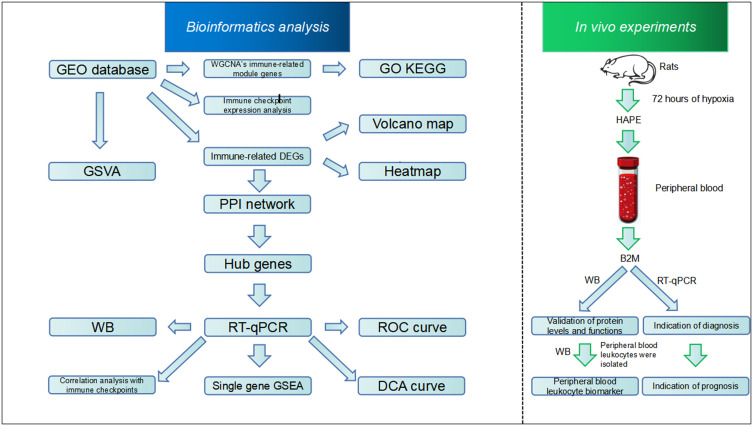
Abstract
Purpose
We aimed to investigate whether peripheral blood biomarkers B2M related to immune response can serve as indicators of HAPE pathophysiological characteristics or disease progression.
Patients and Methods
Bioinformatics technology was used to explore the peripheral blood pathophysiological mechanisms and immune hub genes related to the occurrence of HAPE. The hub gene was verified through animal experiments, and its function and correlation between its expression level and the diagnosis, treatment effect and prognosis of HAPE were explored.
Results
The GSVA results showed that the occurrence of HAPE was related to the down-regulation of immune response pathways by RUNX3 and STING. WGCNA results showed that the peripheral blood immune gene module related to the development of HAPE was related to the decrease of immune function and the increase of immune checkpoint molecule PD-L1 gene expression, and the expression of immune checkpoint genes LILRB2 and SIGLEC15 increased. Cytoscape software, RT-qPCR and WB confirmed that the hub gene B2M is a specific peripheral blood biomarker of HAPE. ROC, DCA, RT-qPCR, HE and Masson results showed that the expression of peripheral blood B2M has the ability to indicate the diagnosis, treatment effect and prognosis of HAPE. The decreased expression of B2M protein in peripheral blood leukocytes may be a marker of HAPE. Single-gene GSEA confirmed that the reduced expression of B2M in peripheral blood may be involved in the down-regulation of the antigen presentation pathway mediated by MHC class I molecules, was positively correlated with the down-regulation of the TNF signaling pathway, and was negatively correlated with the expression of LILRB2 and SIGLEC15.
Conclusion
The occurrence of HAPE may be related to decreased immune function and immune tolerance. Peripheral blood B2M may be involved in the related pathways, its expression level can prompt the diagnosis, treatment and prognosis of HAPE.
Keywords: high altitude pulmonary edema, immune tolerance, biomarker, B2M, prognosis
Graphical Abstract
Introduction
HAPE, a disease characterized by rapid progression and a specific mortality rate, develops at high altitudes. Most researchers believe that inflammation is not its causative factor. They believe that except for high altitude hypoxia factors, genetic susceptibility, sleep, mental factors and other factors are important factors inducing the occurrence of HAPE.1,2 While certain studies maintain the belief that inflammation plays a role in causing diseases,3,4 other studies indicate that the quantity of immune cells in the body decrease notably at high altitudes.5,6 Additionally, signal transduction pathways associated with immune function are hindered, resulting in impaired generation and regulation of immune responses compared to low-altitude conditions. Consequently, this diminished immune capability reduces the body’s ability to combat pathogens. Susceptible to a variety of bacteria and virus.7 Functional inhibition of peripheral blood immune cells can lead to uncontrolled inflammatory responses, which can lead to increased pulmonary vascular permeability and the formation of pulmonary edema;8 at the same time, functionally impaired peripheral blood immune cells can also affect the production and clearance of inflammatory factors,9 further promoting the occurrence of HAPE and aggravating its condition.10 Additional investigation into the molecular mechanisms that trigger or suppress immune activity in the bloodstream will enhance our comprehension of the pathophysiology and therapeutic approaches for HAPE.
HAPE has an acute onset and rapid progress, and the mortality rate may reach 50% without timely treatment.8 Early diagnosis and prompt treatment are crucial for improving prognosis, reducing complications, and decreasing mortality.11 Currently, the conventional diagnostic approach for HAPE involves comprehensive evaluation based on clinical manifestations and medical imaging findings.12 However, early or atypical cases of HAPE pose challenges in terms of diagnosis due to their nonspecific clinical features and chest X-ray results.13 HAPE often presents symptoms such as severe shortness of breath, coughing, coughing up foamy or pink sputum.14 Physical examination of high-altitude pulmonary edema may reveal crackles or rales in the lungs, rapid or irregular heartbeat, and signs of poor oxygenation.15 In the initial stages of HAPE, interstitial edema is predominant, while alveolar edema becomes more prominent in later stages.16 Patients with interstitial pulmonary edema often exhibit atypical moist rales, wheezing, and dyspnea that can lead to misdiagnosis,11 potentially resulting in delayed treatment initiation and poor prognosis. Therefore, there is an urgent need for a sensitive laboratory indicator capable of rapidly screening for early or atypical HAPE cases.17 The non-invasive detection of mRNA expression of biomarkers in peripheral blood is a straightforward and highly sensitive method, extensively employed for early disease diagnosis, treatment evaluation, and prognosis assessment. It holds significant potential in the diagnosis and management of HAPE.18,19 Previous studies showed that the peripheral blood biomarkers Complement C3, haptoglobin (Hp), alpha 1-antitrypsin (α1-AT), apolipoprotein A-I (ApoA-I), apolipoprotein A-IV (ApoA-IV), SULT1A1, and natriuretic peptides (NPs) may play an important role in the diagnosis and prognosis evaluation of HAPE. However, the application of these markers is limited due to their lack of high sensitivity or specificity.20,21 Immune response and inflammatory response are important links in the pathophysiological process of HAPE.22 The relationship between biomarkers related to peripheral immune response and pathophysiological mechanisms or disease progression of HAPE is still unclear and needs further exploration. High-throughput sequencing technology combined with bioinformatics methods and animal experiments been used to detect biomarkers of different diseases and has the ability to enhance the diagnosis of HAPE patients as well as treatment evaluation and prognostic evaluation.23,24 We use bioinformatics to explore new pathophysiological mechanisms of HAPE. Our previous research found that the peripheral blood immune-related biomarker B2M is associated with the occurrence of HAPE. In order to further study the correlation between peripheral blood B2M and the occurrence, development and prognosis of HAPE, we used the immune genes of the GEO data set to conduct differential gene hub gene analysis and protein expression changes to further verify that peripheral blood B2M may be a biomarker of HAPE. Animal experiments were conducted to explore the relationship between its expression level and disease prognosis. The results of this study can provide new theoretical basis for pathophysiological research, early diagnosis, clinical treatment effect and prognosis assessment of HAPE.25
Material and Methods
Gene Set Variation Analysis (GSVA)
GSVA is a bioinformatic-based analysis method that can be used to infer the activity of gene set-enriched pathways in individual samples.26 The enrichment analysis of molecular signaling pathways for each sample in the GSE52209 dataset of the high altitude adaptation group and the HAPE group was performed using the “GSVA” package from the “C2: all canonical pathways” in the MSigDB database. The difference in molecular signaling pathways between the two groups was analyzed using the “limma” package. The results presented in the figure represent the top 10 pathways with up-regulation and down-regulation, ranked based on absolute value of t value from largest to smallest. A pathway with p < 0.05 was considered to have significant differences.
Weighted Gene Co-Expression Network Analysis (WGCNA) Was Used to Study the Peripheral Blood Pathophysiological Mechanism of HAPE
In order to study the immune function of peripheral blood in HAPE, we performed WGCNA on 1896 immune genes in peripheral blood of 14 samples from the high altitude adaptation group and 17 samples from the HAPE in GSE52209 to explore the relationship between immune genes and phenotypes. WGCNA was used to construct a gene co-expression network to explore the relationship between genes and phenotypes.27 We used the gene expression profile, we calculated the Median Absolute Deviation (MAD) of each gene separately, and excluded the top 50% of genes with the smallest MAD. WGCNA was used to construct a scale-free co-expression network. β is a soft threshold parameter that can emphasize strong correlations between genes and penalize weak correlations, and its value is set to 14. The adjacency was transformed into a topological overlap matrix (TOM), which could measure the network connectivity of a Gene defined as the sum of its adjacency with all other Genes for network Gene ration. To classify Genes with similar expression profiles into Gene modules, average linkage hierarchical clustering was conducted according to the TOM-based dissimilarity measure with a minimum size (Gene group) of 30 for the Genes dendrogram. We also merged modules with distance less than 0.25, and finally obtained 8 co-expression modules. It is worth to note that the grey module is considered as a set of genes that cannot be assigned to any module. We used the phenotype and correlation of the module genes to calculate the association between the gene module and the onset of HAPE. “ClusterProfiler” package was used for GO and KEGG analyses.
Dataset Preprocessing and Identification of Immune-Related DEGs
In order to study the differentially expressed genes between the plateau adaptation group and the HAPE group, we searched the GEO database (https://www.ncbi.nlm.nih.gov/geo) and downloaded the microarray matrix data “Miniml” from the data set GSE52209 Format file, including 31 individuals ascended to the 3500m plateau, out of whom 14 successfully acclimatized to the high-altitude environment, while 17 developed HAPE within a span of 48–72 hours. Peripheral blood samples were collected for transcriptome sequencing of peripheral blood. The selection of two data sets, the plateau adaptation group and the HAPE group, can exclude the variable hypoxia that exists in plain and plateau environments. Download the intersection of two sets of data sets, the gene expression profile and the immune gene set (https://www.immport.org), to obtain the immune-related gene expression profile. Use the “Limma” package of R software to identify immune-related Differentially expressed genes (DEGs), then use the “ggplot2” package to generate a volcano plot, and then use the R software’s “pheatmap” package to classify up and down-regulation according to the log2|FC| value of DEGs to create heat maps.
PPI Network Analysis and Hub Gene Identification
The STRING database (https://cn.string-db.org/) was employed for protein-protein interaction (PPI) network analysis, utilizing a confidence score threshold of > 0.4 to identify PPI pairs and then using Cytoscape’s MCODE, MCC, DMNC, MNC and Degree algorithms perform cluster analysis on the protein network of genes. Hub genes were found in the gene cluster with the highest score after the intersection of various algorithms is the hub gene related to HAPE peripheral blood and immunity.
