this, a luciferase reporter construct containing a 12-fold multimerized RBPJκ binding-site (pGa-981-21) (37) was tested in BJAB cells (Fig. 5C). EBNA2 transactivation could be titrated in an almost linear fashion, but coexpression of SMN did not result in an additional stimulation. Furthermore, expression of the RBPJκ binding-deficient EBNA2 322LE mutant resulted in a complete loss of transactivation which, in contrast to the results obtained when testing the LMP1 promoter, could not be rescued by coexpression of SMN. These data suggest that the presence of RBPJκ bound to the promoter is necessary but is not sufficient for coactivation by EBNA2 and SMN.
FIG. 4.
EBNA2-mediated transactivation of the LMP1 promoter involves binding of EBNA2 to DP103. (A) Mapping of the SMN binding site on DP103. 293GP cells were transfected with pSG5 constructs encoding HA-tagged DP103 mutants and WT myc-tagged SMN, as indicated. Cell extracts were immunoprecipitated with anti-HA MAb 3F10 (IP: anti HA and control 2) or irrelevant MAb 9C2 (controls 1 and 3) and analyzed by SDS–10% PAGE and Western blotting. Precipitated HA-tagged DP103 mutants were detected by using anti-HA MAb 3F10 (WB: anti HA), coprecipitated myc-tagged WT SMN by using anti-myc MAb 9E10 (WB: anti myc). Panels designated input represent ca. 10% of unprecipitated extracts. The positions of the molecular mass markers (in kilodaltons) are indicated on the left side of each panel. Deletion of aa 665 to 824 of DP103 (ΔC159) abolished coprecipitation of myc-tagged SMN. (B) Schematic representation of the DP103 and EBNA2 mutants tested. (C) Coexpression of SMN rescued impaired transactivation of the −327/+40 LMP1 promoter luciferase construct by the DP103 binding-deficient mutant EBNA2 Δ121-216 and the RBPJκ binding-deficient mutant EBNA2 322LE. Assays were performed as described for Fig. 1B. Graphs represent the mean values of three independent experiments performed in duplicate (±SEM).
FIG. 5.
The presence of RBPJκ is essential but not sufficient for coactivation by EBNA2 and SMN. (A) Schematic map of positions −327 to +40 of the LMP1 promoter and the different luciferase constructs tested. Shaded boxes represent the positions of cellular transcription factor binding sites, and black boxes represent the positions RBPJκ binding sites relative to the transcription start site (arrow). (B) Deletion mutants of the LMP1 promoter were tested for responsiveness to EBNA2 and coexpressed HA-tagged SMN in BJAB cells, as indicated. Deletion of the RBPJκ binding sites abolished EBNA2 transactivation and coactivation by SMN. (C) No coactivation of a multimerized RBPJκ site by SMN and increasing amounts of WT EBNA2 (1, 4, and 10 μg) or by SMN and RBPJκ binding-deficient EBNA2 322LE. (B and C) Assays were performed as described for Fig. 1B. Graphs represent the mean values of three independent experiments performed in duplicate (±SEM).
