Abstract
Pancreatic ductal adenocarcinoma (PDAC) patients’ express higher levels of the orphan Nuclear Receptor 4A2 (NR4A2, NURR1) compared to normal pancreas and NR4A2 is a prognostic factor for patient survival. Knockdown of NR4A2 by RNA interference (RNAi) inhibited cell proliferation, invasion, and migration. RNA sequencing performed in NR4A2(+/+) and NR4A2(-/-) MiaPaCa2 cells demonstrated that NR4A2 played a significant role in cellular metabolism. Human antigen R (HuR) and isocitrate dehydrogenase 1 (IDH1) were identified as NR4A2 target genes. HuR is a pro-oncogenic RNA binding protein and silencing of HuR by RNAi significantly downregulated expression of NR4A2. Expression of HuR and IDH1 were significantly downregulated after treatment with NR4A2 inverse agonist, 1,1-bis(3’-indolyl)-1-(p-chlorophenyl)methane resulting in significant inhibition of tumor growth in an athymic nude mouse xenograft model. This study demonstrates that NR4A2 and HuR regulate genes and signaling pathways that enhance tumorigenesis and targeting NR4A2 and HuR expression with an NR4A2 inverse agonist represents a novel regimen for treating PDAC.
Keywords: Pancreatic cancer, NR4A2, HuR, IDH1, NR4A2 antagonist treatment
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is one of the leading causes of cancer deaths, with five-year and one-year survival rates of 5-7 and 20% respectively [1]. PDAC is lethal due to poor clinical prognosis, late-stage diagnosis with surgical abscission being the only curative therapy (for <20% patients) [2,3]. The common symptoms of PDAC including loss of weight, abdominal pain, nausea, and vomiting resemble symptoms of other diseases [4] and this often results in late-stage diagnosis. Out of the >350,000 people that are diagnosed annually with pancreatic cancer worldwide, more than 340,000 die from the disease [2]. Treatments of various solid cancers including pancreatic cancer have improved [1] however, this has not significantly impacted the high mortality rates observed for PDAC patients. Despite the increasing data on genetic alterations and impairment of signaling pathways that lead to pancreatic tumorigenesis, this has not resulted in development of more effective therapies [5].
The orphan nuclear receptor 4A (NR4A) sub-family members of NR4A1 (Nur77), NR4A2 (Nurr1) and NR4A3 (Nor1) are stress/inflammation-induced immediate-early genes that play key functional roles in maintaining cellular homeostasis and in pathophysiology [6]. It has previously been reported that offspring from the crossing of NR4A1-/- and NR4A3-/- rapidly develop symptoms of acute myeloid leukemia and both receptors exhibit tumor suppressor-like activities in blood-derived cancers [7]. In contrast there is extensive data in solid tumors that both NR4A1 and NR4A2 are pro-oncogenic and exhibit tumor promoter-like activity [8]; NR4A3 has not been extensively studied in solid tumors but there is evidence for tumor suppressor activity or lack of any effects [9,10]. Research in our laboratory has focused on the pro-oncogenic role of NR4A1 and NR4A2 in solid tumors and development of ligands for these receptors that act as inverse agonists that inhibit these receptors. Functional assays based on receptor knockdown studies show that both NR4A1 and NR4A2 play a role in solid tumor-derived cancer cell proliferation, survival, migration and invasion and regulation of genes and gene products that mediate these responses [6]. For example, NR4A1 regulates genes that maintain low cellular redox levels and knockdown of NR4A1 or treatment with bis-indole derived (C-DIM) NR4A1 inverse agonists increased ROS and ROS-dependent stress and inhibited mTOR signaling [11-15]. In pancreatic cancer cells, this response was due, in part, to downregulation of pro-reductant genes thioredoxin domain containing 5 (TXNDC5) and isocitrate dehydrogenase-1 (IDH1) [15]. NR4A2 also regulates glioblastoma and pancreatic cancer cell growth, survival, migration and invasion, and ongoing studies in these cell lines show that NR4A1 and NR4A2 regulate comparable pro-oncogenic pathways and some of the same genes [10,15-17]. Our recent study also showed that NR4A2 was overexpressed in PDAC patients compared to non-tumor tissue and was also a negative prognostic factor for PDAC patient survival and regulated chemoresistance and autophagy in pancreatic cancer cell lines [17].
The RNA binding protein Hu-antigen R (HuR) exhibits many of the same pro-oncogenic functions reported for NR4A2, and also regulate some of the same genes including survivin, β-catenin, Slug, Zeb1 and IDH1 [15,16,18-25]. Since the functional activities of NR4A2 and HuR are comparable, the aim of this study was to investigate possible NR4A2-HuR interactions. Our results show that HuR and NR4A2 interact in pancreatic cancer cells and that NR4A2 regulates HuR expression and vice versa. Moreover, the NR4A2 ligand, 1,1-bis(3’-indolyl)-1-(4-chlorophenyl)methane (C-DIM12) downregulates HuR and thus targeting NR4A2 represents a novel chemotherapeutic approach for targeting HuR in cancer cells.
Materials and methods
Cell lines and transfection with small-interference RNA and plasmids
MiaPaCa2, HPNE and Panc1 pancreatic cancer cell lines were obtained from ATCC (American Type Culture Collection) and were cultured in in DMEM supplemented with 10% FBS (Gibco/Invitrogen), 1% L-glutamine (Gibco/Invitrogen), and 1% penicillin-streptomycin (Invitrogen, Carlsbad, CA) at 37°C in 5% humidified CO2 incubators. CRISPR/Cas9-mediated knockout of NR4A2 in MiaPaCa2 cells was accomplished using guide RNAs targeting NR4A2, fused with CRISPR/Cas9 and GFP protein as previously described [17]. CRISPR Universal Negative Control plasmid (CRISPR06-1EA) were purchased from Sigma-Aldrich (St. Louis, MO). Cells were harvested after 48 hours of transfection and GFP positive cells were single sorted by using a FACS Calibur flow cytometer and cells which did not express NR4A2 were isolated as described [17]. The guide RNA sequences used were: NR4A2-1 (gatcccgggtcgtcccacat), NR4A2-2 (gggcttgtagtaaaccgacc). For all cell cultures experiments the mycoplasma detection kit MycoAlert (Lonza) was performed after each thawing and at least monthly to ensure mycoplasma free cells were used.
Cells were plated at 60% confluency in 6-well plates, and transient transfections were performed using Lipofectamine 3000 (Invitrogen), and Opti-MEM (Invitrogen) according to the manufacturer’s protocol; 48 hours after transfection, cells were treated or analyzed, as described. Small interfering RNA (siRNA) oligos were purchased from Invitrogen and are summarized in Supplementary Table 1.
Cell growth and Annexin V staining assays
Cells were plated in 96-well plates at 1×103 cells per well. After 5 days of incubation, cell growth was measured using Quant-iTTM PicoGreenTM dsDNA assay kit (Invitrogen). To estimate cell death, cells were trypsinized and counted after Trypan blue staining (Invitrogen) with a Hausser bright-line hemocytometer (Fisher Scientific). Annexin V/PI staining was performed using the Dead Cell Apoptosis Kit (ThermoFisher Scientific #V13245, Waltham, MA), according to the manufacturer’s instructions. Staining was measured with an Accuri C6 flow cytometer and analyzed with FlowJo Version 10.2 software.
Immunoblot analysis
Cells were lysed using 1% Triton in TBS containing protease and phosphatase inhibitors. Tumors were lysed using 1X RIPA buffer containing protease and phosphatase inhibitors. Equal amounts of total protein were separated by electrophoresis on a 4-12% Bis-Tris gel and transferred to a PVDF membrane. Blots were blocked in 5% BSA, and then probed with antibodies against anti-NR4A2 (sc-376984, SCB), anti-IDH1 (ab184615, Abcam), anti-HuR (sc-5261 clone 3A2, SCB), anti-Sp1 (ab124804, Abcam), Sp3 (sc-644, SCB), Sp4 (sc-645, SCB) and anti-α-tubulin (21445, Cell Signaling Technology). Chemiluminescent (32106, Thermo Fisher Scientific) signals were captured using a Syngene G-BOX iChemi XT imager.
Chromatin immunoprecipitation assay
Cells were cross-linked for 10 minutes at room temperature by addition of a solution containing 11% formaldehyde in 50 mM HEPES pH 7.4, 100 mM NaCl, 1 mM EDTA pH 8.0, 0.5 mM EGTA pH 8.0, followed by 5 minutes quenching with a solution of 2.5 M glycine. Cells were then washed with PBS (2X), the supernatant was aspirated, and the cell pellet was flash frozen in liquid nitrogen and stored at -80°C until used. Antibody precipitation, reversing crosslinks, sonication and DNA purification were carried out as described [10,17]. The primers are summarized in Supplementary Table 1.
