Abstract
The envelope (Env) protein of Moloney murine leukemia virus is the primary mediator of viral entry. We constructed a large pool of insertion mutations in the env gene and analyzed the fitness of each mutant in completing two critical steps in the virus life cycle: (i) the expression and delivery of the Env protein to the cell surface during virion assembly and (ii) the infectivity of virions displaying the mutant proteins. The majority of the mutants were poorly expressed at the producer cell surface, suggesting folding defects due to the presence of the inserted residues. The mutants with residual infectivity had insertions either in the amino-terminal signal sequence region, two disulfide-bonded loops in the receptor binding domain, discrete regions of the carboxy-terminal region of the surface subunit (SU), or the cytoplasmic tail. Insertions that allowed the mutants to reach the cell surface but not to mediate detectable infection were located within the amino-terminal sequence of the mature Env, within the SU carboxy-terminal region, near putative receptor binding residues, and throughout the fusion peptide. Independent analysis of select mutants in this group allowed more precise identification of the defect in Env function. Mapping of mutant phenotypes to a structural model of the receptor-binding domain provides insights into the protein's functional organization. The high-resolution functional map reported here will be valuable for the engineering of the Env protein for a variety of uses, including gene therapy.
Cell entry by retroviruses is mediated by the virally encoded Env glycoprotein on the virus surface. The Env protein of Moloney murine leukemia virus (MoMLV), the best-characterized mammalian type C retrovirus, is synthesized as a single 85-kDa precursor that includes an amino-terminal signal sequence that directs its insertion into the lumen of the endoplasmic reticulum (ER) (61). During transport to the cell surface, the Env precursor undergoes a series of posttranslational modifications, including glycosylation, oligomerization, disulfide bond formation and cleavage into surface (SU) and transmembrane (TM) subunits (19, 46, 47, 48). At the cell surface, the Env trimer is incorporated into the membrane of the budding virion, where it is further modified by the virally encoded protease, which removes the carboxy-terminal 16 amino acids (the R-peptide) of TM to yield the entry-competent glycoprotein (22, 50, 51, 52).
The entry process is initiated by the specific interaction of the Env protein with the receptor protein MCAT-1, which is located on the surface of a susceptible cell (1). Receptor-bound virion particles are then internalized, probably by clathrin-independent endocytosis (38). By analogy with the influenza hemagglutinin (HA) fusion protein, the reduced pH within the endosome mediates a conformational change in the Moloney Env protein that exposes the fusion peptide at the amino terminus of TM (55). Fusion, initiated by the insertion of the fusion peptide into the endosomal membrane, permits the release of the genome-containing virus core into the cytoplasm of the infected cell (63).
Earlier approaches to defining functionally important elements of the Env protein involved making relatively large-scale alterations in the protein (for example, deletions and chimeras) and then analyzing their effects on different steps of the life cycle (3, 4, 24). These studies have been further refined by analyzing mutants with subtler alterations, such as single residue substitutions or small insertions (2, 9, 11, 20, 45). However, the ability to ascribe function to particular features of Env has been limited by the relatively small number of mutations analyzed in comparison to the large size of the protein. We wanted to approach the problem of functional analysis from a more global perspective, in a manner allowing detailed comparative analysis of the phenotypic consequences of individual mutations. To do this, we utilized genetic footprinting, a comprehensive mutagenesis and selection strategy (54). This approach permits the rapid generation and functional analysis of a large pool of insertion variants of any cloned gene, obviating the need to generate and analyze each mutant individually and therefore greatly expanding the number of mutants which can be included in a single study. Moreover, it allows facile quantitative comparison of the phenotypes of the mutants. We generated a comprehensive pool of insertion variants of the env gene and subjected the pool en masse to two selections for normal Env function: the ability of the mutant Env proteins to (i) be detected at the cell surface of producer cells and (ii) mediate viral infection. By obtaining data for 378 mutants, each with an insertion at a unique site in the env gene, we have been able to ascribe function to previously unmutagenized sequences, thereby extending the results of earlier studies and providing the basis for the more complete functional characterization of the entire protein.
MATERIALS AND METHODS
Plasmids.
pNCA contains the entire wild-type MoMLV genome in a pUC19 backbone. pUC19 P/A was generated by subcloning the PmlI-AflIII fragment of pNCA (nucleotides 6179 to 9038) into pUC19. pBMN-kana is a replication-defective retroviral expression vector in which the ampicillin resistance gene within the pUC19 backbone has been replaced by the kanamycin resistance gene (35; S. M. Rothenberg, unpublished data). The 2,557-nucleotide EcoRI-NheI fragment of pUC19 P/A (which contains the env open reading frame, the polypyrimidine tract, and part of the 3′ U3 region) was subcloned into pBMN-kana after mutagenesis to generate the pool of mutant env vectors. Due to this subcloning step, a proportion of the subcloned fragment, originating from plasmid molecules containing insertions located outside of the env gene in the plasmid backbone, lacked insertions and was therefore wild type.
Mutagenesis.
A 15-nucleotide oligonucleotide was inserted at diverse sites within plasmid pUC19 P/A by using the bacteriophage Mu transposase (MuA) as described previously (36). Briefly, the integration substrate NotI5 was generated by annealing two single-stranded oligonucleotides, NotI5A (5′-TGCGGCCGCGCACGAAAAACGCGAAAGCGTTTCACGATAAATGCGAAAAC-3′) and NotI5B (5′-GTTTTCGCATTTATCGTGAAACGCTTTCGCGTTTTTCGTGCGCGGCCGCA-3′), in 50 mM NaCl. The resulting duplex oligonucleotide contained recognition sites for MuA and the restriction endonuclease NotI. The integration reaction was carried out by incubating the NotI5 substrate, target plasmid pUC19 P/A, and MuA at 30°C for 1 h. The five-nucleotide gaps produced by the concerted integration of two NOT15 oligonucleotides into the target plasmid were repaired by Taq DNA polymerase (AmpliTaq; Perkin-Elmer). The repaired products were then restriction digested with NotI (New England Biolabs) and recircularized by ligation with T4 DNA ligase (New England Biolabs) to generate a pool of 15-nucleotide single-insertion variants of the MoMLV env gene.
Cell lines and antibodies.
293T, Rat1, and XC cell lines were cultured in Dulbecco modified Eagle medium supplemented with 4.5 g of glucose/liter, 10% fetal calf serum, and glutamine. Mouse hybridoma and Jurkat cell lines were cultured in RPMI 1640 supplemented with 10% fetal calf serum and glutamine. Hybridomas 538 and 573, each of which produces an immunoglobulin M (IgM) antibody against Env, and 715, which produces an anti-Env IgG, were a generous gift from Bruce Chesebro, Laboratory of Persistent Viral Disease, Rocky Mountain Laboratories. 83A25, an anti-Env IgG used for the receptor-binding assay, was kindly provided by Leonard Evans, Laboratory of Persistent Viral Disease, Rocky Mountain Laboratories. 293T gag pol, expressing the MoMLV Gag and Pol proteins, and Phoenix-A, expressing MoMLV Gag and Pol and the amphotropic MLV Env protein, were kindly provided by Garry P. Nolan, Stanford University.
Selection for cell surface expression of Env.
Primary virus stocks were generated by calcium phosphate-mediated transfection of Phoenix-A cells with the pool of mutant Env vectors. The supernatant medium containing the resulting viruses was overlaid onto Jurkat cells at a multiplicity of infection (MOI) of ∼0.05. For the isolation of cells expressing mutant envelopes capable of reaching the cell surface, the transduced Jurkat cells were incubated simultaneously with a mixture of supernatants from hybridomas 538, 573, and 715 for 30 min on ice, followed by sequential staining with two goat anti-mouse immunoglobulin-fluorescein isothiocyanate (FITC) conjugates (Caltag), one recognizing the μ chain of IgM and the other recognizing the heavy and light chains of IgG. Jurkat cells were used instead of 293T gag pol cells because pilot experiments had shown that the higher levels of Env expression on the surface of the Jurkat cells, as assayed by reactivity with the anti-Env monoclonal antibodies, allowed better separation of Env-expressing and nonexpressing cells. Stained cells were incubated with anti-FITC paramagnetic Microbeads (Miltenyi), and cells expressing Env at the surface were isolated by magnetic cell sorting according to the manufacturer's protocol. The isolated cells were further purified by fluorescence-activated cell sorting with a FACStar flow cytometer (Becton Dickinson, Shared FACS Facility, Stanford University).
Selection for infectivity.
The same conditions used for the transduction of Jurkat cells were used to transduce a population of 293T gag pol cells at an MOI of ∼0.05. For the actual infectivity selection, the supernatant medium from the transduced 293T gag pol cells was used to infect a population of Rat1 cells.
