Abstract
Although immune checkpoint blockade therapy (ICBT) has revolutionized cancer treatment with good therapeutic response in a number of human cancers, including bladder cancer, many cancers still do not respond to ICBT. Analyzing genetic signatures helps the understanding of underlying biological mechanisms. Here, based on two cohorts of bladder cancer patients receiving ICBT, we identified three novel ICBT-associated signatures in the bladder cancer microenvironment, involving genomic stability, angiogenesis and RNA regulatory, which affect PD-L1 expression and patient response to ICBT. The combinations of these signatures with TMB or PD-L1 expression improved the overall survival prediction efficiency over TMB and PD-L1 expression alone for patients receiving ICBT. Moreover, we utilized two methods to search potential drugs or small-molecules that have an impact on ICBT-associated signatures. This study provides new molecular insight into ICBT response of bladder cancer and has the potential to improve the prediction accuracy for patients to benefit from ICBT.
Keywords: Immune checkpoint blockade, PD-1/PD-L1, tumor microenvironment, molecular signatures, bladder cancer
Introduction
In the last decade, with the mechanistic understanding of immune checkpoints and success in checkpoint blockade using antibodies for the treatment of certain cancers, immunotherapy has become one of the hottest areas in cancer research, with promise of long-lasting therapeutic effect [1]. Currently, however, only a proportion of cancers have a good response to immune checkpoint blockade therapy (ICBT). Now, it has been recognized that the use of molecular signatures has been particularly important, linking diverse experimental systems that dissect the complexity of biological systems using next generation sequencing data of clinical samples in a way that was not previously feasible. The molecular heterogeneity of cancer in individuals, resulting from the acquisition of multiple genetic alterations that contribute to the development of cancer, underlies the discrepancy of immunotherapy. Indeed, this heterogeneity might contribute to the observation of tumor signature, which can often find in multiple cohorts that represent a common biological feature, such as the signature represented breast cancer recurrence in previous study [2,3]. Although the ability to interpret the meaning of the individual genes in these signatures remains a challenge, this does not diminish the power of the signature to characterize biological states [4]. At present, there are various ways that can identify tumor signatures from rich datasets, some of which may be within the modules of co-expressed genes discovery [5]. In the previous study, Wolf et al. used co-expression networks to identify and define 11 modules of genes, which are consistently co-regulated across multiple datasets, as signatures to reflect the biological process such as estrogen signaling, development/differentiation and immune signaling [6]. Clustering approaches can classify a selected set of genes into subsets each of which contains mutually related genes. For instance, Karasinska et al. have used consensus clustering to identify glycolytic and cholesterogenic genes in pancreatic ductal adenocarcinom samples [7].
Bladder urothelial carcinoma (BLCA) is the sixth most common cancer and one of the few cancers with good response to ICBT. However, only 30% advanced/metastatic bladder cancers respond to anti-PD-1/PD-L1 ICBT, and it has been suggested as a good model to study cancer immune response mechanisms in order to improve immunotherapy efficacy [1]. Due to the limited understanding of the tumor signature of BLCA in immunotherapy, improving the efficiency of immunotherapy and designing new treatment strategies for BLCA has continued to be an arduous task. In the current study, we performed weighted gene co-expression networks and consensus clustering analyses of two cohorts of BLCA patients receiving ICBT to discover the potential alterations impacting the outcome of ICBT immunotherapy of BLCA and identified three novel ICBT signatures: ICBT Genomic stability (IGS), ICBT angiogenesis signature (IAS) and ICBT RNA regulatory signature (IRS). They share some interactions and biological processes within molecules and significantly improve the treatment response predictive ability of TMB. The combination of ICBT signatures with TMB can enhance TMB’s ability to distinguish the patients responding to immunotherapy. Furthermore, they diversely affect the prognosis related to PD-L1 expression of patients with ICBT treatment, which may provide us extra potential mechanisms for understanding immunotherapy based on anti-PDL1/PDL1. Additionally, using two methods, silico screening of 2249 drug targets and 1770 compounds and connectivity map (CMap) analysis, we identified PGR, SSTR5 and BDKRB1 as potential target molecules; and PHA.00816795, fasudil and imatinib as potential drugs, which may have an impact on ICBT-associated signatures to improve the efficiency of ICBT therapy.
Material and methods
Study cohort
Three independent cohorts were collected to analyze, including two public cohorts that patients received ICBT, IMvigor210 trial and GSE176307 cohort. Patients did not receive ICBT were collected from the Cancer Genome Atlas (TCGA) BLCA cohort. IMvigor210 trial was a single-arm phase II study investigating the PD-L1 immune checkpoint inhibitors (Atezolizumab, 1200 mg three times weekly) in patients with metastatic bladder urothelial carcinoma [8,9]. All data of IMvigor210 trial were accessed through the IMvigor210CoreBiologies R package (http://research-pub.gene.com/IMvigor210CoreBiologies/). Patients in GSE176307 cohort with advanced urothelial carcinoma had received treatment with at least one dose of ICBT within the University of North Carolina hospital system between January 2014 and June 2018 [10]. All data of GSE176307 cohort had been obtained from Gene Expression Omnibus (GEO) database (GEO accession: GSE176307). In the cohorts received ICBT, both patients who had achieved complete response (CR) and partial response (PR) to ICBT were defined as responders in this study, while patients with stable disease (SD) and progressive disease (PD) were defined as non-responders [11]. The clinicopathological characteristics and genomic data of patients without ICBT in TCGA BLCA cohort were downloaded from https://xenabrowser.net/datapages/.
Identification of tumor ICBT signature
Unsigned gene co-expression networks were constructed by the WGCNA package as used in previous articles [12,13]. To ensure that the results of network construction were reliable, outlier samples were removed. Module identification was accomplished with the dynamic tree cut method by hierarchically clustering genes and highly similar modules were identified by clustering and then merged together with a height cut-off of 0.25. To test the stability of each identified module, module preservation and quality statistics were computed with the modulePreservation function (nPermutations=200) implemented in the WGCNA package by using GSE176307 cohort as test dataset. The correlation between modules and clinical features was evaluated by spearman’s correlation tests to search biologically meaningful modules. The unsupervised clustering was executed by using the ConsensuClusterPlus R package and repeated 1,000 times to ensure the classification stability [14].
Functional enrichment analysis and construction of PPI network
Genes within the ICBT signatures were subjected to over-representation analyses with Gene Ontology (GO) by enrichGO function in clusterProfiler package and the simplify function was performed to reduce redundancy of enriched GO terms [15]. Based on the online database resource STRING (Search Tool for the Retrieval of Interacting Genes/Proteins, http://string-db.org) [16], the protein-protein interaction (PPI) networks required experimental evidence of genes within the ICBT signatures were provided with confidence score cutoff of 900, PPI network was constructed and explored using various interactive visualization methods offered within Network Analyst (https://www.networkanalyst.ca/) [17]. Oncoplot function be performed to visualize the landscape of mutated genes for each ICBT signature by maftools package.
Quality control and processing of single-cell sequencing
CellRanger (version 3.0.0) was used to process the raw data from BioProject PRJNA662018 in GEO database [18], including demultiplex cellular barcodes, map reads to the transcriptome, and down-sample reads. The R package Seurat (version 4.1.1) was used to create “Seurat object” by CreateSeuratObject function for further processing [19], cells with UMI numbers <1,000 or with over 10% mitochondrial-derived UMI counts were considered low-quality cells and were removed. Single cells with over 6,000 genes detected were also filtered out to eliminate potential doublets. Finally, 55,922 single cells remained, and they were applied in downstream analyses. SingleR is an R package for automatic cell-type annotation of single-cell RNA-seq sequencing data and 55,922 cells in present study were annotated based on cell samples with known type labels as reference data sets by SingleR [20]. CellPhoneDB (V3) was used to execute the analysis of cell-cell communication at the molecular level [21].
Cancer cell line cell growth/survival dependent gene analysis
Expression profile data of human cancer cell lines (CCLs) were sourced from the Broad Institute’s Cancer Cell Line Encyclopedia (CCLE) project (https://portals.broadinstitute.org/ccle/) [22]. The CERES score was used to measure the dependency of a particular gene in a specific CCL, where a lower score indicates a higher probability that the gene is crucial for the growth and survival of the given CCL. CERES scores from genome-wide CRISPR knockout screens for 18,333 genes across 739 cell lines were obtained from the dependency map (DepMap) portal (https://depmap.org/portal/).
