Abstract
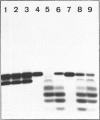
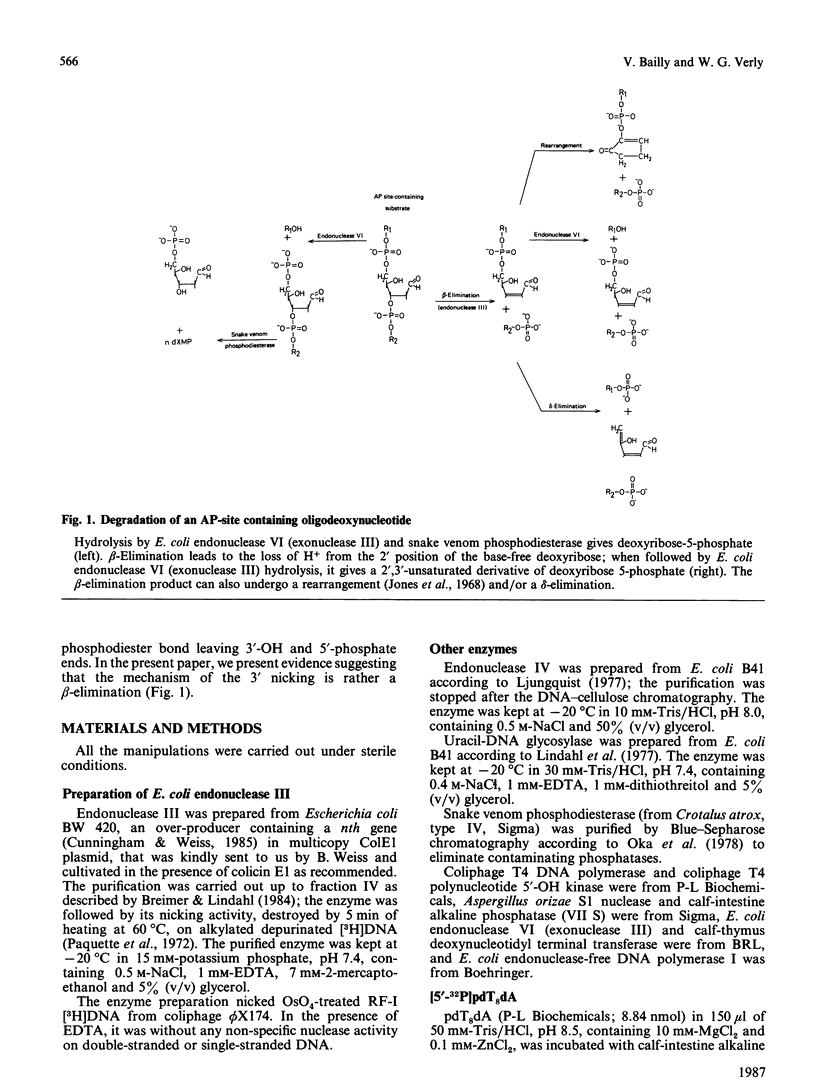
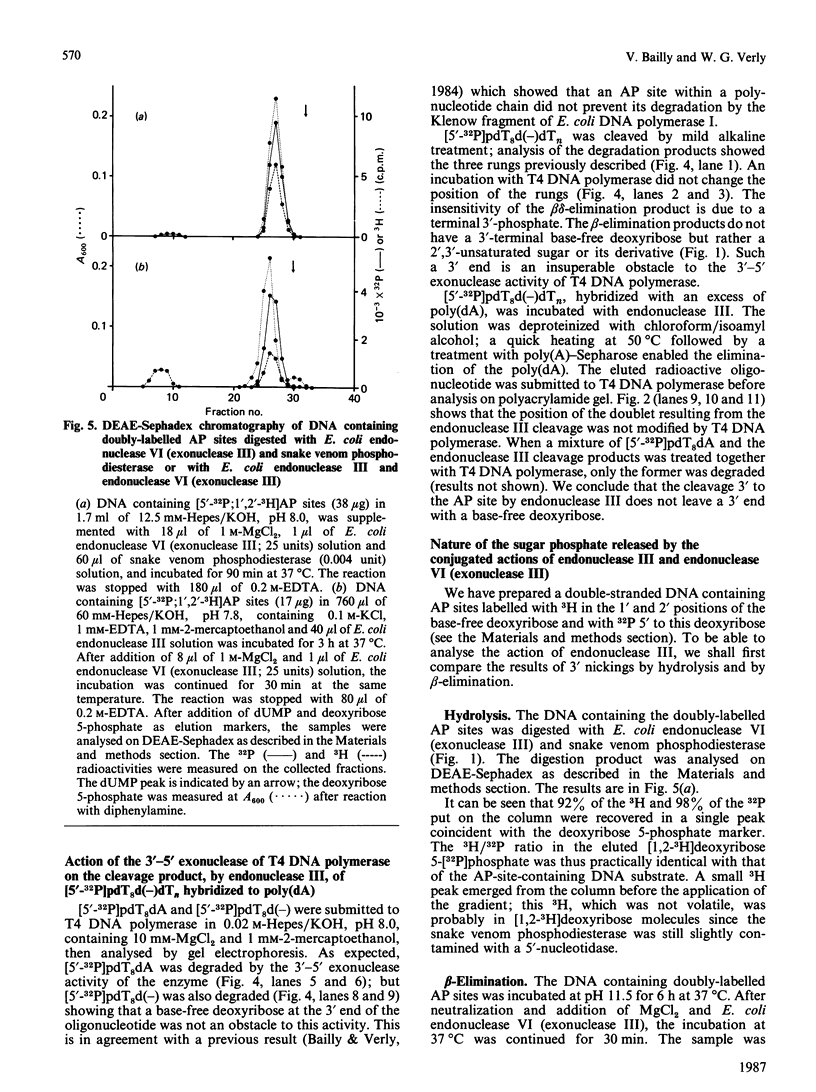
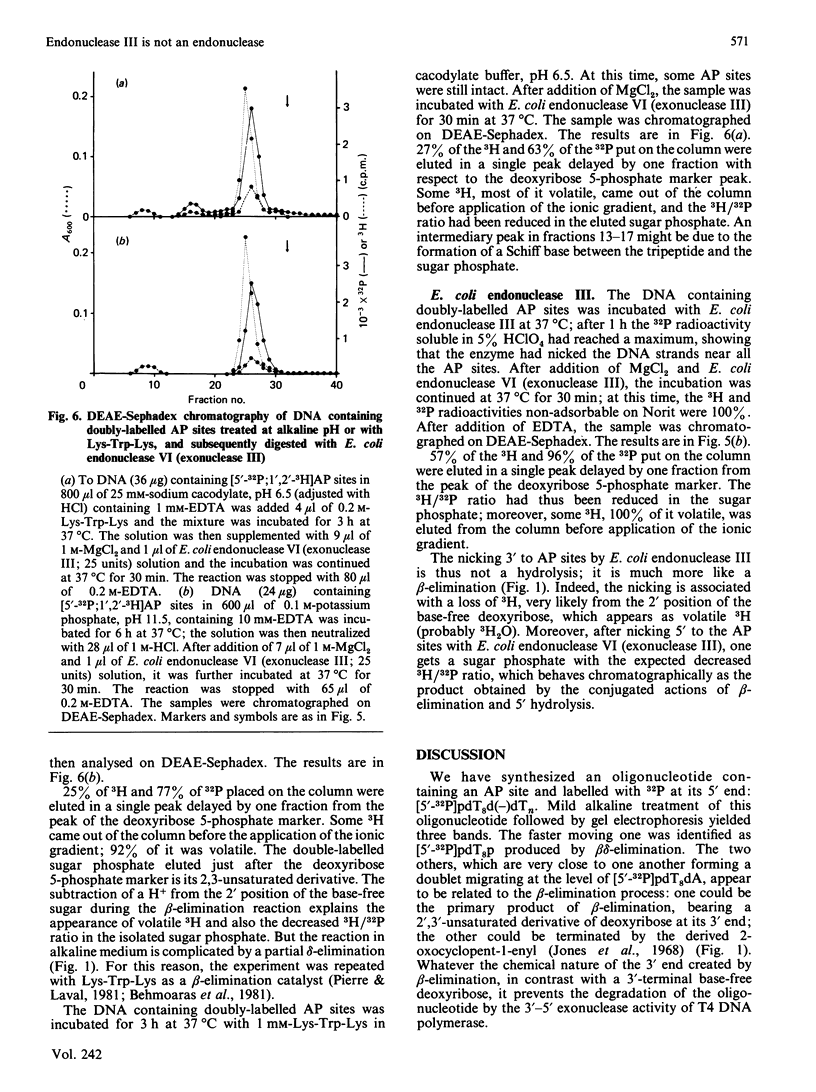
The oligonucleotide [5'-32P]pdT8d(-)dTn, containing an apurinic/apyrimidinic (AP) site [d(-)], yields three radioactive products when incubated at alkaline pH: two of them, forming a doublet approximately at the level of pdT8dA when analysed by polyacrylamide-gel electrophoresis, are the result of the beta-elimination reaction, whereas the third is pdT8p resulting from beta delta-elimination. The incubation of [5'-32P]pdT8d(-)dTn, hybridized with poly(dA), with E. coli endonuclease III yields two radioactive products which have the same electrophoretic behaviour as the doublet obtained by alkaline beta-elimination. The oligonucleotide pdT8d(-) is degraded by the 3'-5' exonuclease activity of T4 DNA polymerase as well as pdT8dA, showing that a base-free deoxyribose at the 3' end is not an obstacle for this activity. The radioactive products from [5'-32P]pdT8d(-)dTn cleaved by alkaline beta-elimination or by E. coli endonuclease III are not degraded by the 3'-5' exonuclease activity of T4 DNA polymerase. When DNA containing AP sites labelled with 32P 5' to the base-free deoxyribose labelled with 3H in the 1' and 2' positions is degraded by E. coli endonuclease VI (exonuclease III) and snake venom phosphodiesterase, the two radionuclides are found exclusively in deoxyribose 5-phosphate and the 3H/32P ratio in this sugar phosphate is the same as in the substrate DNA. When DNA containing these doubly-labelled AP