Boyden chamber invasion assay and migration assay
Cell suspension (50×103 cells in 500 µl 2.5% FBS DMEM) was added on the CorningTM Boyden Invasion chamber with CorningTM Matrigel Matrix (Life Sciences; 354480) disc and 500 µl 10% FBS DMEM was added to the well. Cells were incubated at 37°C/5% CO2 and allowed to invade through the porous membrane. After 24 hours, the discs were taken out of the medium and placed in 500 µl 0.5% crystal violet for 10-15 minutes at room temperature. The discs were washed and dried for 24 hours. For migration assays, cells were seeded at 60-70% confluency in a 6-well plate. After 24 hours, cells were transfected with siNR4A2 or NR4A2 overexpression plasmid using Lipofectamine 2000 (Invitrogen; 11668-027) and Opti-MEM (Gibco; 31985-062). After 24 hours of seeding, the cell monolayers were scratched using micropipette tips and the photographs of the scratch on each plate were taken at 0 and after 48 hours by using EVOS FL microscope and the ratio of migrated cells was analyzed by using the ImageJ software (National Institutes of Health, Bethesda, MD).
Co-immunoprecipitation assay
Co-immunoprecipitation assay was carried using Pierce Co-Immunoprecipitation kit (Pierce; 26149). The AminoLink Plus Coupling Resin provided in the kit was used for coupling the anti-NR4A2 and anti-Sp4 antibodies following the manufacture’s protocol. Protein lysate was extracted from MiaPaCa2 cells using the provided IP Lysis/Wash Buffer and all Co-IP steps were performed according to the manufacture’s protocol. The total cell lysate and Co-IP samples were then used to detect NR4A2 and Sp4 on SDS gel for western blot analysis using the anti-NR4A2 or anti-Sp4 antibodies.
Clonogenic assay
Cells (1000-2000/well) were plated in a 6-well plate. The media was not changed during experiments unless indicated. Upon completion of the experiments, colonies were fixed in reagent containing 80% methanol and stained with 0.5% crystal violet. To determine relative growth, dye was extracted from stained colonies with 10% acetic acid and the associated absorbance measured at 600 nm using a Microplate Reader/Synergy HT BioTek plate reader.
Xenograft tumor growth studies
Athymic nude mice (Female, Nude Foxn-1 nu, 6-8 weeks old) were ordered from EN-VIGO and housed in a pathogen-free environment with vented cages. The mice were housed in Texas A&M University’s animal facility with the standard of AAALAC. The protocol of the study was approved by IACUC (#2020-0138), Texas A&M University. Mice were allowed to acclimatize for 1-2 weeks before the experiment. The animals (10 per treatment group) were fed with standard chow diet and water during the experiment. MiaPaCa2 cells were cultured at a density (4×106) in 10% FBS DMEM. The cells were harvested and resuspended in 100 µl of PBS with Matrigel. Mice (10 per treatment group) were injected with MiaPaCa2 cells (4×106) subcutaneously. In two weeks when tumors grew to a size of 100 mm3, mice were administered C-DIM12 (50 mg/kg) or vehicle control by intraperitoneal injections three times a week for a period of 6 weeks. Some of the mice were observed for their survival and tumor growth after the final injection.
Mice were also injected with wild-type and CRISPR/Cas9-derived NR4A2-knockout MiaPaCa2 cells and tumor growth was evaluated for 4 weeks. Some mice were observed for tumor-free survival after the end of the experiment. Tumor volumes and body weights were determined periodically; a Vernier caliper was used to measure the size of the tumor, and tumor volume was determined by the formula (Volume = Length × Width2/2). At the end of the experiment, the mice were euthanized under deep isoflurane anesthesia. The tumors were resected and stored in 10% neutral buffer formalin for immunohistochemistry or stored at -80°C until processed for protein and RNA analysis.
siRNA and cDNA transfection
MiaPaCa2 cell were seeded on a 6-well plate. After 24 hours the transfection mixture containing siNR4A2 (1 µmol/L)/ siHuR (1 µmol/L) or NR4A2 overexpression plasmid (1 µg) along with Lipofectamine 2000 (Invitrogen; 11668-027) and Opti-MEM (Gibco; 31985-062) was added. After 48 hours, cells were harvested for protein and RNA analysis. Overexpression (NOE) and empty vector were used as previously described [17].
RNP-IP and RT-qPCR
Cells were plated at 50% confluency in 100 mm dishes. The following day, immunoprecipitation was performed using either anti-HuR or IgG control antibodies as previously described [23]. For RT-qPCR, the RNA lysate was extracted from the cultured cells using RNeasy Mini Kit (Qiagen; 74104 and 74106) and cells were harvested according to the manufacture’s protocol. The cDNA was extracted by using High-Capacity cDNA Reverse Transcription Kits as per manufacture’s protocol (Applied Biosystems; 4368813, 4368814, 4374966, and 4374967). The RT-qPCR was performed by using Taqman™ Universal Master Mix II (Thermo Fisher Scientific; 4440038) with probe (NR4A2 probe- Thermo Fisher Scientific; 4331182, ELAVL1 probe- Life Technologies; 4453320, IDH1 probe- Thermo Fisher Scientific; 4351372) and cDNA and was analyzed by using the Bio-Rad CFX Maestro software. Oligonucleotides are summarized in Supplementary Table 1.
RNA sequencing
RNA quality was assessed via the Agilent 2100 Bioanalyzer (Agilent Technologies). Strand-specific RNA-seq library was prepared using NEBNext Ultra II Directional RNA Library Prep Kit (NEB, Ipswich, MA) according to the manufacturer’s protocols. RNA-sequencing was performed using 150-bp paired-end format on a NovaSeq 6000 (Illumina) sequencer. RNA-seq quality was checked by running FastQC, and TrimGalore was used for adapter and quality trimming. Sequence reads were aligned to the hg19 human genome build using the STAR aligning program [26]. Quantification of all genes and their isoforms was performed using FPKM normalized values using Cufflinks v2.2.1, DESeq2 analysis with an adjusted P-value <0.05 was used to get a list of differentially expressed genes [27].
Immunohistochemistry
IHC staining was performed on paraffin embedded tissue sections. Sections were incubated with Histoclear (National Diagnostics; HS-200) for 5 minutes (3X), 100% ethanol (2X) and in 95% ethanol (2X) for 10 minutes each, then with purified water (2X) for 5 minutes. Antigen retrieval was performed by boiling for 10 minutes in 10 mM sodium citrate pH 6.0, followed by washing in water, then 3% hydrogen peroxide for 10 minutes, then water. 1X TBST + 2.5% normal goat serum (Vector; MP-7451) was used as a blocking solution and the following primary antibodies were diluted in Signal Stain Antibody Diluent (Cell Signaling technology; 8112) and incubated overnight at 4°C: Ki67 (Abcam; ab16667), c-Casp3 (Cell Signaling Technology; 9664). Slides were then washed for 5 minutes in 1X TBST (3X) and incubated in Signal Stain Boost IHC Detection Reagent (Cell Signaling Technology; 8114) for 30 minutes at room temperature, followed by washes in 1X TBST (3X) for 5 minutes. The signal was detected using the AEC Peroxidase (HRP) Substrate Kit (Vector; SK-4200) according to manufacturer’s instructions. Slides were counterstained with hematoxylin (Vector; H3401). Coverslips were mounted with Permount Mounting solution (Thermo Fisher Scientific; 00-4958-02). Terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) staining was performed following the In-Situ Apoptosis Detection Kit (Trevigen; 4810-30-K) with 40-fold magnification as described [17].
Statistical analysis
Data were expressed as mean ± standard error of the mean (SEM) of at least three in-dependent experiments. An unpaired, two-tailed student t test was used to determine the differences between groups (*P<0.05; **P<0.01; ***P<0.001). ANOVA test was used for the analysis of tumor measurements among treated groups.
