Abstract
Zeb1, a key epithelial-mesenchymal transition (EMT) regulator, has recently been found to be involved in M2 macrophage polarization in the tumor immune microenvironment, thereby promoting tumor development. However, the underlying mechanism of Zeb1-induced M2 macrophage polarization remains largely unexplored. To identify the potential role of Zeb1 in remodeling the tumor immune microenvironment in breast cancer, we crossed the floxed Zeb1 allele homozygously into PyMT mice to generate PyMT;Zeb1cKO (MMTV-Cre;PyMT;Zeb1fl/fl) mice. We found that the recruitment of M2-type tumor-associated macrophages (TAMs) was significantly reduced in tumors from PyMT;Zeb1cKO mice, and their tumor suppressive effects were weakened. Mechanistically, Zeb1 played a crucial role in transcriptionally promoting the production of Cxcl1 in tumor cells. In turn, Cxcl1 activated the Cxcr2-Jak-Stat3 pathway to induce M2 polarization of TAMs in a paracrine manner, which eventually led to T-cell inactivation and impaired the antitumor immune response in breast cancer. Our results collectively revealed an important role of Zeb1 in remodeling the tumor microenvironment, suggesting a novel therapeutic intervention for the treatment of advanced breast cancer.
Keywords: Breast cancer, Zeb1, Cxcl1, M2 macrophage polarization, antitumor immune response, tumor immune microenvironment
Introduction
The tumor microenvironment (TME) contains numerous types of cells, including immune and nonimmune cells [1,2]. Innate and adaptive immune cells recognize and destroy tumor cells; however, their functions can be modified by tumor cells to establish an immunosuppressive microenvironment that is favorable for tumor progression [3]. Infiltrating immunosuppressive cells include tumor-associated macrophages (TAMs), granulocytic myeloid-derived suppressor cells (G-MDSCs) and monocytic myeloid-derived suppressor cells (M-MDSCs) [4]. Tumorigenesis is regulated by TAMs, which are prominent immune cells in the TME [5,6]. Macrophages can be polarized toward one of two distinct phenotypes based on microenvironmental signals: the classic (M1) phenotype or the alternative (M2) phenotype [7]. TAMs, which predominantly display M2-like properties, play an important role in the crosstalk between cancer cells and the immune system [8,9]. Accordingly, TAMs can produce various factors that contribute to cancer development through sustaining proliferative signaling, evading growth suppression and immune destruction, thus enabling invasion and metastasis, inducing angiogenesis and resisting cell death [10-12]. For example, by producing immunosuppressive molecules such as IL-10 and TGF-β, TAMs suppress effective antitumor immune responses [13,14]. Moreover, TAM-derived IL-35 enhances the proliferation of breast cancer cells and thus accelerates tumorigenesis in patients [15]. Therefore, it is crucial to further elucidate the specific regulatory mechanism of TAM polarization in the immunosuppressive TME, which could be a viable therapeutic target for the treatment of advanced human cancers.
In addition, tumor cells can affect the TME through a process known as TME remodeling or reprogramming. Cancer cells have been shown to modulate the TME through multiple effects on stromal and immune cells, such as juxtacrine, paracrine and endocrine effects [16,17]. Upon juxtacrine signaling, adjacent cells directly interact with each other through ligand-receptor recognition. For example, cancer cells expressing PD-L1 can interfere directly with T cells expressing programmed death protein 1 (PD-1) [18]. Owing to the dependence of juxtacrine signals on membrane proteins and the fact that endocrine signals primarily act on distant tissues, paracrine cytokine secretion is the main mechanism through which cancer cells interact with the TME [19,20]. For instance, to escape from the immune system, breast cancer cells secrete ILT4 to induce T-cell senescence and suppress tumor immunity [21]. Moreover, chemokines such as CCL20, are secreted by cancer cells and stimulate the infiltration and polarization of macrophages [22]. Of note, a growing body of evidence has shown that members of the CXC chemokine family are also involved in the infiltration and polarization of macrophages in several types of human cancers, suggesting their potential as therapeutic targets for cancer treatment [23].
Zinc-finger E-box binding homeobox 1 (Zeb1) has been shown to influence cell fate across a wide range of tissues during development and homeostasis [24,25]. As a transcription factor, Zeb1 binds directly to the promoter region of its target genes, thus activating or suppressing gene expression [26-28]. Various types of human cancers have undergone malignant progression due to the ectopic expression of Zeb1, which is mostly found at the invasive fronts of tumors [29-36]. The presence of Zeb1 in cancer cells confers proinvasive and stem-like properties, which are associated with a relatively poor clinical prognosis [32,36-42]. Nevertheless, whether cancer cell-derived Zeb1 functions through paracrine processes to regulate components of the TME remains largely unclear.
In this study, we generated PyMT;Zeb1cKO (MMTV-Cre;PyMT;Zebfl/fl ) mice and discovered that the proportion of M2-like TAMs in the TME was dramatically decreased by Zeb1 depletion in breast cancer cells through Cxcl1-dependent paracrine action. At the molecular level, we identified Zeb1 as a key activator of Cxcl1 transcription by binding to its promoter. Moreover, the secretion of Cxcl1 from cancer cells with ectopic Zeb1 contributes to the M2-like polarization of TAMs by triggering the Cxcr2-Jak-Stat3 pathway, which eventually results in remodeling of the immunosuppressive TME of breast cancer in vitro and in vivo. Together, our data revealed the important correlation between the dysregulation of cancer cell-derived Cxcl1 and the immunosuppressive TME through a Zeb1-dependent mechanism, highlighting the possibility of identifying new targets and therapeutic strategies for cancer treatment.
Materials and methods
Generation of conditional Zeb1-knockout MMTV-PyMT mice
MMTV-PyMT mice (on the FVB/N background) and mice expressing FVB/N-MMTV-Cre under the control of a mouse mammary tumor virus (MMTV) promotor (on the FVB/N background) were purchased from Jackson Laboratory. MMTV-PyMT mice were crossed with Zeb1fl/fl mice (on the C57BL/6 background) to generate Zeb1fl/fl PyMT+/- mice (PyMT), which were then crossed with MMTV-Cre mice to generate Zeb1fl/fl PyMT+/- Cre +/- mice (PyMT;Zeb1cKO ). The mice were palpated twice a week for tumor initiation and growth. All mice were handled in accordance with protocols approved by the Animal Care and Use Committees of Nankai University.
Allograft tumor model
Cxcl1-/- mice (on the C57BL/6 background) were purchased from GemPharmatech Company. To establish the allograft tumor model, 2×106 primary tumor cells in 100 μl of PBS mixed with 100 μl of Matrigel (#354234; Corning) were subcutaneously implanted into the 4th mammary fat pads of female mice. The mice were monitored for tumor growth every other day according to the animal protocol and were sacrificed at the time of tumor removal for analysis.
Cell culture
Cells were cultured at 37℃ in 5% CO2 in a humidified incubator. MDA-MB-231 cells were cultured in RPMI-1640 containing L-glutamine (#11875093, Gibco) supplemented with 10% (vol/vol) FBS (#A5670701, Gibco), 100 IU/ml penicillin and 100 µg/ml streptomycin (#15070063, Gibco). SUM-159 cells and primary tumor cells were cultured in DMEM containing L-glutamine (#11965092, Gibco) supplemented with 10% (vol/vol) FBS, 100 IU/ml penicillin and 100 µg/ml streptomycin. THP1 cells were cultured in RPMI-1640 medium without calcium nitrate (#PWL034, Meiluncell) and supplemented with 2.05 mM L-glutamine (#A2916801, Gibco).
Peritoneal macrophage (PM) isolation
Eight-week-old male C57BL/6 mice were given 4% thioglycolate medium (#LA4184, Solarbio) by intraperitoneal (i.p.) injection. After 4 days, PMs were obtained by i.p. lavage with 8 ml of ice-cold PBS supplemented with 3% (vol/vol) FBS. The PMs were subsequently pelleted, resuspended in complete RPMI-1640 medium and plated into cell culture dishes. PMs were incubated for 4 h, after which the nonadherent cells were removed by washing twice with PBS.
Fluorescence-activated cell sorting (FACS) analysis
Tumor-infiltrating immune cells were characterized by flow cytometry. Tumors were dissected and incubated in 1 mg/ml collagenase at 37℃ for 1 h to generate a single-cell suspension. For each test, 1×106 cells were stained with fluorescently labeled antibodies for surface marker expression analysis, followed by fixation with 4% paraformaldehyde. The following antibodies were used: Alexa Fluor 647-conjugated anti-mCD206 (#141711, Biolegend), PerCP-conjugated anti-mTGF-β1 (#141409, Biolegend), APC-conjugated anti-hCD206 (#321110, Biolegend) and PE-conjugated anti-hTGF-β1 (#562490, BD Biosciences). The cells were then subjected to FACS, and the results were analyzed via FlowJo software.
Real-time quantitative PCR (qPCR) analysis
RNA was isolated using TRIzol (#19203ES60, Yeasen) according to the manufacturer’s protocol. RNA was transcribed into cDNA using Revert Aid H Minus reverse transcriptase (#RR037A, TaKaRa) according to the manufacturer’s protocol. Gene expression was analyzed using SYBR Green mix (#11184ES25, Yeasen) for real-time qPCR under the following PCR conditions: 15 s at 95℃, 10 s at 60℃, and 5 s at 72℃. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a housekeeping gene to normalize the data. The primer details can be found in Table S1.
