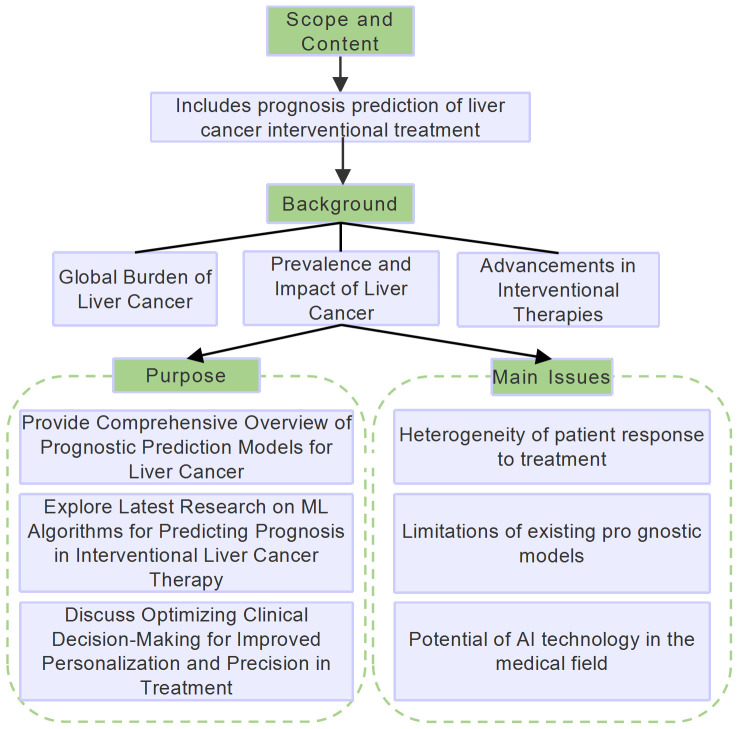
Abstract
The treatment for liver cancer has transitioned from traditional surgical resection to interventional therapies, which have become increasingly popular among patients due to their minimally invasive nature and significant local efficacy. However, with advancements in treatment technologies, accurately assessing patient response and predicting long-term survival has become a crucial research topic. Over the past decade, machine algorithms have made remarkable progress in the medical field, particularly in hepatology and prognosis studies of hepatocellular carcinoma (HCC). Machine algorithms, including deep learning and machine learning, can identify prognostic patterns and trends by analyzing vast amounts of clinical data. Despite significant advancements, several issues remain unresolved in the prognosis prediction of liver cancer using machine algorithms. Key challenges and main controversies include effectively integrating multi-source clinical data to improve prediction accuracy, addressing data privacy and ethical concerns, and enhancing the transparency and interpretability of machine algorithm decision-making processes. This paper aims to systematically review and analyze the current applications and potential of machine algorithms in predicting the prognosis of patients undergoing interventional therapy for liver cancer, providing theoretical and empirical support for future research and clinical practice.
Keywords: Machine algorithms, liver cancer, interventional therapy, prognosis prediction
Introduction
Liver cancer is one of the most common malignant tumors worldwide and ranks third among the leading causes of cancer-related deaths in humans [1]. According to estimates from the International Agency for Research on Cancer (IARC), there were approximately 870,000 new liver cancer cases and 760,000 deaths globally in 2022 [2]. Hepatocellular carcinoma (HCC) is the most common type of liver cancer, accounting for about 80% of all liver cancer cases, followed by intrahepatic cholangiocarcinoma (ICC), which accounts for approximately 15%, with other rare types of liver cancer making up around 5% [2,3]. This review primarily focuses on HCC due to its predominant role in liver cancer; however, we also address ICC and other less common types to provide a comprehensive overview of the current state of prognostic prediction models for liver cancer patients undergoing interventional treatments.
Interventional therapy for liver cancer, such as transcatheter arterial chemoembolization (TACE), is a crucial treatment method that can effectively control tumor growth and extend patient survival across early, intermediate, and advanced stages [4,5]. Interventional treatment strategies for liver cancer are either vascular or non-vascular (Figure 1). Non-vascular interventional techniques include various ablation methods, such as microwave ablation (MWA), radiofrequency ablation (RFA), irreversible electroporation (IRE), cryoablation (CRA), high-intensity focused ultrasound (HIFU), and laser ablation (LSA) [6]. Additionally, radiotherapy, including stereotactic body radiotherapy (SBRT) and iodine-125 seed implantation brachytherapy, is also part of non-vascular interventional therapy. In the vascular interventional treatment domain, transhepatic arterial interventions, particularly TACE and hepatic arterial infusion chemotherapy (HAIC), are mainstream clinical choices due to their significant efficacy. However, patients’ responses and prognoses to interventional treatments vary significantly, with objective response rates (ORRs) ranging from 40% to 80% and overall survival (OS) time varying from 13 to 48 months [7,8] (Figure 1).
Figure 1.
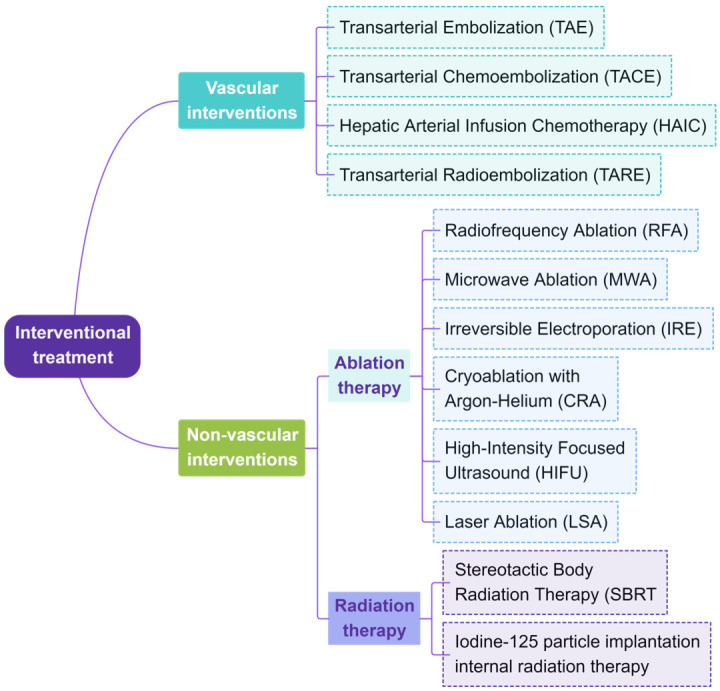
Main interventional treatment methods.
In recent years, with the development of artificial intelligence (AI) technology, machine learning (ML) has been increasingly applied in the medical field, providing new approaches for predicting the prognosis of liver cancer. ML processes large amounts of clinical and imaging data, allowing for advanced feature generation and quantitative radiomics parameter analysis, thus helping to identify hidden patterns in the data [9,10]. Compared to traditional statistical methods, ML algorithms can detect complex patterns in liver cancer treatment data more clearly. These algorithms simulate human learning process, effectively extracting and analyzing key features from multi-source data, such as tumor growth dynamics, changes in imaging performance, and post-treatment response [11,12]. This review aims to explore the latest research advancements of ML algorithms in predicting the prognosis of patients undergoing interventional therapy for liver cancer by summarizing recent key clinical data on interventional treatment outcomes, and discussing how these algorithms can optimize clinical decision-making processes, so as to improve treatment personalization and precision. Flowchart of the study is shown in Figure 2.
Figure 2.
Flowchart of the study.
Overview of liver cancer interventional therapies
Interventional therapy plays a crucial role in managing intermediate and advanced HCC patients, especially for those who are ineligible for surgery or transplantation. Since early-stage HCC often lacks obvious symptoms, most patients are diagnosed at an advanced stage [13]. In the Chinese Guidelines for the Diagnosis and Treatment of Primary Liver Cancer (2022 Edition) [14], interventional therapy is recommended as a treatment strategy spanning from early to late-stage liver cancer, with TACE recognized as one of the most commonly used non-surgical treatments for liver cancer. Similarly, in the internationally recognized Barcelona Clinic Liver Cancer (BCLC) Staging System and Treatment Strategy [15], interventional therapy, particularly ablation therapy, is recommended as the first-line treatment for very early-stage liver cancer (single tumor ≤ 2 cm) in patients not suitable for liver transplantation.
Ablation therapy has been shown to provide efficacy comparable to surgical resection in early-stage liver cancer patients [16]. For intermediate and advanced liver cancer, downstaging conversion therapy strategies, especially local interventional therapy, have played a key role in converting initially unresectable liver cancer into a resectable state [17]. After years of in-depth research, the combination of local interventional therapy and systemic targeted immunotherapy has been proven to significantly improve tumor response rates and conversion resection possibilities, becoming a crucial strategy for downstaging intermediate and advanced liver cancers [18]. A prospective phase II study in China (DoHAICs study, NCT05166772) evaluated the efficacy of HAIC combined with donafenib and sintilimab as a first-line treatment for unresectable liver cancer [19]. The results showed that the ORR of the combined treatment was 78.3%, with a conversion success rate of 65.2%. Recent studies have reported better tumor response rates at 1 month, 3 months, and 6 months following TACE combined with MWA treatment [7].
Although interventional therapy offers diverse treatment options and demonstrates good efficacy for HCC patients, challenges remain in predicting treatment response due to biological heterogeneity of tumors and complications, such as bleeding, liver and kidney function impairment, and ectopic embolization of embolic agents, which are crucial for ensuring patient safety and treatment efficacy [7,20]. These challenges highlight the importance of accurately predicting patient prognosis.