To Determine the Differentially Expressed Genes Between the Plain Normal Group and the Plateau Acclimatization Group and Between the Plateau Acclimatization Group and the Acute Altitude Sickness Group
All transcriptome sequencing data were obtained from the GEO dataset GSE75665, which was the data obtained after taking peripheral blood of normal plain people in the plain and entering the 5300m altitude within 72 hours, and then taking peripheral blood again for transcriptome sequencing. All subjects were informed and consented to participate in the trial. We performed transcriptome sequencing on peripheral blood, which was the plateau adaptation group. Use the “Limma” package of R software to identify immune-related DEGs, then use the “ggplot2” package to generate a volcano plot, and then use the “pheatmap” function of the R package to classify up and down-regulation according to the log2|FC| value of DEGs to create heat maps.
Comparative Analysis of Expression Levels of Immune Checkpoint Genes and Hub Genes
Immune checkpoint is an inhibitory receptor on immune cells that can inhibit T cell function and plays an important role in maintaining autoimmune tolerance and regulating the duration and amplitude of physiological immune responses.28 Use the R software “stats” and “car” packages to select the two-tailed t test to perform statistics on the expression values of common immune checkpoint genes and hub genes in the altitude adaptation group and HAPE group in the GSE52209 data set, and the results were visualized using the “ggplot2” package. The significance standard for the expression difference between the two groups of samples is p<0.05.
Establishment of HAPE Model
24 male SD rats, weighing (200±20) g, were collected from the animal experiment center of Daping Hospital of Army Medical University. Experiment approval was obtained from Army Medical University’s Ethics Committee. The implementation of research completely follows the 3R principle, which means substitution, reduction and optimization. The experimental animals were placed in a hypobaric oxygen chamber (ZS-DY-HP, Zhongshi Technology, China) and rose to a simulated altitude of about 5000m in about 20 minutes. The air pressure was 53.4-54kpa, the oxygen concentration was 17.5%, the humidity was 55%, and the constant temperature was 23-26℃. A light cycle of 12 hours is implemented, while food and water are provided ad libitum. Rats were randomly assigned to two groups: (1) control group (n=12), which did not undergo negative pressure hypoxia treatment; (2) HAPE group (n=12), which was exposed to negative pressure hypoxia for 72 hours. At the end of modeling, air was put into the chamber, and the pressure inside the chamber slowly rose to atmospheric pressure.
Determination of Water Content Changes
The rats were divided into two groups: Untreated plain (Control) group (n=6) and HAPE group (n=6). Immediately after removal from the hypobaric chamber, the animals were sacrificed under anesthesia and intraperitoneally injected with 350 mg/kg 2.5% tribromoethanol. The lung tissue of both lungs was collected to measure the wet weight of the lungs. The lung tissue was placed in a constant-temperature oven at 50°C for 72h until constant weight was determined as lung dry weight. The lung tissue was weighed with a precision balance (BSA224S, Sartorius, Germany) with a minimum graduation value of 0.1 mg and then the lung water content was calculated. The lung water content was determined by expressed as the ratio of wet (W) weight/dry (D) weight.
Evans Blue Staining
Evans blue (10%) was injected into the tail vein of SD rats (n=3) and untreated rats (n=3). About 1 hour later, after the eyes and skin of the rats appear blue, 2.5% tribromoethanol 350mg/kg is injected intraperitoneally. The rats were anesthetized by injection and then perfused through the heart, and then 100 mg of left upper lung tissue was taken. Place the tissue in a 1.5 mL centrifuge tube, add 1 mL of dimethylformamide solution (dimethylformamide: PBS = 1:1), quickly use a tissue homogenizer to homogenize the lung tissue and then centrifuge. Take the supernatant after centrifugation, 1mL was added to a 1.5mL EP tube at a ratio of supernatant: dimethylformamide solution = 3:7, incubate at 37°C for 24 hours, take the supernatant solution, and use an absorption light microplate reader (800TS, BioTEK, USA) to measure the absorbance value (OD value) at 630nm, and at the same time measure the OD value of Evans blue staining with known different concentration gradients, draw a standard curve, and calculate the Evans blue content of the tissue sample to be tested based on the standard curve.
Reverse Transcription-Polymerase Chain Reaction (RT-qPCR)
After rats (control=4; HAPE=4) were modeled, their peripheral blood cells were lysed using TRIzol reagent, and the concentration of RNA was detected using a fluorescence quantification meter. Then use the reverse transcription kit (RC112-01, Vazyme, China) to reverse transcribe the total RNA, add 2 × ChamQ Universal Master Mix (Q711-02, Vazyme, China) to the cDNA sample that needs to be amplified, and use the CFX96 real-time fluorescence quantification system (CFX 96 Touch, Bio-Rad, USA), the relative expression of each sample is the Ct ratio with β-actin as the internal reference. The primer sequences are shown in the table (Table 1).
Table 1.
Primers for RT-qPCR
| Gene | Forward (5’-3’) | Reverse (5’-3’) |
|---|---|---|
| B2M JAK2 ICAM1 PDCD1LG2 β-actin |
GGCTCACACTGAATTCACACC CACCCAATCATGTCTTCCACA AGACCTTGAGAATCTACAACTTTTC CAACCAGAGGGGATGCACTG CTTCCTCCCTGGAGAAGAGC |
ACATGTCTCGGTCCCAGGT AATCATGCCGCCACTGAGC GGGAAGTACCCTGTGAGGTG TTGCGAGCTCAGGTTCAGTA CCACAGGATTCCATACCCAGG |
Isolation of Leukocytes from Peripheral Blood of Rats
Fresh anticoagulated peripheral blood samples were collected, and an appropriate amount of dedicated separation solution was added to the prepared centrifuge tube according to the blood volume, ensuring that the total amount of fluid did not exceed ⅔ of the centrifuge tube volume. During addition, blood was drawn using a Pasteur tube and then slowly and evenly applied to the surface of the separation solution. The centrifuge tube containing the mixture was centrifuged at 1000g centrifugal force for 30min at room temperature using a horizontal rotor. At the end of centrifugation, a distinct layered structure is visible: the top layer is the diluted plasma, followed immediately by a layer of lymphocytes, the granulocyte layer is in the separation solution, and the bottom layer is the erythrocyte layer. Granulocytes in the lymphocyte layer and separation solution were carefully aspirated, and erythrocytes were removed by the appropriate addition of erythrocyte lysate. The supernatant was discarded, and the treated cells were re-suspended in an appropriate volume of solution for use in subsequent experiments.
Western Blotting (WB)
Samples were lysed with RIPA lysis buffer (P0013B, Beyotime, China), total protein concentration was measured by BCA protein assay kit (P0011, Beyotime, China), total protein was electrophoresed through SDS-PAGE and electrotransferred to a PVDF membrane (IPVH00010, Millipore, USA). Then incubate overnight at 4°C in 5% skim milk powder containing primary antibody, and incubate for 2 hours at room temperature in 5% skim milk powder containing HRP-labeled secondary antibody of the corresponding species, using an imaging system (ChemiDoc MP, Bio-Rad, USA). The signal on the membrane was recorded and Fusioncapt software was used to quantify the gel total protein and target band intensity. The dilution ratio used for the primary antibody B2M (GTX112815, Gentex, USA) was 1:1000, and the dilution ratio used for the goat anti-rabbit IgG (LF102, EpiZyme, China) HRP-conjugated antibody was 1:2500.
Single-Gene Gene Set Enrichment Analysis (GSEA)
In order to evaluate the biological functions involved in biomarkers, single-gene GSEA analysis of biomarkers was performed using the expression profile of the data set. NOM p-value<0.05 and FDR q-value<0.25 were considered significant results.
Receiver Operating Characteristic (ROC) Curve Analysis
The “pROC” package was used to analyze the ROC curves of biomarkers in the GSE52209 dataset, and the results were visualized using the “ggplot2” package. Genes with AUC>0.7 were considered relevant for HAPE diagnosis in plateau environments.
Decision Curve Analysis (DCA) Curve
Clinical utility was assessed through DCA using the “rmda” package to help determine the diagnostic value and clinical practical applicability of the nomogram.
Hematoxylin-Eosin Staining (HE) Staining
The modeling process was as described above. After modeling, 3 rats in each group were extracted from the left upper lung tissue under anesthesia. Rinse in PBS solution and fix in 4% paraformaldehyde solution for more than 24 hours, cut, dehydrate, and soak in wax. These tissues were then embedded, cut into 4µm thick sections, deparaffinized, stained with hematoxylin and eosin, dehydrated and sealed. Observations were performed with a microscope (U-HGLGPS IX71, Olympus, Japan).
Masson Stain
Dewax paraffin sections to water. Wash with tap water and distilled water in sequence, use Regaud’s hematoxylin staining solution or Weigert’s hematoxylin semen to stain the nucleus. The staining time is 5–10 minutes, wash thoroughly with water, wash again with distilled water, and use Masson Ponceau acid fuchsin solution to stain the nucleus. The tissue was stained for 5–10 minutes. The sections were soaked with 2% glacial acetic acid aqueous solution for a while and differentiated with 1% phosphomolybdic acid aqueous solution for 3–5 minutes. Without washing, directly use aniline blue or light green solution for dyeing. The dyeing time is 5 minutes. Use 0.2% glacial acetic acid aqueous solution to soak the sections for a while, and finally use 95% alcohol, absolute alcohol and xylene to clear them, and use neutral gum for mounting. Observations were performed with a microscope (U-HGLGPS IX71, Olympus, Japan).
Statistical Analysis
GraphPad Prism 8.0 software was used for graphing and statistical analysis. RT-qPCR and WB statistical graphs are expressed as mean ± standard error of the mean (SEM). Two-tailed T test and Welch test were used as needed, and differences were statistically significant at p<0.05.
