Abstract
Cervical cancer accounts for 10-15% of cancer-related mortality among women globally. Infection with high-risk human papillomavirus (HPV) types constitutes a significant etiological factor in the development of cervical carcinoma. The integration of HPV DNA into the host genome is considered a pivotal event in cervical carcinogenesis. Nevertheless, the precise mechanisms underlying HPV integration and its role in promoting cancer progression remain inadequately understood. Therefore, this study aims to identify potential common denominators at HPV DNA integration sites and to analyze the adjacent cellular sequences. We conducted whole-genome sequencing on 13 primary cervical cancer samples, employing the chromosomal coordinates of 537 breakpoints to assess the statistical overrepresentation of integration sites in relation to various chromatin features. Our analysis, which encompassed all chromosomes, identified several integration hotspots within the human genome, notably at 14q32.2, 10p15, and 2q37. Additionally, our findings indicated a preferential integration of HPV DNA into intragenic and gene-dense regions of human chromosomes. A substantial number of host cellular genes impacted by the integration sites were associated with cancer, including IKZF2, IL26, AHRR, and PDCD6. Furthermore, the cellular genes targeted by integration were enriched in tumor-related terms and pathways, as demonstrated by gene ontology and KEGG analysis. In conclusion, these findings enhance our understanding of HPV integration sites and provide deeper insights into the molecular mechanisms underlying the pathogenesis of cervical carcinoma.
Keywords: HPV, cervical cancer, integration hotspots, WGS
Introduction
Cervical cancer ranks as the second most prevalent cause of cancer-related mortality among women globally, impacting approximately 500,000 individuals annually [1-4]. The development of high-grade cervical neoplasia is predominantly attributed to the integration of the human papillomavirus (HPV) genome into the host chromosome [5,7-11]. Consequently, this integration is recognized as a critical event in the progression of precancerous lesions to malignant cancer [12-18]. Therefore, elucidating the mechanisms underlying viral genome integration is essential for advancing therapeutic strategies for viral infections and the development of gene therapies [18].
Numerous studies have been undertaken to elucidate the preferential sites of DNA tumor virus integration, yielding varying conclusions [8,19-22]. Initially, HPV integration was considered a random process occurring across nearly all chromosomes without specific hotspots [2,17,23,24]. However, emerging evidence indicates that certain genomic regions, such as fragile sites, are preferentially targeted by the virus for integration. These regions have been reported as integration sites with greater frequency than others, thereby supporting the hypothesis that the distribution of integration sites is non-random [7,23-27]. Furthermore, clusters of integration sites have been identified in specific cytogenetic bands, including 3q28 [6,13,28], 8q24 [29-33], and 13q22 [7,24,34,35], which are now commonly referred to as integration hotspots.
However, previous studies were constrained by relatively small sample sizes, and their breakpoints were often biased [13,16,36,37]. The precise identification of small stretches of integration sites amidst a vast background of episomal forms remains a significant technical challenge [19].
Therefore, it is imperative to develop more efficient methodologies to enable comprehensive mapping of HPV integration sites, which is essential for gaining a deeper understanding of cervical carcinogenesis.
In pursuit of this objective, we employed whole-genome sequencing (WGS) to detect and analyze HPV integration in 13 cervical carcinoma samples. Additionally, to elucidate the pathogenic role of integration-targeted cellular genes (ITGs) in cervical carcinogenesis, we aggregated and scrutinized all available integration data for high-risk HPV (HR-HPV) types, focusing on the characteristics of the targeted loci within the human genome by the integration events. Our study provides an objective and comprehensive HPV integration map for cervical carcinomas, identifying novel hotspots and potential mechanisms. Specifically, we conducted an extensive analysis of HPV prevalence and characterized the precise integration sites of HPV DNA in 13 cervical cancer specimens. This research advances current understanding of HPV integration patterns in cervical carcinoma and offers new perspectives on the pathogenesis of cervical cancer.
Materials and methods
Clinical material
Snap-frozen primary cervical samples were collected from 13 treatment-naïve Chinese patients diagnosed with cervical adenocarcinoma, sourced from the Tissue Bank at Shanghai Tenth People’s Hospital, Tongji University. A board-certified pathologist conducted direct visualization to assess tumor characteristics and tissue heterogeneity. Histological analysis confirmed that the tumor sections contained a minimum of 10% tumor cells, ensuring their suitability for viral nucleic acid isolation and subsequent analysis. This study received approval from the institutional review board and ethics committee at Shanghai Jiao Tong University. Informed consent was obtained from the patients for sequencing analyses and data release.
DNA preparation and whole-genome sequencing
Genomic DNA was extracted from frozen tumor tissues utilizing the QIAamp® DNA Mini Kit (Qiagen, Hilden, Germany) in accordance with the manufacturer’s instructions. Subsequent whole-genome sequencing, employing 2×150-bp paired-end reads at a coverage depth of 60×, was conducted using the HiSeq X Ten platform (Illumina Inc., California, USA). The entire experimental procedure adhered strictly to the manufacturer’s protocol. The whole-genome sequencing was executed at CloudHealth Medical Group Ltd., Shanghai, People’s Republic of China.
Reference sequences
As the initial step in our pipeline, we obtained HPV genome data in FASTA format. Specifically, we acquired genomes of 18 high-risk HPV (HR-HPV) types, including types 6, 16, 18, 11, 33, 31, 35, 39, 45, 52, 56, 58, 59, 66, 68, 69, 82, and 83, from the National Center for Biotechnology Information (NCBI) database (accessible at www.ncbi.nlm.nih.gov/). All HPV reference sequences were concatenated to form a multiFASTA sequence (HPV_Ref) using BioPerl modules. For the human genome, we utilized the GRCh37 major release as the reference assembly, available at ftp://ftp.ensembl.org/pub/release-85/fasta/homo_sapiens/.
Filtering abundant sequencing reads
For quality control, we initially filtered low-quality reads and potential PCR duplicates using a custom Perl script. Subsequently, 3’ and 5’ adapters were trimmed employing the Adapter Removal program with default parameters, resulting in high-quality clean reads for further analysis.
HPV-aligned reads detection
Evaluating the HPV-aligned reads was essential for identifying HPV presence in the respective samples. For HPV detection, we indexed the multiFASTA HPV reference file (HPV_Ref) using the BWA aligner, followed by aligning the reads to the indexed genome. The aligned reads were then extracted from the SAM file using a Perl script.
Human-HPV integration loci detection
To identify integration sites, we constructed a multiFASTA reference genome (Homo_HPV_Ref) that included both human and high-risk human papillomavirus (HR-HPV) genomes of 18 types. For potential HPV integration sites within the human genome, we re-mapped selected HPV-aligned reads to this reference using the BWA-MEM program with default settings. The alignment files were then analyzed to identify reads where one mate aligned to a human chromosome and the other to HPV. Subsequently, we excluded reads that perfectly paired-end aligned to the HPV genome with a definitive alignment value (≥ 25) using BWA and retained chimeric read pairs, where part of the read sequence aligned to the human genome and part to the HPV genome.
Validation and assay of HPV integration sites
To ensure the accuracy of the alignment and to precisely identify the integration site as well as the sequence bridging the viral and cellular genomes, all meta-reads containing both viral and cellular sequences underwent further analysis using BLASTn comparisons against the whole-genome database.
Annotation of integration-targeted genes and fragile sites
The integration sites within human chromosomes and HPV genomes, along with the corresponding HPV subtypes, were subsequently parsed and annotated utilizing a gene reference annotation file obtained from the Ensembl genome browser (http://www.ensembl.org/index.html). Genes with transcription start sites located within 50 kilobases of the HPV integration sites were classified as ITGs. Additionally, we investigated the correspondence between HPV breakpoints and documented fragile sites in the human genome, as cataloged in the NCBI database.
Gene functional annotation analysis
For gene functional annotation analysis, we utilized the Database for Annotation, Visualization, and Integrated Discovery (DAVID) (https://david.ncifcrf.gov/), employing Gene Ontology (GO) categories and KEGG Pathways as reference databases.
Results
HPV integration analysis based on the WGS strategy
Leveraging the high throughput capacity of whole genome sequencing (WGS), we devised a multiplex strategy for the determination of human papillomavirus (HPV) integration sites. The analytical workflow is depicted in Figure 1. From a single lane of HiSeq ×10, we generated a total of 106.3 million sequence read pairs. Following data processing, we implemented a cutoff value for the basic alignment score (≥ 25) to pre-select the most promising junction candidates from over 9,000 filtered viral-cellular junction sequences for validation, thereby constituting the site-detection library (Table S1). Furthermore, to precisely identify the breakpoint between GRCh37 and HPV, we extracted all reads mapped to both GRCh37 and HPV and subsequently aligned each read to GRCh37 and HPV using BLASTn with parameters set to a minimum score of 35 and a minimum identity of 85%. Specifically, in instances where two or more paired reads mapped to nearly identical locations (within ±2 base pairs), only one read was retained for analysis. This approach resulted in the identification of 537 distinct integration events, each with associated chromosomal loci information.