Connectivity Map (CMap) analysis
The Connectivity Map (CMap) is a database containing genome-wide transcriptional expression profiles of bioactive small molecules derived from cultured human cell lines, along with pattern-matching algorithms. This combination facilitates the discovery of drug-disease relationships and the elucidation of drug mechanisms of action [23,24]. Genes comprising three novel ICBT signatures were gathered and used for CMap analysis. The “CMAP_gene_signatures.RData” file, containing signatures related to 1,288 compounds, was downloaded from the database (https://www.pmgenomics.ca/bhklab/sites/default/files/downloads) website and utilized to calculate the matching score. The analysis process adhered to the methodology described in previous publications [25,26].
Statistics
All statistical analyses were conducted in R (version 4.1.3). Fisher’s exact test for categorical data and Kruskal-Wallis test or Student’s t test for continuous data when appropriate was used to depict the discrepancy of subgroups. Kaplan-Meier method was used to assess overall survival (OS), whereas log-rank test and Cox proportional hazards regression were applied for the assessment of the prognostic and risk significance. Spearman correlation analysis was performed to investigate the bivariate correlation, Fisher’s r-to-z transformation was used to calculate a value of z that was applied to assess the significance of the difference between two correlation coefficients. Multiple logistic regression analysis was used to establish the ICBT signature and TMB panel. Receiver operating characteristic (ROC) curves were constructed, and area under the ROC curve (AUC) was used to evaluate the diagnostic performance of the selected panel. Statistical significance was defined as P<0.05 unless specified otherwise.
Results
Expression patterns of gene modules in bladder cancer
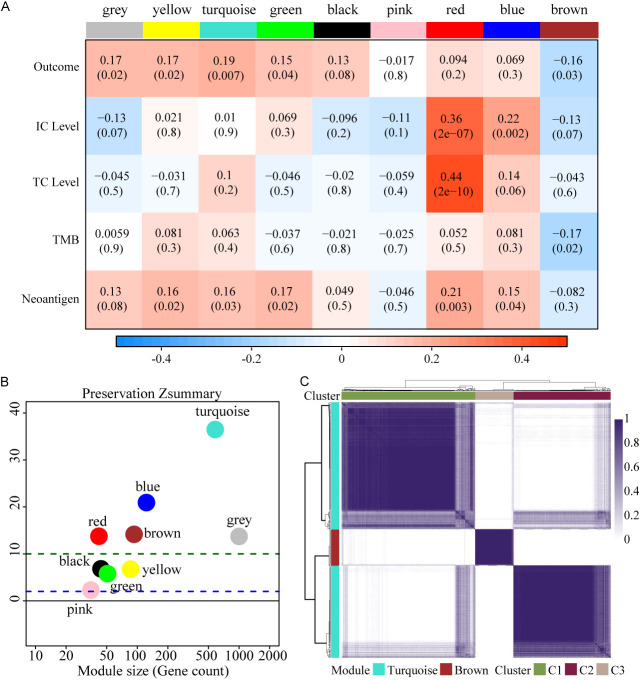
Eight gene modules were identified through weighted correlation network analysis (WGCNA) in IMvigor210 cohort (Figure 1A) and further validated using the GSE176307 dataset. Comparing the gene modules discovered in IMvigor210 cohort with the validation dataset GSE176307 cohort, turquoise, brown, blue and red modules were found to be most stable, whereas the rest modules were not very stable with their Z-summary statistics below 10 (Figure 1B) [27]. Turquoise and brown modules were significantly positive and negative, respectively, correlated with the outcome of immunotherapy. By consensus clustering, turquoise and brown modules show three different expression patterns. The gene set from turquoise module that appears in the C1 subtype was defined as turquoise-A signature, those in C3 subtype were turquoise-B signature. Brown module in C2 subtype was defined as brown signature (Figure 1C). We considered these three signatures as ICBT signatures.
Figure 1.
Expression patterns of gene modules in bladder cancer. A. 8 co-expressed modules were identified. While the “grey” module was reserved for genes identified as not co-expressed. Each row to a feature related to immunotherapy. Each cell contains the correlation and the P-value. The table is color-coded by correlation according to the color legend. B. The Z-summary statistics of the module preservation, the dashed blue and green lines indicate the thresholds Z=2 and Z=10, respectively. These horizontal lines indicate the Z-summary thresholds for strong evidence of conservation (above 10) and for low to moderate evidence of conservation (above 2). C. Consensus clustering solution (k=3) for turquoise and brown module genes in IMvigor210 cohort (n=192).
Tumor stratification by ICBT signatures reveals different relationship between the signatures and PD-L1 expression
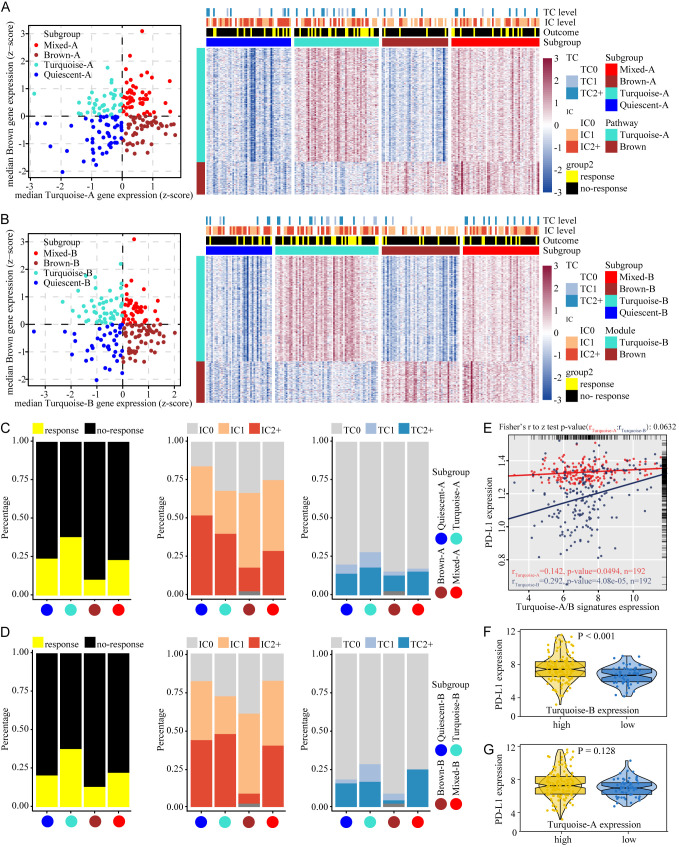
To identify whether the two signatures from turquoise module are discrepant in the stratification of tumors in combination with brown signature, we calculated median expression levels of turquoise-signatures (A or B, respectively) and brown signature genes for each sample, which were used in assigning the samples into one of four groups based on the expression of brown and one of the turquoise signatures (A or B): ‘Quiescent’ (with both signatures at low expression levels), ‘Turquoise’ (with high expression level of a turquoise signature and low expression level of brown signature), ‘Brown’ (with low expression level of a turquoise signature and high expression level of brown signature) and ‘Mixed’ (with both signatures highly expressed). When turquoise-A and brown signature were used in combination, expression levels of turquoise-A and brown signature genes across the subgroups are consistent with expectation of stratification (Figure 2A). Turquoise-B and brown signature were utilized with the same method to complete the stratification of the tumor (Figure 2B).
Figure 2.
Tumor stratification reveals the relationship between ICBT signature and PD-L1 expression. (A) Stratification of tumors based on expression of turquoise-A and brown signature genes. (B) Stratification of tumors based on turquoise-B and brown signature genes. Scatter plot showing median expression levels of turquoise (A or B) and brown signature genes in each tumor sample. Corresponding subgroups were assigned on the basis of the relative expression levels of signature genes. Heatmap depict expression levels of signature genes in each subgroup. (C) The proportion of patients responding to ICBT and the proportion of different IC and TC levels when used turquoise-A and brown signature genes to identify subgroup. (D) The proportion of features when used turquoise-B and brown signature genes to identify subgroup. (E) Correlations between signatures and PD-L1 expression. Red represent turquoise-A signature, blue for turquoise-B signature. (F) PD-L1 expression in high or low expression of turquoise-B signature. (G) PD-L1 expression in high or low expression of turquoise-A signature. ICBT, immune checkpoint blockade therapy; PD-L1, programmed cell death ligand-1; IC, immune cells; TC, tumor cells.