Results
Comparison of GSVA and Immune Checkpoint Gene Expression in the Plateau Adaptation Group and HAPE Group in the Data Set GSE52209
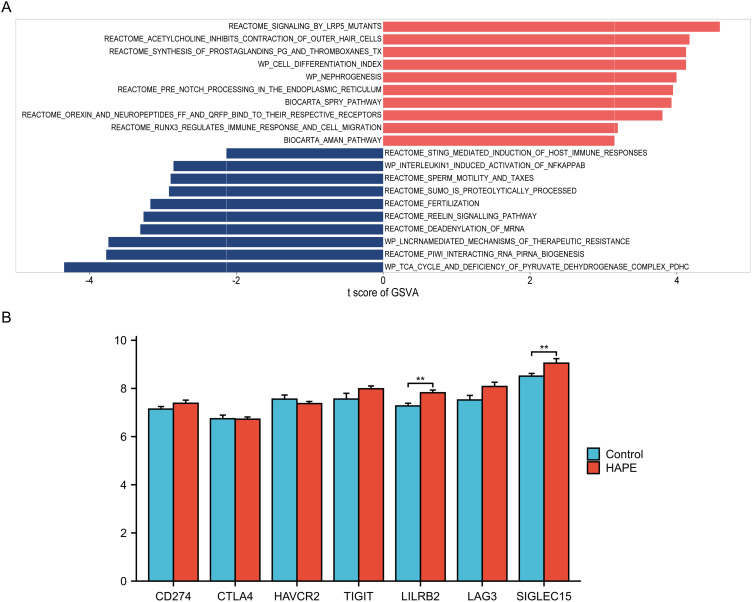
GSVA analysis can calculate the enrichment score of the screened molecular signaling pathways for each patient. The results of differential analysis of molecular signaling pathways obtained from the two groups of samples are displayed in the form of bidirectional bar graphs through the limma package. After analyzing the differences between the two groups of samples, the results of the top 10 up-regulated and down-regulated molecular signaling pathways are shown in the figure The figure shows that RUNX3 regulates the inflammatory response and up-regulation of cell migration pathways, STING-mediated immune host response and leukocyte-mediated Downregulation of the NFKAPPAB activation pathway induced by phytozoin 1 is related to the occurrence of HAPE (Figure 1A, Table S1).
Figure 1.
Comparison of regulatory differences in molecular signaling pathways and immune checkpoint gene expression between the altitude adaptation group and the HAPE group. (A) The vertical axis represents different molecular signaling pathways, and the horizontal axis represents the degree of difference in regulation of molecular signaling pathways between different groups. Up-regulated and down-regulated molecular signaling pathways with absolute t values are marked in red and blue respectively; (B) Comparison of immune checkpoint gene expression between two groups. * refers to p<0.05, ** indicates that the difference between the two groups is more significant, p<0.01.
To further verify whether other factors were responsible for the decreased immune function of the patients, we performed immune checkpoint expression analysis. Comparative results of common immune checkpoint gene expressions showed that the expression levels of LILRB2 and SIGLEC15 in the HAPE group were higher than those in the altitude adaptation group.29 The results indicate that under altitude conditions, patients with HAPE may have developed immune tolerance compared with the altitude adaptation population (Figure 1B).
WGCNA Was Used to Analyze the Gene Modules Related to the Occurrence of HAPE
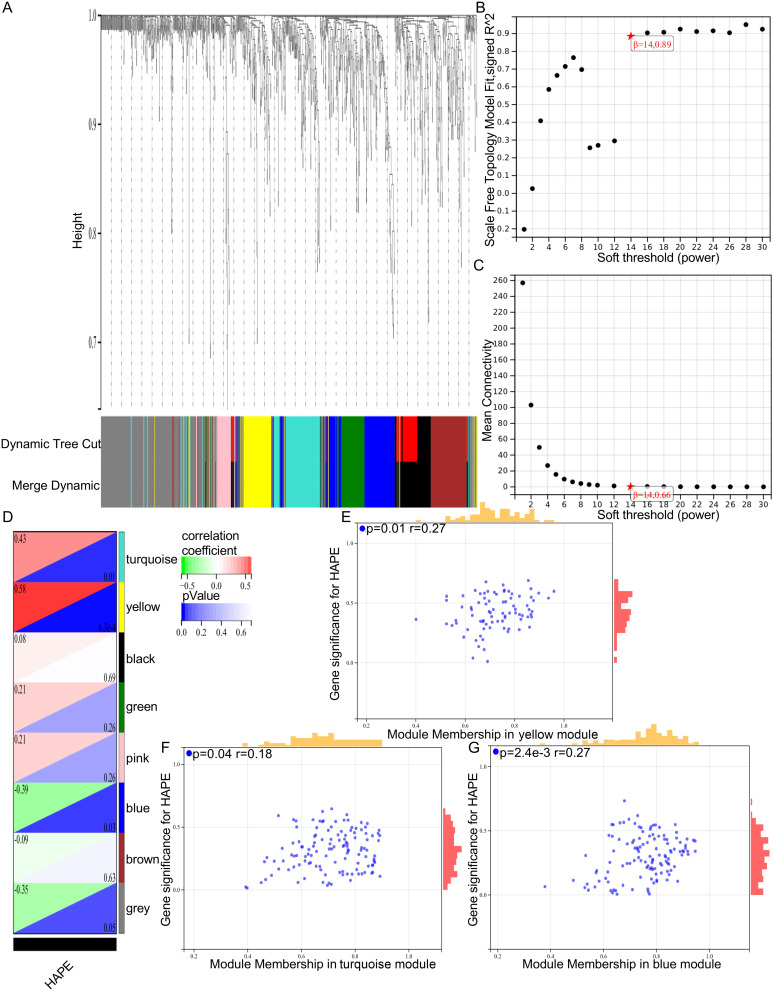
In order to further investigate the immunopathological mechanism of peripheral blood related to the occurrence of HAPE, we performed WGCNA analysis of immune genes on the two sets of GSE52009 datasets (Figure 2A–), and performed correlation analysis of gene modules related to the occurrence of HAPE. The results showed that the yellow, turquoise and blue gene modules were closely related to the occurrence of HAPE, with correlation coefficients of 0.58, 0.43 and -0.39, respectively. The yellow gene module was most associated with the occurrence of HAPE (Figure 2D). The results of correlation map showed that the genes in the three modules were positively correlated with the occurrence of HAPE, and the correlation coefficients were 0.27, 0.18 and 0.27, respectively (Figure 2E–). The above results indicated that the gene module constructed by WGCNA was successful, and the genes in the yellow, turquoise and blue modules were closely related to the occurrence of HAPE.
Figure 2.
Gene modules associated with the development of HAPE identification. (A) Genes are grouped in a dendrogram based on topological overlap. Gene modules corresponding to different colors are shown; (B and C) Network topology analysis with different soft thresholds using the scale-free fit index (left) and mean connectivity (right); (D) Identify 7 modules (non-grey). The yellow module (r = 0.58, P = 6.3e-4), turquoise module (r =0.43, P = 0.01), and blue module (r =−0.39, P =0.03) had the highest correlation; (E–G) Significance and module gene association of HAPE occurrence genes.
Functional Exploration of Immune-Related Module Genes in Peripheral Blood of HAPE
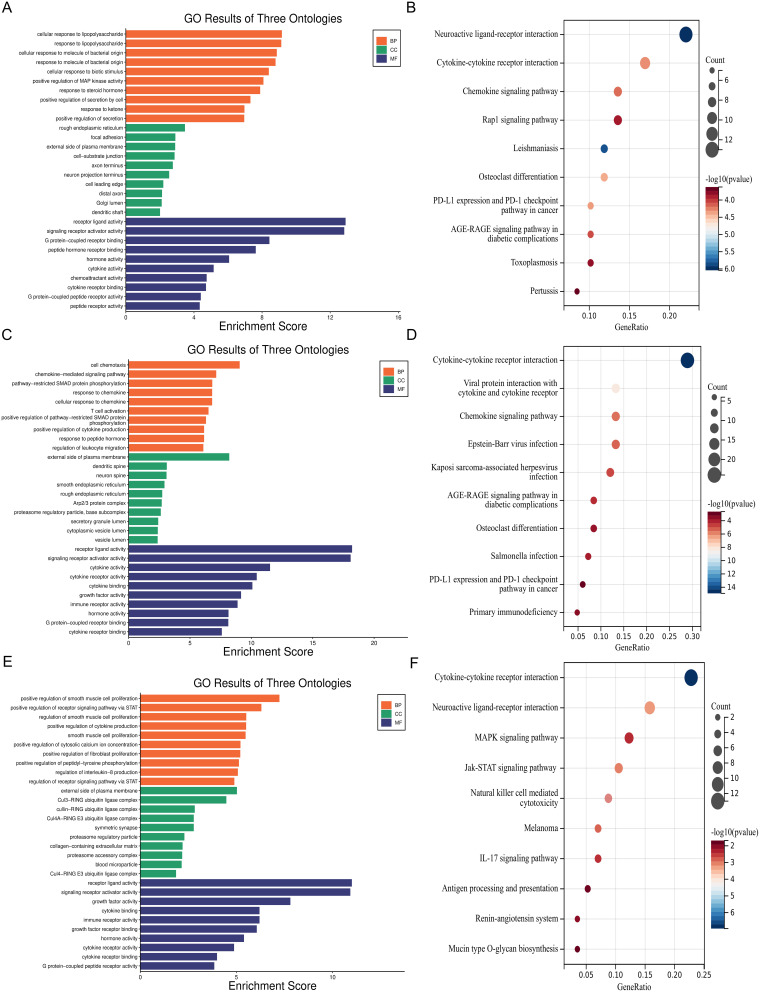
In order to further explore the enrichment function and molecular signaling pathways of module genes. We performed GO and KEGG enrichment analyses for the yellow (Figure 3A and B), turquoise (Figure 3C and D) and blue module genes (Figure 3E and F) separately. According to KEGG enrichment analysis and correlation analysis of gene modules in WGCNA, the occurrence of HAPE may be closely related to primary immune deficiency, increased expression of immune tolerance molecule PD-L1 and down-regulation of antigen processing and presentation.
Figure 3.
GO and KEGG enrichment analysis of immune-related gene modules in peripheral blood of patients with HAPE. (A and B) GO and KEGG enrichment analysis of yellow module genes; (C and D) GO and KEGG enrichment analysis of turquoise module genes; (E and F) GO and KEGG enrichment analysis of blue module genes.