Immunofluorescence and immunohistochemistry
For immunofluorescence, tumor samples were frozen in OCT compound and cut into 5-µm-thick sections. The tissue slices were fixed in absolute methanol for 5 min at -20℃. The antibodies used for murine tissue immunostaining were as follows: anti-F4/80 (#ab6640, Abcam) and anti-CD206 (#ab64693, Abcam). The sections were then incubated with the appropriate fluorophore-conjugated secondary antibodies (Alexa 594- or 488-conjugated, Bioss), and the nuclei were counterstained with DAPI (#C1002, Beyotime). The samples were mounted using fluorescent mounting medium (#36307ES25, Yeasen). All immunofluorescence images were acquired and analyzed using a Leica SPEII confocal laser-scanning microscope (Leica Microsystems). Image acquisition was performed with the same laser power, gain, and offset settings. Multiple independent fields (15-20 for every section; 20× or 40× magnification) of each tumor section were randomly chosen and analyzed for at least three tumors for each experimental condition. Image quantification was performed using NIH ImageJ, and the data are expressed as the fluorescence area.
For immunohistochemistry, paraffin-embedded sections were deparaffinized and treated with antigen retrieval solution. The tissue sections were incubated with endogenous peroxidase blockers and then in blocking buffer (5% goat serum in PBS). The sections were incubated overnight at 4℃ in an antibody mixture (#9028, Origene) containing primary antibodies against Zeb1 (#ab87280, Abcam), Cxcl1 (#12335-1-AP, Proteintech) and CD163 (#ab182422, Abcam), followed by incubation with biotinylated secondary antibodies (#9001, Origene). Standard DAB (#9019, Origene) was used for the detection of HRP activity. The slides were counterstained with hematoxylin, dehydrated and mounted. Microarrays of human breast cancer tissues (HBreD136Su02 and OD-CT-RpBre01-006) were obtained from Shanghai Outdo Biotech Co., Ltd. (Shanghai, China).
Cell migration assay
To analyze macrophage migration, we used a Transwell migration system. The macrophages were plated in the inner wells. Conditional medium (CM) supplemented with 10% FBS was added to the outer wells. Using ImageJ, the migrated cells were counted at 20 h by fixation with 4% formaldehyde and staining with crystal violet.
Chromatin immunoprecipitation (ChIP) and sequential ChIP assays
The ChIP assay was performed using an EZ-ChIP kit (#17-371, Millipore) according to the manufacturer’s instructions. Briefly, the cells were fixed in 37% formaldehyde for 10 min, and the reaction was quenched with 0.125 M glycine for 5 min. The cells were then washed three times with ice-cold PBS and lysed in lysis buffer supplemented with a protease inhibitor cocktail for 20 min on ice. Sonication was performed to fragment the DNA in the cell lysates into 200 to 1,000-bp fragments. After centrifugation, the supernatants were precleared with protein A/G beads at 4℃ for 1 h. As an input control, 2.5% of the sheared chromatin was utilized; the remaining chromatin was incubated overnight in the presence of antibodies against Zeb1 (#21544-1-AP, Proteintech), acetyl-histone H3 (Lys4; #ab176799, Abcam), acetyl-histone H3 (Lys14; #ab52946, Abcam), acetyl-histone H3 (Lys18; #ab177870, Abcam) and an IgG isotope control (#sc-2763, Santa Cruz) at 4℃ under rotation, followed by another incubation with protein A/G beads (#sc-2003, Santa Cruz) for 1 h at 4℃. After one wash with low-salt wash buffer, one wash with high-salt wash buffer, one wash with LiCl wash buffer and two washes with TE buffer, the bead-bound chromatin was eluted twice with elution buffer (1% SDS and 0.1 M NaHCO3). The eluted DNA-protein complexes were incubated with 0.2 M NaCl overnight at 65℃, RNase A for 30 min at 37℃ and proteinase K for 1.5 h at 45℃. The bound DNA was purified using spin columns and then subjected to PCR analysis.
For sequential ChIP assays, after sequential washes, half of the beads were eluted and saved for subsequent assays. Half of the beads were eluted with 2% DTT buffer at 37℃ for 30 min. The supernatants were incubated with an anti-CBP antibody (#7389, Cell Signaling Technology), an anti-p300 antibody (#70088, Cell Signaling Technology) and an IgG isotope control overnight at 4℃, followed by incubation with protein A/G beads for 1 h at 4℃. The chromatin bound to the beads was sequentially washed with low- and high-salt wash buffers, LiCl wash buffer, and TE buffer (twice) and eluted twice with elution buffer. The eluted DNA-protein complexes and the resulting bound DNA were treated as described above. The sequences of the primers used are listed in Table S1.
Immunoblotting and co-immunoprecipitation (Co-IP) assays
RIPA buffer supplemented with protease inhibitor cocktail (#87786, Thermo Fisher) was used to lyse the cells for immunoblot analysis. The cell lysates were subjected to immunoblotting using antibodies against Zeb1 (#21544-1-AP, Proteintech), CBP (#7389, Cell Signaling Technology), p300 (#70088, Cell Signaling Technology) and β-actin (#sc-47778, Santa Cruz), followed by incubation with appropriate HRP-conjugated secondary antibodies.
For the IP assays, cells were harvested and lysed on ice for 30 min in RIPA buffer containing a protease inhibitor cocktail. The sonicated cell lysates were clarified, incubated with antibodies against Zeb1 (#21544-1-AP, Proteintech) and then incubated with precleared protein A/G agarose beads. The immunocomplexes were subjected to immunoblotting using antibodies against CBP, p300 and Zeb1.
Dual-luciferase reporter assay
Fragments of the human Cxcl1 promoter (-1600 ~ +71) and truncated mutants were subcloned and inserted into the pGL3 basic luciferase reporter vector (#E1751, Promega) by PCR. The Cxcl1 reporter constructs and control pRL-TK Renilla luciferase constructs were co-transfected into wild-type or Zeb1-expressing MDA-MB-231 cells. Dual-luciferase reporter assays were performed according to the manufacturer’s instructions (#E1910, Promega).
Enzyme-linked immunosorbent assay (ELISA)
ELISAs were used to measure the concentrations of chemokines secreted by M1 and M2 macrophages. To measure chemokine levels, mouse serum was collected from the orbital plexus and incubated at room temperature for 30 min before being centrifuged for 15 min at 3500 RPM, and the supernatant was collected for analysis. All the samples were processed and analyzed according to the manufacturer’s instructions. The results were obtained using a microplate spectrophotometer.
Statistical analysis
All the data were presented as the mean ± standard error of the mean (SEM). One-way ANOVA with Bonferroni’s test was used for multiple-group analyses, and an unpaired Student’s t-test was used to compare two groups. The P value was calculated by the log-rank test. Differences were considered statistically significant if P < 0.05.
Results
Zeb1 functions in the remodeling of the immunosuppressive TME
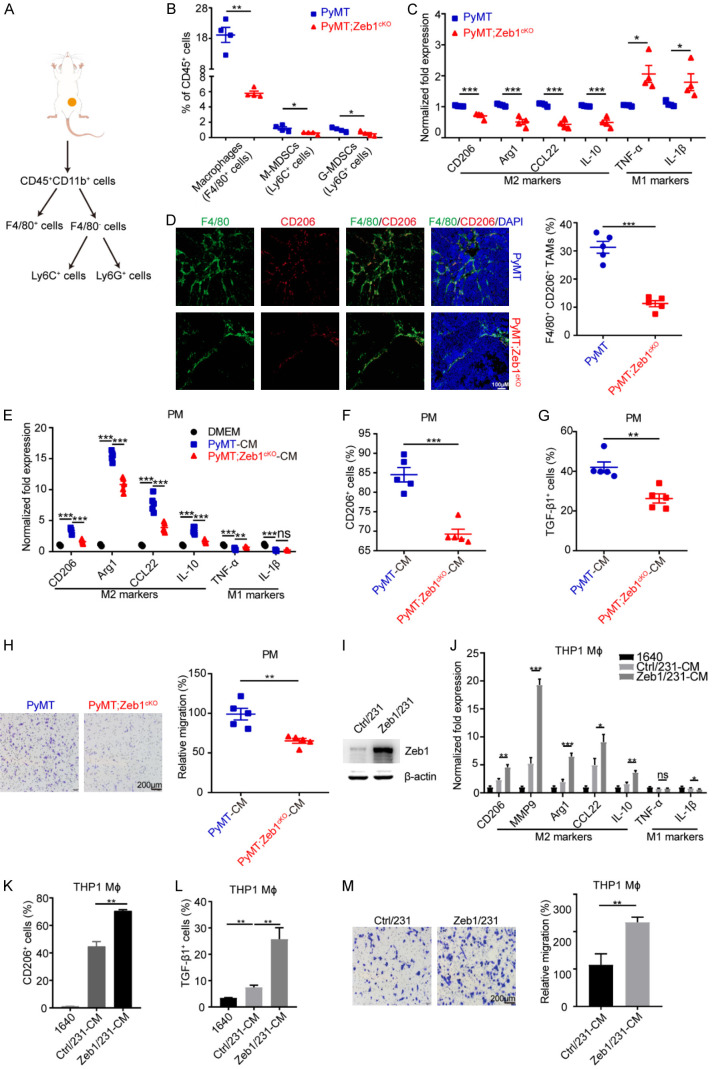
To identify the potential role of Zeb1 in fostering the tumor immune microenvironment, we crossed the floxed Zeb1 allele homozygously into PyMT mice to generate PyMT;Zeb1cKO mice [43] and examined the percentages of TAMs, M-MDSCs and G-MDSCs, which are the main tumor immunosuppressive cells in the TME, using FACS analysis (Figure 1A). The results showed that the proportion of TAMs dramatically decreased upon Zeb1 depletion, whereas those of M-MDSCs and G-MDSCs did not (Figure 1B). Moreover, we found that the expression of M2-like TAM markers, including CD206, Arg1, CCL22 and IL-10, was downregulated in F4/80+ TAMs derived from PyMT;Zeb1cKO tumors; however, the expression of M1-like TAM markers, such as TNF-α and IL-1β, was increased (Figure 1C). Accordingly, the immunofluorescence assay further revealed that tumors with Zeb1 depletion exhibited fewer F4/80+/CD206+ M2-like TAMs in the TME (Figure 1D), highlighting a potential correlation between endogenous Zeb1 dysregulation and the development of an immunosuppressive TME in breast cancer.
