Abstract
Examination of the three-dimensional structure of intact herpes simplex virus type 1 (HSV-1) virions had revealed that the icosahedrally symmetrical interaction between the tegument and capsid involves the pentons but not the hexons (Z. H. Zhou, D. H. Chen, J. Jakana, F. J. Rixon, and W. Chiu, J. Virol. 73:3210–3218, 1999). To account for this, we postulated that the presence of the small capsid protein, VP26, on top of the hexons was masking potential binding sites and preventing tegument attachment. We have now tested this hypothesis by determining the structure of virions lacking VP26. Apart from the obvious absence of VP26 from the capsids, the structures of the VP26 minus and wild-type virions were essentially identical. Notably, they showed the same tegument attachment patterns, thereby demonstrating that VP26 is not responsible for the divergent tegument binding properties of pentons and hexons.
Herpesvirus virions have complex and characteristic multilayered structures in which the capsid is separated from the envelope by a thick layer of protein called the tegument (7, 12). The 125-nm-diameter capsid has the form of a T=16 icosahedron with 12 pentons occupying the icosahedral vertices and 150 hexons forming the faces and edges (11, 18, 23). In herpes simplex virus type 1 (HSV-1), the pentons and hexons contain five and six copies, respectively, of the 150-kDa major capsid protein, VP5 (6, 23). They are surrounded and connected by 320 triplexes, which are heterotrimers formed from two copies of a 34-kDa protein (VP23) and a single copy of a 50-kDa protein (VP19C) (6, 9, 21). Six copies of a 12-kDa protein (VP26) occupy the top of each hexon (16, 22). Although the structure of the capsid is increasingly well understood, little is known about the organization of the tegument and how it interacts with the other components of the virion. Recently, we published the first structural analysis of the intact HSV-1 virion using electron cryomicroscopy and three-dimensional (3D) reconstruction (20). The majority of the tegument mass was not icosahedrally ordered and was not resolved. However, some density attributable to the tegument was seen in close contact with the capsid. The contacts between the capsid and tegument were of limited extent and were confined to the vicinities of the 12 vertices, where they extended from near the tip of the penton to the upper surfaces of the Ta and Tc triplexes. The reason why tegument contacts are restricted to these particular subunits when apparently similar potential contact sites are present on other capsomeres and triplexes was not obvious. A possible reason suggested is that the corresponding binding site near the tip of the penton is occupied in the hexon by VP26, making it unavailable to the tegument. To test this possibility we analyzed the pattern of tegument-capsid interactions in virions that lack VP26.
Generation of VP26 minus virus.
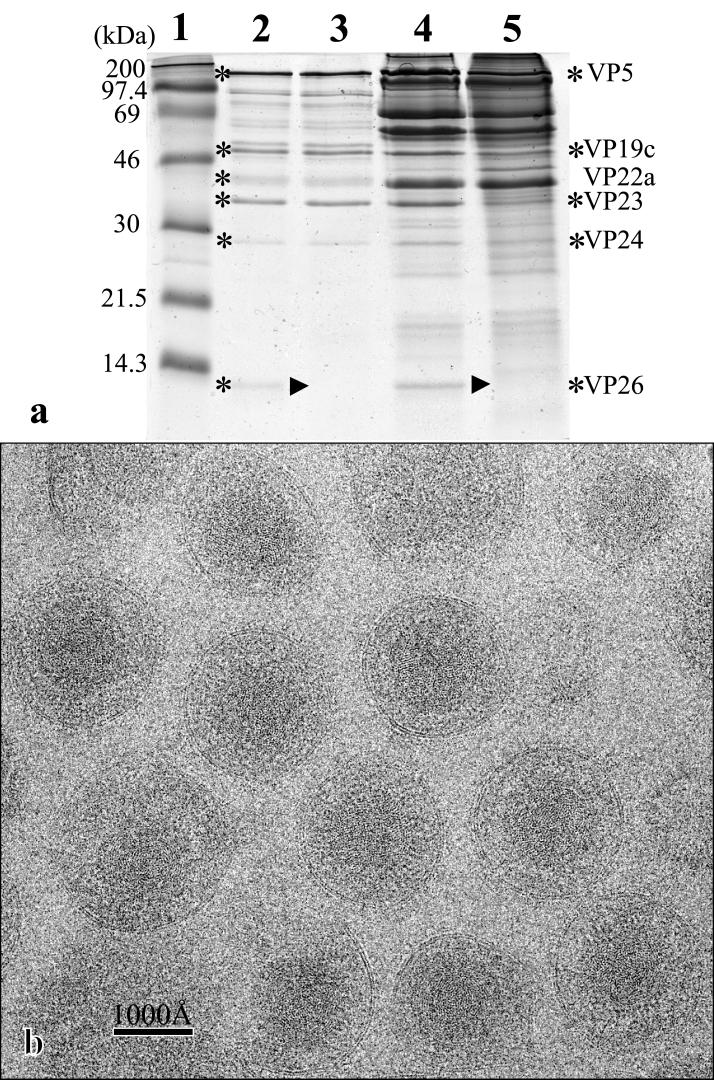