Nucleic acid preparation.
All plasmid DNAs and genomic DNA samples were prepared by using the appropriate DNA purification kits from Qiagen. Initially, the integrated proviral mutants were enriched by restriction digestion of each genomic DNA sample with BglII and NheI (which releases a 2.6-kb proviral fragment containing the env gene), followed by gel isolation of DNA fragments which were 2.0 to 3.0 kb in size. PCR was then performed in a mixture containing 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 625 μM concentrations of each deoxynucleoside triphosphate, 0.25 μM concentrations of each primer, 1 U of Taq DNA polymerase (AmpliTaq; Perkin-Elmer), 0.01 U of Deep Vent DNA polymerase (Deep Vent; New England Biolabs), and 50 ng of purified genomic DNA samples per 50-μl reaction mixture. PCR conditions consisted of 5 min at 95°C, followed by 30 cycles of 30 s at 95°C, 30 s at 53°C, and 3.5 min at 72°C. The PCR primers MoMLV 2347 (5′-GGGATCCATGCATCTCGAGTG-3′) and MoMLV 4510 (5′-CTTTTATTTTATCGTCGACCCTA-3′) were complementary to proviral sequences flanking the env gene. Multiple identical PCRs were pooled to generate the template for the radiolabeled footprinting PCR (see below).
Genetic footprinting.
PCR was carried out by using 25 ng of the preamplified env DNA as a template and two env-specific primers, one radiolabeled and the other biotinylated. Analysis of the entire env gene required separate PCRs with 14 different primer pairs, each of which covered 200 to 300 nucleotides of the env sequence. Primer sequences are available online (http://genome-www.stanford.edu/geneticfootprinting/). PCR conditions consisted of 25 cycles of 30 s at 95°C, 30 s at 57°C, and 1 min at 72°C with the same buffer as used for the initial PCR (without Deep Vent). PCR products containing regions of single-stranded DNA as a result of incomplete extension were removed with single-stranded affinity matrix (Clontech), and the remaining double-stranded products were recovered by using the QiaQuick PCR Purification Kit (Qiagen). The products were further purified by absorption to 100 μl of streptavidin-agarose beads (Sigma) for 1 h at 37°C with gentle rotation. After an extensive washing, each bead-DNA mixture was restriction digested with 100 U of NotI for 20 min at 37°C with gentle rotation. The supernatant containing the released radiolabeled PCR fragments was separated from the beads by centrifugation through a Micro Bio-Spin column (Bio-Rad) and then concentrated by ethanol precipitation. Samples were analyzed by denaturing polyacrylamide gel electrophoresis, and the fragments representing the mutants were visualized with a Molecular Dynamics PhosphorImager. The position of each insertion within the env sequence was determined by comparison of each footprinting gel lane with an env sequencing ladder generated by using wild-type env DNA as a template and the same radiolabeled primer used for the radiolabeled PCR in a dideoxy sequencing reaction.
Quantitation of mutant fitness.
The intensity of the product band representing each mutant was determined with the ImageQuant 5.0 software package (Molecular Dynamics). The relative fitness of each mutant was determined by computing the ratio of the postselection intensity to the preselection intensity for each corresponding band. To normalize the calculated ratio to the activity of the wild-type Env protein, a sample of the PCR, prior to NotI digestion, was analyzed by gel electrophoresis to separate unmutagenized wild-type DNA in the pool from all of the insert-containing mutants, which are 15 nucleotides longer than the wild type because of the presence of the insertion. The ratio of the intensities of the upper (mutant) to the lower (unmutagenized) band was then calculated for each sample. Next, the ratio, i.e., (mutant postselection/nonmutant postselection)/(mutant preselection/nonmutant preselection), was calculated to provide a factor representing the overall fitness of the population of insertion mutants relative to the wild type. Finally, the ratio of the postselection to preselection intensities of each restriction-digested band was multiplied by this region-wide fitness factor to determine the fitness of each mutant relative to the wild type. Two independent selections for cell surface expression and three independent selections for infectivity were carried out, and the results for each mutant were averaged. The average coefficients of variation for all mutants with fitnesses of >10% of the wild-type level were 0.35 and 0.47 for the selections for cell surface expression and infectivity, respectively. A complete graphical display of the data is available online (http://genome-www.stanford.edu/geneticfootprinting/).
Isolation and analysis of individual mutant clones.
A fraction of the mutant vector pool was electroporated into Escherichia coli, and the resulting transformants were isolated on solid media. Select clones were purified and sequenced to identify the exact position and structure of each insertion. The cell surface expression of each mutant was determined by incubating 293T gag pol cells, transfected with each mutant, with the hybridoma supernatant mixture, followed by flow cytometry. Receptor-binding activity was assayed by incubating transfected 293T gag pol cells with NIH 3T3 cells in suspension at 4°C for 1 h, followed by staining of the virus-cell mixture with antibody 83A25 and flow cytometry (28, 40). Fusion activity was determined by coculture of XC cells and transfected 293T gag pol cells, followed by quantitation of the number of visible syncytia with more than four nuclei by light microscopy. Infectivity was determined by cotransfection of each mutant clone with a green fluorescent protein (GFP)-encoding retroviral vector into 293T gag pol cells, followed by transduction of Rat 1 cells with the resulting viral supernatant and determination of the fraction of GFP-positive cells by flow cytometry.
Computer modeling.
All structural models were generated with Swiss-PDB Viewer 3.7, which can be obtained online (http://www.expasy.ch/spdbv/). The MoMLV receptor-binding domain (RBD) was fitted to the coordinates of the Friend MLV RBD by using the threading function in the program. The coordinates of the Friend trimer model were kindly provided by Peter Kim (Whitehead Institute for Biomedical Research).
RESULTS
Construction of a library of insertion variants of the MoMLV env gene.
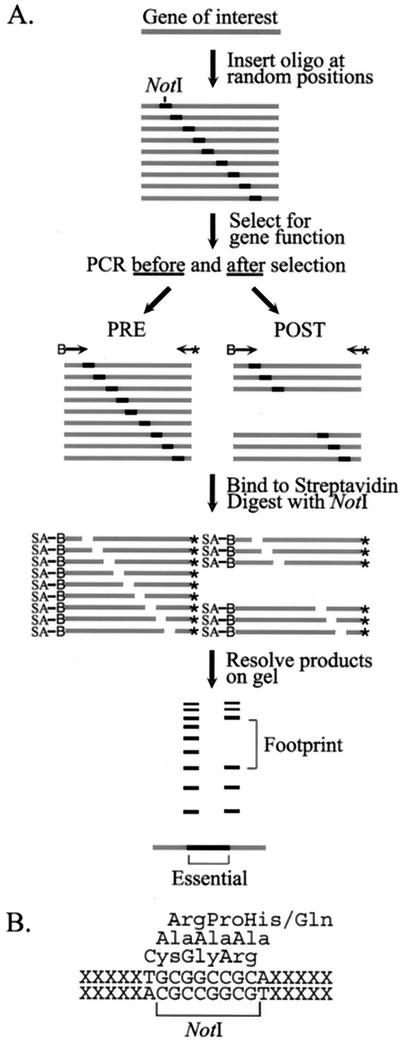
An overview of the genetic footprinting procedure is shown in Fig. 1A. Briefly, a pool of insertion variants of a gene of interest, generated by an in vitro transposition reaction, is subjected to a selection en masse for the normal function of the encoded protein. The resulting pre- and postselection pools are analyzed by a PCR approach that generates a population of fragments, each representing a different mutant in the pool. The size of each fragment reflects the position of the insertion in the gene sequence. Comparison of the pre- and postselection samples after gel electrophoresis reveals the footprints, i.e., regions of the postselection lane in which the product bands representing mutants are depleted relative to the preselection lane, corresponding to functionally essential regions of the gene.
FIG. 1.
Overview of genetic footprinting. (A) Selection and analysis. A pool of single insertion variants of a gene is subjected to a selection for gene function. The resulting pre- and postselection pools are analyzed by a PCR approach that generates a uniquely sized fragment for each mutant, reflecting the position of the insertion in the gene sequence. Polyacrylamide gel electrophoresis of the fragments generates a footprint, corresponding to an essential region of the gene in which insertions decrease function. Symbols: B→, biotinylated primer; ←✽, radiolabeled primer; SA, streptavidin-agarose. (B) Sequence of the insertion site. Duplication of the target sequence (XXXXX) results in the net in-frame insertion of 15 nucleotides. The identity of the inserted amino acids depends on the relative frame of the insert within the target gene sequence.
