Abstract
Colorectal cancer (CRC) is a lethal malignancy and a leading cause of cancer-related mortality worldwide. Chromosomal instability (CIN) is a key driver of genomic instability in CRC and is characterized by aneuploidy and somatic copy-number alterations. This study aimed to predict CIN in CRC using histological data from whole slide images (WSIs). CRC samples from TCGA were analyzed, with tumor regions segmented into tiles and nuclei for feature extraction using convolutional neural network (CNN) and morphologic analysis. Binary classification models were developed to distinguish high and low aneuploidy scores (AS) based on slide-level features. The analysis included 313 patients with 315 WSIs, resulting in over 350,000 tumor tiles and nearly 2.7 million tumor cell nuclei. The ResNet18-SSL model, pre-trained on histopathological images, demonstrated superior accuracy in tile-based AS prediction, while DenseNet121 excelled in nucleus-based prediction. Combining CNN-based and morphological features enhanced the classification accuracy of nucleus-based predictions. Additionally, significant correlations were observed between morphological features and copy-number signatures. Unsupervised clustering of nuclear features revealed that distinct groups are significantly correlated with CIN and TP53 mutations. This study underscores the potential of histological features from WSIs to predict CIN in CRC samples. Nuclear feature analysis, combined with deep-learning techniques, offers a robust method for CIN prediction, highlighting the importance of further research into the relationships between histological and molecular phenotypes.
Keywords: Colorectal cancer, chromosomal instability, digital pathology, deep learning, nucleus
Introduction
Colorectal cancer (CRC) is the second leading cause of cancer-related deaths globally, and its incidence in young patients is rising [1,2]. Traditionally, pathologic diagnoses of CRC, conducted via biopsy or resection specimens, have relied on hematoxylin and eosin (H&E) stained slides, which are essential for tumor staging and guiding therapeutic decisions. In recent years, molecular characteristics such as microsatellite instability (MSI), tumor mutation burden (TMB), CpG island methylator phenotype (CIMP), and chromosomal instability (CIN) have emerged as critical prognostic indicators and therapeutic targets [3-6]. Among the molecular phenotypes, CIN is characterized by persistent loss and gain of chromosomes at high grades [7]. CIN in CRC is marked by pronounced aneuploidy and frequent somatic copy-number alterations (SCNA) [8]. Notably, mutations in genes such as APC, TP53, KRAS, SMAD4, SOX9, and FBXW7 significantly contribute to CIN in CRC [8].
CIN is a hallmark of various human malignancies, and is often associated with tumor initiation, progression, metastasis, prognosis, and therapeutic resistance [7]. Therefore, predicting CIN enables the identification of tumors with aggressive features, including those that exhibit resistance to standard and immune-based therapies. This approach can guide treatment decisions, enabling more personalized and potentially effective therapeutic approaches. Furthermore, this approach can help to select more intensive treatments or explore novel targeted therapies. Nevertheless, the patterns of SCNA associated with CIN vary across different tumor types, including CRC, posing challenges in analyzing CIN across different molecular subtypes within CRC and other types of carcinomas. To address these challenges, Taylor et al. proposed a pan-cancer chromosome arm-level scoring system that applicable to 33 cancer types within in The Cancer Genome Atlas (TCGA) [9]. Similarly, Drews et al. developed a copy-number signature system that considers diverse causes of CIN, such as mitotic errors, replication stress, homologous recombination deficiency, telomere crisis, and breakage fusion bridge cycles, as well as its consequences across a broad spectrum of cancers [10]. Despite these advancements, a comprehensive model integrating histological data and CIN measurements in CRC is still lacking.
Recent progress in digital pathology and artificial intelligence have revealed previously elusive relationships between histological images, molecular phenotypes, and patient outcome [11]. These techniques facilitate the prediction of molecular features from whole slide images (WSIs) of malignancies with far greater accuracy than tranditional visual assessment [12].
There are largely two methods for feature extraction and biomarker prediction from WSIs [13]. The first approach involves segmenting WSIs into small patches, extracting features from each patch, and aggregating them later. For example, previous studies have successfully employed histological models, based on segmented tiles, to predict molecular features of CRC, including MSI status [12,14-18]. The tile-based prediction of CIN has also been explored in breast cancer [19]. The second approach is focusing on single-cell analysis, where individual cells are segmented, classified, and analyzed for feature extraction. This method has been used to predict CIN in prostate, lung, and head and neck cancers [13,20]. In the this study, we aimed to use TCGA data of CRC (TCGA-CRC) to predict diverse aspects of CIN based on histology, applying both tile-based and nuclear-based methods to determine the optimal model for predicting CIN in CRC.
Material and methods
Datasets
Images of formalin-fixed paraffin-embedded H&E stained CRC samples were obtained from TCGA database. WSIs of colon adenocarcinoma (TCGA-COAD) and rectal adenocarcinoma (TCGA-READ) at 40× magnification were selected. The aneuploidy score (AS), determined by the number of arms altered (either amplified or deleted) in each sample, and whole-genome doubling (WGD) were obtained from Taylor et al. [9]. Detectable CIN and copy-number signatures were obtained from Drews et al. [10]. We used survival data adopted from Liu et al. [21]. Clinical, mutation, and fraction genome altered data were obtained from cBioPortal for Cancer Genomics [22].