In the two subgroups established by turquoise-A/B and brown signature, the proportion of patients responding to ICBT was consistent, ‘Turquoise’ group was the highest, ‘Brown’ group was the lowest, and ‘Quiescent’ and ‘Mixed’ were moderate (Figure 2C and 2D left). The alterations of the proportion of IC (PD-L1 expression on immune cells) and TC (PD-L1 expression on tumor cells) levels was more significant between ‘Turquoise’ and ‘Brown’ group (IC level: Figure 2C and 2D midst; TC level: Figure 2C and 2D right), we speculated that this change was determinate by the correlation turquoise-A/B and PDL1 expression. To verify our speculation, we performed correlation tests. Turquoise-B signature showed a higher correlation with PD-L1 expression (rturquoise-B=0.292, P=4.08e-05) compared to Turquoise-A signature (rturquoise-A=0.142, P=0.0494), the discrepancy of correlations was statistically significant (Test rturquoise-B and rturquoise-A, P=0.0632) (Figure 2E). PD-L1 expression was higher in the subgroup with high turquoise-B signature expression than in low turquoise-B signature expression subgroup (Figure 2F, P<0.001). However, there was no difference of PD-L1 expression between the high/low expression subgroups of turquoise-A signature (Figure 2G, P=0.128). Above results indicate that although turquoise-A and turquoise-B signature were derived from the same gene module, only one signature, turquoise-B signature, affects PD-L1 expression, suggesting the intrinsic functional differences between the two signatures from the same gene module.
Functional and molecular characteristics of ICBT signatures
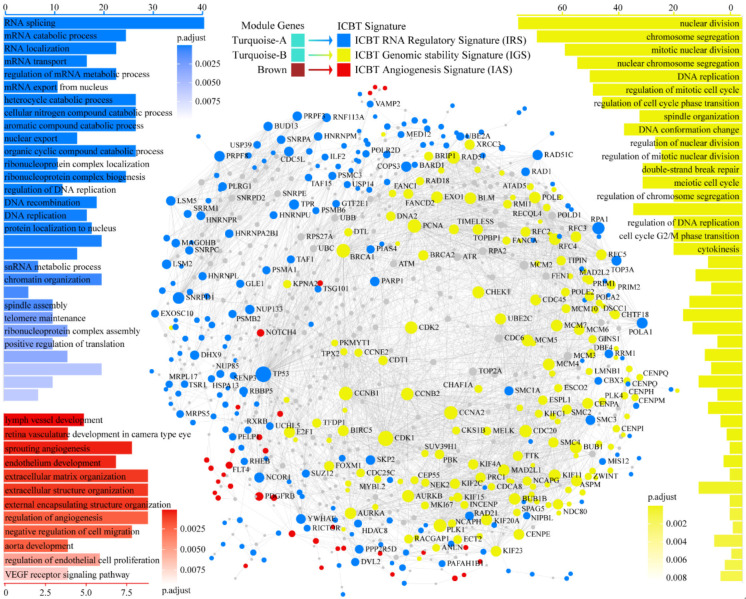
The characteristics of ICBT signatures were shown in Figure 3. GO functional enrichment analyses for ICBT signatures showed that turquoise-A signature is mainly related to the biological processes of regulation of RNA, such as RNA splicing, mRNA catabolic process, RNA localization and mRNA transport, which is an RNA regulatory signal. Therefore, we defined turquoise-A signature as ICBT RNA regulatory Signature (IRS). The biological processes involved in Turquoise-B signature mainly include nuclear division, chromosome segregation, mitotic nuclear division, DNA replication and regulation of mitotic cell cycle, suggesting that turquoise-B signature is associated with potential genomic instability. We defined turquoise-B signature as ICBT Genomic stability Signature (IGS). Brown signature is an angiogenesis signal, involving lymph vessel development, sprouting angiogenesis, endothelium development and extracellular matrix organization. We defined it as ICBT Angiogenesis Signature (IAS). PPI networks analysis revealed the interaction pattern within the three ICBT signatures. There are a wide range of interactions within independent ICBT signatures, and IRS has an intensive mutual regulation effect on IGS and IAS, while there is less interaction between IGS and IAS.
Figure 3.
Functional and molecular characteristics of ICBT signatures. Blue represent IRS, yellow for IGS, red for IAS. In the PPI network, each node is an independent protein, and the lines represent the interaction between the nodes. The size of a node is determined by the number of edges connected to the node. The position of a node is determined by the distance between the node and all the interacting nodes around it, meaning that proteins in the network tend to interact with neighboring proteins. ICBT, immune checkpoint blockade therapy; IRS, ICBT RNA regulatory signature; IGS, ICBT genomic stability signature; IAS, ICBT angiogenesis signature; PPI, Protein-protein interaction.
ICBT signatures improve the prediction value of TMB or PD-L1 ICBT response and patient overall survival in BLCA
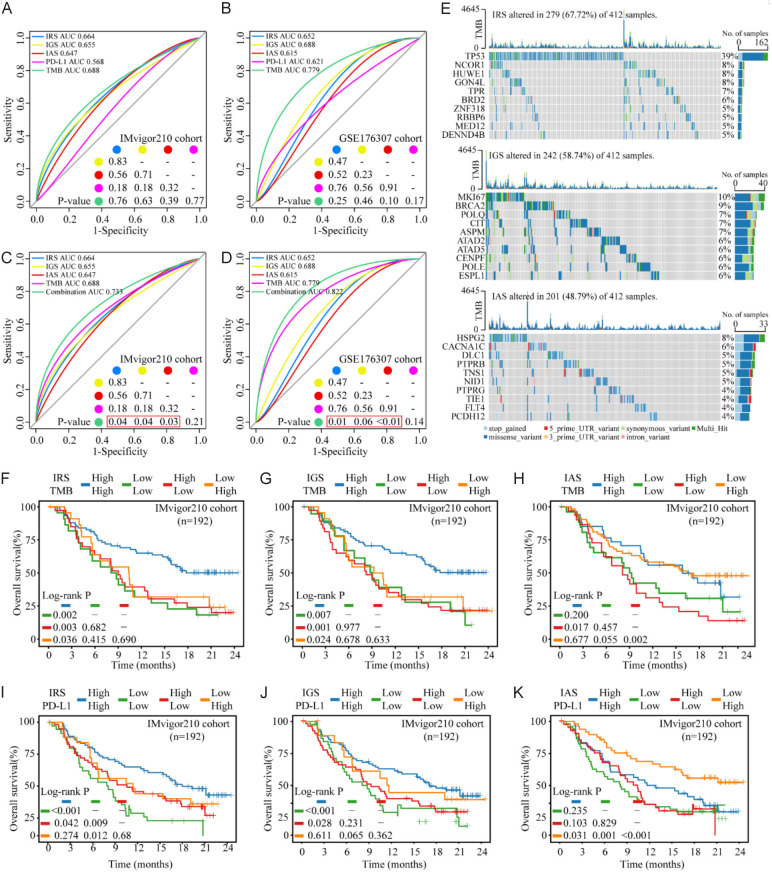
In order to check the ability of ICBT signatures in improving the prediction value of TMB or PD-L1 ICBT response, we calculated the predictive power of ICBT signatures, TMB, and PD-L1 expression for patients receiving ICBT. All features had certain values in predicting ICBT response, with TMB of the highest area under the curve (AUC) and PD-L1 the lowest AUC in both cohorts, except slightly higher AUC of PD-L1 (0.621) than IAS (0.615) in the GSE176307 cohort. However, there were not statistical differences between these AUCs (Figure 4A and 4B). The combination of all the three ICBT signatures and TMB increased the prediction value from AUC 0.688 and 0.779 for TMB to 0.733 and 0.822 for the combination in the IMvigor210 and GSE176307 cohort respectively (Figure 4C and 4D). We also visualized the landscape of the top 10 most frequently mutated genes for each ICBT signature (Figure 4E).
Figure 4.
ICBT signatures and TMB or PD-L1 panel predicts response and benefit of ICBT in BLCA. (A and B) Predictive power of TMB, PD-L1, and ICBT signatures used independently for response of ICBT in the IMvigor210 cohort (A) and GSE176307 cohort (B). (C and D) The combination of ICBT signatures and TMB could significantly improve the predictive ability of ICBT signatures for the response of ICBT in the IMvigor210 cohort (C) and GSE176307 cohort (D). (E) The landscape of the top 10 most frequently mutated genes for each ICBT signature. (F-H) Kaplan-Meier survival curves of IRS/IGS/IAS and TMB panel for OS of patients with ICBT from IMvigor210 cohort. (I-K) Kaplan-Meier survival curves of IRS/IGS/IAS and PD-L1 panel for OS of patients with ICBT from IMvigor210 cohort. ICBT, immune checkpoint blockade therapy; TMB, tumor mutation burden; BLCA, bladder urothelial carcinoma; PD-L1, programmed cell death ligand-1; IRS, ICBT RNA regulatory signature; IGS, ICBT genomic stability signature; IAS, ICBT angiogenesis signature; OS, overall survival.