Figure 1.
Zeb1 regulates the M2-like polarization of TAMs. (A) Flow chart of immunosuppressive cell dissociation. (B) Flow cytometry analysis of immunosuppressive cells in breast cancer tissues (n = 4 for both the PyMT group and the PyMT;Zeb1cKO group). (C) Relative mRNA levels of M1- and M2-TAM markers in F4/80+ macrophages sorted from breast cancer tissues (n = 4 for both the PyMT group and the PyMT;Zeb1cKO group). (D) Immunofluorescence staining for F4/80 and CD206 in breast cancer tissues (n = 5 for both the PyMT group and the PyMT;Zeb1cKO group). (E) Relative mRNA levels of M1- and M2-TAM markers in peritoneal macrophages treated with CM from primary cancer cells (n = 5 for both the PyMT group and the PyMT;Zeb1cKO group). (F, G) Flow cytometry analysis of CD206+ (F) and TGF-β1+ (G) cells in peritoneal macrophages treated with CM from primary cancer cells (n = 5 for both the PyMT group and the PyMT;Zeb1cKO group). (H) Analysis of the migration of peritoneal macrophages treated with CM from primary cancer cells using the Transwell assay (n = 5 for both the PyMT group and the PyMT;Zeb1cKO group). (I) Western blotting analysis of Zeb1 expression in Zeb1-expressing MDA-MB-231 cells. (J) Relative mRNA levels of M1- and M2-TAM markers in THP1 macrophages treated with CM from Zeb1-expressing MDA-MB-231 cells. (K, L) Flow cytometry analysis of CD206+ (K) and TGF-β1+ (L) THP1 macrophages treated with CM from Zeb1-expressing MDA-MB-231 cells. (M) Transwell assay analysis of the migration of THP1 macrophages treated with CM from Zeb1-expressing MDA-MB-231 cells. The indicated P values were calculated using two-tailed unpaired Student’s t-tests. The data are presented as the mean ± SEM in (B-H). The dots represent individual samples in (B-H). The data are representative of four (B, C), five (D-H) or three (J-M) independent experiments. Source data are provided as a source data file.
Next, we extracted PMs from C57BL/6 mice, followed by treatment with CM from the primary tumor cells of PyMT;Zeb1cKO and PyMT mice. The analysis of qPCR revealed that the expression of M2-like TAM markers (CD206, Arg1, CCL22 and IL-10) was reduced in PMs treated with CM from PyMT;Zeb1cKO tumor cells, whereas the expression of M1-like TAM markers (TNF-α and IL-1β) was not markedly altered (Figure 1E). Consistently, the inhibition of M2-like phenotypes was further confirmed by FACS analysis, showing that the percentages of CD206+ (Figure 1F) and TGF-β1+ (Figure 1G) cells were reduced in PMs treated with CM from PyMT;Zeb1cKO tumor cells compared with those in the PyMT control. Transwell assays also demonstrated that cell migration was strongly impeded in PMs by treatment with CM from PyMT;Zeb1cKO tumor cells (Figure 1H).
In addition, we established gain- (Figure 1I) and loss-of-function (Figure S1A) of Zeb1 in MDA-MB-231 human breast cancer cells. The results revealed that the phenotypes of M2-like TAM polarization were markedly increased in PMA-primed THP1 cells cultured in CM from Zeb1-expressing MDA-MB-231 cells (Figure 1J-M), whereas these effects were weakened by Zeb1 depletion (Figure S1B-E). These experiments were also performed in SUM-159 cells and similar results were obtained (Figures S2 and S3). Together, our observations demonstrated that Zeb1-dependent paracrine action is involved in M2-like TAM polarization, which results in the development of an immunosuppressive TME in breast cancer.
Zeb1 transcriptionally activates Cxcl1
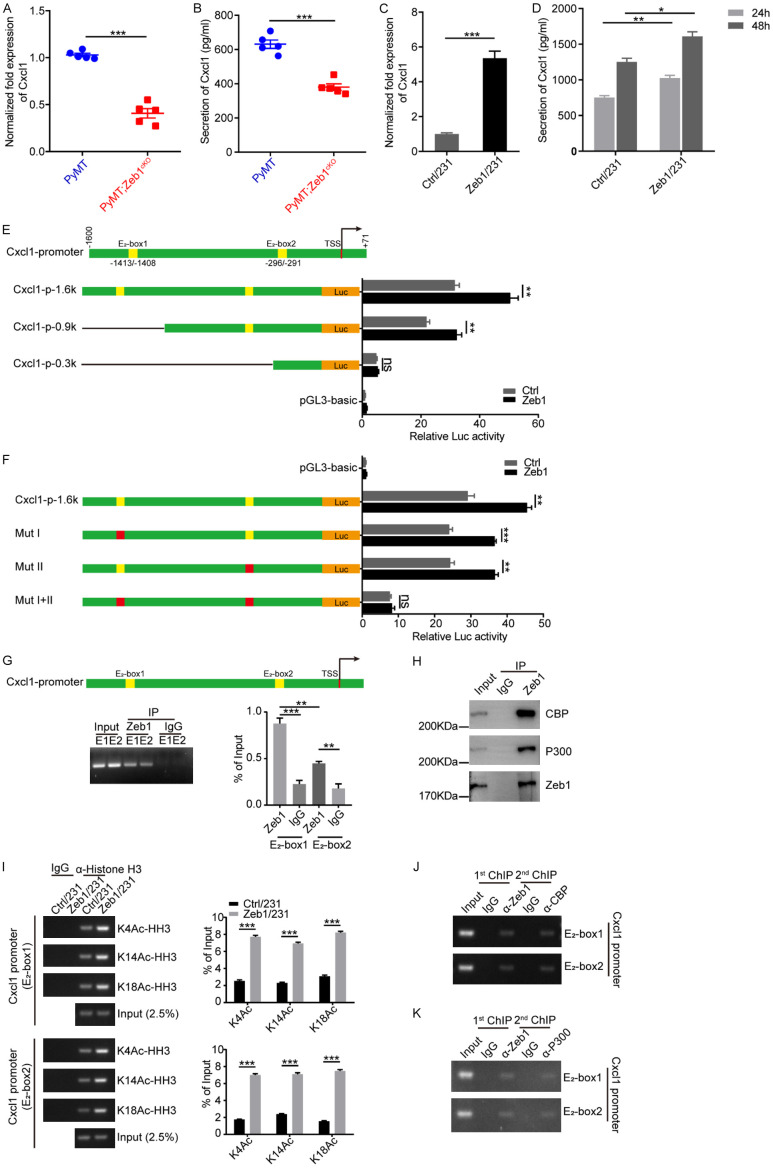
Consequently, we investigated the molecular mechanism by which Zeb1 regulated M2-like TAM polarization. For this purpose, primary tumor cells were isolated from PyMT;Zeb1cKO and PyMT mice for RNA sequencing. The results demonstrated that the expression of various paracrine factors was reduced by the loss of Zeb1 expression in PyMT;Zeb1cKO tumor cells, especially with significant alterations in the expression of CCL5, IL23a, and Cxcl1 (Figure S4A). Taken together with the RNA sequencing analysis of MDA-MB-231 cells with Zeb1 knockdown, the mRNA level of Cxcl1 was consistently decreased (Figure S4B), suggesting that Zeb1 might promote M2-like TAM polarization by modulating Cxcl1 production. Indeed, qPCR (Figure 2A) and ELISA (Figure 2B) further confirmed that Zeb1 depletion strongly decreased Cxcl1 expression at both the mRNA and protein levels in PyMT;Zeb1cKO tumor cells, which was further validated in MDA-MB-231 cells with Zeb1 knockdown (Figure S5). In contrast, we demonstrated that MDA-MB-231 cells with ectopic Zeb1 expression presented increased mRNA expression (Figure 2C) and protein secretion (Figure 2D) of Cxcl1. These experiments were also performed in SUM-159 cells and similar results were obtained (Figures S6 and S7).
Figure 2.
Zeb1 transcriptionally activates Cxcl1. (A) Relative mRNA levels of Cxcl1 in breast cancer tissues (n = 5 for both the PyMT group and the PyMT;Zeb1cKO group). (B) Analysis of the Cxcl1 concentration in CM by ELISA (n = 5 for both the PyMT group and the PyMT;Zeb1cKO group). (C) Relative mRNA levels of Cxcl1 in Zeb1-expressing MDA-MB-231 cells. (D) Analysis of the Cxcl1 concentration in CM from Zeb1-expressing MDA-MB-231 cells via ELISA. (E) Luciferase assays of the wild-type (-1600/+71) and truncated Cxcl1 promoters in Zeb1-expressing MDA-MB-231 cells. (F) Luciferase assays of the wild-type Cxcl1 promoter (-1600/+71) and E2 box element-mutated Cxcl1 promoters in Zeb1-expressing MDA-MB-231 cells. (G) ChIP assays showing the recruitment of Zeb1 to E2 box elements in the endogenous Cxcl1 promoter in MDA-MB-231 cells. (H) Co-IP analysis of the interaction between ZEB1 and P300 or CBP in MDA-MB-231 cells. (I) ChIP assays showing the recruitment of K4-, K14- and K18-acetylated α-histone H3 to E2-box elements in the endogenous Cxcl1 promoter in Zeb1-expressing MDA-MB-231 cells. (J, K) ChIP-on-ChIP analysis showing the co-occupation of E2-box elements in the endogenous Cxcl1 promoter by Zeb1 with either CBP (J) or P300 (K) in MDA-MB-231 cells. The indicated P values were calculated using two-tailed unpaired Student’s t-tests. The data are presented as the mean ± SEM in (A, B). The dots represent individual samples in (A, B). The data are representative of five (A, B) or three (C-K) independent experiments. Source data are provided as a source data file.