DISCUSSION
This study demonstrates a functional cooperation of EBNA2 and SMN as a novel mechanism involved in EBNA2-mediated transactivation of the viral LMP1 promoter. Initially identifying SMN as a DP103-interacting protein in the yeast two-hybrid system, we further established this interaction in B cells (Fig. 1A) and showed that SMN can coactivate the LMP1 promoter in the presence of EBNA2, whereas DP103 exhibited a slightly negative influence on EBNA2-mediated transactivation (Fig. 1B). Since this coactivation by EBNA2 and SMN was detectable in different B-cell lines in both luciferase assays and by an increased LMP1 protein level in vivo (Fig. 2B), we assume that this effect is due to an enhanced transcriptional activation, resulting in increased amounts of luciferase or LMP1 transcripts, respectively. In particular, we could exclude in control experiments possible side effects of overexpressed SMN on splicing efficiency, levels of EBNA2 expression, and the viability of the transfected cells. To dissect the mechanism underlying the coactivation by EBNA2 and SMN, we mapped the binding sites of both DP103 and SMN in coimmunoprecipitation experiments from transfected mammalian 293 GP cells and in the yeast two-hybrid system. Thereby, we could extend the results of Charroux et al. (2), who demonstrated that deletion of the SMN exon 7 reduced binding to DP103 (there named Gemin3), since we showed that only deletion of both SMN exons 6 and 7 completely abolished the interaction with DP103 (Fig. 3A), suggesting that exon 6 is the main binding site for DP103 on SMN. Furthermore, we mapped the interacting domain of SMN on DP103 to the C terminus (aa 665 to 824) of DP103 (Fig. 4A) by testing several mutants deleting different portions of DP103. In contrast, Charroux et al. defined the binding site (aa 456 to 547) by showing that an in vitro-translated myc-tagged 179-aa DP103 polypeptide (Gemin3 ΔN368ΔC277) bound to recombinant glutathione S-transferase (GST)–SMN in GST pulldown experiments, whereas two mutants with deletions of large parts of DP103 (ΔC328 and ΔN548) did not (2). The possible existence of multiple interaction sites with different affinities and also the different systems used or the varying design of the mutants tested could account for the disparate results. Testing different SMN mutants on EBNA2-mediated transactivation, we found indeed that the loss of binding to DP103 released SMN ΔEx6/7 from nuclear gems and/or coiled bodies (Fig. 3D and E) and resulted in an enhanced coactivation (Fig. 3C). Consistent with these findings, DP103 binding-deficient EBNA2 Δ121-216 was impaired in transactivation and could be rescued to WT levels by coexpression of SMN (Fig. 4C). This led us to the hypothesis that EBNA2 targets DP103 to release transcriptionally active SMN, since both EBNA2 and SMN bind to the C terminus of DP103. Furthermore, immunofluorescence experiments suggested that EBNA2 can target nuclear gems or coiled bodies, thereby releasing SMN (Fig. 2C and D), since the localization of endogenous SMN changed upon expression of EGFP-EBNA2 into a speckled pattern. A similar distribution of EBNA2 and corepressors of RBPJκ in experiments with Vero cells has been described by Zhang et al. (44). Recently, Strasswimmer et al. (36) reported that SMN interacts with the bovine papillomavirus E2 transactivator, resulting in an enhanced transactivation of a viral E2-responsive promoter, although the mechanisms leading to this effect have not been further elucidated. Although similar mechanisms underlying SMN-mediated coactivation within the RNA polymerase II transcription complex could be assumed, it should be considered that EBNA2, in contrast to bovine papillomavirus E2, neither directly binds to SMN nor directly binds to DNA. Furthermore, the SMA-derived SMN mutants (Y272C, E134K, and ΔEx7) did not show a dominant-negative effect on transactivation of the LMP1 promoter, whereas SMN ΔEx7 severely impaired bovine papillomavirus E2-mediated transactivation (36). Thus far, we cannot determine where within the promoter-bound RNA polymerase II transcription complex SMN exerts its coactivational function. Although it seems very likely that a direct interaction of SMN with the CTD of RNA polymerase II, as reported by Pellizzoni et al. (30), is involved, a direct binding of SMN to the promoter or DNA-bound transcription factors also seems possible. In particular, we could show that the presence of RBPJκ bound to the promoter was essential for coactivation (Fig. 5B), but a multimerized RBPJκ binding site could not be coactivated (Fig. 5C), indicating that SMN is somehow able to bridge the interaction of EBNA2 and RBPJκ. At this point, we also cannot determine whether EBNA2, by targeting DP103 to release SMN, otherwise changes the composition or binding affinities of the subnuclear spliceosomal complexes containing both proteins (3) and whether these changes could also affect transcription. Finally, different SMN-containing nuclear complexes, as proposed by Meister et al. (25), could be involved in the coactivation reported here. These complexes could determine alternative functions of SMN in either transcriptional activation or posttranscriptional processing, supporting the idea of a transcriptosome (8) tightly coupling both functions. Regarding EBV-induced transformation of resting human B lymphocytes, which is closely linked to LMP1 expression, our data suggest that the proposed displacement of endogenous SMN from endogenous DP103 by EBNA2 is involved in, but not absolutely required for, EBNA2-mediated transactivation and transformation. It has been reported in a different context that the deletion of aa 143 to 230 or of aa 112 to 142, encompassing the DP103 binding site on EBNA2 (aa 121 to 216), severely impaired but did not completely abolish LMP1 transactivation and viral transformation (5, 40).
