Abstract
The synthesis of retroviral DNA is initiated near the 5′ end of the RNA. DNA synthesis is transferred from the 5′ end to the 3′ end of viral RNA in an RNase H-dependent step. In the case of human immunodeficiency virus type 1 (HIV-1) (and certain other retroviruses that have complex secondary structures at the ends of the viral RNA), there is the possibility that DNA synthesis can lead to a self-priming event that would block viral replication. The extent of RNase H cleavage must be sufficient to allow the strand transfer reaction to occur, but not so extensive that self-priming occurs. We have used a series of model RNA substrates, with and without a 5′ cap, to investigate the rules governing RNase H cleavage at the 5′ end of the HIV-1 genome. These in vitro RNase H cleavage reactions produce an RNA fragment of the size needed to block self-priming but still allow strand transfer. The cleavages seen in vitro can be understood in light of the structure of HIV-1 reverse transcriptase in a complex with an RNA/DNA substrate.
The retroviral genome is single-stranded RNA. When the virus infects a susceptible cell, the core enters the cytoplasm and genomic RNA is converted into linear double-stranded DNA by the viral enzyme reverse transcriptase (RT) (reviewed in references 3, 11, and 19). RT has two enzymatic activities: a DNA polymerase that can copy either an RNA or a DNA template, and RNase H, which will cleave RNA if, and only if, it is part of an RNA/DNA duplex. These two activities collaborate in the conversion of the RNA genome into DNA. RT, like many other DNA polymerases, requires both a template and a primer. Reverse transcription is initiated from a host tRNA base-paired to the viral genome at the primer binding site near the 5′ end of the viral genome. First-strand DNA synthesis creates an RNA/DNA duplex; this duplex is a substrate for RNase H.
Because the primer binding site is near the 5′ end of the RNA genome, DNA synthesis rapidly reaches the end of the RNA. This DNA is called minus-strand strong-stop DNA (−sssDNA). DNA synthesis is then transferred to the 3′ end of the RNA genome. This transfer reaction requires that RNase H degrade the 5′ end of the RNA genome. The 5′ ends of the genomes of several complex retroviruses (human immunodeficiency virus type 1 [HIV-1], HIV-2, and human T-cell leukemia virus type 2) can form complex secondary structures; in the absence of the complementary RNA, the newly synthesized −sssDNAs of these retroviruses can fold into complex structures. Some of these complex DNA structures have the potential to self-prime. Self-priming is prevented in vivo; the process appears to involve both the nucleocapsid (NC) protein and a piece of viral RNA from the 5′ end of the genome (5, 7, 10). The available data, most of which were obtained with in vitro reactions using model substrates, suggest that a residual piece from the 5′ end of the RNA genome prevents self-priming. To block self-priming, the residual RNA must be long enough so that when it is annealed to −sssDNA, the RNA is able to prevent self-priming; however, the residual RNA/DNA duplex must be small enough that it does not block the strand transfer reaction (5, 8).
In a previous study, HIV-1 RT was allowed to copy in vitro an RNA whose sequence matched the 5′ end of the HIV-1 genome (5). The 5′ end of the RNA was relatively resistant to RNA degradation; RNase H cleavage of the 5′ end of the RNA produced a product approximately 14 bases long. This RNA has an important role in the strand transfer reaction; in the presence of NC, the RNA remains hybridized to viral DNA and prevents self-priming. However, the residual 5′ RNA is small enough that it does not block the first-strand transfer reaction. Because the strand transfer reaction is a critical step in reverse transcription, we have used additional model substrates to investigate the RNase H cleavages of RNA/DNA duplexes that mimic the substrates created when reverse transcription reaches the 5′ end of the genome. In particular, we wanted to examine whether 5′ or 3′ extensions of the DNA strand would affect RNA cleavage and whether capping of the viral RNA would alter RNase H activity. A 5′ DNA extension has a negative effect on RNase H cleavage, and a 3′ extension has a positive effect. In some cases, a 5′ cap modestly affects the specificity of RNase H cleavage. The results can be explained in terms of the structure of HIV-1 RT in a complex with an RNA/DNA duplex (17).
RNase H cleavage of short RNAs whose sequence matches the 5′ end of the HIV-1 genome.