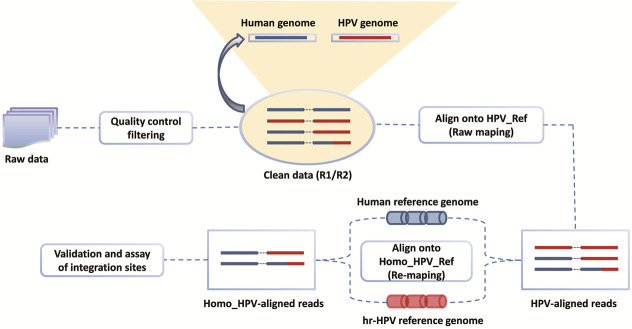
Figure 1.
Overall workflow. The pipeline includes the workflow of the experiment and the bioinformatic processes. In the process, raw data is initially filtered and then raw-mapped. Next, chimeric paired-end reads are selected for re-mapping of paired-end reads. HPV-aligned reads undergo re-mapping to locate the HPV integration sites. An in-house program is used to perform the paired-end reads assembly.
Position information of HPV integration into human genome and disruption in viral genome
The increased number of integration sites allowed for a comprehensive investigation of their distribution across the entire genome. For this analysis, all integration sites were standardized to reference “chromosome bands”. To mitigate bias arising from variations in chromosome band lengths, each “chromosome band” was represented by a ratio reflecting the proportion of the human genome it encompasses. Consequently, “densities” were calculated to assess the frequencies of integration hotspots, defined as the counts of validated integration sites within each band, divided by the corresponding band ratio.
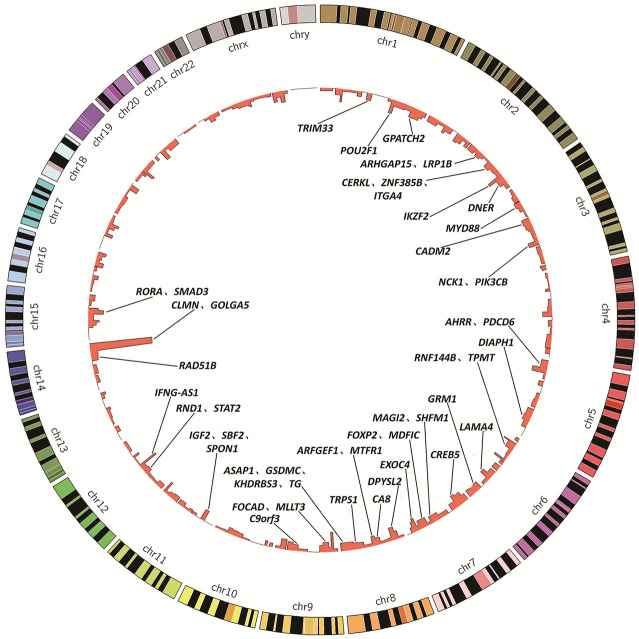
The distribution of HPV integration sites was observed across the entire genome, encompassing all chromosomes (Figures 2, S1). Specific locations of each cellular integration site are delineated in Table S2. The most frequently observed integration locus was a region approximately 17,138 kb in size on chromosome 14q32, which contained between 1 and 36 HPV integration sites identified across multiple samples (S1-T, S2-T, S3-T, S4-T and S7-T) (Figure S2). Additionally, substantial evidence was found for recurrent integration in other chromosomal hotspots, notably within the cytogenetic bands 12q15 (density = 24.44), 9p23 (density = 23.81), 2q34 (density = 19.66), and 15q22 (density = 18.43) (Table S3).
Figure 2.
Location of integration sites in the human genome. Human chromosomes (1-22, X, Y) are arranged around the circle. The innermost ring shows HPV integration sites, stacking multiple events that occur at the same location.
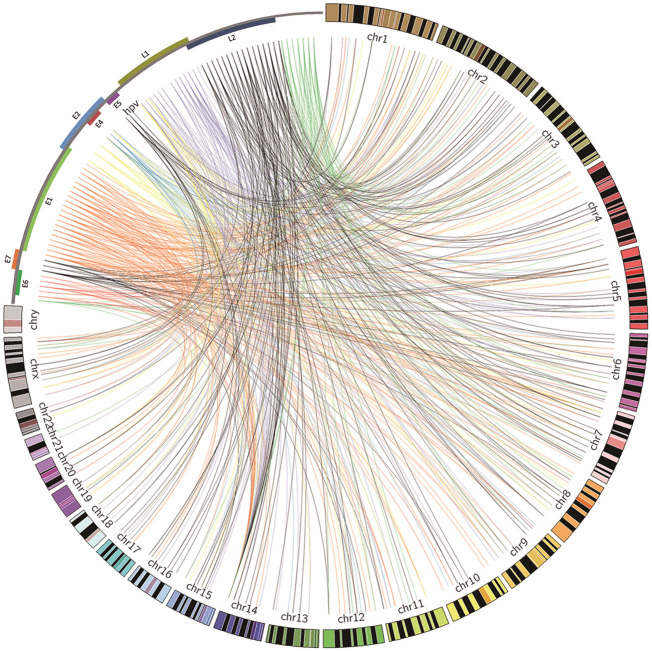
Furthermore, our observations revealed that HPV insertional breakpoints were concentrated in specific chromosomal regions. The genomic distances between distinct viral integration sites within a “cluster”, defined as containing three or more unique HPV integration sites, extended up to 1.5 Mb. We conducted a meticulous analysis of these clusters and categorized them into three distinct groups. In the first group, each cytogenetic band exhibited three or more HPV integration sites per sample, suggesting the likelihood of integration in a concatenated array (Figure S3). For example, HPV integrant clusters mapped to 2q34 in S2-T (n = 4), 1q31.3 in S3-T (n = 3), and 8p21.3 in S3-T (n = 4). In the second group, some integrant clusters arose from at least three individual cases, and they likely represented authentic hotspots, such as 14q32 in S1-T (n = 3), S2-T (n = 2), S3-T (n = 36), and S4-T (n = 1). In the last group, the clusters showed both of the aforementioned properties, such as 7p21.3 in S3-T (n = 3), S4-T (n = 1), and S7-T (n = 1) (Table S4). Collectively, sequence analysis of the junctions showed that the sites of viral gene disruption occurred broadly across the HPV genome (Figure 3).
Figure 3.
Distribution of integration sites across the HPV genome and human genomes. Integration breakpoints are shown for the HPV-positive tumors. Breakpoint colors correspond to HPV genes where an integration event occurred. The HPV and human genomes are drawn at different scales (created in Circos39).
Densities were also adopted herein to reduce the bias caused by length differences between different HPV open reading frames (ORFs). Disruptions were significantly more frequent in the L1 gene (n = 109, density = 29.11), followed by the E1 gene (n = 152, density = 25.25), E5 gene (n = 27, density = 8.50), and the E4 gene (n = 29, density = 7.99) (Table S5).
Characterization of integration hotspots
To investigate the characteristics of HPV integration into the human genome, recurrent “hotspots” of HPV integration (No. of integration sites > 3; density > the average score of those identified integration bands [7.28]) in the host genome were analyzed.
HPV integrations occur within or near cellular genes
The genomic regions corresponding to HPV integration hotspots were subjected to further analysis for the presence of known genes (Table S6). Genes directly disrupted by HPV integration, as well as those with transcription start sites within 50 kb of the integration breakpoints, were classified as integration target genes (ITGs), in accordance with previous reports. Out of the 263 integration events identified within the 57 chromosomal hotspots analyzed, 203 events were found to occur either within a gene region or in close proximity to a gene region (Table 1).
Table 1.