Using this approach, we generated a pool of insertion variants of the MoMLV env gene. Due to the transposition mechanism and the length of the introduced sequence, each mutant should contain a net in-frame insertion of five amino acids. However, the actual identity of the amino acids encoded by the inserted sequence depends on its reading frame within the env sequence (Fig. 1B). Sequencing analysis of 80 individually isolated mutant clones demonstrated that nearly 90% contained a single insertion with the expected sequence at a unique position in the env gene (data not shown).
Genetic selections for Env functions.
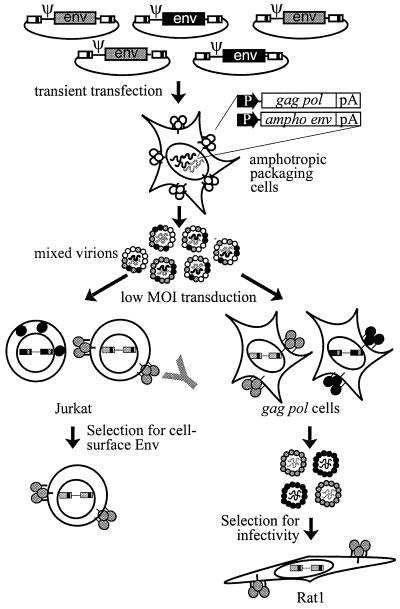
We wanted to deliver each mutant in the pool into individual packaging cells so that the function of each could be assessed without complementation by wild-type virus introduced into the same cell. However, transfection methods typically create precipitated complexes of DNA, which allows each transfected cell to take up multiple vector molecules. This permits nonfunctional mutants to be complemented in trans by functional mutants if both types are present in the same cell. To circumvent this problem, the pool of vectors was first transfected into Phoenix-A cells, which constitutively express the Moloney Gag and Pol proteins as well as the amphotropic Env protein, each from a stably integrated defective viral genome which lacks a packaging signal (Fig. 2). The amphotropic Env protein recognizes a cellular receptor, Pit-2, which is distinct from the receptor for MoMLV Env, MCAT-1 (1, 33). Therefore, although the virions produced by the transfected Phoenix-A cells could have contained multiple mutant MoMLV Env proteins, each should have incorporated the amphotropic Env protein (but not mRNA) as well. Next, the supernatant from the transfected cells was used to transduce Pit-2-positive, MCAT-1-negative cells at an MOI of ∼0.05. This strategy permits the entire pool to be introduced into cells, independently of the function of each mutant in the pool, while decreasing the chance of complementation by minimizing the number of cells transduced by more than one virus.
FIG. 2.
Selections for Env function. The pool of mutant retroviral vectors was transiently transfected into amphotropic packaging cells. The resulting virus was used to transduce, at a low MOI, either Jurkat T cells or 293T gag pol cells, which express the receptor for amphotropic virus but not for MoMLV. Jurkat cells expressing cell surface Env were isolated on the basis of their reactivity with a pool of anti-Env antibodies. Infectious mutants were selected by transducing Rat1 cells, which express the receptor for MoMLV, with the virus produced by the gag pol cells. Symbols:  , vectors;
, vectors;  , viral genomes; ○, Env proteins;
, viral genomes; ○, Env proteins;  , proviruses. Functional mutants at each stage are colored gray; nonfunctional mutants are colored black. The amphotropic Env protein is colored white.
, proviruses. Functional mutants at each stage are colored gray; nonfunctional mutants are colored black. The amphotropic Env protein is colored white.  , Anti-Env antibody.
, Anti-Env antibody.
To assess the ability of the mutants to reach the producer cell surface, Jurkat T cells were used as targets (Fig. 2). Cells in which the introduced Env variants could reach the cell surface were isolated on the basis of their reactivity with a pool of three anti-Env monoclonal antibodies. Multiple antibodies were used in order to minimize the effect on the analysis of the disruption of any single antibody-binding epitope by a particular insertion. Genomic DNA from the initially transduced Jurkat cells was prepared for the preselection sample of the integrated mutants, while genomic DNA from the antibody-isolated cells was collected for the postselection sample.
A similar strategy was used to determine the effects of the insertions on viral infection. The supernatant from the initial transfected Phoenix-A cells was overlaid onto 293T gag pol cells, which constitutively express the MoMLV Gag and Pol proteins as well as Pit-2 but not MCAT-1 or any other Env protein. The introduction of wild-type env DNA into these cells results in the release of infectious virus particles into the supernatant. For the actual selection, a second transduction was carried out with the supernatant from the transduced 293T gag pol cells as the source of the viral variants and a population of Rat1 cells, which constitutively express the rat homologue of MCAT-1, as the targets. Genomic DNA from the 293T gag pol cells was prepared for the preselection sample, while genomic DNA from the infected Rat 1 cells was prepared for the postselection sample.
Analysis and normalization.
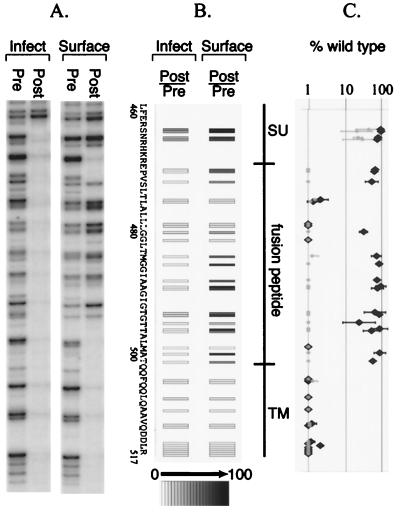
Figure 3 shows representative footprinting gels for the samples from each selection. Each mutant in the pool is represented by a fragment of unique size in the preselection sample, representing the position of the insertion in the env sequence. The nonrandom transposition by MuA has two important consequences for the resulting mutant pool (43). First, although the mutant pool is comprehensive, the actual number of unique mutants that we could reliably detect was less than the maximum number possible if each internucleotide position in the gene was used equally as a target. Second, the frequency of each mutant in the pool varies, reflected in the varying intensities of the bands representing each mutant in the preselection lanes.
FIG. 3.
Representation of the footprinting data. (A) Footprinting gels for a group of mutants from one infectivity selection (Infect) and one cell surface selection (Surface). (B) Display of mutant fitness. For each mutant, a fitness value in each selection is determined by computing the ratio of the postselection: preselection intensities of each mutant fragment and then normalizing to the wild type (see Materials and Methods). Each mutant is then represented by a rectangle, mapped to the position of the insertion within the amino acid sequence of the Env protein, and assigned a gray scale value (white for 0% wild type, black for 100% wild type) for the mean fitness of replicate selections. (C) Graphical representation of mutant fitness. One standard deviation around the mean fitness values for the replicated selections is shown. Black, cell surface selection; gray, infectivity.
The recovery of each mutant after selection was determined by computing the ratio of the intensities of each mutant fragment in the post- and preselection lanes. Importantly, the presence of wild-type DNA in the starting pool allowed the intensity ratio to be expressed as the fraction activity of the wild-type Env protein (see Materials and Methods).
Of 378 mutants we analyzed, 232 (61%) were poorly detected at the cell surface and did not mediate infection at greater than 10% of the wild-type levels. A total of 99 mutants (26%) that reached the cell surface also retained significant infectivity, while 42 (11%) reached the cell surface but were noninfectious. Five mutants survived the infectivity selection despite their apparent absence at the cell surface.
Insertions within the signal sequence.
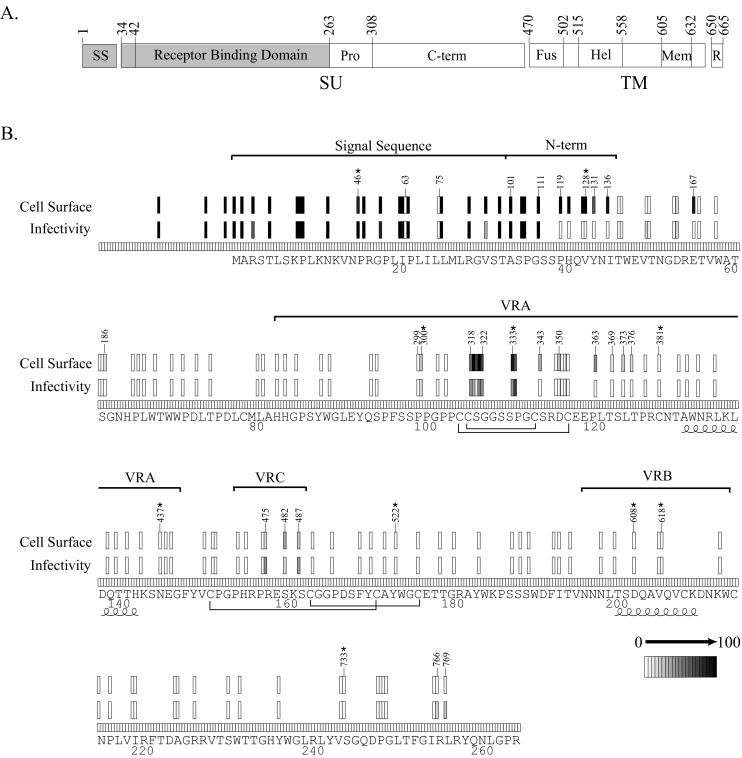
The Env signal sequence is required for the Env protein to enter the secretory pathway and reach the cell surface, where it is incorporated into budding virions (25). Interestingly, all but two of the mutants with insertions in the Env signal sequence demonstrated significant activity in both selections (Fig. 4). Individual analysis of mutant 46 confirmed the phenotype observed by genetic footprinting (Fig. 5). Our results are consistent with previous studies of secreted and transmembrane proteins, which have demonstrated that the signal sequence can tolerate a variety of mutations, including single amino acid substitutions and even complete replacement by random sequences (29, 30, 31). The two exceptions were mutants 63 and 75, which were poorly detected at the cell surface. Their insertions are predicted to introduce the residues Arg-Pro-His within a centrally located hydrophobic stretch of residues (residues 18 to 28). Previous studies have shown that disruption of the continuity of these residues by the introduction of charged amino acids can block secretion (26).