Next, to test whether Zeb1 transcriptionally regulates the gene expression of Cxcl1, we constructed a luciferase reporter plasmid containing an approximately -1600/+71 region of the wild-type human Cxcl1 promoter (Figure 2E), which has two canonical E2-box elements (CAGGTG) at positions -1413/-1408 and -296/-291 for the recruitment of Zeb1 [44]. A luciferase assay indicated that Zeb1 overexpression increased the activity of the wild-type Cxcl1 promoter in MDA-MB-231 cells (Figure 2E). Deletion or site-directed mutagenesis of either E2 box did not markedly affect Zeb1-induced transcriptional activation of the Cxcl1 promoter, whereas simultaneous deletion or mutation of both E2 boxes completely abolished this effect (Figure 2E and 2F). Notably, quantitative ChIP analysis demonstrated that endogenous Zeb1 was able to be recruited to both E2-box elements in the promoter of Cxcl1 (Figure 2G). Given that transcription factors might activate target genes by recruiting coactivators to promote histone acetylation at the promoters of these genes [45], we performed a Co-IP assay and found that endogenous Zeb1 was physically associated with the histone acetyltransferases CREB-binding protein (CBP) and p300 in MDA-MB-231 cells (Figure 2H). Consequently, Zeb1 overexpression led to a marked increase in histone acetylation, including histone H3 lysine 4 acetylation (H3K4Ac), H3K14Ac and H3K18Ac in the promoter of Cxcl1 in an E2 box-dependent manner (Figure 2I). ChIP-on-ChIP analysis further indicated that Zeb1, CBP and p300 co-occupied the promoter of Cxcl1 (Figure 2J and 2K), suggesting that CBP/p300 interacts with Zeb1 in the promoter of Cxcl1 and thus induces its transcriptional activation in breast cancer cells.
Zeb1-induced Cxcl1 production contributes to M2-like TAM polarization
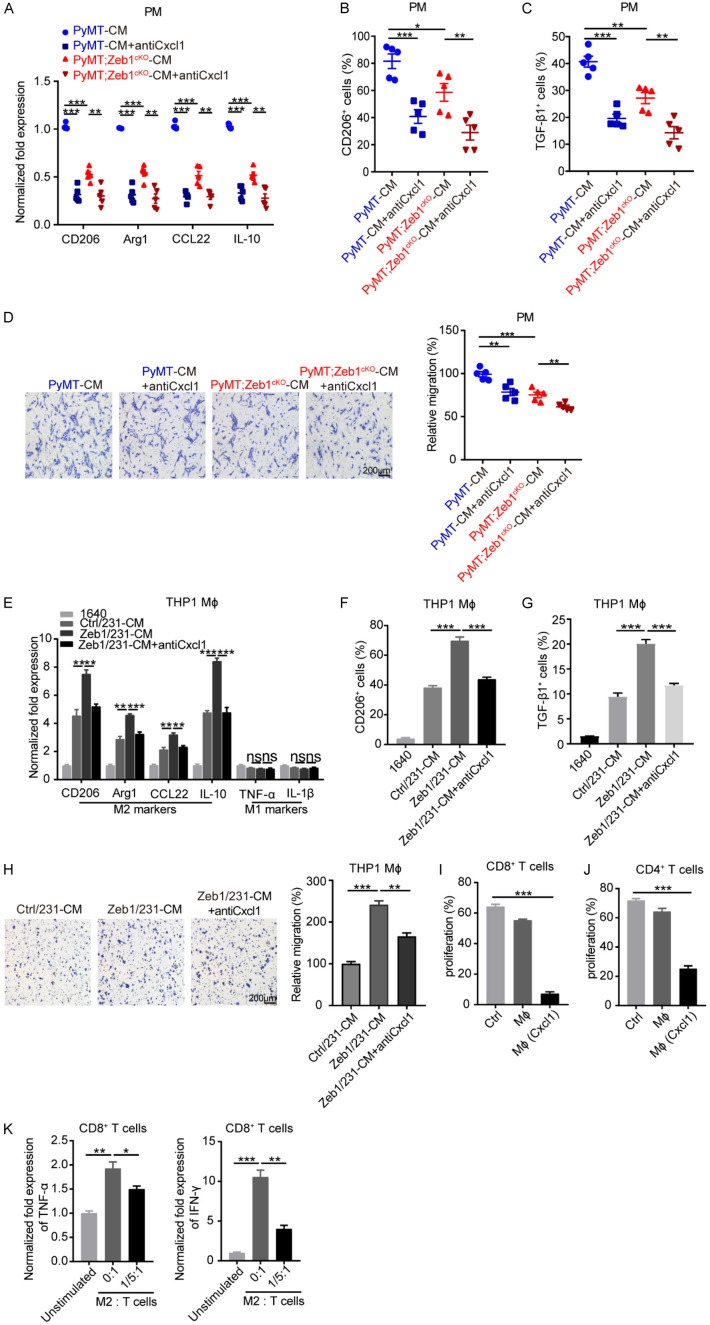
Subsequently, we sought to validate whether cancer cell-derived Cxcl1 can modulate the M2-like polarization of TAMs. Thus, PMs were treated with CM from PyMT;Zeb1cKO or PyMT tumor cells in the presence or absence of a Cxcl1 neutralizing antibody. The results of qPCR (Figure 3A), FACS (Figure 3B and 3C) and Transwell (Figure 3D) assays demonstrated that preincubation with the Cxcl1-neutralizing antibody significantly blocked M2-like TAM polarization, including M2-related marker expression and cell migration, in PMs after treatment with CM from PyMT tumor cells; however, these effects were attenuated in PMs incubated with CM from PyMT;Zeb1cKO tumor cells. We also performed these experiments in MDA-MB-231 (Figure 3E-H) and SUM-159 (Figure S8) cells overexpressing Zeb1, confirming that Cxcl1 derived from breast cancer cells with ectopic Zeb1 expression predominantly contributes to M2-like TAM polarization.
Figure 3.
Zeb1-induced Cxcl1 production contributes to M2-like TAM polarization. (A) Relative mRNA levels of M2-TAM markers in peritoneal macrophages treated with CM from primary cancer cells in the presence of a Cxcl1-neutralizing antibody (n = 5 for both the PyMT group and the PyMT;Zeb1cKO group). (B, C) Flow cytometry analysis of CD206+ (B) and TGF-β1+ (C) cells in peritoneal macrophages treated with CM from primary cancer cells in the presence of a Cxcl1-neutralizing antibody (n = 5 for both the PyMT group and the PyMT;Zeb1cKO group). (D) Transwell assay analysis of the migration of peritoneal macrophages treated with CM from primary cancer cells in the presence of a Cxcl1-neutralizing antibody (n = 5 for both the PyMT group and the PyMT;Zeb1cKO group). (E) Relative mRNA levels of M1- and M2-TAM markers in THP1 macrophages treated with CM from Zeb1-expressing MDA-MB-231 cells in the presence of a Cxcl1-neutralizing antibody. (F, G) Flow cytometry analysis of CD206+ (F) and TGF-β1+ (G) THP1 macrophages treated with CM from Zeb1-expressing MDA-MB-231 cells in the presence of a Cxcl1-neutralizing antibody. (H) Analysis of the migration of THP1 macrophages treated with CM from Zeb1-expressing MDA-MB-231 cells in the presence of a Cxcl1-neutralizing antibody via the Transwell assay. (I, J) Flow cytometry analysis of CFSE+ cells among CD8+ (I) and CD4+ (J) T cells co-cultured with peritoneal macrophages treated with rmCxcl1. (K) Relative mRNA levels of TNF-α and IFN-γ in CD8+ T cells co-cultured with peritoneal macrophages treated with rmCxcl1. The indicated P values were calculated using two-tailed unpaired Student’s t-tests. The data are presented as the mean ± SEM in (A-H). The dots represent individual samples in (A-D). The data are representative of five (A-D) or three (E-K) independent experiments. Source data are provided as a source data file.
To further examine the immunosuppressive effects of M2-like TAMs induced by Cxcl1, we stimulated CD8+ and CD4+ T cells with anti-CD3/CD28 antibodies and recombinant mouse IL2 (rmIL-2) followed by coculture with PMs in the presence or absence of recombinant mouse Cxcl1 (rmCxcl1). The results of FACS analysis showed that the proliferation of both activated CD8+ (Figure 3I) and CD4+ (Figure 3J) T cells were strongly inhibited by rmCxcl1-pretreated PMs. Similarly, the mRNA levels of cytotoxic TNF-α and IFN-γ, two cytokines with documented immunostimulatory and antitumor activity [46,47], were also decreased in activated CD8+ T cells cocultured with PMs in the presence of rmCxcl1 (Figure 3K). Together, these observations demonstrated a Zeb1-Cxcl1-dependent paracrine mechanism that is involved in the development of an immunosuppressive TME.