Results
NR4A2 expression plays a key role in proliferation, invasion, and migration of pancreatic cancer cells
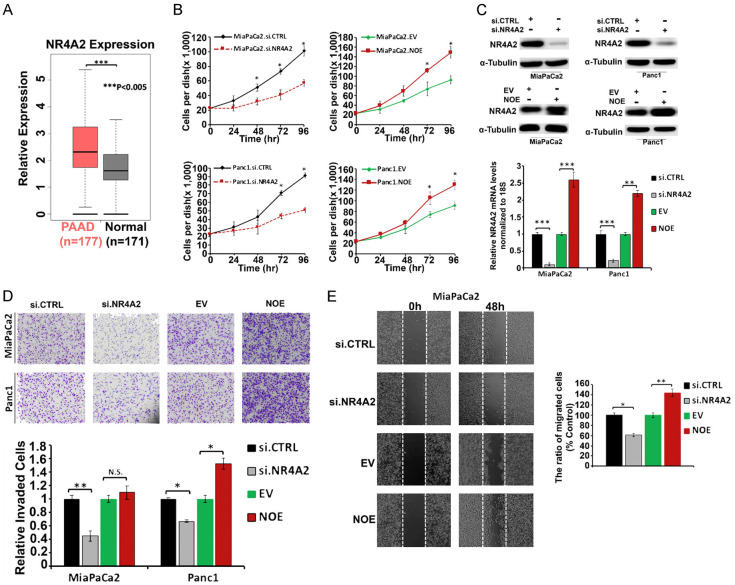
Analysis of the TCGA database showed that NR4A2 was highly expressed in tissues from PDAC cancer patients compared to normal donors, and the high expression of NR4A2 (Figure 1A) in tumors corresponded to poor patient survival (Ρ<0.05) [17]. The functional effects of NR4A2 were determined by gene silencing or overexpression (OE). Figure 1B shows that silencing of NR4A2 by RNA interference resulted a significant decrease in proliferation of MiaPaCa2 and Panc1 pancreatic cancer cells after treatment for 72-96 hours whereas transfection of these cells with an NR4A2 overexpression plasmid significantly increased proliferation of both cell lines after 72 and 96 hours (Figure 1B). Figure 1C illustrates the effects of knockdown/overexpression on NR4A2 levels in MiaPaCa2 and Panc1 cells. Results obtained in Boyden chamber assay showed that NR4A2 silencing (Figure 1D) reduced invasion by 55% in MiaPaCa2 and by 40% in Panc1 cells (Figure 1D). Overexpression of NR4A2 increased the invasion in Panc1 cells however the effects on invasion of MiaPaCa2 cells was not significant (Figure 1D). In the wound healing assay, NR4A2 silencing by RNA interference resulted in a significant decrease in migration whereas NR4A2 overexpression resulted in an increase in migration in Panc1 cells (Figure 1E). These data demonstrate that NR4A2 plays a significant pro-oncogenic role in pancreatic cancer cell proliferation, invasion and migration and these results are comparable to those previously observed after NR4A2 knockdown in pancreatic cancer and glioblastoma cells [10,17].
Figure 1.
NR4A2 plays an important role in pancreatic cancer cell survival and migration. A. Expression of NR4A2 in PDAC patient’s tissues as compared to that in normal pancreas tissues, *P<0.05 (data extracted from TCGA database). B. MiaPaCa2 and Panc1 cell proliferation after RNA silencing of NR4A2 (siNR4A2) and overexpression of NR4A2 (NOE) compared with controls (siCTRL and EV) was determined over time. C. Western blot and RT-qPCR analysis for NR4A2 expression in MiaPaCa2 and Panc1 cells after NR4A2 overexpression or knockdown. D. MiaPaCa2 and Panc1 cell images (4× magnification) and quantification of invaded cell numbers in Boyden chamber assay. E. MiaPaCa2 cell migration Images (4× magnification) and quantification of migrated cells in the scratch assay after NR4A1 knockdown or overexpression (NOE) in MiaPaCa2 cells. Quantitated results are means ± SE for at least 3 replicate determinations; significance, *P<0.05, **P<0.01, ***P<0.001.
NR4A2 regulates HuR and IDH1 expression in pancreatic cancer cells
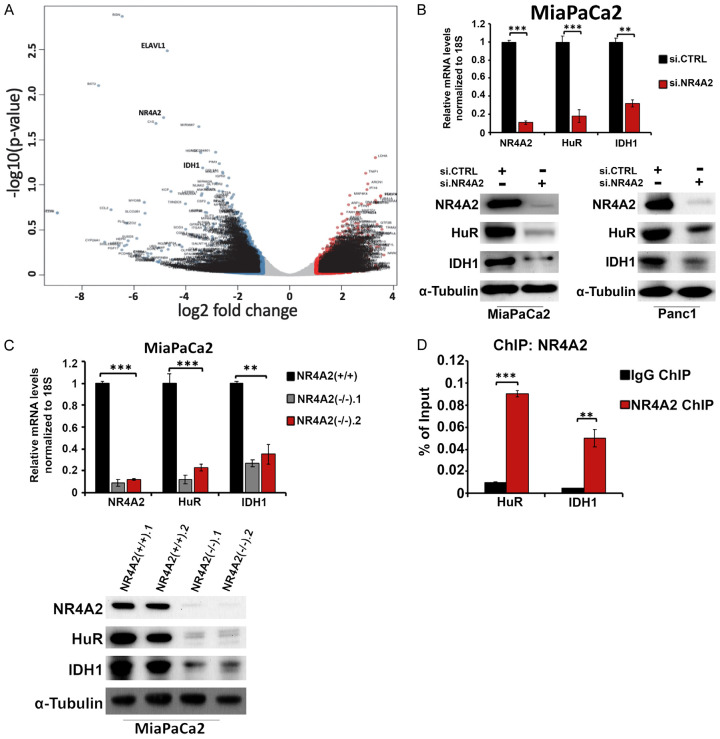
To identify genes regulated by NR4A2 we performed RNA sequencing (RNA-Seq) in MiaPaCa2 cells. NR4A2 expression was deleted by gene editing using CRISPR/Cas9 system as previously described [14]. Figure 2A illustrates a volcano plot of the results and subsequent data analysis showed that loss of NR4A2 was associated with downregulation of IDH1 and HuR. According to Metabolite Set Enrichment Analysis pathways and gene associated with the tricarboxylic acid cycle and NADPH synthesis pathways were also affected by NR4A2 knockdown (Supplementary Figure 1A and 1B) and these are currently being investigated. Based on results of previous studies showing that NR4A1 and HuR regulated IDH1 expression [17,20], we focused mechanistic analysis of both the HuR and IDH1 genes which were significantly downregulated in MiaPaCa2 cells after knockdown of NR4A2 by RNA interference. HuR encodes an RNA-binding protein that maintains mRNA stability and IDH1 encodes an enzyme that catalyzes the reversible conversion of isocitrate and α-ketoglutarate, with the production of NADPH [18-25,28]. To validate our RNA sequencing data, we showed that NR4A2, HuR and IDH1 levels were decreased in MiaPaCa2 cells after NR4A2 knockdown by RNA interference (Figure 2B). The western blot data was consistent with the RT-qPCR results and showed that NR4A2 knockdown decreased expression of HuR and IDH1 proteins in MiaPaca2 and Panc1 cells (Figure 2B). The western blot and RT-qPCR analysis of wild-type (NR4A2+/+) and 2 different NR4A2 knockdown (NR4A2-/-) (by RNA interference) cell lines also showed a significant downregulation of HuR and IDH1 mRNA and protein expression in the knockout cell lines (Figure 2C). Interactions of NR4A2 with the HuR and IDH1 gene promoters was carried out by ChIP analysis in MiaPaCa2 cells. Immunoprecipitation with antibodies showed enrichment of NR4A2 on the HuR and IDH1 promoters (Figure 2D) and these results demonstrate that NR4A2 regulates expression of HuR and IDH1 in MiaPaCa2 cells.
Figure 2.
NR4A2 regulates HuR (ELAVL1) and IDH1 in pancreatic cancer cells. A. RNA sequencing was performed in MiaPaCa2 cells (gene edited by CRISPR-Cas9) [14] and MiaPaCa2 wildtype. Volcano plot showing significant changes in gene expression in NR4A2(-/-) cells as compared to NR4A2(+/+) cells. B. Western blot analysis and RT-qPCR analysis of HuR and IDH1 after silencing of NR4A2. C. Western blot and RT-qPCR analysis of HuR and IDH1 expression after CRISPR knockout of NR4A2 in MiaPaCa2 cells. D. ChIP analysis in MiaPaCa2 for HuR and IDH1 (HuR and IDH1 promoter) bound to NR4A2 protein relative to IgG as determined by RT-qPCR. Quantitated results are means ± SE for at least 3 replicate determinations; significance, **P<0.01, ***P<0.001.