We further investigated the impact of ICBT signatures on the survival outcome of patients received ICBT. Using Kaplan-Meier analysis of combined IRS expression with TMB in the IMvigor210 cohort, we noted that the IRShighTMBhigh subgroup have the best prognosis compared to other three groups (Figure 4F, compared to IRSlowTMBlow subgroup P=0.002, compared to IRShighTMBlow subgroup P=0.003, compared to IRSlowTMBhigh subgroup P=0.036). Similar data and feature were observed for the combination of IGS expression and TMB (Figure 4G, IGShighTMBhigh subgroup compared to IGSlowTMBlow subgroup P=0.007, compared to IGShighTMBlow subgroup P=0.001, compared to IGSlowTMBhigh subgroup P=0.024). In both analyses, only the group with both high relevant ICBT signature and TMB had significant improved overall survival over the other groups, while the other three groups had a similar survival rate, suggesting that both TMB and the ICBT signatures should be considered for patient outcome prediction under ICBT. However, adding high IAS expression to TMB high group did not improved OS compared to TMB high only patients (Figure 4H), therefore, no additional prediction value over TMB.
We also investigated if these signatures can be used to improve the prediction value of PD-L1 for ICBT efficiency. As expected, in both IMvigor210 and GSE176307 cohort, high PD-L1 expression was significantly associated with improved OS (Figure S1A, P_IMvigor210<0.001, P_GSE176307=0.03). We examined the combined effect of IRS panel and PD-L1 expression on patients’ OS benefit. In the IMvigor210 cohort, patients in IRShighPD-L1high subgroup had a significantly better OS than IRSlowPD-L1low subgroup (Figure 4I, P_IMvigor210<0.001). However, there was no difference observed between IRShighPD-L1high and IRSlowPD-L1low subgroup in the GSE176307 cohort (Figure S1C, P_GSE176307=0.713), potentially due to small number of sample number in this cohort, therefore high change of false message. The long-term survival rates of combination of IRS and PD-L1 expression (Both synchronized high or low expression) compared to using PD-L1 alone, obtained the improvement after about eleven months of ICBT (Figure S1B, blue and green line). We speculated that this was due to the delayed clinical effect of immunotherapy [28]. These results suggest that both IRS and IGS combining with TMB had significant predictive value for the survival benefit of ICBT in BLCA.
Next, we combined IGS, the signature shown above as significant correlation with PD-L1 expression status. IGShighPD-L1high subgroup had a significantly better OS than IGSlowPD-L1low subgroup in two cohort (Figure 4J, P_IMvigor210<0.001; Figure S1D, P_GSE176307=0.02). Moreover, considering the relation between IGS and genomic instability, we validated the impact of the IGS subgroup on adjuvant chemotherapy (ACT) for patients in TCGA cohort. In the subgroup with high expression of IGS, patients with ACT had a significant survival benefit compared with patients without ACT (Figure S1F, top), however, there was no benefit of OS between patients with and without ACT in the subgroup with low expression of IGS (Figure S1F, bottom). Finally, we found IASlowPD-L1high subgroup had a significantly better OS than other subgroups (Figure 4K, IASlowPD-L1high subgroup compared to IAShighPD-L1high subgroup P=0.031, compared to IASlowPD-L1low subgroup P=0.001, compared to IAShighPD-L1low subgroup P<0.001). We only found the trend of better OS to IASlowPD-L1high subgroup in GSE176307 cohort (Figure S1E) with a limited cohort size. The performance of long-term survival rates using the combination of IAS and PD-L1 expression (IASlowPD-L1high compared to IAShighPD-L1low group) compared to using PD-L1 alone was similar to IGS above, but an ascent was observed after 17 months of treatment (Figure S1B, red and green line). Furthermore, to identify whether the effect of IAS was consistent with the known angiogenic signal pathways, we tested the effect with PD-L1 expression panel on ICBT benefit of patient in vascular endothelial growth factors (VEGF) pathway (Kyoto encyclopedia of genes and genomes, KEGG) [29] and angiogenesis signature (Bindea et al.) [30]. VEGF pathway showed an opposite effect in the two cohorts, lacking consistency. The influence of angiogenesis signature on the ICBT benefit prediction value of PD-L1 expression was consistent with IAS, but AngiolowPD-L1high subgroup couldn’t indicate the trend toward improved immunotherapy response rate, which was a special characteristic of IAS (Figure S1G and S1H).
Expression pattern of ICBT signatures in different cell types within the cancer lesion
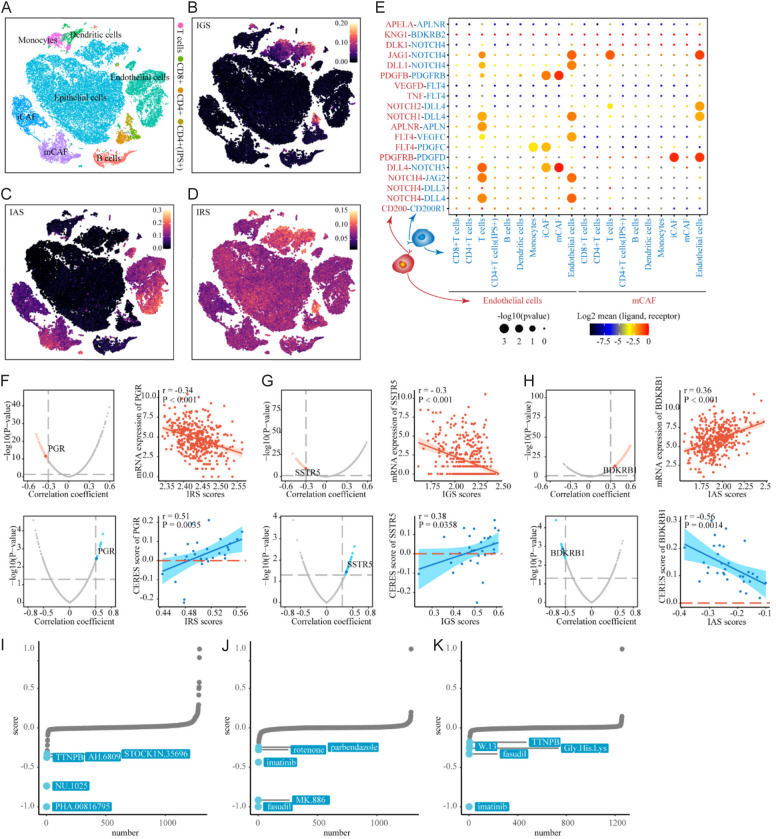
To determine which cell population in BLCA lesion produced the expression pattern of ICBT signatures, we used scRNA-seq data from BioProject PRJNA662018 in GEO database, control and removal of the batch effect between patients (Figure S2A-D, see “Methods”). 55,922 cells were clustered into six major clusters by annotating cell types with ‘HumanPrimaryCellAtlasData’ [31] of singleR: epithelial cells, endothelial cells, cancer-associated fibroblasts (CAF), T cells, B cells and myeloid cells (Figure S2E). Immune cells and CAF were further annotated by using ‘MonacoImmuneData’ [32-34] from singleR as: CD8+ T cells, CD4+ T cells, monocytes, dendritic cells and B cells, inflammatory cancer-associated fibroblasts (iCAF) and myo-cancer-associated fibroblasts (mCAF) (Figure 5A). We noted that IGS was not extensively activated in BLCA cells, and there was a specific cluster of bladder cancer epithelial cells, which were not clustered with the main epithelial cell cluster, with significant activation of IGS (Figure 5B). At the same time, IGS was also activated in a specific cluster of CD4+ T cells, which was defined as ‘CD4+ T cell (IGS+)’. IAS was significantly activated only in endothelial cells, iCAFs and mCAFs (Figure 5C), whereas VEGF pathway was activated in all cell types (Figure S2F) and angiogenesis signature was activated only in endothelial cells (Figure S2G). Consistent with our speculation, IRS was a broad RNA regulatory signal that was activated in all cell types, especially the IGS activated clusters, which may be due to both of them derived from one co-expression module in WGCNA (Figure 5D). Using CellphoneDB, we investigated the cell-cell interaction network between the cells with activated IAS and immune cell based on the genes from IAS. Notably, cells with activated IAS showed the most interactions inside of them, mainly endothelial cells, iCAF and mCAF. For other cell types, endothelial cells had the most interaction with T cells expected cytotoxic T cell (CTL), mainly based on the NOTCH family and corresponding ligands, suggesting a possible pathway of angiogenesis and immune regulation in bladder cancer (Figure 5E).
Figure 5.