Tumor tile selection, nuclear segmentation, and nuclear extraction
Tumor regions within the WSIs were identified using manual annotations by Loeffler and Kather (available at dx.doi.org/10.5281/zenodo.5320076). The tumor areas were segmented into non-overlapping 512 × 512-pixel tiles, each with a spatial resolution of 1.0 μm/pixel. The tiles were normalized using the Macenko method [23]. A pathologist (YK) reviewed the non-tumorous tiles. To extract tumor cell nuclei, the tumor areas were manually annotated into small clusters to minimize the inclusion of endothelial cells, fibroblasts, and immune cells. Annotation was performed in QuPath v0.4.3 [24]. The StarDist model, which is compatible with QuPath, was used to detect cell nuclei within the annotated areas [25]. Tumor and non-tumor cells were classified using a random tree classifier built into QuPath. After non-tumor cells were removed, we generated 100 × 100-pixel images with a single central nucleus to extract features from each nucleus.
Convolutional neural network (CNN)-based and morphological feature extraction
Deep features from tumor patches were extracted using pre-trained CNNs. We employed DenseNet121 [26], ResNet18 [27], and VGG11 [28] models, pre-trained on ImageNet [29], as well as ResNet18 model pre-trained on histopathological images using self-supervised learning (SSL) techniques [30], trained on 400,000 histopathological tiles of various organs from public databases. The feature dimensions were 1,000 for ImageNet-pretrained features and 512 for SSL-based features.
In addition to deep features, 15 morphological features associated with size, shape, and intensity were detected for each tumor nucleus, as described by Abel et al. [20]. These features included measurements of area, major and minor axis lengths, perimeter, circularity, eccentricity, solidity, and the mean and standard deviation (SD) of the pixel grayscale intensity, pixel saturation, and pixel A and B channels in the LAB color space.
Feature aggregation for tiles and nuclei
Tile-level features were aggregated into slide-level features using max pooling [19]. The slide-level tile deep feature was derived from the maximum value within each feature. Therefore, slide-level tile deep features have the same dimensions as tile-level deep features, enabling the derivation of model prediction scores at the tile-level. Once the AS prediction model has been trained using slide-level tile deep features, the prediction scores for individual tiles were derived by feeding the tile-level deep features into the trained model.
To transform the nuclei-level features into slide-level features, the mean and SD of each feature were calculated from all nuclei on a single slide. This processes yielded 30 slide-level nuclear morphological features, such as the mean major axis length and SD of circularity. Similarly, the mean and SD of the deep features of the cell nuclei were calculated, resulting in either 1,024 or 2,000 slide-level nuclear deep features, depending on whether the feature extractor was based on SSL or was pre-trained on ImageNet.
AS prediction models
We developed binary classification models to distinguish between high (AS-H, AS > 10) and low (AS-L, AS ≤ 10) in WSIs using different slide-level features, including those derived from tile-level or nuclei-level feature extractors. The classification models are built on multi-layer perceptrons (MLPs) with four layers: an input layer, two hidden layers, and an output layer. The number of nodes in the input layer was determined by the slide-level features. Each of the two hidden layers contained 512 nodes, followed by 100 nodes in the output layer and a single node for the final output.
Each model was trained and evaluated using a five-fold cross-validation approach. We used the StratifiedKFold function in scikit-learn to randomly divide the dataset into five stratified folds. One of the folds was assigned as the test set, while the remaining four folds were used for training (80%) and validation (20%). The process was iterated for each of the five folds, resulting in five outcomes per model. The average and SD of the results were used to determine the overall cross-validation performance.
The models were trained to minimize the binary cross-entropy loss function. The weights of the MLP were initialized using the He initialization method [31]. Stochastic gradient descent (SGD) optimizer was used to train network weights with a learning rate of 0.00005. The training was stopped early if the validation loss did not decrease within 200 epochs. Other parameter settings in the training process were as follows: maximum epochs, 2,000; batch size, 8; momentum, 0.9; and activation function, rectified linear unit (RELU) function. All experiments were conducted using Python language (v.3.8) and an NVIDIA A6000 GPU. The models were implemented using PyTorch v.1.12 [32].
Nuclear classification in tumor tiles
Cell components within tiles were classified using Hover-Net on the PanNuke dataset [33] which categorized the cells into five groups: neoplastic, connective, non-neoplastic epithelia, necrotic, and inflammatory [34]. AS prediction scores were predicted at the tile-level by feeding individual tile features into trained MLP models, as mentioned previously. The density of tumor cells within each slide was calculated as the number of neoplastic cells divided by the number of tiles. The tumor-to-immune cell ratio was calculated by dividing the number of neoplastic cells by the number of inflammatory cells.
Unsupervised clustering
The clustering induced by morphological features was investigated by using the uniform manifold approximation and projection (UMAP) method to map the slide-level nuclei morphological features onto a 2D space [35]. In the UMAP space, the hierarchical density-based clustering of applications with noise (HDBSCAN) was applied to obtain cluster labels, with the minimum cluster size set to 10 [36].