VP26 is encoded by gene UL35 (8). Sequences downstream from the UL35 ORF were amplified using the primers GACAGGATCCTGAGGCCCGGGGAGTTCCTTCTGG and GACAAAGCTTCAGACCCTGTATGTCTCTGACG. The PCR product was digested with BamHI and HindIII, using the sites built into the primers (underlined), and ligated into BamHI/HindIII-digested pCMV10 (13) to give pUL35construct1. The open reading frame (ORF) for the green fluorescent protein (GFP) was isolated from plasmid pGFPemd-b [R] basic (Packard Bioscience) by digestion with KpnI and BamHI and ligated into KpnI/BamHI-digested pUL35construct1 to give pUL35construct2. Sequences upstream of the UL35 ORF were amplified by PCR using the primers GACAGAATTCCCGAGCAGGCTATTACCCGTCGC and GACAGGTACCATGGGGATCCGAGGTCGGGAAGCGATATGGGGGTGTCG. The PCR product was digested with EcoRI and NcoI, using the sites built into the primers (unlabelled), and ligated into EcoRI/NcoI-digested ppUL35construct2 to give pUL35construct3. pUL35construct3 contains HSV-1 sequences from 70186 to 71385 (5) with an internal deletion from 70557 to 70901 that removes the entire UL35 ORF and replaces it with the GFP ORF. BHK cells were transfected with pUL35construct3 and superinfected 5 h later with 1 PFU of HSV-1 strain 17 per cell. After incubation at 37°C for 48 h, the infected-cell medium was harvested and the virus was titrated and grown in medium containing 1% carboxymethyl cellulose. Plaques were examined using a Nikon Microphot-SA fluorescence microscope. Fluorescing plaques were picked and subjected to three further rounds of purification. A single plaque isolate was selected and designated dmVP26-minus. To confirm that the VP26 protein was missing, wild-type and dmVP26-minus capsids and virions were prepared as described previously (14, 23) and their protein contents were analyzed by electrophoresis through a polyacrylamide gel (Fig. 1a). In both capsids and virions, the 12-kDa VP26 protein is absent from the dmVP26-minus profile.
FIG. 1.
(a) Sodium dodecyl sulfate-polyacrylamide gel of purified wild-type B capsids (lane 2) and virions (lane 4), dmVP26-minus capsids (lane 3) and virions (lane 5), and standard size markers (lane 1). The locations of the capsid proteins in the capsid and virion profiles are indicated (∗) to the left of lane 2 and to the right of lane 5, respectively. The arrowheads indicate the expected position of the missing VP26 protein in lanes 3 and 5. (b) Electron cryomicroscopy (400 kV) images of ice-embedded dmVP26-minus virions.
Icosahedral reconstruction.
dmVP26-minus virions were purified through Ficoll gradients (14). The purified virions were embedded in vitreous ice suspended across holes in holey carbon grids (10) (Fig. 1b). Electron microscopy, digitizing of the images, 3D reconstruction, and correction of the contrast transfer function were carried out as described previously (20). A 3D reconstruction to an effective resolution of 17 Å was obtained by merging 918 particles selected from 33 micrographs. The structures shown here are truncated to a lower resolution of 20 Å in order to allow direct comparison with the previously published wild-type HSV-1 virion reconstruction (20).