Overview of machine algorithms
Machine algorithms, such as ML and deep learning, have shown significant achievements and application potential in medical prognosis prediction. ML technology can automatically learn and identify patterns from historical data, while deep learning simulates the human brain’s working mechanisms, using complex algorithms to process and analyze data.
Logistic regression (LR), known for its simple model and high computational efficiency, is widely used in liver cancer diagnosis, prediction, and prognosis assessment [21,22]. Support vector machines (SVM) are powerful classification algorithms that find optimal hyperplanes in the feature space to differentiate between categories. In medical image processing, SVMs are often combined with feature extraction techniques [23]. Random forest (RF) algorithms improve classification accuracy and evaluate the importance of different clinical features by constructing multiple decision tree models [24]. Although the k-nearest neighbors (KNN) algorithm has limitations in computational efficiency, its simplicity and intuitive nature still make it suitable for classification tasks in small patient datasets [25]. Naive Bayes simplifies computation by assuming feature independence, making it useful for handling gene expression data with many features and helping identify relevant genetic markers [26]. Gradient boosting decision trees (GBDT) construct models through iterative optimization, accurately evaluating and guiding the latest model with annotations generated from privileged information [27].
Deep learning algorithms, such as convolutional neural networks (CNN), recurrent neural networks (RNN), and deep neural networks (DNN), automatically extract features in medical image analysis, improving the accuracy of liver cancer diagnosis and metastasis prediction [28,29]. Wu et al. [30] proposed a phase difference network (PDN), utilizing phase difference and multi-head self-attention mechanisms to distinguish HCC and ICC from four-phase CT images, showing better performance than traditional deep learning methods. The Transformer, a neural network model based on self-attention mechanisms, consists of multiple encoders and decoders. Tang et al. [31] developed a hybrid model combining graph neural networks and Transformer, which effectively utilized global context information from whole-slide images, significantly improving the accuracy and clinical value of HCC prognosis assessment.
Generative adversarial networks (GANs) demonstrate significant potential in medical image enhancement and data generation. The 2021 AI data challenge successfully showcased GANs’ ability to generate numerous rare malignant tumor MRI images from a few real MRI samples, with qualitative and quantitative evaluations confirming its effectiveness [32]. Bayesian networks, with their advantages in handling uncertainty and causal relationship modeling, provide deeper insights into liver cancer prediction and treatment. Cai et al. [33] identified portal vein tumor thrombus as the most important predictor of survival time in HCC patients after liver resection using Bayesian networks and importance measurement methods.
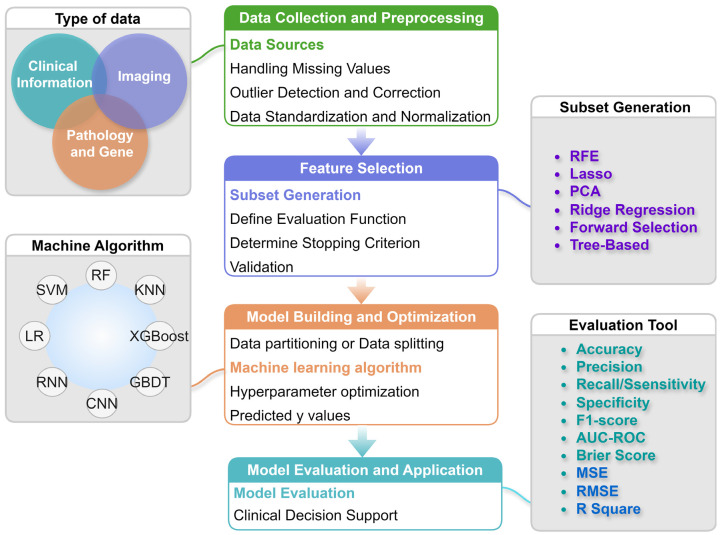
The clinical application process of ML includes clinical data acquisition and preprocessing, feature selection, model training and tuning, model diagnosis, multi-model fusion, and deploying validated models into clinical practice to assist in diagnosis and treatment decisions (Figure 3). In clinical applications, the ML process must particularly focus on data representativeness and diversity, ensuring the model generalizability to different patient groups while considering interpretability, allowing doctors to understand and trust its outputs.
Figure 3.
Flow of the machine learning algorithm. LR: Logistic Regression; SVM: Support Vector Machine; RF: Random Forest; KNN: K-Nearest Neighbors; XGBoost: eXtreme Gradient Boosting; GBDT: Gradient Boosting Decision Tree; CNN: Convolutional Neural Network; RNN: Recurrent Neural Network; RFE: Recursive Feature Elimination; PCA: Principal Component Analysis; AUC-ROC: Area Under the Receiver Operating Characteristic Curve; MSE: Mean Squared Error; RMSE: Root Mean Squared Error.
Application of ML algorithms in predicting prognosis for liver cancer patients undergoing interventional therapies
TACE
Many HCC patients are diagnosed at intermediate or advanced stages, missing the optimal treatment window. TACE is the gold-standard treatment for intermediate HCC patients [34,35], but patients’ responses to TACE vary significantly, and not all benefit from it [36]. Therefore, developing models to predict the efficacy of TACE is crucial.
Previous studies have constructed various ML models using clinical and radiological variables to predict outcomes for HCC patients post-TACE. Spleen volume (SV) is an undervalued, automatically retrievable imaging biomarker. Muller et al. [37] used CNN algorithms to automatically assess SV and found that high SV correlated significantly with reduced survival in liver cancer patients. SV is also a strong predictor of hepatic decompensation post-TACE. Bartnik et al. [38] analyzed radiomics features from multiple organs of interest in liver cancer patients using deep learning, showing that their radiomics model outperformed traditional clinical models in predicting progression-free survival (PFS) after TACE. These studies highlight the importance of non-tumor regional features in clinical prediction. Bernatz et al. [39] analyzed CT images after three consecutive TACE procedures, combining radiomics features with clinical mHAP-II scores. Their RF model achieved an AUC of 0.70 and an accuracy of 0.72 at the lesion level, an AUC of 0.62 at the patient level, and a C-index of 0.67 for OS prediction, demonstrating potential in improving TACE response prediction. Dong et al. [40] selected clinical data from patients receiving their first TACE for unresectable liver cancer, identified three features (portal vein tumor thrombus type, albumin level, and intrahepatic tumor distribution) using the LASSO algorithm, and built various prognostic models (XGBoost, decision tree, SVM, RF, KNN, and ANN). Among them, the RF model performed best, with an AUC of 0.802, accuracy of 0.784, sensitivity of 0.904, and specificity of 0.480. Ma et al. [41] compared different machine algorithms for predicting responses in unresectable HCC patients receiving lenvatinib combined with TACE, finding that SVM and RF models performed best in accuracy and AUC. The RF model reached an AUC of 0.91, indicating high predictive accuracy. Peng et al. [42] emphasized the high accuracy of radiomics conventional ML and deep learning models in preoperative TACE response prediction (deep learning model AUC = 0.972, integrated model AUC = 0.994). Combining these models with clinical variables offers a novel and accurate method for predicting treatment responses in intermediate-stage liver cancer patients. Zhang et al. [43] developed and validated a fully automated deep learning framework to predict TACE response in real time for HCC patients. Overall, these studies demonstrate the potential of ML models in predicting TACE response and the importance of non-tumor regional features and automated imaging analysis. Relevant studies are summarized in Table 1.
Table 1.
ML-based prognostic model characteristics of HCC patients after TACE
| Author | ML Algorithms | Prediction Targets | Key Predictors | Main Results/Performance Indicators | Model Validation and Interpretability | Other Important Findings |
|---|---|---|---|---|---|---|
| Muller 2022 [32] | CNN | OS, PFS, TTUP | SV | Significant correlation between SV and survival rates | Internal validation, S⊘rensen Dice score, Bland-Altman plot | Spleen volume significantly correlates with risk of liver dysfunction after TACE |
| Bartnik 2024 [33] | DL, RSF, COX | OS, PFS | Tumor VOI and non-tumor VOI | OS: C-index range 0.616 to 0.640. PFS: C-index 0.713 | Cross-validation, XAI | Multiple VOI features extracted from CT images, overcoming manual segmentation limitations |
| Bernatz 2023 [34] | RF | TACE response, OS | Radiomic features and clinical mHAP-II score | Lesion-level AUC 0.70, Accuracy 0.72; Patient-level AUC 0.62; C-index 0.67 | Reliability and redundancy analysis | Supports the potential of lipid deposition as an imaging biomarker |
| Dong 2021 [35] | XGBoost, Decision Tree, SVM, RF, KNN, ANN | Early treatment response post first cTACE | Portal vein tumor thrombus type, Albumin level, Tumor distribution in liver | RF model performed best, AUC 0.802, Accuracy 0.784, Sensitivity 0.904, Specificity 0.480 | 5-fold cross-validation | Portal vein tumor thrombus type is the most important factor for predicting response to first cTACE treatment |
| Ma 2023 [36] | CART, AdaBoost, XGBoost, RF, SVM | Response to combination therapy (lenvatinib + TACE) | K, LDL, D-D, Red blood cells, ALT, ALB, Mono, Tumor size, TG, and Age | RF model AUC 0.91, SVM and RF performed best | SHAP algorithm enhanced model interpretability | Lower serum K, older age, higher BMI, and larger tumor size correlate with better efficacy of combination therapy |
| Peng 2021 [37] | Linear model, LR, SVM, GBM, RF, DL | TACE treatment response | Tumor size | DL model AUC 0.972, Integrated model AUC 0.994 | Multicenter data validated model robustness | Tumor size significantly correlates with initial treatment response, while AFP levels do not |
| Zhang 2022 [38] | ResNet18 and Multilayer Perceptron | TACE treatment response | DSA video information, Demographics, and liver function parameters | Accuracy rates on internal and external validation sets were 78.2% and 75.1% respectively | Internal and external validation | Predictive model performance using segmentation results as input is slightly lower than using true segmentation results, but not significantly |
ML: Machine Learning; CNN: Convolutional Neural Network; OS: Overall Survival; PFS: Progression-Free Survival; TTUP: Time to Tumor Progression; SV: Segmentation Volume; TACE: Transarterial Chemoembolization; VOI: Volume of Interest; DL: Deep Learning; RSF: Random Survival Forest; COX: Cox Proportional Hazards Model; RF: Random Forest; AUC: Area Under the Curve; mHAP-II: Modified Hepatoma Arterial Embolization Prognostic Score; SVM: Support Vector Machine; KNN: k-Nearest Neighbors; GBM: Gradient Boosting Machine; LR: Logistic Regression; DL: Deep Learning (used in the context of the algorithm name); AFP: Alpha-Fetoprotein; ALT: Alanine Aminotransferase; ALB: Albumin; Mono: Monocytes; TG: Triglyceride; BMI: Body Mass Index; DSA: Digital Subtraction Angiography; AUC: Area Under the Receiver Operating Characteristic Curve; XAI: Explainable Artificial Intelligence; SHAP: SHapley Additive exPlanations.