Statistical analysis
Spearman’s rank correlation was used to examine the relationship between morphological features and the activities of copy-number signatures (CX1-CX17). The correlation between clusters and clinical and molecular features was evaluated using Pearson’s chi-square test. Kaplan-Meier analysis with a log-rank test was used to calculate overall and disease-free survival. p values < 0.05 were considered as statistically significant.
Results
Tissue annotation, segmentation, and nuclear extraction
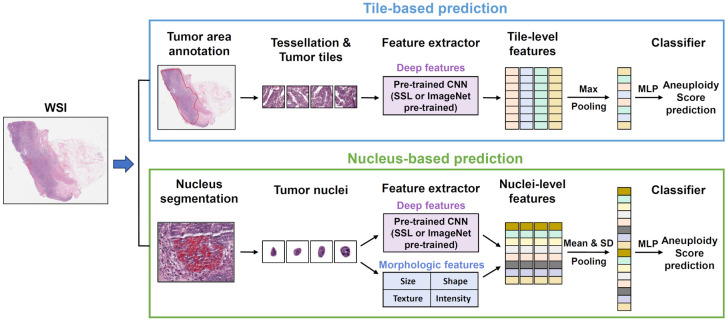
An overview of tumor annotation, segmentation, and model development process is summarized in Figure 1. Slides that StarDist failed to properly detect the nuclei were excluded from the anlaysis. In cases of mucinous adenocarcinoma with signet ring cell component, StarDist annotated entire cells instead of just the nuclei. After excluding inadequate cases and slides, the final cohort comprised 313 patients with 315 WSIs, yielding 353,712 tumor tiles and 2,693,097 cell nuclei. On average, this corresponded to approximately 1,000 tiles and 8,500 cell nuclei per slide.
Figure 1.
Overview of workflow for tile-based (upper portion) and nucleus-based (lower portion) aneuploidy score (AS) prediction model development from whole-slide images (WSIs). CNN, convolutional neural network; MLP, multi-layer perceptron; SSL, self-supervised learning.
Model comparisons and predictions
The performance of various pre-trained models was evaluated (Table 1). The ResNet18-SSL model exhibited superior accuracy and area under the receiver operating characteristic curve (AUROC) in tile-based AS prediction, while DenseNet121 performed optimally in nucleus-based prediction. However, ResNet18-SSL outperformed DenseNet121 in terms of AUROC (Supplementary Figure 1). When deep and morphological features were combined in nucleus-based models, the classification accuracy improved compared to using either feature alone (Table 2). Models predicting WGD and fraction genome altered using combined features achieved modest accuracy (Supplementary Table 1).
Table 1.
Comparison of deep learning models for predicting AS
| Source | Backbone | Accuracy (SD) | AUROC (SD) | F1 (SD) |
|---|---|---|---|---|
| Tile | ResNet18 - ImageNet | 0.638 (0.034) | 0.661 (0.033) | 0.720 (0.042) |
| Tile | ResNet18 - SSL | 0.686 (0.038) | 0.742 (0.023) | 0.735 (0.040) |
| Tile | Vgg11 - ImageNet | 0.590 (0.017) | 0.629 (0.043) | 0.686 (0.030) |
| Tile | DenseNet121 - ImageNet | 0.670 (0.063) | 0.717 (0.061) | 0.738 (0.051) |
| Nucleus | Resnet18 - ImageNet | 0.667 (0.039) | 0.752 (0.058) | 0.720 (0.048) |
| Nucleus | Resnet18 - SSL | 0.673 (0.051) | 0.736 (0.060) | 0.730 (0.058) |
| Nucleus | Vgg11 - ImageNet | 0.670 (0.031) | 0.714 (0.043) | 0.734 (0.045) |
| Nucleus | DenseNet121 - ImageNet | 0.717 (0.059) | 0.740 (0.060) | 0.758 (0.053) |
Table 2.
Nucleus-based model with and without feature integration
| Accuracy (SD) | AUROC (SD) | F1 (SD) | |
|---|---|---|---|
| Morphological features (M) | 0.689 (0.048) | 0.737 (0.072) | 0.741 (0.054) |
| Deep features (D) | 0.717 (0.059) | 0.740 (0.060) | 0.758 (0.053) |
| M+D | 0.721 (0.053) | 0.750 (0.054) | 0.766 (0.050) |
Impact of MSI on model performance
MSI is a critical molecular feature associated with CRC histology [14-18]. Among 42 MSI-high CRC cases, AS 0 was most common (17 cases, 40.5%), accounting for 68% of AS 0 cases analyzed (17 out of 25). Therefore, the effect of MSI-high cases on AS prediction was investigated by comparing models that included and excluded AS 0 cases for both tile- and nucleus-based predictions (Table 3). Tile-based models exhibited a steep decline in accuracy and AUROC, whereas the nucleus-based models showed limited changes. Both tile- and nucleus-based models were affected when all MSI-high cases were excluded, likely due to severe class imbalance (Supplementary Tables 2 and 3).
Table 3.