FIG. 4.
Genetic footprinting of the Env signal peptide and RBD. (A) Schematic diagram of the Env protein. (B) Phenotypic map of the mutants. The shaded area in panel A has been enlarged to permit the visualization of the phenotype of each mutant and the position of each insertion as in Fig. 3. Regions discussed in the text are labeled. Select mutants are named according to the nucleotide position of the insertions. Each mutant tested in individual assays is denoted by an asterisk. Disulfide-bonded cysteine residues in VRA and VRC are bracketed. The alpha-helices in VRA and VRB are represented by curled lines.
FIG. 5.
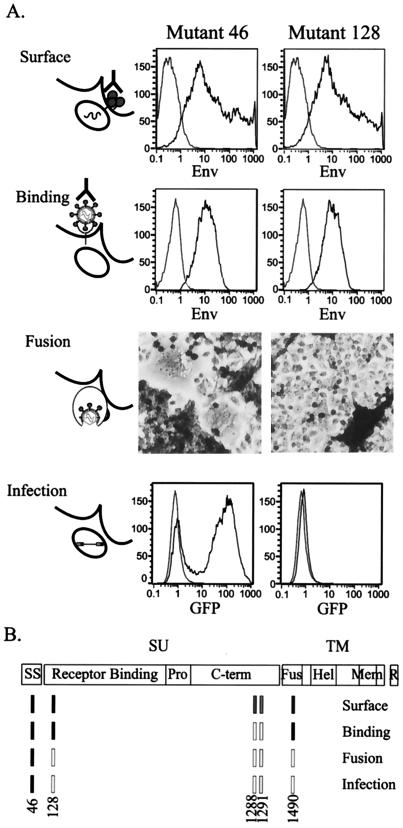
Individual analysis of mutant clones. (A) A mutant from the SU amino-terminal region is deficient in fusion. Mutant 46, a wild-type mutant with an insertion in the signal sequence, and mutant 128, from the SU amino-terminal region identified by genetic footprinting, were tested in individual assays for Env function as described in Materials and Methods. Although mutant 128 could efficiently reach the cell surface and bind the receptor, no syncytia were observed in the XC fusion assay. As expected, mutant 128 was also noninfectious. In the histogram plots, gray lines indicate mock-transfected cells, and black lines indicate the mutants. (B) Summary of noninfectious mutants that reach the producer cell surface. Mutant phenotypes are colored as in Fig. 3.
The amino terminus of mature Env.
Ten insertion mutations within the amino-terminal 13 residues of Env, formed after signal peptide removal, were characterized (Fig. 4). Mutants 101, 105, 106, and 111, with insertions within the sequence encoding the first four residues of the mature Env protein (residues 34 to 37), were equivalent to the wild type in both selections. However, six mutants (119, 122, 127, 128, 131, and 136), with insertions that disrupt the sequence of residues 40 to 46, displayed detectable cell surface expression but poor infectivity. We further characterized the activity of one of these mutants, mutant 128, in individual assays for specific Env functions (Fig. 5). Though this mutant retained significant receptor binding activity, it lacked cell-cell fusion activity. Therefore, a specific defect in fusion appears to account for the loss of infectivity. Consistent with our results, substitution or deletion of the histidine at position 41 or the deletion of residues 34 to 40 or 34 to 41 has previously been shown to abrogate fusion activity (2).
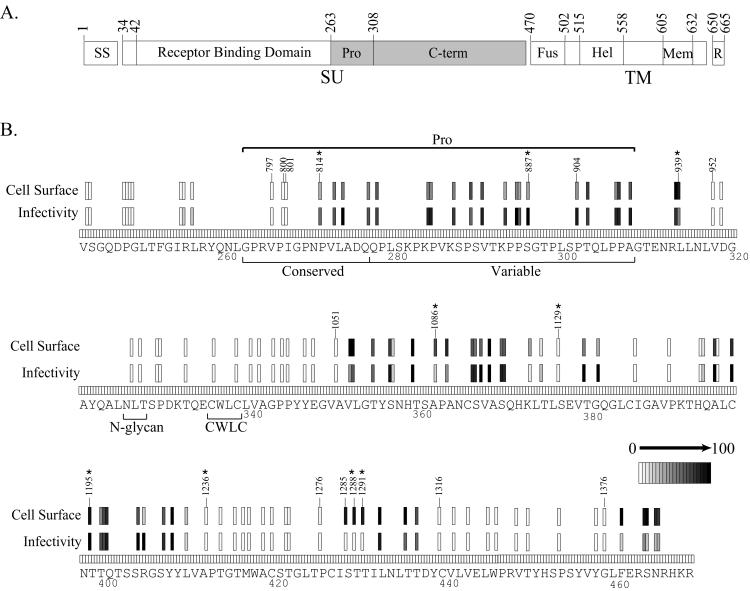
RBD.
We characterized 117 insertions within the MoMLV RBD (Fig. 4, residues 42 to 262). Only 19 mutants were detected at the cell surface at levels at least 10% that of the wild-type protein. Of these, 17 had insertions in either of two regions (variable region A [VRA] and VRC), which vary in sequence and length among the Env proteins of viruses with different tropisms (4, 16). The other two mutants (167 and 186), which were noninfectious despite appreciable cell surface expression, contained insertions in a more conserved region between the amino terminus and VRA.
Seven VRA mutants that reached the cell surface were still infectious (mutants 300, 318, 319, 321, 322, 333, and 334). All but one (mutant 300) contained insertions within a stretch of nine residues (106 to 114) that form a disulfide-bonded surface loop in the RBD of the closely related Friend virus ecotropic envelope protein (16). The insertion in mutant 300 was also located on the protein surface. VRA mutants 343, 350, 363, 373, and 376 were detected at the cell surface at >10% of wild-type levels but were deficient in infectivity. Interestingly, all carried insertions near potential receptor-binding residues defined by mutagenesis studies (2, 11, 40, 45). Infectious VRC mutants 475, 482, and 487 also possessed insertions within a disulfide-bonded loop. None of the insertions in the viable mutants within these loops resulted in the alteration of the cysteine residues involved in the formation of the disulfide bonds.
