Abstract
A comprehensive genetic diversity analysis of 87 Passiflora germplasm accessions domesticated and cultivated for several years in the karst region of Guizhou, China, was conducted utilizing simple sequence repeat (SSR) fluorescent markers. These Passiflora species, renowned for their culinary and medicinal value, could bring significant economic and ecological benefits to the region. This study aimed to assess the genetic resources of these species and facilitate the selection of superior cultivars adapted to the karst environment. Our analysis revealed an abundance of SSR loci within the Passiflora transcriptome, with single-base repeats being the most prevalent type. Through rigorous primer screening and amplification, we successfully identified 27 SSR primer pairs exhibiting robust polymorphisms. Further interrogation at eight microsatellite loci revealed 68 alleles, underscoring the high level of genetic diversity present in the cultivated accessions. The average expected heterozygosity was 0.202, with the ssr18 locus exhibiting the highest value of 0.768, indicating significant genetic variation. The mean polymorphic information content (PIC) of 0.657 indicates the informativeness of these SSR markers. Comparative analyses of the cultivated and potential wild progenitors revealed distinct genetic variations among the different Passiflora types. Genetic structure and clustering analyses of the 87 accessions revealed seven distinct groups, suggesting gene flow and similarities among the resources. Notably, a DNA fingerprinting system was established using eight SSR primer pairs, effectively distinguishing the selected cultivars that had adapted to the karst mountainous region. This study not only deepens our understanding of Passiflora genetic resources in the karst environment but also provides a valuable reference for conservation, genetic improvement, and cultivar selection. The rich genetic diversity of the Passiflora germplasm underscores their potential for sustainable utilization in breeding programs aimed at enhancing the economic and ecological viability of these valuable plant species.
Keywords: Passiflora, SSR fluorescent markers, genetic diversity analysis, fingerprinting
1. Introduction
The genus Passiflora is the largest genus within the expansive Passifloraceae family, encompassing more than 520 species globally. The vast majority of these species are native to the Americas, particularly countries such as Colombia, Brazil, Ecuador, and Peru, while a few are dispersed throughout other tropical and subtropical regions, such as Southeast Asia, Australia, and New Zealand [1,2,3]. The Passiflora species, commonly referred to as passionflower, have been widely cultivated in tropical and subtropical regions worldwide because of their multifaceted benefits, including edibility, medicinal properties, and ornamental appeal [4]. In terms of edibility, approximately 60 species of Passiflora are considered palatable, with notable examples including P. edulis, P. edulis f. flavicarpa, P. ligularis, and P. quadrangularis. In terms of medicinal importance, numerous Passiflora plants boast a rich history of use in traditional folk medicines in several American and European nations, where they have been employed as remedies for a diverse array of neurological conditions [5]. Active compounds extracted from the leaves, fruits, peels, and seeds of these plants exhibit a range of beneficial effects, including sedative, antioxidant, anti-inflammatory, anxiolytic, and anticancer effects [6,7,8].
Molecular markers such as AFLP, SSR, and SNP provide polymorphic information at the DNA level, demonstrating high accuracy and precision, particularly suitable for situations requiring the precise differentiation of germplasm resources and varieties. These molecular markers are not influenced by environmental factors and can stably reflect genetic variations, serving as essential tools for genetic analysis and variety improvement by breeders and researchers [9]. The use of simple sequence repeat (SSR) molecular marker technology, on the other hand, involves the amplification of targeted DNA fragments and the detection of products through electrophoresis. These markers consist of 1–6 nucleotide repeat units, and variations in the number of these repeat units contribute to the genetic diversity among the cultivars. SSR marker technology follows the rules of Mendelian codominant inheritance, exhibits excellent stability and repeatability, possesses abundant polymorphic loci, provides large amounts of information, and requires minimal DNA amounts. SSR technology has been extensively employed in various applications, such as DNA fingerprinting, kinship analysis, and genetic diversity studies. Its merits include codominant inheritance, abundant polymorphic loci, and low DNA requirements [10,11]. Specifically, SSR fluorescent labelling via capillary electrophoresis technology has been applied in plants, including pistachio [12], chestnut [13], upland cotton [14], plum [15], broomcorn millet [16,17], sweet potato [18], and apricot [19], primarily for analysing the genetic diversity of germplasm resources, constructing genetic maps, gene marking and mapping, cultivar identification, and parentage analysis. The International Union for the Protection of New Varieties of Plants (UPOV) recommends the combined use of SSR and SNP markers as the preferred method for constructing DNA fingerprints (UPOV 2007) [20].
Currently, the application of SSR molecular marker technology in the Passiflora species focuses primarily on genetic diversity analysis, kinship studies, and the conservation of germplasm resources. Santos et al. pioneered the identification of 669 SSR loci in a cultivated Passiflora species via the BAC-end method [21]. Araya et al. subsequently leveraged partial sequences from the genome sequencing of cultivated Passiflora species to develop 816 pairs of SSR primers targeting functional genomic regions. Among these, 57 SSR primer pairs were selected and validated in 79 samples of both wild and cultivated Passiflora species, revealing polymorphisms in 42 primer pairs [22]. Furthermore, da Costa et al. utilized cDNA sequences from suppression subtractive hybridization (SSH) libraries to devise 42 SSR primers, with 34 demonstrating high transferability between yellow/purple-fruited Passiflora and Passiflora coccinea [23]. Research has shown that the escaped habitats of Passiflora in karst peak-cluster depression regions are complex and diverse, where the Passiflora species coexist with companion species in a mutually beneficial symbiosis, adapting well to the environment while exhibiting rapid growth and high biomass production. In Guizhou’s karst region, Passiflora cultivation has emerged as a local industry. The growth of Passiflora in rocky desertification areas can improve soil porosity, increase aeration and water permeability, lower ambient temperatures, and increase humidity, resulting in significant cooling and humidifying effects during hot weather, thereby contributing positively to the ecosystem [24]. Selecting and breeding suitable Passiflora varieties for karst regions can provide both economic and ecological benefits. This study employed SSR fluorescent labeling via capillary electrophoresis technology to conduct a genetic diversity analysis of 87 Passiflora germplasm accessions domesticated and cultivated over several years in Guizhou’s karst region and Xishuangbanna, China. By leveraging the SSR markers developed from transcriptome sequencing, we aimed to elucidate the kinship relationships, population genetic structures, and genetic backgrounds of diverse Passiflora germplasm resources. This work provides a solid foundation for the selection, breeding, application, preservation, and molecular marker-assisted breeding of suitable Passiflora varieties in the karst regions.
2. Results
2.1. Characterization of SSR Loci in the Passionfruit Transcriptome
Through an analysis of the passionfruit transcriptome, the transcriptome was sequenced from the purple-fruited Passiflora from the Guizhou Province; we identified 27,303 SSR loci within 86,880 unigenes, exhibiting a frequency of 31.43%, with an average of one SSR locus per 4.6 kb. Notably, 4517 unigenes contained more than one SSR locus (Table 1). The SSR repertoire in the passionfruit transcriptome was diverse, encompassing all repeat types from mono- to hexanucleotide repeats, albeit with significant variations in abundance among the different repeat types. Mononucleotide repeats were the most abundant, accounting for 63.7% of all SSR loci, with A/T repeats being the dominant type. Dinucleotide repeats constituted 21.0% of the total, dominated by AG/CT repeats. Trinucleotide repeats constituted 14.0% of the SSR loci, with AAG/CTT repeats being prevalent. Tetranucleotide repeats accounted for 0.9% of the total, with AAAT/ATTT repeats being the most numerous. Pentanucleotide and hexanucleotide repeats were less frequent, with proportions of 0.3% and 0.1%, respectively.
Table 1.
SSR Locus information from the passionfruit transcriptome.
| Repeats | 5 | 6 | 7 | 8 | 9 | 10–15 | 16–20 | >21 | Total |
|---|---|---|---|---|---|---|---|---|---|
| Mono-motif | (17,401) | ||||||||
| A/T | 13,911 | 2780 | 579 | 17,270 | |||||
| C/G | 93 | 3 | 35 | 131 | |||||
| Din-motif | (5738) | ||||||||
| AC/GT | 256 | 148 | 113 | 44 | 80 | 641 | |||
| AG/CT | 1110 | 639 | 465 | 446 | 454 | 1 | 3115 | ||
| AT/AT | 615 | 423 | 313 | 284 | 336 | 1971 | |||
| CG/CG | 11 | 11 | |||||||
| Tri-motif | (3808) | ||||||||
| AAC/GTT | 175 | 35 | 31 | 2 | 1 | 244 | |||
| AAG/CTT | 589 | 262 | 148 | 7 | 1006 | ||||
| AAT/ATT | 157 | 72 | 57 | 8 | 294 | ||||
| ACC/GGT | 221 | 87 | 65 | 9 | 1 | 378 | |||
| ACG/CGT | 55 | 37 | 20 | 11 | 123 | ||||
| ACT/AGT | 31 | 14 | 4 | 2 | 51 | ||||
| AGC/CTG | 290 | 128 | 45 | 4 | 467 | ||||
| AGG/CCT | 399 | 145 | 123 | 3 | 670 | ||||
| ATC/ATG | 270 | 98 | 65 | 6 | 439 | ||||
| CCG/CGG | 89 | 31 | 13 | 3 | 136 | ||||
| Tetra-motif | 216 | 24 | 6 | 249 | |||||
| Penta-motif | 57 | 4 | 7 | 1 | 69 | ||||
| Hexa-motif | 7 | 23 | 4 | 2 | 2 | 38 | |||
| Total number of unigenes examined | 86,880 | ||||||||
| Total size of examined sequences (bp) | 125,855,003 | ||||||||
| Total number of identified SSRs | 27,303 | ||||||||
| Number of SSR containing unigenes | 21,578 | ||||||||
| Number of unigenes containing more than 1 SSR | 4517 | ||||||||
| Number of SSRs present in compound formation | 1427 | ||||||||
The repeat number is a crucial factor influencing SSR polymorphism. The repeat numbers of SSRs in the passionfruit transcriptome ranged from 5 to 24, with 10 repeats being the most abundant. Among the different repeat types, mononucleotide repeats were predominant (10 repeats), followed by dinucleotide repeats (6 repeats), and trinucleotide, tetranucleotide, and pentanucleotide repeats (5 repeats). Interestingly, hexanucleotide repeats showed a higher abundance (6 repeats). The number of SSR loci for mononucleotide to pentanucleotide repeats decreased with increasing repeat number, whereas for hexanucleotide repeats, the repeat number increased from 5 to 6.