Prediction of AS with and without AS 0 cases
| AS 0 cases | Accuracy (SD) | AUROC (SD) | F1 (SD) | |
|---|---|---|---|---|
| Tile-based Model | Include | 0.686 (0.038) | 0.742 (0.023) | 0.735 (0.040) |
| Exclude | 0.628 (0.040) | 0.651 (0.085) | 0.734 (0.043) | |
| Nuclei-based Model | Include | 0.721 (0.053) | 0.750 (0.054) | 0.766 (0.050) |
| Exclude | 0.714 (0.045) | 0.732 (0.082) | 0.787 (0.037) |
Effect of tumor cell density and tumor-to-immune cell ratio on tile-based prediction
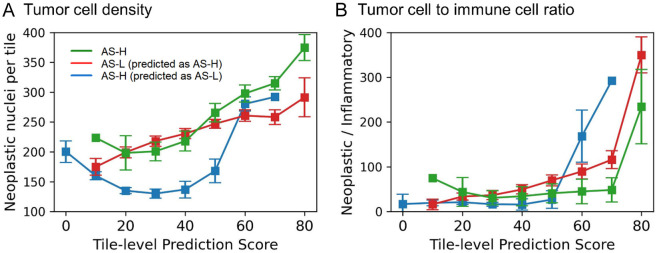
In analyzing mislabeled cases in tile-based models, we hypothesized that AS-H cases misclassified as AS-L would exhibit lower tumor cellularity, while mislabeled AS-L cases predicted as AS-H would display higher tumor cellularity. To assess this, Hover-Net was employed to label the cell components within the tiles. AS prediction scores at the tile-level were generated by feeding individual tile features into trained MLP models. Consistent with the hypothesis, cases incorrectly predicted as AS-L exhibited significantly lower tumor cell densities than those of both misclassified and accurately predicted AS-H cases (Figure 2A; Supplementary Table 4). Interestingly, tiles with a higher probability of AS had a higher tumor cell density than those with a lower probability of AS (Figure 2A). Another potential confounding factor we considered was the tumor-to-immune cell ratio. However, this ratio did not show a marked difference between the incorrectly predicted AS-H and AS-L cases (Figure 2B).
Figure 2.
Influence of tumor cell density and tumor cell to immune cell ratio in tile-level AS prediction. A: Tumor cell density of three cases representing AS-H (green line), AS-L incorrectly predicted as AS-H (red line), and AS-H incorrectly predicted as AS-L (blue line), each aligned with the prediction scores of their respective tiles (0-80%). B: Ratio between tumor cells and immune cells. Three cases are displayed according to tile-level prediction scores. AS-H, high aneuploidy score; AS-L, low aneuploidy score.
Key parameters in morphological features
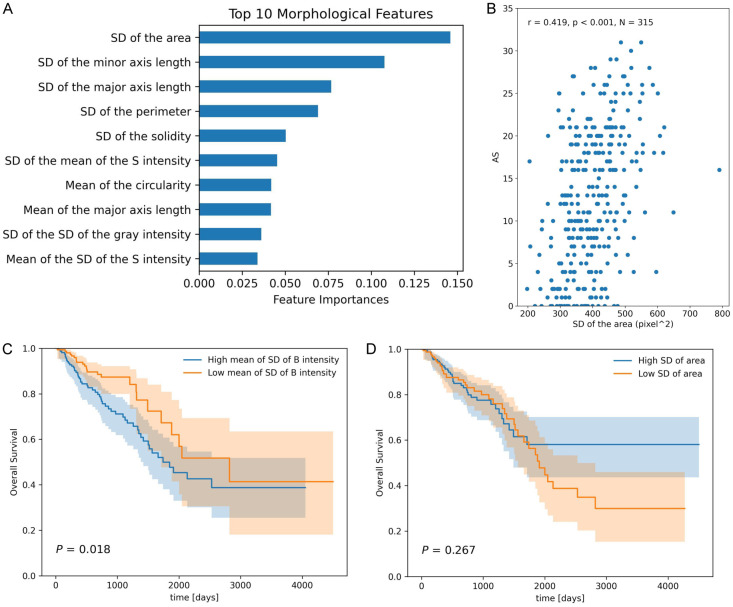
To determine the most important morphological parameters for predicting AS, feature importance was extracted using a random forest model. Out of the 30 evaluated parameters, the SD of the tumor nuclear area ranked highest (Figure 3A) and showed a significant correlation with AS (Spearman r = 0.419, P < 0.001, Figure 3B). Other parameters with high importance included SD of the minor axis length, SD of the major axis length, and SD of the perimeter, all of which reflect variability in nuclear size. Additionally, we explored the relationship between morphological features and patient survival. Univariate Cox proportional hazard models fitted to each morphological feature revealed that the mean SD of B channel intensity in the LAB color space was strongly associated with both overall and disease-free survival (Supplementary Table 5). Patients were stratified into two groups based on the median value of the feature, and Kaplan-Meier analysis indicated that a lower mean SD of B channel intensity significantly correlated with improved overall survival (Figure 3C). However, the SD of the area, despite being the most important feature for predicting AS, was not a prognostic factor (Figure 3D). Notably, AS showed no association with survival in this cohort of 313 cases (P = 0.149).
Figure 3.
Nuclear morphology with feature importance and its correlation with survival. A: Nuclear morphological features with top feature importance. B: Correlation between SD of tumor cell area and AS. C: Overall survival according to mean SD of B intensity. D: Overall survival according to SD of tumor cell area.