To measure the ability of the RNase H of HIV-1 to cleave RNA sequences from the 5′ end of the HIV-1 genome, a series of short synthetic RNAs were chemically synthesized and gel purified by Oligos, Etc., Inc. (Wilsonville, Ore.). The lyophilized RNAs were dissolved in diethylpyrocarbonate-treated water and aliquoted in Eppendorf tubes stored at −80°C. For each experiment, one tube was taken out of the freezer, thawed at room temperature, and immediately used in a labeling reaction. The RNA was 5′ labeled in vitro using T4 polynucleotide kinase from Boehringer Mannheim and r-32P from Amersham Pharmacia Biotech (Piscataway, N.J.). The labeled RNA was purified using Quick Spin Columns-G25 Sephadex (Boehringer Mannheim). All of the RNAs shared a common 5′ end; the RNAs differed in length (10, 12, or 16 bases long) (Fig. 1). To create RNase H substrates, the 5′ ends of the RNAs were radioactively labeled using [32P]ATP and T4 polynucleotide kinase. The labeled RNAs were hybridized to cDNA oligonucleotides. DNA oligonucleotides were synthesized by BioServe Biotechnologies (Laurel, Md.). The lyophilized oligonucleotides were dissolved in diethylpyrocarbonate-treated water and stored at −20°C.
FIG. 1.
Sequences of the RNA and DNA oligonucleotides. The sequence of the 5′ end of the HIV-1 genome is shown at top. The asterisk denotes a 5′ cap structure. The RNA oligonucleotides are shown beneath the HIV-1 RNA. The DNA oligonucleotides are shown at bottom. The DNA oligonucleotides are shown from 3′ to 5′ to make the comparisons with the RNA oligonucleotides easier.
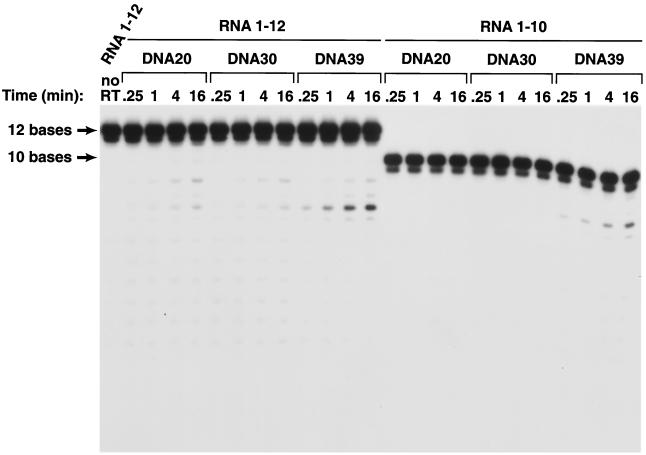
When HIV-1 RT reaches the end of the HIV-1 genome, the 5′ end of the viral RNA is base-paired with newly synthesized DNA. RNase H digestion leaves approximately 14 bases from the 5′ end of the genome and a long minus-strand DNA whose 5′ end extends well beyond the 3′ end of the residual piece of viral RNA. To ask whether a 5′ DNA extension affected RNase H cleavage, DNA oligonucleotides 20 and 30 bases long were prepared (Fig. 1) and hybridized to the 10-, 12-, and 16-base-long RNAs. The resulting RNA/DNA duplexes were used as substrates for in vitro RNase H digestion assays with purified HIV-1 RT. The open reading frame encoding wild-type HIV-1 RT was cloned into a plasmid similar to p6HRT-PROT (1, 2, 13). The plasmid is based on the expression vector pT5m, and it was introduced into the Escherichia coli strain BL21(DE3)(pLysE) (2, 13, 16, 18). After induction with isopropyl β-D-thiogalactopyranoside, the plasmid expresses both the p66 form of HIV-1 RT and HIV-1 protease. Approximately 50% of the overexpressed p66 RT is converted to the p51 form by HIV-1 protease, and p66/p51 heterodimers accumulate in E. coli. The p66/p51 heterodimers were purified by metal chelate chromatography (2, 13, 14).