2.2. Screening of SSR Primers
A total of 50 loci were randomly selected for primer design, and six randomly chosen passionfruit samples were used to screen the 50 pairs of SSR primers. As a result, 27 primer pairs were identified that exhibited a distinct and robust amplification of major bands (Table 2). Among these primer pairs, eight displayed stable amplification with clear major bands and exhibited good polymorphism. The primer numbers for these eight selected primers were SSR1, SSR2, SSR3, SSR18, SSR30, SSR32, SSR34, and SSR39, and the amplification patterns of the two samples in this study using eight core primers are shown in Figure S1.
Table 2.
The 27 SSR primers that amplified distinct major bands.
| Name | SSR Motif | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|---|
| SSR1 | (GGAGTT)6 | GGAGGAGGGCTTCGAGGAGT | TGCAGCCACCACGTCTACTG |
| SSR2 | (CTGATA)5 | TGGAAGATGGTTGAAGCAAAGGGT | AGGCATCCATGTCAGATTGCA |
| SSR3 | (GAACTG)5 | TGATGCTGCGTACGGCTCAC | CTTCCTGACGCAGAGCGGAG |
| SSR5 | (AGCTGG)6 | TCCCCACCAGACTCTGCACC | CAGTGCCAGTTCCAGTGCCA |
| SSR6 | (CACTAC)5 | GAAATCCTTCCACCTGGG | CAACAACTTTGCCTGCTT |
| SSR8 | (AGATT)7 | GAGAATGGCACAATGAGA | TAGAGGTGGAGCGGATTA |
| SSR9 | (CATAG)5 | TTTATACTTCTTCGCCATCA | GGTCAGTTTCCAGGGTGC |
| SSR12 | (CTGGG)5 | TGGAGCATCTTTGTTGAG | GGTCGGATAGTAAATAGGG |
| SSR14 | (ATCCA)5 | CGCCACAATCTAACTGCT | TCCACCTCATTAACCCTC |
| SSR17 | (ACCAT)5 | CACTGACCTCTGCGACTA | GTAAGAATGCTCCCGACA |
| SSR18 | (AGATT)7 | GGGCATCGCAGAGTAACAG | GCCGTCCCATACCCAAGT |
| SSR21 | (TGCT)5 | GGCTGCCTCATAGACCAC | GAAAGTTACAAACCGACA |
| SSR24 | (TGAT)5 | CAGATAAGATTCTTCCGCACTA | ATGGGAAATGGAGGCTGT |
| SSR26 | (GATA)5 | TGTGATGAAGCACCCTGAA | GAGCGGTATAAGTTGTCTGG |
| SSR27 | (TGTC)5 | TTTTCTTACAGGCTTTATTGCG | ACTCAGCGGTTGGTTTGC |
| SSR28 | (TATT)5 | ACGTTATGTTGGGTGCTA | TTGTTCGGTACAGAGGAG |
| SSR29 | (TGTT)5 | ATACGCCCAATTAAGGAG | GTCGGTGATGGTTAGCAG |
| SSR30 | (GATA)5 | TGCCTGTGATGAAGCACCCTG | AATTTGCCGCGGTTGTGCTG |
| SSR31 | (GATA)5 | CAGAGACGGCCTGCAGATGG | GGAAGTGTTGGCACTTGCGT |
| SSR32 | (TTCT)5 | GTCACCGAACTGCCCAACCA | GTCTCGCCTCGGCTTCTGAC |
| SSR34 | (ACCA)5 | GTACCACCATCGGCCTGTCG | GGGCGTTGTCCGGATGAGAG |
| SSR36 | (TATT)5 | AGCGACTTCTGCTGTGTGGT | TGGATGTTCGTGGATGCCCT |
| SSR38 | (TCAT)5 | CCTCAGACAATCACTGGCGCT | GGCAGAATCAATCGTCGTGCG |
| SSR39 | (CATC)5 | GCCAAAGAGTGGGAGTCGGG | CAAGCGTTGGGCGTGGAGTA |
| SSR46 | (TCCT)5 | GATCGGCGGACAAAGAAGGA | TTTCACCTGCCTCTTGCCTT |
| SSR47 | (TCCT)5 | TGCACGAGCCATACAGAGACG | ACAGTGGGAACAAGGATGGCG |
| SSR48 | (GATA)5 | CGCTGACCGGCCTTTCAAGA | AGTCCTGCTTCCCCGGGATT |
2.3. SSR Polymorphic Variation
In the genetic assessment of 87 Passiflora germplasm resources utilizing eight core primers, a total of 68 alleles were detected, ranging from 4 alleles for SSR30 to 15 alleles for SSR39, with an average of 8.625 alleles per primer. Each sample contributed either two identical (monomorphic, single peak) or distinct (polymorphic, double peak) alleles at each locus. The effective number of alleles (Ne) varied from 1.977 for SSR34 to 4.724 for SSR18, with an average of 3.100. The observed heterozygosity (Ho) ranged from 0.079 for SSR2 to 0.449 for SSR18, with a mean value of 0.235. In contrast, the expected heterozygosity (He) ranged from 0.494 for SSR34 to 0.788 for SSR18, with an average value of 0.650. The Shannon diversity index (I), which quantifies the uncertainty associated with predicting the identity of an allele from a randomly chosen individual, ranged from 1.021 for SSR34 to 1.786 for SSR32, with a mean of 1.410. Notably, the polymorphic information content (PIC), a measure of the informativeness of a genetic marker, exceeded 0.250 for all the primers, ranging from 0.460 for SSR34 to 0.762 for SSR18, with an average of 0.614 (Table 3). These results demonstrate the high level of genetic diversity and potential utility of the selected SSR markers in genetic studies of Passiflora species.
Table 3.
Polymorphism analysis of SSR primers at 8 loci.
| Primer | Na | Ne | I | Ho | He | PIC |
|---|---|---|---|---|---|---|
| ssr1 | 5.000 | 2.183 | 1.105 | 0.244 | 0.542 | 0.512 |
| ssr2 | 7.000 | 2.730 | 1.311 | 0.079 | 0.634 | 0.595 |
| ssr3 | 6.000 | 3.645 | 1.460 | 0.127 | 0.726 | 0.684 |
| ssr18 | 10.000 | 4.724 | 1.814 | 0.449 | 0.788 | 0.762 |
| ssr30 | 4.000 | 2.363 | 1.062 | 0.423 | 0.577 | 0.523 |
| ssr32 | 15.000 | 3.928 | 1.786 | 0.229 | 0.745 | 0.711 |
| ssr34 | 7.000 | 1.977 | 1.021 | 0.171 | 0.494 | 0.460 |
| ssr39 | 15.000 | 3.248 | 1.723 | 0.161 | 0.692 | 0.670 |
| Mean | 8.625 | 3.100 | 1.410 | 0.235 | 0.650 | 0.614 |
2.4. Analysis of Genetic Diversity among Passiflora Germplasm Resources
This study analysed 87 germplasm resources belonging to 22 species and their cultivars within the genus Passiflora. The range of the average number of alleles (Na) per population varied from 0.750 in P. xishuangbannaensis to 2.750 in P. edulis. Similarly, the average effective number of alleles (Ne) also showed a significant variation, ranging from 0.750 in P. xishuangbannaensis to 1.880 in P. edulis. Notably, both P. miniata Xishuangbanna Red and P. serrulata presented a value of zero for average expected heterozygosity (He), Shannon’s diversity index (I), and observed heterozygosity (Ho). In contrast, the remaining 22 species and their cultivars presented a broader range of genetic diversity indices, with the He values ranging from 0.055 for P. miniata to 0.436 for P. edulis, the I values varying from 0.087 for P. caerulea to 0.701 for P. edulis, and the Ho values ranging from 0.063 for P. miniata to 0.563 for P. ‘Lady Margaret’ (Table 4).
Table 4.