NR4A2 regulation of HuR and IDH1 through specificity protein (Sp) transcription factors in pancreatic cancer cells
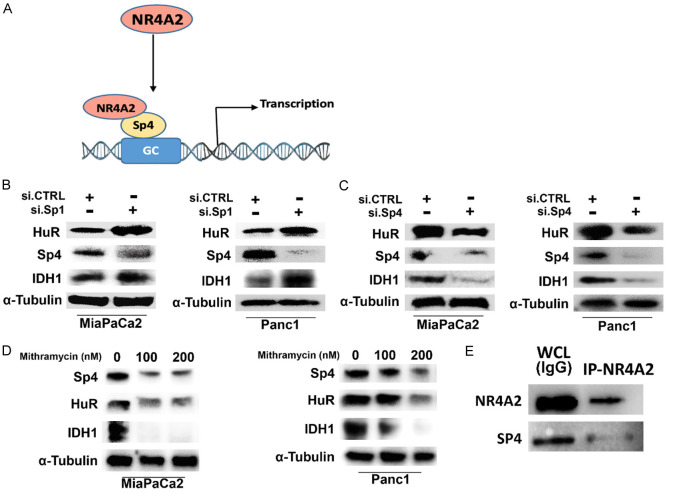
There is evidence that NR4A2 and NR4A1 regulate similar functions and genes in pancreatic and other cancer cell lines and previous studies show that NR4A1 acts as a ligand-dependent co-activator of several Sp regulated genes including survivin, PAX3-FOX01, β1- and β3-integrins and α5- and α6-integrins [16,29-32]. Mechanism studies showed that these genes were regulated by NR4A1/Sp1 or NR4A1/Sp4 binding to GC-rich gene promoters. Both the HuR and IDH1 gene promoters contain GC-rich Sp binding sites and in the absence of cognate NR4A2 binding sequences we hypothesize that HuR and IDH1 may also be NR4A2/Sp1- or NR4A2/Sp4-regulated genes (Figure 3A). Knockdown of Sp1 in MiaPaCa2 cells was minimal (Figure 3B); similar results were observed with another oligonucleotide (data not shown) and levels of HuR and IDH1 were not decreased. Surprisingly, in Panc1 cells Sp1 silencing decreased Sp1 but increased levels of IDH1 and HuR. The reason for increased expression of IDH1 and HuR is not known however the results indicate that both genes are not regulated by NR4A2/Sp1. In contrast, knockdown of Sp4 in MiaPaCa2 and Panc1 cells decreased levels of Sp1, IDH1 and HuR gene products (Figure 3C) suggesting that NR4A2/Sp4 plays a role in regulation of IDH1 and HuR in MiaPaCa2 and Panc1 cells. This mechanism is further supported by the observation that mithramycin, which blocks binding of Sp1 and Sp4 to GC-rich sites [33] also decreased levels of Sp4, HuR and IDH1 in MiaPaCa2 and Panc1 cells (Figure 3D). These results are consistent with previous studies on NR4A1/Sp4 regulation of other genes [12,29-32]. We also observed in co-immunoprecipitation studies that NR4A2 antibodies co-immunoprecipitated both NR4A2 and Sp4 (Figure 3E); similar results were observed for Sp4 antibodies, however the NR4A2 band was very weak (data not shown).
Figure 3.
NR4A2 regulated expression of HuR and IDH1 involves interactions with Sp transcription factors. (A) Schematic representation of NR4A2 binding to Sp4 and regulating gene transcription. (B) Expression of gene products after RNA silencing of Sp1, (C) Sp4 and (D) treatment with mithramycin in pancreatic cancer cell lines (MiaPaCa2 and Panc1). (E) Co-immunoprecipitation of NR4A2 and Sp4 using NR4A2 antibodies and subsequent western blot analysis as outlined in the methods. Sp4 antibodies also co-immunoprecipitated Sp4 and NR4A2 however the NR4A2 band was weak (data not shown).
Bis-indole derived C-DIM12/NR4A2 inverse agonist inhibits pancreatic cancer cell proliferation and migration
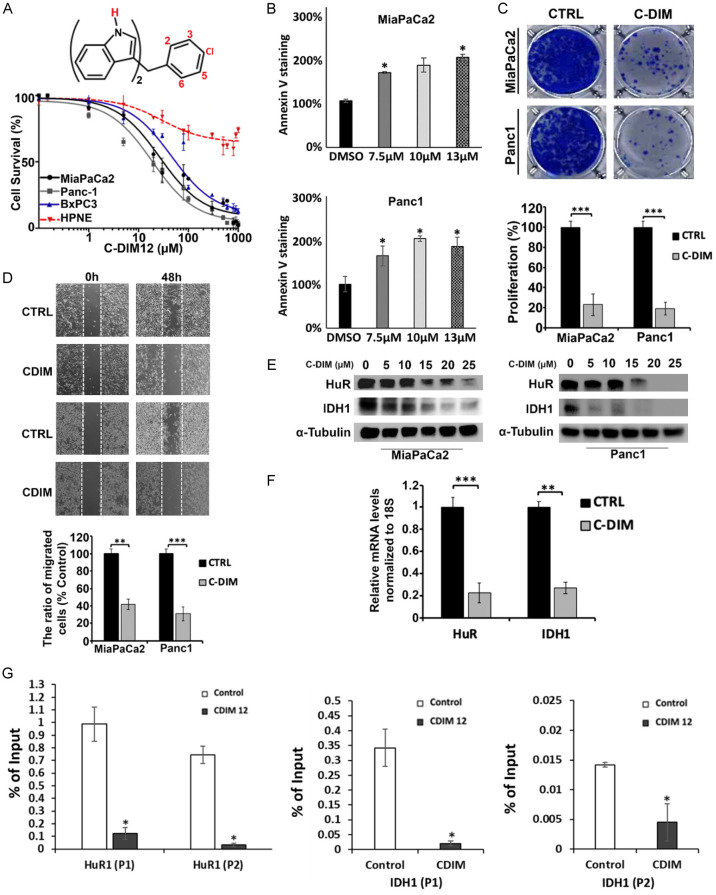
Previous studies showed that 1,1-bis(3’-indolyl)-1-(ρ-substituted phenyl) methane (C-DIM) analogs inhibited growth and induce apoptosis in cancer cell lines by acting as NR4A1 or NR4A2 inverse agonists [7]. Specifically, 1,1-bis(3’-indolyl)-1-(ρ-chlorophenyl) methane (DIM-C-ρPhCl or C-DIM12) acts as an NR4A2 inverse agonist in cancer cells [10,17,34] and treatment of MiaPaCa2, Panc1, BxPC3 pancreatic cancer cells and HPNE normal pancreatic ductal cells with C-DIM12 (1-1000 µM) preferentially decreased % survival in the cancer cell lines in a dose-dependent manner whereas this decrease was attenuated in non-transformed HPNE cells (Figure 4A). We also observed that treatment with 15 µM DIM12 induced MiaPaCa2 and Panc1 cell death as indicated by increased Annexin V staining (Figure 4B). A soft agar clonogenic assay in MiaPaCa2 and Panc1 cells showed that C-DIM12 (15 µM) inhibited anchorage-independent growth over a period of 4 weeks (Figure 4C) and in a scratch assay 15 µM C-DIM12 decreased migration of MiaPaCa2 and Panc1 cells by 60-70% over a period of 48 hours (Figure 4D). Western blot analysis showed that C-DIM12 (5-25 µM) downregulated expression of NR4A2 targets, HuR and IDH1 proteins in MiaPaCa2 and Panc1 cells (Figure 4E) and mRNA in Panc1 cells (Figure 4F). Moreover, we show in Figure 4G that treatment with C-DIM12 decreases NR4A2 interaction with the HuR and IDH1 gene promoter (2 primer sets for each gene) in a ChIP assay. These results demonstrate that the NR4A2 inverse agonist C-DIM12 inactivates NR4A2- dependent transcriptional activity and the effects of this compound in terms of function and gene expression (HuR and IDH1) overlaps those observed after NR4A2 knockdown (Figures 1, 2 and 3).
Figure 4.
C-DIM12 inhibits cell proliferation and migration and downregulates expression of HuR and IDH1. (A) The structure of DIM-pPhCl (C-DIM12) and percentage of cell survival after treatment with 0-1000 μM of C-DIM12 for 24 hours. (B) MiaPaCa2 and Panc1 cells were treated with 15 μM C-DIM12 for 24 hours and apoptosis was measured by Annexin V staining (images were taken at 4× magnification). (C) Soft agar colony formation assay was determined in pancreatic cancer cells treated with 15 μM C-DIM12 for 4 weeks. (D) Cell migration images and quantification of migrated cells in a scratch assay after treatment of pancreatic cancer cells with C-DIM12 for 0 or 48 hours. (E) Western blot and (F) RT-qPCR analysis of HuR and IDH1 levels in pancreatic cancer cells after treatment with C-DIM12 for 24 hours. (G) ChIP assay results showing that C-DIM12 decreases interaction of NR4A2 with the HuR and IDH1 gene promoter (2 primer sets used for each gene). Quantitated results are means ± SE for at least 3 replicate determinations; significance, *P<0.05, **P<0.01, ***P<0.001.