sites is degraded by alkaline treatment or with Lys-Trp-Lys, followed by E. coli endonuclease VI (exonuclease III), some 3H is found in a volatile compound (probably 3H2O) whereas the 3H/32P ratio is decreased in the resulting sugar phosphate which has a chromatographic behaviour different from that of deoxyribose 5-phosphate. Treatment of the DNA containing doubly-labelled AP sites with E. coli endonuclease III, then with E. coli endonuclease VI (exonuclease III), also results in the loss of 3H and the formation of a sugar phosphate with a lower 3H/32P ratio that behaves chromatographically as the beta-elimination product digested with E. coli endonuclease VI (exonuclease III). From these data, we conclude that E. coli endonuclease III cleaves the phosphodiester bond 3' to the AP site, but that the cleavage is not a hydrolysis leaving a base-free deoxyribose at the 3' end as it has been so far assumed. The cleavage might be the result of a beta-elimination analogous to the one produced by an alkaline pH or Lys-Trp-Lys. Thus it would seem that E. coli 'endonuclease III' is, after all, not an endonuclease.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bailly V., Verly W. G. The excision of AP sites by the 3'-5' exonuclease activity of the Klenow fragment of Escherichia coli DNA polymerase I. FEBS Lett. 1984 Dec 10;178(2):223–227. doi: 10.1016/0014-5793(84)80605-5. [DOI] [PubMed] [Google Scholar]
- Behmoaras T., Toulmé J. J., Hélène C. A tryptophan-containing peptide recognizes and cleaves DNA at apurinic sites. Nature. 1981 Aug 27;292(5826):858–859. doi: 10.1038/292858a0. [DOI] [PubMed] [Google Scholar]