In summary, our results suggest that transcriptionally inactive SMN is complexed to DP103 in a pool of spliceosomal proteins, most likely within nuclear gems and/or coiled bodies. In this scenario EBNA2, by targeting DP103, releases a transcriptionally active form of SMN that might interact with parts of the transcription machinery, e.g., the CTD of RNA polymerase II (30), thereby regulating transactivation of the viral LMP1 promoter by EBNA2.
ACKNOWLEDGMENTS
This work was supported by a grant of the Deutsche Forschungsgemeinschaft (DFG) through Sonderforschungsbereich 399 (Projekt B1).
We thank G. Laux and U. Zimber-Strobl (GSF, Munich, Germany) for generously providing plasmids and U. Fischer (MPI für Biochemie, Martinsried, Germany) for providing antibody 7B10. We are grateful to A. Schmid and I. Schultz (Institut für Physiologie, Homburg, Germany) and N. Schuster and K. Krieglstein (Institut für Anatomie, Homburg, Germany) for help with confocal microscopy and to P. Sommer (Institut Pasteur, Paris, France) for helpful discussions.
REFERENCES
- 1.Campbell L, Hunter C M D, Mohagheg P, Tinsley J M, Brasch M A, Davies K E. Direct interaction of SMN with dp103, a putative RNA helicase: a role for SMN in transcription regulation? Hum Mol Genet. 2000;9:1093–1100. doi: 10.1093/hmg/9.7.1093. [DOI] [PubMed] [Google Scholar]
- 2.Charroux B, Pellizzoni L, Perkinson R A, Shevchenko A, Mann M, Dreyfuss G. Gemin3: a novel DEAD box protein that interacts with SMN, the spinal muscular atrophy gene product, and is a component of gems. J Cell Biol. 1999;147:1181–1194. doi: 10.1083/jcb.147.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Charroux B, Pellizzoni L, Perkinson R A, Yong J, Shevchenko A, Mann M, Dreyfuss G. Gemin4. A novel component of the SMN complex that is found in both gems and nucleoli. J Cell Biol. 2000;148:1177–1186. doi: 10.1083/jcb.148.6.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clermont O, Burlet P, Cruad C, Bertrandy S, Melki J, Munnich A, Lefevbre S. Mutation analysis of the SMN gene in undeleted SMA patients. Am J Hum Genet. 1997;61:A329. [Google Scholar]
- 5.Cohen J I, Wang F, Kieff E. Epstein-Barr virus nuclear protein 2 mutations define essential domains for transformation and transactivation. J Virol. 1991;65:2545–2554. doi: 10.1128/jvi.65.5.2545-2554.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fischer N, Voss M D, Mueller Lantzsch N, Grässer F A. A potential NES of the Epstein-Barr virus nuclear antigen 1 (EBNA1) does not confer shuttling. FEBS Lett. 1999;447:311–314. doi: 10.1016/s0014-5793(99)00313-0. [DOI] [PubMed] [Google Scholar]
- 7.Fischer U, Liu Q, Dreyfuss G. The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell. 1997;90:1023–1029. doi: 10.1016/s0092-8674(00)80368-2. [DOI] [PubMed] [Google Scholar]
- 8.Gall J G, Bellini M, Wu Z, Murphy C. Assembly of the nuclear processing and transcription machinery: Cajal bodies (coiled bodies) and transcriptosomes. Mol Biol Cell. 1999;10:4385–4402. doi: 10.1091/mbc.10.12.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grässer F A, Sauder C, Haiss P, Hille A, König S, Göttel S, Kremmer E, Leinenbach H P, Zeppezauer M, Mueller Lantzsch N. Detection of proteins associated with the Epstein-Barr virus nuclear antigen 2: EBNA2A binds to histone H1 and unknown cellular proteins of 130, 110, 105, and 95 kDa. Virology. 1993;195:550–560. doi: 10.1006/viro.1993.1406. [DOI] [PubMed] [Google Scholar]