PPI Network Analysis and Hub Gene Identification of Plateau Adaptation Group, HAPE Group and Immune-Related DEGs in the Data Set GSE52209
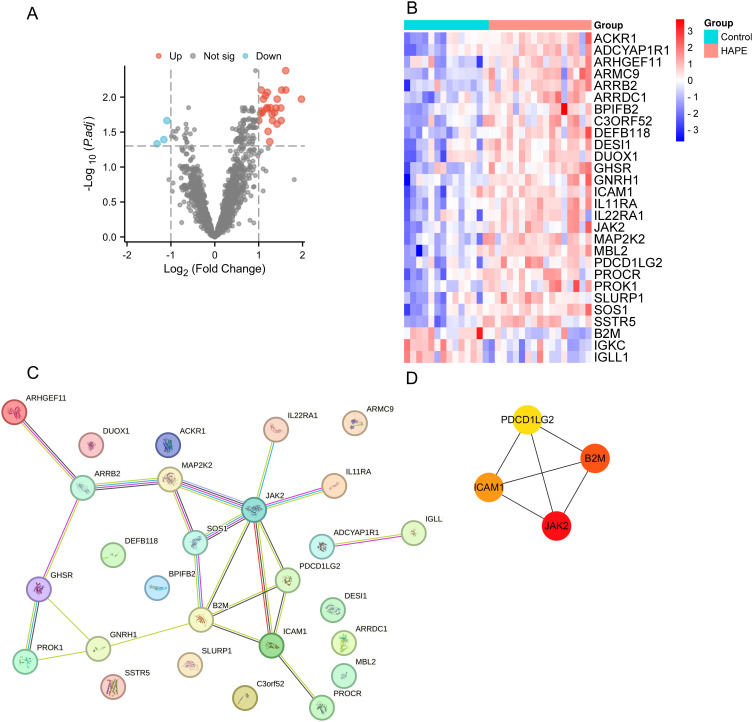
In order to explore peripheral blood immune biomarkers related to the occurrence and development of HAPE, the data set genes were intersected with the import database immune gene data set to obtain 1896 immune genes. We analyzed and found that there are a total of 28 immune genes in the data set. DEGs, of which 25 DEGs were up-regulated and 3 DEGs were down-regulated (Table 2, Figure 4A and B).
Table 2.
Identification of Immune-Related DEGs in the Plateau Adaptation Group and HAPE Group in the Data Set GSE52209
| DEGs | Gene Terms |
|---|---|
| Up-regulated | DESI1 SSTR5 IL11RA MBL2 ICAM1 ARMC9 ARRB2 PROCR ACKR1 IL22RA1 ARHGEF11 C3ORF52 MAP2K2 BPIFB2 DUOX1 PDCD1LG2 ARRDC1 JAK2 GHSR SOS1 ADCYAP1R1 SLURP1 DEFB118 GNRH1 PROK1 |
| Down-regulated | B2M IGKC IGLL1 |
Figure 4.
PPI network analysis and hub gene identification of immune-related DEGs in the plateau adaptation group and HAPE group in the data set GSE52209. (A) Volcano plot of DEGs; (B) DEGs heat map shows all up-regulated and down-regulated DEGs. The expression levels of DEGs are represented by color shades, with blue representing down-regulation and red representing up-regulation; (C) PPI network of DEGs; (D) hub genes that make up the PPI network.
Protein-protein interaction (PPI) network analysis of immune-related DEGs in peripheral blood was performed using the STRING website (https://cn.string-db.org/) (Figure 4C). We use Cytoscape software to determine the B2M, JAK2, ICAM1, PDCD1LG2 is HAPE peripheral blood and immune related DEGs hub genes (Figure 4D).
Effects of Hypoxic Plateau Conditions on Hub Gene Expression
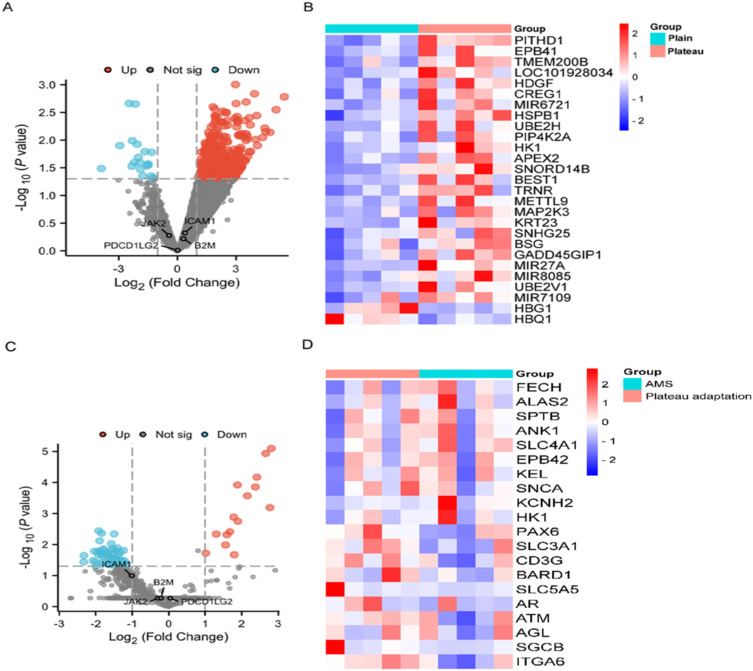
In order to verify whether peripheral blood B2M, JAK2, ICAM1 and PDCD1LG2 gene expression change under plateau hypoxic conditions, we used the normal plain group (n=5) and plateau adaptation group (n=5) data sets in the GSE75665 data set to conduct DEGs analysis to verify peripheral blood B2M, JAK2, ICAM1 and PDCD1LG2 gene expression changes (Figure 5A and B, Table 3). The results show that the above four genes are not DEGs, and their expression changes are not significant, indicating that the plateau hypoxic environment has no effect on the expression of B2M, JAK2, ICAM1 and PDCD1LG2 in peripheral blood.
Figure 5.
Plateau conditions have no effect on hub gene expression. (A) Volcano plot of DEGs in normal plain group and plateau adaptation group in GSE75665 volcano plot of DEGs in normal plain group and plateau adaptation group in GSE75665; (B) Heat map of DEGs in normal plain group and plateau adaptation group in GSE75665. The difference between the two groups of samples is significant at p<0.05; (C) Volcano plot of DEGs in GSE75665 for the plateau adaptation and acute altitude response groups; (D) Heat map of top 20 differentially expressed genes in GSE75665 according to the degree of difference between high altitude acclimatization group and acute altitude sickness group. The difference between the two groups of samples was significant, p<0.05.
Table 3.
DEGs of the Plain Normal Group and Plateau Adaptation Group in the GSE75665 Data Set
| DEGS | Gene terms |
|---|---|
| Upregulated | PITHD1 EPB41 TMEM200B LOC101928034 hDGF CREG1 MIR6721 hSPB1 UBE2H PIP4K2A HK1 APEX2 SNORD14B BEST1 TRNR METTL9 MAP2K3 KRT23 SNHG25 BSG GADD45GIP1 MIR27A MIR8085 UBE2V1 |
| Downregulated | MIR7109 hBG1 hBQ1 |
Hub Gene Can Be Used for Rapid Screening of HAPE at High Altitude
According to the results of bioinformatics analysis in the figure, B2M, JAK2, ICAM1 and PDCD1LG2 in GSE75665 are not DEGs in the high altitude adaptation group and the acute mountain sickness group, and cannot indicate acute altitude response under high altitude conditions. They have certain specificity as biomarkers of HAPE (Figure 5C and D, Table 4).
Table 4.
DEGs of the Plateau Adaptation Group and the AMS Group in the GSE75665 Data Set
| DEGs | Gene Terms |
|---|---|
| Up-regulated | FECH ALAS2 SPTB ANK1 SLC4A1 EPB42 KEL SNCA KCNH2 hK1 CA2 hMBS TPM1 PNP UROD |
| Down-regulated | PAX6 SLC3A1 CD3G BARD1 SLC5A5 AR ATM AGL SGCB ITGA6 UBE3A CHM HPRT1 MTR SLC26A2 GUCY2D PEX1 PEX2 SLC12A3 BLM XPA GK ROM1 MSH2 CACNA1A ATXN7 BRCA2 ATRX CD40LG PKD2 PRNP ATP7A FH DLD SMN1 C3 CLCN5 MSH6 LYST NF1 ERCC6 AGA ABCB4 RB1 IFNGR1 ERCC3 ADSL ATXN1 OCRL TPMT MYO5A COL4A4 APC INSR EPHX1 ABCC2 NME1 PDHA1 PCCA SHH CYBB HSD17B4 |
Abbreviations: HAPE, high altitude pulmonary edema; GEO, gene expression omnibus; DEGs, differentially expressed genes; GSVA, gene set variation analysis; RUNX3, runt-related transcription factor 3; STING, stimulator of interferon genes; PPI, protein-protein interaction;
RT-qPCR, reverse transcription-polymerase chain reaction; WB, Western blotting; ROC, receiver operating characteristic; DCA, decision curve analysis; GSEA, gene set enrichment analysis; HE, hematoxylin-eosin; SEM, standard error of the mean; LILRB2, recombinant leukocyte immunoglobulin like receptor subfamily B, member 2; SIGLEC15, sialic acid-binding Ig-like lectin 15; B2M, beta-2 microglobulin; JAK2, Janus kinase 2; ICAM-1, Intercellular cell adhesion molecule-1; PDCD1LG2, programmed cell death 1 ligand 2; AMS, Acute mountain sickness; PD-L1, programmed cell death 1 ligand 1; MHC I, major histocompatibility complex class 1; Hp, haptoglobin; α1-AT, alpha-1-antitrypsin; ApoA-1, apolipoprotein A-I; ApoA-IV, apolipoprotein A-IV; NPs, natriuretic peptides; SULT1A1, sulfotransferase 1A1.
HAPE in Rats Model of Identification
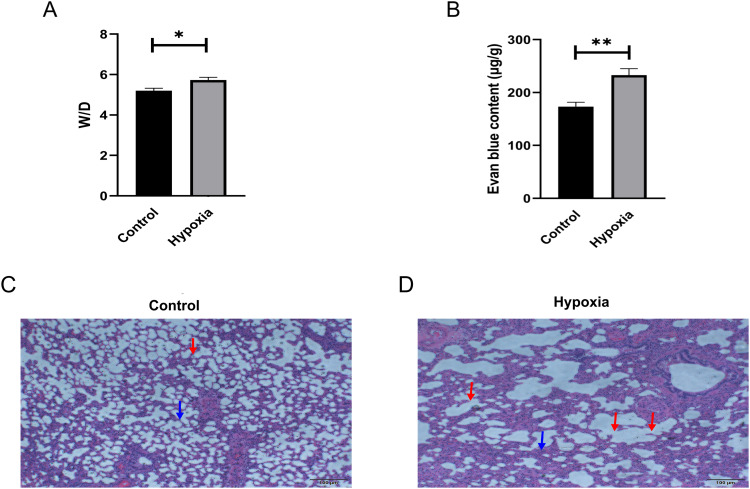
Their study showed that HAPE can occur in SD rats for 2–3 days in a hypobaric chamber that simulates an altitude of 5000 to 6000m.30,31 We are going to SD rats under low pressure in the chamber for 72 hours. We found that the water content changed through the statistical graph (Figure 6A). It is worth noting that HAPE modeling was successful. Because the investigators believed that increased vascular permeability in lung tissue was the cause of HAPE, we used Evans blue staining to detect pulmonary vascular permeability. The results showed that the absorbance value (OD) at 630nm of lung tissue samples from HAPE rats was significantly increased compared with plain normal rats (Figure 6B), and pulmonary vascular permeability was increased in HAPE rats. To further evaluate the modeling of HAPE, identification with histological staining was performed. HE staining showed congestion of alveolar wall capillaries, thickening of alveolar septum, destruction of alveolar structure and edema in the HAPE model group (Figure 6C and D).