- Breimer L. H., Lindahl T. DNA glycosylase activities for thymine residues damaged by ring saturation, fragmentation, or ring contraction are functions of endonuclease III in Escherichia coli. J Biol Chem. 1984 May 10;259(9):5543–5548. [PubMed] [Google Scholar]
- Clements J. E., Rogers S. G., Weiss B. A DNase for apurinic/apyrimidinic sites associated with exonuclease III of Hemophilus influenzae. J Biol Chem. 1978 May 10;253(9):2990–2999. [PubMed] [Google Scholar]
- Cunningham R. P., Weiss B. Endonuclease III (nth) mutants of Escherichia coli. Proc Natl Acad Sci U S A. 1985 Jan;82(2):474–478. doi: 10.1073/pnas.82.2.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demple B., Linn S. DNA N-glycosylases and UV repair. Nature. 1980 Sep 18;287(5779):203–208. doi: 10.1038/287203a0. [DOI] [PubMed] [Google Scholar]
- Doetsch P. W., Helland D. E., Haseltine W. A. Mechanism of action of a mammalian DNA repair endonuclease. Biochemistry. 1986 Apr 22;25(8):2212–2220. doi: 10.1021/bi00356a054. [DOI] [PubMed] [Google Scholar]
- Gates F. T., Linn S. Endonuclease from Escherichia coli that acts specifically upon duplex DNA damaged by ultraviolet light, osmium tetroxide, acid, or x-rays. J Biol Chem. 1977 May 10;252(9):2802–2807. [PubMed] [Google Scholar]
- Grossman L., Grafstrom R. AP sites and AP endonucleases. Biochimie. 1982 Aug-Sep;64(8-9):577–580. doi: 10.1016/s0300-9084(82)80090-4. [DOI] [PubMed] [Google Scholar]
- Katcher H. L., Wallace S. S. Characterization of the Escherichia coli X-ray endonuclease, endonuclease III. Biochemistry. 1983 Aug 16;22(17):4071–4081. doi: 10.1021/bi00286a013. [DOI] [PubMed] [Google Scholar]