- 10.Grundhoff A T, Kremmer E, Tureci O, Glieden A, Gindorf C, Atz J, Mueller Lantzsch N, Schubach W H, Grässer F A. Characterization of DP103, a novel DEAD box protein that binds to the Epstein-Barr virus nuclear proteins EBNA2 and EBNA3C. J Biol Chem. 1999;274:19136–19144. doi: 10.1074/jbc.274.27.19136. [DOI] [PubMed] [Google Scholar]
- 11.Hannus S, Buhler D, Romano M, Seraphin B, Fischer U. The Schizosaccharomyces pombe protein Yab8p and a novel factor, Yip1p, share structural and functional similarity with the spinal muscular atrophy-associated proteins SMN and SIP1. Hum Mol Genet. 2000;9:663–674. doi: 10.1093/hmg/9.5.663. [DOI] [PubMed] [Google Scholar]
- 12.Henkel T, Ling P D, Hayward S D, Peterson M G. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein Jκ. Science. 1994;265:92–95. doi: 10.1126/science.8016657. [DOI] [PubMed] [Google Scholar]
- 13.Iwahashi H, Eguchi Y, Yasuhara N, Hanafusa T, Matsuzawa Y, Tsujimoto Y. Synergistic anti-apoptotic activity between Bcl-2 and SMN implicated in spinal muscular atrophy. Nature. 1997;390:413–417. doi: 10.1038/37144. [DOI] [PubMed] [Google Scholar]
- 14.Jayachandra S, Low K G, Thlick A E, Yu J, Ling P D, Chang Y, Moore P S. Three unrelated viral transforming proteins (vIRF, EBNA2, and E1A) induce the MYC oncogene through the interferon-responsive PRF element by using different transcription coadaptors. Proc Natl Acad Sci USA. 1999;96:11566–11571. doi: 10.1073/pnas.96.20.11566. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Kieff E. Epstein-Barr virus and its replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2343–2396. [Google Scholar]
- 16.Kremmer E, Kranz B, Hille A, Klein K, Eulitz M, Hoffmann-Fezer G, Feiden W, Herrmann K, Delecluse H-J, Delsol G, Bornkamm G W, Mueller-Lantzsch N, Grässer F A. Rat monoclonal antibodies differentiating between the Epstein-Barr virus nuclear antigens 2A (EBNA2A) and 2B (EBNA2B) Virology. 1995;208:336–342. doi: 10.1006/viro.1995.1157. [DOI] [PubMed] [Google Scholar]
- 17.Laux G, Adam B, Strobl L J, Moreau-Gachelin F. The Spi-1/PU.1 and Spi-B ets family transcription factors and the recombination signal binding protein RBP-Jκ interact with an Epstein-Barr virus nuclear antigen 2 responsive cis-element. EMBO J. 1994;13:5624–5632. doi: 10.1002/j.1460-2075.1994.tb06900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laux G, Dugrillon F, Eckert C, Adam B, Zimber Strobl U, Bornkamm G W. Identification and characterization of an Epstein-Barr virus nuclear antigen 2-responsive cis element in the bidirectional promoter region of latent membrane protein and terminal protein 2 genes. J Virol. 1994;68:6947–6958. doi: 10.1128/jvi.68.11.6947-6958.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 20.Ling P D, Hayward S D. Contribution of conserved amino acids in mediating the interaction between EBNA2 and CBF1/RBPJκ. J Virol. 1995;69:1944–1950. doi: 10.1128/jvi.69.3.1944-1950.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Q, Dreyfus G. A novel nuclear structure containing the survival of motor neurons protein. EMBO J. 1996;15:3555–3565. [PMC free article] [PubMed] [Google Scholar]