Nuclear morphology and copy-number signatures
Among the 315 analyzed slides, 220 had detectable CIN and were assigned activity value for 17 copy-number signatures (CX1-CX17). Among them, AS exhibited the strongest correlation with, CX6 (Spearman’s r = 0.420, P < 0.001), which is characterized by whole-arm and chromosomal changes potentially resulting from chromosome missegregation due to defective mitosis, followed by CX8 and CX4 (Table 4). An inverse correlation with CX11 and CX12. Various other copy-number signatures were also significantly correlated with diverse morphological features of the tumor nuclei, suggesting that differences in nuclear morphology may reflect specific pattern of change within CIN (Supplementary Table 6).
Table 4.
Correlation between AS and copy-number signatures
| Signature | Spearman’s r | P value | Putative cause according to Drews et al. [10] |
|---|---|---|---|
| CX4 | 0.254 | < 0.001 | PI3K-AKT-mediated toleration of whole-genome duplication |
| CX6 | 0.420 | < 0.001 | Chromosome missegregation via defective mitosis |
| CX8 | 0.348 | < 0.001 | Replication stress |
| CX11 | -0.168 | 0.012 | Replication stress |
| CX12 | -0.153 | 0.023 | Unknown |
Clustering of nuclear features
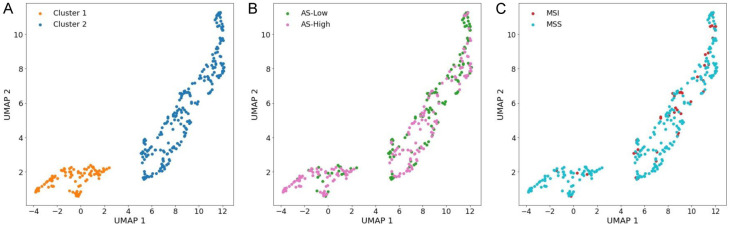
Unsupervised clustering of morphological features revealed two clusters in UMAP (Figure 4A). Cluster 1 comprised predominantly of AS-H/microsatellite stable (MSS) cases, while cluster 2 included most AS-L and MSI-high cases (Figure 4B and 4C). However, clustering did not have significant prognostic implications for overall survival (P = 0.809). A notable correlation was observed between the clusters and clinical and molecular features, particularly with MSI status (P = 0.002) and AS (P < 0.001). No significant associations were found with other clinical parameters, such as pTNM, pT, pN, pM stages, or sex (Supplementary Table 7). Notably, while BRAF mutations, which are often associated with MSI, exhibited no significant association with the clusters (P = 0.136), TP53 mutations, which are related to CIN, demonstrated a significant correlation (P = 0.047).
Figure 4.
Unsupervised clustering based on nuclear morphology. (A) Clustering of nuclear morphological features. Clusters are depicted according to AS (B) or MSI status (C). MSI, microsatellite instability; MSS, microsatellite stable.
Discussion
Tumor aneuploidy, a nearly universal feature of human malignancies [9], can predict poor prognosis following immunotherapy across multiple cancer types [37]. Additionally, AS-H tumors can be used to stratify patients with worse survival in samples with a low tumor mutational burden. However, CIN is not limited to aneuploidy alone; it encompasses a broader range of genomic alterations [38]. Copy-number signatures account for various causes and changes in patterns associated with CIN [10]. These signatures have predicted platinum sensitivity in ovarian, esophageal, and breast cancer. Therefore, our prediction of CIN using AS and copy-number signatures based on histology could inform the development of personalized treatment plans for patients with CRC.
In breast cancer and CRC, CIN has been predicted using tile-based models [12,19]. Notably, tiles predictive of high CIN tend to have larger neoplastic cell nuclei than those of the tiles predictive of low CIN, highlighting the ongoing importance of nuclear components in predicting CIN. This supports our approach of using nuclear features as a more effective method for predicting CIN compared to tile images alone. Previous studies have predicted MSI-high with great accuracy using tile-based models [12,14,39,40]. Tiles that predict MSI-high have been associated with tumor-infiltrating lymphocytes and poorly differentiated morphology, whereas tiles that predict MSS show well-differentiated tumor morphology. These findings align with our current results, demonstrating the effectiveness of tile-based models for predicting molecular features influenced by tumor cell density, including MSI.
Due to the large number of cell nuclei in the WSIs, significant storage and computing resources are required to segment all nuclei and extract features. Abel et al. extracted every nucleus from each WSI to investigate genomic instability [20]. Although capturing all cell nuclei may provide a more precise representation of tumor microenvironments than sampling regions of interest (ROIs), it can also lead to overexposure to unwanted tissue regions, such as normal mucosa and stroma without malignancy. Instead, we opted to select tumor nucleus-rich ROIs, extracting approximately 8,000 cell nuclei per slide, focusing on specific cell types in clinical settings for future deployment.
Our analysis revealed that the ResNet18-SSL model, pre-trained using histopathological images, outperformed models using the ImageNet features in the tile-based prediction. However, there was no similar improvement in the nucleus-based model, suggesting that pre-training on histological image may not provide an optimal representation for smaller nucleus images. Therefore, future studies should focus on developing SSL techniques on large-scale nuclear images to generate nucleus-specific models that could facilitate single-cell analysis. Furthermore, deep features exhibited better performance than that of morphological features; however, morphological features were more interpretable than “black-box” deep features in the nucleus-based model. Future research should aim to enhance the performance of deep features by leveraging advanced SSL techniques and minimizing performance discrepancies between deep and morphological features by identifying and incorporating additional interpretable morphological features.