Figure 6.
Identification of HAPE rat model. (A) Lung water content was measured by lung wet weight (W)/lung dry weight (D) (n=6); (B) Pulmonary vascular permeability measured by Evans blue staining of lung tissue in the two groups (n=3). Statistical plot data are shown as Mean±SEM, and the significance level for the comparison of the two groups is *p<0.05 and ** p<0.01; (C and D) HE staining images of plain normal rats and hypoxic rats (n=3), Scale bar: 200μm. The blue arrow points to the thickening of the alveolar septum, and the red arrow points to the structural destruction of the alveolar wall.
HAPE Hub Gene Expression and Animal Experiment Verification
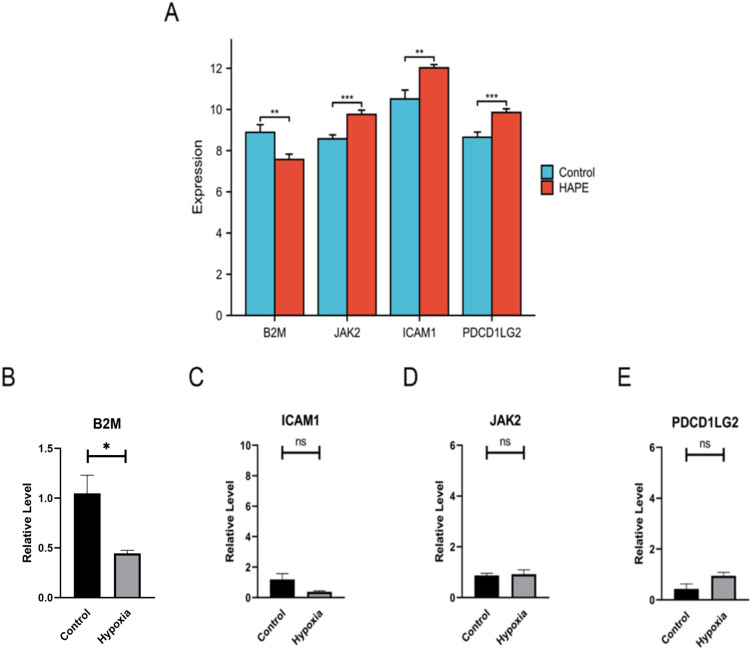
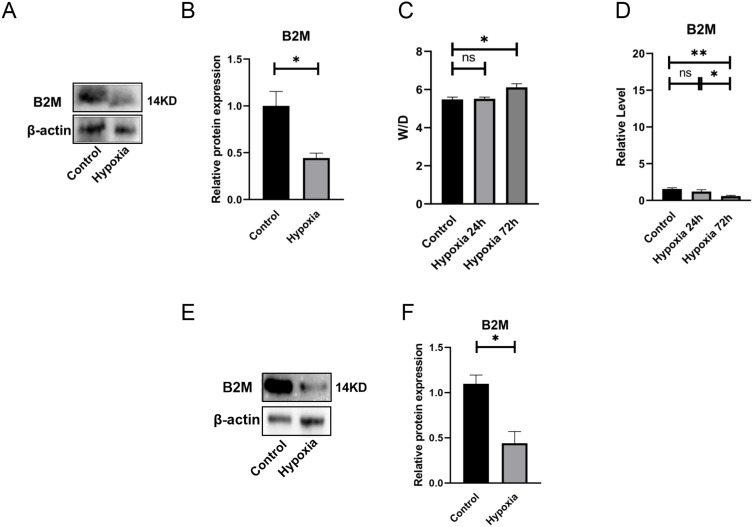
To compare the expression levels of core genes JAK2, B2M, ICAM1 and PDCD1LG2 in the peripheral blood of the GSE52209 high altitude adaptation group and the HAPE group, the “PheatMap” package of R software was used to analyze and compare the expression levels of JAK2, B2M, ICAM1 and PDCD1LG2 in the two groups. The results showed that the expression level of B2M in HAPE patients was significantly lower than that in high altitude adaptation group, and the expression levels of JAK2, ICAM1 and PDCD1LG2 were significantly higher than those in high altitude adaptation group (Figure 7A). Combined with the above results, plateau conditions did not affect the expression of B2M, JAK2, ICAM1 and PDCD1LG2, and B2M, JAK2, ICAM1 and PDCD1LG2 may serve as biomarkers suggestive of HAPE in plain areas. The results in the figure show that the change trend of RT-qPCR relative expression of B2M under hypoxic conditions is consistent with the results of bioinformatics analysis. The relative expression levels of JAK2, ICAM1 and PDCD1LG2 did not change significantly, which was inconsistent with the results of bioinformatics analysis. B2M may be used as a biomarker for HAPE in plain areas (Figure 7B–).
Figure 7.
Validation of B2M mRNA level in peripheral blood in rat model of HAPE. (A) Expression of hub genes in the high altitude adaptation group and HAPE group in the GSE52209 dataset; (B–E) RT-qPCR relative expression level determination of hub genes after plain and HAPE modeling (n=4). Ns indicates that the difference between the two groups is not significant, * refers to p<0.05, ** indicates that the difference between the two groups is more significant, p<0.01.
Effects of Hypoxia on B2M Expression in Peripheral Blood of Rats and Verification of B2M Protein Expression in Animal Models
In order to verify whether the changes in the relative expression level of B2M protein are consistent with the relative expression change trends of RT-qPCR, we conducted WB experiments. The experimental results showed that compared with the normal plain group, the expression of B2M protein in peripheral blood of HAPE was reduced (Figure 8A and B). The relative expression changes were consistent with the RT-qPCR results (Figure 7B). Previous studies have shown that β-actin can be used as an internal reference gene for RT-qPCR under hypoxic conditions, and its expression will not change under hypoxia.32 Studies have shown that plasma RNA content in peripheral blood is scarce, and β-actin can be used as a reference gene for RT-qPCR experiments in peripheral blood of rats.33
Figure 8.
Validation of biomarker B2M protein levels. (A and B) B2M WB experimental determination of peripheral blood in plain and HAPE model rats (n=3). β-actin is the internal reference protein (Loading Control), which is used as a reference for the relative ratio in WB experimental statistics. The statistical chart shows the statistical results of each group of rat samples (n=3) after modeling; (C) Changes in the water content of the lungs of rats in a hypobaric chamber for 24h and 72h (n=6); (D) Changes in B2M expression in peripheral blood of rats exposed to hypobaric chamber for 24h and 72h (n=4); (E and F) B2M WB assay was used to determine peripheral blood leukocytes in plain and HAPE model rats. β-actin is the internal reference protein (Loading Control), which is used as a reference for the relative ratio in WB experimental statistics. The statistical chart shows the statistical results of each group of rat samples (n=3) after modeling. Ns indicates that the difference between the two groups is not significant, * refers to p<0.05, ** indicates that the difference between the two groups is more significant, p<0.01.
In order to verify the relative expression of B2M in peripheral blood of rats under hypoxia, the relative expression values of B2M in peripheral blood samples of rats were measured by RT-qPCR at 24h and 72h of hypoxia. The W/D ratio of bilateral lung tissue showed that the rats did not develop HAPE after 24 hours of hypoxia (Figure 8C and D). We can use the comparison results of the relative expression values of hub gene RT-qPCR genes between plain normal rats and HAPE rats to verify the comparison results between plain normal humans and patients with HAPE.
Evaluation of B2M Protein Expression in Peripheral Blood Leukocytes of Rats with HAPE
Blood mainly contains leukocytes, erythrocytes, platelets and plasma, and peripheral blood plasma RNA content is scarce, erythrocytes and platelets cannot carry out gene transcription, and RNA expression may not change.34,35 Therefore, we hypothesized that B2M is mainly expressed in peripheral blood leukocytes, and leukocytes were isolated from peripheral blood of rats for WB detection immediately after modeling. The results showed that the relative expression level of B2M protein in leukocytes decreased after the occurrence of HAPE (Figure 8E and F), and the decrease in the relative expression level of B2M protein in peripheral blood leukocytes may be closely related to the occurrence of HAPE.
Analysis of the Function of B2M in Peripheral Blood of HAPE and Its Correlation with Immune Checkpoint Genes
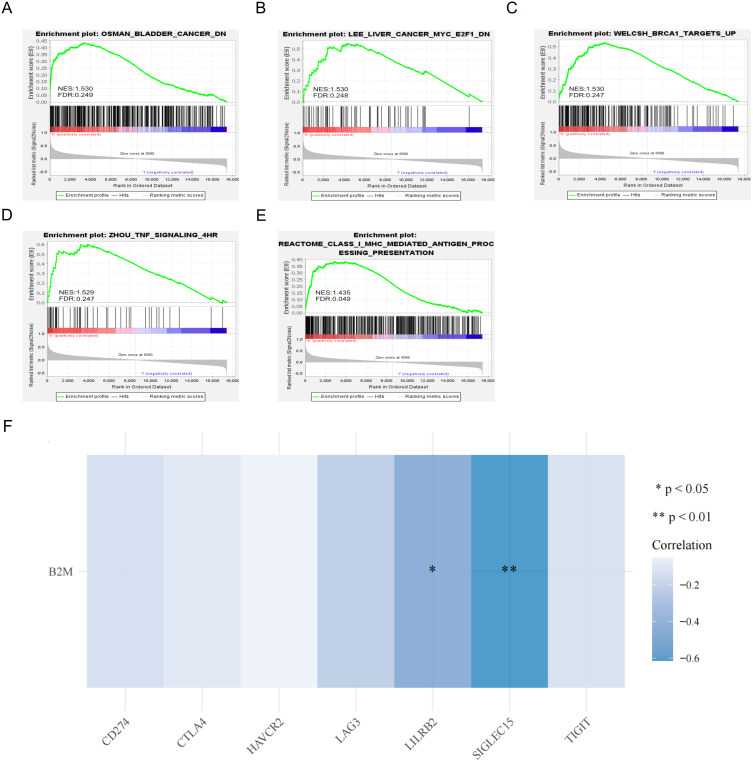
In order to study the molecular function of B2M in peripheral blood and its participation in signaling pathways, we performed a single-gene GSEA enrichment analysis of the GSE52209 data set. The results showed that the expression level of B2M is positively correlated with the TNF signaling pathway and the antigen presentation pathway mediated by MHC class I molecules (Figure 9A–). The results of correlation analysis between B2M and common immune checkpoint genes showed that B2M was negatively correlated with the expression of LILRB2 and SIGLEC15 genes (Figure 9F).