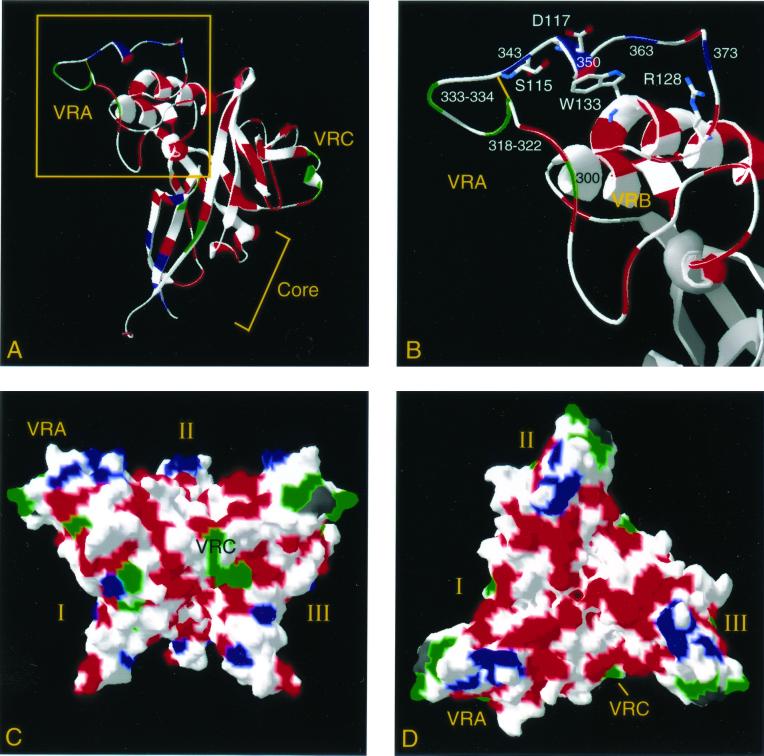
In order to visualize their context in the folded protein, we mapped the location and phenotype of each mutant onto a model of the structure of the MoMLV RBD (Fig. 6A) (16). The model consists of a beta-sandwich core domain, formed from conserved residues, and a helical subdomain with large extended loops formed by the variable regions. As described above, the insertions in the infectious mutants (green) map primarily to two disulfide-bonded loops, one each in VRA and VRC, which in the structure are located on the protein surface. The majority of the mutants that failed to reach the cell surface (red) carried insertions within secondary structural elements that make up the core of the molecule. The VRA insertions that inhibited infection without affecting delivery to the cell surface (blue) were located near residues whose substitution diminishes receptor binding (Fig. 6B).
FIG. 6.
Mutant phenotypes mapped to a model of the RBD. The MoMLV RBD model was generated from the crystal structure of the closely related Friend MLV domain (see Materials and Methods). Each residue is colored according to the fitness of insertions within its sequence as follows: green, >10% viability in both selections; blue, >10% viability in the cell surface selection but <10% infectivity; red, <10% viability in both selections; white, no insertion. (A) Ribbon diagram of the RBD monomer. (B) The region boxed in panel A has been enlarged to show the positions of insertions in the vicinity of the receptor-binding surface. The side chains of previously identified receptor contacting residues are displayed and labeled with the name and position of each residue. Viable mutants are numbered. (C) Molecular surface model of the Friend RBD trimer, viewed from the side. Each monomer is labeled I, II, or III. The VRA and VRC regions of monomer I are indicated. The Ala-Ala dipeptide absent in the Moloney VRA region is colored in dark gray. (D) Same as panel C, but viewed down the trimer threefold axis.
The mutations were also mapped onto a model of the biologically active trimeric form of the Friend RBD (Fig. 6C and D) (15, 16). Most infectious mutants localize to surface-exposed residues, particularly at the apices of the trimer defined by the VRA region of one monomer and the VRC loop of the neighboring trimer. The noninfectious mutants that reached the cell surface neatly define the putative receptor-binding surface, one for each lobe of the trimer. The majority of the mutations that disrupted cell surface expression are located within the highly conserved core or affect regions of the variable loops that have previously been shown to be important for proper folding or assembly (16, 40).
PRR.
The phenotypes of insertions within the proline-rich region (PRR) agreed well with previous studies (34, 62). Three mutants with insertions within the more highly conserved amino-terminal sequence were poorly detected at the cell surface, while the remaining insertions throughout the more variable carboxy-terminal sequence were well tolerated (Fig. 7).
FIG. 7.
Genetic footprinting of the SU PRR and carboxy-terminal domain. Domains and select mutants are named as in Fig. 4.
The carboxy-terminal region of SU.
We characterized the effects of 85 insertions within the highly conserved MoMLV SU C-terminal domain (Fig. 7). The majority of mutants fell into two classes: complete loss of function (e.g., no cell surface detection or infectivity) or nearly wild-type function. Furthermore, since mutants with similar phenotypes tended to cluster together in the primary amino acid sequence, we were able to infer functions of discrete sequences. None of the mutants with insertions in the sequence encoding residues 318 to 348 could be detected at the cell surface. Interestingly, residues 326 to 328 constitute a highly conserved glycosylation site. Substituting for the glycosylated Asn prevents glycan addition and causes the intracellular accumulation of the unprocessed Env protein, suggesting a folding defect (18). This segment also contains a highly conserved motif at residues 336 to 339 containing two cysteine residues, one of which has been proposed to join SU to TM by a labile disulfide bond (49). Substitution of the corresponding cysteines in the reticuloendotheliosis retrovirus Env leads to impaired cell surface delivery and altered intracellular processing, again suggesting a folding defect (23). Since it is possible that the epitopes recognized by the antibodies used for cell surface detection are located within SU (B. Chesebro and L. Evans, unpublished data), the loss of SU as a result of a weakened SU-TM interaction could also account for the observed phenotype.
Two other discrete subdomains were identified in the C-terminal region. Two previously reported insertion mutations within the sequence defined by mutants 1236 through 1276 resulted in decreased infectivity and increased shedding of SU (21). The release of SU from TM could explain the lack of cell surface detection observed for the mutants in this region in our study. The remaining subdomain, defined by mutants 1316 to 1376, forms part of a motif (residues 440 to 469) that in the human immunodeficiency virus type 1 SU protein (GP120) was suggested to interact with the TM subunit (GP41) on the basis of computer modeling and antibody binding studies, again suggesting a possible role for this region in SU-TM association (53).
Three mutants (1285, 1288, and 1291), with insertions within the sequence encoding residues 428 to 431, were detected at the cell surface but lacked infectivity. Mutants 1288 and 1291 did not demonstrate receptor binding or fusion activities in our assays despite significant cell surface levels of each (Fig. 5), suggesting a defect in Env incorporation into virions or receptor binding. However, neither reacted with the 83A25 antibody used for the receptor binding assay when expressed at the cell surface, even though each reacted well with the hybridoma supernatants used for the cell surface selection (data not shown). Therefore, it is possible that the disruption of the 83A25 binding epitope in these mutants accounts for the apparent defect in receptor binding.
MoMLV fusion peptide.
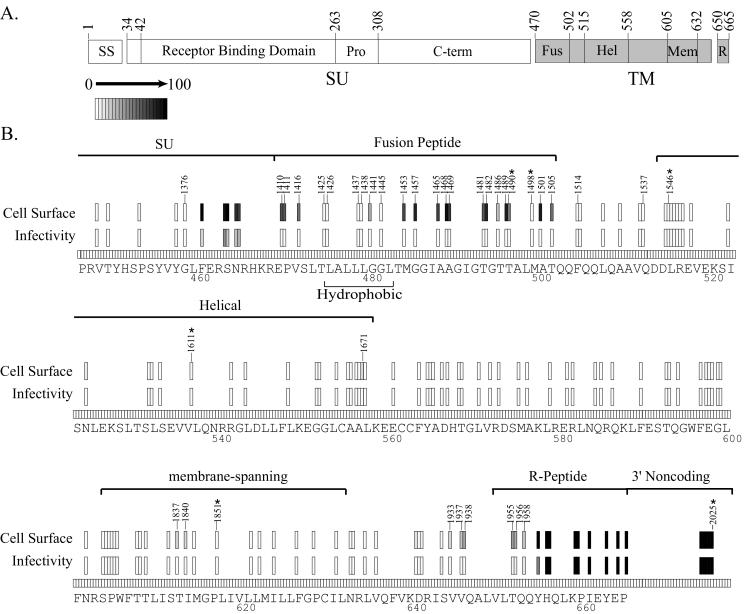
We analyzed 22 insertion variants of the MoMLV fusion peptide (Fig. 8). Sixteen permitted cell surface detection but eliminated infectivity. One of these, mutant 1490, was found to have a specific fusion defect upon individual analysis, a finding consistent with function of the fusion peptide in mediating membrane fusion (Fig. 5). Five of the mutants with severely reduced cell surface detection and infectivity had insertions within a central hydrophobic stretch (residues L476 to L483) in which the subsitution of select residues diminishes both surface expression and incorporation of Env into virions and eliminates infectivity (67). Our results support a role for the hydrophobic stretch in the proper processing of Env, as has been suggested for a similar region in the human immunodeficiency virus type 1 fusion peptide (13).
FIG. 8.
Summary of insertions in the Moloney TM subunit. Domains and select mutants are named as in Fig. 4.
Remainder of the TM subunit.
Most of the remaining insertions in the extracellular portion of TM had severe effects on Env detection at the producer cell surface (Fig. 8). These included six mutants with insertions between the fusion peptide and the helical domain (residues 503 to 514). Two previously characterized insertion mutations within this region (between residues 507 to 508 and residues 511 to 512) completely abrogated infectivity and fusion activity, although their cell surface expression was not assayed (21). Of 26 helical-region mutants, 25 displayed the same phenotype. Since the helical region is the primary mediator of Env oligomerization, it is possible that oligomer stability was disrupted by the presence of the insertions. As is known for the G protein of vesicular stomatitis virus, disruption of oligomerization would presumably cause the Env mutants to become trapped within the ER, thereby preventing their cell surface expression (12). Since the influenza virus HA helical region is also involved in the fusion mechanism, the lone noninfectious mutant in the Env helical region that retained appreciable cell surface expression (i.e., mutant 1671) may be defective in fusion (5, 6, 7, 64). All of the membrane-spanning domain mutants demonstrated significantly diminished cell surface expression and complete lack of infectivity, supporting its function in proper membrane association.