Expression pattern of ICBT signatures among cells in tumor microenvironment and identification of ICBT signatures-related drug targets. (A) tSNE plot of single cells profiled in the presenting study colored by major cell types. (B-D) tSNE plot of the expression pattern of IGS in the single cells profiled (B); IAS in the single cells profiled (C); IRS in the single cells profiled (D). (E) Bubble plots show ligand-receptor pairs of genes from IAS between clusters with activated IAS and other immune cell groups; (F) Red volcano plot (left) and scatter plots (right) of Spearman’s correlations and significance between IRS score and mRNA expression of drug targets. Red dots indicate the significant positive correlations (P<0.05, and Spearman’s r absolute value >0.3). Blue volcano plot (left) and scatter plots (right) of Spearman’s correlations and significance between IRS score and CERES score of drug targets. Blue dots indicate the significant negative correlations (P<0.05, and Spearman’s r absolute value >0.3); (G) IGS; (H) IAS; (I) CMap analysis of IRS, the lower the score of compounds, the more likely the drug is to reverse the molecular characteristics of the disease; (J) CMap analysis of IGS; (K) CMap analysis of IAS. ICBT, immune checkpoint blockade therapy; IRS, ICBT RNA regulatory signature; IGS, ICBT genomic stability signature; IAS, ICBT angiogenesis signature.
Identification of potential drug and small-molecules targets for ICBT signatures
To identify potential targets for improving ICBT based on ICBT signature, we collected target information for 6,125 compounds and performed a two-step analysis to identify candidate targets. Firstly, the correlation coefficient between the expression level of druggable mRNA and ICBT signatures was calculated. Targets were identified with a correlation coefficient greater than an absolute value of 0.30 (P<0.05). Next, by conducting a correlation analysis between CERES scores and ICBT signature scores based on bladder cancer cell lines, we further identified poor prognosis-dependent targets, defined as Spearman’s r absolute value >0.5 and P<0.05 (Figure 5F-H). The final targets identified were PGR (for IRS) and BDKRB1 (for IAS), with the corresponding compounds listed in Table S1. We did not find significant targets for IGS based on the above criteria. When we downregulated the cutoff of Spearman’s r absolute value to 0.3, SSTR5 was identified. This analysis suggests that inhibiting these three genes in ICBT signature patients could achieve favorable treatment outcome in ICBT. Several hypotheses linking therapeutic compounds to new disease signature have been experimentally validated using CMap analysis [35-37]. In CMap analysis, we identify that PHA.00816795 as potential synergist of IRS-mediated (Figure 5I) and fasudil as potential synergist of IGS-mediated improvement of ICBT efficacy (Figure 5J), while Imatinib as potential inhibitors of IAS-mediated poor outcomes of ICBT (Figure 5K).
Discussion
Treatment of bladder urothelial carcinoma, especially muscle invasive bladder urothelial carcinoma is still a big problem. Anti-immune checkpoint therapies, in particular anti-PD-1/PD-L1, benefit only less than 30% of patients with advanced disease [1]. New targets or combined therapy strategies are still waiting to be discovered, and such development could be enhanced by a better understanding of the tumor signature of bladder urothelial carcinoma. In the present study, we identified three novel signatures related to ICBT response in bladder urothelial carcinoma: IGS, representing a special state of genomic stability; IAS, an angiogenesis signal; IRS, an RNA regulatory signal.
The extracellular immunopathogenic PD-L1/PD-1 pathway described in cancer immune checkpoint therapy is represented by PD-L1 expressed on the surface of cancer or stromal cells, which binds to PD-1 expressed on the surface of immune cells, leading to signaling downstream of PD-1 to suppress anti-tumor immunity [38,39]. Our study found that IGS, associated to DNA duplication, cell division and cell cycle transition, was only positive in a special subgroup of BLCA epithelial cells significantly discrepancy from the majority of BLCA epithelial cells. Tumor endogenous PD-L1 also plays an important role in cancer [40], while IGS was significantly correlated with PD-L1 expression in tumor. IGS, representing genomic instability usually companies an increase of neoantigens, which will activate PD-L1 for cancer cell to survive [41]. Moreover, previous studies have shown that tumor PD-L1 expression changes with cell cycle, which has the highest expression in M/G1 phase and a sharp decrease in G1/S phase [42]. These correlations may explain that the changes in PD-L1 expression are associated with IGS, that only cells in a special cell state (IGS+) would upregulated the expression of PD-L1. It has also been reported in melanoma and ovarian cancer studies that tumor PD-L1 promoted cell proliferation [43], which may take more activation of IGS. In our study, it is yet to be investigated if IGS leads to the up-regulation of cell cycle-dependent PD-L1 expression or the increased expression of PD-L1 activates IGS. Moreover, our study found that the subgroup of patients with high IGS activation could benefit from adjuvant chemotherapy, whereas the subgroup with low IGS expression level could not, although the in-depth mechanism is not yet clear. Finally, as expected, combination of IGS panel and TMB or PD-L1 significantly distinguished a patient subgroup with high sensitivity of ICBT. We speculated the increased prediction value of TMB or PD-L1 for ICBT in the IGS positive subgroup might be due to the cell proliferation activity and high genomic instability caused by IGS activation.
In addition to the small subpopulation of epithelial cells, the other cells with significantly activated IGS were a cluster of CD4+ T cells and a cluster of dendritic cells, both of them participate in the signal transduction of antigen recognition by T cell antigen receptor [44,45]. At present, the research of immunotherapeutic targets mainly focuses on CD8+ T cells. However, in tumor immunity, CD4+ T cells can activate CD8+ T cells through a variety of mechanisms to evolve into cytotoxic T lymphocytes, while maintaining and enhancing the antitumor response of CTL [46], especially in bladder cancer. The enrichment level of CD4+ T cells were higher than CD8+ T cells, indicating better tumor-specific states in bladder cancer. Moreover, CD4+ T cells can directly induce apoptosis of bladder cancer cells in vitro [47]. On the other hand, CD4+ T cell activation depends on subpopulation of dendritic cells [48]. The specific activation of IGS in above cell types raises the suspicion that this state of changing genome occurring simultaneously in multiple cell types shares a common feature, an antitumor immunity activated program blocked by PD-1/PD-L1 immune checkpoint, potentially involving CD4+ T cell proliferation and functional recovery [47,49], which has rarely been reported in bladder cancer previously.
Overall, IGS is a novel ICBT genomic instability signature that correlates with tumor PD-L1 expression of tumor and it only exists in the specific classes of bladder cancer epithelial cells, CD4+ T cells and dendritic cells. This finding provides further understanding of the impact of bladder cancer heterogeneity on immune checkpoint therapy.
IAS represents a class of gene signature related to tumor angiogenesis. Unlike traditional tumor angiogenesis signature, which is only expressed in bladder cancer cells and endothelial cells, IAS was activated in tumor microenvironment non-immune cells, including endothelial cells, iCAF and mCAF. Previous studies have shown that tumor vascular endothelial cells can be activated by overexpress PD-L1. The inhibition of a series of immune functions of CD8+ T lymphocytes and PD-L1 on tumor vascular endothelial cells promotes tumor genesis, progression and metastasis [50,51]. However, these studies only focus on tumor vascular endothelial cells. IAS was also activated in CAFs, which has been reported to affect the tumor intravasation in microenvironment [52].
We demonstrated that IASlowPD-L1high subgroup showed a trend toward improved immunotherapy response rate compared to other states of expression. Tumor vessel and immune microenvironment interact mutually [53]. CAFs can be differentiated from epithelial cells, endothelial cells, smooth muscle cells and bone marrow mesenchymal stem cells in tumor and participate in the shape of the tumor extracellular matrix. The secretion of a large amount of collagen and fibronectin can promote the solidification of tumor extracellular matrix, inhibit the infiltration of immune cells recruited by chemokines secreted from tumor tissues and impede the penetration of anti-tumor drugs to reduce the effect of tumor treatment. In addition, CAFs are also directly involved in the regulation of tumor vascular network [54-56]. As IAS is activated in both endothelial cells and CAFs, IAS, representing tumor vascular regulation, might be contributed by both of these cell types. By investigating the cell-cell interaction network among the cell types using CellphoneDB, we also found that IAS in endothelial cells and CAFs mainly interacts with other cell types through the ligands and corresponding receptors of NOTCH family and platelet-derived growth factor (PDGF) family. In addition, IAS were also activated in a few non-cytotoxic T cells and interacted with endothelial cells through the ligands and corresponding receptors of NOTCH family. This provides evidence that angiogenesis, as represented by IAS, affects the immune microenvironment. Taken together, IAS is a characteristic angiogenic signature in bladder urothelial carcinoma, involving the co-regulation of endothelial cells and CAFs.