HuR regulates NR4A2 expression
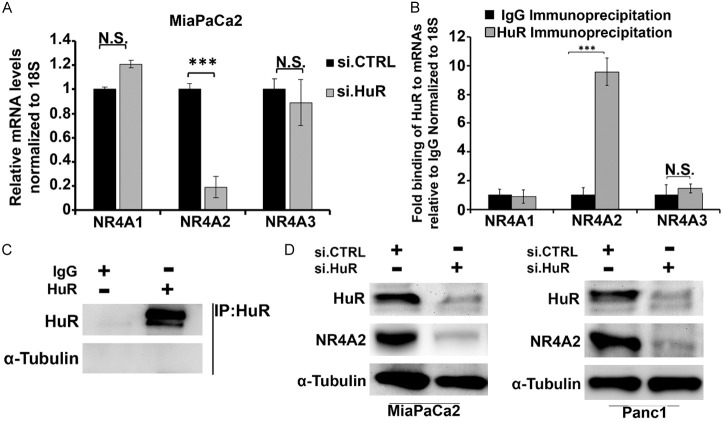
Since HuR also plays an important role in regulation of gene expression, we investigated the effects of HuR silencing on expression of NR4A family members NR4A1, NR4A2 or NR4A3 and observed that NR4A2 but not NR4A1 and NR4A3 expression were affected by HuR knockdown (Figure 5A). Immunoprecipitation of MiaPaCa2 cell lysate with HuR antibodies showed NR4A2 but not NR4A1 or NR4A3 mRNAs were enriched in the precipitate (Figure 5B) and the antibody specificity is indicated in Figure 5C. This confirms that among NR4A proteins NR4A2 preferentially is targeted by and interacts with HuR. Knockdown of HuR by RNAi also significantly decreased NR4A2 protein in MiaPaCa2 and Panc1 cells (Figure 5D). Bioinformatics analysis shows that there are 2-4 HuR binding sites on the 3’UTR of NR4A2 however studies in MiaPaCa2 cells with the 3’-UTR of NR4A2 did not reveal any functional interactions in HuR with the NR4A2- 3’-UTR and the mechanism of NR4A2 regulation by HuR are currently being investigated.
Figure 5.
HuR regulates the expression of NR4A2. (A) Relative mRNA levels of NR4A1, NR4A2 and NR4A3 in MiaPaCa2 cells after transfection with siCTRL or siHuR. (B) Immunoprecipitation analysis in MiaPaCa2 for NR4As mRNAs bound to HuR protein relative to IgG and (C) preferential immunoprecipitation of HuR by HuR antibodies and not IgG. (D) Western blot analysis of HuR and NR4A2 expression in MiaPaCa2 and Panc1 cells transfected with siCTRL (control/non-specific) and siNR4A2. Quantitated results are means ± SE for at least 3 replicate determinations; significance, ***P<0.001.
NR4A2 knockout suppresses tumor growth in vivo
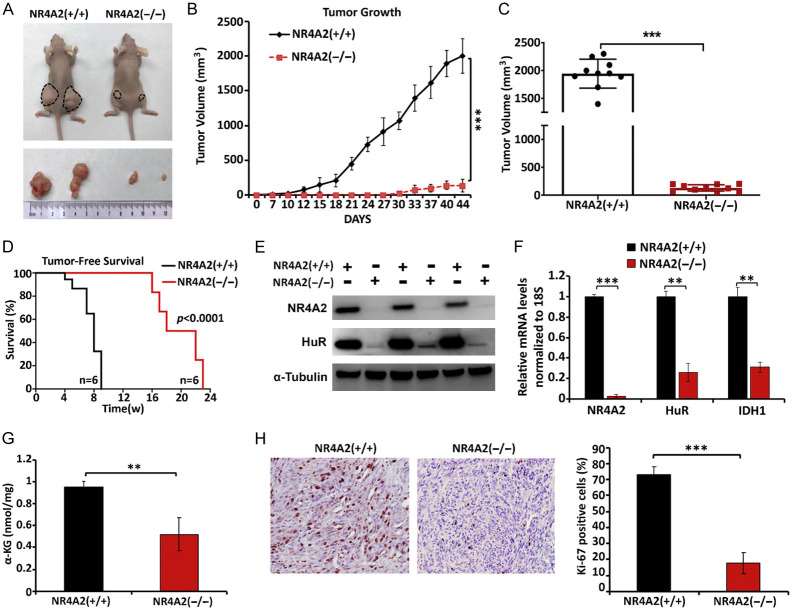
To determine the effect of NR4A2 expression on pancreatic tumor growth in vivo, we used female athymic nude mice bearing wild-type and CRISPR/Cas9-derived NR4A2-knockout MiaPaCa2 cells as xenografts. Mice bearing NR4A2 knockout cells exhibited a 12.6-fold decrease in tumor volume (150 mm3) compared to tumors derived from wild type cells (1200 mm3) (Figure 6A-C). At the end of the experiment, a second group of mice were monitored until tumors reached a size of approximately 2000 mm3 and the results show that loss of NR4A2 significantly enhanced tumor free survival (Figure 6D). Immunoblot analysis of the tumor lysates validated NR4A2 knockdown in NR4A2(-/-) group vs NR4A2(+/+) group and this was accompanied by downregulation of HuR which is consistent with our in vitro data (Figures 6E and 2D). Results of NR4A2 knockdown also decreased expression of HuR and IDH1 mRNA in tumors (Figure 6F) and this was also accompanied by decreased expression of α-ketoglutarate (αK6) (Figure 6G) which is dependent on IDH1- mediated conversion of isocitrate to α-ketoglutarate. Immunohistochemical analysis of the Ki-67, marker for cell proliferation showed that knockdown of NR4A2 also decreased the percentage of Ki-67 positive cells (Figure 6H) indicating that NR4A2 play a role in tumor growth.
Figure 6.
NR4A2 knockout inhibits tumor growth in vivo. (A) Mice were injected with wild-type and CRISPR/Cas9-derived NR4A2 knockout MiaPaCa2 cells and tumor growth was monitored for 6 weeks. Images represents the tumor size at the end of the experiment. (B) Tumor growth associated with wild-type and NR4A2 knockout MiaPaCa2 cells as xenografts and (C) tumor volumes determined 6 weeks after initial injection. (D) Mice with tumors were examined for tumor free survival until the tumors were palpable. (E) Western blot analysis of HuR expression in tumors derived from wild-type (NR4A2+/+) and knockout (NR4A2-/-) MiaPaCa2 cells as xenografts. (F) Relative mRNA levels of NR4A2, HuR and IDH1 normalized to mRNA levels of 18S and (G) α-ketoglutarate levels in tumors derived from wild-type and NR4A2 knockout MiaPaCa2 cells. (H) Immunohistochemistry images and Percentage analysis of Ki-67 in 10% neutral buffer formalin fixed and paraffin embedded (FFPE) tissue sections of tumors derived from wild-type and NR4A2 knockout MiaPaCa2 cell xenografts. Quantitated results are means ± SE for at least 3 replicate determinations; significance, **P<0.01, ***P<0.001.