Five mutants (1933, 1937, 1938, 1955, and 1956) with insertions near the R-peptide cleavage site were expressed at the cell surface but could not mediate detectable infection. Since residues flanking the actual cleavage site can influence the efficiency of processing by the viral protease, these insertions may have inhibited the removal of the R-peptide, secondarily inhibiting fusion (41, 42, 50, 51). Consistent with this mechanism, mutants lacking the residues encompassing the insertions in these mutants demonstrate decreased R-peptide cleavage and infectivity (27, 66). Insertions within the remainder of the R-peptide sequence were generally well tolerated, as was a previously reported insertion mutation in the same sequence (58).
DISCUSSION
The experiments described here were designed to determine the effects of a comprehensive set of mutations in the MoMLV Env protein on two critical steps of the virus life cycle: (i) the transport of the Env protein to the producer cell surface during virus assembly and (ii) its capacity to mediate infection. The time and effort required to generate and analyze each mutant individually would have precluded the systematic, quantitative analysis of so many mutants in a single study. Therefore, we devised a strategy that permitted the construction and analysis of all of the mutants simultaneously, as a single pool, yet still obtained quantitative information about the function of each mutant in the pool. An initial transduction with amphotropic virus allowed us to introduce the mutants into individual cells independently of their function. Carrying out this transduction at an MOI of ∼0.05 meant that <5% of the transduced cells were infected by more than one virus, if we assume that the individual transduction events followed a Poisson distribution. Therefore, the chance that a functional mutant could complement a nonfunctional one was minimized.
Most insertion mutations impaired cell surface expression. A total of 61% of the mutants were detected at the producer cell surface at <10% of the wild-type level. Since ER retention is a common feature of misfolded proteins in the secretory pathway, improper folding could have prevented most of the mutants from reaching the producer cell surface (14). Most of the RBD mutants that were poorly detected at the producer cell surface carried insertions in secondary structural elements whose tight packing is likely critical for the overall conformation of the entire domain (Fig. 6). Even in the less-conserved variable regions, all of the insertions in either of the two alpha-helices in VRA and VRB prevented cell surface detection. In the crystal structure, these helices are intimately associated with each other and with residues in the extended loop of VRA. These interactions may be necessary for the proper assembly of the RBD (16, 40).
Since improper folding could also influence the accessibility of epitopes recognized by the antibodies we used to detect cell-surface expression, it is possible that some of the insertions blocked detection of the mutants protein rather than cell surface expression. Our use of a pool of three different monoclonal antibodies against Env was intended to minimize the likelihood of such an event.
We identified three discrete segments of the C-terminal region of SU in which insertions severely decreased the level of the Env protein detected at the producer cell surface (Fig. 7). Based on prior studies, two mechanisms for the observed phenotypes are most plausible. First, the phenotypes could be due to defective folding, as discussed above for the RBD mutants. Second, the insertions could have weakened the association between SU and TM. Such mutants might have reached the producer cell surface but so efficiently shed the SU subunit that they were not recognized by the hybridoma mixture. One of the segments identified contains a highly conserved CWLC motif (residues 336 to 339) proposed to link SU to TM by a disulfide bond (49). The observation that insertions in the SU C-terminal domain can weaken SU-TM stability without altering the sequence of this motif could explain the phenotype we observed for mutants with insertions outside of the motif (21).
Five mutants retained more than 10% of wild-type infectivity despite their seeming absence from the producer cell surface in our assays. It is possible that differences in the sensitivities of the two assays could have permitted better detection of infectivity than cell surface expression. However, it is also possible that insertions that decrease the overall levels of cell surface expression could nevertheless independently enhance the infectivity of the individual mutant protein molecules that manage to reach the surface, through positive effects on steps in the life cycle which occur downstream of cell surface expression.
Fusion-defective mutants.
Insertions within discrete sequences of the SU amino terminus and the fusion peptide prevented infection without significantly reducing the delivery of the mutant proteins to the producer cell surface (Fig. 4 and 8). Furthermore, specific mutants from each region, when analyzed in greater detail, were found to have fusion defects (Fig. 5).
Previous studies have suggested that the length of the fusion peptide could be an important determinant of its function. The insertion of a single alanine residue between the first and second residues of the influenza virus HA fusion peptide abrogated cell-cell fusion at a step following fusion peptide association with a target membrane (57). In addition, although removal of the amino-terminal residue prevents fusion, the restoration of the normal length of the fusion peptide by the insertion of a leucine residue downstream restores fusion activity (44). The fusion defects of these mutants may therefore be due to the increased length of the fusion peptide caused by the insertion of five residues.
Less is known about how the amino terminus of SU can influence the fusion process. Recent studies have demonstrated that substituting His41 in a conserved PHQV motif abolishes fusion (2, 37). Our results demonstrated that insertions in the sequence encoding residues 40 to 46 abolished infectivity. Detailed analysis of one of these mutants (mutant 128) identified a defect in fusion (Fig. 5). Residues near the amino terminus of the HA1 subunit of influenza virus HA influence fusion (56). In particular, substituting His17 in the amino-terminal region of HA1 destabilizes the neutral pH location of the fusion peptide at the amino terminus of HA2 (equivalent to MoMLV TM) and permits fusion to occur at a higher than normal pH (10). In the crystal structure of HA at neutral pH, His17 forms part of a surface of ionizable residues that interact with the fusion peptide at pH 7.0 (but not at the lower pH at which fusion takes place) (8, 56, 64). The insertions in the amino terminus of MoMLV SU may similarly destabilize an interaction with the fusion peptide, allowing a premature switch to the fusogenic conformation. Further support for a functional interaction between the N-terminal and downstream sequences comes from a recent study of chimeras between Moloney and amphotropic Env proteins. Substituting the amphotropic envelope N-terminal residues with the 17 residues at the N terminus of the Moloney protein enhanced the infectivity of a chimeric Env protein containing the amphotropic RBD joined with the C terminus of the Moloney protein (extending from the PRR through the R-peptide) but not of the wild-type amphotropic protein lacking the downstream C-terminal Moloney sequences (39).
Viable insertions within the RBD localize to surface loops.
Only 19 RBD mutants were detected at the cell surface, most with insertions in the variable regions (Fig. 4). Five VRA mutants, which were noninfectious despite reaching the producer cell surface, clustered in a region of VRA that, in the Friend envelope structure, constitutes the putative receptor-binding surface (Fig. 6B) (16). Although the insertions in our mutants do not actually alter the sequence of the key receptor-binding residues as previously defined by mutagenesis, they could block access of the receptor to the key residues or change the three-dimensional relationship of these residues.
Six mutations in VRA that did not abolish infectivity localized to the disulfide-bonded loop formed by residues 106 to 114 (Fig. 6A and B). In a recent study, insertions in the same loop had minimal effects on infectivity (65). Further support for the relative lack of functional importance of this loop comes from our observation that the most significant difference between the otherwise highly homologous Friend and Moloney RBD sequences occurs in this loop, where the Friend domain contains an insertion of two contiguous Ala residues not present in MoMLV (data not shown). Three mutants with insertions in a disulfide-bonded loop in VRC also retained significant infectivity (Fig. 6A), in agreement with previous studies (11, 65).
Implications for vector design and gene therapy.
The methods and results described in this study have important implications for the development of Env protein variants with novel receptor specificities, for use in retroviral vector design and gene therapy approaches. Prior attempts to retarget Env, which have relied on the preexisting knowledge about the Env protein's functional organization, have met with limited success, in part because only one or a few receptor-binding ligands have been inserted at a limited number of sites in the protein (32, 59, 60, 65). The results presented here point to several sites in Env that can be tested as candidates for insertion of novel ligand-binding domains. Alternatively, the NotI site that was introduced in our library provides a convenient means for constructing a library of Env variants in which alternative ligand binding domains are inserted en masse at this site in our library. Selecting the resulting library for the ability to infect cells with the targeted receptor, but lacking the ecotropic receptor, may allow identification of Env variants with altered cell tropisms.
ACKNOWLEDGMENTS
We thank Jon Pollack for critical reading of the manuscript and Joe DeRisi for software support.