Integration targeted genes in hotspots and relationship with tumors
| Hotspots | Integrated times | Fragile sites | ITGs in the hotspot and relationship with tumors | |||
|---|---|---|---|---|---|---|
|
|
|
|||||
| HPV16 | HPV18 | Other types | Y | N | ||
| 14q32 | 43 | - | - | - | CLMN c, GOLGA5 c, CYP46A1, DDX24, DYNC1H1 | RPL3P4 d, AL162151.3 d, HSP90AA1 c, IFI27L1 c, AB019438.66, CTD-2240H23.2, HHIPL1, HOMER2P1, IFI27, IFI27L2, IGHV1-67, IGHV1-68, IGHV1-69, IGHV2-70, IGHV3-38, IGHV3-41, IGHV3-42, IGHV3-43, IGHV3-63, IGHV3-64, IGHV3-65, IGHV3-66, IGHV3-71, IGHV3-72, IGHV4-39, IGHV7-40, IGHVII-40-1, IGHVII-43-1, IGHVII-44-2, IGHVII-62-1, IGHVII-65-1, IGHVII-67-1, IGHVIII-38-1, IGHVIII-44, IGHVIII-67-2, IGHVIII-67-3, IGHVIII-67-4, IGHVIV-44-1, LINC00221, OTUB2, RN7SL472P, RP11-1017G21.4, RP11-543C4.3, RP11-725G5.3, SLC20A1P1 |
| 12q15 | 3 | - | - | - | IFNG-AS1 c,d, IL26 d, DYRK2, IFNG, IL22 | MDM1, RP11-335O4.1, RP11-335O4.3, RP11-444B24.2 |
| 9p23 | 4 | - | - | - | PES1P2, RP11-23D5.1, RP11-284P20.3, RPL3P11 | |
| 2q34 | - | - | 4 | FRA2I | IKZF2 c,d | RP11-105N14.1 d, RP11-105N14.3 d, AC079610.1 c |
| 15q22 | 5 | - | - | FRA15A a | RORA c,d, SMAD3 c, CA12 | FAM81A c, RP11-219B17.1 c, RP11-321G12.1 c, AAGAB, AC007950.1, CTD-2501E16.1, CTD-2501E16.2, NARG2, RP11-342M21.2, RP11-356M20.1, RP11-356M20.2, RPS24P16, Y_RNA |
| 5p14 | 6 | - | - | FRA5E a | - | CDH18 c,d, CTD-2061E9.1, HSPD1P15, MSNP1, RNU6-909P |
| 1q24 | 2 | - | 2 | FRA1G | POU2F1 c, UCK2 | LINC00970 c,d, RPL29P7 d, SUMO1P2 d, RP11-525G13.2, RP11-52A20.2, RP11-7G12.2 |
| 7q33 | 3 | - | - | FRA7H | EXOC4 c,d | AC083875.1, AC083875.2, AC091736.1 |
| 8p21 | 5 | - | - | - | DPYSL2 c, U3 | AC100802.3 c, RP11-108E14.1 c, AC021613.1, RP11-1G11.2, RP11-404E12.1, RPL30P9, SLC18A1 |
| 7p15 | 4 | - | - | - | CREB5 c,d, EVX1, NPY | AC005105.2 d, RPL35P4 c, AC004485.3, EVX1-AS, RP1-170O19.17, snoU13 |
| 7p21 | 6 | - | - | FRA7B | THSD7A c,d, PHF14 c, AC004538.3, AC004543.2, TMEM106B | |
| 6q21 | 4 | - | - | FRA16B b, FRA16C | LAMA4 c, SLC22A16, TUBE1, U3, WISP3 | HS3ST5 c, LRP3-399L15.3 c, CTA-331P3.1, FAM229B, RN7SL617P, RP1-142L7.5, RP1-249H1.2 |
| Xq27 | 4 | - | - | FRAXD, FRAXA a,b | HNRNPCP10, RNU6-382P, RP11-434J24.2, RP11-434J24.3, RP3-406C18.1 | |
| 11p15 | 4 | - | - | FRA11C, FRA11I b | IGF2 c, SBF2 c, SPON1 c, INS, INS-IGF2, MIR483 | RIC3 c, RP11-1H15.2 c, AC132217.4, IGF2-AS, MIR4686, RNA5SP332, RP11-379P15.1, TH, TUB |
| 3q22 | 4 | - | - | - | NCK1 c, PIK3CB c | COL6A4P2 c, IL20RB c, TMCC1 c, AC083799.1, AC130888.1, ENPP7P3, IL20RB-AS1, RNU6-1142P, RP11-85F14.1, RP11-93K22.14, TMCC1-AS1, Y_RNA |
| 1q41 | 4 | - | - | FRA1H | GPATCH2 c, KCNK2, TLR5 | AURKAPS1, MORF4L1P1, RAB3GAP2, RP11-239E10.2, RP11-302I18.1, RP11-323K10.1, XRCC6P3 |
| 20p11 | 4 | - | - | FRA20A a,b | - | DZANK1 c, RALGAPA2 c, RP1-122P22.2 c, AL121761.1, AL121761.2, FAM182B, GCNT1P1, LINC00851, MIR3192, POLR3F, RNA5SP476, RP13-401N8.1, RPL12L3, VN1R108P, ZNF337 |
| 1q42 | 5 | - | - | FRA1H a | ABCB10, B3GALNT2, EGLN1, EXOC8, SPRTN | NVL c, TBCE c, BTNL10, CNIH4, GNPAT, HIST3H2A, HIST3H2BA, HIST3H2BB, HMGN2P19, MIR4666A, RNA5SP78, RNA5SP80, RNF187, RNU4-21P, RNU6-1008P, RP11-293G6__A.2, RP11-293G6__A.3, RP11-365O16.5, snoU13, TAF5L, URB2 |
| 9q33 | 5 | - | - | FRA9B b, FRA9E | CRB2, STOM | ASTN2 c, BRINP1 c, RP11-162D16.2 c, GGTA1P, RN7SKP125, RP11-787B4.2, STRBP |
| Xq25 | 2 | - | 1 | - | U3 | - |
| 8q13 | 3 | - | - | - | ARFGEF1 c, MTFR1 c, PDE7A | CPA6, RP11-707M3.3, RP11-7F18.2 |
| 9p21 | 5 | - | - | FRA9A a,b, FRA9C a | FOCAD c, MLLT3 c | LINGO2 c, MIR4474, RP11-321L2.2, RP11-32I2.1, RP11-73E6.2, SNORA30 |
| 4q22 | 4 | - | - | - | PPM1K | CCSER1 c, RP11-10L7.1 c, HERC6, Y_RNA |
| 2p12 | 3 | - | - | FRA2E | - | LRRTM4 c, AC073628.1, RNU6-812P, RNU6-827P |
| 7q31 | 7 | - | - | FRA7F, FRA7G a | FOXP2 c, MDFIC c, MET | RP11-328J2.1 c, SLC13A1 c, AC006159.3, AC006926.1, RP11-500M10.1, RP11-95P9.1 |
| 18q22 | 4 | - | - | FRA18B, FRA18C | TSHZ1 | CDH19 c, RP11-638L3.1, RP11-659F24.1, ZADH2 |
| 6p22 | 3 | - | 2 | FRA6A b, FRA6C a | ZNF322, RNF144B c, TPMT c, KDM1B | RP11-457M11.2 d, VN1R14P d, ABT1, NHLRC1, RP11-457M11.5, RP11-457M11.6, snoU13 |
| 9q31 | 4 | - | - | FRA9B b, FRA9E | - | RNA5SP293, RP11-380I20.2, RP11-436F21.1, RPL36AP6 |
| 16q21 | 3 | - | - | FRA16B b, FRA16C | - | AC012322.1 c, LINC00922 c, RP11-351A20.1, RP11-744D14.1 |
| 2q36 | 3 | - | - | - | DNER c | SPHKAP c, AC007559.1 |
| 8q24 | 9 | - | - | FRA8C a, FRA8E a,b, FRA8D a | ASAP1 c, GSDMC c, KHDRBS3 c, TG c, ZFAT | DENND3 c, RP11-1082L8.3 c, RP11-30J20.1 c, RP11-369K17.1 c, AC083843.1, AC131568.1, CTD-2182N23.1, CTD-2342N23.3, LINC00964, MAPRE1P1, MIR4662B, RNU6-1255P, RP11-1082L8.4, RP11-274M4.1, RP11-809O17.1, SLC45A4, SNORA12 |
| 3p12 | 4 | - | - | - | CADM2 c,d | CADM2-AS2 c, RP11-260O18.1 c, RPL7AP23 |
| 2q31 | 4 | - | - | FRA2G a | CERKL c, ZNF385B c, ITGA4 c | AC013410.1, AC068706.1, AC068706.2, AC073069.2, MTX2 |
| 2p11 | 3 | - | - | FRA2A b | RPIA | ANKRD36BP2 c, AC096579.1, AC096579.13, AC128677.4, MIR4436A |
| 6q24 | 3 | - | - | - | GRM1 c,d | FUNDC2P3 |
| 7q21 | 6 | - | - | FRA7J, FRA7E a | MAGI2 c, SHFM1 c | ANKIB1 c, MTERF c, AC007566.11, AC073958.2, AC092013.1, RP11-682N22.1 |
| 5q31 | 4 | - | - | FRA5C a | DIAPH1 c, SLC22A5 | AC005592.2 c,d, AC116366.5 c, C5orf56 c, AC116366.6, CTD-2024I7.13, PCDHGA1, PCDHGA10, PCDHGA11, PCDHGA12, PCDHGA2, PCDHGA3, PCDHGA4, PCDHGA5, PCDHGA6, PCDHGA7, PCDHGA8, PCDHGA9, PCDHGB1, PCDHGB2, PCDHGB3, PCDHGB4, PCDHGB6, PCDHGB7, PCDHGC3, PCDHGC4, PCDHGC5, RN7SL68P, Y_RNA |
| 14q31 | 3 | - | - | - | - | CTD-2128A3.2 c, RP11-526N18.1 c, CTD-2128A3.3, EML5, LINC00911, PTPN21, RNU4-22P, RP11-507K2.2, RP11-507K2.3, ZC3H14 |
| 8q12 | 3 | - | - | - | CA8 c,d | YTHDF3 c, RN7SL135P, RP11-16E18.1, RP11-16E18.3, YTHDF3-AS1 |
| 1p13 | 3 | - | - | FRA1E | TRIM33 c | NHLH2 c, NTNG1 c, AC114491.1, EIF2S2P5, HNRNPA1P43, PKMP1, RP11-270C12.3, RP4-591B8.2, Y_RNA |
| 7p14 | 4 | - | - | FRA7C a | INHBA, INHBA-AS1 | AC005027.3 c,d, AOAH c, AC007349.4, RP11-85E16.1 |
| 5p15 | 5 | - | - | - | AHRR c,d, PDCD6 c,d, MRPL36, NDUFS6, SDHA | CTD-2143L24.1 c, TAS2R1 c, CTD-2001E22.1, CTD-2083E4.5, CTD-2083E4.6, CTD-2228K2.1, CTD-2228K2.2 |
| 14q24 | 3 | - | - | FRA14C a | RAD51B c | C14orf166B c, ANGEL1, CTD-2566J3.1, DLST, PROX2, RN7SKP17, RN7SL706P, RP11-316E14.2, RP11-316E14.6, RP11-488C13.1, RPS6KL1, YLPM1 |
| 8q23 | 3 | - | - | FRA8C, FRA8E b | TRPS1 c | RP11-790J24.1 c, TMEM74 c, RP11-25P11.2, RP11-790J24.2, RP11-946L20.1, RP11-946L20.2 |
| 5q23 | 4 | - | - | FRA5C | - | AC004769.1, AC093267.1, CTD-2334D19.1, RP11-166A12.1, snoU13, SNX2 |