NR4A2 inverse agonist has a potent effect on pancreatic cancer cells, in-vivo
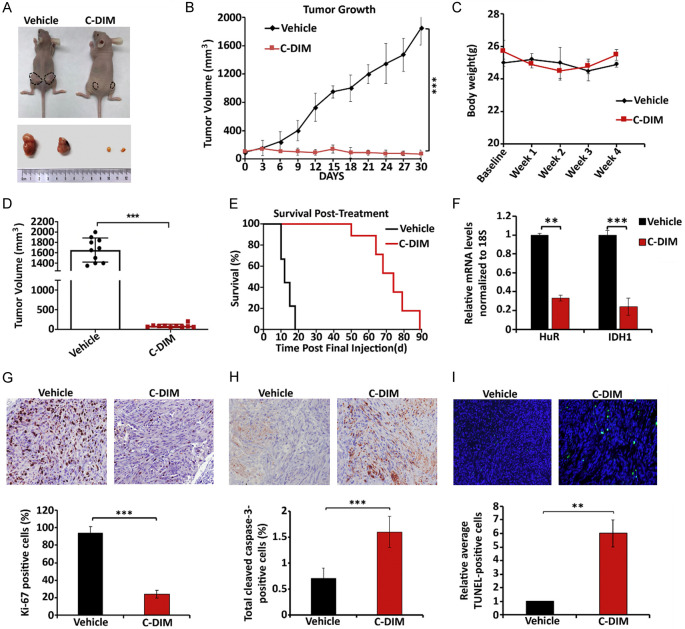
We also investigated effects of the NR4A2 inverse agonist C-DIM12 on pancreatic tumors in an athymic nude mouse model. Mice bearing MiaPaCa2 cells as xenografts were injected (ip) with C-DIM12 (50 mg/kg) and vehicle (corn oil) three times a week. Over the course of the experiment, C-DIM12 significantly decreased tumor growth (Figure 7A and 7B) but did not affect body weight (Figure 7C); organ weights were also unchanged and pathology was unchanged. C-DIM12 also decreased tumor volumes (Figure 7D) and in a longer-term survival experiment where the drug treatment was terminated after 30 days, mice in the treatment group exhibited longer survival than the control mice (Figure 7E). We did not observe any changes in organ weights or organ damage, and this is consistent with results of previous studies with this compound [10,17,34-36]. Treatment with C-DIM12 downregulated expression of HuR and IDH1 (Figure 7F) and immunohistochemical analysis showed that C-DIM12-dependent inactivation of NR4A2 decreased expression of Ki-67 (Figure 7G). Tumors from the C-DIM12 treated mice exhibited an increased percentage of cleaved Caspase-3 positive cells (Figure 7H) and enhanced TUNEL staining (Figure 7I) indicating that the tumor growth inhibiting effects of C-DIM12 were also due to induced apoptosis. Results for the NR4A2 antagonist C-DIM12 paralleled those observed after NR4A2 knockdown in MiaPaCa2 cells (Figure 6).
Figure 7.
C-DIM12 inhibits tumor growth in vivo. (A) Mice were injected with MiaPaCa2 cells and were treated with vehicle and C-DIM12 (50 mg/kg) when tumors reached the size of 100 mm3. Images represents the tumor size at the end of the experiment. (B) Tumor growth curve for MiaPaCa2 xenografts in nude mice, treated with 50 mg/kg C-DIM12 or vehicle control 3 times a week and (C) body weight changes in nude mice, treated with 50 mg/kg C-DIM or vehicle control 3 times a week over a period of 4 weeks and (D) scatter plots showing tumor volumes in treated and control group determined 6 weeks after initial injection. (E) Survival was evaluated after the last injection of C-DIM12. (F) Relative HuR and IDH1 mRNA levels in tumors derived from MiaPaCa2 cell xenografts (vehicle control and C-DIM12 treated). (G) Percentage of Ki-67 positive cells in tumors from vehicle control and C-DIM12 treated mice bearing MiaPaCa2 cells xenografts and Ki-67 positive cells in tumors. (H) Percentage of caspase-3 positive cells in vehicle control and C-DIM12 treated mice and (I) relative average TUNEL-positive cells in vehicle control and C-DIM treated mice. Quantitated results are means ± SE for at least 3 replicate determinations; significance, *P<0.05, **P<0.01, ***P<0.001.
Discussion
PDAC is one of the deadliest tumors and is typically diagnosed in the later stages of tumor development and treatment options in terms of surgery and chemotherapy have limited effectiveness [1,4]. The sequence of oncogene expression and mutations of tumor suppressor genes that lead to formation and metastasis of pancreatic tumors has been determined and some of these genetic alterations and their functions have been confirmed in mouse models [38-40]. However, clinical development of specific agents that target pro-oncogenic factors expressed in pancreatic cancer has not been robust and despite its limited effectiveness gemcitabine remains the first line treatment for pancreatic cancer. It has previously been demonstrated that the orphan nuclear receptor NR4A1 is highly expressed in multiple solid tumors including pancreatic cancer and NR4A1 knockdown results in decrease tumor growth, survival migration and invasion (rev. in 8). There is also evidence that NR4A2 exhibits comparable pro-oncogenic functions in solid tumor-derived cell lines [41-49] and a recent report shows that NR4A2 regulates growth, survival, migration, and invasion of patient derived glioblastoma cells [10].
Analysis of genomic data showed that NR4A2 is more highly expressed in PDAC vs normal pancreas and patients with high expression of NR4A2 exhibit decreased overall survival (Figure 1A). Knockdown and overexpression of NR4A2 in pancreatic cancer cells demonstrate that NR4A2 is pro-oncogenic as observed in glioblastoma and other cell lines [10,41-49] and these results complement our recent study on the role of NR4A2 in pancreatic cancer [17]. NR4A1 and NR4A2 are primarily pro-oncogenic factors in solid tumors whereas results of breeding of NR4A1+/- with NR4A3-/- mice show that NR4A1 is a tumor suppressor in blood-derived tumors [7] and the reasons for differences in the roles of NR4A sub-family members in blood-derived and solid tumors is unknown. However, in pancreatic cancer cells both NR4A2 and NR4A1 exhibit pro-oncogenic activity and regulate cell growth survival and invasion/migration and ligands for both receptors inhibit these responses. Thus, both NR4A2 and NR4A1 ligands that act as receptor inhibitors or inverse agonist in pancreatic cancer cells represent novel mechanism-based anticancer agents with potential for clinical applications for treating patients overexpressing the orphan receptors.
NR4A2 regulated genes that dictate the pro-oncogenic functions of this receptor include genes associated with autophagy [17] and changes in gene expression after knockdown of NR4A2 were further analyzed in MiaPaCa2 cells. Metabolite Set Enrichment Analysis indicated that NR4A2 regulated genes are involved in multiple pathways including the tricarboxylic acid cycle. Previous studies reported that both HuR and IDH1 were highly expressed in pancreatic tumors and regulation of IDH1 by HuR plays a role in pancreatic cancer cell survival [5,18,20,49,50]. Results of RNAseq show that both IDH1 and HuR are decreased after NR4A2 knockdown, and we further investigated the mechanisms of NR4A2 regulation of HuR/IDH1 and the potential for targeting both pro-oncogenic factors with the NR4A2 inverse agonist C-DIM12. The results of RNA interference studies show that NR4A2 directly regulates HuR and IDH1 gene expression through interactions of NR4A2 with their respective promoters (Figure 2). Moreover, treatment of pancreatic cancer cells with C-DIM12 mimicked the effect of NR4A2 knockdown and decreased interaction of NR4A1 with the HuR and IDH1 promoters (Figure 4) demonstrating that the NR4A2 ligands can be used to simultaneously target both HuR and IDH1 in pancreatic cancers. These results complement previous studies in pancreatic cancer cells showing the NR4A1 also regulated IDH1 expression and an NR4A1 inverse agonist decreased expression of this gene [16]. Mechanistic studies show that NR4A1 regulation of several genes in pancreatic and other cancer cell lines involves NR4A1/Sp interactions at GC-rich promoter sites where NR4A1 acts as a nuclear cofactor of Sp1 or Sp4 [12,16,29-32]. This type of gene regulation has been observed for many other nuclear receptors which enhance Sp (Sp1 or Sp4) regulated gene expression by targeting cis-acting GC-rich elements in target gene promoters [51]. Results of Sp knockdown studies and treatments with mithramycin that blocks Sp-DNA interactions confirm that NR4A2/Sp4 regulates expression of HuR and IDH1 by direct interactions with GC-rich sequences in the HuR and IDH1 promoters (Figure 3). Thus, IDH1 expression is inhibited directly by the NR4A2 inverse agonist and indirectly by parallel downregulation of HuR. Kang and coworkers previously showed the NF-κB regulated expression of HuR in gastric cancer cells [52] whereas C-DIM12/NR4A1 inhibited NF-κB in neuronal cell lines [53]. In this study we did not investigate the role of NF-κB in the NR4A2-dependent inhibition of HuR expression in pancreatic cancer cells. Differences in the effects of NF-κB on regulation of HuR could be due to several possibilities including cell context dependent variability of nuclear factors and chromatin structure and these will be investigated in future studies.