Tumor cell-derived Cxcl1 is predominantly responsible for M2-like TAM polarization in the TME
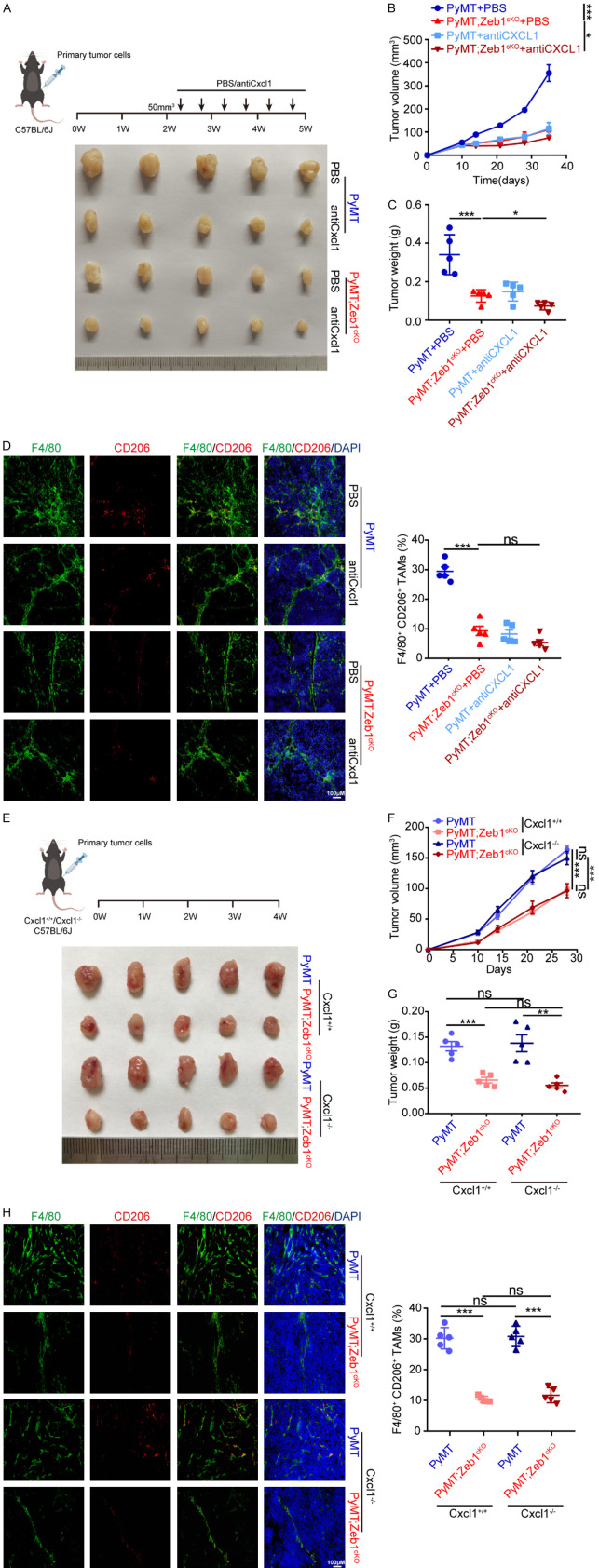
In addition, to determine the role of the Zeb1-Cxcl1 axis in tumor progression in vivo, we established allograft models in female C57BL/6 mice using breast cancer cells from PyMT and PyMT;Zeb1cKO mice, followed by treatment with a Cxcl1-neutralizing antibody (Figure 4A). Compared with that of PyMT allografts, the growth of allografts established with PyMT;Zeb1cKO tumor cells was significantly reduced. Importantly, the addition of Cxcl1-neutralizing antibody resulted in obvious tumor regression in PyMT allografts, whereas these effects were not as evident in allografts established with PyMT;Zeb1cKO tumor cells (Figure 4B, 4C). Consistently, the immunofluorescence assay further revealed a decrease in the percentage of F4/80+/CD206+ M2-like TAMs in tumors from PyMT allografts upon Cxcl1-neutralizing antibody treatment, which was strongly attenuated by Zeb1 depletion in PyMT;Zeb1cKO tumors (Figure 4D). These results indicated that breast cancer cell-derived Zeb1 promotes M2-like TAM polarization via a Cxcl1-dependent mechanism.
Figure 4.
Tumor cell-derived Cxcl1 is predominantly responsible for M2-TAM polarization in the TME. (A) Representative image of tumors dissected from a xenograft mouse model treated with a Cxcl1-neutralizing antibody. (B, C) The volume (B) and weight (C) of tumors generated by parental primary tumor cell injection into the mammary fat pads of C57BL/6 mice. (D) Immunofluorescence staining for F4/80 and CD206 in breast cancer tissues (n = 5 for the PyMT group, the PyMT+anti-Cxcl1 group, the PyMT;Zeb1cKO group, and the PyMT;Zeb1cKO+anti-Cxcl1 group). (E) Representative image of tumors dissected from xenograft mice model with or without Cxcl1-neutralizing antibody treatment. (F, G) The volume (F) and weight (G) of tumors generated by parental primary tumor cell injection into the mammary fat pads of C57BL/6 mice with or without Cxcl1-neutralizing antibody treatment. (H) Immunofluorescence staining for F4/80 and CD206 in breast cancer tissues (n = 5 for the PyMT group, the PyMT;Zeb1cKO group, the PyMT+Cxcl1-/- group, and the PyMT;Zeb1cKO+Cxcl1-/- group). The indicated P values were calculated using two-tailed unpaired Student’s t-tests. The data are presented as the means ± SEMs in (B-D) and (F-H). The dots represent individual samples in (C-D) and (G-H). The data are representative of five (A-H) independent experiments. Source data are provided as a source data file.
To further determine whether Cxcl1 from tumor cells plays a key role in Zeb1-regulated tumor growth in vivo, we constructed a breast cancer allograft model using female Cxcl1+/+ and Cxcl1-/- C57BL/6 mice with primary tumor cells from PyMT;Zeb1cKO and PyMT mice. Notably, the loss of Zeb1 markedly inhibited tumor growth in allografts established with PyMT;Zeb1cKO tumor cells compared with that established with PyMT tumor cells, regardless of Cxcl1 expression in the recipient mice (Figure 4E-G). Taken together with the immunofluorescence results showing that tumors from both Cxcl1+/+ and Cxcl1-/- mice with Zeb1 depletion exhibited decreased numbers of F4/80+/CD206+ M2-like TAMs (Figure 4H), these observations revealed that Zeb1-induced Cxcl1 production from breast cancer cells predominantly contributes to tumor development.
Cxcl1 induces M2-like TAM polarization via Jak-Stat3 signaling
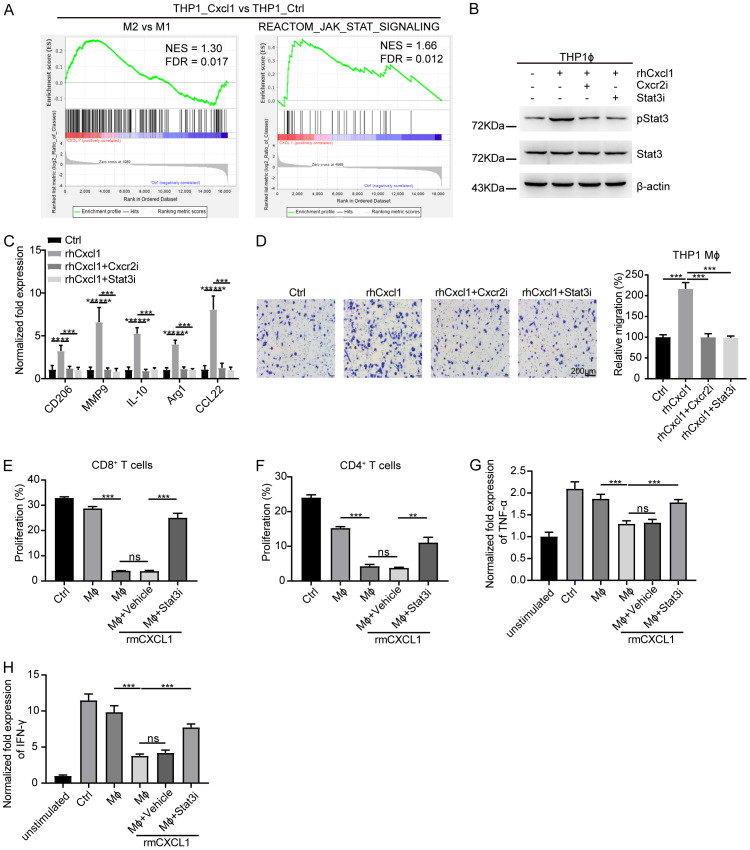
To further identify the potential mechanism involved in Cxcl1-induced M2-like TAM polarization, we isolated total RNA from PMA-primed THP1 cells by treatment with rhCxcl1 and performed RNA sequencing. The gene set enrichment analysis (GSEA) confirmed that rhCxcl1 treatment was significantly associated with the M2-like TAM signature and Jak-Stat signature (Figure 5A). Previous reports have shown that Jak2/Stat3 activation modulates the signal transduction of Cxcr2, a chemokine receptor for Cxcl1, to support M2-like TAM polarization [48]. Hence, western blotting analysis revealed that the expression of activated Stat3 increased in PMA-primed THP1 cells treated with rhCxcl1, whereas this effect was reversed by the addition of either the Cxcr2 inhibitor Elubrixin or the Stat3 inhibitor Stattic (Figure 5B). Similarly, qPCR (Figure 5C) and Transwell (Figure 5D) assays revealed that the expression of M2 markers and cell migration increased in PMA-primed THP1 cells treated with rhCxcl1; however, these effects were abolished by treatment with Elubrixin and Stattic, respectively.
Figure 5.
Cxcl1 induces M2-type TAM polarization via Jak-Stat3 signaling. (A) GSEA of transcriptome data from THP1_Cxcl1 vs. THP1 cells showing enrichment of gene signatures associated with M2-like polarization and the Jak-Stat pathway in THP1_Cxcl1 cells. (B) Western blotting analysis of Stat3 and phosphorylated Stat3 (pStat3) levels in THP1 macrophages treated with rhCxcl1 in the presence of either the Cxcr2 or Stat3 inhibitor. (C) Relative mRNA levels of M2-TAM markers in THP1 macrophages treated with rhCxcl1 in the presence of either the Cxcr2 or Stat3 inhibitor. (D) Transwell migration assay of THP1 macrophages treated with rhCxcl1 in the presence of either the Cxcr2 or Stat3 inhibitor. (E, F) Flow cytometry analysis of CFSE+ cells among CD8+ (E) and CD4+ (F) T cells cocultured with peritoneal macrophages treated with rmCxcl1 in the presence of Stat3 inhibitor. (G, H) Relative mRNA levels of TNF-α and IFN-γ in CD8+ T cells cocultured with peritoneal macrophages treated with rmCxcl1 in the presence of Stat3 inhibitor. The indicated P values were calculated using two-tailed unpaired Student’s t-tests. The data are presented as the mean ± SEM in (C-H). Source data are provided as a source data file.