TACE models typically combine clinical, radiological, and radiomics features, with non-tumor regional features (such as spleen volume) serving as important predictors. These models employ deep learning and conventional ML algorithms, with RF models showing excellent performance in several studies. AUC values range from 0.62 to 0.994, with most studies reporting AUCs above 0.8. Key predictive features include portal vein tumor thrombus, albumin levels, and intrahepatic tumor distribution. Automated imaging analysis and deep learning frameworks demonstrate significant potential, offering new perspectives for predicting TACE efficacy.
HAIC
HAIC involves catheterizing the hepatic artery supplying the tumor and continuously infusing chemotherapeutic agents. It is advantageous for advanced liver cancer with portal vein tumor thrombus, arterio-venous fistula, and poorly vascularized liver metastases [35].
Recent advances show that combining deep learning with radiomics features significantly improves the predicted accuracy of HAIC treatment response. Xu et al. [44] developed the DLRN model, integrating deep learning, radiomics features, and key clinical variables, achieving high accuracy in training, internal, and external validation cohorts (AUCs of 0.988, 0.915, and 0.896, respectively). The model also predicted survival based on treatment response, with the median OS in the response group significantly higher than in the non-response group. Quan et al. [45] used the InceptionV4 CNN model with preoperative MRI data and clinical factors (HAIC cycle count, tumor thrombus, neutrophil-lymphocyte ratio, and gamma-glutamyltransferase), achieving an AUC of 0.871 in the training cohort and 0.826 in the internal validation cohort. Another retrospective study used a combination model of MRI radiomics and ALBI score to predict HAIC treatment response, providing a nomogram to assess PFS [46]. Patients with high scores had a median PFS of 6.0 months, significantly shorter than 9.0 months in low-score patients. He et al. [47] further explored radiomics features extracted from dual-phase contrast-enhanced CT (CECT), combined with clinical variables and MTM subtypes, and established a multi-task deep learning radiomics (MDLR) model to provide accurate HAIC prognostic risk stratification for HCC patients. Relevant studies are summarized in Table 2.
Table 2.
ML-based prognostic model characteristics of HCC patients after HAIC
| Author | ML Algorithms | Prediction Target | Key Predictive Factors | Main Results/Performance Metrics | Model Validation & Interpretability | Other Important Findings |
|---|---|---|---|---|---|---|
| Xu 2022 [39] | DL, XGBoost | OR | APE, RVI, R score, DL score | AUC in training set = 0.988, internal validation set AUC = 0.915, external validation set AUC = 0.896 | Internal and external validation | Radiological parameters (APE and RVI) may predict the efficacy of HAIC better than clinical characteristics |
| Quan 2024 [40] | InceptionV4-CNN | HAIC response | MRI data, HAIC cycles, cancer thrombus, NLR | AUC in training cohort = 0.871, internal validation cohort AUC = 0.826 | Cross-validation and independent validation, CAM used for visualization | Age, HAIC cycle number, tumor thrombus, extrahepatic spread, and AST level are independent predictors |
| Zhao 2023 [41] | LR | PFS | Radiomic score (Radscore) and ALBI score | Combined model AUC in training and validation sets are 0.79 and 0.75, respectively | Internal validation | NA |
| He 2023 [42] | MDLR | Post-HAIC patient prognosis | CECT radiomic features, portal vein cancer thrombus, HAIC response, HAIC cycles | AUC for survival prediction model in internal and external validation sets are 0.87 and 0.83 | Internal and external validation | Tumor burden and distribution as well as tumor microenvironment features are associated with prognosis |
XGBoost: Extreme Gradient Boosting; OR: Objective Response; APE: Asymmetry of Parenchymal Enhancement; RVI: Reduction in Viable Tumor on Initial; R score: Radiographic Response Score; DL score: Deep Learning Score; HAIC: Hepatic Arterial Infusion Chemotherapy; MRI: Magnetic Resonance Imaging; NLR: Neutrophil-to-Lymphocyte Ratio; CAM: Class Activation Mapping; PFS: Progression-Free Survival; Radscore: Radiomic Score; ALBI: Albumin-Bilirubin Grade; MDLR: Multitask Deep Learning Radiomics; CECT: Contrast-Enhanced Computed Tomography; AST: Aspartate Aminotransferase; NA: Not Available.
HAIC models often integrate deep learning, radiomics features, and clinical variables, commonly using pre-treatment MRI and CT images for feature extraction. These models achieve AUC values ranging from 0.826 to 0.988, with key predictive features including HAIC cycle count, tumor thrombus, neutrophil-lymphocyte ratio, and gamma-glutamyltransferase. MDLR models exhibit high accuracy, capable of predicting both treatment response and survival outcomes, providing personalized prognostic assessment tools for HAIC treatment.
TARE
Transarterial radioembolization (TARE), also known as selective internal radiation therapy (SIRT), involves injecting the radioactive isotope yttrium-90 (90Y) to treat liver cancer.
Roll et al. [48] extracted and analyzed radiomics features from pre-TARE CT images of patients with colorectal cancer liver metastases. Two independent radiomics features (energy and maximum correlation coefficient) reflected tumor heterogeneity. Their multivariate LR model successfully distinguished high-risk from low-risk patients, with an AUC of 0.75, providing a new prognostic assessment tool. Ince et al. [49] found that ML models (SVM, LR, RF, LightGBM) combining radiomics features from pre-treatment contrast-enhanced MRI and clinical data significantly improved TARE response prediction. Kobe et al. [50] used features from pre-TARE CBCT images, achieving high sensitivity (94.2%) and moderate specificity (67.7%) with a multi-layer perceptron ANN in an external test set. Marinelli et al. [51] collected baseline and early post-TARE MRI of HCC patients, using semi-automatic segmentation to extract radiomics features. Their XGBoost model showed high accuracy (AUC = 0.89) in an independent validation cohort, particularly outperforming models using only clinical parameters and conventional imaging features in predicting complete response. Balli et al. [52] combined dynamic MRI radiomics scores with clinical features using LASSO feature selection and LR to build a radiomics model. Triple cross-validation optimized parameters, with the model predicting response to 90Y TARE in intrahepatic cholangiocarcinoma patients, showing that responders had significantly lower radiomics scores. Axial T2W with fat suppression sequence achieved an AUC of 0.839, indicating high predictive accuracy. Aujay et al. [53] compared the European Association for the Study of the Liver (EASL) criteria, using radiomics combined with MRI data to assess treatment response in patients with locally advanced HCC undergoing TARE. They found that long emphasis, short axis length, surface area, and grayscale non-uniformity in arterial phase images could accurately predict early treatment response, demonstrating the potential of radiomics combined with LR in predicting TARE efficacy. Relevant studies are summarized in Table 3.
Table 3.
ML-based prognostic model characteristics of HCC patients after TARE
| Author | ML Algorithms | Prediction Target | Key Predictive Factors | Main Results/Performance Metrics | Model Validation & Interpretability | Other Important Findings |
|---|---|---|---|---|---|---|
| Roll 2024 [43] | Multivariate LR | Treatment Response and Survival Outcome | Energy, Maximal Correlation Coefficient | AUC: 0.75 | Feature selection with Boruta algorithm | Radiomic analysis can quantify tumor heterogeneity, including blood supply, cellular vitality, density, and fibrosis |
| Ince 2023 [44] | SVM, LR, RF, LightGBM | Treatment Response | 8 Radiomic features, 4 Clinical features | AUC: 0.88-0.94 | 5-fold cross-validation | Age and preoperative total bilirubin level significantly correlate with TARE treatment response |
| Kobe 2021 [45] | Multilayer Perceptron, ANN | Disease Control (PR/SD) and PD | 104 Texture Analysis Features from CBCT, 15 features after selection | AUC: 0.85, Sensitivity 94.2%, Specificity 67.7% | 10-fold cross-validation | NA |
| Marinelli 2023 [46] | XGBoost | Treatment Response at 4-6 Months Post-Treatment | Radiomic features from baseline and early post-treatment (1-2 months) MRI images | AUC: 0.89 | NA | Combined baseline and early follow-up MRI radiomic data better predict patient treatment response |
| Balli 2024 [47] | LASSO, LR | Radiological Response at 6 Months Post-Treatment | Rad-score, Bifurcation Lesions | AUC: 0.696-0.880 | DeLong test, NRI, IDI | First study to use MRI radiomics to predict TARE treatment response in ICC patients |
| Aujay 2022 [48] | LR | Treatment Response | Longitudinal Emphasis, Minor Axis Length, Surface Area, and Gray Level Non-uniformity | AUC: 1 | Cross-validation | Heterogeneity parameters in arterial and portal venous phase images before and after treatment not significantly different between responders and non-responders |
LightGBM: A gradient boosting framework that uses tree-based learning; ANN: Artificial Neural Network; PR: Partial Response; SD: Stable Disease; PD: Progressive Disease; CBCT: Cone Beam Computed Tomography; ICC: Intrahepatic Cholangiocarcinoma; TARE: Transarterial Radioembolization; NRI: Net Reclassification Improvement; IDI: Integrated Discrimination Improvement.