Genetic diversity analysis of Passiflora species and their cultivars.
| Species (Cultivars) | Na | Ne | I | Ho | He | Number |
|---|---|---|---|---|---|---|
| P. altebilobata | 1.250 | 1.250 | 0.260 | 0.375 | 0.188 | 1 |
| P. edulis | 2.750 | 1.889 | 0.701 | 0.270 | 0.436 | 20 |
| P. edulis f. flavicarpa | 2.375 | 1.642 | 0.556 | 0.174 | 0.326 | 8 |
| P.trifasciata | 1.375 | 1.375 | 0.347 | 0.500 | 0.250 | 1 |
| P. wilsonii | 1.000 | 1.000 | 0.173 | 0.250 | 0.125 | 1 |
| P. xishuangbannaensis | 0.750 | 0.750 | 0.087 | 0.125 | 0.063 | 1 |
| P.’Lady Margaret’ | 1.875 | 1.783 | 0.547 | 0.563 | 0.375 | 2 |
| P. amethystina | 1.375 | 1.325 | 0.244 | 0.313 | 0.172 | 2 |
| P. caerulea | 1.125 | 1.125 | 0.087 | 0.125 | 0.063 | 1 |
| P. incarnata × P. laurifiolia | 2.375 | 1.880 | 0.666 | 0.406 | 0.410 | 4 |
| P. foetida | 1.875 | 1.700 | 0.462 | 0.169 | 0.290 | 5 |
| P. incarnata | 1.500 | 1.500 | 0.347 | 0.375 | 0.250 | 2 |
| P. ligularis | 2.000 | 1.382 | 0.436 | 0.117 | 0.269 | 5 |
| P. miniata | 1.500 | 1.079 | 0.113 | 0.063 | 0.055 | 10 |
| P. miniata Xishuangbanna Red | 0.875 | 0.875 | 0.000 | 0.000 | 0.000 | 1 |
| P. miniata × P. serrulata | 1.375 | 1.350 | 0.253 | 0.333 | 0.181 | 6 |
| P. morifolia | 1.500 | 1.371 | 0.414 | 0.188 | 0.262 | 3 |
| P. quadrangularis | 1.375 | 1.196 | 0.192 | 0.083 | 0.125 | 3 |
| P. serrulata | 1.000 | 1.000 | 0.000 | 0.000 | 0.000 | 1 |
| P. suberosa | 2.125 | 1.791 | 0.621 | 0.390 | 0.388 | 6 |
| P. violacea ‘victoria’ | 1.250 | 1.250 | 0.173 | 0.250 | 0.125 | 2 |
| P. yucatanensis | 1.250 | 1.167 | 0.260 | 0.188 | 0.156 | 2 |
2.5. Genetic Structures of Passiflora Germplasm Resources
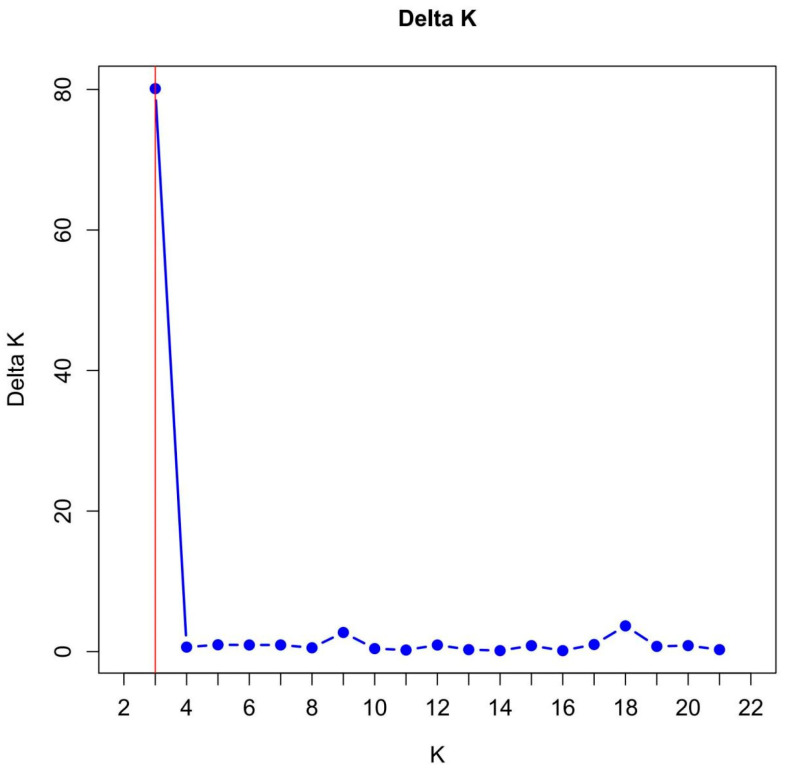
The genetic structure of 87 passionfruit (Passiflora spp.) germplasm accessions was analysed on the basis of SSR molecular marker data. The analysis was performed by setting the number of clusters (K) ranging from 1 to 22, with 15 repetitions for each K value. The optimal K value was determined using the ΔK, which peaked at K = 3 (Figure 1). Notably, there was a distinct increase in ΔK at K = 9, suggesting the incorporation of a new subpopulation within the tested materials, indicating that K = 9 represents a crucial subdivision point (Figure 1).
Figure 1.
Graphical depiction of the relationship between K and Δk.
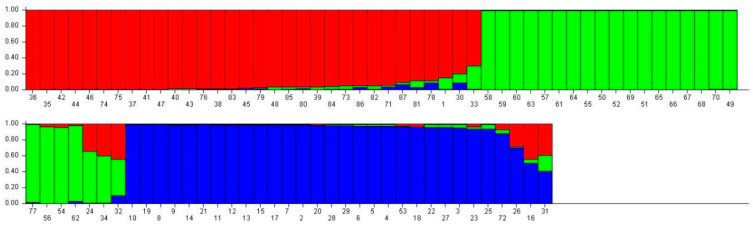
When K = 3, the 87 accessions were divided into three distinct groups (Figure 2), implying three potential genetic origins. The red group consisted primarily of species such as P. amethystina, P. foetida, P. quadrangularis, P. caerulea, P. incarnata × P. laurifolia, P. incarnata, P. suberosa, P. violacea ‘victoria’, P. morifolia, P. yucatanensis, P. altebilobata, P. trifasciata, and P. ‘Lady Margaret’. The green group was dominated by P. miniata, P. miniata Xishuangbanna Red, P. serrulata, P. ligularis, P. miniata × P. serrulata, and P. serrulata. Finally, the blue group was composed mainly of cultivated varieties of P. edulis, P. edulis f. flavicarpa, and their hybrids.
Figure 2.
Population genetic structure of 87 Passiflora germplasm accessions (K = 3).
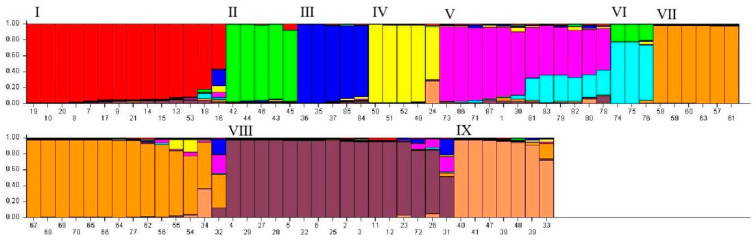
At K = 9, a more detailed subdivision was observed, with the 87 accessions classified into nine groups (Figure 3), suggesting nine potential genetic origins. Groups 1 and 8 were dominated by P. edulis, P. edulis f. flavicarpa, and their hybrids. Group 2 contained only P. foetida, whereas Group 3 was characterized by P. hystina, P. caerulea, and P. violacea ‘victoria’. Group 4 was predominantly P. ligularis and P. edulis f. flavicarpa “mi zhuan”, with the latter’s primary genetic contribution coming from Groups 4 and 8. Group 5 was composed of P. morifolia, P. yucatanensis, P. alitebilobata, P. trifiata, and P. suberosa, with P. suberosa’s major gene flow originating from Groups 5 and 6. Group 6 exclusively consisted of P. quadrangularis, with gene flow from Groups 2 and 6. Group 7 was dominated by P. miniata, P. miniata × P. serrulata, P. miniata Xishuangbanna Red, P. serrulata, P. ‘Lady Margaret’, and P. xishuangbannaensis, with the latter exhibiting the most complex genetic admixture from Groups 3, 5, 7, and 8. Finally, Group 9 was mainly composed of P. incarnata × P. laurifolia, P. incarnata, and P. ‘Lady Margaret’. The mixed composition of species within each group suggests that these varieties do not exhibit significant differences in their most primitive population classifications. To better understand the evolutionary patterns among different populations, tracing the origins of some varieties back to their more primitive habitats is necessary.
Figure 3.
Population genetic structure of 87 Passiflora germplasm accessions (K = 9).
In the STRUCTURE population structure analysis, we assessed the association of individuals with different genetic clusters based on their Q values, which represent the probability of an individual belonging to a particular cluster. A Q value ≥0.6 was considered indicative of a significant genetic contribution from that cluster. For the 87 Passiflora germplasm accessions, the distribution of Q values across clusters is presented in Table 4. Results show that 79 accessions (90.8% of the total) had Q values >0.6 in at least one cluster, indicating strong genetic affiliations with at least one cluster. Furthermore, 65.5% and 56.3% of the accessions had Q values ≥0.8 and ≥0.9, respectively, in at least one cluster, reflecting their close genetic relationships with specific clusters. Nevertheless, all the accessions likely contain genetic variations from the other clusters to some extent, highlighting the genetic diversity within the germplasm collection (Table 5).