Consequently, we cocultured CD8+ or CD4+ T cells with Cxcl1-pretreated PMs in the presence or absence of Stattic. FACS analysis showed that Cxcl1-pretreated PMs significantly inhibited the proliferation of CD8+ (Figure 5E) and CD4+ (Figure 5F) T cells, while these effects were blocked by Stattic. In consistent, the results of qPCR demonstrated that the expression of TNF-α (Figure 5G) and IFN-γ (Figure 5H) was markedly decreased in CD8+ T cells cocultured with PMs in the presence of Cxcl1, which was further attenuated upon Stattic treatment. Taken together, our observations revealed that Cxcl1 promotes M2-like TAM polarization by activating Cxcr2-Jak-Stat3 signaling.
The expression of Zeb1 and Cxcl1 is positively correlated with M2-like TAM polarization in breast cancer patients
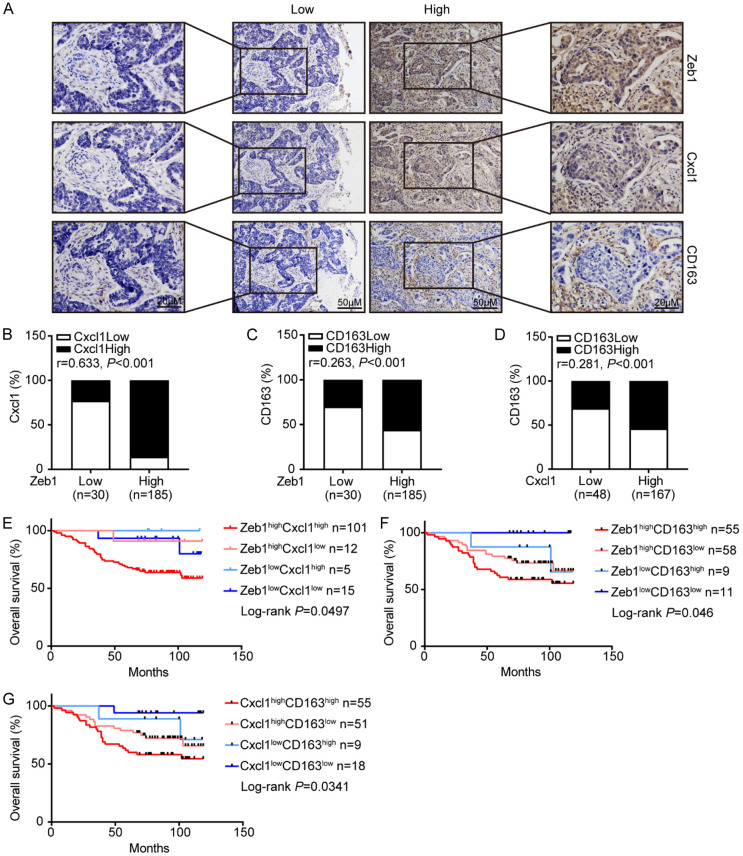
To further examine the pathological associations among Zeb1, Cxcl1 and M2-like TAM polarization, we performed immunohistochemical staining for Zeb1, Cxcl1 and CD163 (an M2-like TAM marker) in human primary breast cancer samples from 215 patients. As shown in Figure 6A, the subjects were divided into two groups based on their Zeb1 expression score. The results demonstrated strong positive correlations among the expressions of Zeb1, Cxcl1 and CD163 (Figure 6B-D). Notably, survival rate analysis indicated that cancer patients with concomitantly high expression of Zeb1, Cxcl1 and CD163 in their tumors had shorter overall survival than those with low Zeb1, Cxcl1 and CD163 expression (Figure 6E-G). Taken together, these observations revealed that aberrant function of the Zeb1-Cxcl1 axis might contribute to M2-like TAM polarization in the TME, which could be used to predict poor clinical outcomes in breast cancer patients.
Figure 6.
Zeb1 and Cxcl1 expression is positively correlated with M2-like TAM polarization in breast cancer patients. A. Immunohistochemistry staining images of Zeb1, Cxcl1 and CD163 from serial sections of the same tumor from two distinct cases. B-D. Positive associations between the expression of Zeb1, Cxcl1 and CD163 in 215 breast cancer samples. E-G. The overall survival of patients with high or low expression of Zeb1, Cxcl1 or CD163, as determined by Kaplan-Meier analysis. The indicated P values were calculated using the log-rank test. Source data are provided as a source data file.
Discussion
TAMs, which display M2-like properties, are thought to play a pivotal role in the crosstalk between cancer cells and their TME. Identifying the oncogenic signaling pathways that instruct cancer cells to educate TAMs could lead to the development of improved antineoplastic therapies. According to our findings, breast cancer cell-derived Zeb1 contributes to the M2-like polarization of TAMs through the paracrine action of Cxcl1, thereby suppressing the anticancer immune response and facilitating cancer progression. Mechanistically, the binding of Zeb1 to the Cxcl1 promoter leads to the transcriptional activation of Cxcl1 in breast cancer cells, which subsequently triggers Cxcr2-Jak-Stat3 signaling and thus induces the M2-like polarization of TAMs. Therefore, we have demonstrated an alternative mechanism of Zeb1-mediated Cxcl1 production, which is involved in reprogramming of the immunosuppressive TME to drive breast tumorigenesis.
Several mechanisms have been established to contribute to the ability of Zeb1 to promote cancer progression [32,36]. Furthermore, our results concerning the occurrence of Zeb1-dependent TME reprogramming revealed that Zeb1 has regulatory potential beyond its effect on EMT and is also linked to additional oncogenic processes, including the M2-like polarization of TAMs. At the molecular level, we found that Zeb1 promotes M2 polarization of macrophages to facilitate the immune escape of breast cancer cells by inducing the paracrine production of Cxcl1. Consistent with our hypothesis, Zeb1 is also involved in M2 macrophage polarization in ovarian cancer, which is mediated through a specific CCL18-Zeb1-M-CSF feedback loop that stimulates tumor-macrophage interplay [48]. This discrepancy may be attributable to the different approaches used for screening: in that study, a cytokine array with CM was used to screen out M-CSF, whereas in our investigation, Cxcl1 was screened by transcriptome sequencing with primary PyMT tumor cells in combination with MDA-MB-231 cells. Moreover, we previously identified that through the secretion of lactate, an aerobic glycolytic metabolite, Zeb1 confers M2 macrophage polarization in breast cancer [49]. These findings collectively shed new light on the important role of cancer cell-intrinsic Zeb1 in TME reprogramming via paracrine action. In this process, Zeb1 most likely facilitates the recruitment of immunosuppressive cells by regulating the expression of specific chemokines such as Cxcl1 in breast, colorectal and pancreatic cancers [50-53]. Indeed, we also found that knockout of Zeb1 reduced the proportion of immunosuppressive M-MDSCs and G-MDSCs in tumors from PyMT;Zeb1cKO mice.
A protumoral TME is formed when immunosuppressive cells are attracted to chemokine-releasing macrophages or cancer cells [50,52,54]. In particular, protumoral M2-like TAMs tend to accumulate in Cxcl1-rich areas of the TME, and this process contributes to tumorigenesis [54,55]. However, exactly how Cxcl1 educates the M2-like TAMs has remained largely unclear. In this study, we proposed a potential mechanism by which Zeb1-induced Cxcl1 expression potentiates the protumoral function of M2-like TAMs through binding to the Cxcr2 receptor on macrophages and subsequently triggering the Jak2-Stat3 signaling pathway. In consistent, as the cognate receptor for Cxcl chemokines, Cxcr2 also confers the M2-like TAM phenotypes in glioblastoma and colorectal cancer [47,51]. Notably, our present study identifying the pivotal role of Zeb1-Cxcl1-induced activation of Stat3 signaling in TAMs might offer additional approaches for targeting immunosuppressive TAMs in breast cancer.
Interestingly, as a product of macrophages, Cxcl1 also recruits neutrophils to establish an immunosuppressive environment and thus promotes cancer metastasis [51]. Moreover, these findings indicate that Cxcl1 secreted by macrophages might promote their own M2 polarization to form positive feedback in the TME. Indeed, our results showed that knockout of host Cxcl1 did not significantly affect tumor growth or the proportion of M2 macrophages, which does not rule out the potential role of macrophage-derived Cxcl1 in the polarization of M2-like TAMs.
There are several limitations in this study. To validate our mechanism clinically, we examined the expression of Zeb1, Cxcl1 and CD163 in human breast cancer samples via immunohistochemical staining. However, we detected the total expression of Cxcl1 in tumor tissues rather than the secreted Cxcl1 level because of technological limitations. Although we measured the secretion of Cxcl1 from PyMT;Zeb1cKO primary tumor cells and human breast cancer cell lines with aberrant Zeb1 expression, Cxcl1 production in the blood serum of breast cancer patients should be further validated.
In summary, our findings demonstrated that breast cancer cells with ectopic Zeb1 produxce elevated Cxcl1, thus conferring immunosuppressive properties to tumors in a paracrine-dependent manner. Importantly, our study identified potential therapeutic approaches that disrupt Zeb1-induced M2-like polarization of TAMs and tumorigenesis, which ultimately leads to improved cancer outcomes in patients with aggressive breast cancer.
Acknowledgements
This work was supported by grants from the International Science and Technology Cooperation Project of China (No. 2022YFE0133300) and the National Natural Science Foundation of China (No. 82172801 and No. 82472870).