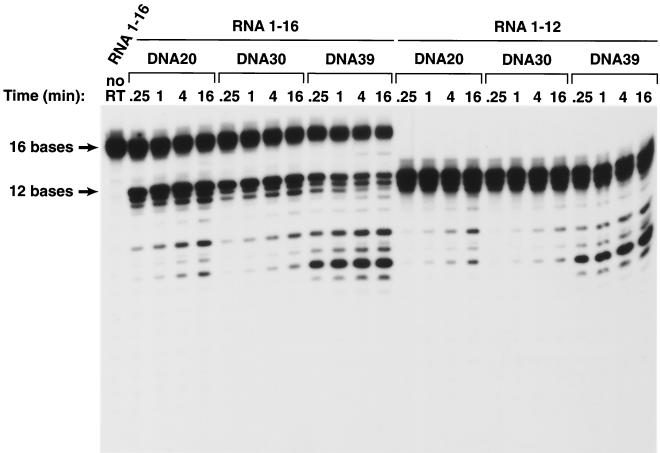
If the RNA in the resulting duplexes was 10 bases long, there was no measurable cleavage with either the DNA 20 or DNA 30 substrate (Fig. 2). The 12-base RNA was cleaved poorly when hybridized to either DNA 20 or DNA 30 (Fig. 2 and Fig. 3). Hybrids prepared with the 16-base RNA were much better RNase H substrates. Hybrids prepared with the 12-base RNA and DNA 20 or DNA 30 were preferentially cleaved at a position approximately 8 bases from their 5′ ends; the 16-base RNA was preferentially cleaved either 13 or 14 bases from its 5′ end; there were secondary cleavages approximately 8 and 10 bases from the 5′ end (Fig. 2 and Fig. 3). Making the 5′ DNA extension 10 bases longer (using DNA 30 instead of DNA 20 to make the duplex) reduced the efficiency of the cleavage of substrates prepared with either the 12-base-long RNAs or the 16-base-long RNAs. RT can bind to the RNA/DNA duplex in either of two orientations; from the enzyme's point of view, the RNA strand can be either the primer or the template strand. For RNase H digestion to occur, RT must bind the heteroduplex such that the RNA strand is the template. If there is a relatively long 5′ DNA extension, this might favor a binding mode for HIV-1 RT in which the RNA strand is the primer, not the template. We believe this explains the observation that substrates prepared with DNA 30 are cleaved less well than those prepared with DNA 20.
FIG. 2.
Cleavage of short RNA oligonucleotides hybridized to various DNA oligonucleotides. The sequences of the RNA and DNA oligonucleotides are given in Fig. 1. The RNA oligonucleotides were 5′ end labeled with T4 polynucleotide kinase. About 20,000 cpm of the 32P-labeled RNA or capped RNA templates were hybridized to approximately 20 ng of the individual oligonucleotides as described above in the presence of 50 mM Tris-HCl (pH 8.0)–50 mM NaCl–5 mM MgCl2–2.0 mM dithiothreitol–100 μg of acetylated bovine serum albumin/ml–10 mM 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate. The mixtures of RNA and oligonucleotides were heated to 70°C for 10 min and then slowly cooled to room temperature. The reactions were initiated by adding 50 ng of purified wild-type HIV-1 RT and MgCl2 to a final concentration of 5 mM, in a final volume of 12 μl, and were then incubated at 37°C. Samples were removed at 0.25, 1, 4, and 16 min, and the reactions were terminated by adding 2× RNA loading buffer. The products were heat denatured and separated on a denaturing 15% polyacrylamide–7 M urea gel in Tris-borate-EDTA buffer at 1,600 V for approximately 90 min (7). The gel was dried and autoradiographed.
FIG. 3.
Cleavage of RNA oligonucleotides hybridized to various DNA oligonucleotides by the RNase H of HIV-1 RT. The assay is similar to the assay shown in Fig. 2, except that RNA 1-16 was used. The autoradiogram was exposed for a longer time in the experiment shown here. To simplify the comparison of the data in Fig. 2 and 3, RNA 1-12 was used in the experiments presented in both figures.