Table 5.
Distribution of the Q values of the nine groups.
| Group | Number of Varieties Group | Number of Varieties | |||
|---|---|---|---|---|---|
| Q < 0.6 | Q ≥ 0.6 | Q ≥ 0.8 | Q ≥ 0.9 | ||
| 1 | 14 | 1 | 13 | 13 | 12 |
| 2 | 5 | 0 | 5 | 5 | 4 |
| 3 | 5 | 0 | 5 | 5 | 4 |
| 4 | 5 | 0 | 5 | 4 | 4 |
| 5 | 12 | 4 | 8 | 5 | 3 |
| 6 | 3 | 0 | 3 | 0 | 0 |
| 7 | 20 | 2 | 18 | 17 | 16 |
| 8 | 16 | 1 | 15 | 2 | 0 |
| 9 | 7 | 0 | 7 | 6 | 6 |
| Total | 87 | 8 | 79 | 57 | 49 |
2.6. Cluster Analysis of Passiflora Germplasm Resources
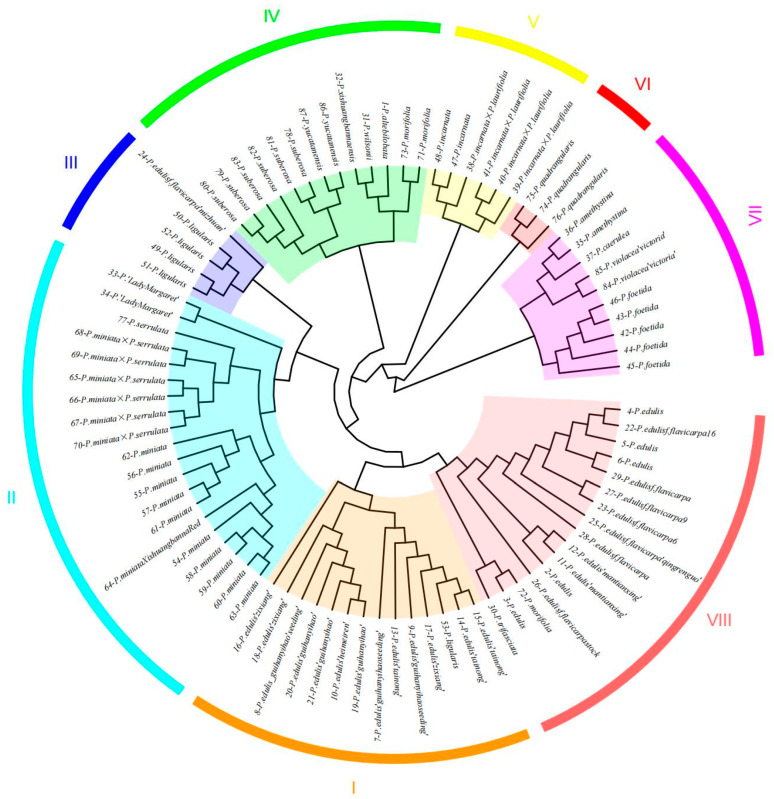
A cluster analysis was conducted on 87 Passiflora germplasm resources on the basis of Nei’s genetic distance. The results indicated that eight pairs of core primers were able to distinguish these 87 Passiflora germplasm resources, which were subsequently grouped into eight distinct clusters (Figure 4). Cluster 1 consisted primarily of cultivated varieties of P. edulis. Cluster 2 was composed mainly of P. miniata × P. serrulata, P. miniata Xishuangbanna Red, P. ‘Lady Margaret’, and P. serrulata. Cluster 3 was dominated by P. edulis f. flavicarpa ‘mizhuan’ and P. ligularis. Cluster 4 included P. yucatanensis, P. morifolia, P. altebilobata, P. wilsonii, and P. xishuangbannaensis. Cluster 5 was composed primarily of P. incarnata × P. laurifolia and P. incarnata. Cluster 6 was composed exclusively of P. quadrangularis. Cluster 7 included P. caerulea, P. amethystina, P. foetida, and P. violacea ‘victoria’. Finally, Cluster 8 was composed primarily of cultivated varieties of P. edulis, P. edulis f. flavicarpa, and P. trifasciata.
Figure 4.
UPGMA clustering dendrogram of 87 Passiflora germplasm accessions.
2.7. Construction of Fingerprint Profiles for Passiflora Germplasm Resources
The eight primer pairs were ranked in descending order on the basis of their PIC values presented in Table 3. The resulting order was as follows: ssr18, ssr3, ssr32, ssr2, ssr39, ssr30, ssr1, and ssr34. The corresponding codes for each fragment amplified by these primers are provided in Table 6. By utilizing the primer order and fragment sizes of the amplified bands (where ‘0’ signifies the absence of a discernible peak), alphabetically arranged fingerprint profiles were constructed for each germplasm resource (Table 7). These fingerprint profiles serve as robust tools for identifying and differentiating Passiflora germplasm resources, meeting the standards for scientific reporting in international journals.
Table 6.
Letter codes for primer fragment sizes.
| Primer Name | Code | Size (bp) | Primer Name | Code | Size (bp) | Primer Name | Code | Size (bp) |
|---|---|---|---|---|---|---|---|---|
| SSR1 | 87 | A | 366 | F | 224 | O | ||
| 93 | B | 367 | G | SSR34 | 130 | A | ||
| 99 | C | 372 | H | 198 | B | |||
| 105 | D | 377 | I | 202 | C | |||
| 117 | E | 387 | J | 206 | D | |||
| SSR2 | 175 | A | SSR30 | 248 | A | 209 | E | |
| 193 | B | 252 | B | 210 | F | |||
| 199 | C | 256 | C | 322 | G | |||
| 203 | D | 260 | D | SSR39 | 239 | A | ||
| 205 | E | SSR32 | 144 | A | 243 | B | ||
| 211 | F | 156 | B | 251 | C | |||
| 307 | G | 160 | C | 259 | D | |||
| SSR3 | 220 | A | 164 | D | 267 | E | ||
| 226 | B | 168 | E | 275 | F | |||
| 232 | C | 172 | F | 279 | G | |||
| 238 | D | 176 | G | 283 | H | |||
| 244 | E | 180 | H | 287 | I | |||
| 250 | F | 184 | I | 291 | J | |||
| SSR18 | 305 | A | 188 | J | 295 | K | ||
| 342 | B | 192 | K | 299 | L | |||
| 347 | C | 196 | L | 307 | M | |||
| 357 | D | 204 | M | 315 | N | |||
| 362 | E | 212 | N | 319 | O |
Table 7.
Fingerprint profiles of 87 germplasm accessions of the genus Passiflora.