TARE models utilize pre-treatment imaging (CT, MRI, CBCT) for radiomics feature extraction, with the combination of radiomics features and clinical data significantly improving prediction accuracy. These models employ various ML algorithms, including SVM, LR, RF, LightGBM, and XGBoost, with AUC values ranging from 0.75 to 0.89. The models can predict both overall response and complete response, with key radiomics features reflecting tumor heterogeneity. These models offer new perspectives for predicting TARE treatment efficacy, aiding in personalized treatment decisions.
RFA
Treatment guidelines from BCLC, CNLC, and NCCN emphasize RFA as the preferred method for early HCC with small tumors. Studies indicate that the five-year survival rate for RFA-treated patients ranges from 26% to 56.7%, and the five-year disease-free survival rate is between 15% and 28.7% [54-56]. However, the high recurrence rate, rapid tumor growth, and invasiveness post-recurrence remain to be clinical concerns [57].
Recent research explores ML in predicting outcomes for HCC patients undergoing RFA. One study used the XGBoost algorithm on multidimensional data from patients receiving localized RFA between 2018 and 2022 [58]. Their model achieved an accuracy of 78.9% and an AUC of 0.80 in an independent validation set. Tong et al. [59] compared five algorithms (LR, decision tree, GBDT, RF, GBM) in predicting overall mortality post-RFA, and identified platelet count, Alpha-Fetoprotein (AFP), age, tumor size, and total bilirubin as key prognostic factors. GBM was found to have the highest accuracy (0.681), indicating the potential and differences of various algorithms in predicting HCC patient prognosis. Sato et al. [60] developed a transformer-based ML model analyzing data from 1778 treatment-naïve HCC patients undergoing RFA, aiming to improve the prediction of OS. This model used clinical and pathological features, evaluated by Harrel’s c-index, showing superior discrimination compared to traditional deep learning models. Results indicated the transformer’s high discriminative ability in external validation cohorts and its capacity to provide personalized cumulative recurrence prediction curves. Another study analyzed 898 early-stage HCC patients using Lasso and Cox regression analysis to identify independent risk factors like age, gender, BCLC stage, tumor size, globulin, and γ-glutamyl transpeptidase [61]. The nomogram, validated by C-index, ROC, calibration, and decision curve analysis (DCA), showed excellent discrimination, consistency, and clinical utility. RFA treatment showed potential in improving long-term survival for solitary HCC patients with tumor diameters ≤ 5 cm. He et al. [62] revealed the effectiveness of RFA in improving 5-year OS and cancer-specific survival (CSS) rates compared to radiotherapy, chemotherapy, and blank control groups by analyzing data from the SEER database. Further Cox regression analysis and the development of the XGBoost model identified key prognostic factors such as age, race, marital status, grade, cirrhosis, tumor size, and AFP level, and constructed a valid predictive model. The XGBoost model demonstrated good predictive performance in the validation cohort through ROC curve, calibration plot, and DCA, providing a personalized CSS predictive tool for patients with isolated HCC with a diameter of less than 5 cm. The relevant studies are listed in Table 4.
Table 4.
ML-based prognostic model characteristics of HCC patients after RFA
| Author | ML Algorithms | Prediction Target | Key Predictive Factors | Main Results/Performance Metrics | Model Validation & Interpretability | Other Important Findings |
|---|---|---|---|---|---|---|
| Hamed 2024 [53] | XGBoost | Disease control at 12 months | Child-Pugh score, WBC count, Heparan concentration, Diabetes, Hypertension, Tumor size, AFP | Accuracy and AUC are 78.9% and 0.80, respectively | Internal validation | 1-year survival and local control rates are 94.6% and 61.3%, respectively |
| Tong 2021 [54] | RF, LR, LightGBM, GBDT, Decision Tree | Total mortality | PLT, AFP, Age, Tumor size, Total bilirubin | GBDT has the highest accuracy (0.681), precision (0.721), AUC: 0.714 | Internal validation | NA |
| Sato 2024 [55] | Transformer-based ML model (SurvTRACE) | OS | Age, Gender, Number and size of tumors, Liver function indicators (Albumin, Total bilirubin, AST, ALT), Tumor markers (AFP, DCP), Hepatitis virus infection status, Platelet count, Prothrombin time | C-index of 0.69 | Internal and external validation | 5-year and 10-year survival rates are 63.7% and 30.4%, respectively. 1-year, 3-year, and 5-year local tumor recurrence rates are 1.7%, 5.3%, and 6.5%, respectively |
| Zhang 2024 [56] | Lasso regression and Cox regression analysis | RFS | Age, Gender, BCLC stage, Tumor size, Glob, γ-GT | AUC for 1-year, 3-year, and 5-year RFS are 0.721, 0.756, and 0.779, respectively | Internal validation | NA |
| He 2023 [57] | LR, SVM, RF, KNN, XGBoost | CSS | Liver cirrhosis, Tumor size, AFP level, Age, Marital status | XGBoost model’s AUC for predicting 1-year, 3-year, and 5-year CSS are 0.88, 0.81, and 0.79, respectively | Internal validation | Compared to other treatment modalities, RFA shows better performance in improving OS and CSS for patients with single HCC ≤ 5 cm, but still lower than hepatectomy |
RFA: Radiofrequency Ablation; RF: Random Forest; PLT: Platelet Count; DCP: Des gamma Carboxyprothrombin; RFS: Recurrence-Free Survival; BCLC: Barcelona Clinic Liver Cancer; Glob: Globulin; γ-GT: γ-Glutamyl Transpeptidase; CSS: Cancer-Specific Survival; c-index: Concordance Index; AFP: Alpha-Fetoprotein.
RFA models utilize multidimensional clinical and pathological data, comparing various algorithms including XGBoost, LR, decision tree, GBDT, RF, and GBM. AUC values range from 0.68 to 0.80, with key prognostic factors including platelet count, AFP, age, tumor size, and total bilirubin. Transformer-based models show promise in predicting OS. These models can predict both overall mortality and cancer-specific survival, providing powerful tools for prognostic assessment following RFA treatment.
MWA
MWA is a commonly used ablation method, particularly for tumors with diameters ranging from 3 to 5 cm. It has been shown to have high efficacy and ablation efficiency. Compared to radiofrequency ablation (RFA), MWA significantly shortens the procedure time and is less sensitive to the heat-sink effect of blood flow, thereby reducing the risk of incomplete ablation in the treatment of larger tumors [55]. However, despite its advantages in ablation efficiency, MWA does not show significant differences in local efficacy, complication rates, or long-term survival outcomes compared to RFA [63].
In a study predicting local tumor progression (LTP) in early-stage HCC patients post-MWA, Ren et al. [64] analyzed the clinicopathological data and ablation parameters of 607 untreated early HCC patients. They developed predictive models using four ML algorithms, including CatBoost, RF, XGBoost, and LR. Among these models, the CatBoost algorithm, which combined nine key variables - tumor number, albumin and alpha-fetoprotein levels, tumor size, age, and international normalized ratio - exhibited the highest predictive accuracy (AUC: 0.898). The study suggested that precise ablation planning and personalized treatment based on these predictive factors could significantly reduce the risk of LTP, thereby improving the success rate of MWA in treating early-stage HCC.
Another study [65] employed ML techniques to predict early recurrence (ER) risk by analyzing the clinical data of 1,574 early HCC patients who underwent MWA. This study constructed ML models, including RF, support vector machine (SVM), and XGBoost, and enhanced their interpretability using SHAP and LIME algorithms. The XGBoost model performed best in predicting ER, accurately identifying key risk factors such as tumor number, platelet count, and alpha-fetoprotein level. Their XGBoost-based prediction system is available online, providing clinicians with a practical tool (http://114.251.235.51:8001/).
In the study on the predictability of LTP after MWA in colorectal cancer liver metastases, clinical data and MRI radiomic features were analyzed to develop two combined models [66]. Model 2, which incorporated T2 fat-suppressed and early arterial phase T1 fat-suppressed features, showed better performance in predicting LTP, with an AUC of 0.981. These highly accurate models offer new perspectives for clinical practice, but the studies also emphasize the need for further large-scale research to validate the generalizability and reliability of these models. Related research is summarized in Table 5.
Table 5.
