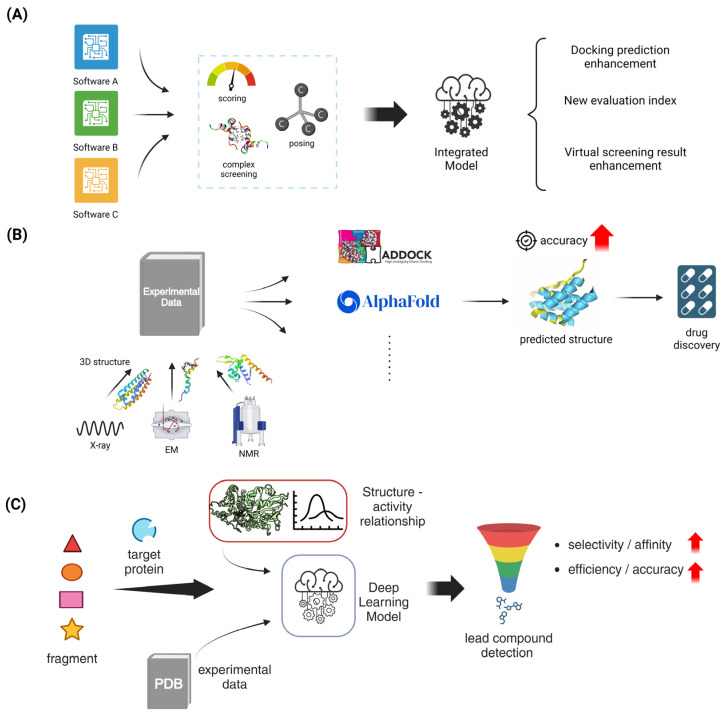
Figure 4.
This figure highlights various approaches that enhance the accuracy and reliability of drug discovery processes by integrating computational models, experimental data, and deep learning methods. It showcases how combining these elements can improve prediction performance, structural accuracy, and lead compound optimization. (A) A model integrating output data from various software improves prediction performance, generates new evaluation metrics, and provides more reliable information during the virtual screening stage. Input parameters include docking scores, molecular (or component) poses, and representations of complexes. (B) Experimental data-based libraries enable the use of various software tools. These libraries compile 3D structures obtained through methods such as X-ray crystallography, electron microscopy, and NMR spectroscopy. By leveraging actual data, software like AlphaFold and HADDOCK can achieve highly accurate structural predictions, ultimately contributing to the drug development process. (C) A deep learning model for simulating the binding of lead compound candidates to target proteins can achieve superior performance by integrating structure-activity relationship data with experimental data. Experimental data can be sourced from databases like PDB, which mainly include data obtained from X-ray crystallography, electron microscopy, and NMR spectroscopy. Ultimately, the integrated deep learning model enhances selectivity and affinity during the lead compound optimization stage, improving efficiency and accuracy at every step.