| Sample | Species (Cultivars) | SSR Fingerprinting |
|---|---|---|
| 1-P. altebilobata | P. altebilobata | BICCACBBFL00CCDD |
| 2-P. edulis | P. edulis | IIDDJLCFLL00CCDD |
| 3-P. edulis | P. edulis | II00JJCCLLCCCCDD |
| 4-P. edulis | P. edulis | BIDDJJCCLLACCCDD |
| 5-P. edulis | P. edulis | IIDDJJCCLLCCCCDD |
| 6-P. edulis | P. edulis | IIDDJLCCLLCCCCDD |
| 7-P. edulis “gui han yi hao” (seeding) | P. edulis | GIDFJL00LLCDCEFF |
| 8-P. edulis “gui han yi hao” (seeding) | P. edulis | IIFFJJFFLLCDEEFF |
| 9-P. edulis “gui han yi hao” (seeding) | P. edulis | GIDDJJFFLLCDCCFF |
| 10-P. edulis “hei mei ren” | P. edulis | GIFFJJFFLLDDEEFF |
| 11-P. edulis “man tian xing” | P. edulis | IIDDJJFFLLAACCDD |
| 12-P. edulis “man tian xing” | P. edulis | IIDDJJFFLLAACCDD |
| 13-P. edulis “tai nong” | P. edulis | 00DDJLFFLLCDCEDF |
| 14-P. edulis “tai nong” | P. edulis | GIDDJJFFLLCDCEDF |
| 15-P. edulis “tai nong” | P. edulis | GIDDJJFFLLCDCEDF |
| 16-P. edulis “zi xiang” | P. edulis | GIDFIIFFKKCDACDF |
| 17-P. edulis “zi xiang” | P. edulis | GIDFJLFFLLCDCEDF |
| 18-P. edulis “zi xiang” | P. edulis | ABFFJLFFLLCDEEDF |
| 19-P. edulis “gui han yi hao” | P. edulis | GGFFJJFFLLDDEEFF |
| 20-P. edulis “gui han yi hao” | P. edulis | FFFFJJFFLLDDEEFF |
| 21-P. edulis “gui han yi hao” | P. edulis | GGFFJJCFLLDDEEFF |
| 22-P. edulis f. flavicarpa “16#” | P. edulis f. flavicarpa | BIDDJJCCLLCCCCDD |
| 23-P. edulis f. flavicarpa “6#” | P. edulis f. flavicarpa | BBDDJJCCLLACACDD |
| 24-P. edulis f. flavicarpa “mi zhuan” | P. edulis f. flavicarpa | EECCIICCMMCCACDF |
| 25-P. edulis f. flavicarpa “qing ren guo” | P. edulis f. flavicarpa | BBDDLLCCLLCCCCDD |
| 26-P. edulis f. flavicarpa (stock) | P. edulis f. flavicarpa | BCEEJJCCLLAACCDD |
| 27-P. edulis f. flavicarpa”9# “ | P. edulis f. flavicarpa | BBDDJJCCLLACCCDD |
| 28-P. edulis f. flavicarpa | P. edulis f. flavicarpa | IIDDJLCCLLACCC00 |
| 29-P. edulis f. flavicarpa | P. edulis f. flavicarpa | IIDDJLCCLLACCCDD |
| 30-P. trifasciata | P.trifasciata | GI00IJACCDCCCCDD |
| 31-P. wilsonii | P. wilsonii | BI00IJBBLL00CCDD |
| 32-P. xishuangbannaensis | P. xishuangbannaensis | BH0000BBLL00CCDD |
| 33-P. ‘Lady Margaret’ | P. ‘Lady Margaret’ | BIACNNCCHLCCACCC |
| 34-P. ‘Lady Margaret’ | P. ‘Lady Margaret’ | BGACINCCHLCCACDD |
| 35-P. amethystina | P. amethystina | DGBBGGBBIIDDABAD |
| 36-P. amethystina | P. amethystina | DGBBGGBBIIDDABAA |
| 37-P. caerulea | P. caerulea | DDBBIJBBJJDDBBDD |
| 38-P. incarnata × P. laurifiolia | P. incarnata × P. laurifiolia | BGACMMEELLBCAA00 |
| 39-P. incarnata × P. laurifiolia | P. incarnata × P. laurifiolia | EECEIICCLLBDABCC |
| 40-P. incarnata × P. laurifiolia | P. incarnata × P. laurifiolia | EECEOOCCLLBDABCC |
| 41-P. incarnata × P. laurifiolia | P. incarnata × P. laurifiolia | EECEOOCCELBDABCC |
| 42-P. foetida | P. foetida | 00BBHHDDNNCCBB00 |
| 43-P. foetida | P. foetida | AIBBHHCCNNCCBB00 |
| 44-P. foetida | P. foetida | AABBHH00NNCDBBDD |
| 45-P. foetida | P. foetida | GIBBHHGGLNCCBBFF |
| 46-P. foetida | P. foetida | GIBBHH00NNCDBB00 |
| 47-P. incarnata | P. incarnata | EECC00EEOOBCACCC |
| 48-P. incarnata | P. incarnata | BBCCHMEEOOBCACCC |
| 49-P. ligularis | P. ligularis | IICCII00MMCCCC00 |
| 50-P. ligularis | P. ligularis | IICCIICCMMCCCCDD |
| 51-P. ligularis | P. ligularis | IICCIICCMMCCCC00 |
| 52-P. ligularis | P. ligularis | IICCIICCMMCCCCDD |
| 53-P. ligularis | P. ligularis | GIDDJJFFLLCDCEDE |
| 54-P. miniata | P. miniata | BICCIICCEHCCCCDD |
| 55-P. miniata | P. miniata | BICCIICCHHCCCC00 |
| 56-P. miniata | P. miniata | BJCCIICCHHCCCC00 |
| 57-P. miniata | P. miniata | BBCCIICCHHCCCC00 |
| 58-P. miniata | P. miniata | BBCCIICCHHCCCCDD |
| 59-P. miniata | P. miniata | BBCCIICCHHCCCCDD |
| 60-P. miniata | P. miniata | BBCCIICCHHCCCCDD |
| 61-P. miniata | P. miniata | BBCCIICCHHCCCC00 |
| 62-P. miniata | P. miniata | BGCCIICCHHCCCC00 |
| 63-P. miniata | P. miniata | BBCCIICCHHCCCCDD |
| 64-P. miniata Xishuangbanna Red | P. miniata Xishuangbanna Red | 00CCIICCHHCCCCDD |
| 65-P. miniata × P. serrulata | P. miniata × P. serrulata | BHCCIICCHLCDCCDD |
| 66-P. miniata × P. serrulata | P. miniata × P. serrulata | BHCCIICCHLCDCCDD |
| 67-P. miniata × P. serrulata | P. miniata × P. serrulata | BHCCIICCHLCDCCDD |
| 68-P. miniata × P. serrulata | P. miniata × P. serrulata | HHCCIICCHLCDCCDD |
| 69-P. miniata × P. serrulata | P. miniata × P. serrulata | HHCCIICCHLCDCCDD |
| 70-P. miniata × P. serrulata | P. miniata × P. serrulata | BHCCIICCHLCDCCDD |
| 71-P. morifolia | P. morifolia | 0000DJBBAACDCCDD |
| 72-P. morifolia | P. morifolia | 0000JJBCLLCCCCDD |
| 73-P. morifolia | P. morifolia | 0000BBBCBB00CCDD |
| 74-P. quadrangularis | P. quadrangularis | AAFFGGAALLBBDDDD |
| 75-P. quadrangularis | P. quadrangularis | AAFFGGAALLBBDDDD |
| 76-P. quadrangularis | P. quadrangularis | AAFFIIAALLBCDDBD |
| 77-P. serrulata | P. serrulata | HHCCIICCLLDDCCDD |
| 78-P. suberosa | P. suberosa | II00IJ00GLBBCCDG |
| 79-P. suberosa | P. suberosa | 00000000GG0000DG |
| 80-P. suberosa | P. suberosa | 00000000GG00AC00 |
| 81-P. suberosa | P. suberosa | JJ00IJ00GGCCCC00 |
| 87-P. suberosa | P. suberosa | AI00DIACGG00CC00 |
| 83-P. suberosa | P. suberosa | 0000EE00GG00CCGG |
| 84-P. violacea ‘victoria’ | P. violacea ‘victoria’ | DI00IIBBLLCCAB00 |
| 85-P. violacea ‘victoria’ | P. violacea ‘victoria’ | DIBBIIBBLLCCABDD |
| 86-P. yucatanensis | P. yucatanensis | II00BK00DDCDCC00 |
| 87-P. yucatanensis | P. yucatanensis | II00FF00DDACCCDD |
3. Discussion
3.1. Characterization of Passiflora SSR Loci and Primer Screening
In this study, 86,880 unigenes were identified from the Passiflora transcriptome, resulting in the identification of 27,303 SSR loci, with an SSR frequency of 31.43%. Research on microsatellite markers developed for the Passiflora species has often detected low polymorphism, with SSR frequencies ranging from 15% to 24.7% [21]. The frequency and occurrence of SSRs are influenced by factors such as species, taxonomic groups, SSR databases and development tools, and SSR search criteria [23,25,26].
In many plants, dinucleotide and trinucleotide repeats are the primary repeat types, albeit with differing dominant motifs [27,28]. In the present study, mononucleotide, dinucleotide, and trinucleotide repeats were predominant in the Passiflora transcriptome, accounting for 63.72%, 20.40%, and 14.28% of the total SSRs, respectively. In contrast, tetranucleotide, pentanucleotide, and hexanucleotide repeats constituted a smaller proportion (1.6%). Among the SSR motifs in Passiflora, A/T was the most abundant among the mononucleotide repeats, AG/CT dominated among the dinucleotide repeats, and AAG/CTT was the most prevalent among the trinucleotide repeats, all of which are consistent with the observation by Meng et al., who reported that AAG/CTT is a common motif in dicotyledonous plants. Additionally, the scarcity of CG/GC, a common feature in most dicotyledonous plants, was also observed in Passiflora, which presented the lowest CG/GC content [29].
The polymorphism level of the SSR molecular markers is a crucial criterion for assessing their usability, and SSR length is a major factor influencing polymorphism. SSRs with lengths ≥20 bp exhibit high polymorphism, whereas those between 12 and 20 bp in length display moderate polymorphism, and those shorter than 12 bp have extremely low polymorphism [30]. In this study, 50 SSR primers were designed on the basis of randomly selected loci, and six Passiflora samples were used to screen these 50 primer pairs. A total of 27 primer pairs produced clear and distinct primary bands, among which 8 were further selected for their stable amplification, clear bands, and polymorphism. Studies have shown that microsatellite markers developed for Passiflora edulis var. flavicarpa can be utilized for the genetic analysis of other Passiflora species. Silva et al. examined the transferability of 31 previously designed SSR primers between the wild and cultivated Passiflora species and reported high transferability for several SSR markers, indicating the feasibility of establishing universal SSR markers for both wild and cultivated Passiflora species. These markers can be applied to studies on the genetic diversity, identification, and exploitation of Passiflora germplasm resources, providing polymorphic molecular markers for further investigations of the genetic variation patterns of Passiflora germplasm resources [31].
3.2. Analysis of Genetic Diversity in Passiflora Germplasm Resources
Genetic diversity serves as a crucial indicator in the study of plant adaptability to environmental changes, with low genetic diversity leading to species decline in nature [32,33]. The similarity between the observed and expected heterozygosity values can also indicate the level of genetic diversity within a population, with more similar values indicating a greater genetic diversity [13]. Among the population genetic parameters, He is commonly used to measure the genetic diversity of a group. A higher He value reflects lower genetic uniformity and richer genetic diversity. In this study, the average He value of the samples of the 87 Passiflora species was 0.202, with the highest He value of 0.768 observed at the ssr18 locus, indicating substantial genetic diversity among the tested Passiflora samples.