ML-based prognostic model characteristics of HCC patients after MWA
| Author | ML Algorithms | Prediction Target | Key Predictive Factors | Main Results/Performance Metrics | Model Validation & Interpretability | Other Important Findings |
|---|---|---|---|---|---|---|
| Ren 2023 [59] | CatBoost, SVM, RF, LR | LTP | Number of tumors, Albumin, AFP, Tumor size, Age, INR | Best performance by CatBoost model, AUC of 0.898 | Internal and external validation | NA |
| An 2022 [60] | LR, RF, SVM, XGBoost | ER | Tumor number, Platelets, AFP, Comorbidity score, WBC, ChE, PT, Neutrophils, Etiology | Best performance by XGBoost model, AUC 0.74 (internal) and 0.76 (external) | Internal and external validation, SHAP and LIME algorithms for model explanation | NA |
| Shahveranova 2023 [61] | LR | LTP | Preoperative extrahepatic metastasis, Tumor size, CA 19-9 | Combined Model 2 (clinical data and Phase 2 radiomic features) has the highest discriminative performance for LTP prediction (AUC 0.981) | NA | LTP group patients have significantly higher radiomic scores in both MRI phases (Phase 1 and Phase 2) |
CatBoost: A machine learning algorithm based on decision trees; LTP: Local Tumor Progression; INR: International Normalized Ratio; ER: Early Recurrence; ChE: Cholinesterase; PT: Prothrombin Time; WBC: White Blood Cell Count; CRLM: Colorectal Liver Metastasis; CA 19-9: A tumor marker for gastrointestinal malignancies; LIME: Local Interpretable Model-agnostic Explanations; AFP: Alpha-Fetoprotein.
MWA models focus primarily on predicting LTP and ER, utilizing various ML algorithms including CatBoost, RF, XGBoost, and LR. AUC values range from 0.898 to 0.981, with key predictive features including tumor number, albumin levels, AFP, tumor size, and platelet count. These models combine clinical data with MRI radiomics features, with some studies providing online prediction tools for clinical use, offering important references for prognostic assessment and personalized treatment following MWA.
Overall, these ML-based prognostic models for HCC patients undergoing interventional therapies share common characteristics. They typically combine clinical, radiological, and radiomics features to improve prediction accuracy. Various ML algorithms are applied, with RF and XGBoost frequently performing well. Pre-treatment imaging (CT, MRI) is commonly used for feature extraction. These models can predict various outcomes, including treatment response, survival, and recurrence. AUC values generally range from 0.7 to 0.9, indicating good to excellent predictive performance. Furthermore, there is an increasing trend towards using deep learning and multi-task learning approaches, providing more precise and personalized tools for prognostic assessment following HCC interventional treatments.
In summary, DL and ML models have demonstrated high accuracy in predicting outcomes for various liver cancer treatments. For instance, in PFS prediction, models by Bartnik et al. [38] and Quan et al. [45] outperformed traditional clinical models. In predicting treatment response, studies by Muller et al. [37] and Peng et al. [42] highlighted the advantages of radiomics combined with deep learning. For OS prediction, models by Ma et al. [41] and Sato et al. [60] showed strong predictive capabilities. Additionally, in LTP prediction, the models by Ren et al. [64] and those incorporating MRI radiomics features [66] demonstrated exceptionally high accuracy (Table 6).
Table 6.
Predictive accuracy of machine algorithm models for liver cancer outcomes
| Prediction Target | Author | Machine algorithm | AUC/C-index |
|---|---|---|---|
| PFS | Bartnik [38] | Deep Learning | - |
| PFS | Quan [45] | Deep Learning | 0.826 |
| Treatment Response | Muller [37] | CNN | - |
| Treatment Response | Peng [42] | Deep Learning | 0.994 |
| Treatment Response | Xu [39] | DLRN | 0.988 |
| CR Prediction | Marinelli [46] | XGBoost | 0.89 |
| OS | Ma [41] | RF | 0.91 |
| OS | Sato [60] | Transformer Model | - |
| LTP | Ren [64] | CatBoost | 0.898 |
| LTP | Shahveranova [66] | - | 0.981 |
PFS: Progression-Free Survival; LTP: Local Tumor Progression; CNN: Convolutional Neural Network.
Conclusion
Currently, the application of ML algorithms in predicting the prognosis of interventional therapy for liver cancer focuses on two main areas. The first is the prognostic evaluation based on imaging features, such as analyzing CT and MRI imaging data to identify tumor size, morphology, and vascular characteristics, thereby predicting treatment efficacy and recurrence risk [67]. For instance, deep learning algorithms have demonstrated remarkable ability in analyzing liver cancer CT images, accurately identifying tumor boundaries and vascular invasion [68-70]. The second area involves integrating multidimensional clinical information of patients, including age, gender, tumor stage, and liver function, to construct complex prognostic models that help determine the optimal treatment plan.
We found that models integrating clinical, imaging, and radiomics features exhibit superior predictive accuracy. Although RF and XGBoost algorithms perform well in many cases, researchers are exploring a range of algorithms from traditional ML to advanced deep learning methods. Imaging, particularly pre-treatment CT and MRI, plays a crucial role in feature extraction across most models. These models predict not only treatment response but also survival and recurrence risk, demonstrating good to excellent predictive capabilities, with AUC values typically ranging from 0.7 to 0.9. As deep learning and multi-task learning approaches become more prevalent, prediction accuracy and personalization are continually improving.
However, despite the significant advances in applying ML to HCC and ICC interventional treatments, there are areas needing further investigation. For instance, many studies rely on single-center data, so multi-center, large-scale studies are necessary to validate models across different patient populations. Additionally, while certain features (e.g., tumor characteristics, liver function markers) consistently emerge as important predictors across various interventions, a deeper understanding of their biological basis is needed.
With increasing model complexity, ensuring interpretability is crucial for clinical adoption. Techniques such as SHAP and LIME, used in some studies, should be more widely implemented. Moreover, despite promising results, integrating these models into clinical decision-making remains a challenge. Future research should focus on developing user-friendly interfaces and decision support tools.
Most current models emphasize short-term outcomes, so developing models that can predict long-term outcomes and account for changes in patient status over time is a crucial next step. With the growing use of combination therapies, models capable of predicting outcomes for these complex treatment regimens will be increasingly valuable.
Furthermore, the high accuracy of personalized treatment plans highlights the potential for highly individualized therapy. Future research should concentrate on developing dynamic models that can adapt recommendations as patient conditions evolve. For example, Zhang et al.’s [43] automated deep learning framework for TACE response prediction points to the possibility of real-time treatment outcome prediction, potentially allowing immediate adjustments during the procedure.
In the realm of liver cancer prognosis prediction, ML algorithms face core challenges including algorithm bias, data quality, and ethical considerations (Figure 4). Algorithm bias primarily manifests as models potentially overfitting specific datasets, impacting their applicability across diverse patient populations [71]. Addressing this issue requires rigorous cross-validation and generalization capability assessments to ensure model robustness. On the data front, diversity and consistency of data are crucial for enhancing model performance [72]. Promoting multicenter studies and establishing unified data standards can help expand dataset scope and improve model generalizability. Additionally, algorithm transparency and fairness are critical ethical considerations, necessitating that model decision processes be interpretable and unbiased, with effective oversight mechanisms in place [71,73-75].
Figure 4.
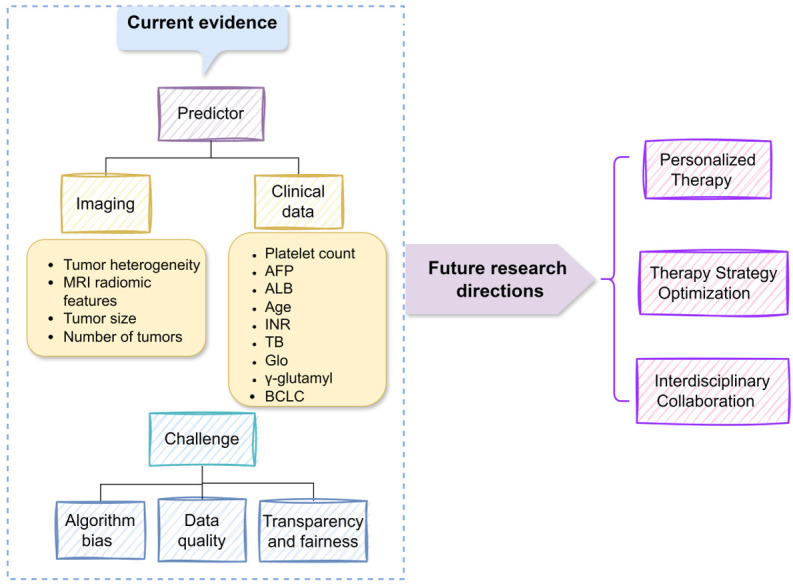
Summary of current evidence and future challenges for prognostic indicators in interventional therapy for liver cancer. AFP: Alpha-Fetoprotein; ALB: Albumin; INR: International Normalized Ratio; BCLC: Barcelona Clinic Liver Cancer.
Looking ahead, liver cancer treatment will become more personalized and precise. The integration of high-resolution imaging technologies and advanced ML algorithms will foster the development of intelligent decision support systems based on imaging features. Furthermore, emerging therapies such as targeted therapies and immunotherapies, along with in-depth research into biomarkers and molecular mechanisms, will drive the optimization of liver cancer treatment strategies, improving treatment outcomes and patient quality of life. Interdisciplinary collaboration and in-depth research are essential for continued progress in this field.
Disclosure of conflict of interest
None.