This study has certain limitations as well as advantages over previous studies. First, while we used only 15 morphological features from the nucleus, other methods, such as CellProfiler, extract over 100 morphological features [41]. Xia et al. conducted a prognostic prediction study for CRC using morphological features of nucleus extracted from WSIs using CellProfiler [42]. Their approach of using the Lasso-Cox model identified seemingly obscure factors such as ‘Median_Identifyeosinpromarycytoplasm_Texture_Entropy_maskosingray_3_01_256’ as a significant predictor of survival. This might be due to a lack of pipeline to exclude tumor-infiltrating lymphocytes and tumor-associated macrophages within the tumor area. Meanwhile, another study suggested that variations in nuclear shape and intensity - indicators of nuclear pleomorphism - are independent prognostic factors in squamous cell carcinoma and adenocarcinoma of the lung [43]. The results of the present study also indicate that nuclear anisokaryosis, a clearly interpretable factor, is correlated with CIN, and that variation in nuclear intensity could be a putative prognostic marker.
Second, this study did not include an external validation set. This is due to the lack of available AS data in the external CRC WSI cohort datasets. Future studies are needed to explore the generalizability of the trained model. Nonetheless, a recent study has shown a similar pattern between SD of the tumor nuclei area and AS in breast cancer, lung adenocarcinoma, and prostate adenocarcinoma [20], indicating that our findings represent a common phenomenon across multiple cancer types. In another study using WSI of CRC, CIN and genomic stability were predicted using tiles with a higher AUROC (0.83) than that in the present study [12]. However, this dichotomous category specific to TCGA-CRC do not directly match with AS; suggesting that our methods may have broader applicability.
Conclusion
In conclusion, the present study used diverse histological features to predict CIN in patients with CRC. The nuclear features of CRC tumor cells demonstrate a robust association with CIN, along with its putative initiator TP53 mutation. Our study has highlighted various directions for future research into the molecular relationships in CRC and development of targeted treatments and personalized management strategies.
Acknowledgements
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (RS-2023-00238446).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73:17–48. doi: 10.3322/caac.21763. [DOI] [PubMed] [Google Scholar]
- 3.Singh MP, Rai S, Pandey A, Singh NK, Srivastava S. Molecular subtypes of colorectal cancer: an emerging therapeutic opportunity for personalized medicine. Genes Dis. 2019;8:133–145. doi: 10.1016/j.gendis.2019.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Y, Ma Y, Wu Z, Zeng F, Song B, Zhang Y, Li J, Lui S, Wu M. Tumor mutational burden predicting the efficacy of immune checkpoint inhibitors in colorectal cancer: a systematic review and meta-analysis. Front Immunol. 2021;12:751407. doi: 10.3389/fimmu.2021.751407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang X, Zhang W, Cao P. Advances in CpG island methylator phenotype colorectal cancer therapies. Front Oncol. 2021;11:629390. doi: 10.3389/fonc.2021.629390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pino MS, Chung DC. The chromosomal instability pathway in colon cancer. Gastroenterology. 2010;138:2059–2072. doi: 10.1053/j.gastro.2009.12.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tijhuis AE, Johnson SC, McClelland SE. The emerging links between chromosomal instability (CIN), metastasis, inflammation and tumour immunity. Mol Cytogenet. 2019;12:17. doi: 10.1186/s13039-019-0429-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y, Sethi NS, Hinoue T, Schneider BG, Cherniack AD, Sanchez-Vega F, Seoane JA, Farshidfar F, Bowlby R, Islam M, Kim J, Chatila W, Akbani R, Kanchi RS, Rabkin CS, Willis JE, Wang KK, McCall SJ, Mishra L, Ojesina AI, Bullman S, Pedamallu CS, Lazar AJ, Sakai R Cancer Genome Atlas Research Network. Thorsson V, Bass AJ, Laird PW. Comparative molecular analysis of gastrointestinal adenocarcinomas. Cancer Cell. 2018;33:721–735. e8. doi: 10.1016/j.ccell.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taylor AM, Shih J, Ha G, Gao GF, Zhang X, Berger AC, Schumacher SE, Wang C, Hu H, Liu J, Lazar AJ Cancer Genome Atlas Research Network. Cherniack AD, Beroukhim R, Meyerson M. Genomic and functional approaches to understanding cancer aneuploidy. Cancer Cell. 2018;33:676–689. e3. doi: 10.1016/j.ccell.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drews RM, Hernando B, Tarabichi M, Haase K, Lesluyes T, Smith PS, Morrill Gavarro L, Couturier DL, Liu L, Schneider M, Brenton JD, Van Loo P, Macintyre G, Markowetz F. A pan-cancer compendium of chromosomal instability. Nature. 