Although a 5′ DNA extension appeared to decrease the efficiency of RNase H cleavage, a 3′ DNA extension substantially enhanced RNase H cleavage, particularly for substrates prepared with the shorter RNAs. The 10-base-long RNA, which was not detectably cleaved when hybridized to DNA 20 or DNA 30, could be cleaved (slowly) if hybridized to DNA 39. There was a considerable enhancement of the cleavage of the RNA if the duplexes were prepared with either the 12-base-long or the 16-base long RNA and DNA 39. The effect seemed to be primarily on the rate of cleavage rather than on the specificity of cleavage. The observation that the 9-base 3′ extension on DNA 39 affects the efficiency of cleavage without having an obvious effect on the specificity of cleavage suggests that the 3′ extension is more likely to affect the strength of the binding of the substrate to HIV-1 RT than to alter the way the substrate is bound. However, this argument does not rule out the possibility that it is the mode of binding that is affected. The capped RNA templates were labeled and synthesized in vitro using the Capped RNA synthesis kit from Ambion, Inc. (Austin, Tex.). The DNA oligonucleotides 5′GAGAGACCTCCCTATAGTGAGTCGTTTTAAATT3′, 5′AATTTAATACGACTCACTATAGGGAGGTCTCTC3′, 5′GTCTAACCAGAGAGACCTCCCTATAGTGAGTCGTATTAAATT3′, and 5′AAT T TAATACGAC TCAC TATAGGGAGG TC TC TC TGGTTAGAC3′ were synthesized and gel purified by Gibco BRL (Rockville, Md.). Each pair of the DNA oligonucleotides was first annealed by heating in 10 mM Tris-HCl (pH 8.0)–1 mM EDTA–100 mM NaCl to 100°C and then slowly cooled to room temperature. The annealed oligonucleotides were used as templates for RNA synthesis and [α-32P]UTP labeling in vitro using the capped RNA synthesis kit mMESSAGE mMACHINE from Ambion, Inc. and [α-32P]UTP from Amersham Pharmacia Biotech. The capped RNA synthesis protocol from Ambion was used to synthesize the RNA, except that a pulse-chase labeling procedure was used instead of direct pulse labeling during the capped RNA synthesis. The transcription reaction was first mixed with RNA loading buffer and heated to 100°C for 3 min, and RNAs of appropriate size were purified on a 20% polyacrylamide gel. The gel slice containing labeled full-length RNA was cut out from the gel, soaked in RNA extraction buffer with a final concentration of 100 μg of proteinase K/ml overnight at 4°C, and purified by phenol-chloroform extraction and ethanol precipitation. The RNA was resuspended in water and used in the annealing reaction.
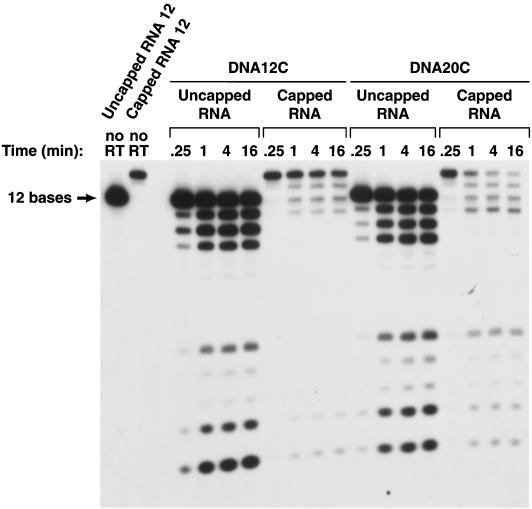
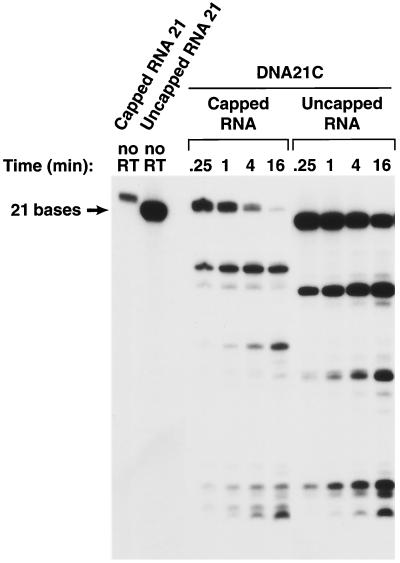
