Abstract
Anaplastic thyroid cancer (ATC) is the most aggressive thyroid cancer, with very limited treatment options. Mutations of p53 are associated with lethal outcomes of ATC. In this study, we tested the hypothesis that wild type p53 (WTp53) mitigates its aggressive progression. We used human 8505C cells (from human ATC tumors) as a model, harboring a BRAFV600E mutation and single of mutated p53C742G allele. We exogenously expressed WTp53 or mutant p53C742G into 8505C cells (8505C-WTp53 or 8505C-MTp53, respectively). The expressed WTp53 inhibited cell proliferation, decreased cell migration, and induced apoptosis via induction of proapoptotic WTp53 target BAX and PUMA genes in vitro. Mouse xenograft studies showed suppression of tumors induced by 8505C-WTp53 but not by 8505C-MTp53 cells. Consistent with in vitro findings, WTp53 inhibited proliferation of tumor cells, evidenced by decreased proliferation marker Ki-67 in tumors. WTp53 also induced apoptosis in xenograft tumors as shown by increased cleaved caspase-3 proteins and pro-apoptotic regulators, BAX and PUMA. Single cells RNA-sequencing (scRNA-seq) of tumors induced by 8505C, 8505C-WTp53, and 8505C-MTp53 cells demonstrated differential expression gene (DEG) patterns between 8505C-WTp53 and 8505C tumors. DEGs analysis identified alteration of multiple pathways, leading to attenuating the oncogenic actions of mutant p53. The discovery of the suppression of TNFα via NFκB pathway topped the pathways list, resulting in subduing the deleterious inflammatory responses caused by mutant p53. Our findings that exogenously expressed WTp53 could counter act the oncogenic actions of p53 has heightened the feasibility of using CRISPR/Cas9 genome editing to modify the p53 alleles for potential treatment of ATC.
Keywords: Anaplastic thyroid cancer, tumor suppressor p53, mutant p53, proliferation, apoptosis, thyroid carcinogenesis, single cell RNA-seq
Introduction
Thyroid cancers in humans consist of an array of several different histologic and biologic types (papillary, follicular, medullary, clear cell, anaplastic, and Hürthle cell), but the majority of clinically important human thyroid cancers are of the papillary (PTC) and follicular (FTC) types. FTC and PTC are highly differentiated and have a relatively good prognosis. The 5-year relative survival rate for regional papillary thyroid cancer is 99%. For regional follicular cancer, the rate is 98%, and for regional medullary cancer, it is 92%. However, for regional anaplastic thyroid cancer, the rate is 11% [1] and ATC patients rarely survived beyond 1 year after diagnosis [2,3].
The molecular genetics underlying thyroid carcinogenesis have been studied extensively. The known genetic changes causing PTC are RET chromosomal rearrangement or mutations of RAS or BRAF proto-oncogenes which lead to constitutive activation of the mitogen-activated protein kinase (MAPK) pathway to drive cancer progression [4]. While in FTC, he known genetic mutations are PAX8/PPARγ rearrangements and mutations in the RAS and PIK3CA gene [5,6]. As a result, identification of these genetic lesions and elucidation of their aberrant signaling in PTC and FTC have facilitated the development of targeted therapies to provide more options in the treatment of patients with differentiated thyroid cancer.
However, a recent study has shown that genetic alterations in ATC are more complex than those of the differentiated thyroid cancers. In addition to the frequent mutations in the BRAF and RAS oncogenes, a higher frequency of mutations in TP53, TERT promoter, PI3K/AKT/mTOR pathway effectors, SWI/SNF subunits, and histone methyltransferases have also been identified [7]. TP53 mutations, specifically, have been observed in 73% of human tumors identified and tend to occur at the late stage of ATC progression [3]. These observations suggest that occurrence of TP53 mutations could be a key reason for the poor outcomes of ATC patients. Currently, the underlying events resulting in frequent mutations of TP53 in late stages of carcinogenesis is not known. However, the high frequency of TP53 mutations provided an opportunity to test the hypothesis that the deleterious actions of p53 mutants could be counteracted by exogenously introduced WTp53. Toward this aim, we exogenously expressed either WTp53 or the mutated p53C742G (MTp53) in a human ATC cell line (8505C cells), which harbors both a BRAFV600E mutation and a single mutated p53C742G allele (MTp53). Extensive molecular characterization of 8505C cells stably expressing WTp53 (8505C-WTp53) showed that WTp53 attenuated oncogenic phenotypes by decreasing proliferation, inducing apoptosis, and weakening migration capacity of tumor cells in vitro, and suppressing tumor growth in vivo in xenograft models. Transcriptomic analysis from single cell RNA sequencing (scRNA-seq) uncovered multiple pathways that could be responsible for the reversal of the cancer phenotype of 8505C cells. Importantly, we also found that the exogenously expressed WTp53 induced the re-expression of the SLC5A5 gene [encoding sodium (Na)-iodide symporter; NIS], which should be amendable for improving the radio-iodide treatment efficacy of ATC patients.