2022;606:976–983. doi: 10.1038/s41586-022-04789-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Litjens G, Sánchez CI, Timofeeva N, Hermsen M, Nagtegaal I, Kovacs I, Hulsbergen-van de Kaa C, Bult P, van Ginneken B, van der Laak J. Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci Rep. 2016;6:26286. doi: 10.1038/srep26286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bilal M, Raza SEA, Azam A, Graham S, Ilyas M, Cree IA, Snead D, Minhas F, Rajpoot NM. Development and validation of a weakly supervised deep learning framework to predict the status of molecular pathways and key mutations in colorectal cancer from routine histology images: a retrospective study. Lancet Digit Health. 2021;3:e763–e772. doi: 10.1016/S2589-7500(21)00180-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu F, Wang X, Sali R, Li R. Single-cell heterogeneity-aware transformer-guided multiple instance learning for cancer aneuploidy prediction from whole slide histopathology images. IEEE J Biomed Health Inform. 2023 doi: 10.1109/JBHI.2023.3262454. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Echle A, Grabsch HI, Quirke P, van den Brandt PA, West NP, Hutchins GGA, Heij LR, Tan X, Richman SD, Krause J, Alwers E, Jenniskens J, Offermans K, Gray R, Brenner H, Chang-Claude J, Trautwein C, Pearson AT, Boor P, Luedde T, Gaisa NT, Hoffmeister M, Kather JN. Clinical-grade detection of microsatellite instability in colorectal tumors by deep learning. Gastroenterology. 2020;159:1406–1416. e11. doi: 10.1053/j.gastro.2020.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kather JN, Pearson AT, Halama N, Jäger D, Krause J, Loosen SH, Marx A, Boor P, Tacke F, Neumann UP, Grabsch HI, Yoshikawa T, Brenner H, Chang-Claude J, Hoffmeister M, Trautwein C, Luedde T. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat Med. 2019;25:1054–1056. doi: 10.1038/s41591-019-0462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamashita R, Long J, Longacre T, Peng L, Berry G, Martin B, Higgins J, Rubin DL, Shen J. Deep learning model for the prediction of microsatellite instability in colorectal cancer: a diagnostic study. Lancet Oncol. 2021;22:132–141. doi: 10.1016/S1470-2045(20)30535-0. [DOI] [PubMed] [Google Scholar]
- 17.Tsai PC, Lee TH, Kuo KC, Su FY, Lee TM, Marostica E, Ugai T, Zhao M, Lau MC, Vayrynen JP, Giannakis M, Takashima Y, Kahaki SM, Wu K, Song M, Meyerhardt JA, Chan AT, Chiang JH, Nowak J, Ogino S, Yu KH. Histopathology images predict multi-omics aberrations and prognoses in colorectal cancer patients. Nat Commun. 2023;14:2102. doi: 10.1038/s41467-023-37179-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wagner SJ, Reisenbuchler D, West NP, Niehues JM, Zhu J, Foersch S, Veldhuizen GP, Quirke P, Grabsch HI, van den Brandt PA, Hutchins GGA, Richman SD, Yuan T, Langer R, Jenniskens JCA, Offermans K, Mueller W, Gray R, Gruber SB, Greenson JK, Rennert G, Bonner JD, Schmolze D, Jonnagaddala J, Hawkins NJ, Ward RL, Morton D, Seymour M, Magill L, Nowak M, Hay J, Koelzer VH, Church DN TransSCOT consortium. Matek C, Geppert C, Peng C, Zhi C, Ouyang X, James JA, Loughrey MB, Salto-Tellez M, Brenner H, Hoffmeister M, Truhn D, Schnabel JA, Boxberg M, Peng T, Kather JN. Transformer-based biomarker prediction from colorectal cancer histology: a large-scale multicentric study. Cancer Cell. 2023;41:1650–1661. e1654. doi: 10.1016/j.ccell.2023.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu Z, Verma A, Naveed U, Bakhoum SF, Khosravi P, Elemento O. Deep learning predicts chromosomal instability from histopathology images. iScience. 2021;24:102394. doi: 10.1016/j.isci.2021.102394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abel J, Jain S, Rajan D, Padigela H, Leidal K, Prakash A, Conway J, Nercessian M, Kirkup C, Javed SA, Egger R, Trotter B, Gerardin Y, Brosnan-Cashman JA, Dhoot A, Montalto MC, Wapinski I, Khosla A, Drage MG, Yu L, Taylor-Weiner A. Cell-type-specific nuclear morphology predicts genomic instability and prognosis in multiple cancer types. bioRxiv. 2023 2023.2005.2015.539600. [Google Scholar]
- 21.Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee AV, Omberg L, Wolf DM, Shriver CD, Thorsson V Cancer Genome Atlas Research Network. Hu H. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173:400–416. e411. doi: 10.1016/j.cell.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macenko M, Niethammer M, Marron JS, Borland D, Woosley JT, Guan XJ, Schmitt C, Thomas NE. A method for normalizing histology slides for quantitative analysis. IEEE International Symposium on Biomedical Imaging: From Nano to Macro. 2009;1-2:1107–1110. [Google Scholar]