Previous studies have characterized the important functions of HuR in pancreatic cancer which include a role in enhancing cell growth, migration, and invasion [5,18,20,52]. Since these functions overlap with those observed for NR4A2 in this study we hypothesized that the functions of HuR may be due, in part, to regulation of NR4A2 expression. This hypothesis was confirmed by studies showing that HuR specifically regulates and interacts with NR4A2 but not NR4A1 or NR4A3 expression in pancreatic cancer cells (Figure 5) however the mechanisms of HuR regulation of NR4A2 were not determined and are currently being investigated. In this study we also demonstrate the crosstalk between NR4A2 and HuR and that drugs targeting either gene represent a unique approach for treating pancreatic cancer. These results complement our recent study which showed that NR4A2 regulated autophagy through the autophagic genes ATG7 and ATG12, and enhanced gemcitabine resistance and these could be reversed by knockdown of NR4A2 or treatment with C-DIM12 [17]. Thus, we have further demonstrated the efficacy of targeting NR4A2 with the NR4A2 inverse agonist C-DIM12 which is a potent inhibitor of pancreatic tumor growth (Figure 7). Our results in this study and our previous report showing that C-DIM12 decreased gemcitabine resistance in pancreatic cancer cells [17] suggest that the bis-indole derived NR4A2 ligands in combination with gemcitabine may be a highly effective combination therapy for treating pancreatic cancer patients.
Acknowledgements
We would like to thank the support of AgriLife Research and the Syd Kyle Chair endowment is gratefully acknowledged. This research was funded by the department of defense; DOD-W81XWH-18-0592 (M. Zarei) and the National Institutes of Health P30-ES029067 (S. Safe). All experiments involving mice were approved by Texas A&M University’s Animal Care and Use Committee (#2020-0138).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Adamska A, Domenichini A, Falasca M. Pancreatic ductal adenocarcinoma: current and evolving therapies. Int J Mol Sci. 2017;18:1338. doi: 10.3390/ijms18071338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Foucher ED, Ghigo C, Chouaib S, Galon J, Iovanna J, Olive D. Pancreatic ductal adenocarcinoma: a strong imbalance of good and bad immunological cops in the tumor microenvironment. Front Immunol. 2018;9:1044. doi: 10.3389/fimmu.2018.01044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grant TJ, Hua K, Singh A. Molecular pathogenesis of pancreatic cancer. Prog Mol Biol Transl Sci. 2016;144:241–75. doi: 10.1016/bs.pmbts.2016.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oberstein PE, Olive KP. Pancreatic cancer: why is it so hard to treat? Therap Adv Gastroenterol. 2013;6:321–37. doi: 10.1177/1756283X13478680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jimbo M, Blanco FF, Huang YH, Telonis AG, Screnci BA, Cosma GL, Alexeev V, Gonye GE, Yeo CJ, Sawicki JA, Winter JM, Brody JR. Targeting the mRNA-binding protein HuR impairs malignant characteristics of pancreatic ductal adenocarcinoma cells. Oncotarget. 2015;6:27312–31. doi: 10.18632/oncotarget.4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pearen MA, Muscat GE. Minireview: nuclear hormone receptor 4A signaling: implications for metabolic disease. Mol Endocrinol. 2010;24:1891–903. doi: 10.1210/me.2010-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mullican SE, Zhang S, Konopleva M, Ruvolo V, Andreeff M, Milbrandt J, Conneely OM. Abrogation of nuclear receptors Nr4a3 and Nr4a1 leads to development of acute myeloid leukemia. Nat Med. 2007;13:730–5. doi: 10.1038/nm1579. [DOI] [PubMed] [Google Scholar]
- 8.Safe S, Karki K. The paradoxical roles of orphan nuclear receptor 4A (NR4A) in cancer. Mol Cancer Res. 2021;19:180–91. doi: 10.1158/1541-7786.MCR-20-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fedorova O, Petukhov A, Daks A, Shuvalov O, Leonova T, Vasileva E, Aksenov N, Melino G, Barlev NA. Orphan receptor NR4A3 is a novel target of p53 that contributes to apoptosis. Oncogene. 2019;38:2108–2122. doi: 10.1038/s41388-018-0566-8. [DOI] [PubMed] [Google Scholar]
- 10.Karki K, Li X, Jin UH, Mohankumar K, Zarei M, Michelhaugh SK, Mittal S, Tjalkens R, Safe S. Nuclear receptor 4A2 (NR4A2) is a druggable target for glioblastomas. J Neurooncol. 2020;146:25–39. doi: 10.1007/s11060-019-03349-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mohankumar K, Li X, Sridharan S, Karki K, Safe S. Nuclear receptor 4A1 (NR4A1) antagonists induce ROS-dependent inhibition of mTOR signaling in endometrial cancer. Gynecol Oncol. 2019;154:218–27. doi: 10.1016/j.ygyno.2019.04.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lacey A, Hedrick E, Li X, Patel K, Doddapaneni R, Singh M, Safe S. Nuclear receptor 4A1 (NR4A1) as a drug target for treating rhabdomyosarcoma (RMS) Oncotarget. 2016;7:31257–69. doi: 10.18632/oncotarget.9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hedrick E, Lee SO, Kim G, Abdelrahim M, Jin UH, Safe S, Abudayyeh A. Nuclear receptor 4A1 (NR4A1) as a drug target for renal cell adenocarcinoma. PLoS One. 2015;10:e0128308. doi: 10.1371/journal.pone.0128308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hedrick E, Lee SO, Doddapaneni R, Singh M, Safe S. Nuclear receptor 4A1 as a drug target for breast cancer chemotherapy. Endocr Relat Cancer. 2015;22:831–40. doi: 10.1530/ERC-15-0063. [DOI] [PubMed] [Google Scholar]
- 15.Lee SO, Jin UH, Kang JH, Kim SB, Guthrie AS, Sreevalsan S, Lee JS, Safe S. The orphan nuclear receptor NR4A1 (Nur77) regulates oxidative and endoplasmic reticulum stress in pancreatic cancer cells. Mol Cancer Res. 2014;12:527–38. doi: 10.1158/1541-7786.MCR-13-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee SO, Abdelrahim M, Yoon K, Chintharlapalli S, Papineni S, Kim K, Wang H, Safe S. Inactivation of the orphan nuclear receptor TR3/Nur77 inhibits pancreatic cancer cell and tumor growth. Cancer Res. 2010;70:6824–36. doi: 10.1158/0008-5472.CAN-10-1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zarei M, Shrestha R, Johnson S, Yu Z, Karki K, Vaziri-Gohar A, Epps J, Du H, Suva L, Zarei M, Safe S. Nuclear receptor 4A2 (NR4A2/NURR1) regulates autophagy and chemoresistance in pancreatic ductal adenocarcinoma. Cancer Res Commun. 2021;1:65–78. doi: 10.1158/2767-9764.CRC-21-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brody JR, Dixon DA. Complex HuR function in pancreatic cancer cells. Wiley Interdiscip Rev RNA. 2018;9:e1469. doi: 10.1002/wrna.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J, Guo Y, Chu H, Guan Y, Bi J, Wang B. Multiple functions of the RNA-binding protein HuR in cancer progression, treatment responses and prognosis. Int J Mol Sci. 2013;14:10015–41. doi: 10.3390/ijms140510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zarei M, Lal S, Parker SJ, Nevler A, Vaziri-Gohar A, Dukleska K, Mambelli-Lisboa NC, Moffat C, Blanco FF, Chand SN, Jimbo M, Cozzitorto JA, Jiang W, Yeo CJ, Londin ER, Seifert EL, Metallo CM, Brody JR, Winter JM. Posttranscriptional upregulation of IDH1 by HuR establishes a powerful survival phenotype in pancreatic cancer cells. Cancer Res. 2017;77:4460–71. doi: 10.1158/0008-5472.CAN-17-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lukosiute-Urboniene A, Jasukaitiene A, Silkuniene G, Barauskas V, Gulbinas A, Dambrauskas Z. Human antigen R mediated post-transcriptional regulation of inhibitors of apoptosis proteins in pancreatic cancer. World J Gastroenterol. 2019;25:205–19. doi: 10.3748/wjg.v25.i2.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong R, Chen P, Polireddy K, Wu X, Wang T, Ramesh R, Dixon DA, Xu L, Aubé J, Chen Q. An RNA-binding protein, Hu-antigen R, in pancreatic cancer epithelial to mesenchymal transition, metastasis, and cancer stem cells. Mol Cancer Ther. 2020;19:2267–77. doi: 10.1158/1535-7163.MCT-19-0822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shrestha R, Mohankumar K, Safe S. Bis-indole derived nuclear receptor 4A1 (NR4A1) antagonists inhibit TGFβ-induced invasion of embryonal rhabdomyosarcoma cells. Am J Cancer Res. 2020;10:2495–509. [PMC free article] [PubMed] [Google Scholar]