| 3p22 | 3 | - | - | - | MYD88 c, DLEC1, U8 | Y_RNA d, AC018359.1 c, ULK4 c, AC123023.1, ACAA1, ATP6V0E1P2, OXSR1 |
| 2q14 | 4 | - | - | FRA2B b, FRA2F | CNTNAP c, RN7SKP102 | |
| 12q13 | 3 | - | - | FRA12A a,b | RND1 c, STAT2 c, CNPY2, EIF4B, IL23A, KRT18, KRT8 | AC107016.1, AC107016.2, APOF, CACNB3, CCDC65, DDX23, PAN2, RNU6-600P, RNU7-40P, RP11-302B13.1, RP11-302B13.5, RP11-348M3.2, RP11-977G19.10, RP11-977G19.11, RP11-977G19.12 |
| 22q12 | 3 | - | - | FRA22B | - | RP5-1119A7.14 c, BX470187.1, CTA-929C8.8, FOXRED2, LL22NC03-86D4.1, RPS15AP38, TXN2, Y_RNA |
| 2p24 | 3 | - | - | FRA2C a | - | NBAS c, AC008069.2, RN7SKP168, RP11-32P22.1 |
| 2q22 | 3 | - | - | FRA2F, FRA2K a | ARHGAP15 c, LRP1B c | AC068287.1, AC092652.1, AC093084.1, RNU6-904P, RP11-570L15.1, RP11-570L15.2 |
| 6q25 | 3 | - | - | FRA6E | TFB1M | CLDN20 |
| 6q14 | 3 | - | - | FRA6D | - | FILIP1, KRT18P64, RNU1-34P, RP11-30P6.1, RP11-379B8.1, RP1-161C16.1, RP11-801I18.1, RPL26P20 |
| 1p34 | 3 | - | - | FRA1B | MUTYH | TESK2 c, HPDL, RP11-291L19.1, RP11-329N22.1, RP11-422J8.1, snoU13, TOE1 |
| 2p25 | 3 | - | - | FRA2C | - | FAM110C, AC007463.2 |
| 9q22 | 3 | - | - | FRA9D | C9orf3 c, SPTLC1 | LINC00475, MIR2278, MTND4P15, RP11-100G15.10, RP11-100G15.2, RP11-100G15.3, RP11-100G15.4, RP11-100G15.5, RP11-100G15.7, RP11-23B15.1, RP11-49O14.3, RP11-546O6.4, snoU13, SPTLC1 |
| 6q16 | 3 | - | - | FRA6G, FRA6F | - | RP3-463P15.1 |
Abbreviations: HPV, human papillomavirus; ITGs, integration-targeted cellular genes; Y: Yes; means that these genes had been reported to be tumor-related based on NCBI database; N: no; means that there was no report about the relationship between this gene and tumors.
Underlined type indicates integrations that occurred within a fragile site.
Rare fragile sites are shown in bold.
Underlined Italics indicate genes directly targeted.
The bold genes were recurrently integration targeted genes.
In this study, a total of 516 integration target genes (ITGs) were identified and subsequently analyzed. These genes encompassed various categories, including protein-coding genes, processed pseudogenes, microRNAs (miRNAs), and others. Notably, approximately 45.81% (93 out of 203) of the confirmed HPV integration sites within hotspots were situated within coding genes, and 73 cellular DNA breakpoints were located in processed transcripts.
In addition to these 516 ITGs, 29 recurrently targeted host genes (RTGs) were also analyzed. Y_RNA was the gene most frequently affected by viral integration (8 events), followed by small nucleolar RNA U13 (snoU13, 6 events), AL162151.3 (4 events) and RPL3P4 (4 events). Furthermore, four genes were affected three times, while 21 recurrent target genes (RTGs) were affected twice within the analyzed dataset. Additionally, we deemed it pertinent to investigate the implications of these ITGs as functionally cancer-related genes. Consistent with the observation that genes located in chromosomal hotspots are associated with tumors, 14 out of the 29 RTGs have also been reported to be tumor-related (Table 2). This finding suggests that HPV DNA fragments preferentially integrate into genomic hotspots where tumor-related genes are situated.
Table 2.
Recurrent targeted genes and relationship with tumors
| Gene symbol | Band | Integrated times | Cancer related | Summary | Reference (PMID) |
|---|---|---|---|---|---|
| Y_RNA | 15q22, 3q22, 4q22, 5q31, 1p13, 3p22, 22q12 | 8 | N | - | - |
| snoU13 | 7p15, 1q42, 6p22, 5q23, 1p34, 9q22 | 6 | N | - | - |
| AL162151.3 | 14q32 | 4 | N | - | - |
| RPL3P4 | 14q32 | 4 | N | - | - |
| IKZF2 | 2q34 | 3 | Y | Functions pivotally in T-cell differentiation and activation. | NCBI gene/Ref (23600753) |
| RP11-105N14.1 | 2q34 | 3 | N | - | - |
| RP11-105N14.3 | 2q34 | 3 | N | - | - |
| U3 | 8p21, 6q21, Xq25 | 3 | Y | Non-conventional regulatory functions of U3 (or fragments derived from it) in mRNA metabolism. | NCBI gene/Ref (27517747) |
| IFNG-AS1 | 12q15 | 2 | Y | Mutations in this gene are associated with an increased susceptibility to viral, bacterial and parasitic infections and to several autoimmune diseases. | NCBI gene/Ref (28600289) |
| IL26 | 12q15 | 2 | Y | The encoded protein is thought to contribute to the transformed phenotype of T cells after infection by herpesvirus samimiri. | NCBI gene/Ref (23704922) |
| RORA | 15q22 | 2 | Y | The encoded protein has been shown to interact with NM23-1, the product of a tumor metastasis suppressor candidate gene. | NCBI gene/Ref (22104449) |
| CDH18 | 5p14 | 2 | Y | - | - |
| LINC00970 | 1q24 | 2 | N | - | - |
| RPL29P7 | 1q24 | 2 | N | - | - |
| SUMO1P2 | 1q24 | 2 | N | - | - |
| EXOC4 | 7q33 | 2 | Y | The encoded protein is found to interact with the actin cytoskeletal remodeling and vesicle transport machinery. | NCBI gene/Ref (23207790) |
| CREB5 | 7p15 | 2 | Y | This gene binds to the cAMP response element and activates transcription. | UniProt/Ref (25076032) |
| AC005105.2 | 7p15 | 2 | N | - | - |
| THSD7A | 7p21 | 2 | Y | The encoded protein appears to interact with integrin αVβ3 and paxillin to inhibit endothelial cell migration and tube formation. | NCBI gene/Ref (28035718) |
| RP11-457M11.2 | 6p22 | 2 | N | - | - |
| VN1R14P | 6p22 | 2 | N | - | - |
| ZNF322 | 6p22 | 2 | Y | The gene may regulate transcriptional activation in MAPK signaling pathways. | NCBI gene/Ref (15555580) |
| CADM2 | 3p12 | 2 | Y | Adhesion molecule that engages in homo- and heterophilic interactions with the other nectin-like family members, leading to cell aggregation. | UniProt/Ref (23643812) |
| GRM1 | 6q24 | 2 | Y | This gene may be associated with many disease states, including schizophrenia, bipolar disorder, depression, and breast cancer. | NCBI gene/Ref (27458247) |
| AC005592.2 | 5q31 | 2 | N | - | - |
| CA8 | 8q12 | 2 | Y | Polymorphisms in this gene are associated with osteoporosis, and overexpression of this gene in osteosarcoma cells suggests an oncogenic role. | NCBI gene/Ref (26711783) |
| AC005027.3 | 7p14 | 2 | N | - | - |
| AHRR | 5p15 | 2 | Y | The protein encoded by this gene is involved in regulation of cell growth and differentiation. | NCBI gene/Ref (16755028) |
| PDCD6 | 5p15 | 2 | Y | May mediate Ca2+-regulated signals along the death pathway: interaction with DAPK1 can accelerate apoptotic cell death by increasing caspase-3 activity. | NCBI gene/Ref (25362542) |
Abbreviations: Y: Yes; means that these genes had been reported to be tumor-related based on NCBI database; N: no; means that there was no report about the relationship between this gene and tumors.