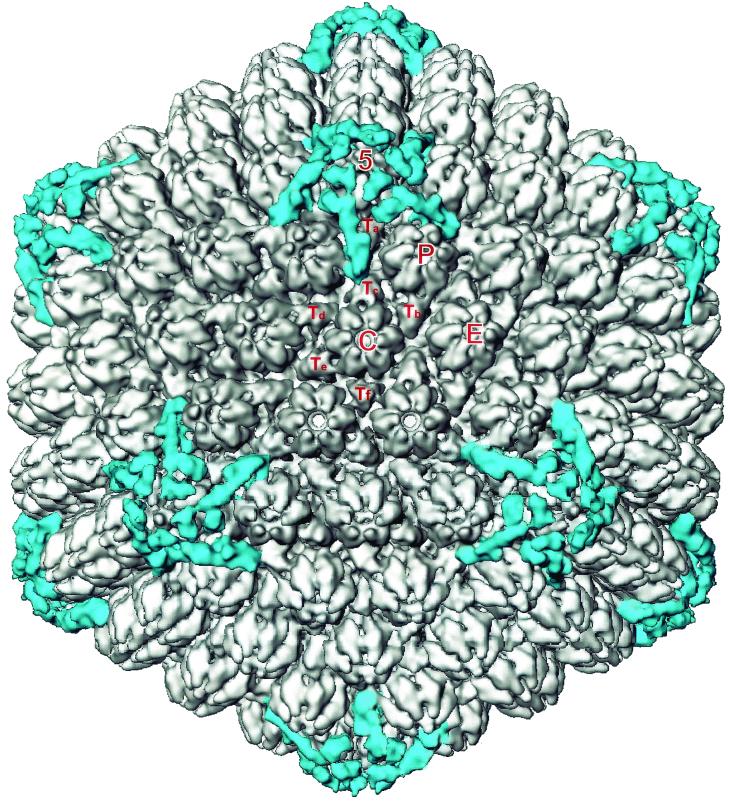
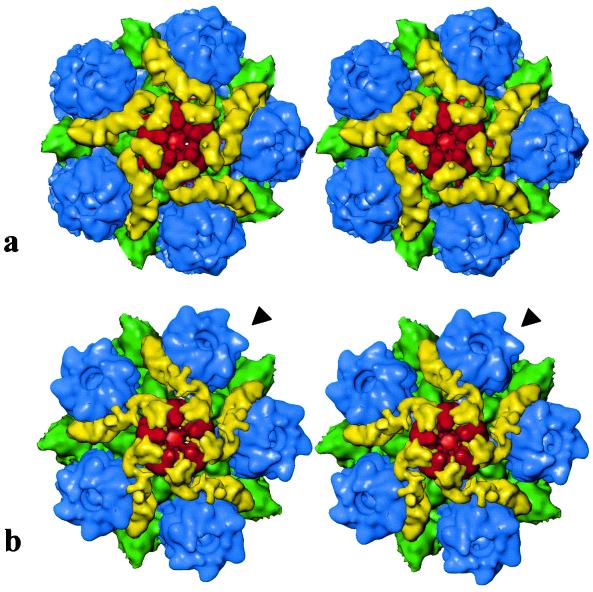
As had been found earlier for the wild-type virions, the bulk of the envelope and tegument material has no icosahedral symmetry and appeared as unconnected masses surrounding the capsid (data not shown). At a density threshold of 1.2ς these unconnected masses disappear, exposing the underlying capsid (Fig. 2). As was seen previously with wild-type virions (20), detectable tegument material is confined to the vicinities of the icosahedral fivefold vertices and there is no suggestion of additional sites of tegument binding. The only obvious difference between the wild-type and dmVP26-minus capsids was seen at the top of each hexon, where a ring of density, corresponding to the six copies of VP26, is missing from the dmVP26-minus map (Fig. 3). Apart from this, the hexons appear to be unchanged, and comparison of the pentons revealed virtual identity between the wild-type and dmVP26-minus structures. In both cases the attached tegument material is in the form of five convoluted ribbons of density, each extending radially outwards from the upper domains of neighboring penton subunits and making contact with the peripentonal triplex (Ta), the P hexon, and triplex Tc (Fig. 3). Another feature distinguishing virion capsids and B capsids noted previously is the closure of the penton channel, and this is also a feature of the dmVP26-minus reconstruction (Fig. 3).
FIG. 2.
Visualization of icosahedrally ordered tegument in dmVP26-minus virions. The dmVP26-minus virion reconstruction is displayed at 1.2 standard deviations above mean density. At this density threshold, only the icosahedrally ordered tegument densities are visible. To generate this map, the five copies of tegument density around one penton were first computationally isolated. These densities were then replicated and rotated around the icosahedral symmetry axes to the positions of the 11 other vertices to obtain a map containing only tegument densities (blue). This map was then superimposed back onto the dmVP26-minus virion reconstruction (gray). The unique structural components of the capsid are labeled 5 (penton), P, C, E, and Ta-Tf (icosahedrally unrelated hexons and triplexes, respectively).
FIG. 3.
(a) Stereo pair of a computationally isolated portion of the dmVP26-minus map shown in Fig. 2 that includes the penton (red), P hexons (blue), triplexes Ta and Tc (green), and the icosahedrally ordered tegument densities (yellow). (b) Stereo pair of the equivalent portion of the wild-type HSV-1 map of Zhou et al. (20). The arrowhead points to one of the six horn-shaped VP26 densities on the top of each hexon. These densities are missing from the equivalent positions on the dmVP26-minus map.