- 24.Bankhead P, Loughrey MB, Fernandez JA, Dombrowski Y, McArt DG, Dunne PD, McQuaid S, Gray RT, Murray LJ, Coleman HG, James JA, Salto-Tellez M, Hamilton PW. QuPath: open source software for digital pathology image analysis. Sci Rep. 2017;7:16878. doi: 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmidt U, Weigert M, Broaddus C, Myers G. Cell detection with star-convex polygons. Medical Image Computing and Computer Assisted Intervention - Miccai 2018, Pt Ii. 2018;11071:265–273. [Google Scholar]
- 26.Huang G, Liu Z, Van Der Maaten L, Weinberger KQ. Densely connected convolutional networks. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017:4700–4708. [Google Scholar]
- 27.He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016:770–778. [Google Scholar]
- 28.Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556 2014. [Google Scholar]
- 29.Deng J, Dong W, Socher R, Li LJ, Li K, Li FF. ImageNet: a large-scale hierarchical image database. IEEE Conference on Computer Vision and Pattern Recognition. 2009;1-4:248–255. [Google Scholar]
- 30.Chen T, Kornblith S, Norouzi M, Hinton G. A simple framework for contrastive learning of visual representations. International Conference on Machine Learning. 2020;119:1597–1607. [Google Scholar]
- 31.He KM, Zhang XY, Ren SQ, Sun J. Delving deep into rectifiers: surpassing human-level performance on ImageNet classification. IEEE International Conference on Computer Vision (ICCV) 2015:1026–1034. [Google Scholar]
- 32.Paszke A, Gross S, Massa F, Lerer A, Bradbury J, Chanan G, Killeen T, Lin ZM, Gimelshein N, Antiga L, Desmaison A, Köpf A, Yang E, DeVito Z, Raison M, Tejani A, Chilamkurthy S, Steiner B, Fang L, Bai JJ, Chintala S. PyTorch: an imperative style, high-performance deep learning library. Advances in Neural Information Processing Systems. 2019:32. [Google Scholar]
- 33.Gamper J, Koohbanani NA, Benes K, Graham S, Jahanifar M, Khurram SA, Azam A, Hewitt K, Rajpoot N. Pannuke dataset extension, insights and baselines. arXiv preprint arXiv:2003.10778 2020. [Google Scholar]
- 34.Graham S, Vu QD, Raza SEA, Azam A, Tsang YW, Kwak JT, Rajpoot N. Hover-Net: simultaneous segmentation and classification of nuclei in multi-tissue histology images. Med Image Anal. 2019;58:101563. doi: 10.1016/j.media.2019.101563. [DOI] [PubMed] [Google Scholar]
- 35.McInnes L, Healy J, Melville J. Umap: uniform manifold approximation and projection for dimension reduction. arXiv preprint arXiv:1802.03426 2018. [Google Scholar]
- 36.McInnes L, Healy J, Astels S. hdbscan: hierarchical density based clustering. J Open Source Softw. 2017;2:205. [Google Scholar]
- 37.Spurr LF, Weichselbaum RR, Pitroda SP. Tumor aneuploidy predicts survival following immunotherapy across multiple cancers. Nat Genet. 2022;54:1782–1785. doi: 10.1038/s41588-022-01235-4. [DOI] [PubMed] [Google Scholar]
- 38.Potapova TA, Zhu J, Li R. Aneuploidy and chromosomal instability: a vicious cycle driving cellular evolution and cancer genome chaos. Cancer Metastasis Rev. 2013;32:377–389. doi: 10.1007/s10555-013-9436-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Niehues JM, Quirke P, West NP, Grabsch HI, van Treeck M, Schirris Y, Veldhuizen GP, Hutchins GGA, Richman SD, Foersch S, Brinker TJ, Fukuoka J, Bychkov A, Uegami W, Truhn D, Brenner H, Brobeil A, Hoffmeister M, Kather JN. Generalizable biomarker prediction from cancer pathology slides with self-supervised deep learning: a retrospective multi-centric study. Cell Rep Med. 2023;4:100980. doi: 10.1016/j.xcrm.2023.100980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saillard C, Dubois R, Tchita O, Loiseau N, Garcia T, Adriansen A, Carpentier S, Reyre J, Enea D, von Loga K, Kamoun A, Rossat S, Wiscart C, Sefta M, Auffret M, Guillou L, Fouillet A, Kather JN, Svrcek M. Validation of MSIntuit as an AI-based pre-screening tool for MSI detection from colorectal cancer histology slides. Nat Commun. 2023;14:6695. doi: 10.1038/s41467-023-42453-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stirling DR, Swain-Bowden MJ, Lucas AM, Carpenter AE, Cimini BA, Goodman A. CellProfiler 4: improvements in speed, utility and usability. BMC Bioinformatics. 2021;22:433. doi: 10.1186/s12859-021-04344-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiao X, Wang Z, Kong Y, Lu H. Deep learning-based morphological feature analysis and the prognostic association study in colon adenocarcinoma histopathological images. Front Oncol. 2023;13:1081529. doi: 10.3389/fonc.2023.1081529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu C, Bera K, Wang X, Prasanna P, Xu J, Janowczyk A, Beig N, Yang M, Fu P, Lewis J, Choi H, Schmid RA, Berezowska S, Schalper K, Rimm D, Velcheti V, Madabhushi A. A prognostic model for overall survival of patients with early-stage non-small cell lung cancer: a multicentre, retrospective study. Lancet Digit Health. 2020;2:e594–e606. doi: 10.1016/s2589-7500(20)30225-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.