- 22.Lorson C L, Strasswimmer J, Yao J M, Baleja J D, Hahnen E, Wirth B, Le T, Burghes A H, Androphy E J. SMN oligomerization defect correlates with spinal muscular atrophy severity. Nat Genet. 1998;19:63–66. doi: 10.1038/ng0598-63. [DOI] [PubMed] [Google Scholar]
- 23.Mann K P, Staunton D, Thorley-Lawson D A. Epstein-Barr virus-encoded protein found in plasma membranes of transformed cells. J Virol. 1985;55:710–720. doi: 10.1128/jvi.55.3.710-720.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McAndrew P E, Parsons D W, Simard L R, Rochette C, Ray P N, Mendell J R, Prior T W, Burghes A H. Identification of proximal spinal muscular atrophy carriers and patients by analysis of SMNT and SMNC gene copy number. Am J Hum Genet. 1997;60:1411–1422. doi: 10.1086/515465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meister G, Bühler D, Laggerbauer B, Zobawa M, Lottspeich F, Fischer U. Characterization of a nuclear 20S complex containing the survival of motor neurons (SMN) protein and a specific subset of spliceosomal Sm proteins. Hum Mol Genet. 2000;9:1977–1986. doi: 10.1093/hmg/9.13.1977. [DOI] [PubMed] [Google Scholar]
- 26.Mohaghegh P, Rodrigues N R, Owen N, Ponting C P, Le T T, Burghes A H, Davies K E. Analysis of mutations in the tudor domain of the survival motor neuron protein SMN. Eur J Hum Genet. 1999;7:519–525. doi: 10.1038/sj.ejhg.5200346. [DOI] [PubMed] [Google Scholar]
- 27.Ou Q, Mouillet J F, Yan X, Dorn C, Crawford P A, Sadovsky Y. The DEAD box protein DP103 is a regulator of steroidogenic factor-1. Mol Endocrinol. 2001;15:69–79. doi: 10.1210/mend.15.1.0580. [DOI] [PubMed] [Google Scholar]
- 28.Owen N, Doe C L, Mellor J, Davies K E. Characterization of the Schizosaccharomyces pombe orthologue of the human survival motor neuron (SMN) protein. Hum Mol Genet. 2000;9:675–684. doi: 10.1093/hmg/9.5.675. [DOI] [PubMed] [Google Scholar]
- 29.Pause A, Sonenberg N. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 1992;11:2643–2654. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pellizzoni L, Charroux B, Rappsilber J, Mann M, Dreyfuss G. A functional interaction between the survival motor neuron complex and RNA polymerase II. J Cell Biol. 2001;152:75–85. doi: 10.1083/jcb.152.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pellizzoni L, Kataoka N, Charroux B, Dreyfuss G. A novel function for SMN, the spinal muscular atrophy disease gene product, in pre-mRNA splicing. Cell. 1998;95:615–624. doi: 10.1016/s0092-8674(00)81632-3. [DOI] [PubMed] [Google Scholar]
- 32.Phizicky E M, Fields S. Protein-protein interactions: methods for detection and analysis. Microbiol Rev. 1995;59:94–123. doi: 10.1128/mr.59.1.94-123.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rickinson A B, Kieff E. Epstein-Barr virus. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2397–2446. [Google Scholar]
- 34.Sauder C, Gotzinger N, Schubach W H, Horvath G C, Kremmer E, Krebs A, König S, Zimber Strobl U, Mueller-Lantzsch N, Grässer F A. Mutational analysis of the Epstein-Barr virus nuclear antigen 2 by far-Western blotting and DNA-binding studies. J Gen Virol. 1996;77:991–996. doi: 10.1099/0022-1317-77-5-991. [DOI] [PubMed] [Google Scholar]