References
- 1.Sidali S, Trepo E, Sutter O, Nault JC. New concepts in the treatment of hepatocellular carcinoma. United European Gastroenterol J. 2022;10:765–774. doi: 10.1002/ueg2.12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Global cancer burden growing, amidst mounting need for services. World Health Organization; 2024. [PMC free article] [PubMed] [Google Scholar]
- 3.Rumgay H, Ferlay J, de Martel C, Georges D, Ibrahim AS, Zheng R, Wei W, Lemmens VEPP, Soerjomataram I. Global, regional and national burden of primary liver cancer by subtype. Eur J Cancer. 2022;161:108–118. doi: 10.1016/j.ejca.2021.11.023. [DOI] [PubMed] [Google Scholar]
- 4.Cassinotto C, Nogue E, Morell M, Panaro F, Molinari N, Guiu B. Changing trends in hepatocellular carcinoma management: results from a nationwide database in the last decade. Eur J Cancer. 2021;146:48–55. doi: 10.1016/j.ejca.2021.01.009. [DOI] [PubMed] [Google Scholar]
- 5.Duan R, Gong F, Wang Y, Huang C, Wu J, Hu L, Liu M, Qiu S, Lu L, Lin Y. Transarterial chemoembolization (TACE) plus tyrosine kinase inhibitors versus TACE in patients with hepatocellular carcinoma: a systematic review and meta-analysis. World J Surg Oncol. 2023;21:120. doi: 10.1186/s12957-023-02961-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Childs A, Meyer T. Hepatocellular Carcinoma: Treatment. 2019. pp. 703–714. [Google Scholar]
- 7.Ji J, Yang W, Shi HB, Liu S, Zhou WZ. Transcatheter arterial chemoembolization alone versus combined with microwave ablation for recurrent small hepatocellular carcinoma after resection: a retrospective comparative study. BMC Gastroenterol. 2022;22:321. doi: 10.1186/s12876-022-02387-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo Y, Ren Y, Chen L, Sun T, Zhang W, Sun B, Zhu L, Xiong F, Zheng C. Transarterial chemoembolization combined with camrelizumab for recurrent hepatocellular carcinoma. BMC Cancer. 2022;22:270. doi: 10.1186/s12885-022-09325-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mokrane FZ, Lu L, Vavasseur A, Otal P, Peron JM, Luk L, Yang H, Ammari S, Saenger Y, Rousseau H, Zhao B, Schwartz LH, Dercle L. Radiomics machine-learning signature for diagnosis of hepatocellular carcinoma in cirrhotic patients with indeterminate liver nodules. Eur Radiol. 2020;30:558–570. doi: 10.1007/s00330-019-06347-w. [DOI] [PubMed] [Google Scholar]
- 10.Zou ZM, Chang DH, Liu H, Xiao YD. Current updates in machine learning in the prediction of therapeutic outcome of hepatocellular carcinoma: what should we know? Insights Imaging. 2021;12:31. doi: 10.1186/s13244-021-00977-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong B, Zhang H, Duan Y, Yao S, Chen Y, Zhang C. Development of a machine learning-based model to predict prognosis of alpha-fetoprotein-positive hepatocellular carcinoma. J Transl Med. 2024;22:455. doi: 10.1186/s12967-024-05203-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calderaro J, Zigutyte L, Truhn D, Jaffe A, Kather JN. Artificial intelligence in liver cancer - new tools for research and patient management. Nat Rev Gastroenterol Hepatol. 2024;21:585–599. doi: 10.1038/s41575-024-00919-y. [DOI] [PubMed] [Google Scholar]
- 13.Demir T, Lee SS, Kaseb AO. Systemic therapy of liver cancer. Adv Cancer Res. 2021;149:257–294. doi: 10.1016/bs.acr.2020.12.001. [DOI] [PubMed] [Google Scholar]
- 14.Zhou J, Sun H, Wang Z, Cong W, Zeng M, Zhou W, Bie P, Liu L, Wen T, Kuang M, Han G, Yan Z, Wang M, Liu R, Lu L, Ren Z, Zeng Z, Liang P, Liang C, Chen M, Yan F, Wang W, Hou J, Ji Y, Yun J, Bai X, Cai D, Chen W, Chen Y, Cheng W, Cheng S, Dai C, Guo W, Guo Y, Hua B, Huang X, Jia W, Li Q, Li T, Li X, Li Y, Li Y, Liang J, Ling C, Liu T, Liu X, Lu S, Lv G, Mao Y, Meng Z, Peng T, Ren W, Shi H, Shi G, Shi M, Song T, Tao K, Wang J, Wang K, Wang L, Wang W, Wang X, Wang Z, Xiang B, Xing B, Xu J, Yang J, Yang J, Yang Y, Yang Y, Ye S, Yin Z, Zeng Y, Zhang B, Zhang B, Zhang L, Zhang S, Zhang T, Zhang Y, Zhao M, Zhao Y, Zheng H, Zhou L, Zhu J, Zhu K, Liu R, Shi Y, Xiao Y, Zhang L, Yang C, Wu Z, Dai Z, Chen M, Cai J, Wang W, Cai X, Li Q, Shen F, Qin S, Teng G, Dong J, Fan J. Guidelines for the diagnosis and treatment of primary liver cancer (2022 Edition) Liver Cancer. 2023;12:405–444. doi: 10.1159/000530495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reig M, Forner A, Rimola J, Ferrer-Fabrega J, Burrel M, Garcia-Criado A, Kelley RK, Galle PR, Mazzaferro V, Salem R, Sangro B, Singal AG, Vogel A, Fuster J, Ayuso C, Bruix J. BCLC strategy for prognosis prediction and treatment recommendation: the 2022 update. J Hepatol. 2022;76:681–693. doi: 10.1016/j.jhep.2021.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang B, Xi X, Yu H, Jiang H, Liang Z, Smayi A, Wu B, Yang Y. Evaluation of the effectiveness of surgical resection and ablation for the treatment of early-stage hepatocellular carcinoma: a retrospective cohort study. Cancer Rep (Hoboken) 2024;7:e2030. doi: 10.1002/cnr2.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karachaliou GS, Dimitrokallis N, Moris DP. Downstaging strategies for unresectable hepatocellular carcinoma. World J Gastroenterol. 2024;30:2731–2733. doi: 10.3748/wjg.v30.i20.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li M, Bhoori S, Mehta N, Mazzaferro V. Immunotherapy for hepatocellular carcinoma: the next evolution in expanding access to liver transplantation. J Hepatol. 2024;81:743–755. doi: 10.1016/j.jhep.2024.05.037. [DOI] [PubMed] [Google Scholar]
- 19.Zhang W, Gao W, Liu C, Li G, Zhang Q. Donafenib combined with hepatic artery infusion chemotherapy (HAIC) and sintilimab for unresectable hepatocellular carcinoma (uHCC): a prospective, single-arm phase II trial (DoHAICs study) J. Clin. Oncol. 2023;41:e16165. [Google Scholar]
- 20.Della Corte A, Rimini M, Steidler S, Palumbo D, Ratti F, Aldrighetti L, Cascinu S, Casadei-Gardini A, De Cobelli F. Combined loco-regional and systemic treatment strategies for hepatocellular carcinoma: from basics to new developments. Cardiovasc Intervent Radiol. 2023;46:175–186. doi: 10.1007/s00270-022-03327-4. [DOI] [PubMed] [Google Scholar]
- 21.Ksiazek W, Gandor M, Plawiak P. Comparison of various approaches to combine logistic regression with genetic algorithms in survival prediction of hepatocellular carcinoma. Comput Biol Med. 2021;134:104431. doi: 10.1016/j.compbiomed.2021.104431. [DOI] [PubMed] [Google Scholar]
- 22.Jin Z, Chen L, Zhong B, Zhou H, Zhu H, Zhou H, Song J, Guo J, Zhu X, Ji J, Ni C, Teng G. Machine-learning analysis of contrast-enhanced computed tomography radiomics predicts patients with hepatocellular carcinoma who are unsuitable for initial transarterial chemoembolization monotherapy: a multicenter study. Transl Oncol. 2021;14:101034. doi: 10.1016/j.tranon.2021.101034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chowdhary CL, Acharjya DP. Segmentation and feature extraction in medical imaging: a systematic review. Procedia Comput Sci. 2020;167:26–36. [Google Scholar]
- 24.Svetnik V, Liaw A, Tong C, Culberson JC, Sheridan RP, Feuston BP. Random forest: a classification and regression tool for compound classification and QSAR modeling. J Chem Inf Comput Sci. 2003;43:1947–1958. doi: 10.1021/ci034160g. [DOI] [PubMed] [Google Scholar]
- 25.Zhang S, Li X, Zong M, Zhu X, Wang R. Efficient kNN classification with different numbers of nearest neighbors. IEEE Trans Neural Netw Learn Syst. 2018;29:1774–1785. doi: 10.1109/TNNLS.2017.2673241. [DOI] [PubMed] [Google Scholar]
- 26.Ziemski M, Wisanwanichthan T, Bokulich NA, Kaehler BD. Beating naive bayes at taxonomic classification of 16S rRNA gene sequences. Front Microbiol. 2021;12:644487. doi: 10.3389/fmicb.2021.644487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li X, Du B, Zhang Y, Xu C, Tao D. Iterative privileged learning. IEEE Trans Neural Netw Learn Syst. 2020;31:2805–2817. doi: 10.1109/TNNLS.2018.2889906. [DOI] [PubMed] [Google Scholar]
- 28.Said D, Carbonell G, Stocker D, Hectors S, Vietti-Violi N, Bane O, Chin X, Schwartz M, Tabrizian P, Lewis S, Greenspan H, Jegou S, Schiratti JB, Jehanno P, Taouli B. Semiautomated segmentation of hepatocellular carcinoma tumors with MRI using convolutional neural networks. Eur Radiol. 2023;33:6020–6032. doi: 10.1007/s00330-023-09613-0. [DOI] [PubMed] [Google Scholar]