IRS is a signature related to RNA regulation, involving in biological processes such as RNA splicing, mRNA catabolic process, RNA localization and mRNA transport. IRS is widely activated in bladder urothelial carcinoma, including all cell types, especially in IGS-activated cells, which may be explained by that IRS and IGS were derived from the same module in the gene co-expression network. Like IGS, we found that combination of IRS panel and TMB or PD-L1 significantly distinguished a patient subgroup with high sensitiveness of ICBT, but the combination with PD-L1 was inconsistent, which needs validation in larger sample size cohort. Generally, tumor cells were thought to teem with multiple RNA regulatory program activation. IRS may be the signature that describes the magnitude of cellular mobilization of RNA regulatory programs after patients receive immune checkpoint therapy.
Most importantly, these ICBT signatures improved the ICBT outcome prediction value of the currently used biomarkers TMB and PD-L1 expression. Theoretically, high PD-L1 expression should be an accurate marker for receiving anti-PD-1/PD-L1 ICBT. However, all the relevant clinical trials have shown that PD-L1 expression has limited predictive value and TMB has later been identified as a better prediction biomarker [57,58]. The combination of both IGS and IRS with either TMB or PD-L1 expression improved their prediction efficiency of OS for patients receiving ICBT that patients with both high level of these ICBT signature and high TMB or PD-L1 expression had much better survival benefit than other patient groups. The striking finding showed that, in the combined IGS or IRS analysis with TMB, only patients with both high immune signature and TMB had significantly better chance of benefiting from ICBT (double OS rate than any other groups at 24-month following up period), whilst the OS of the other three groups of patients were similar. Therefore, both signatures are promising ICBT response prediction biomarkers in combination with TMB. IAS has limited impact on the ICBT response prediction efficiency when combined with TMB. We considered that potential reason was low gene mutation frequency in IAS signature compared to IRS/IGS. While patients with low IAS and high PP-L1 expression had over 50% chance of survival under the ICBT, significantly better than any other groups of patients in this combined analysis and was similar to those with both high TMB and IGS or IRS in different KM analysis. If should be further determined in large sample cohort(s) which of these combinations will perform the best in predicting ICBT outcome and if the combination of all these predicting factors will be significantly better than these two factor combinations.
Based on the ICBT signatures we identified, we explored potential molecular targets and compounds to improve the efficacy of immunotherapy. PGR (Progesterone Receptor) is one of the molecules with loss of expression in triple negative breast cancer (TNBC), which is more likely to benefit from immunotherapy [59]. In bladder cancer, PGR is considered as a biomarker for the bad prognosis of bladder cancer by several studies [60,61]. Weather targeting PGR can benefit immunotherapy of bladder cancer warrant further investigations. SSTR5 (Somatostatin Receptor 5) is one of five G-protein coupled receptors that sense the peptide hormone somatostatin. SSTR5 inhibitors have potential in the treatment of type 2 diabetes, but the research in immunotherapy is still insufficient [62]. SSTR3 in this family is a known prognostic marker in bladder cancer [63], but further studies on SSTR5 is required to establish its roles in bladder cancer. BDKRB1 (Bradykinin Receptor B1) has been reported to affect migration and invasion of malignant glioblastoma cells and to limit encephalitogenic T lymphocyte recruitment to the central nervous system [64]. However, data on BDKRB1 in immunotherapy and bladder cancer are very limited. PHA.00816795, fasudil, and imatinib are considered the most likely compounds in CMap analysis to reverse tumor state of ICBT signatures (Table S2). Further experimental validation to confirm their potential safety and efficacy are warranted.
There are limitations of our study. Our study collected immunotherapy patient data form IMvigor210 and GSE176307 cohorts, where both are composed of metastatic BLCA patients, but single-cell and TCGA data are mainly from non-metastatic cases. Further investigations are required to determine whether ICBT signatures are affected by the difference between advanced and early disease. Finally, the biological mechanisms of the involvement of certain genes of ICBT signatures, such as SRSF10 and CCDC59, are yet to be experimentally confirmed.
Conclusions
In conclusion, our work highlights three novel ICBT signatures and describes the expression pattern of ICBT signatures among cells in the BLCA lesion. The combination of IGS or IRS panel with TMB and the combination of IAS with PD-L1 expression enhanced the prognostic value in term of OS for patients receiving ICBT and may have great value in patient selection for ICBT. This study also provides clues to understand the role of ICBT signatures, which promotes further mechanism discovery in BLCA progression and response to therapies.
Acknowledgements
This study was sponsored by the National Natural Science Foundation of China (Grant No. 82373077, U1904162).
Disclosure of conflict of interest
None.
Abbreviations
- AUC
area under the ROC curve
- BLCA
bladder urothelial carcinoma
- CR
complete response
- CTL
cytotoxic T cell
- IAS
ICBT angiogenesis signature
- ICBT
Immune checkpoint blockade therapy
- IGS
ICBT genomic stability signature
- IRS
ICBT RNA regulatory signature
- KEGG
Kyoto encyclopedia of genes and genomes
- OS
overall survival
- PD-1
programmed death 1
- PD-L1
programmed cell death ligand 1
- PD
progressive disease
- PPI
protein-protein interaction
- PR
partial response
- ROC
receiver operating characteristic
- SD
table disease
- TCGA
The Cancer Genome Atlas
- TMB
Tumor mutational burden
- VEGF
vascular endothelial growth factors
- WGCNA
weighted gene co-expression network analysis
Figures S1 and S2
Table S1
Table S2
References
- 1.Song D, Powles T, Shi L, Zhang L, Ingersoll MA, Lu YJ. Bladder cancer, a unique model to understand cancer immunity and develop immunotherapy approaches. J Pathol. 2019;249:151–165. doi: 10.1002/path.5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ein-Dor L, Kela I, Getz G, Givol D, Domany E. Outcome signature genes in breast cancer: is there a unique set? Bioinformatics. 2005;21:171–178. doi: 10.1093/bioinformatics/bth469. [DOI] [PubMed] [Google Scholar]
- 3.Fan C, Oh DS, Wessels L, Weigelt B, Nuyten DS, Nobel AB, van’t Veer LJ, Perou CM. Concordance among gene-expression-based predictors for breast cancer. N Engl J Med. 2006;355:560–569. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- 4.Nevins JR, Potti A. Mining gene expression profiles: expression signatures as cancer phenotypes. Nat Rev Genet. 2007;8:601–609. doi: 10.1038/nrg2137. [DOI] [PubMed] [Google Scholar]
- 5.Segal E, Friedman N, Kaminski N, Regev A, Koller D. From signatures to models: understanding cancer using microarrays. Nat Genet. 2005;37(Suppl):S38–45. doi: 10.1038/ng1561. [DOI] [PubMed] [Google Scholar]
- 6.Wolf DM, Lenburg ME, Yau C, Boudreau A, van’t Veer LJ. Gene co-expression modules as clinically relevant hallmarks of breast cancer diversity. PLoS One. 2014;9:e88309. doi: 10.1371/journal.pone.0088309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karasinska JM, Topham JT, Kalloger SE, Jang GH, Denroche RE, Culibrk L, Williamson LM, Wong HL, Lee MKC, O’Kane GM, Moore RA, Mungall AJ, Moore MJ, Warren C, Metcalfe A, Notta F, Knox JJ, Gallinger S, Laskin J, Marra MA, Jones SJM, Renouf DJ, Schaeffer DF. Altered gene expression along the glycolysis-cholesterol synthesis axis is associated with outcome in pancreatic cancer. Clin Cancer Res. 2020;26:135–146. doi: 10.1158/1078-0432.CCR-19-1543. [DOI] [PubMed] [Google Scholar]