The consensus T7 promoter sequence was added to the capped model RNA, and the resulting RNA was not an exact match of the 5′ end of HIV-1 genomic RNA (Fig. 1). Two capped RNAs were made, capped RNA 12 and capped RNA 21. The corresponding uncapped RNAs were also synthesized (uncapped RNA 12 and uncapped RNA 21). cDNA oligonucleotides were synthesized (DNA 12C and DNA 21C). In addition, a DNA was prepared that was complementary to capped RNA 12 but, when hybridized, created a 3′ extension (DNA 20C) (Fig. 1). The uncapped RNA 12 differs from RNA 12 in sequence (Fig. 1) and appears to be a better RNase H substrate when hybridized to DNA 12C or DNA 20C. The susceptibilities of the corresponding capped RNA and uncapped RNAs to the RNase H of HIV-1 RT were compared. If the shorter capped RNA substrate (RNA 12C) was hybridized to the corresponding DNA (DNA 12C), the capped RNA was less susceptible to cleavage, particularly in the center of the RNA (near position 6), than the corresponding uncapped RNA. This difference in susceptibility to cleavage was not absolute; when the capped RNA 12 and uncapped RNA 12 were hybridized to DNA 20C, creating a substrate with a 3′ primer extension, both the capped and uncapped versions of RNA 12C could be cleaved near the middle of the RNA (Fig. 4). If the RNA/DNA duplexes were longer, capped and uncapped RNA 21 hybridized to DNA 21C, and there were no obvious differences in the patterns of RNase H cleavage (Fig. 5).
FIG. 4.
Effects of a 5′ cap on cleavage of an RNA oligonucleotide by the RNase H of HIV-1 RT. A 12-base-long RNA was synthesized in vitro that either had, or did not have, a 5′ cap. The RNAs were labeled by including [α32P]UTP in the synthesis mix. The full-length RNAs were purified by gel electrophoresis and hybridized to DNA oligonucleotides. The resulting heteroduplexes were incubated with HIV-1 RT for various lengths of time (0.25 to 16 min), and the reaction products were fractionated on a 15% polyacrylamide–7 M urea gel.
FIG. 5.
Effects of a 5′ cap on the cleavage of an RNA oligonucleotide by the RNase H of HIV-1 RT. The experiment shown in this figure is similar to the experiment shown in Fig. 4, except that the RNA is 21 bases long instead of 12 bases long.
Although the RNase H of HIV-1 RT does not selectively bind to specific nucleic acid sequences, it is able to cleave certain substrates with considerable precision. These precise cleavages are important; RNase H cleavages define the ends of the linear viral DNA that are the substrates for integration (see references 3, 11, and 19 for reviews). In addition, it appears that the first-strand transfer requires that the residual 5′ RNA left by RNase H be a specific size. This 5′ RNA must be large enough so that in the presence of NC, it can block self-priming of −sssDNA; however, the RNA must be small enough to allow the transfer of −sssDNA to the 3′ end of viral RNA (5, 8).
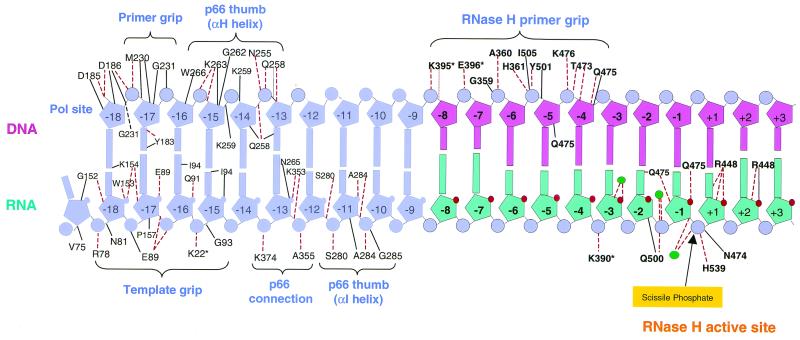
We have tried to determine how the RNase H of HIV-1 RT cleaves model substrates that are related to those generated when HIV-1 RT copies the 5′ end of the viral genome and to understand why HIV-1 RT makes the cleavages it does. Fortunately, the structure of the complex between HIV-1 RT and an RNA/DNA heteroduplex (17) is quite helpful in interpreting the cleavage data (Fig. 6).
FIG. 6.