- 29.Albaradei S, Thafar M, Alsaedi A, Van Neste C, Gojobori T, Essack M, Gao X. Machine learning and deep learning methods that use omics data for metastasis prediction. Comput Struct Biotechnol J. 2021;19:5008–5018. doi: 10.1016/j.csbj.2021.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu Y, Chen G, Feng Z, Cui H, Rao F, Ni Y, Huang Z, Zhu W. Phase difference network for efficient differentiation of hepatic tumors with multi-phase CT. Annu Int Conf IEEE Eng Med Biol Soc. 2023;2023:1–5. doi: 10.1109/EMBC40787.2023.10340090. [DOI] [PubMed] [Google Scholar]
- 31.Tang L, Diao S, Li C, He M, Ru K, Qin W. Global contextual representation via graph-transformer fusion for hepatocellular carcinoma prognosis in whole-slide images. Comput Med Imaging Graph. 2024;115:102378. doi: 10.1016/j.compmedimag.2024.102378. [DOI] [PubMed] [Google Scholar]
- 32.Mule S, Lawrance L, Belkouchi Y, Vilgrain V, Lewin M, Trillaud H, Hoeffel C, Laurent V, Ammari S, Morand E, Faucoz O, Tenenhaus A, Cotten A, Meder JF, Talbot H, Luciani A, Lassau N. Generative adversarial networks (GAN)-based data augmentation of rare liver cancers: the SFR 2021 artificial intelligence data challenge. Diagn Interv Imaging. 2023;104:43–48. doi: 10.1016/j.diii.2022.09.005. [DOI] [PubMed] [Google Scholar]
- 33.Cai ZQ, Si SB, Chen C, Zhao Y, Ma YY, Wang L, Geng ZM. Analysis of prognostic factors for survival after hepatectomy for hepatocellular carcinoma based on a bayesian network. PLoS One. 2015;10:e0120805. doi: 10.1371/journal.pone.0120805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho Y, Choi JW, Kwon H, Kim KY, Lee BC, Chu HH, Lee DH, Lee HA, Kim GM, Oh JS, Hyun D, Lee IJ, Rhim H Research Committee of the Korean Liver Cancer Association. Transarterial chemoembolization for hepatocellular carcinoma: 2023 expert consensus-based practical recommendations of the Korean Liver Cancer Association. Clin Mol Hepatol. 2023;29:521–541. doi: 10.3350/cmh.2023.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clinical Guidelines Committee of Chinese College of Interventionalists. Chinese clinical practice guidelines for transarterial chemoembolization of hepatocellular carcinoma (2023 edition) Zhonghua Yi Xue Za Zhi. 2023;103:2674–2694. doi: 10.3760/cma.j.cn112137-20230630-01114. [DOI] [PubMed] [Google Scholar]
- 36.Mendez Romero A, van der Holt B, Willemssen FEJA, de Man RA, Heijmen BJM, Habraken S, Westerveld H, van Delden OM, Klumpen HJ, Tjwa ETTL, Braam PM, Jenniskens SFM, Vanwolleghem T, Weytjens R, D’Archambeau O, de Vos-Geelen J, Buijsen J, van der Leij C, den Toom W, Sprengers D, IJzermans JNM, Moelker A. Transarterial chemoembolization with drug-eluting beads versus stereotactic body radiation therapy for hepatocellular carcinoma: outcomes from a multicenter, randomized, phase 2 trial (the TRENDY trial) Int J Radiat Oncol Biol Phys. 2023;117:45–52. doi: 10.1016/j.ijrobp.2023.03.064. [DOI] [PubMed] [Google Scholar]
- 37.Muller L, Kloeckner R, Mahringer-Kunz A, Stoehr F, Duber C, Arnhold G, Gairing SJ, Foerster F, Weinmann A, Galle PR, Mittler J, Pinto Dos Santos D, Hahn F. Fully automated AI-based splenic segmentation for predicting survival and estimating the risk of hepatic decompensation in TACE patients with HCC. Eur Radiol. 2022;32:6302–6313. doi: 10.1007/s00330-022-08737-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bartnik K, Krzyzinski M, Bartczak T, Korzeniowski K, Lamparski K, Wroblewski T, Grat M, Holowko W, Mech K, Lisowska J, Januszewicz M, Biecek P. A novel radiomics approach for predicting TACE outcomes in hepatocellular carcinoma patients using deep learning for multi-organ segmentation. Sci Rep. 2024;14:14779. doi: 10.1038/s41598-024-65630-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bernatz S, Elenberger O, Ackermann J, Lenga L, Martin SS, Scholtz JE, Koch V, Grunewald LD, Herrmann Y, Kinzler MN, Stehle A, Koch I, Zeuzem S, Bankov K, Doering C, Reis H, Flinner N, Schulze F, Wild PJ, Hammerstingl R, Eichler K, Gruber-Rouh T, Vogl TJ, Dos Santos DP, Mahmoudi S. CT-radiomics and clinical risk scores for response and overall survival prognostication in TACE HCC patients. Sci Rep. 2023;13:533. doi: 10.1038/s41598-023-27714-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dong Z, Lin Y, Lin F, Luo X, Lin Z, Zhang Y, Li L, Li ZP, Feng ST, Cai H, Peng Z. Prediction of early treatment response to initial conventional transarterial chemoembolization therapy for hepatocellular carcinoma by machine-learning model based on computed tomography. J Hepatocell Carcinoma. 2021;8:1473–1484. doi: 10.2147/JHC.S334674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma J, Bo Z, Zhao Z, Yang J, Yang Y, Li H, Yang Y, Wang J, Su Q, Wang J, Chen K, Yu Z, Wang Y, Chen G. Machine learning to predict the response to lenvatinib combined with transarterial chemoembolization for unresectable hepatocellular carcinoma. Cancers (Basel) 2023;15:625. doi: 10.3390/cancers15030625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peng J, Huang J, Huang G, Zhang J. Predicting the initial treatment response to transarterial chemoembolization in intermediate-stage hepatocellular carcinoma by the integration of radiomics and deep learning. Front Oncol. 2021;11:730282. doi: 10.3389/fonc.2021.730282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang L, Jiang Y, Jin Z, Jiang W, Zhang B, Wang C, Wu L, Chen L, Chen Q, Liu S, You J, Mo X, Liu J, Xiong Z, Huang T, Yang L, Wan X, Wen G, Han XG, Fan W, Zhang S. Real-time automatic prediction of treatment response to transcatheter arterial chemoembolization in patients with hepatocellular carcinoma using deep learning based on digital subtraction angiography videos. Cancer Imaging. 2022;22:23. doi: 10.1186/s40644-022-00457-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu Z, An C, Shi F, Ren H, Li Y, Chen S, Dou J, Wang Y, Yan S, Lu J, Chen H. Automatic prediction of hepatic arterial infusion chemotherapy response in advanced hepatocellular carcinoma with deep learning radiomic nomogram. Eur Radiol. 2023;33:9038–9051. doi: 10.1007/s00330-023-09953-x. [DOI] [PubMed] [Google Scholar]
- 45.Quan B, Li J, Mi H, Li M, Liu W, Yao F, Chen R, Shan Y, Xu P, Ren Z, Yin X. Development and preliminary validation of a novel convolutional neural network model for predicting treatment response in patients with unresectable hepatocellular carcinoma receiving hepatic arterial infusion chemotherapy. J Imaging Inform Med. 2024;37:1282–1296. doi: 10.1007/s10278-024-01003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao Y, Huang F, Liu S, Jian L, Xia X, Lin H, Liu J. Prediction of therapeutic response of unresectable hepatocellular carcinoma to hepatic arterial infusion chemotherapy based on pretherapeutic MRI radiomics and Albumin-Bilirubin score. J Cancer Res Clin Oncol. 2023;149:5181–5192. doi: 10.1007/s00432-022-04467-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He X, Li K, Wei R, Zuo M, Yao W, Zheng Z, He X, Fu Y, Li C, An C, Liu W. A multitask deep learning radiomics model for predicting the macrotrabecular-massive subtype and prognosis of hepatocellular carcinoma after hepatic arterial infusion chemotherapy. Radiol Med. 2023;128:1508–1520. doi: 10.1007/s11547-023-01719-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roll W, Masthoff M, Kohler M, Rahbar K, Stegger L, Ventura D, Morgul H, Trebicka J, Schafers M, Heindel W, Wildgruber M, Schindler P. Radiomics-based prediction model for outcome of radioembolization in metastatic colorectal cancer. Cardiovasc Intervent Radiol. 2024;47:462–471. doi: 10.1007/s00270-024-03680-6. [DOI] [PubMed] [Google Scholar]
- 49.Ince O, Onder H, Gencturk M, Cebeci H, Golzarian J, Young S. Prediction of response of hepatocellular carcinoma to radioembolization: machine learning using preprocedural clinical factors and mr imaging radiomics. J Vasc Interv Radiol. 2023;34:235–243. e3. doi: 10.1016/j.jvir.2022.11.004. [DOI] [PubMed] [Google Scholar]