The tissue samples were obtained with written informed consent from each patient.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Ring SS, Cupovic J, Onder L, Lutge M, Perez-Shibayama C, Gil-Cruz C, Scandella E, De Martin A, Morbe U, Hartmann F, Wenger R, Spiegl M, Besse A, Bonilla WV, Stemeseder F, Schmidt S, Orlinger KK, Krebs P, Ludewig B, Flatz L. Viral vector-mediated reprogramming of the fibroblastic tumor stroma sustains curative melanoma treatment. Nat Commun. 2021;12:4734. doi: 10.1038/s41467-021-25057-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou Z, Wu H, Yang R, Xu A, Zhang Q, Dong J, Qian C, Sun M. GSH depletion liposome adjuvant for augmenting the photothermal immunotherapy of breast cancer. Sci Adv. 2020;6:eabc4373. doi: 10.1126/sciadv.abc4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mao X, Tey SK, Yeung CLS, Kwong EML, Fung YME, Chung CYS, Mak LY, Wong DKH, Yuen MF, Ho JCM, Pang H, Wong MP, Leung CO, Lee TKW, Ma V, Cho WC, Cao P, Xu X, Gao Y, Yam JWP. Nidogen 1-enriched extracellular vesicles facilitate extrahepatic metastasis of liver cancer by activating pulmonary fibroblasts to secrete tumor necrosis factor receptor 1. Adv Sci (Weinh) 2020;7:2002157. doi: 10.1002/advs.202002157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Vlerken-Ysla L, Tyurina YY, Kagan VE, Gabrilovich DI. Functional states of myeloid cells in cancer. Cancer Cell. 2023;41:490–504. doi: 10.1016/j.ccell.2023.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allouch A, Voisin L, Zhang Y, Raza SQ, Lecluse Y, Calvo J, Selimoglu-Buet D, de Botton S, Louache F, Pflumio F, Solary E, Perfettini JL. CDKN1A is a target for phagocytosis-mediated cellular immunotherapy in acute leukemia. Nat Commun. 2022;13:6739. doi: 10.1038/s41467-022-34548-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moeini P, Niedzwiedzka-Rystwej P. Tumor-associated macrophages: combination of therapies, the approach to improve cancer treatment. Int J Mol Sci. 2021;22:7239. doi: 10.3390/ijms22137239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orecchioni M, Ghosheh Y, Pramod AB, Ley K. Corrigendum: macrophage polarization: different gene signatures in M1(LPS+) vs. classically and M2(LPS-) vs. alternatively activated macrophages. Front Immunol. 2020;11:234. doi: 10.3389/fimmu.2020.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao H, Guo Y, Li B, Li X, Wang Y, Han S, Cheng D, Shuai X. M2-like tumor-associated macrophage-targeted codelivery of STAT6 inhibitor and ikkbeta siRNA induces M2-to-M1 repolarization for cancer immunotherapy with low immune side effects. ACS Cent Sci. 2020;6:1208–1222. doi: 10.1021/acscentsci.9b01235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Locati M, Curtale G, Mantovani A. Diversity, mechanisms, and significance of macrophage plasticity. Annu Rev Pathol. 2020;15:123–147. doi: 10.1146/annurev-pathmechdis-012418-012718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engblom C, Pfirschke C, Pittet MJ. The role of myeloid cells in cancer therapies. Nat Rev Cancer. 2016;16:447–462. doi: 10.1038/nrc.2016.54. [DOI] [PubMed] [Google Scholar]
- 11.Bejarano L, Jordao MJC, Joyce JA. Therapeutic targeting of the tumor microenvironment. Cancer Discov. 2021;11:933–959. doi: 10.1158/2159-8290.CD-20-1808. [DOI] [PubMed] [Google Scholar]
- 12.Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21:309–322. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y, Cao X. Immunosuppressive cells in tumor immune escape and metastasis. J Mol Med (Berl) 2016;94:509–522. doi: 10.1007/s00109-015-1376-x. [DOI] [PubMed] [Google Scholar]
- 14.Jayasingam SD, Citartan M, Thang TH, Mat Zin AA, Ang KC, Ch’ng ES. Evaluating the polarization of tumor-associated macrophages into M1 and M2 phenotypes in human cancer tissue: technicalities and challenges in routine clinical practice. Front Oncol. 2020;9:1512. doi: 10.3389/fonc.2019.01512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruiz-Torres SJ, Bourn JR, Benight NM, Hunt BG, Lester C, Waltz SE. Macrophage-mediated RON signaling supports breast cancer growth and progression through modulation of IL-35. Oncogene. 2022;41:321–333. doi: 10.1038/s41388-021-02091-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horwitz SM, Koch R, Porcu P, Oki Y, Moskowitz A, Perez M, Myskowski P, Officer A, Jaffe JD, Morrow SN, Allen K, Douglas M, Stern H, Sweeney J, Kelly P, Kelly V, Aster JC, Weaver D, Foss FM, Weinstock DM. Activity of the PI3K-delta, gamma inhibitor duvelisib in a phase 1 trial and preclinical models of T-cell lymphoma. Blood. 2018;131:888–898. doi: 10.1182/blood-2017-08-802470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Weverwijk A, de Visser KE. Mechanisms driving the immunoregulatory function of cancer cells. Nat Rev Cancer. 2023;23:193–215. doi: 10.1038/s41568-022-00544-4. [DOI] [PubMed] [Google Scholar]
- 18.Casey SC, Tong L, Li Y, Do R, Walz S, Fitzgerald KN, Gouw AM, Baylot V, Gutgemann I, Eilers M, Felsher DW. MYC regulates the antitumor immune response through CD47 and PD-L1. Science. 2016;352:227–231. doi: 10.1126/science.aac9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merlino G, Miodini P, Callari M, D’Aiuto F, Cappelletti V, Daidone MG. Prognostic and functional role of subtype-specific tumor-stroma interaction in breast cancer. Mol Oncol. 2017;11:1399–1412. doi: 10.1002/1878-0261.12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Z, Wang X, Zhang T, Su L, Liu B, Zhu Z, Li C. LncRNA MALAT1 promotes gastric cancer progression via inhibiting autophagic flux and inducing fibroblast activation. Cell Death Dis. 2021;12:368. doi: 10.1038/s41419-021-03645-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao A, Liu X, Lin W, Wang J, Wang S, Si F, Huang L, Zhao Y, Sun Y, Peng G. Tumor-derived ILT4 induces T cell senescence and suppresses tumor immunity. J Immunother Cancer. 2021;9:e001536. doi: 10.1136/jitc-2020-001536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu C, Fan L, Lin Y, Shen W, Qi Y, Zhang Y, Chen Z, Wang L, Long Y, Hou T, Si J, Chen S. Fusobacterium nucleatum promotes colorectal cancer metastasis through miR-1322/CCL20 axis and M2 polarization. Gut Microbes. 2021;13:1980347. doi: 10.1080/19490976.2021.1980347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsai CF, Chen GW, Chen YC, Shen CK, Lu DY, Yang LY, Chen JH, Yeh WL. Regulatory effects of quercetin on M1/M2 macrophage polarization and oxidative/antioxidative balance. Nutrients. 2021;14:67. doi: 10.3390/nu14010067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta B, Errington AC, Jimenez-Pascual A, Eftychidis V, Brabletz S, Stemmler MP, Brabletz T, Petrik D, Siebzehnrubl FA. The transcription factor ZEB1 regulates stem cell self-renewal and cell fate in the adult hippocampus. Cell Rep. 2021;36:109588. doi: 10.1016/j.celrep.2021.109588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J, Wencker M, Marliac Q, Berton A, Hasan U, Schneider R, Laubreton D, Cherrier DE, Mathieu AL, Rey A, Jiang W, Caramel J, Genestier L, Marcais A, Marvel J, Ghavi-Helm Y, Walzer T. Zeb1 represses TCR signaling, promotes the proliferation of T cell progenitors and is essential for NK1.1(+) T cell development. Cell Mol Immunol. 2021;18:2140–2152. doi: 10.1038/s41423-020-0459-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim EJ, Kim JS, Lee S, Cheon I, Kim SR, Ko YH, Kang K, Tan X, Kurie JM, Ahn YH. ZEB1-regulated lnc-Nr2f1 promotes the migration and invasion of lung adenocarcinoma cells. Cancer Lett. 2022;533:215601. doi: 10.1016/j.canlet.2022.215601. [DOI] [PubMed] [Google Scholar]
- 27.Han X, Long Y, Duan X, Liu Z, Hu X, Zhou J, Li N, Wang Y, Qin J. ZEB1 induces ROS generation through directly promoting MCT4 transcription to facilitate breast cancer. Exp Cell Res. 2022;412:113044. doi: 10.1016/j.yexcr.2022.113044. [DOI] [PubMed] [Google Scholar]
- 28.Han X, Duan X, Liu Z, Long Y, Liu C, Zhou J, Li N, Qin J, Wang Y. ZEB1 directly inhibits GPX4 transcription contributing to ROS accumulation in breast cancer cells. Breast Cancer Res Treat. 2021;188:329–342. doi: 10.1007/s10549-021-06301-9. [DOI] [PubMed] [Google Scholar]
- 29.Chaffer CL, Marjanovic ND, Lee T, Bell G, Kleer CG, Reinhardt F, D’Alessio AC, Young RA, Weinberg RA. Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell. 2013;154:61–74. doi: 10.1016/j.cell.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krebs AM, Mitschke J, Lasierra Losada M, Schmalhofer O, Boerries M, Busch H, Boettcher M, Mougiakakos D, Reichardt W, Bronsert P, Brunton VG, Pilarsky C, Winkler TH, Brabletz S, Stemmler MP, Brabletz T. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat Cell Biol. 2017;19:518–529. doi: 10.1038/ncb3513. [DOI] [PubMed] [Google Scholar]