- 35.Schrank B, Gotz R, Gunnersen J M, Ure J M, Toyka K V, Smith A G, Sendtner M. Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos. Proc Natl Acad Sci USA. 1997;94:9920–9925. doi: 10.1073/pnas.94.18.9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strasswimmer J, Lorson C L, Breiding D E, Chen J J, Le T, Burghes A H, Androphy E J. Identification of survival motor neuron as a transcriptional activator-binding protein. Hum Mol Genet. 1999;8:1219–1226. doi: 10.1093/hmg/8.7.1219. [DOI] [PubMed] [Google Scholar]
- 37.Strobl L J, Höfelmayr H, Stein C, Marschall G, Brielmeier M, Laux G, Bornkamm G W, Zimber Strobl U. Both Epstein-Barr viral nuclear antigen 2 (EBNA2) and activated Notch1 transactivate genes by interacting with the cellular protein RBP-Jκ. Immunobiology. 1997;198:299–306. doi: 10.1016/s0171-2985(97)80050-2. [DOI] [PubMed] [Google Scholar]
- 38.Tong X, Drapkin R, Reinberg D, Kieff E. The 62- and 80-kDa subunits of transcription factor IIH mediate the interaction with Epstein-Barr virus nuclear protein 2. Proc Natl Acad Sci USA. 1995;92:3259–3263. doi: 10.1073/pnas.92.8.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tong X, Wang F, Thut C J, Kieff E. The Epstein-Barr virus nuclear protein 2 acidic domain can interact with TFIIB, TAF40, and RPA70 but not with TATA-binding protein. J Virol. 1995;69:585–588. doi: 10.1128/jvi.69.1.585-588.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tong X, Yalamanchili R, Harada S, Kieff E. The EBNA-2 arginine-glycine domain is critical but not essential for B-lymphocyte growth transformation; the rest of region 3 lacks essential interactive domains. J Virol. 1994;68:6188–6197. doi: 10.1128/jvi.68.10.6188-6197.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang L, Grossman S R, Kieff E. Epstein-Barr virus nuclear protein 2 interacts with p300, CBP, and PCAF histone acetyltransferases in activation of the LMP1 promoter. Proc Natl Acad Sci USA. 2000;97:430–435. doi: 10.1073/pnas.97.1.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu D Y, Krumm A, Schubach W H. Promotor-specific targeting of human SWi-SNF complex by Epstein-Barr virus nuclear protein 2. J Virol. 2000;74:8893–8903. doi: 10.1128/jvi.74.19.8893-8903.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu D Y, Krumm A, Schubach W H. Epstein-Barr virus nuclear protein 2 (EBNA2) binds to a component of the human SNF-SWI complex, hSNF5/Ini1. J Virol. 1996;70:6020–6028. doi: 10.1128/jvi.70.9.6020-6028.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang J, Chen H, Weinmaster G, Hayward S D. Epstein-Barr virus BamHI-A rightward transcript-encoded RPMS protein interacts with the CBF1-associated corepressor CIR to negatively regulate the activity of EBNA2 and NotchIC. J Virol. 2001;75:2946–2956. doi: 10.1128/JVI.75.6.2946-2956.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zimber-Strobl U, Kremmer E, Grässer F, Marschall G, Laux G, Bornkamm G W. The Epstein-Barr virus nuclear antigen 2 interacts with an EBNA2 responsive cis-element of the terminal protein 1 gene promoter. EMBO J. 1993;12:167–175. doi: 10.1002/j.1460-2075.1993.tb05642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zimber-Strobl U, Strobl L J, Meitinger C, Hinrichs R, Sakai T, Furukawa T, Honjo T, Bornkamm G W. Epstein-Barr virus nuclear antigen 2 exerts its transactivating function through interaction with recombination signal binding protein RBP-Jκ, the homologue of Drosophila Suppressor of Hairless. EMBO J. 1994;13:4973–4982. doi: 10.1002/j.1460-2075.1994.tb06824.x. [DOI] [PMC free article] [PubMed] [Google Scholar]