- 8.Necchi A, Joseph RW, Loriot Y, Hoffman-Censits J, Perez-Gracia JL, Petrylak DP, Derleth CL, Tayama D, Zhu Q, Ding B, Kaiser C, Rosenberg JE. Atezolizumab in platinum-treated locally advanced or metastatic urothelial carcinoma: post-progression outcomes from the phase II IMvigor210 study. Ann Oncol. 2017;28:3044–3050. doi: 10.1093/annonc/mdx518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mariathasan S, Turley SJ, Nickles D, Castiglioni A, Yuen K, Wang Y, Kadel EE III, Koeppen H, Astarita JL, Cubas R, Jhunjhunwala S, Banchereau R, Yang Y, Guan Y, Chalouni C, Ziai J, Şenbabaoğlu Y, Santoro S, Sheinson D, Hung J, Giltnane JM, Pierce AA, Mesh K, Lianoglou S, Riegler J, Carano RAD, Eriksson P, Höglund M, Somarriba L, Halligan DL, van der Heijden MS, Loriot Y, Rosenberg JE, Fong L, Mellman I, Chen DS, Green M, Derleth C, Fine GD, Hegde PS, Bourgon R, Powles T. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature. 2018;554:544–548. doi: 10.1038/nature25501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rose TL, Weir WH, Mayhew GM, Shibata Y, Eulitt P, Uronis JM, Zhou M, Nielsen M, Smith AB, Woods M, Hayward MC, Salazar AH, Milowsky MI, Wobker SE, McGinty K, Millburn MV, Eisner JR, Kim WY. Fibroblast growth factor receptor 3 alterations and response to immune checkpoint inhibition in metastatic urothelial cancer: a real world experience. Br J Cancer. 2021;125:1251–1260. doi: 10.1038/s41416-021-01488-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosenberg JE, Hoffman-Censits J, Powles T, van der Heijden MS, Balar AV, Necchi A, Dawson N, O’Donnell PH, Balmanoukian A, Loriot Y, Srinivas S, Retz MM, Grivas P, Joseph RW, Galsky MD, Fleming MT, Petrylak DP, Perez-Gracia JL, Burris HA, Castellano D, Canil C, Bellmunt J, Bajorin D, Nickles D, Bourgon R, Frampton GM, Cui N, Mariathasan S, Abidoye O, Fine GD, Dreicer R. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet. 2016;387:1909–1920. doi: 10.1016/S0140-6736(16)00561-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu Y, Huang Y, Li C, Ou S, Xu C, Kang Z. Clinical value of M1 macrophage-related genes identification in bladder urothelial carcinoma and in vitro validation. Front Genet. 2022;13:1047004. doi: 10.3389/fgene.2022.1047004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou G, Soufan O, Ewald J, Hancock REW, Basu N, Xia J. NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019;47:W234–W241. doi: 10.1093/nar/gkz240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Z, Zhou L, Liu L, Hou Y, Xiong M, Yang Y, Hu J, Chen K. Single-cell RNA sequencing highlights the role of inflammatory cancer-associated fibroblasts in bladder urothelial carcinoma. Nat Commun. 2020;11:5077. doi: 10.1038/s41467-020-18916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, Chak S, Naikawadi RP, Wolters PJ, Abate AR, Butte AJ, Bhattacharya M. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol. 2019;20:163–172. doi: 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc. 2020;15:1484–1506. doi: 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- 22.Ghandi M, Huang FW, Jané-Valbuena J, Kryukov GV, Lo CC, McDonald ER 3rd, Barretina J, Gelfand ET, Bielski CM, Li H, Hu K, Andreev-Drakhlin AY, Kim J, Hess JM, Haas BJ, Aguet F, Weir BA, Rothberg MV, Paolella BR, Lawrence MS, Akbani R, Lu Y, Tiv HL, Gokhale PC, de Weck A, Mansour AA, Oh C, Shih J, Hadi K, Rosen Y, Bistline J, Venkatesan K, Reddy A, Sonkin D, Liu M, Lehar J, Korn JM, Porter DA, Jones MD, Golji J, Caponigro G, Taylor JE, Dunning CM, Creech AL, Warren AC, McFarland JM, Zamanighomi M, Kauffmann A, Stransky N, Imielinski M, Maruvka YE, Cherniack AD, Tsherniak A, Vazquez F, Jaffe JD, Lane AA, Weinstock DM, Johannessen CM, Morrissey MP, Stegmeier F, Schlegel R, Hahn WC, Getz G, Mills GB, Boehm JS, Golub TR, Garraway LA, Sellers WR. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature. 2019;569:503–508. doi: 10.1038/s41586-019-1186-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, Reich M, Hieronymus H, Wei G, Armstrong SA, Haggarty SJ, Clemons PA, Wei R, Carr SA, Lander ES, Golub TR. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 24.Qu XA, Rajpal DK. Applications of Connectivity Map in drug discovery and development. Drug Discov Today. 2012;17:1289–1298. doi: 10.1016/j.drudis.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 25.Malta TM, Sokolov A, Gentles AJ, Burzykowski T, Poisson L, Weinstein JN, Kamińska B, Huelsken J, Omberg L, Gevaert O, Colaprico A, Czerwińska P, Mazurek S, Mishra L, Heyn H, Krasnitz A, Godwin AK, Lazar AJ Cancer Genome Atlas Research Network. Stuart JM, Hoadley KA, Laird PW, Noushmehr H, Wiznerowicz M. Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell. 2018;173:338–354. e315. doi: 10.1016/j.cell.2018.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang C, Zhang H, Chen M, Wang S, Qian R, Zhang L, Huang X, Wang J, Liu Z, Qin W, Wang C, Hang H, Wang H. A survey of optimal strategy for signature-based drug repositioning and an application to liver cancer. Elife. 2022;11:e71880. doi: 10.7554/eLife.71880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langfelder P, Luo R, Oldham MC, Horvath S. Is my network module preserved and reproducible? PLoS Comput Biol. 2011;7:e1001057. doi: 10.1371/journal.pcbi.1001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu X, Meng J, Su L, Jiang L, Wang H, Zhu J, Huang M, Cheng W, Xu L, Ruan X, Yeh S, Liang C, Yan F. Multi-omics consensus ensemble refines the classification of muscle-invasive bladder cancer with stratified prognosis, tumour microenvironment and distinct sensitivity to frontline therapies. Clin Transl Med. 2021;11:e601. doi: 10.1002/ctm2.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, Angell H, Fredriksen T, Lafontaine L, Berger A, Bruneval P, Fridman WH, Becker C, Pagès F, Speicher MR, Trajanoski Z, Galon J. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 31.Schmiedel BJ, Singh D, Madrigal A, Valdovino-Gonzalez AG, White BM, Zapardiel-Gonzalo J, Ha B, Altay G, Greenbaum JA, McVicker G, Seumois G, Rao A, Kronenberg M, Peters B, Vijayanand P. Impact of genetic polymorphisms on human immune cell gene expression. Cell. 2018;175:1701–1715. e1716. doi: 10.1016/j.cell.2018.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu W, Monaco G, Wong EH, Tan WLW, Kared H, Simoni Y, Tan SW, How WZY, Tan CTY, Lee BTK, Carbajo D, K G S, Low ICH, Mok EWH, Foo S, Lum J, Tey HL, Tan WP, Poidinger M, Newell E, Ng TP, Foo R, Akbar AN, Fülöp T, Larbi A. Mapping of γ/δ T cells reveals Vδ2+ T cells resistance to senescence. EBioMedicine. 2019;39:44–58. doi: 10.1016/j.ebiom.2018.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monaco G, Lee B, Xu W, Mustafah S, Hwang YY, Carré C, Burdin N, Visan L, Ceccarelli M, Poidinger M, Zippelius A, Pedro de Magalhães J, Larbi A. RNA-Seq signatures normalized by mRNA abundance allow absolute deconvolution of human immune cell types. Cell Rep. 2019;26:1627–1640. e1627. doi: 10.1016/j.celrep.2019.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang EY, Mao T, Klein J, Dai Y, Huck JD, Jaycox JR, Liu F, Zhou T, Israelow B, Wong P, Coppi A, Lucas C, Silva J, Oh JE, Song E, Perotti ES, Zheng NS, Fischer S, Campbell M, Fournier JB, Wyllie AL, Vogels CBF, Ott IM, Kalinich CC, Petrone ME, Watkins AE Yale IMPACT Team. Dela Cruz C, Farhadian SF, Schulz WL, Ma S, Grubaugh ND, Ko AI, Iwasaki A, Ring AM. Diverse functional autoantibodies in patients with COVID-19. Nature. 2021;595:283–288. doi: 10.1038/s41586-021-03631-y. [DOI] [PubMed] [Google Scholar]