Contacts between HIV-1 RT and an RNase H substrate. The DNA (primer) strand is shown above, and the RNA (template) strand is below. Some of the interaction elements of HIV-1 RT are indicated (RNase H primer grip, p66 thumb, p66 thumb αI, p66 thumb αH, p66 connection template grip, and primer grip). The RNase H active site and the scissile phosphate are indicated. The contacts made by individual amino acids are shown. A dotted line connecting the nucleic acid to an amino acid indicates a hydrogen bond, and a solid line indicates another type of interaction. The figure was prepared based on the structure of an RNA/DNA duplex bound to HIV-1 RT (17).
Extending the 5′ end of the primer (DNA) strand diminishes RNase H cleavage. The structure of HIV-1 RT in a complex with an RNA/DNA duplex does not show any significant interactions between RNase H and the primer strand beyond the cleavage site in the RNA. However, there might be some interactions that are not revealed in the structure; the electron density for the nucleic acid is weak past the RNase H active site. For RNase H cleavage to occur, the substrate must, in most cases, be an RNA/DNA duplex in the immediate vicinity of the RNase H active site; however, in the available structure, there are no obvious interactions between RNase H and the RNA (template strand) beyond two bases 3′ of the scissile phosphate. This interpretation of the structural data is supported by the biochemical analyses: that portion of the RNA/DNA duplex more than a few base pairs beyond (3′ on the RNA) the RNase H active site is not important for RNase H cleavage. It would appear that the primary effect of increasing the length of the 5′ primer (DNA) extension is to alter the binding mode with RT. There is no absolute requirement for RT to bind to a nucleic acid such that the RNA strand is the template and the DNA strand is the primer. If the substrate has a long 5′ DNA extension, RT can bind in the configuration in which the DNA strand is the template and the RNA strand is the primer. The competition from the second binding mode for RT would lead to a reduction in the RNase H cleavage of substrates that have long 5′ DNA extensions.
The preferred mode of binding of HIV-1 RT with nucleic acid substrates in which the double-stranded portion is at least 18 bp long places the 3′ end of the primer strand at the polymerase active site (4, 6, 9, 12, 15, 17, 20). If the template strand nucleic acid substrate has a 5′ extension, this extension can interact with the fingers subdomain of the p66 subunit. If a nucleic acid substrate has one end blunt and the other with a 5′ template extension, RT preferentially binds so that the extended template interacts with the p66 fingers, whether that template is RNA or DNA.
The isolated RNase H domain of HIV-1 RT is properly folded, but it does not cleave an RNA/DNA duplex. The problem is that the isolated RNase H domain does not bind to its substrate well enough to cleave it (reviewed in reference 11). Several elements of the polymerase domain are involved in binding an RNase H substrate. A network of amino acids in the RNase H domain and the connection subdomain called the RNase H primer grip interacts with the nucleic acid (17). The nucleic acid also interacts with the fingers, thumb, and palm subdomains. The requirement for these elements of the polymerase subdomain in the binding of an RNase H substrate determines the minimal RNA/DNA duplex that can be cleaved by the RNase H of HIV-1 RT. The cleavage data we have obtained suggest that if the 5′ end of the RNA template exactly matches the 5′ end of the HIV-1 genome, the double-stranded region must be 12 bases long (or longer) in order to see even minimal cleavage; a 10-base duplex is not cleaved, and a 16-base duplex is a much better substrate. The structure suggests that the binding site composed of RNase H itself and the RNase H primer grip contacts 11 base pairs (from the phosphate between positions −9 and −8 from the cleavage site or the primer strand to the ribose at +2 on the template strand). We did not explicitly test an 11-base substrate; however, cleavage is quite inefficient with a 12-bp blunt-ended duplex derived from the 5′ end of the HIV-1 genome. The more efficient cleavage of the uncapped RNA 12 substrates suggests that nucleic acid sequence or structure is important in determining interactions with RNase H. For substrates that exactly match the 5′ end of the HIV-1 genome, efficient RNase H cleavage requires interactions in the polymerase domain beyond the RNase H primer grip. The 16-mer substrate is cleaved efficiently 13 or 14 bases from the 5′ end of the RNA. This would place the end of the αI and αH helix of the duplex in a position where it can interact with the thumb of p66 and with the p66 connection subdomain; apparently this is sufficient to allow reasonably efficient cleavage (Fig. 6).