HPV integration within fragile sites
Furthermore, we investigated all integration loci at hotspots for the presence of fragile sites, which are genomic regions susceptible to chromosomal breaks that facilitate the integration of foreign DNA. Utilizing the NCBI fragile site map viewer, we identified a significant correlation between fragile sites and HPV integration sites. Notably, 56.65% (149 out of 263) of the integration events within these hotspots were situated in or near a fragile site. Specifically, 53 integration sites were located within common or rare fragile sites, while 96 sites were within a 5 Mb proximity to a common or rare fragile site (Table S7). The remaining integration sites did not exhibit any association with fragile sites.
Furthermore, a comparative analysis was conducted between all integration sites and the mapped fragile sites available in the database (Table S8). In this study, we reanalyzed all integration loci in relation to the mapped fragile sites. The combined data revealed a significant correlation between fragile sites and HPV integration sites. Specifically, 54.75% of the 537 integration sites were found to target fragile sites, and this percentage is likely an underestimate given that not all fragile sites have been mapped to date.
Functional analysis of genes involved with HPV integration hotspots
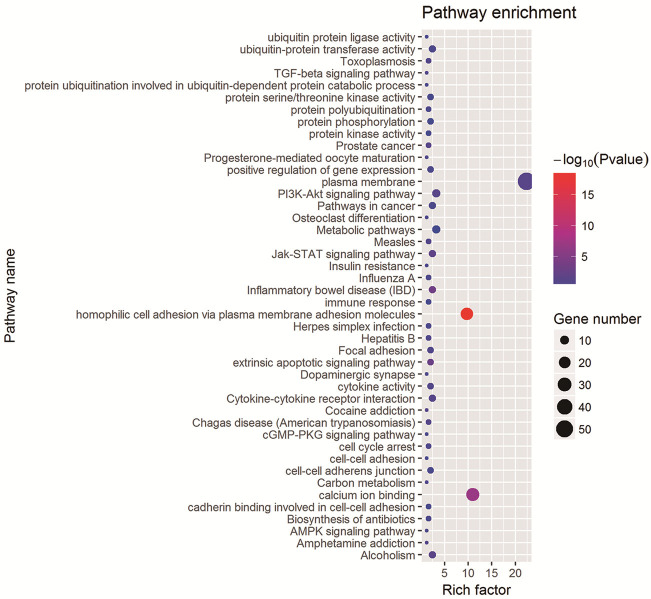
To elucidate the potential roles of ITGs in HPV-related cervical cancer, a functional annotation analysis was conducted using the DAVID web service. Out of 516 ITGs, 100 were identified through DAVID analysis. Gene Ontology (GO) analysis indicated significant enrichment in four specific terms: “homophilic cell adhesion via plasma membrane adhesion molecules”, “extrinsic apoptotic signaling pathway”, “calcium ion binding”, and “plasma membrane” (P < 0.05; Figure 4; Table S9). Additionally, KEGG pathway annotation analysis revealed significant clustering in three pathways: “Jak-STAT signaling pathway”, “Inflammatory bowel disease (IBD)”, and “Alcoholism” (P < 0.05; Figure 4).
Figure 4.
Functional annotation analysis of ITGs. Summary of significant pathways of ITGs in the discovery.
Microhomology among the viral and human genomes
The identification of reads spanning the insertion site enabled the determination of integration breakpoints with single-nucleotide precision. A total of 130 integration sites, characterized by defined recombination sites observed in this study, were collected for further analysis. Upon alignment to the reference viral genome and the human genome, three distinct patterns of host-virus integration sequences were identified: direct ligation, insertion of unaligned nucleotides, and overlapping (Table S8).
Approximately 9.23% of the 130 viral-cellular junction sequences were found to occur via direct ligation. Due to the presence of certain nucleotides that could not be aligned to a specific genomic sequence within both viral and cellular sequences, these nucleotides were classified as insertions of unaligned nucleotides, constituting approximately 11.54% of the total 130 integration junction sequences.
The most common pattern observed among the validated viral-cellular sequences was overlapping, characterized by nucleotides shared between the viral and cellular genomes. Notably, 79.23% of these 130 junctions were located in regions exhibiting microhomology, ranging from 1 to 21 base pairs, between the viral and human genomes. For example, a 4-base sequence similarity was observed between the viral L1 gene and the human gene at the human-viral interface. These 4-base segments were identical in both the HPV-derived portion of the L1 gene and the corresponding human-derived region at the human-viral boundary.
Discussion
In the current study, an innovative multiplex strategy was developed for the detection of HPV integration breakpoints, employing 13 cervical carcinoma samples to identify integration sites with varying frequencies. The methodology incorporated a tailored targeted sequencing protocol that capitalizes on the high-throughput capabilities of next-generation DNA sequencing. The identification of 537 validated HPV integration sites underscores the effectiveness of this technique. Moreover, this approach demonstrates the potential to reveal over 50% of previously undetected low-frequency integration sites, thereby surpassing existing methods [38,39]. The restricted coverage and validated reads of the novel sites imply that HPV integration may occur less frequently in tumor cells at these locations. Consequently, our methodology presents several advantages over previously reported approaches. Unlike the commonly utilized PCR method [20,37,40,41], our tool facilitates the detection of integration sites on a genome-wide scale. Moreover, although both our strategy and the HPV-capture approach [19,41] employ massively parallel sequencing platforms, our current method exhibits substantially greater efficiency in identifying viral integration.
Additionally, this methodology is applicable for identifying breakpoints in a range of viruses, including hepatitis B virus and human immunodeficiency virus. Furthermore, it is a cost-effective technique that delivers rapid results and is capable of detecting numerous integration breakpoints previously identified by other methods. Moreover, the integration sites identified by this approach exhibit higher validation rates, underscoring its specificity in detecting viral integration breakpoints.
A primary limitation of the genome-wide HPV integration methodology is its dependence on targeting only previously identified viruses with established genome references for integration identification. Furthermore, a significant technical challenge is presented by the substantial proportion of data comprised of human-genome reads and free-virus reads. These constraints could potentially be alleviated through the implementation of Next-Next-generation sequencing technology, which provides longer reads that may improve the characterization of HPV insertion sequences and enhance validation rates. Furthermore, our objective is to identify additional integration breakpoints while minimizing the quantity of sequencing data necessary. To this end, we have developed a methodology that exhibits high specificity and sensitivity in detecting HPV integration breakpoints. This method is applicable to screening viral integration in large sample cohorts, thereby enabling a systematic investigation of its association with disease etiology and tumorigenesis in a comprehensive and unbiased manner.
Utilizing the integration site analysis in conjunction with the novel multiplex strategy on these 13 cervical cancer samples will facilitate a comprehensive mapping of HPV integration sites, surpassing the scope of currently available data. Consistent with findings from other studies, HPV integration into the host genome is not entirely random but exhibits a preference for specific chromosomal locations. In our analysis, we validated several previously identified hotspots of HPV integration, including 2q34 [8,42], 8q24 [27,42-45], 15q22 [27,44], and 9p23 [3,42], and identified some new hotspots such as 1q24, 7q33, 7p15, and 3q22. These clusters may signify regions of high genomic instability that are particularly vulnerable to HPV integration or may represent loci containing genes crucial for the development of cervical tumors [23,35,43,45,46]. The greatest number of integration events was observed at 14q32, a substantial chromosomal region rich in ITGs, which has been previously identified as a hotspot for HPV integration [5,28,29]. Additionally, region 8q24 is another well-established hotspot for HPV integration which contains the MYC gene (alias c-MYC), FRA8C, FRA8D and FRA8E [31,34,45].
Moreover, the large-scale analysis undertaken in this study allowed us to conclude that HPV DNAs prefer to integrate into intragenic and gene-dense regions. Analysis of the chromosomal hotspot ITGs in this cohort revealed that some of the genes disrupted by HPV integration are involved in tumor development in other cancer entities (e.g., IFNG-AS1 [47], IL26 [48], RORA [49], AHRR [50] and PDCD6 [51]). Among those identified ITGs, Y_RNA and IKZF2 are illustrative. The cellular gene Y_RNA, located in multiple chromosomal hotspots 15q22, 3q22, 4q22, 5q31, 1p13, 3p22 and 22q12, was the gene most recurrently targeted by viral integration. Previous research has indicated that Y RNAs can function as repressors of Ro60 and as initiation factors for DNA replication [52,53]. Additionally, Y RNAs are overexpressed in certain human tumors and are essential for cell proliferation [54]. Furthermore, small, microRNA-sized breakdown products of Y RNAs may play a role in autoimmunity and other pathological conditions [55]. The IKZF2 gene, located at the fourth most common HPV integration site, 2q34, encodes a member of the Ikaros family of zinc-finger proteins, which are believed to be crucial for T-cell differentiation and activation [56]. Given that all three integration events identified in this cohort were located within the intragenic region of IKZF2, it would be pertinent to investigate whether this gene is frequently mutated in cervical cancers.
The analysis of RTGs identified in this large-scale study further substantiates our hypothesis. Our functional annotation found that most of the ITGs were enriched in tumor-related terms and pathways, including the GO terms of “extrinsic apoptotic signaling pathway” and “cytokine activity” and the KEGG terms of “Cytokine-cytokine receptor interaction”, “Jak-STAT signaling pathway”, and “PI3K-Akt signaling pathway” which also exhibited high “reactive” pathway score in a recent study [57].