- 31.Viswanathan VS, Ryan MJ, Dhruv HD, Gill S, Eichhoff OM, Seashore-Ludlow B, Kaffenberger SD, Eaton JK, Shimada K, Aguirre AJ, Viswanathan SR, Chattopadhyay S, Tamayo P, Yang WS, Rees MG, Chen S, Boskovic ZV, Javaid S, Huang C, Wu X, Tseng YY, Roider EM, Gao D, Cleary JM, Wolpin BM, Mesirov JP, Haber DA, Engelman JA, Boehm JS, Kotz JD, Hon CS, Chen Y, Hahn WC, Levesque MP, Doench JG, Berens ME, Shamji AF, Clemons PA, Stockwell BR, Schreiber SL. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature. 2017;547:453–457. doi: 10.1038/nature23007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caramel J, Ligier M, Puisieux A. Pleiotropic roles for ZEB1 in cancer. Cancer Res. 2018;78:30–35. doi: 10.1158/0008-5472.CAN-17-2476. [DOI] [PubMed] [Google Scholar]
- 33.de Barrios O, Sanchez-Moral L, Cortes M, Ninfali C, Profitos-Peleja N, Martinez-Campanario MC, Siles L, Del Campo R, Fernandez-Acenero MJ, Darling DS, Castells A, Maurel J, Salas A, Dean DC, Postigo A. ZEB1 promotes inflammation and progression towards inflammation-driven carcinoma through repression of the DNA repair glycosylase MPG in epithelial cells. Gut. 2019;68:2129–2141. doi: 10.1136/gutjnl-2018-317294. [DOI] [PubMed] [Google Scholar]
- 34.Fu R, Han CF, Ni T, Di L, Liu LJ, Lv WC, Bi YR, Jiang N, He Y, Li HM, Wang S, Xie H, Chen BA, Wang XS, Weiss SJ, Lu T, Guo QL, Wu ZQ. A ZEB1/p53 signaling axis in stromal fibroblasts promotes mammary epithelial tumours. Nat Commun. 2019;10:3210. doi: 10.1038/s41467-019-11278-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kroger C, Afeyan A, Mraz J, Eaton EN, Reinhardt F, Khodor YL, Thiru P, Bierie B, Ye X, Burge CB, Weinberg RA. Acquisition of a hybrid E/M state is essential for tumorigenicity of basal breast cancer cells. Proc Natl Acad Sci U S A. 2019;116:7353–7362. doi: 10.1073/pnas.1812876116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu HT, Zhong HT, Li GW, Shen JX, Ye QQ, Zhang ML, Liu J. Oncogenic functions of the EMT-related transcription factor ZEB1 in breast cancer. J Transl Med. 2020;18:51. doi: 10.1186/s12967-020-02240-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E, Dirbas FM, Somlo G, Pera RA, Lao K, Clarke MF. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D, zur Hausen A, Brunton VG, Morton J, Sansom O, Schuler J, Stemmler MP, Herzberger C, Hopt U, Keck T, Brabletz S, Brabletz T. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 39.Chao CH, Chang CC, Wu MJ, Ko HW, Wang D, Hung MC, Yang JY, Chang CJ. MicroRNA-205 signaling regulates mammary stem cell fate and tumorigenesis. J Clin Invest. 2014;124:3093–3106. doi: 10.1172/JCI73351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morel AP, Ginestier C, Pommier RM, Cabaud O, Ruiz E, Wicinski J, Devouassoux-Shisheboran M, Combaret V, Finetti P, Chassot C, Pinatel C, Fauvet F, Saintigny P, Thomas E, Moyret-Lalle C, Lachuer J, Despras E, Jauffret JL, Bertucci F, Guitton J, Wierinckx A, Wang Q, Radosevic-Robin N, Penault-Llorca F, Cox DG, Hollande F, Ansieau S, Caramel J, Birnbaum D, Vigneron AM, Tissier A, Charafe-Jauffret E, Puisieux A. A stemness-related ZEB1-MSRB3 axis governs cellular pliancy and breast cancer genome stability. Nat Med. 2017;23:568–578. doi: 10.1038/nm.4323. [DOI] [PubMed] [Google Scholar]
- 41.Castano Z, San Juan BP, Spiegel A, Pant A, DeCristo MJ, Laszewski T, Ubellacker JM, Janssen SR, Dongre A, Reinhardt F, Henderson A, Del Rio AG, Gifford AM, Herbert ZT, Hutchinson JN, Weinberg RA, Chaffer CL, McAllister SS. IL-1beta inflammatory response driven by primary breast cancer prevents metastasis-initiating cell colonization. Nat Cell Biol. 2018;20:1084–1097. doi: 10.1038/s41556-018-0173-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang H, Zhou C, Zhang Z, Wang Q, Wei H, Shi W, Li J, Wang Z, Ou Y, Wang W, Wang H, Zhang Q, Sun W, Sun P, Yang S. Jagged1-Notch1-deployed tumor perivascular niche promotes breast cancer stem cell phenotype through Zeb1. Nat Commun. 2020;11:5129. doi: 10.1038/s41467-020-18860-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin EY, Jones JG, Li P, Zhu L, Whitney KD, Muller WJ, Pollard JW. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am J Pathol. 2003;163:2113–2126. doi: 10.1016/S0002-9440(10)63568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lacher MD, Shiina M, Chang P, Keller D, Tiirikainen MI, Korn WM. ZEB1 limits adenoviral infectability by transcriptionally repressing the coxsackie virus and adenovirus receptor. Mol Cancer. 2011;10:91. doi: 10.1186/1476-4598-10-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hsieh WC, Sutter BM, Ruess H, Barnes SD, Malladi VS, Tu BP. Glucose starvation induces a switch in the histone acetylome for activation of gluconeogenic and fat metabolism genes. Mol Cell. 2022;82:60–74. e65. doi: 10.1016/j.molcel.2021.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruegg C, Yilmaz A, Bieler G, Bamat J, Chaubert P, Lejeune FJ. Evidence for the involvement of endothelial cell integrin alphaVbeta3 in the disruption of the tumor vasculature induced by TNF and IFN-gamma. Nat Med. 1998;4:408–414. doi: 10.1038/nm0498-408. [DOI] [PubMed] [Google Scholar]
- 47.Yuan W, Zhang Q, Gu D, Lu C, Dixit D, Gimple RC, Gao Y, Gao J, Li D, Shan D, Hu L, Li L, Li Y, Ci S, You H, Yan L, Chen K, Zhao N, Xu C, Lan J, Liu D, Zhang J, Shi Z, Wu Q, Yang K, Zhao L, Qiu Z, Lv D, Gao W, Yang H, Lin F, Wang Q, Man J, Li C, Tao W, Agnihotri S, Qian X, Mack SC, Zhang N, You Y, Rich JN, Sun G, Wang X. Dual role of CXCL8 in maintaining the mesenchymal state of glioblastoma stem cells and M2-like tumor-associated macrophages. Clin Cancer Res. 2023;29:3779–3792. doi: 10.1158/1078-0432.CCR-22-3273. [DOI] [PubMed] [Google Scholar]
- 48.Long L, Hu Y, Long T, Lu X, Tuo Y, Li Y, Ke Z. Tumor-associated macrophages induced spheroid formation by CCL18-ZEB1-M-CSF feedback loop to promote transcoelomic metastasis of ovarian cancer. J Immunother Cancer. 2021;9:e003973. doi: 10.1136/jitc-2021-003973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang H, Wei H, Wang H, Wang Z, Li J, Ou Y, Xiao X, Wang W, Chang A, Sun W, Zhao L, Yang S. Zeb1-induced metabolic reprogramming of glycolysis is essential for macrophage polarization in breast cancer. Cell Death Dis. 2022;13:206. doi: 10.1038/s41419-022-04632-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu ZY, Zheng M, Li YM, Fan XY, Wang JC, Li ZC, Yang HJ, Yu JM, Cui J, Jiang JL, Tang J, Chen ZN. RIP3 promotes colitis-associated colorectal cancer by controlling tumor cell proliferation and CXCL1-induced immune suppression. Theranostics. 2019;9:3659–3673. doi: 10.7150/thno.32126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen H, Pan Y, Zhou Q, Liang C, Wong CC, Zhou Y, Huang D, Liu W, Zhai J, Gou H, Su H, Zhang X, Xu H, Wang Y, Kang W, Kei Wu WK, Yu J. METTL3 inhibits antitumor immunity by targeting m(6)A-BHLHE41-CXCL1/CXCR2 axis to promote colorectal cancer. Gastroenterology. 2022;163:891–907. doi: 10.1053/j.gastro.2022.06.024. [DOI] [PubMed] [Google Scholar]
- 52.Kemp SB, Carpenter ES, Steele NG, Donahue KL, Nwosu ZC, Pacheco A, Velez-Delgado A, Menjivar RE, Lima F, The S, Espinoza CE, Brown K, Long D, Lyssiotis CA, Rao A, Zhang Y, Pasca di Magliano M, Crawford HC. Apolipoprotein E promotes immune suppression in pancreatic cancer through NF-kappaB-mediated production of CXCL1. Cancer Res. 2021;81:4305–4318. doi: 10.1158/0008-5472.CAN-20-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hao M, Huang B, Wu R, Peng Z, Luo KQ. The interaction between macrophages and triple-negative breast cancer cells induces ROS-mediated interleukin 1alpha expression to enhance tumorigenesis and metastasis. Adv Sci (Weinh) 2023;10:e2302857. doi: 10.1002/advs.202302857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seifert L, Werba G, Tiwari S, Giao Ly NN, Alothman S, Alqunaibit D, Avanzi A, Barilla R, Daley D, Greco SH, Torres-Hernandez A, Pergamo M, Ochi A, Zambirinis CP, Pansari M, Rendon M, Tippens D, Hundeyin M, Mani VR, Hajdu C, Engle D, Miller G. The necrosome promotes pancreatic oncogenesis via CXCL1 and Mincle-induced immune suppression. Nature. 2016;532:245–249. doi: 10.1038/nature17403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu H, Jiang N, Li J, Jin Q, Jin J, Guo J, Wei X, Wang X, Yao L, Meng D, Zhi X. Tumor cell SPTBN1 inhibits M2 polarization of macrophages by suppressing CXCL1 expression. J Cell Physiol. 2024;239:97–111. doi: 10.1002/jcp.31146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.