Figure 9.
B2M single-gene GSEA analysis and correlation analysis with common immune checkpoint genes. (A–E) Single-gene GSEA analysis of B2M in the GSE52209 data set. The significant difference was normal p<0.05, FDR<0.25; (F) Heat map of correlation between B2M and common immune checkpoint genes.
Evaluation of the Correlation Between Peripheral Blood B2M and the Diagnosis and Prognosis of HAPE
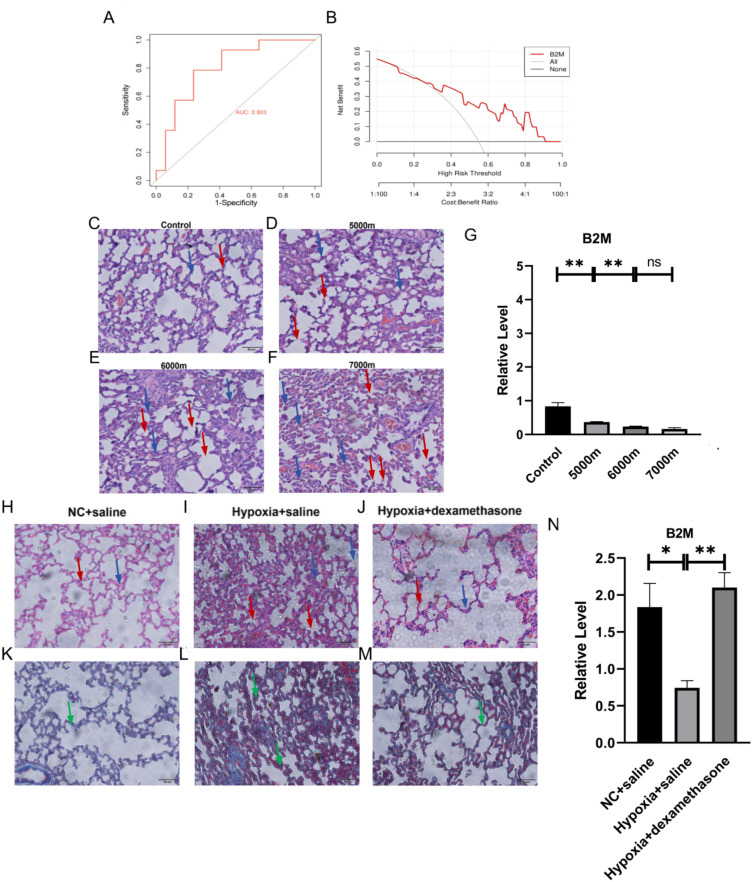
In order to evaluate the diagnostic utility of B2M in diagnosing HAPE in a high altitude environment, because HAPE samples are limited, we performed an internal ROC curve analysis of the data set. ROC curve analysis is mostly used for the verification and evaluation of diagnostic and prognostic markers, and calculated the area under curve (AUC), the AUC of the hub gene B2M is 0.803>0.7 (Figure 10A), which has high diagnostic value. According to our research results, there is no difference in the expression of B2M between plain and plateau adapted populations. Therefore, B2M can be used as a peripheral blood biomarker of HAPE with high diagnostic value in plateau environment. The ordinate of the DCA curve represents the net benefit. The net benefit of B2M diagnosis is much greater than the full positive extreme curve, which proves that significantly reduced B2M expression has better diagnostic performance for high altitude pulmonary edema and has better benefits in clinical diagnosis (Figure 10B).
Figure 10.
Evaluation of the correlation between peripheral blood B2M and high altitude pulmonary edema diagnosis and prognosis (A) ROC curve analysis of hub genes, AUC: area under the curve, significant difference criterion is AUC>0.7; (B) DCA curve analysis The diagnostic value of clinical HAPE, the ordinate represents the net benefit; (C–F) are the HE staining of lung tissue in the blank control group, 5000m model group, 6000m model group and 7000m model group respectively (n=3). The blue arrow points to the thickening of the alveolar septum, and the red arrow points to the structural destruction of the alveolar wall; (G) Comparison of B2M in Relative expression of RT-qPCR in peripheral blood of HAPE models at 5000m, 6000m and 7000m altitude (n=4); (H–J) HE of NC+normal saline group, hypoxia+normal saline group, hypoxia+dexamethasone group respectively Staining situation (n=3); (K–M) Masson staining situation of NC+normal saline group, hypoxia+normal saline group and hypoxia+dexamethasone group (n=3) respectively. Green arrows point to collagen fiber proliferation; (N) Comparison at around 5000m altitude relative expression level of B2M RT-qPCR in peripheral blood after 4 hours of dexamethasone treatment after establishing HAPE model (n=4). Ns indicates that the difference between the two groups is not significant, * refers to p<0.05, ** indicates that the difference between the two groups is more significant, p<0.01.
To evaluate the correlation between B2M and HAPE prognosis, that is, the changes in B2M expression as the severity of HAPE increases. HAPE model (n=3) was constructed by simulating altitudes of 5000m, 6000m and 7000m for 72 hours in a hypobaric oxygen chamber. The upper left lung tissue was taken for HE staining and the results are as shown below. The figure shows that as the altitude increases, the degree of edema in HAPE gradually increases, and the degree of alveolar septal thickening gradually increases. The severity of HAPE increases with the gradual increase in the simulated altitude of the hypobaric oxygen chamber (Figure 10C–). In order to evaluate the correlation between the expression of B2M and the severity of HAPE, we used SD rats in a hypobaric oxygen chamber to simulate altitudes of about 5000m, 6000m and 7000m for 72 hours to construct a HAPE model, and collected peripheral blood for RT-qPCR of B2M. As shown below, the relative expression of B2M at 6000m was significantly lower than that at 5000m, and the relative expression at 7000m was lower than that at 6000m. The results show that B2M gradually decreases with increasing altitude, and B2M is closely related to the severity of HAPE (Figure 10G).
In order to evaluate the correlation between B2M and the therapeutic effect of HAPE, we first evaluated the lung tissue damage of HAPE after treatment. SD rats were used to simulate an altitude of 5000m for 72 hours in a hypobaric oxygen chamber to construct a HAPE model. 4 hours later, 0.5mg/kg dexamethasone sodium phosphate injection (H41021255, Suicheng Pharmaceutical Industry, China) dissolved in 1mL saline was injected via the tail vein. The left upper lung tissue was taken for HE and Masson staining. Dexamethasone can reduce HAPE capillaries. Permeability, the function of reducing fluid leakage from the lungs, has been used as a treatment for HAPE. From the HE staining results in the figure, it can be seen that the alveolar septum in the hypoxia+normal saline group is thickened and accompanied by edema. The Masson staining results show that the degree of fibrosis is increased. In the hypoxia+dexamethasone group, alveolar septal thickening, edema, and fibrosis were reduced, indicating that dexamethasone can delay the progression of the disease and may have a better therapeutic effect on HAPE (Figure 10H–). In order to evaluate the correlation between the relative expression level of B2M and the therapeutic effect of HAPE, we took the peripheral blood of the above three groups of rats for B2M RT-qPCR detection. Currently, there are no reports on the direct and indirect effects of dexamethasone on B2M. Dexamethasone can be used for Evaluate the correlation between B2M and HAPE treatment effects. The result figure shows that the relative expression of B2M in the hypoxia+normal saline group is reduced compared with the NC+normal saline group and the hypoxia+dexamethasone group compared with the hypoxia+normal saline group, and the difference is significant. The results show that the relative expression of B2M increased after HAPE treatment, B2M is closely related to the therapeutic effect of HAPE (Figure 10N).
Discussion
We used the plateau adaptation group and HAPE group transcriptome data sets for GSVA analysis. The results showed that compared with the plateau adaptation group, the HAPE group had an up-regulated RUNX3-regulated immune response pathway, and down-regulated STING-mediated host immune response induction and IL-1-induced NF-κB activation pathways. Studies have shown that RUNX3 has the effect of suppressing immune function. Downregulation or mutation of RUNX3 is related to the occurrence of psoriasis and rheumatoid arthritis.36,37 RUNX3 can inhibit the killing effect of CD8+ T cells, and STING can restore the reduced function.38 The nuclear factor NF-κB pathway is considered a classic pro-inflammatory signaling pathway.39 It can be seen from the GSVA results that the occurrence of HAPE may be related to the decline of immune response function and the downregulation of IL-1-induced NF-κB activation pathway. The expression analysis of immune checkpoint genes in the two groups combined with the above results shows that the occurrence of HAPE may be related to immune tolerance. LILRB2 is a classic immune tolerance receptor molecule expressed by many leukocyte subpopulations, especially monocytes. Studies have shown that increased LILRB2 receptor levels can recognize a variety of MHC class I molecules and transduce inhibitory signals,40 the results of this study indicate that LILRB2’s expression is increased in HAPE patients, which may inhibit the antigen presentation ability mediated by MHC class I molecules. The expression of PD-L1 was elevated in the enrichment analysis of genes in the yellow and turquoise modules that were positively correlated with the occurrence of HAPE by WGCNA analysis. PD-L1 is a common tumor immune checkpoint and also plays an important role in non-tumor diseases. The PD-1/PD-L1 co-inhibitory pathway plays a key role in immune responses and autoimmunity by regulating T cell activity. Excessive activity and high expression of this pathway can suppress T cell and immune function. Studies have shown that it also plays an important role in the occurrence and development of cardiovascular and pulmonary diseases.41,42 This result was consistent with the primary immune deficiency pathway enriched by the turquoise module genes.