Role of VP26 in capsid-tegument interaction.
In an earlier paper (20), it was suggested that the presence of VP26 in the hexons might explain why tegument binds only to the vicinity of the penton despite the presence elsewhere in the capsid of superficially similar arrangements of capsomeres and triplexes. This suggestion was based on the observation that the region of VP5, to which the tegument binds on the penton, is already occupied on the hexons by VP26. The near identity described here between the patterns of tegument-capsid interaction seen in the presence or absence of VP26 clearly eliminates the possibility that accessibility of the binding site is controlling tegument attachment. The most likely explanation now seems to lie in inherent differences in the nature of the upper domains of the penton and hexon subunits, as evidenced by the differential binding patterns of certain monoclonal antibodies (17). Interestingly, the 3D structure of human cytomegalovirus (HCMV) virions has recently been determined and shows a very different pattern of tegument-capsid interaction, with tegument binding extending to all of the hexons and triplexes as well as the pentons (3). A similar pattern was described previously for simian cytomegalovirus (SCMV) cytoplasmic B capsids (15).
The tegument proteins involved in these interactions have not yet been positively identified for HSV-1, HCMV, or SCMV. Comparisons between the protein profiles and 3D structures of SCMV nuclear and cytoplasmic B capsids led Trus et al. (15) to propose the 119-kDa basic phosphoprotein and the 69-kDa upper matrix protein as possible candidates. Neither of these two proteins has a counterpart in HSV-1 (2). However, the SCMV cytoplasmic B capsid protein profile also contained the 205-kDa high-molecular-size protein, which is the SCMV counterpart of VP1–3 in HSV-1 (15). Previously, we speculated that VP1–3 was the most likely candidate for the HSV-1 penton-associated tegument protein (20). This raises the intriguing possibility that the association between tegument and penton is mediated through homologous proteins in HSV-1 (VP1–3) and CMV (the high-molecular-weight protein), while that between tegument and hexon, seen only in HCMV and SCMV, represents an unrelated association (presumably involving basic phosphoprotein and upper matrix protein) that has not been conserved between the two virus subfamilies. It has recently been demonstrated that in HCMV the small capsid protein is essential for virus growth (1), unlike its counterpart (VP26) in HSV-1 (this paper and reference 4). This led to the suggestion that the small capsid protein is involved in the interaction between the tegument and hexon (1). If true, this would support the conjecture by Wingfield et al. (19) that the differential interaction of the small capsid protein with hexons and pentons might serve to expand the range of functional binding sites on the capsid surface. Further information on the nature of these various interactions and their roles in the virus life cycle awaits the accurate identification of the proteins involved.
Acknowledgments
This research was supported by NIH grants (RO1AI38469 and P41RR02250).
REFERENCES
- 1.Borst E M, Mathys S, Wagner M, Muranyi W, Messerle M. Genetic evidence of an essential role for cytomegalovirus small capsid protein in viral growth. J Virol. 2001;75:1450–1458. doi: 10.1128/JVI.75.3.1450-1458.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chee M S, Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Horsnell T, Hutchison C A, Kouzarides T, Martignetti J A, Preddie E, Satchwell S C, Tomlinson P, Weston K M, Barrell B G. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 3.Chen D H, Jiang H, Lee M, Liu F Y, Zhou Z H. Three-dimensional visualization of tegument/capsid interactions in the intact human cytomegalovirus. Virology. 1999;260:10–16. doi: 10.1006/viro.1999.9791. [DOI] [PubMed] [Google Scholar]
- 4.Desai P, DeLuca N A, Person S. Herpes simplex virus type 1 VP26 is not essential for replication in cell culture but influences production of infectious virus in the nervous system of infected mice. Virology. 1998;247:115–124. doi: 10.1006/viro.1998.9230. [DOI] [PubMed] [Google Scholar]