This work was supported by NIH grant HG00983 and the Howard Hughes Medical Institute. S.M.R. and R.A.C. were supported by PHS grant CA09302 from the National Cancer Institute. L.C.L. was supported in part by the Medical Scientist Training Program, University of California-San Francisco. P.O.B. is an associate investigator of the Howard Hughes Medical Institute and Professor of Biochemistry at Stanford University.
REFERENCES
- 1.Albritton L M, Tseng L, Scadden D, Cunningham J M. A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell. 1989;57:659–666. doi: 10.1016/0092-8674(89)90134-7. [DOI] [PubMed] [Google Scholar]
- 2.Bae Y, Kingsman S M, Kingsman A J. Functional dissection of the Moloney murine leukemia virus envelope protein gp70. J Virol. 1997;71:2092–2099. doi: 10.1128/jvi.71.3.2092-2099.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Battini J L, Danos O, Heard J M. Receptor-binding domain of murine leukemia virus envelope glycoproteins. J Virol. 1995;69:713–719. doi: 10.1128/jvi.69.2.713-719.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Battini J L, Heard J M, Danos O. Receptor choice determinants in the envelope glycoproteins of amphotropic, xenotropic, and polytropic murine leukemia viruses. J Virol. 1992;66:1468–1475. doi: 10.1128/jvi.66.3.1468-1475.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bullough P A, Hughson F M, Skehel J J, Wiley D C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 6.Carr C M, Chaudhry C, Kim P S. Influenza hemagglutinin is spring-loaded by a metastable native conformation. Proc Natl Acad Sci USA. 1997;94:14306–14313. doi: 10.1073/pnas.94.26.14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carr C M, Kim P S. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- 8.Chen J, Lee K H, Steinhauer D A, Stevens D J, Skehel J J, Wiley D C. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell. 1998;95:409–417. doi: 10.1016/s0092-8674(00)81771-7. [DOI] [PubMed] [Google Scholar]
- 9.Colicelli J, Lobel L I, Goff S P. A temperature-sensitive mutation constructed by “linker insertion”mutagenesis. Mol Gen Genet. 1985;199:537–539. doi: 10.1007/BF00330771. [DOI] [PubMed] [Google Scholar]
- 10.Daniels R S, Downie J C, Hay A J, Knossow M, Skehel J J, Wang M L, Wiley D C. Fusion mutants of the influenza virus hemagglutinin glycoprotein. Cell. 1985;40:431–439. doi: 10.1016/0092-8674(85)90157-6. [DOI] [PubMed] [Google Scholar]
- 11.Davey R A, Zuo Y, Cunningham J M. Identification of a receptor-binding pocket on the envelope protein of friend murine leukemia virus. J Virol. 1999;73:3758–3763. doi: 10.1128/jvi.73.5.3758-3763.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Silva A M, Balch W E, Helenius A. Quality control in the endoplasmic reticulum: folding and misfolding of vesicular stomatitis virus G protein in cells and in vitro. J Cell Biol. 1990;111:857–866. doi: 10.1083/jcb.111.3.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delahunty M D, Rhee I, Freed E O, Bonifacino J S. Mutational analysis of the fusion peptide of the human immunodeficiency virus type 1: identification of critical glycine residues. Virology. 1996;218:94–102. doi: 10.1006/viro.1996.0169. [DOI] [PubMed] [Google Scholar]
- 14.Doms R W, Lamb R A, Rose J K, Helenius A. Folding and assembly of viral membrane proteins. Virology. 1993;193:545–562. doi: 10.1006/viro.1993.1164. [DOI] [PubMed] [Google Scholar]
- 15.Fass D. Ph.D. dissertation. Cambridge: Massachussetts Institute of Technology; 1997. [Google Scholar]
- 16.Fass D, Davey R A, Hamson C A, Kim P S, Cunningham J M, Berger J M. Structure of a murine leukemia virus receptor-binding glycoprotein at 2.0 angstrom resolution. Science. 1997;277:1662–1666. doi: 10.1126/science.277.5332.1662. [DOI] [PubMed] [Google Scholar]
- 17.Fass D, Harrison S C, Kim P S. Retrovirus envelope domain at 1.7 angstrom resolution. Nat Struct Biol. 1996;3:465–469. doi: 10.1038/nsb0596-465. [DOI] [PubMed] [Google Scholar]
- 18.Felkner R H, Roth M J. Mutational analysis of the N-linked glycosylation sites of the SU envelope protein of Moloney murine leukemia virus. J Virol. 1992;66:4258–4264. doi: 10.1128/jvi.66.7.4258-4264.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freed E O, Risser R. The role of envelope glycoprotein processing in murine leukemia virus infection. J Virol. 1987;61:2852–2856. doi: 10.1128/jvi.61.9.2852-2856.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goff S P, Prasad V R. Linker insertion mutagenesis as probe of structure-function relationships. Methods Enzymol. 1991;208:586–603. doi: 10.1016/0076-6879(91)08030-l. [DOI] [PubMed] [Google Scholar]
- 21.Gray K D, Roth M J. Mutational analysis of the envelope gene of Moloney murine leukemia virus. J Virol. 1993;67:3489–3496. doi: 10.1128/jvi.67.6.3489-3496.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Green N, Shinnick T M, Witte O, Ponticelli A, Sutcliffe J G, Lerner R A. Sequence-specific antibodies show that maturation of Moloney leukemia virus envelope polyprotein involves removal of a COOH-terminal peptide. Proc Natl Acad Sci USA. 1981;78:6023–6027. doi: 10.1073/pnas.78.10.6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gu J, Parthasarathi S, Varela-Echavarria A, Ron Y, Dougherty J P. Mutations of conserved cysteine residues in the CWLC motif of the oncoretrovirus SU protein affect maturation and translocation. Virology. 1995;206:885–893. doi: 10.1006/viro.1995.1011. [DOI] [PubMed] [Google Scholar]
- 24.Heard J M, Danos O. An amino-terminal fragment of the Friend murine leukemia virus envelope glycoprotein binds the ecotropic receptor. J Virol. 1991;65:4026–4032. doi: 10.1128/jvi.65.8.4026-4032.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Izard J W, Kendall D A. Signal peptides: exquisitely designed transport promoters. Mol Microbiol. 1994;13:765–773. doi: 10.1111/j.1365-2958.1994.tb00469.x. [DOI] [PubMed] [Google Scholar]
- 26.Izard J W, Rusch S L, Kendall D A. The amino-terminal charge and core region hydrophobicity interdependently contribute to the function of signal sequences. J Biol Chem. 1996;271:21579–21582. doi: 10.1074/jbc.271.35.21579. [DOI] [PubMed] [Google Scholar]
- 27.Januszeski M M, Cannon P M, Chen D, Rozenberg Y, Anderson W F. Functional analysis of the cytoplasmic tail of Moloney murine leukemia virus envelope protein. J Virol. 1997;71:3613–3619. doi: 10.1128/jvi.71.5.3613-3619.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kadan M J, Sturm S, Anderson W F, Eglitis M A. Detection of receptor-specific murine leukemia virus binding to cells by immunofluorescence analysis. J Virol. 1992;66:2281–2287. doi: 10.1128/jvi.66.4.2281-2287.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaiser C A, Botstein D. Efficiency and diversity of protein localization by random signal sequences. Mol Cell Biol. 1990;10:3163–3173. doi: 10.1128/mcb.10.6.3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaiser C A, Botstein D. Secretion-defective mutations in the signal sequence for Saccharomyces cerevisiae invertase. Mol Cell Biol. 1986;6:2382–2391. doi: 10.1128/mcb.6.7.2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaiser C A, Preuss D, Grisafi P, Botstein D. Many random sequences functionally replace the secretion signal sequence of yeast invertase. Science. 1987;235:312–317. doi: 10.1126/science.3541205. [DOI] [PubMed] [Google Scholar]
- 32.Kasahara N, Dozy A M, Kan Y W. Tissue-specific targeting of retroviral vectors through ligand-receptor interactions. Science. 1994;266:1373–1376. doi: 10.1126/science.7973726. [DOI] [PubMed] [Google Scholar]
- 33.Kavanaugh M P, Kabat D. Identification and characterization of a widely expressed phosphate transporter/retrovirus receptor family. Kidney Int. 1996;49:959–963. doi: 10.1038/ki.1996.135. [DOI] [PubMed] [Google Scholar]
- 34.Kayman S C, Park H, Saxon M, Pinter A. The hypervariable domain of the murine leukemia virus surface protein tolerates large insertions and deletions, enabling development of a retroviral particle display system. J Virol. 1999;73:1802–1808. doi: 10.1128/jvi.73.3.1802-1808.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kinsella T M, Nolan G P. Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum Gene Ther. 1996;7:1405–1413. doi: 10.1089/hum.1996.7.12-1405. [DOI] [PubMed] [Google Scholar]