The Ne value is a significant indicator of genetic variation within a population. An Ne value close to the absolute number of alleles detected suggests a uniform distribution of alleles, whereas a large discrepancy indicates uneven distribution [34,35,36]. In this study, the number of alleles and Ne detected by the eight core primers varied among the 87 Passiflora germplasm resources, indicating an uneven distribution of alleles among the tested samples.
The PIC is used to quantify the degree of polymorphism at a locus. A PIC value greater than 0.5 indicates a high degree of polymorphism, whereas values between 0.25 and 0.50 suggest reasonable polymorphism, and values below 0.25 indicate low polymorphism [37]. Silva et al. utilized 31 previously designed SSR primers to study the transferability of SSR markers between wild and cultivated Passiflora species, reporting PIC values ranging from 0.26 to 0.70, with an average of 0.46 [31].
Previous studies have employed SSR markers to assess genetic variation in 51 Colombian commercial yellow Passiflora accessions (102 individuals), reporting a PIC of 0.74 and Ho and He values of 0.52 and 0.78, respectively [38]. Using RAPD and ISSR markers, studies on 20 passionfruit resources from different provinces in Southern Vietnam reported PIC values ranging from 0.85 to 0.88 [39]. Analysis of 70 purple Passiflora accessions via AFLP and SSR markers revealed low genetic diversity [40]. A study on 127 Passiflora species samples from Brazil and Colombia, including 85 commercial P. edulis samples and 42 samples from 13 wild species, reported average Ho and He values of 0.43 (ranging from 0.01 to 0.77) and 0.50 (ranging from 0.01 to 0.86), respectively, for P. edulis [41]. A study of 79 Passiflora species with 18 SSRs yielded an average PIC value of 0.60, ranging from 0.46 to 0.77 [22].
Collectively, these findings demonstrate the effectiveness of SSR markers in assessing genetic diversity and characterizing Passiflora germplasm resources, contributing to efforts to harness the full potential of Passiflora for agricultural, medicinal, and ornamental applications.
3.3. Genetic Structure and Cluster Analysis of Passiflora Germplasm Resources
Genetic structure analysis and cluster analysis were performed on 87 Passiflora germplasm resources. The results revealed that these resources could be categorized into eight to nine distinct groups. Both analyses categorized the cultivated Passiflora species P. edulis, P. edulis f. flavicarpa, and their hybrids into two main groups, one primarily comprising the yellow-skinned varieties and the other consisting primarily of the purple-skinned varieties. This finding aligns with those of the previous studies on Fujian passionfruit [42] and Brazilian passionfruit [22]. Notably, the genetic variation among passionfruit cultivars and lines was substantial, with no clear correlation observed between the geographical origins of the tested samples and their genetic differences.
Species and varieties with red flowers, such as P. miniata, P. miniata Xishuangbanna Red, and P.‘Lady Margaret’ were grouped into one cluster, whereas those with purple flowers, including P. caerulea, P. amethystina, and P. violacea ‘victoria’, constituted another distinct cluster. Additionally, the hybrid P. incarnata × P. laurifolia and the species P. incarnata itself formed a separate group, which is consistent with the findings of Anderson et al. [34]. The introduction of P. incarnata (or a related species) to the karst regions for cultivation has demonstrated strong stress tolerance, and studies have suggested that it is one of the species most similar to P. edulis in terms of valuable traits such as cold tolerance and phenotypic diversity (e.g., flowering time, flower colour, fruit size, and fertility), despite its inferior fruit quality.
In the cluster analysis of this study, most species clustered with those belonging to the same subgenus. Specifically, P. morifolia and P. trifasciata, both belonging to the Decaloba subgenus, were grouped with P. edulis and P. edulis f. flavicarpa (of the Passiflora subgenus). This outcome may be attributed to the limited number of molecular marker primers used in this study, which failed to distinguish P. morifolia and P. trifasciata from other species. P. ‘Lady Margaret’ is the fanciful name given to an F1 progeny resulting from an interspecific artificial cross between P. coccinea and P. incarnata in 1991. Phenotypically, P. ‘Lady Margaret’ exhibits greater similarity to P. coccinea [43]. The failure of P. ‘Lady Margaret’ to cluster with P. incarnata could stem from genetic differences; despite the hybridization between P. coccinea and P. incarnata, the hybrid offspring may have inherited diverse genetic traits from both parents. These traits manifest as distinct amplification patterns or polymorphic loci in SSR molecular markers, thereby contributing to a genetic divergence between the hybrid and either parent. This divergence is sufficient to prevent P. ‘Lady Margaret’ from grouping with P. incarnata in the cluster analysis. Additionally, genetic recombination, a crucial mechanism in biological evolution, may have occurred during the hybridization process. In the cross between P. coccinea and P. incarnata, genetic recombination could have led to novel genetic combinations in the offspring. These novel combinations might significantly differentiate the hybrid from either parent in terms of SSR molecular markers. Similarly, in the study by Anderson et al., P. incarnata × P. edulis and P. edulis × P. incarnata did not cluster with P. incarnata and displayed relatively distant genetic relationships [44].
3.4. Construction of DNA Fingerprints for Passiflora Germplasm Resources
The construction of DNA fingerprints, characterized by multiple loci, high variability, and simple yet stable inheritance, is considered the simplest and most effective approach for cultivar identification. The higher the PIC value of a primer is, the more capable the primer is of distinguishing cultivar-specific traits, making it suitable for use as a core primer in constructing DNA fingerprinting profiles [37]. In this study, eight pairs of SSR primers were employed to establish the DNA fingerprinting profiles for 87 Passiflora germplasm resources, encompassing 22 species (cultivars) within the genus. Notably, these eight SSR primers were fully capable of differentiating the 22 species (cultivars), and the constructed fingerprinting profiles also discriminated between some species (cultivars) that have been introduced and cultivated for extended periods in the karst mountains of Guizhou. Traditional methods of cultivar identification on the basis of phenotypic traits are often influenced by a combination of environmental factors. In contrast, DNA fingerprinting directly reveals the genetic differences among the cultivars at the DNA level, offering greater precision, polymorphism, and stability than phenotype-based identification does, and it is unaffected by environmental conditions or the plant’s growth status. These findings provide a direct and effective theoretical foundation for cultivar improvement and conservation [45].
4. Materials and Methods
4.1. Experimental Materials
The experimental materials consisted of 87 Passiflora germplasm resources that have been cultivated and domesticated for many years in the Passiflora Germplasm Nursery in Kedu Town, Pingtang County, Guizhou Province (25°44′ N, 106°48′ E, 884 m above sea level), and the Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences (21°56′ N, 101°16′ E, 570 m above sea level). Table S1 presents a summary of these materials. For transcriptome sequencing, the material of interest was purple-fruited Passiflora from Pingtang County, Guizhou Province. The aboveground parts (stems, leaves, flowers, and fruits) of three individual plants were harvested, immediately frozen on dry ice, and transported for storage in an ultralow temperature freezer at −80 °C. RNA was extracted separately from stems, leaves, flowers, and fruits, and then equal amounts were pooled for subsequent transcriptome sequencing.
4.2. SSR Locus Mining and Primer Design
The SSR loci within the Passiflora transcriptome data were identified and analysed using MISA v2.1 (http://pgrc.ipk-gatersleben.de/misa/misa.html (accessed on 1 October 2020)) with the following parameters: minimum repeat numbers of 8, 5, 4, 3, 2, and 2 for mono-, di-, tri-, tetra-, penta-, and hexanucleotide motifs, respectively. Fifty SSR loci were randomly selected, and primers were designed using Primer5.0. The primers were synthesized by GenScript Biotech Corporation (Nanjing, China), dried to powder, diluted to 10 μmol/L, and stored at −20 °C for later use.
4.3. Extraction and Analysis of DNA from Passiflora
Genomic DNA was extracted from Passiflora samples via the BioTeke Plant Genomic DNA Extraction Kit. The concentration and purity of the DNA were measured via a Thermo Scientific NanoDrop 2000C spectrophotometer (Thermo Scientific, Waltham, MA, USA), and the quality of the DNA was preliminarily assessed on the basis of the OD values obtained. The DNA was diluted to a concentration of 30 ng/μL and stored at 4 °C for future use.
4.4. PCR System and Program Setup
The PCR mixture had a total volume of 10 μL, containing 1 μL of 10 × buffer, 0.4 μL of 25 mmol/L MgCl2, 1 μL of 2.5 mmol/L dNTPs, 0.3 μL each of 0.2 μmol/L forward and reverse SSR primers, 0.12 μL of Taq DNA polymerase (TAKARA), 1.5 μL of DNA template, and 5.38 μL of ddH2O. The PCR cycling conditions were as follows: initial denaturation at 94 °C for 5 min, followed by 30 cycles of denaturation at 94 °C for 30 s, annealing at 55–58 °C for 30 s, and extension at 72 °C for 30 s, with a final extension at 72 °C for 3 min.