- 5.McGeoch D J, Dalrymple M A, Davison A J, Dolan A, Frame M C, McNab D, Perry L J, Scott J E, Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988;69:1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 6.Newcomb W W, Trus B L, Booy F P, Steven A C, Wall J S, Brown J C. Structure of the herpes simplex virus capsid: molecular composition of the pentons and the triplexes. J Mol Biol. 1993;232:499–511. doi: 10.1006/jmbi.1993.1406. [DOI] [PubMed] [Google Scholar]
- 7.Rixon F J. Structure and assembly of herpesviruses. Semin Virol. 1993;4:135–144. [Google Scholar]
- 8.Rixon F J, Davison M D, Davison A J. Identification of the genes encoding two capsid proteins of herpes simplex virus type 1 by direct amino acid sequencing. J Gen Virol. 1990;71:1211–1214. doi: 10.1099/0022-1317-71-5-1211. [DOI] [PubMed] [Google Scholar]
- 9.Saad A, Zhou Z H, Jakana J, Chiu W, Rixon F J. Roles of triplex and scaffolding proteins in herpes simplex virus type 1 capsid formation suggested by structures of recombinant particles. J Virol. 1999;73:6821–6830. doi: 10.1128/jvi.73.8.6821-6830.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schrag J D, Prasad B V V, Rixon F J, Chiu W. Three-dimensional structure of the HSV-1 nucleocapsid. Cell. 1989;56:651–660. doi: 10.1016/0092-8674(89)90587-4. [DOI] [PubMed] [Google Scholar]
- 11.Steven A C, Roberts C R, Hay J, Bisher M E, Pun T, Trus B L. Hexavalent capsomers of herpes simplex virus type-2: symmetry, shape, dimensions, and oligomeric status. J Virol. 1986;57:578–584. doi: 10.1128/jvi.57.2.578-584.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steven A C, Spear P G. Herpesvirus capsid assembly and envelopment. In: Chiu W, Burnett R M, Garcea R, editors. Structural biology of viruses. New York, N.Y: Oxford University Press; 1997. pp. 312–351. [Google Scholar]
- 13.Stow N D, Hammarsten O, Arbuckle M I, Elias P. Inhibition of herpes-simplex virus type-1 DNA-replication by mutant forms of the origin-binding protein. Virology. 1993;196:413–418. doi: 10.1006/viro.1993.1496. [DOI] [PubMed] [Google Scholar]
- 14.Szilagyi J F, Cunningham C. Identification and characterization of a novel non-infectious herpes simplex virus-related particle. J Gen Virol. 1991;72:661–668. doi: 10.1099/0022-1317-72-3-661. [DOI] [PubMed] [Google Scholar]
- 15.Trus B L, Gibson W, Cheng N Q, Steven A C. Capsid structure of simian cytomegalovirus from cryoelectron microscopy: evidence for tegument attachment sites. J Virol. 1999;73:2181–2192. doi: 10.1128/jvi.73.3.2181-2192.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trus B L, Homa F L, Booy F P, Newcomb W W, Thomsen D R, Cheng N, Brown J C, Steven A C. Herpes simplex virus capsids assembled in insect cells infected with recombinant baculoviruses: structural authenticity and localization of VP26. J Virol. 1995;69:7362–7366. doi: 10.1128/jvi.69.11.7362-7366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trus B L, Newcomb W W, Booy F P, Brown J C, Steven A C. Distinct monoclonal antibodies separately label the hexons or the pentons of herpes simplex virus capsid. Proc Natl Acad Sci USA. 1992;89:11508–11512. doi: 10.1073/pnas.89.23.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wildy P, Russell W C, Horne R W. The morphology of herpes virus. Virology. 1960;12:204–222. doi: 10.1016/0042-6822(60)90195-1. [DOI] [PubMed] [Google Scholar]
- 19.Wingfield P T, Stahl S J, Thomsen D R, Homa F L, Booy F P, Trus B L, Steven A C. Hexon-only binding of VP26 reflects differences between the hexon and penton conformations of VP5, the major capsid protein of herpes simplex virus. J Virol. 1997;71:8955–8961. doi: 10.1128/jvi.71.12.8955-8961.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou Z H, Chen D H, Jakana J, Rixon F J, Chiu W. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J Virol. 1999;73:3210–3218. doi: 10.1128/jvi.73.4.3210-3218.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Z H, Dougherty M, Jakana J, He J, Rixon F J, Chiu W. Seeing the herpesvirus capsid at 8.5 Å. Science. 2000;288:877–880. doi: 10.1126/science.288.5467.877. [DOI] [PubMed] [Google Scholar]
- 22.Zhou Z H, He J, Jakana J, Tatman J D, Rixon F J, Chiu W. Assembly of VP26 in herpes simplex virus-1 inferred from structures of wild-type and recombinant capsids. Nat Struct Biol. 1995;2:1026–1030. doi: 10.1038/nsb1195-1026. [DOI] [PubMed] [Google Scholar]
- 23.Zhou Z H, Prasad B V V, Jakana J, Rixon F J, Chiu W. Protein subunit structures in the herpes simplex virus A-capsid determined from 400 kV spot-scan electron cryomicroscopy. J Mol Biol. 1994;242:456–469. doi: 10.1006/jmbi.1994.1594. [DOI] [PubMed] [Google Scholar]