- 35.Chang M, Smith S, Thorpe A, Barratt MJ, Karim F. Evaluation of phenoxybenzamine in the CFA model of pain following gene expression studies and connectivity mapping. Mol Pain. 2010;6:56. doi: 10.1186/1744-8069-6-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Claerhout S, Lim JY, Choi W, Park YY, Kim K, Kim SB, Lee JS, Mills GB, Cho JY. Gene expression signature analysis identifies vorinostat as a candidate therapy for gastric cancer. PLoS One. 2011;6:e24662. doi: 10.1371/journal.pone.0024662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dudley JT, Sirota M, Shenoy M, Pai RK, Roedder S, Chiang AP, Morgan AA, Sarwal MM, Pasricha PJ, Butte AJ. Computational repositioning of the anticonvulsant topiramate for inflammatory bowel disease. Sci Transl Med. 2011;3:96ra76. doi: 10.1126/scitranslmed.3002648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zou W, Wolchok JD, Chen L. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: mechanisms, response biomarkers, and combinations. Sci Transl Med. 2016;8:328rv4. doi: 10.1126/scitranslmed.aad7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Topalian SL, Taube JM, Anders RA, Pardoll DM. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer. 2016;16:275–287. doi: 10.1038/nrc.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kornepati AVR, Vadlamudi RK, Curiel TJ. Programmed death ligand 1 signals in cancer cells. Nat Rev Cancer. 2022;22:174–189. doi: 10.1038/s41568-021-00431-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McGranahan N, Furness AJ, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, Jamal-Hanjani M, Wilson GA, Birkbak NJ, Hiley CT, Watkins TB, Shafi S, Murugaesu N, Mitter R, Akarca AU, Linares J, Marafioti T, Henry JY, Van Allen EM, Miao D, Schilling B, Schadendorf D, Garraway LA, Makarov V, Rizvi NA, Snyder A, Hellmann MD, Merghoub T, Wolchok JD, Shukla SA, Wu CJ, Peggs KS, Chan TA, Hadrup SR, Quezada SA, Swanton C. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351:1463–1469. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang J, Bu X, Wang H, Zhu Y, Geng Y, Nihira NT, Tan Y, Ci Y, Wu F, Dai X, Guo J, Huang YH, Fan C, Ren S, Sun Y, Freeman GJ, Sicinski P, Wei W. Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature. 2018;553:91–95. doi: 10.1038/nature25015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gupta HB, Clark CA, Yuan B, Sareddy G, Pandeswara S, Padron AS, Hurez V, Conejo-Garcia J, Vadlamudi R, Li R, Curiel TJ. Tumor cell-intrinsic PD-L1 promotes tumor-initiating cell generation and functions in melanoma and ovarian cancer. Signal Transduct Target Ther. 2016;1:16030. doi: 10.1038/sigtrans.2016.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murray JS, Madri J, Tite J, Carding SR, Bottomly K. MHC control of CD4+ T cell subset activation. J Exp Med. 1989;170:2135–2140. doi: 10.1084/jem.170.6.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bauer CA, Kim EY, Marangoni F, Carrizosa E, Claudio NM, Mempel TR. Dynamic Treg interactions with intratumoral APCs promote local CTL dysfunction. J Clin Invest. 2014;124:2425–2440. doi: 10.1172/JCI66375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laidlaw BJ, Craft JE, Kaech SM. The multifaceted role of CD4(+) T cells in CD8(+) T cell memory. Nat Rev Immunol. 2016;16:102–111. doi: 10.1038/nri.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oh DY, Kwek SS, Raju SS, Li T, McCarthy E, Chow E, Aran D, Ilano A, Pai CS, Rancan C, Allaire K, Burra A, Sun Y, Spitzer MH, Mangul S, Porten S, Meng MV, Friedlander TW, Ye CJ, Fong L. Intratumoral CD4(+) T cells mediate anti-tumor cytotoxicity in human bladder cancer. Cell. 2020;181:1612–1625. e1613. doi: 10.1016/j.cell.2020.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Binnewies M, Mujal AM, Pollack JL, Combes AJ, Hardison EA, Barry KC, Tsui J, Ruhland MK, Kersten K, Abushawish MA, Spasic M, Giurintano JP, Chan V, Daud AI, Ha P, Ye CJ, Roberts EW, Krummel MF. Unleashing type-2 dendritic cells to drive protective antitumor CD4(+) T cell immunity. Cell. 2019;177:556–571. e516. doi: 10.1016/j.cell.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tracy SI, Venkatesh H, Hekim C, Heltemes-Harris LM, Knutson TP, Bachanova V, Farrar MA. Combining nilotinib and PD-L1 blockade reverses CD4+ T-cell dysfunction and prevents relapse in acute B-cell leukemia. Blood. 2022;140:335–348. doi: 10.1182/blood.2021015341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodig N, Ryan T, Allen JA, Pang H, Grabie N, Chernova T, Greenfield EA, Liang SC, Sharpe AH, Lichtman AH, Freeman GJ. Endothelial expression of PD-L1 and PD-L2 down-regulates CD8+ T cell activation and cytolysis. Eur J Immunol. 2003;33:3117–3126. doi: 10.1002/eji.200324270. [DOI] [PubMed] [Google Scholar]
- 51.Campesato LF, Merghoub T. Antiangiogenic therapy and immune checkpoint blockade go hand in hand. Ann Transl Med. 2017;5:497. doi: 10.21037/atm.2017.10.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng H, An M, Luo Y, Diao X, Zhong W, Pang M, Lin Y, Chen J, Li Y, Kong Y, Zhao Y, Yin Y, Ai L, Huang J, Chen C, Lin T. PDGFRα(+)ITGA11(+) fibroblasts foster early-stage cancer lymphovascular invasion and lymphatic metastasis via ITGA11-SELE interplay. Cancer Cell. 2024;42:682–700. e612. doi: 10.1016/j.ccell.2024.02.002. [DOI] [PubMed] [Google Scholar]
- 53.Tian L, Goldstein A, Wang H, Ching Lo H, Sun Kim I, Welte T, Sheng K, Dobrolecki LE, Zhang X, Putluri N, Phung TL, Mani SA, Stossi F, Sreekumar A, Mancini MA, Decker WK, Zong C, Lewis MT, Zhang XH. Mutual regulation of tumour vessel normalization and immunostimulatory reprogramming. Nature. 2017;544:250–254. doi: 10.1038/nature21724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen X, Song E. Turning foes to friends: targeting cancer-associated fibroblasts. Nat Rev Drug Discov. 2019;18:99–115. doi: 10.1038/s41573-018-0004-1. [DOI] [PubMed] [Google Scholar]
- 55.Mao X, Xu J, Wang W, Liang C, Hua J, Liu J, Zhang B, Meng Q, Yu X, Shi S. Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: new findings and future perspectives. Mol Cancer. 2021;20:131. doi: 10.1186/s12943-021-01428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li C, Teixeira AF, Zhu HJ, Ten Dijke P. Cancer associated-fibroblast-derived exosomes in cancer progression. Mol Cancer. 2021;20:154. doi: 10.1186/s12943-021-01463-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, Leming PD, Spigel DR, Antonia SJ, Horn L, Drake CG, Pardoll DM, Chen L, Sharfman WH, Anders RA, Taube JM, McMiller TL, Xu H, Korman AJ, Jure-Kunkel M, Agrawal S, McDonald D, Kollia GD, Gupta A, Wigginton JM, Sznol M. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goodman AM, Kato S, Bazhenova L, Patel SP, Frampton GM, Miller V, Stephens PJ, Daniels GA, Kurzrock R. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther. 2017;16:2598–2608. doi: 10.1158/1535-7163.MCT-17-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kwapisz D. Pembrolizumab and atezolizumab in triple-negative breast cancer. Cancer Immunol Immunother. 2021;70:607–617. doi: 10.1007/s00262-020-02736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kriegmair MC, Wirtz RM, Worst TS, Breyer J, Ritter M, Keck B, Boehmer C, Otto W, Eckstein M, Weis CA, Hartmann A, Bolenz C, Erben P. Prognostic value of molecular breast cancer subtypes based on Her2, ESR1, PGR and Ki67 mRNA-expression in muscle invasive bladder cancer. Transl Oncol. 2018;11:467–476. doi: 10.1016/j.tranon.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang JP, Leng JY, Zhang RK, Zhang L, Zhang B, Jiang WY, Tong L. Functional analysis of gene expression profiling-based prediction in bladder cancer. Oncol Lett. 2018;15:8417–8423. doi: 10.3892/ol.2018.8370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu W, Shao PP, Liang GB, Bawiec J, He J, Aster SD, Wu M, Chicchi G, Wang J, Tsao KL, Shang J, Salituro G, Zhou YP, Li C, Akiyama TE, Metzger DE, Murphy BA, Howard AD, Weber AE, Duffy JL. Discovery and pharmacology of a novel somatostatin subtype 5 (SSTR5) antagonist: synergy with DPP-4 inhibition. ACS Med Chem Lett. 2018;9:1082–1087. doi: 10.1021/acsmedchemlett.8b00305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maas M, Mayer L, Hennenlotter J, Stühler V, Walz S, Scharpf M, Kühs U, Neumann T, Stenzl A, Todenhöfer T. Prognostic impact of somatostatin receptor expression in advanced bladder cancer. Urol Oncol. 2020;38:935.e17–935.e28. doi: 10.1016/j.urolonc.2020.07.005. [DOI] [PubMed] [Google Scholar]
- 64.Sun DP, Lee YW, Chen JT, Lin YW, Chen RM. The Bradykinin-BDKRB1 axis regulates aquaporin 4 gene expression and consequential migration and invasion of malignant glioblastoma cells via a Ca(2+)-MEK1-ERK1/2-NF-κB mechanism. Cancers (Basel) 2020;12:667. doi: 10.3390/cancers12030667. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.