We used PPI network analysis and Cytoscape software to explore the hub genes of immune genes in the data set, and obtained the intersection of peripheral blood and immune-related hub genes JAK2, B2M, ICAM1 and PDCD1LG2. Then we established a HAPE model in a hypobaric oxygen chamber and used biological Informatics analysis and RT-qPCR and WB were used to verify the expression differences of the hub genes between the two groups, and the peripheral blood biomarker B2M related to HAPE and immunity was obtained. We used the single-gene GSEA analysis of the data set to explore the function of B2M, the molecular pathways involved and its regulation. Combined with the above experimental results, the single-gene GSEA analysis results of the data set show that the hub gene B2M may be involved in antigen presentation mediated by MHC class I molecules. The expression level of B2M was positively correlated with the regulation direction of TNF signaling pathway. Previous studies have confirmed that B2M protein is a component of MHC class I antigen-presenting molecules. MHC class I molecules are mainly distributed on all nucleated cells and can present endogenous antigens to CD8+ T cells and NK cells for elimination.43 The protein expression level of B2M can indicate the antigen-presenting ability of peripheral blood cells and is a molecule that describes the immune function status. The TNF pathway is a classic inflammatory pathway,44 which may be down-regulated after immune cell function is reduced or immune tolerance occurs. HLA-I encodes MHC-I,45 B2M mutation may lead to the impairment of HLA expression and function, thereby reducing the expression level of HLA-I, reducing the infiltration of T lymphocytes in tumor tissues, and weakening the host’s ability to recognize and eliminate EBV+ tumor cells, thereby inducing T lymphocyte immune tolerance.46 Combined with the previous GSVA analysis results, it was shown that compared with the plain control group, the inflammation level in the HAPE group was reduced, the antigen presentation ability of peripheral blood cells may be reduced, and immune tolerance may occur.
Studies have reported that the clinical manifestations of AMS are similar to those of early and atypical HAPE, such as headache, nausea and palpitation, which are difficult to differentiate.47 Therefore, HAPE should be differentiated from AMS first. We used ROC and DCA curves to evaluate the diagnostic value of B2M for HAPE in plateau environments. We used the GSE75665 transcriptome dataset to explore the effect of high altitude environment on the expression level of B2M in human peripheral blood and the DEGs in high altitude adaptation group and AMS group. Combined with animal experimental studies showed that that high altitude environment did not affect the expression of B2M in peripheral blood. B2M was not a DEGs between the high altitude adaptation group and the AMS group. The expression level of B2M can identify AMS and HAPE, which may be used as an indicator for rapid screening of HAPE in the high altitude environment. We used HE and Masson staining to evaluate the severity of HAPE animal models at different altitudes and the therapeutic effect of dexamethasone, and the RT-qPCR B2M expression level is used to assess the correlation between the degree of B2M expression and the therapeutic effect and prognosis of HAPE. The results showed that B2M may be used as a peripheral blood biomarker to indicate the treatment and prognosis of HAPE, and can better indicate the severity, disease development and treatment effect of HAPE. Abnormal expression of B2M genes in peripheral blood plays an important role in the diagnosis of various immune system diseases. For example, abnormal expression of B2M genes in peripheral blood is closely related to the occurrence and development of autoimmune diseases (such as rheumatoid arthritis, systemic lupus erythematosus, etc.) and immune-related tumors (such as lymphoma, myeloma, etc).48–51 B2M protein expression can indicate the severity of heart failure and is positively correlated with the severity of heart failure.52 Previous studies have shown that HAPE can cause acute left heart failure, but our study results show that B2M levels are reduced in HAPE patients. Our results demonstrate that B2M is specific as a biomarker related to the occurrence of HAPE. Peripheral blood B2M genes also have extensive application value in disease treatment. According to the expression level of B2M genes, the patient’s response to certain treatments can be predicted. In tumor treatment, detecting the expression levels of B2M genes in peripheral blood can help select appropriate immunotherapy regimens and evaluate patients’ tolerance and efficacy of treatment.53 Peripheral blood B2M genes also play a key role in disease prognosis assessment. By detecting the expression changes of B2M genes during treatment, the treatment effect can be evaluated and the patient’s prognosis can be predicted. Studies have shown that the expression level of B2M genes in peripheral blood is closely related to the survival rate and recurrence rate of tumor patients.54 At present, there are few biomarkers that indicate the prognosis and treatment effect of HAPE, and they lack specificity; reduced B2M expression has rarely been reported in studies indicating the diagnosis, treatment effect and prognosis of other diseases. The number, subsets and functional status of peripheral blood leukocytes can help clinicians judge the development trend of diseases and select appropriate treatment methods.55 Some studies have shown that peripheral blood leukocyte biomarkers can predict the occurrence and development of a variety of diseases.55,56 With the in-depth understanding of peripheral blood leukocyte biomarkers, we will provide more possibilities for the realization of precision medicine. B2M may be a sensitive and specific biomarker for HAPE. Experimental results show that it has high clinical practice value. Its value in indicating the diagnosis and prognosis of HAPE needs further verification in clinical trials.
This study also has many limitations. Due to the limitation of disease types, the sample size of GEO transcriptome can be found is small, and the peripheral blood biomarker B2M obtained after differential analysis needs more clinical samples to verify. The indicator effect of peripheral blood B2M on HAPE has been verified by animal experiments, and further clinical trials are needed to verify it. This study initially explored the indicator role of B2M in the occurrence, severity, and treatment effect of HAPE, and the mechanism needs to be further explored.
Conclusion
The occurrence of HAPE may be related to decreased immune function and immune tolerance. B2M can be used as a biomarker for peripheral blood in HAPE patients. The decreased expression of B2M in peripheral blood of HAPE patients is positively correlated with the downregulation of the antigen presentation pathway mediated by MHC class I molecules. The down-regulation of B2M expression in peripheral blood may be related to the decline of immune function and immune tolerance in patients with HAPE, and has suggestive value for the diagnosis, treatment effect and prognosis of HAPE. Animal experiment results show that the decreased expression level of B2M in the peripheral blood of rats indicates the occurrence of HAPE. Its expression level is negatively correlated with the severity of HAPE and positively correlated with the therapeutic effect of HAPE in rats. The reduced expression of B2M protein in rat peripheral blood leukocytes may be a marker of HAPE in rats. The results of this study can provide new theoretical basis for pathophysiological research, clinical treatment effect and prognosis assessment of HAPE.
Acknowledgments
The assistance rendered by the Editor and anonymous reviewers, along with their valuable comments and suggestions, significantly enhanced the quality of the manuscript for which we are sincerely grateful. The researchers who uploaded the datasets in the GEO database deserve our gratitude.
Funding Statement
This study was supported by the National Natural Science Foundation of China (No.82372529).
Data Sharing Statement
The data set used in this study was obtained from the GEO database, specifically from two sources: GSE52209 (for acclimatization to high altitude group and HAPE group) and GSE75665 (for plain normal group, plateau adapted group and AMS group).
Ethical Standards Disclosure
The Ethics Committee of Army Medical University granted ethical approval for this study (Approval Number: AMUWEC20211773). The implementation of research completely follows the 3R principle, which means substitution, reduction and optimization.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas. All authors drafted, revised or reviewed the article, and agreed on the journal in which the article will be submitted, gave final approval for the version to be published. All authors agree to be accountable for all aspects of the work.
Disclosure
Absence of conflicting interests is declared by the authors.