- 24.Li X, Lee SO, Safe S. Structure-dependent activation of NR4A2 (Nurr1) by 1,1-bis(3’-indolyl)-1-(aromatic)methane analogs in pancreatic cancer cells. Biochem Pharmacol. 2012;83:1445–55. doi: 10.1016/j.bcp.2012.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burkhart RA, Pineda DM, Chand SN, Romeo C, Londin ER, Karoly ED, Cozzitorto JA, Rigoutsos I, Yeo CJ, Brody JR, Winter JM. HuR is a post-transcriptional regulator of core metabolic enzymes in pancreatic cancer. RNA Biol. 2013;10:1312–23. doi: 10.4161/rna.25274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, Jewell CM, Johnson ZR, Irvine DJ, Guarente L, Kelleher JK, Vander Heiden MG, Iliopoulos O, Stephanopoulos G. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2011;481:380–4. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lacey A, Rodrigues-Hoffman A, Safe S. PAX3-FOXO1A expression in rhabdomyosarcoma is driven by the targetable nuclear receptor NR4A1. Cancer Res. 2017;77:732–41. doi: 10.1158/0008-5472.CAN-16-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hedrick E, Lee SO, Doddapaneni R, Singh M, Safe S. NR4A1 antagonists inhibit β1-integrin-dependent breast cancer cell migration. Mol Cell Biol. 2016;36:1383–94. doi: 10.1128/MCB.00912-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karki K, Wright GA, Mohankumar K, Jin UH, Zhang XH, Safe S. A bis-indole-derived NR4A1 antagonist induces PD-L1 degradation and enhances antitumor immunity. Cancer Res. 2020;80:1011–23. doi: 10.1158/0008-5472.CAN-19-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hedrick E, Li X, Safe S. Penfluridol represses integrin expression in breast cancer through induction of reactive oxygen species and downregulation of Sp transcription factors. Mol Cancer Ther. 2017;16:205–16. doi: 10.1158/1535-7163.MCT-16-0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rao M, Atay SM, Shukla V, Hong Y, Upham T, Ripley RT, Hong JA, Zhang M, Reardon E, Fetsch P, Miettinen M, Li X, Peer CJ, Sissung T, Figg WD, De Rienzo A, Bueno R, Schrump DS. Mithramycin depletes specificity protein 1 and activates p53 to mediate senescence and apoptosis of malignant pleural mesothelioma cells. Clin Cancer Res. 2016;22:1197–210. doi: 10.1158/1078-0432.CCR-14-3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Inamoto T, Papineni S, Chintharlapalli S, Cho SD, Safe S, Kamat AM. 1,1-Bis(3’-indolyl)-1-(p-chlorophenyl)methane activates the orphan nuclear receptor Nurr1 and inhibits bladder cancer growth. Mol Cancer Ther. 2008;7:3825–33. doi: 10.1158/1535-7163.MCT-08-0730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hammond SL, Popichak KA, Li X, Hunt LG, Richman EH, Damale PU, Chong EKP, Backos DS, Safe S, Tjalkens RB. The Nurr1 Ligand,1,1-bis(3’-Indolyl)-1-(p-Chlorophenyl)Methane, modulates Glial Reactivity and is neuroprotective in MPTP-induced Parkinsonism. J Pharmacol Exp Ther. 2018;365:636–51. doi: 10.1124/jpet.117.246389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Miranda BR, Popichak KA, Hammond SL, Miller JA, Safe S, Tjalkens RB. Novel para-phenyl substituted diindolylmethanes protect against MPTP neurotoxicity and suppress glial activation in a mouse model of Parkinson’s disease. Toxicol Sci. 2015;143:360–73. doi: 10.1093/toxsci/kfu236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chatterjee S, Walsh EN, Yan AL, Giese KP, Safe S, Abel T. Pharmacological activation of Nr4a rescues age-associated memory decline. Neurobiol Aging. 2020;85:140–4. doi: 10.1016/j.neurobiolaging.2019.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mizrahi JD, Surana R, Valle JW, Shroff RT. Pancreatic cancer. Lancet. 2020;395:2008–20. doi: 10.1016/S0140-6736(20)30974-0. [DOI] [PubMed] [Google Scholar]
- 39.Pérez-Mancera PA, Guerra C, Barbacid M, Tuveson DA. What we have learned about pancreatic cancer from mouse models. Gastroenterology. 2012;142:1079–92. doi: 10.1053/j.gastro.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 40.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, Hong SM, Fu B, Lin MT, Calhoun ES, Kamiyama M, Walter K, Nikolskaya T, Nikolsky Y, Hartigan J, Smith DR, Hidalgo M, Leach SD, Klein AP, Jaffee EM, Goggins M, Maitra A, Iacobuzio-Donahue C, Eshleman JR, Kern SE, Hruban RH, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–6. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han Y, Cai H, Ma L, Ding Y, Tan X, Liu Y, Su T, Yu Y, Chang W, Zhang H, Fu C, Cao G. Nuclear orphan receptor NR4A2 confers chemoresistance and predicts unfavorable prognosis of colorectal carcinoma patients who received postoperative chemotherapy. Eur J Cancer. 2013;49:3420–30. doi: 10.1016/j.ejca.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 42.Beard JA, Tenga A, Chen T. The interplay of NR4A receptors and the oncogene-tumor suppressor networks in cancer. Cell Signal. 2015;27:257–66. doi: 10.1016/j.cellsig.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun L, Liu M, Sun GC, Yang X, Qian Q, Feng S, Mackey LV, Coy DH. Notch signaling activation in cervical cancer cells induces cell growth arrest with the involvement of the nuclear receptor NR4A2. J Cancer. 2016;7:1388–95. doi: 10.7150/jca.15274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Llopis S, Singleton B, Duplessis T, Carrier L, Rowan B, Williams C. Dichotomous roles for the orphan nuclear receptor NURR1 in breast cancer. BMC Cancer. 2013;13:139. doi: 10.1186/1471-2407-13-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ke N, Claassen G, Yu DH, Albers A, Fan W, Tan P, Grifman M, Hu X, Defife K, Nguy V, Meyhack B, Brachat A, Wong-Staal F, Li QX. Nuclear hormone receptor NR4A2 is involved in cell transformation and apoptosis. Cancer Res. 2004;64:8208–12. doi: 10.1158/0008-5472.CAN-04-2134. [DOI] [PubMed] [Google Scholar]
- 46.Li X, Tai HH. Activation of thromboxane A(2) receptors induces orphan nuclear receptor Nurr1 expression and stimulates cell proliferation in human lung cancer cells. Carcinogenesis. 2009;30:1606–13. doi: 10.1093/carcin/bgp161. [DOI] [PubMed] [Google Scholar]
- 47.Han Y, Cai H, Ma L, Ding Y, Tan X, Chang W, Guan W, Liu Y, Shen Q, Yu Y, Zhang H, Cao G. Expression of orphan nuclear receptor NR4A2 in gastric cancer cells confers chemoresistance and predicts an unfavorable postoperative survival of gastric cancer patients with chemotherapy. Cancer. 2013;119:3436–45. doi: 10.1002/cncr.28228. [DOI] [PubMed] [Google Scholar]
- 48.Zhu B, Sun L, Luo W, Li M, Coy DH, Yu L, Yu W. Activated Notch signaling augments cell growth in hepatocellular carcinoma via up-regulating the nuclear receptor NR4A2. Oncotarget. 2017;8:23289–302. doi: 10.18632/oncotarget.15576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Han YF, Cao GW. Role of nuclear receptor NR4A2 in gastrointestinal inflammation and cancers. World J Gastroenterol. 2012;18:6865–73. doi: 10.3748/wjg.v18.i47.6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schultz CW, Preet R, Dhir T, Dixon DA, Brody JR. Understanding and targeting the disease-related RNA binding protein human antigen R (HuR) Wiley Interdiscip Rev RNA. 2020;11:e1581. doi: 10.1002/wrna.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Safe S, Kim K. Non-classical genomic estrogen receptor (ER)/specificity protein and ER/activating protein-1 signaling pathways. J Mol Endocrinol. 2008;41:263–75. doi: 10.1677/JME-08-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kang MJ, Ryu BK, Lee MG, Han J, Lee JH, Ha TK, Byun DS, Chae KS, Lee BH, Chun HS, Lee KY, Kim HJ, Chi SG. NF-κB activates transcription of the RNA-binding factor HuR, via PI3K-AKT signaling, to promote gastric tumorigenesis. Gastroenterology. 2008;135:2030–42. e1–3. doi: 10.1053/j.gastro.2008.08.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.