Consistent with our findings, a recent report [5] similarly demonstrated that ITGs were enriched in tumor-associated KEGG pathways. The analysis conducted in this study provides compelling evidence that dysregulation of ITGs plays a significant role in cervical carcinogenesis.
To characterize the viral integration pattern, we annotated 537 breakpoints within HR-HPV genomes. In agreement with the observations of Rusan et al. [12] and other cervical cancer studies [5,19,58], the breakpoints were distributed broadly across the viral genome. Notably, L1 emerged as the most frequently disrupted gene in our study, which contrasts with previous reports [5,8,58]. Disruption of the L1 gene may result in defects in virion formation and transmission, leading to the elimination of the majority of cervical cells with viral disruption sites in this gene in cases of severe neoplasia [26,58]. Furthermore, the HPV breakpoints in the validated viral-cellular DNA junctions demonstrated a statistically significant preferential distribution within the E1 open reading frame (ORF), consistent with findings from previous studies [12,46]. Integration within the E1 region is predicted to separate the E2 gene from the HPV promoter, thereby likely reducing the expression of the downstream E2 gene [46,59]. Reduced expression of E2 has been documented to facilitate the overexpression of the E6 and E7 oncoproteins, thereby accelerating the progression of cervical lesions [3,15,17,22,45]. Consistent with previous studies [12,29,36] indicating that integrated HPV retains intact copies of the oncogenes E6 and E7, our findings reveal that breakpoints rarely occur within these regions of the viral genome. This observation may highlight the critical role of sustained expression of these genes in the maintenance of malignancy [12,37,58,59]. Thus, our data, derived from 537 integration sites, robustly suggest that the L1 and E1 ORFs are primary targets for linkage to cellular sequences. This insight is instrumental for the efficient detection of integrated HPV genomes, which predominantly target this viral regulatory region.
In our analysis of the dataset, we noted a notable increase in microhomologies at integration breakpoints between the human and HPV genome. This observation implies that various microhomology-mediated DNA repair pathways may have been involved in facilitating the integration process, potentially serving as crucial mechanisms in HPV integration. Furthermore, recurrent sites of HPV insertion were observed at the chromosomal level across various samples, consistently exhibiting identical nucleotide sequences at the viral-cellular junctions. It is plausible that multiple mechanisms of HPV integration are operative, potentially involving regions of microhomology or specific DNA sequences that facilitate the ligation of host and viral sequences in certain instances. This hypothesis warrants systematic investigation in future studies.
Acknowledgements
We express our gratitude to all of the patients who contributed tissue samples for this study. This study was supported partly by grants from Shanghai Natural Science Foundation (18ZR1423900), Project Foundation of Taizhou School of Clinical Medicine, Nanjing Medical University (TZKY20220204 and TZKY20220205), Nantong Health Science and Technology Project (MS2022053), Nantong Basic Science and Technology Program (JC22022027), and Nantong University Clinical Medicine Special Project (2022JZ014).
Informed consent was obtained from the patients for sequencing analyses and data release.
Disclosure of conflict of interest
None.
Abbreviations
- HR-HPV
high-risk human papillomavirus
- ITGs
integration-targeted cellular genes
- LCR
long control region
- NCBI
National Center for Biotechnology Information
- ORFs
HPV open reading frames
- RTGs
recurrently targeted host genes
- WGS
whole genome sequencing
Tables S1, S2, S6, S9 and Figures S1-S3
Table S3
Table S4
Table S5
Table S7
Table S8
References
- 1.Lu X, Lin Q, Lin M, Duan P, Ye L, Chen J, Chen X, Zhang L, Xue X. Multiple-integrations of HPV16 genome and altered transcription of viral oncogenes and cellular genes are associated with the development of cervical cancer. PLoS One. 2014;9:e97588. doi: 10.1371/journal.pone.0097588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Serrao E, Cherepanov P, Engelman AN. Amplification, next-generation sequencing, and genomic DNA mapping of retroviral integration sites. J Vis Exp. 2016:53840. doi: 10.3791/53840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu T, Ferber MJ, Cheung TH, Chung TK, Wong YF, Smith DI. The role of viral integration in the development of cervical cancer. Cancer Genet Cytogenet. 2005;158:27–34. doi: 10.1016/j.cancergencyto.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 4.Katerji M, Duerksen-Hughes PJ. DNA damage in cancer development: special implications in viral oncogenesis. Am J Cancer Res. 2021;11:3956–3979. [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang R, Shen C, Zhao L, Wang J, McCrae M, Chen X, Lu F. Dysregulation of host cellular genes targeted by human papillomavirus (HPV) integration contributes to HPV-related cervical carcinogenesis. Int J Cancer. 2016;138:1163–1174. doi: 10.1002/ijc.29872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi Y, Li L, Hu Z, Li S, Wang S, Liu J, Wu C, He L, Zhou J, Li Z, Hu T, Chen Y, Jia Y, Wang S, Wu L, Cheng X, Yang Z, Yang R, Li X, Huang K, Zhang Q, Zhou H, Tang F, Chen Z, Shen J, Jiang J, Ding H, Xing H, Zhang S, Qu P, Song X, Lin Z, Deng D, Xi L, Lv W, Han X, Tao G, Yan L, Han Z, Li Z, Miao X, Pan S, Shen Y, Wang H, Liu D, Gong E, Li Z, Zhou L, Luan X, Wang C, Song Q, Wu S, Xu H, Shen J, Qiang F, Ma G, Liu L, Chen X, Liu J, Wu J, Shen Y, Wen Y, Chu M, Yu J, Hu X, Fan Y, He H, Jiang Y, Lei Z, Liu C, Chen J, Zhang Y, Yi C, Chen S, Li W, Wang D, Wang Z, Di W, Shen K, Lin D, Shen H, Feng Y, Xie X, Ma D. A genome-wide association study identifies two new cervical cancer susceptibility loci at 4q12 and 17q12. Nat Genet. 2013;45:918–922. doi: 10.1038/ng.2687. [DOI] [PubMed] [Google Scholar]
- 7.Bodelon C, Vinokurova S, Sampson JN, den Boon JA, Walker JL, Horswill MA, Korthauer K, Schiffman M, Sherman ME, Zuna RE, Mitchell J, Zhang X, Boland JF, Chaturvedi AK, Dunn ST, Newton MA, Ahlquist P, Wang SS, Wentzensen N. Chromosomal copy number alterations and HPV integration in cervical precancer and invasive cancer. Carcinogenesis. 2016;37:188–196. doi: 10.1093/carcin/bgv171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wentzensen N, Vinokurova S, von Knebel Doeberitz M. Systematic review of genomic integration sites of human papillomavirus genomes in epithelial dysplasia and invasive cancer of the female lower genital tract. Cancer Res. 2004;64:3878–3884. doi: 10.1158/0008-5472.CAN-04-0009. [DOI] [PubMed] [Google Scholar]
- 9.Asiaf A, Ahmad ST, Mohammad SO, Zargar MA. Review of the current knowledge on the epidemiology, pathogenesis, and prevention of human papillomavirus infection. Eur J Cancer Prev. 2014;23:206–224. doi: 10.1097/CEJ.0b013e328364f273. [DOI] [PubMed] [Google Scholar]
- 10.Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S. Human papillomavirus and cervical cancer. Lancet. 2007;370:890–907. doi: 10.1016/S0140-6736(07)61416-0. [DOI] [PubMed] [Google Scholar]
- 11.Doorbar J, Quint W, Banks L, Bravo IG, Stoler M, Broker TR, Stanley MA. The biology and life-cycle of human papillomaviruses. Vaccine. 2012;30(Suppl 5):F55–70. doi: 10.1016/j.vaccine.2012.06.083. [DOI] [PubMed] [Google Scholar]
- 12.Rusan M, Li YY, Hammerman PS. Genomic landscape of human papillomavirus-associated cancers. Clin Cancer Res. 2015;21:2009–2019. doi: 10.1158/1078-0432.CCR-14-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dall KL, Scarpini CG, Roberts I, Winder DM, Stanley MA, Muralidhar B, Herdman MT, Pett MR, Coleman N. Characterization of naturally occurring HPV16 integration sites isolated from cervical keratinocytes under noncompetitive conditions. Cancer Res. 2008;68:8249–8259. doi: 10.1158/0008-5472.CAN-08-1741. [DOI] [PubMed] [Google Scholar]
- 14.Luft F, Klaes R, Nees M, Dürst M, Heilmann V, Melsheimer P, von Knebel Doeberitz M. Detection of integrated papillomavirus sequences by ligation-mediated PCR (DIPS-PCR) and molecular characterization in cervical cancer cells. Int J Cancer. 2001;92:9–17. [PubMed] [Google Scholar]