- Lindahl T., Ljungquist S., Siegert W., Nyberg B., Sperens B. DNA N-glycosidases: properties of uracil-DNA glycosidase from Escherichia coli. J Biol Chem. 1977 May 25;252(10):3286–3294. [PubMed] [Google Scholar]
- Ljungquist S. A new endonuclease from Escherichia coli acting at apurinic sites in DNA. J Biol Chem. 1977 May 10;252(9):2808–2814. [PubMed] [Google Scholar]
- Oka J., Ueda K., Hayaishi O. Snake venom phosphodiesterase: simple purification with Blue Sepharose and its application to poly(ADP-ribose) study. Biochem Biophys Res Commun. 1978 Feb 28;80(4):841–848. doi: 10.1016/0006-291x(78)91321-9. [DOI] [PubMed] [Google Scholar]
- Paquette Y., Crine P., Verly W. G. Properties of the endonuclease for depurinated DNA from Escherichia coli. Can J Biochem. 1972 Nov;50(11):1199–1209. doi: 10.1139/o72-163. [DOI] [PubMed] [Google Scholar]
- Pierre J., Laval J. Specific nicking of DNA at apurinic sites by peptides containing aromatic residues. J Biol Chem. 1981 Oct 25;256(20):10217–10220. [PubMed] [Google Scholar]
- Radman M. An endonuclease from Escherichia coli that introduces single polynucleotide chain scissions in ultraviolet-irradiated DNA. J Biol Chem. 1976 Mar 10;251(5):1438–1445. [PubMed] [Google Scholar]
- Warner H. R., Demple B. F., Deutsch W. A., Kane C. M., Linn S. Apurinic/apyrimidinic endonucleases in repair of pyrimidine dimers and other lesions in DNA. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4602–4606. doi: 10.1073/pnas.77.8.4602. [DOI] [PMC free article] [PubMed] [Google Scholar]