4.5. Polyacrylamide Gel Electrophoresis
The PCR amplicon lengths were determined via 8% polyacrylamide gels. Electrophoresis was performed in a DYCZ-30A vertical electrophoresis tank (Beijing Liuyi Instrument Factory, Beijing, China) with a 50 bp marker as the standard molecular weight marker. Electrophoresis was conducted at 120 V and 350 mA for 120 min. The PCR products were sent to Sangon Biotech (Shanghai) Co., Ltd. Shanghai, China, for capillary electrophoresis analysis via an ABI 3730XL DNA Analyser. The internal size standard used was GS-500 LIZ.
4.6. Data Statistics and Analysis
4.6.1. Polymorphism Analysis of Microsatellite Loci
The polymorphisms of the eight developed microsatellite loci were evaluated via GenAlEx 6.51b2 [46]. Key polymorphism indices, including the number of alleles (Na), the number of effective alleles (Ne), expected heterozygosity (He), observed heterozygosity (Ho), and Shannon’s diversity index (I), were obtained.
4.6.2. Analysis of Genetic Variation within Populations
Genetic variation within the populations was also analysed via GenAlEx 6.51b2 [46]. Population genetic diversity, which measures the allelic and genotypic composition of a given population, is characterized by the Na, Ne, He, Ho, and I values. The polymorphism information content (PIC) was calculated via PowerMarker 3.25. Population structure was analysed with STRUCTURE 2.2.4 [29], employing the admixture model and allele frequency correlation model.
4.6.3. Analysis of Population Genetic Structure and Clustering
A genetic dendrogram was constructed in MEGA 6.06 [28] on the basis of pairwise Nei’s standard genetic distances calculated via PowerMarker 3.25. A clustering heatmap was generated using R language code to visualise the clustering of the 87 Passiflora samples. STRUCTURE v2.3.4 was used to infer the population structure, employing the admixture model and allele frequency correlation model with 106 iterations of the Monte Carlo Markov chain (MCMC) algorithm. We tested K values ranging from 1 to 22, with 15 runs for each K. The optimal K value (number of subpopulations) was determined via the ΔK method using STRUCTURE HARVESTER (https://lmme.ac.cn/StructureSelector/ (accessed on 29 May 2024)), where the highest ΔK indicates the best-fit number of subpopulations.
4.6.4. Selection of Core Primers and Construction of Fingerprint Maps
Core primers were selected on the basis of their PIC to establish fingerprint maps. Fragments amplified by different primers were numbered according to their length. A two-letter code represents each primer, and “0” indicates the absence of a clear band for that primer in a particular variety. On the basis of the order of the primers and the sizes of the amplified fragments, an alphabetically arranged fingerprint map was constructed for each genotype.
5. Conclusions
This study evaluated the genetic diversity of the Passiflora species which has been domesticated and cultivated for several years in the karst region of Guizhou, China, providing crucial theoretical and technical support for the classification, collection, conservation, elite breeding, and molecular marker-assisted breeding of Passiflora genetic resources. Through primer screening and the genetic diversity analysis of the SSR loci, a series of stable and highly polymorphic SSR primers were developed, and an in-depth investigation of the genetic structure of Passiflora species was conducted. The results revealed a wealth of genetic diversity and distinct genetic structures among the Passiflora resources. There was a certain degree of gene flow among the different populations, while some populations presented high genetic differentiation. Fingerprint maps were constructed to distinguish selected species (cultivars) that have been introduced and cultivated for years in the karst mountains. Genetic differences and relationships among the different germplasm resources were identified. These classifications offer important references and guidance for the conservation, utilization, and elite breeding of Passiflora genetic resources. This study provided in-depth insights into the genetic diversity, genetic structure, and phylogenetic relationships of Passiflora species, providing solid scientific evidence and technical support for the conservation and utilization of their germplasm resources. Future research could further expand the sample range, delve deeper into genetic information, and conduct comprehensive analyses incorporating phenotypic data to elucidate the genetic characteristics and evolutionary history of Passiflora species, thereby offering deeper insights and guidance for the conservation and utilization of their germplasm resources.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms251910815/s1.
Author Contributions
Conceptualization, A.L.; Software, Y.G.; Validation, G.C.; Formal Analysis, P.X.; Resources, M.X.; Writing—Original Draft, F.W.; Project Administration, A.L. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article and Supplementary Materials.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This research was funded by the Science and Technology Project of Guizhou Province (Qian-Ke-He Platform Talents [2021]5624; Qian-Ke-He Support [2021]General 228), the National Natural Science Foundation of China (31960576).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Dhawan K., Dhawan S., Sharma A. Passiflora: A review update. J. Ethnopharmacol. 2004;94:1–23. doi: 10.1016/j.jep.2004.02.023. [DOI] [PubMed] [Google Scholar]
- 2.Ocampo J., Salazar A., Ceballos N., Lopez W. Passiflora: Genetic, Grafting and Biotechnology Approaches. Nova Science Publishers; Hauppauge, NY, USA: 2021. pp. 2–3. [Google Scholar]
- 3.Cerqueira-Silva C.B.M., Faleiro F.G., Jesus O.N.D., Santos E.S.L.D., Souza A.P.D. The Genetic Diversity, Conservation, and Use of Passion Fruit (Passiflora spp.) Springer International Publishing; New York, NY, USA: 2016. [Google Scholar]
- 4.Kugler E.E., King L.A. Passiflora: Passionflowers of the World. Timber Press; Portland, OR, USA: 2004. A Brief History of the Passionflower. [Google Scholar]
- 5.Deng J., Zhou Y., Bai M., Li H., Li L. Anxiolytic and sedative activities of Passiflora edulis f. flavicarpa. J. Ethnopharmacol. 2010;128:148–153. doi: 10.1016/j.jep.2009.12.043. [DOI] [PubMed] [Google Scholar]
- 6.Foudah A.I., Alam P., Kamal Y.T., Alqasoumi S.I., Alqarni M.H., Ross S.A., Yusufoglu H.S. Development and validation of a high-performance thin-layer chromatographic method for the quantitative analysis of vitexin in Passiflora foetida herbal formulations. Saudi Pharm. J. 2019;27:1157–1163. doi: 10.1016/j.jsps.2019.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.García-Ruiz A., Girones-Vilaplana A., León P., Moreno D.A., Stinco C.M., Meléndez-Martínez A.J., Ruales J. Banana Passion Fruit (Passiflora mollissima (Kunth) L.H. Bailey): Microencapsulation, Phytochemical Composition and Antioxidant Capacity. Molecules. 2017;22:85. doi: 10.3390/molecules22010085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gadioli I.L., da Cunha M.D.S.B., de Carvalho M.V.O., Costa A.M., Pineli L.D.L.D.O. A systematic review on phenolic compounds in Passiflora plants: Exploring biodiversity for food, nutrition, and popular medicine. Crit. Rev. Food. Sci. Nutr. 2017;58:785–807. doi: 10.1080/10408398.2016.1224805. [DOI] [PubMed] [Google Scholar]
- 9.Alcaraz M.L., Hormaza J.I. Molecular characterization and genetic diversity in an avocado collection of cultivars and local Spanish genotypes using SSRs. Hereditas. 2010;144:244–253. doi: 10.1111/j.2007.0018-0661.02019x. [DOI] [PubMed] [Google Scholar]
- 10.Sima T., Thohirah L.A., Mohd Y., Mohamed H., Mahbod S., Parisa A., Redmond S. Mining and Development of Novel SSR Markers Using Next Generation Sequencing (NGS) Data in Plants. Molecules. 2018;23:399. doi: 10.3390/molecules23020399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nadeem M.A., Nawaz M.A., Shahid M.Q., Doğan Y., Comertpay G., Yıldız M., Baloch F.S. DNA molecular markers in plant breeding: Current status and recent advancements in genomic selection and genome editing. Biotechnol. Biotechnol. Equip. 2018;32:261–285. doi: 10.1080/13102818.2017.1400401. [DOI] [Google Scholar]
- 12.Karci H., Paizila A., Guney M., Zhaanbaev M., Kafkas S. Revealing genetic diversity and population structure in Pistachio (Pistacia vera L.) by SSR markers. Genet. Resour. Crop Evol. 2022;69:2875–2887. doi: 10.1007/s10722-022-01410-w. [DOI] [Google Scholar]