References
- 1.Nussbaumer-Ochsner Y, Schuepfer N, Ursprung J, et al. Sleep and breathing in high altitude pulmonary edema susceptible subjects at 4559 meters. Sleep. 2012;35:1413–1421. doi: 10.5665/sleep.2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Swenson ER, Maggiorini M, Mongovin S, et al. Pathogenesis of high-altitude pulmonary edema: inflammation is not an etiologic factor. JAMA. 2002;287:2228–2235. doi: 10.1001/jama.287.17.2228 [DOI] [PubMed] [Google Scholar]
- 3.Kandel RS, Mishra R, Gautam J, et al. Patchy vasoconstriction versus inflammation: a debate in the pathogenesis of high altitude pulmonary edema. Cureus. 2020;12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schoene RB, Bracker MD. High-Altitude Pulmonary Edema: the Disguised Killer. Phys Sportsmed. 1988;16:102–114. doi: 10.1080/00913847.1988.11709575 [DOI] [PubMed] [Google Scholar]
- 5.Krishnan S, Stearman RS, Zeng L, et al. Transcriptomic modifications in developmental cardiopulmonary adaptations to chronic hypoxia using a murine model of simulated high-altitude exposure. Am J Physiol Lung Cell Mol Physiol. 2020;319:L456–L470. doi: 10.1152/ajplung.00487.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khanna K, Mishra KP, Ganju L, et al. High-altitude-induced alterations in gut-immune axis: a review. Intern revi immu. 2018;37:119–126. doi: 10.1080/08830185.2017.1407763 [DOI] [PubMed] [Google Scholar]
- 7.Ali M, Choudhary R, Rabyang S, et al. Harsh environmental stressors of high altitude on pathogens susceptibility. Genomic Surveill Pandemic Preparedne. 2023:357–373. [Google Scholar]
- 8.Malla DK, Singh S, Basnet Y, et al. Endotracheal Intubation at 3,600 Meters Above Sea Level for High-Altitude Pulmonary Edema Followed by Helicopter Evacuation in Nepal. Air Medical Journal. 2023;42:58–60. doi: 10.1016/j.amj.2022.11.001 [DOI] [PubMed] [Google Scholar]
- 9.Gao YM, Xu G, Wang B, et al. Cytokine storm syndrome in coronavirus disease 2019: a narrative review. J Internal Med. 2021;289:147–161. doi: 10.1111/joim.13144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Betjes MG. Immune cell dysfunction and inflammation in end-stage renal disease. Nat Rev Nephrol. 2013;9:255–265. doi: 10.1038/nrneph.2013.44 [DOI] [PubMed] [Google Scholar]
- 11.Zhou Q. Standardization of methods for early diagnosis and on-site treatment of high-altitude pulmonary edema. Pulmonary Medicine. 2011;2011:1–7. doi: 10.1155/2011/190648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pennardt A. High-altitude pulmonary edema: diagnosis, prevention, and treatment. Curr Sports Med Rep. 2013;12:115–119. doi: 10.1249/JSR.0b013e318287713b [DOI] [PubMed] [Google Scholar]
- 13.Gluecker T, Capasso P, Schnyder P, et al. Clinical and radiologic features of pulmonary edema. Radiographics. 1999;19:1507–1531. doi: 10.1148/radiographics.19.6.g99no211507 [DOI] [PubMed] [Google Scholar]
- 14.Zubieta-Calleja GR, Zubieta-DeUrioste N, de Jesús Montelongo F, et al. Morphological and functional findings in COVID-19 lung disease as compared to Pneumonia, ARDS, and High-Altitude Pulmonary Edema. Respir Physiol Neurobiol. 2023;309:104000. doi: 10.1016/j.resp.2022.104000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korzeniewski K, Nitsch-Osuch A, Guzek A, Juszczak D. High altitude pulmonary edema in mountain climbers. Respir Physiol Neurobiol. 2015;209:33–38. doi: 10.1016/j.resp.2014.09.023 [DOI] [PubMed] [Google Scholar]
- 16.CHAIT A. Interstitial pulmonary edema. Circulation. 1972;45:1323–1330. doi: 10.1161/01.CIR.45.6.1323 [DOI] [PubMed] [Google Scholar]
- 17.Ansary TM, Hossain MR, Kamiya K, et al. Inflammatory Molecules Associated with Ultraviolet Radiation-Mediated Skin Aging. Int J Mol Sci. 2021. 22. doi: 10.3390/ijms23010022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng L, Li J, Zhang R. Current research status of blood biomarkers in Alzheimer’s disease: diagnosis and prognosis. Ageing Res Rev. 2021;72:101492. doi: 10.1016/j.arr.2021.101492 [DOI] [PubMed] [Google Scholar]
- 19.Mohr S, Liew C. The peripheral-blood transcriptome: new insights into disease and risk assessment. Trends Mol Med. 2007;13:422–432. doi: 10.1016/j.molmed.2007.08.003 [DOI] [PubMed] [Google Scholar]
- 20.Zhang YY, Duan RF, Cui WY, et al. Proteomic identification of human serum biomarkers associated with high altitude pulmonary edema. Zhongguo Ying Yong Sheng Li Xue Za Zhi. 2013;29:501–507. [PubMed] [Google Scholar]
- 21.Bhargava YAAN. The proteome of Hypobaric Induced Hypoxic Lung: insights from Temporal Proteomic Profiling for Biomarker Discovery. Sci Rep. 2015;5:undefined. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woods AMAC. Cardiac biomarkers at high altitude. High Alt Med Biol. 2014;15:4:undefined. [DOI] [PubMed] [Google Scholar]
- 23.Ahmad Y, Shukla D, Garg I, et al. Identification of haptoglobin and apolipoprotein AI as biomarkers for high altitude pulmonary edema. Funct Integrat Genomics. 2011;11:407–417. doi: 10.1007/s10142-011-0234-3 [DOI] [PubMed] [Google Scholar]
- 24.Janvilisri T, Suzuki H, Scaria J, et al. High-Throughput Screening for Biomarker Discovery. Dis. Markers. 2015;2015:108064. doi: 10.1155/2015/108064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuan M, Hu X, Xing W, et al. B2M is a Biomarker Associated With Immune Infiltration In High Altitude Pulmonary Edema. Comb Chem High Throughput Screen. 2024;27:168–185. doi: 10.2174/1386207326666230510095840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 2013;14:7. doi: 10.1186/1471-2105-14-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qin S, Xu L, Yi M, Yu S, Wu K, Luo S. Novel immune checkpoint targets: moving beyond PD-1 and CTLA-4. Mol Cancer. 2019;18:1–14. doi: 10.1186/s12943-019-1091-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qian Y, Yang T, Liang H, et al. Myeloid checkpoints for cancer immunotherapy. Chin J Cancer Res. 2022;34:460. doi: 10.21147/j.issn.1000-9604.2022.05.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang C, Yan M, Jiang H, et al. Protective effects of puerarin on acute lung and cerebrum injury induced by hypobaric hypoxia via the regulation of aquaporin (AQP) via NF-κB signaling pathway. Int Immunopharmacol. 2016;40:300–309. doi: 10.1016/j.intimp.2016.09.010 [DOI] [PubMed] [Google Scholar]
- 31.Liu Z, Cui H, Zhao L, et al. Effects of Plateau Air Quality and Low Oxygen Content on Platelet mtDNA Methylation and High Altitude Pulmonary Edema. Aerosol Air Qual. Res. 2021;21:210222. doi: 10.4209/aaqr.210222 [DOI] [Google Scholar]
- 32.Foldager CB, Munir S, Ulrik-Vinther M, et al. Validation of suitable house keeping genes for hypoxia-cultured human chondrocytes. BMC Mol. Biol. 2009;10:1–8. doi: 10.1186/1471-2199-10-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schultz-Thater E, Frey DM, Margelli D, et al. Whole blood assessment of antigen specific cellular immune response by real time quantitative PCR: a versatile monitoring and discovery tool. J Transl Med. 2008;6(1):1–9. doi: 10.1186/1479-5876-6-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu Y, A EE, Kittelsrud J, et al. Lipid-lowering effect of berberine in human subjects and rats. Phytomedicine. 2012;19(10):861–867. doi: 10.1016/j.phymed.2012.05.009 [DOI] [PubMed] [Google Scholar]
- 35.Wang K, Yuan Y, Cho J, et al. Comparing the MicroRNA spectrum between serum and plasma. PLoS One. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Helms C, Cao L, Krueger JG, et al. A putative RUNX1 binding site variant between SLC9A3R1 and NAT9 is associated with susceptibility to psoriasis. Nat Genet. 2003;35:349–356. doi: 10.1038/ng1268 [DOI] [PubMed] [Google Scholar]
- 37.Tokuhiro S, Yamada R, Chang X, et al. An intronic SNP in a RUNX1 binding site of SLC22A4, encoding an organic cation transporter, is associated with rheumatoid arthritis. Nat Genet. 2003;35:341–348. doi: 10.1038/ng1267 [DOI] [PubMed] [Google Scholar]
- 38.Huang Y, Qin G, Cui T, et al. A bimetallic nanoplatform for STING activation and CRISPR/Cas mediated depletion of the methionine transporter in cancer cells restores anti-tumor immune responses. Nat Commun. 2023;14:4647. doi: 10.1038/s41467-023-40345-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lawrence T. The nuclear factor NF-κB pathway in inflammation. Cold Spring Harbor Perspect Biol. 2009;1:a001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anderson KJ, Allen RL. Regulation of T‐cell immunity by leucocyte immunoglobulin‐like receptors: innate immune receptors for self on antigen‐presenting cells. Immunology. 2009;127:8–17. doi: 10.1111/j.1365-2567.2009.03097.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun Y, Li L, Wu Y, et al. PD-1/PD-L1 in cardiovascular disease. Clin Chim Acta. 2020;505:26–30. doi: 10.1016/j.cca.2020.02.019 [DOI] [PubMed] [Google Scholar]
- 42.Bala-Hampton JE, Bazzell AF, Dains JE. Clinical Management of Pneumonitis in Patients Receiving Anti-PD-1/PD-L1 Therapy. J Adv Pract Oncol. 2018;9:422–428. [PMC free article] [PubMed] [Google Scholar]
- 43.Kyrysyuk O, Wucherpfennig KW. Designing cancer immunotherapies that engage T cells and NK cells. Ann Rev Immunol. 2023;41:17–38. doi: 10.1146/annurev-immunol-101921-044122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mackay F, Ambrose C. The TNF family members BAFF and April: the growing complexity. Cytokine Growth Factor Rev. 2003;14:311–324. doi: 10.1016/S1359-6101(03)00023-6 [DOI] [PubMed] [Google Scholar]
- 45.Hazini A, Fisher K, Seymour L. Deregulation of HLA-I in cancer and its central importance for immunotherapy. J ImmunoThera Canc. 2021;9:e002899. doi: 10.1136/jitc-2021-002899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.He M, Liu B, Tang G, et al. B2M mutation paves the way for immune tolerance in pathogenesis of Epstein-Barr virus positive diffuse large B-cell lymphomas. J Cancer. 2022;13:3615–3622. doi: 10.7150/jca.75813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kurtzman RA, Caruso JL. High-Altitude Illness Death Investigation. Acad Forens Pathol. 2018;8:83–97. doi: 10.23907/2018.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shklovskaya E, Rizos H. MHC class I deficiency in solid tumors and therapeutic strategies to overcome it. Int J Mol Sci. 2021;22:6741. doi: 10.3390/ijms22136741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McPhee CG, Sproule TJ, Shin D, et al. MHC class I family proteins retard systemic lupus erythematosus autoimmunity and B cell lymphomagenesis. J Immunol. 2011;187:4695–4704. doi: 10.4049/jimmunol.1101776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Furie R, Werth VP, Merola JF, et al. Monoclonal antibody targeting BDCA2 ameliorates skin lesions in systemic lupus erythematosus. J Clin Invest. 2019;129:1359–1371. doi: 10.1172/JCI124466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu-Monette ZY, Zhang M, Li J, et al. PD-1/PD-L1 blockade: have we found the key to unleash the antitumor immune response? Front Immunol. 2017;8:1597. doi: 10.3389/fimmu.2017.01597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brioschi M, Gianazza E, Agostoni P, et al. Multiplexed MRM-Based Proteomics Identified Multiple Biomarkers of Disease Severity in Human Heart Failure. Int J Mol Sci. 2021;22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yilmaz A, Cui H, Caligiuri MA, et al. Chimeric antigen receptor-engineered natural killer cells for cancer immunotherapy. J Hematol Oncol. 2020;13:1–22. doi: 10.1186/s13045-020-00998-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin R, Fogarty CE, Ma B, et al. Identification of ferroptosis genes in immune infiltration and prognosis in thyroid papillary carcinoma using network analysis. BMC Genomics. 2021;22:576. doi: 10.1186/s12864-021-07895-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.C AD, W SC, M CC, et al. A framework for the development and interpretation of different sepsis definitions and clinical criteria. Crit Care Med. 2016;44(3):e113–e121. doi: 10.1097/CCM.0000000000001730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang L, Aakre JA, Jiang R, et al. Methylation markers for small cell lung cancer in peripheral blood leukocyte DNA. J Thorac Oncol. 2010;5:778–785. doi: 10.1097/JTO.0b013e3181d6e0b3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data set used in this study was obtained from the GEO database, specifically from two sources: GSE52209 (for acclimatization to high altitude group and HAPE group) and GSE75665 (for plain normal group, plateau adapted group and AMS group).