Although extending the primer strand 5′ (beyond RNase H) has no positive effect on RNase H cleavage, extending the 3′ end of the primer strand does enhance RNase H cleavage. This suggests that a 3′ primer extension, like a 5′ template extension, can interact with the polymerase domain. In some sense this should not be surprising. In addition to the interactions with an extended template, which have already been discussed, there are elements in the fingers, thumb, and palm of the p66 subunit that interact with both the primer and the template strand of a double-stranded nucleic acid (Fig. 6). If the duplex region of an RNA/DNA nucleic acid substrate is relatively short (between 12 and 16 bases long), the nucleic acid can bind in a configuration such that the double-stranded portion contacts the RNase H active site. This interaction is enhanced by either a single-stranded 5′ template extension or a single-stranded 3′ primer extension. Again, this makes sense in terms of the structure of HIV-1 RT. Presumably an extended single-stranded template is still able to interact with the template grip, and similarly, the extended primer can still interact with the primer grip.
Since there can be interactions with a 5′ template extension, we also examined whether a 5′ cap structure would affect the site of cleavage. In doing these experiments we focused primarily on model substrates designed to resemble the structure that arises when RT copies the 5′ end of the viral genome. The most relevant substrates were those in which the 5′ end of the RNA was fully base-paired (except for the cap). If the 5′ end of the RNA is fully base-paired and the duplex is short, only modest differences are seen in the pattern of RNase H digestion whether the duplex is 12 or 21 bp long. The differences seen with a short duplex (12 bases) suggest that the cap may interfere with RT binding rather than enhancing it. This inhibition can be relieved if there is an 8-base 3′ extension on the primer (DNA) strand. These observations can be explained in terms of the structure. When the duplex is short, adding the cap does not enhance binding because there are no interactions between the template (RNA) and the protein between positions −4 and −10 measured from the RNase H active site. There are interactions on the primer strand, through the RNase H primer grip; however, the cap could disrupt the end of the duplex and reduce these interactions. Extending the 3′ end of the primer strand beyond the RNase H primer grip to the polymerase primer grip could overcome this problem. If the duplex region is sufficiently large (here, 21 bases), there are sufficient interactions between RT and the nucleic acid that the cap doesn't have a significant effect on RNase H cleavage. Alternatively, because the cap is somewhat flexible, it could have unique interactions with RT that are difficult to predict from the available structures. Whatever model ultimately proves to be correct, the effects of a 5′ cap on RNase H cleavage are relatively modest.
Acknowledgments
We are grateful to Pat Clark and Peter Frank for preparing purified HIV-1 RT and to Hilda Marusiodis for preparing the manuscript.
S.G.S. was supported by an NIH-NIAID NRSA fellowship (A1 09578). The research in E.A.'s laboratory was supported by an NIH Merit Award (A1 27690). Research in S.H.H.'s laboratory is supported by the National Cancer Institute and the National Institute of General Medical Sciences.
REFERENCES
- 1.Boyer P L, Ferris A L, Hughes S H. Cassette mutagenesis of the reverse transcriptase of human immunodeficiency virus type 1. J Virol. 1992;66:1031–1039. doi: 10.1128/jvi.66.2.1031-1039.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyer P L, Ding J, Arnold E, Hughes S H. Subunit specificity of mutations that confer resistance to nonnucleoside inhibitors in human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother. 1994;38:1909–1914. doi: 10.1128/aac.38.9.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 4.Ding J, Das K, Hsiou Y, Sarafianos S G, Clark A D, Jr, Jacobo-Molina A, Tantillo C, Hughes S H, Arnold E. Structure and functional implications of the polymerase active site region in a complex of HIV-1 RT with a double-stranded DNA template-primer and an antibody Fab fragment at 2.8 D resolution. J Mol Biol. 1998;284:1095–1111. doi: 10.1006/jmbi.1998.2208. [DOI] [PubMed] [Google Scholar]