- 50.Kobe A, Zgraggen J, Messmer F, Puippe G, Sartoretti T, Alkadhi H, Pfammatter T, Mannil M. Prediction of treatment response to transarterial radioembolization of liver metastases: radiomics analysis of pre-treatment cone-beam CT: a proof of concept study. Eur J Radiol Open. 2021;8:100375. doi: 10.1016/j.ejro.2021.100375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marinelli B, Chen M, Stocker D, Charles D, Radell J, Lee JY, Fauveau V, Bello-Martinez R, Kim E, Taouli B. Early prediction of response of hepatocellular carcinoma to Yttrium-90 radiation segmentectomy using a machine learning MR imaging radiomic approach. J Vasc Interv Radiol. 2023;34:1794–1801. e2. doi: 10.1016/j.jvir.2023.06.023. [DOI] [PubMed] [Google Scholar]
- 52.Ballı HT, Piskin FC, Puren Yücel S, Sozutok S, Ozgul D, Aikimbaev K. Predictability of the radiological response to Yttrium-90 transarterial radioembolization by dynamic magnetic resonance imaging-based radiomics analysis in patients with intrahepatic cholangiocarcinoma. Diagn Interv Radiol. 2024;30:193–199. doi: 10.4274/dir.2023.222025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aujay G, Etchegaray C, Blanc JF, Lapuyade B, Papadopoulos P, Pey MA, Bordenave L, Trillaud H, Saut O, Pinaquy JB. Comparison of MRI-based response criteria and radiomics for the prediction of early response to transarterial radioembolization in patients with hepatocellular carcinoma. Diagn Interv Imaging. 2022;103:360–366. doi: 10.1016/j.diii.2022.01.009. [DOI] [PubMed] [Google Scholar]
- 54.Xiong Y, Zhang Y, Hu C. Radiofrequency ablation versus microwave ablation for hepatocellular carcinoma with cirrhosis: a propensity score analysis. Transl Cancer Res. 2024;13:1807–1820. doi: 10.21037/tcr-23-1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chong CCN, Lee KF, Cheung SYS, Chu CCM, Fong AKW, Wong J, Hui JWY, Fung AKY, Lok HT, Lo EYJ, Chan SL, Yu SCH, Ng KKC, Lai PBS. Prospective double-blinded randomized controlled trial of microwave versus radiofrequency ablation for hepatocellular carcinoma (McRFA trial) HPB (Oxford) 2020;22:1121–1127. doi: 10.1016/j.hpb.2020.01.008. [DOI] [PubMed] [Google Scholar]
- 56.Chen JJ, Jin ZC, Zhong BY, Fan W, Zhang WH, Luo B, Wang YQ, Teng GJ, Zhu HD. Locoregional therapies for hepatocellular carcinoma: the current status and future perspectives. United European Gastroenterol J. 2024;12:226–239. doi: 10.1002/ueg2.12554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang Y, Chen Y, Ye F, Cao X, Xin Y, Wang Y, Lei Y, Li X, Feng D, Zhou X, Fan Q. Late recurrence of hepatocellular carcinoma after radiofrequency ablation: a multicenter study of risk factors, patterns, and survival. Eur Radiol. 2021;31:3053–3064. doi: 10.1007/s00330-020-07460-x. [DOI] [PubMed] [Google Scholar]
- 58.Hamed AA, Muhammed A, Abdelbary EAM, Elsharkawy RM, Ali MA. Can machine learning predict favorable outcome after radiofrequency ablation of hepatocellular carcinoma? JCO Clin Cancer Inform. 2024;8:e2300216. doi: 10.1200/CCI.23.00216. [DOI] [PubMed] [Google Scholar]
- 59.Tong J, Liu P, Ji M, Wang Y, Xue Q, Yang JJ, Zhou CM. Machine learning can predict total death after radiofrequency ablation in liver cancer patients. Clin Med Insights Oncol. 2021;15:11795549211000017. doi: 10.1177/11795549211000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sato M, Moriyama M, Fukumoto T, Yamada T, Wake T, Nakagomi R, Nakatsuka T, Minami T, Uchino K, Enooku K, Nakagawa H, Shiina S, Koike K, Fujishiro M, Tateishi R. Development of a transformer model for predicting the prognosis of patients with hepatocellular carcinoma after radiofrequency ablation. Hepatol Int. 2024;18:131–137. doi: 10.1007/s12072-023-10585-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang H, Sheng S, Qiao W, Sun Y, Jin R. Nomogram built based on machine learning to predict recurrence in early-stage hepatocellular carcinoma patients treated with ablation. Front Oncol. 2024;14:1395329. doi: 10.3389/fonc.2024.1395329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.He Q, Xiong Y, Xia P, Yang X, Yu Y, Chen Z. Efficacy of radiofrequency ablation for solitary hepatocellular carcinoma 5 cm or smaller and construction of prognostic model by machine learning: a retrospective cohort study. Research Square; 2023. [Google Scholar]
- 63.Glassberg MB, Ghosh S, Clymer JW, Qadeer RA, Ferko NC, Sadeghirad B, Wright GW, Amaral JF. Microwave ablation compared with radiofrequency ablation for treatment of hepatocellular carcinoma and liver metastases: a systematic review and meta-analysis. Onco Targets Ther. 2019;12:6407–6438. doi: 10.2147/OTT.S204340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ren H, An C, Fu W, Wu J, Yao W, Yu J, Liang P. Prediction of local tumor progression after microwave ablation for early-stage hepatocellular carcinoma with machine learning. J Cancer Res Ther. 2023;19:978–987. doi: 10.4103/jcrt.jcrt_319_23. [DOI] [PubMed] [Google Scholar]
- 65.An C, Yang H, Yu X, Han ZY, Cheng Z, Liu F, Dou J, Li B, Li Y, Li Y, Yu J, Liang P. A machine learning model based on health records for predicting recurrence after microwave ablation of hepatocellular carcinoma. J Hepatocell Carcinoma. 2022;9:671–684. doi: 10.2147/JHC.S358197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shahveranova A, Balli HT, Aikimbaev K, Piskin FC, Sozutok S, Yucel SP. Prediction of local tumor progression after microwave ablation in colorectal carcinoma liver metastases patients by MRI radiomics and clinical characteristics-based combined model: preliminary results. Cardiovasc Intervent Radiol. 2023;46:713–725. doi: 10.1007/s00270-023-03454-6. [DOI] [PubMed] [Google Scholar]
- 67.Jiang Y, Liang X, Han Z, Wang W, Xi S, Li T, Chen C, Yuan Q, Li N, Yu J, Xie Y, Xu Y, Zhou Z, Poultsides GA, Li G, Li R. Radiographical assessment of tumour stroma and treatment outcomes using deep learning: a retrospective, multicohort study. Lancet Digit Health. 2021;3:e371–e382. doi: 10.1016/S2589-7500(21)00065-0. [DOI] [PubMed] [Google Scholar]
- 68.Huang H, Xie Y, Wang G, Zhang L, Zhou W. DLNLF-net: denoised local and non-local deep features fusion network for malignancy characterization of hepatocellular carcinoma. Comput Methods Programs Biomed. 2022;227:107201. doi: 10.1016/j.cmpb.2022.107201. [DOI] [PubMed] [Google Scholar]
- 69.Kucukkaya AS, Zeevi T, Chai NX, Raju R, Haider SP, Elbanan M, Petukhova-Greenstein A, Lin M, Onofrey J, Nowak M, Cooper K, Thomas E, Santana J, Gebauer B, Mulligan D, Staib L, Batra R, Chapiro J. Predicting tumor recurrence on baseline MR imaging in patients with early-stage hepatocellular carcinoma using deep machine learning. Sci Rep. 2023;13:7579. doi: 10.1038/s41598-023-34439-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xia T, Zhao B, Li B, Lei Y, Song Y, Wang Y, Tang T, Ju S. MRI-based radiomics and deep learning in biological characteristics and prognosis of hepatocellular carcinoma: opportunities and challenges. J Magn Reson Imaging. 2024;59:767–783. doi: 10.1002/jmri.28982. [DOI] [PubMed] [Google Scholar]
- 71.Daneshjou R, Smith MP, Sun MD, Rotemberg V, Zou J. Lack of transparency and potential bias in artificial intelligence data sets and algorithms: a scoping review. JAMA Dermatol. 2021;157:1362–1369. doi: 10.1001/jamadermatol.2021.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Corti C, Cobanaj M, Dee EC, Criscitiello C, Tolaney SM, Celi LA, Curigliano G. Artificial intelligence in cancer research and precision medicine: applications, limitations and priorities to drive transformation in the delivery of equitable and unbiased care. Cancer Treat Rev. 2023;112:102498. doi: 10.1016/j.ctrv.2022.102498. [DOI] [PubMed] [Google Scholar]
- 73.Saadat A, Siddiqui T, Taseen S, Mughal S. Revolutionising impacts of artificial intelligence on health care system and its related medical in-transparencies. Ann Biomed Eng. 2024;52:1546–1548. doi: 10.1007/s10439-023-03343-6. [DOI] [PubMed] [Google Scholar]
- 74.Pecqueux M, Riediger C, Distler M, Oehme F, Bork U, Kolbinger FR, Schoffski O, van Wijngaarden P, Weitz J, Schweipert J, Kahlert C. The use and future perspective of Artificial Intelligence-a survey among German surgeons. Front Public Health. 2022;10:982335. doi: 10.3389/fpubh.2022.982335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hantel A, Walsh TP, Marron JM, Kehl KL, Sharp R, Van Allen E, Abel GA. Perspectives of oncologists on the ethical implications of using artificial intelligence for cancer care. JAMA Netw Open. 2024;7:e244077. doi: 10.1001/jamanetworkopen.2024.4077. [DOI] [PMC free article] [PubMed] [Google Scholar]