- 15.Matovina M, Sabol I, Grubisić G, Gasperov NM, Grce M. Identification of human papillomavirus type 16 integration sites in high-grade precancerous cervical lesions. Gynecol Oncol. 2009;113:120–127. doi: 10.1016/j.ygyno.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 16.Pett MR, Alazawi WO, Roberts I, Dowen S, Smith DI, Stanley MA, Coleman N. Acquisition of high-level chromosomal instability is associated with integration of human papillomavirus type 16 in cervical keratinocytes. Cancer Res. 2004;64:1359–1368. doi: 10.1158/0008-5472.can-03-3214. [DOI] [PubMed] [Google Scholar]
- 17.Vojtechova Z, Sabol I, Salakova M, Turek L, Grega M, Smahelova J, Vencalek O, Lukesova E, Klozar J, Tachezy R. Analysis of the integration of human papillomaviruses in head and neck tumours in relation to patients’ prognosis. Int J Cancer. 2016;138:386–395. doi: 10.1002/ijc.29712. [DOI] [PubMed] [Google Scholar]
- 18.Liu CY, Li F, Zeng Y, Tang MZ, Huang Y, Li JT, Zhong RG. Infection and integration of high-risk human papillomavirus in HPV-associated cancer cells. Med Oncol. 2015;32:109. doi: 10.1007/s12032-015-0560-8. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y, Lu Z, Xu R, Ke Y. Comprehensive mapping of the human papillomavirus (HPV) DNA integration sites in cervical carcinomas by HPV capture technology. Oncotarget. 2016;7:5852–5864. doi: 10.18632/oncotarget.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chandrani P, Kulkarni V, Iyer P, Upadhyay P, Chaubal R, Das P, Mulherkar R, Singh R, Dutt A. NGS-based approach to determine the presence of HPV and their sites of integration in human cancer genome. Br J Cancer. 2015;112:1958–1965. doi: 10.1038/bjc.2015.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen D, Enroth S, Ivansson E, Gyllensten U. Pathway analysis of cervical cancer genome-wide association study highlights the MHC region and pathways involved in response to infection. Hum Mol Genet. 2014;23:6047–6060. doi: 10.1093/hmg/ddu304. [DOI] [PubMed] [Google Scholar]
- 22.Liang WS, Aldrich J, Nasser S, Kurdoglu A, Phillips L, Reiman R, McDonald J, Izatt T, Christoforides A, Baker A, Craig C, Egan JB, Chase DM, Farley JH, Bryce AH, Stewart AK, Borad MJ, Carpten JD, Craig DW, Monk BJ. Simultaneous characterization of somatic events and HPV-18 integration in a metastatic cervical carcinoma patient using DNA and RNA sequencing. Int J Gynecol Cancer. 2014;24:329–338. doi: 10.1097/IGC.0000000000000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Das P, Thomas A, Mahantshetty U, Shrivastava SK, Deodhar K, Mulherkar R. HPV genotyping and site of viral integration in cervical cancers in indian women. PLoS One. 2012;7:e41012. doi: 10.1371/journal.pone.0041012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Christiansen IK, Sandve GK, Schmitz M, Dürst M, Hovig E. Transcriptionally active regions are the preferred targets for chromosomal HPV integration in cervical carcinogenesis. PLoS One. 2015;10:e0119566. doi: 10.1371/journal.pone.0119566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ojesina AI, Lichtenstein L, Freeman SS, Pedamallu CS, Imaz-Rosshandler I, Pugh TJ, Cherniack AD, Ambrogio L, Cibulskis K, Bertelsen B, Romero-Cordoba S, Treviño V, Vazquez-Santillan K, Guadarrama AS, Wright AA, Rosenberg MW, Duke F, Kaplan B, Wang R, Nickerson E, Walline HM, Lawrence MS, Stewart C, Carter SL, McKenna A, Rodriguez-Sanchez IP, Espinosa-Castilla M, Woie K, Bjorge L, Wik E, Halle MK, Hoivik EA, Krakstad C, Gabiño NB, Gómez-Macías GS, Valdez-Chapa LD, Garza-Rodríguez ML, Maytorena G, Vazquez J, Rodea C, Cravioto A, Cortes ML, Greulich H, Crum CP, Neuberg DS, Hidalgo-Miranda A, Escareno CR, Akslen LA, Carey TE, Vintermyr OK, Gabriel SB, Barrera-Saldaña HA, Melendez-Zajgla J, Getz G, Salvesen HB, Meyerson M. Landscape of genomic alterations in cervical carcinomas. Nature. 2014;506:371–375. doi: 10.1038/nature12881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, Yang Y, Zhang R, Cai Y, Yang X, Wang Z, Li Y, Cheng X, Ye X, Xiang Y, Zhu B. Preferential sites for the integration and disruption of human papillomavirus 16 in cervical lesions. J Clin Virol. 2013;56:342–347. doi: 10.1016/j.jcv.2012.12.014. [DOI] [PubMed] [Google Scholar]
- 27.Schmitz M, Driesch C, Jansen L, Runnebaum IB, Dürst M. Non-random integration of the HPV genome in cervical cancer. PLoS One. 2012;7:e39632. doi: 10.1371/journal.pone.0039632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nambaru L, Meenakumari B, Swaminathan R, Rajkumar T. Prognostic significance of HPV physical status and integration sites in cervical cancer. Asian Pac J Cancer Prev. 2009;10:355–360. [PubMed] [Google Scholar]
- 29.Popescu NC. Genetic alterations in cancer as a result of breakage at fragile sites. Cancer Lett. 2003;192:1–17. doi: 10.1016/s0304-3835(02)00596-7. [DOI] [PubMed] [Google Scholar]
- 30.Diao MK, Liu CY, Liu HW, Li JT, Li F, Mehryar MM, Wang YJ, Zhan SB, Zhou YB, Zhong RG, Zeng Y. Integrated HPV genomes tend to integrate in gene desert areas in the CaSki, HeLa, and SiHa cervical cancer cell lines. Life Sci. 2015;127:46–52. doi: 10.1016/j.lfs.2015.01.039. [DOI] [PubMed] [Google Scholar]
- 31.Ferber MJ, Eilers P, Schuuring E, Fenton JA, Fleuren GJ, Kenter G, Szuhai K, Smith DI, Raap AK, Brink AA. Positioning of cervical carcinoma and Burkitt lymphoma translocation breakpoints with respect to the human papillomavirus integration cluster in FRA8C at 8q24.13. Cancer Genet Cytogenet. 2004;154:1–9. doi: 10.1016/j.cancergencyto.2004.01.028. [DOI] [PubMed] [Google Scholar]
- 32.Brink AA, Wiegant JC, Szuhai K, Tanke HJ, Kenter GG, Fleuren GJ, Schuuring E, Raap AK. Simultaneous mapping of human papillomavirus integration sites and molecular karyotyping in short-term cultures of cervical carcinomas by using 49-color combined binary ratio labeling fluorescence in situ hybridization. Cancer Genet Cytogenet. 2002;134:145–150. doi: 10.1016/s0165-4608(01)00620-3. [DOI] [PubMed] [Google Scholar]
- 33.Adey A, Burton JN, Kitzman JO, Hiatt JB, Lewis AP, Martin BK, Qiu R, Lee C, Shendure J. The haplotype-resolved genome and epigenome of the aneuploid HeLa cancer cell line. Nature. 2013;500:207–211. doi: 10.1038/nature12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmitz M, Driesch C, Beer-Grondke K, Jansen L, Runnebaum IB, Dürst M. Loss of gene function as a consequence of human papillomavirus DNA integration. Int J Cancer. 2012;131:E593–602. doi: 10.1002/ijc.27433. [DOI] [PubMed] [Google Scholar]
- 35.Ferber MJ, Thorland EC, Brink AA, Rapp AK, Phillips LA, McGovern R, Gostout BS, Cheung TH, Chung TK, Fu WY, Smith DI. Preferential integration of human papillomavirus type 18 near the c-myc locus in cervical carcinoma. Oncogene. 2003;22:7233–7242. doi: 10.1038/sj.onc.1207006. [DOI] [PubMed] [Google Scholar]
- 36.Hu Z, Zhu D, Wang W, Li W, Jia W, Zeng X, Ding W, Yu L, Wang X, Wang L, Shen H, Zhang C, Liu H, Liu X, Zhao Y, Fang X, Li S, Chen W, Tang T, Fu A, Wang Z, Chen G, Gao Q, Li S, Xi L, Wang C, Liao S, Ma X, Wu P, Li K, Wang S, Zhou J, Wang J, Xu X, Wang H, Ma D. Genome-wide profiling of HPV integration in cervical cancer identifies clustered genomic hot spots and a potential microhomology-mediated integration mechanism. Nat Genet. 2015;47:158–163. doi: 10.1038/ng.3178. [DOI] [PubMed] [Google Scholar]
- 37.Thorland EC, Myers SL, Persing DH, Sarkar G, McGovern RM, Gostout BS, Smith DI. Human papillomavirus type 16 integrations in cervical tumors frequently occur in common fragile sites. Cancer Res. 2000;60:5916–5921. [PubMed] [Google Scholar]
- 38.Chen D, Gaborieau V, Zhao Y, Chabrier A, Wang H, Waterboer T, Zaridze D, Lissowska J, Rudnai P, Fabianova E, Bencko V, Janout V, Foretova L, Mates IN, Szeszenia-Dabrowska N, Boffetta P, Pawlita M, Lathrop M, Gyllensten U, Brennan P, McKay JD. A systematic investigation of the contribution of genetic variation within the MHC region to HPV seropositivity. Hum Mol Genet. 2015;24:2681–2688. doi: 10.1093/hmg/ddv015. [DOI] [PubMed] [Google Scholar]