- 5.Driscoll M D, Golinelli M-P, Hughes S H. In vitro analysis of human immunodeficiency virus type 1 minus-strand strong-stop DNA synthesis and genomic RNA processing. J Virol. 2001;75:672–686. doi: 10.1128/JVI.75.2.672-686.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furfine E S, Reardon J E. Reverse-transcriptase-RNase H from human immunodeficiency virus: relationship of the DNA polymerase and RNA hydrolysis activities. J Biol Chem. 1991;266:406–412. [PubMed] [Google Scholar]
- 7.Gao H-Q, Boyer P L, Arnold E, Hughes S H. Effects of mutations in the polymerase domain on the polymerase, RNase H and strand transfer activities of human immunodeficiency virus type 1 reverse transcriptase. J Mol Biol. 1998;277:559–572. doi: 10.1006/jmbi.1998.1624. [DOI] [PubMed] [Google Scholar]
- 8.Golinelli M-P, Hughes S H. Self-priming of retroviral minus-strand strong-stop DNAs. Virology. 2001;285:278–290. doi: 10.1006/viro.2001.0970. [DOI] [PubMed] [Google Scholar]
- 9.Gopalakrishnan V, Peliska J A, Benkovic S J. Human immunodeficiency virus type 1 reverse transcriptase: spatial and temporal relationship between the polymerase and RNase H activities. Proc Natl Acad Sci USA. 1992;89:10763–10767. doi: 10.1073/pnas.89.22.10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo J, Henderson L E, Bess J, Kane B, Levin J G. Human immunodeficiency virus type 1 nucleocapsid protein promotes efficient strand transfer and specific viral DNA synthesis by inhibiting TAR-dependent self-priming from minus-strand strong-stop DNA. J Virol. 1997;71:5178–5188. doi: 10.1128/jvi.71.7.5178-5188.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hughes S H, Arnold E, Hostomsky Z. RNase H of retroviral reverse transcriptase. In: Toulmé J J, Crouch R J, editors. Ribonucleases H. Paris, France: INSERM Editions; 1998. pp. 195–224. [Google Scholar]
- 12.Jacobo-Molina A, Ding J, Nanni R G, Clark A D, Jr, Lu X, Tantillo C, Williams R L, Kamer G, Ferris A L, Clark P, Hizi A, Hughes S H, Arnold E. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 Å resolution shows bent DNA. Proc Natl Acad Sci USA. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le Grice S F, Grüninger-Leitch F. Rapid purification of homodimer and heterodimer HIV-1 reverse transcriptase by metal chelate affinity chromatography. Eur J Biochem. 1990;187:307–314. doi: 10.1111/j.1432-1033.1990.tb15306.x. [DOI] [PubMed] [Google Scholar]
- 14.Le Grice S F, Naas T, Wohlgensinger B, Schatz O. Subunit-selective mutagenesis indicates minimal polymerase activity in heterodimer-associated p51 HIV-1 reverse transcriptase. EMBO J. 1991;10:3905–3911. doi: 10.1002/j.1460-2075.1991.tb04960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Post K, Guo J, Kalman E, Uchida T, Crouch R J, Le Grice S F, Fay P J, Bambara R A. A large deletion in the connection subdomain of murine leukemia virus reverse transcriptase or replacement of the RNase H domain with Escherichia coli RNase H results in altered polymerase and RNase H activities. Biochemistry. 1993;32:5508–5517. doi: 10.1021/bi00072a004. [DOI] [PubMed] [Google Scholar]
- 16.Rosenberg A H, Lade B N, Chui D S, Lin S W, Dunn J J, Studier F W. Vectors for selective expression of cloned DNAs by T7 RNA polymerase. Gene. 1987;56:125–135. doi: 10.1016/0378-1119(87)90165-x. [DOI] [PubMed] [Google Scholar]
- 17.Sarafianos S G, Das K, Tantillo C, Clark A D, Jr, Ding J, Whitcomb J M, Boyer P L, Hughes S H, Arnold E E. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J. 2001;20:1449–1461. doi: 10.1093/emboj/20.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 19.Whitcomb J M, Hughes S H. Retroviral reverse transcription and integration: progress and problems. Annu Rev Cell Biol. 1992;7:275–306. doi: 10.1146/annurev.cb.08.110192.001423. [DOI] [PubMed] [Google Scholar]
- 20.Wöhrl B M, Moelling K. Interactin of HIV-1 ribonuclease H with polypurine tract containing RNA-DNA hybrids. Biochemistry. 1990;29:10141–10147. doi: 10.1021/bi00496a001. [DOI] [PubMed] [Google Scholar]