- 13.Nie X.H., Wang Z.H., Liu N.W., Song L., Cao Q.Q. Fingerprinting 146 Chinese chestnut (Castanea mollissima Blume) accessions and selecting a core collection using SSR markers. J. Integr. Agric. 2021;20:1277–1286. doi: 10.1016/S2095-3119(20)63400-1. [DOI] [Google Scholar]
- 14.Zhao S.-Q., Liang B., Li L.-B., Pang C.-Y., Su J.-J., Song M.-Z., Wei H.-L., Wang H.-T., Fan S.-L., Yu S.-X. Genetic Diversity Analysis of Early-maturity Related Germplasm in Upland Cotton (Gossypium hirsutum L.) Based on SSR Markers. J. Plant Genet. Resour. 2016;17:599–606. doi: 10.13430/j.cnki.jpgr.2016.04.003. [DOI] [Google Scholar]
- 15.Wang R., Li X., Zhang W., Ou J., Fang C., Song Q., Zhou H. SSR analysis and fingerprint construction to evaluate the genetic diversity of medicinal plum varieties. J. Plant Biochem. Biotechnol. 2022;31:1–11. doi: 10.1007/s13562-021-00681-1. [DOI] [Google Scholar]
- 16.Liu M., Xu Y., He J., Zhang S., Wang Y., Lu P. Genetic Diversity and Population Structure of Broomcorn Millet (Panicum miliaceum L.) Cultivars and Landraces in China Based on Microsatellite Markers. Int. J. Mol. Sci. 2016;17:370. doi: 10.3390/ijms17030370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kou S.J., Huo A.H., Fu G.Q., Ji J.J., Wang Y., Zuo Z.X., Liu M.X., Lu P. Genetic diversity and population genetic structure of Chinese broomcorn millet analyzed using fluorescent SSR markers. Sci. Agric. Sin. 2019;52:1475–1487. [Google Scholar]
- 18.Meng Y.S., Ning Z.H.A.O., Hui L.I., Hong Z.H.A.I., He S.Z., Liu Q.C. SSR fingerprinting of 203 sweetpotato (Ipomoea batatas (L.) Lam.) varieties. J. Agric. Sci. 2018;17:86–93. doi: 10.1016/S2095-3119(17)61687-3. [DOI] [Google Scholar]
- 19.Corrado G., Forlani M., Rao R., Basile B. Diversity and Relationships among Neglected Apricot (Prunus armeniaca L.) Landraces Using Morphological Traits and SSR Markers: Implications for Agro-Biodiversity Conservation. Plants. 2021;10:1341. doi: 10.3390/plants10071341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Button P. New developments in the International Union for the Protection of New Varieties of Plants (UPOV) J. Psychiatr. Ment. Health Nurs. 2007;12:464–471. doi: 10.17660/ActaHortic.2006.714.22. [DOI] [Google Scholar]
- 21.Santos A.A., Penha H.A., Bellec A., Munhoz C.D.F., Pedrosa-Harand A., Berges H., Carneiro Vieira M.L. Begin at the beginning: A BAC-end view of the passion fruit (Passiflora) genome. BMC Genom. 2014;15:816. doi: 10.1186/1471-2164-15-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Susan A., Martins A.M., Junqueira N.T., Costa A.M., Faleiro F.G., Ferreira M.E. Microsatellite marker development by partial sequencing of the sour passion fruit genome (Passiflora edulis Sims) BMC Genom. 2017;18:549. doi: 10.1186/s12864-017-3881-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Da Costa Z.P., Munhoz C.D.F., Vieira M.L.C. Report on the development of putative functional SSR and SNP markers in passion fruits. BMC Res. Notes. 2017;10:445. doi: 10.1186/s13104-017-2771-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong W.P., Li A.D., Zhang J.L., Long X.Q. Characteristics of the habitat and niche of naturalized passionflowers in karst peak-cluster depressions. Zhejiang Agric. Sci. 2019;31:7. [Google Scholar]
- 25.Srivastava S., Avvaru A.K., Sowpati D.T., Mishra R.K. Patterns of microsatellite distribution across eukaryotic genomes. BMC Genom. 2019;20:153. doi: 10.1186/s12864-019-5516-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Q., Su X., Ma H., Du K., Yang M., Chen B., Fu S., Fu T., Xiang C., Zhao Q. Development of genic SSR marker resources from RNA-seq data in Camellia japonica and their application in the genus Camellia. Sci. Rep. 2021;11:9919. doi: 10.1038/s41598-021-89350-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kong Q. Ph.D. Thesis. Huazhong Agricultural University; Wuhan, China: 2006. Identification, Development, and Utilization of EST-SSR Markers in the Cucumis Genus Based on Public Sequence Databases. [Google Scholar]
- 28.Zhu L., Wu H., Li H., Tang H., Yang L. Short Tandem Repeats in plants: Genomic distribution and function prediction. Electron. J. Biotechnol. 2020;50:37–44. doi: 10.1016/j.ejbt.2020.12.003. [DOI] [Google Scholar]
- 29.Meng M., Tang W., Liu J., Huang S.X., Yu J.D., Liu F.F., Lin L., Zhang X., Liu Y.S. Development of EST-SSR molecular markers based on the transcriptome sequence of Actinidia chinensis ‘Hongyang’. Chin. J. Appl. Environ. Biol. 2014;20:7 [Google Scholar]
- 30.Turudic A., Liber Z., Grdisa M., Jakse J., Varga F., Poljak I., Satovic Z. Dig-up Primers: A Pipeline for Identification of Polymorphic Microsatellites Loci within Assemblies of Related Species. Int. J. Mol. Sci. 2024;25:3169. doi: 10.3390/ijms25063169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Silva M.A.A., Souza M.M., Silva G.S., Melo C.A.F., Corrêa R.X., Araújo I.S., Conceição L.D. Analysis of transferability of microsatellite primers (SSR) in wild Passiflora species and intraspecific genetic diversity in Passiflora alata. Genet. Mol. Res. GMR. 2014;13:5908–5918. doi: 10.4238/2014.August.7.6. [DOI] [PubMed] [Google Scholar]
- 32.Rao V.R., Hodgkin T. Genetic diversity and conservation and utilization of plant genetic resources. Plant Cell Tissue Organ Cult. 2002;68:1–19. [Google Scholar]
- 33.Xu M., Li A., Teng Y., Sun Z., Xu M. Exploring the adaptive mechanism of Passiflora edulis in karst areas via an integrative analysis of nutrient elements and transcriptional profiles. BMC Plant Biol. 2019;19:185. doi: 10.1186/s12870-019-1797-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma Z.S., Li L., Zhang Y.P. Defining Individual-Level Genetic Diversity and Similarity Profiles. Sci. Rep. 2020;10:5805. doi: 10.1038/s41598-020-62362-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hedrick P. Large variance in reproductive success and the Ne/N ratio. Evolution. 2005;59:1596. [PubMed] [Google Scholar]
- 36.Wang J., Santiago E., Caballero A. Prediction and estimation of effective population size. Heredity. 2016;117:193–206. doi: 10.1038/hdy.2016.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lemos Serrote C.M.R.L. Determining the Polymorphism Information Content of a molecular marker. Gene Int. J. Focus. Gene Cloning Gene Struct. Funct. 2020;726:144175. doi: 10.1016/j.gene.2019.144175. [DOI] [PubMed] [Google Scholar]
- 38.Ocampo J., Acosta-Barón N., Hernández-Fernández J. Variability and genetic structure of yellow passion fruit (Passiflora edulis f. flavicarpa Degener) in Colombia using microsatellite DNA markers. Agron. Colomb. 2017;35:135–149. doi: 10.15446/agron.colomb.v35n2.59973. [DOI] [Google Scholar]
- 39.Ho V.T., Ngo T.K.A., Phan T.H.T., Tran T.K.P. Genetic diversity among passion fruit (Passiflora edulis) accessions of southern Vietnam using RAPD and ISSR markers. SABRAO J. Breed. Genet. 2021;53:1–14. [Google Scholar]
- 40.Ortiz D.C., Bohórquez A., Duque M.C., Tohme J., Cuéllar D., Mosquera Vásquez T. Evaluating purple passion fruit (Passiflora edulis Sims f. edulis) genetic variability in individuals from commercial plantations in Colombia. Genet. Resour. Crop Evol. 2012;59:1089–1099. doi: 10.1007/s10722-011-9745-y. [DOI] [Google Scholar]
- 41.Cerqueira-Silva C.B.M., Jesus O.N., Santos E.S.L., Corrêa R.X., Souza A.P. Genetic Breeding and Diversity of the Genus Passiflora: Progress and Perspectives in Molecular and Genetic Studies. Int. J. Mol. Sci. 2014;15:14122–14152. doi: 10.3390/ijms150814122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei X.Q., Xu L., Zhang X.J., Yu D., Chen Z.F., Zhang L.M., Chen C.Z., Xu J.H. Analysis of SSR information in the transcriptome of wax apple and development of molecular markers. Acta Hortic. Sin. 2018;45:11. [Google Scholar]
- 43.Aizza L.C.B., Sawaya A.C.H.F., Dornelas M.C. Identification of anthocyanins in the corona of two species of Passiflora and their hybrid by ultra-high performance chromatography with electrospray ionization tandem mass spectrometry (UHPLC-ESI-MS/MS)—ScienceDirect. Biochem. Syst. Ecol. 2019;85:60–67. doi: 10.1016/j.bse.2019.05.003. [DOI] [Google Scholar]
- 44.Anderson J.D., Vidal R.F., Brym M., Stafne E.T., Resende M.F.R., Viana A.P., Chambers A.H. Genotyping-by-sequencing of passion fruit (Passiflora spp.) generates genomic resources for breeding and systematics. Genet. Resour. Crop Evol. 2022;69:2769–2786. doi: 10.1007/s10722-022-01397-4. [DOI] [Google Scholar]
- 45.Xue J.H., Jiang L., Ma X.L., Bing Y.H., Zhao S.C., Ma K.P. Construction of DNA fingerprint of lotus varieties. Biodivers. Sci. 2016;24:9 [Google Scholar]
- 46.Peakall R., Smouse P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics. 2012 doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are contained within the article and Supplementary Materials.