- 39.Kazemian M, Ren M, Lin JX, Liao W, Spolski R, Leonard WJ. Possible human papillomavirus 38 contamination of endometrial cancer RNA sequencing samples in The Cancer Genome Atlas Database. J Virol. 2015;89:8967–8973. doi: 10.1128/JVI.00822-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miura K, Mishima H, Kinoshita A, Hayashida C, Abe S, Tokunaga K, Masuzaki H, Yoshiura K. Genome-wide association study of HPV-associated cervical cancer in Japanese women. J Med Virol. 2014;86:1153–1158. doi: 10.1002/jmv.23943. [DOI] [PubMed] [Google Scholar]
- 41.Dutta S, Chakraborty C, Dutta AK, Mandal RK, Roychoudhury S, Basu P, Panda CK. Physical and methylation status of human papillomavirus 16 in asymptomatic cervical infections changes with malignant transformation. J Clin Pathol. 2015;68:206–211. doi: 10.1136/jclinpath-2014-202611. [DOI] [PubMed] [Google Scholar]
- 42.Peter M, Stransky N, Couturier J, Hupé P, Barillot E, de Cremoux P, Cottu P, Radvanyi F, Sastre-Garau X. Frequent genomic structural alterations at HPV insertion sites in cervical carcinoma. J Pathol. 2010;221:320–330. doi: 10.1002/path.2713. [DOI] [PubMed] [Google Scholar]
- 43.Thorland EC, Myers SL, Gostout BS, Smith DI. Common fragile sites are preferential targets for HPV16 integrations in cervical tumors. Oncogene. 2003;22:1225–1237. doi: 10.1038/sj.onc.1206170. [DOI] [PubMed] [Google Scholar]
- 44.Doolittle-Hall JM, Cunningham Glasspoole DL, Seaman WT, Webster-Cyriaque J. Meta-analysis of DNA tumor-viral integration site selection indicates a role for repeats, gene expression and epigenetics. Cancers (Basel) 2015;7:2217–2235. doi: 10.3390/cancers7040887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kraus I, Driesch C, Vinokurova S, Hovig E, Schneider A, von Knebel Doeberitz M, Dürst M. The majority of viral-cellular fusion transcripts in cervical carcinomas cotranscribe cellular sequences of known or predicted genes. Cancer Res. 2008;68:2514–2522. doi: 10.1158/0008-5472.CAN-07-2776. [DOI] [PubMed] [Google Scholar]
- 46.Parfenov M, Pedamallu CS, Gehlenborg N, Freeman SS, Danilova L, Bristow CA, Lee S, Hadjipanayis AG, Ivanova EV, Wilkerson MD, Protopopov A, Yang L, Seth S, Song X, Tang J, Ren X, Zhang J, Pantazi A, Santoso N, Xu AW, Mahadeshwar H, Wheeler DA, Haddad RI, Jung J, Ojesina AI, Issaeva N, Yarbrough WG, Hayes DN, Grandis JR, El-Naggar AK, Meyerson M, Park PJ, Chin L, Seidman JG, Hammerman PS, Kucherlapati R Cancer Genome Atlas Network. Characterization of HPV and host genome interactions in primary head and neck cancers. Proc Natl Acad Sci U S A. 2014;111:15544–15549. doi: 10.1073/pnas.1416074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spurlock CF 3rd, Shaginurova G, Tossberg JT, Hester JD, Chapman N, Guo Y, Crooke PS 3rd, Aune TM. Profiles of long noncoding RNAs in human naive and memory T cells. J Immunol. 2017;199:547–558. doi: 10.4049/jimmunol.1700232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cammayo-Fletcher PLT, Flores RA, Nguyen BT, Altanzul B, Fernandez-Colorado CP, Kim WH, Devi RM, Kim S, Min W. Identification of critical immune regulators and potential interactions of IL-26 in riemerella anatipestifer-infected ducks by transcriptome analysis and profiling. Microorganisms. 2024;12:973. doi: 10.3390/microorganisms12050973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cai Y, Chen L, Liu X, Yao W, Hou W. GmNF-YC4 delays soybean flowering and maturation by directly repressing GmFT2a and GmFT5a expression. J Integr Plant Biol. 2024;66:1370–1384. doi: 10.1111/jipb.13668. [DOI] [PubMed] [Google Scholar]
- 50.Ishihara Y, Tsuji M, Vogel CFA. Suppressive effects of aryl-hydrocarbon receptor repressor on adipocyte differentiation in 3T3-L1 cells. Arch Biochem Biophys. 2018;642:75–80. doi: 10.1016/j.abb.2018.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou B, Bai P, Xue H, Zhang Z, Shi S, Zhang K, Wang Y, Wang K, Quan Y, Song Y, Zhang L. Single nucleotide polymorphisms in PDCD6 gene are associated with the development of cervical squamous cell carcinoma. Fam Cancer. 2015;14:1–8. doi: 10.1007/s10689-014-9767-7. [DOI] [PubMed] [Google Scholar]
- 52.Sim S, Weinberg DE, Fuchs G, Choi K, Chung J, Wolin SL. The subcellular distribution of an RNA quality control protein, the Ro autoantigen, is regulated by noncoding Y RNA binding. Mol Biol Cell. 2009;20:1555–1564. doi: 10.1091/mbc.E08-11-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang AT, Langley AR, Christov CP, Kheir E, Shafee T, Gardiner TJ, Krude T. Dynamic interaction of Y RNAs with chromatin and initiation proteins during human DNA replication. J Cell Sci. 2011;124:2058–2069. doi: 10.1242/jcs.086561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Christov CP, Trivier E, Krude T. Noncoding human Y RNAs are overexpressed in tumours and required for cell proliferation. Br J Cancer. 2008;98:981–988. doi: 10.1038/sj.bjc.6604254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Verhagen AP, Pruijn GJ. Are the Ro RNP-associated Y RNAs concealing microRNAs? Y RNA-derived miRNAs may be involved in autoimmunity. Bioessays. 2011;33:674–682. doi: 10.1002/bies.201100048. [DOI] [PubMed] [Google Scholar]
- 56.Zhao S, Liu W, Li Y, Liu P, Li S, Dou D, Wang Y, Yang R, Xiang R, Liu F. Alternative splice variants modulates dominant-negative function of helios in T-cell leukemia. PLoS One. 2016;11:e0163328. doi: 10.1371/journal.pone.0163328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cancer Genome Atlas Research Network; Albert Einstein College of Medicine; Analytical Biological Services; Barretos Cancer Hospital; Baylor College of Medicine; Beckman Research Institute of City of Hope; Buck Institute for Research on Aging; Canada’s Michael Smith Genome Sciences Centre; Harvard Medical School; Helen F. Graham Cancer Center & Research Institute at Christiana Care Health Services; HudsonAlpha Institute for Biotechnology; ILSbio, LLC; Indiana University School of Medicine; Institute of Human Virology; Institute for Systems Biology; International Genomics Consortium; Leidos Biomedical; Massachusetts General Hospital; McDonnell Genome Institute at Washington University; Medical College of Wisconsin; Medical University of South Carolina; Memorial Sloan Kettering Cancer Center; Montefiore Medical Center; NantOmics; National Cancer Institute; National Hospital, Abuja, Nigeria; National Human Genome Research Institute; National Institute of Environmental Health Sciences; National Institute on Deafness & Other Communication Disorders; Ontario Tumour Bank, London Health Sciences Centre; Ontario Tumour Bank, Ontario Institute for Cancer Research; Ontario Tumour Bank, The Ottawa Hospital; Oregon Health & Science University; Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center; SRA International; St Joseph’s Candler Health System; Eli & Edythe L. Broad Institute of Massachusetts Institute of Technology & Harvard University; Research Institute at Nationwide Children’s Hospital; Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins University; University of Bergen; University of Texas MD Anderson Cancer Center; University of Abuja Teaching Hospital; University of Alabama at Birmingham; University of California, Irvine; University of California Santa Cruz; University of Kansas Medical Center; University of Lausanne; University of New Mexico Health Sciences Center; University of North Carolina at Chapel Hill; University of Oklahoma Health Sciences Center; University of Pittsburgh; University of São Paulo, Ribeir ão Preto Medical School; University of Southern California; University of Washington; University of Wisconsin School of Medicine & Public Health; Van Andel Research Institute; Washington University in St Louis. Integrated genomic and molecular characterization of cervical cancer. Nature. 2017;543:378–384. doi: 10.1038/nature21386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Z, Liu C, Liu W, Lv X, Hu T, Yang F, Yang W, He L, Huang X. Long-read sequencing reveals the structural complexity of genomic integration of HPV DNA in cervical cancer cell lines. BMC Genomics. 2024;25:198. doi: 10.1186/s12864-024-10101-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu L, Majerciak V, Lobanov A, Mirza S, Band V, Liu H, Cam M, Hughes SH, Lowy DR, Zheng ZM. HPV oncogenes expressed from only one of multiple integrated HPV DNA copies drive clonal cell expansion in cervical cancer. mBio. 2024;15:e0072924. doi: 10.1128/mbio.00729-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.