- 36.Laurent L C, Olsen M N, Crowley R A, Savilahti H, Brown P O. Functional characterization of the human immunodeficiency virus type 1 genome by genetic footprinting. J Virol. 2000;74:2760–2769. doi: 10.1128/jvi.74.6.2760-2769.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lavillette D, Ruggieri A, Russell S J, Cosset F L. Activation of a cell entry pathway common to type C mammalian retroviruses by soluble envelope fragments. J Virol. 2000;74:295–304. doi: 10.1128/jvi.74.1.295-304.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee S, Zhao Y, Anderson W F. Receptor-mediated Moloney murine leukemia virus entry can occur independently of the clathrin-coated-pit-mediated endocytic pathway. J Virol. 1999;73:5994–6005. doi: 10.1128/jvi.73.7.5994-6005.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu C W, Roth M J. Functional characterization of the N termini of murine leukemia virus envelope proteins. J Virol. 2001;75:4357–4366. doi: 10.1128/JVI.75.9.4357-4366.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.MacKrell A J, Soong N W, Curtis C M, Anderson W F. Identification of a subdomain in the Moloney murine leukemia virus envelope protein involved in receptor binding. J Virol. 1996;70:1768–1774. doi: 10.1128/jvi.70.3.1768-1774.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Menendez-Arias L, Weber I T, Oroszlan S. Mutational analysis of the substrate binding pocket of murine leukemia virus protease and comparison with human immunodeficiency virus proteases. J Biol Chem. 1995;270:29162–29168. doi: 10.1074/jbc.270.49.29162. [DOI] [PubMed] [Google Scholar]
- 42.Menendez-Arias L, Weber I T, Soss J, Harrison R W, Gotte D, Oroszlan S. Kinetic and modeling studies of subsites S4–S3′ of Moloney murine leukemia virus protease. J Biol Chem. 1994;269:16795–16801. [PubMed] [Google Scholar]
- 43.Mizuuchi M, Mizuuchi K. Target site selection in transposition of phage Mu. Cold Spring Harbor Symp Quant Biol. 1993;58:515–523. doi: 10.1101/sqb.1993.058.01.058. [DOI] [PubMed] [Google Scholar]
- 44.Orlich M, Rott R. Thermolysin activation mutants with changes in the fusogenic region of an influenza virus hemagglutinin. J Virol. 1994;68:7537–7539. doi: 10.1128/jvi.68.11.7537-7539.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Panda B R, Kingsman S M, Kingsman A J. Mutational analysis of the putative receptor-binding domain of Moloney murine leukemia virus glycoprotein gp70. Virology. 2000;273:90–100. doi: 10.1006/viro.2000.0397. [DOI] [PubMed] [Google Scholar]
- 46.Pinter A, Fleissner E. The presence of disulfide-linked gp70–p15(E) complexes in AKR murine leukemia virus. Virology. 1977;83:417–422. doi: 10.1016/0042-6822(77)90187-8. [DOI] [PubMed] [Google Scholar]
- 47.Pinter A, Honnen W J. Topography of murine leukemia virus envelope proteins: characterization of transmembrane components. J Virol. 1983;46:1056–1060. doi: 10.1128/jvi.46.3.1056-1060.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pinter A, Honnen W J, Li J S. Studies with inhibitors of oligosaccharide processing indicate a functional role for complex sugars in the transport and proteolysis of Friend mink cell focus-inducing murine leukemia virus envelope proteins. Virology. 1984;136:196–210. doi: 10.1016/0042-6822(84)90259-9. [DOI] [PubMed] [Google Scholar]
- 49.Pinter A, Kopelman R, Li Z, Kayman S C, Sanders D A. Localization of the labile disulfide bond between SU and TM of the murine leukemia virus envelope protein complex to a highly conserved CWLC motif in SU that resembles the active-site sequence of thiol-disulfide exchange enzymes. J Virol. 1997;71:8073–8077. doi: 10.1128/jvi.71.10.8073-8077.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ragheb J A, Anderson W F. pH-independent murine leukemia virus ecotropic envelope-mediated cell fusion: implications for the role of the R peptide and p12E TM in viral entry. J Virol. 1994;68:3220–3231. doi: 10.1128/jvi.68.5.3220-3231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rein A, Mirro J, Haynes J G, Ernst S M, Nagashima K. Function of the cytoplasmic domain of a retroviral transmembrane protein: p15E-p2E cleavage activates the membrane fusion capability of the murine leukemia virus Env protein. J Virol. 1994;68:1773–1781. doi: 10.1128/jvi.68.3.1773-1781.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schultz A, Rein A. Maturation of murine leukemia virus Env proteins in the absence of other viral proteins. Virology. 1985;145:335–339. doi: 10.1016/0042-6822(85)90168-0. [DOI] [PubMed] [Google Scholar]
- 53.Schulz T F, Jameson B A, Lopalco L, Siccardi A G, Weiss R A, Moore J P. Conserved structural features in the interaction between retroviral surface and transmembrane glycoproteins? AIDS Res Hum Retrovir. 1992;8:1571–1580. doi: 10.1089/aid.1992.8.1571. [DOI] [PubMed] [Google Scholar]
- 54.Singh I R, Crowley R A, Brown P O. High-resolution functional mapping of a cloned gene by genetic footprinting. Proc Natl Acad Sci USA. 1997;94:1304–1309. doi: 10.1073/pnas.94.4.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Skehel J J, Bizebard T, Bullough P A, Hughson F M, Knossow M, Steinhauer D A, Wharton S A, Wiley D C. Membrane fusion by influenza hemagglutinin. Cold Spring Harbor Symp Quant Biol. 1995;60:573–580. doi: 10.1101/sqb.1995.060.01.061. [DOI] [PubMed] [Google Scholar]
- 56.Steinhauer D A. Role of hemagglutinin cleavage for the pathogenicity of influenza virus. Virology. 1999;258:1–20. doi: 10.1006/viro.1999.9716. [DOI] [PubMed] [Google Scholar]
- 57.Steinhauer D A, Wharton S A, Skehel J J, Wiley D C. Studies of the membrane fusion activities of fusion peptide mutants of influenza virus hemagglutinin. J Virol. 1995;69:6643–6651. doi: 10.1128/jvi.69.11.6643-6651.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thomas A, Gray K D, Roth M J. Analysis of mutations within the cytoplasmic domain of the Moloney murine leukemia virus transmembrane protein. Virology. 1997;227:305–313. doi: 10.1006/viro.1996.8333. [DOI] [PubMed] [Google Scholar]
- 59.Valsesia-Wittmann S, Drynda A, Deleage G, Aumailley M, Heard J M, Danos O, Verdier G, Cosset F L. Modifications in the binding domain of avian retrovirus envelope protein to redirect the host range of retroviral vectors. J Virol. 1994;68:4609–4619. doi: 10.1128/jvi.68.7.4609-4619.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Valsesia-Wittmann S, Morling F J, Hatziioannou T, Russell S J, Cosset F L. Receptor co-operation in retrovirus entry: recruitment of an auxiliary entry mechanism after retargeted binding. EMBO J. 1997;16:1214–1223. doi: 10.1093/emboj/16.6.1214. . (Erratum, 16:4153.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Walter P, Johnson A E. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu Rev Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- 62.Weimin Wu B, Cannon P M, Gordon E M, Hall F L, Anderson W F. Characterization of the proline-rich region of murine leukemia virus envelope protein. J Virol. 1998;72:5383–5391. doi: 10.1128/jvi.72.7.5383-5391.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.White J M. Membrane fusion: the influenza paradigm. Cold Spring Harbor Symp Quant Biol. 1995;60:581–588. doi: 10.1101/sqb.1995.060.01.062. [DOI] [PubMed] [Google Scholar]
- 64.Wilson I A, Skehel J J, Wiley D C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 65.Wu B W, Lu J, Gallaher T K, Anderson W F, Cannon P M. Identification of regions in the Moloney murine leukemia virus SU protein that tolerate the insertion of an integrin-binding peptide. Virology. 2000;269:7–17. doi: 10.1006/viro.2000.0201. [DOI] [PubMed] [Google Scholar]
- 66.Yang C, Compans R W. Analysis of the murine leukemia virus R peptide: delineation of the molecular determinants which are important for its fusion inhibition activity. J Virol. 1997;71:8490–8496. doi: 10.1128/jvi.71.11.8490-8496.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhu N L, Cannon P M, Chen D, Anderson W F. Mutational analysis of the fusion peptide of Moloney murine leukemia virus transmembrane protein p15E. J Virol. 1998;72:1632–1639. doi: 10.1128/jvi.72.2.1632-1639.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]