Materials and methods
Preparation of 8505C cell stably expressing WTp53 or MTp53 in 8505C cells
The human ATC cell line 8505C used in this study was described previously [21]. To generate 8505C cells stably expressing either WTp53 or MTp53, lentivirus was prepared using plasmids encoding WTp53 and MTp53 cDNAs, respectively. DNA sequence corresponding to WT TP53 was obtained using the UCSC Genome Browser under accession number ENST00000269305.9. Wild type TP53 and mutant TP53 (R248G) cDNA sequences were then synthesized using Twist Biosciences and subsequently cloned into pGMC00020 (Addgene 195304) using isothermal assembly (PMID: 19363495) [WT-p53 (pMC0749), MTp53 (pMC0750), respectively]. Plasmids were verified using Sanger sequencing. To generate lentivirus containing WT TP53 and mutant p53 cDNA, HEK293T cells were seeded (1.8 × 107 cells on 15 cm plates) and were transfected with plasmids pMC0749 (WT-TP53), or pMC0750 (mutant TP53) together with psPAX2, and pMD2.G (both from ADDGENE) using Lipofectamine 3000 (Invitrogen, Carlsbad, CA). psPAX2 and pMD2.G were obtained from Addgene. After 24 hours, the medium was replaced and after 24 hours, media was collected, filtered, and concentrated by Lenti-X concentrator solution (Takara, Otsu, Japan). The final mixed solution was then centrifuged at 1500 g for 45 min at 4°C and the pellet was then resuspended in phosphate-buffered saline (PBS). The concentrated lentivirus for both WTp53 and MTp53 was then used to transduce parental 8505C cells to generate 8505C-WTp53 and 8505C-MTp53 cells. The transduced 8505C cells were then treated with 20 μg/ml of blasticidin (Thermo Fisher Scientific, Waltham, MA, USA) for 5-7 days for selection of cells expressing either WTp53 or MTp53.
Confocal imaging of localization of p53 and NIS in cells
Cells (8505C, 8505C-WTp53, 8505C-MTp53) were seeded onto poly-L-lysine-coated coverslips at 3 × 105 cells/per coverslip, and cultured cells were grown until reaching 90% confluency. Surface membranes were stained with MemBrite® Fix 640/660 (Biotium, Fremont, CA) for 10 min, according to manufacturer’s instructions, and then fixed in 10% neutral buffered formalin. Endogenous peroxidase activity was inhibited by incubation for 30 min in 0.3% H2O2 in 0.01 M Tris, and non-specific binding was reduced by blocking in normal goat serum at room temperature. Cells on coverslips were incubated with anti-NIS antibodies (hNIS 331 Abs, 1:200 dilution), anti-p53 antibodies (60283-1-Ig Abs, 1:200 dilution) at 4°C. They then went through several washing with 1 × PBS and were incubated in a 1:200 dilution of Goat anti-Rabbit IgG Alexa Flor488 (A11034, ThermoFisher), Goat anti-mouse IgG Alexa Flor 568 (A11004, ThermoFisher) for 1 hour in a light-protected chamber at room temperature. Coverslips were counterstained with 4, 6-diamidino-2-phenylindole dihydrochloride (DAPI) and mounted on slides using ProLong Diamond Antifade mount reagent (Thermo Fisher Scientific) with DAPI nuclear staining. Confocal images were collected using a Nikon SoRa spinning disk microscope equipped with a 60 × Apo TIRF oil immersion objective lens (N.A. 1.49) and Photometrics BSI sCMOS camera. The Nikon Elements image analysis software (v.5.41) was used to adjust the brightness and contrast to be consistent for all images in the dataset.
In vivo mouse xenograft study
All animal experiments were performed under protocols approved by the National Cancer Institute Animal Care and Use Committee. For xenograft studies, 6- to 8-week-old female athymic nude mice were used as previously described [22]. Cell preparations of 8505C, 8505C-WTp53, and 505C-MTp53 (5 × 106 cells) were mixed in 200 μl 50% Matrigel basement membrane matrix and subcutaneously inoculated into the right flank of mice. We then monitored the frequency of tumor initiation and tumor growth for about 12 weeks. When the longest diameter of the biggest tumor exceeded 20 mm, we simultaneously euthanized all the mice to dissect the tumors for further analyses.
Flow cytometry analysis
Apoptosis assays for all three ATC cell lines, 8505C, 8505C-WTp53, and 505C-MTp53 cells, were carried out as described previously [22]. The list of antibodies used in the study is included in Supplementary Table 1.
To assay for reactive oxygen species (ROS) activity, we treated 8505C, 8505C-WTp53, and 8505C-MTp53 cells, with 10 μM of 2’,7’-Dichlorodihydrofluorescein diacetate (H2DCFDA) for 30 minutes at 37°C in the dark. Briefly, H2DCFDA diffuses into the cytoplasm, and subsequently cellular esterases remove the acetate groups from the compound to form a non-fluorescent moiety that can be oxidized by intracellular ROS to form the fluorescent products [2’,7’-dichlorofluorescein (DCF)] which was measured by flow cytometry (BD FACS CantoII). The data analysis was done by FlowJo software 10.5.3.
Immunohistochemistry
Tissues from a xenografted tumor were fixed in 10% neutral buffered formalin and subsequently embedded in paraffin. Five-micrometer-thick sections were prepared and stained with hematoxylin and eosin. IHC was performed as previously described [23]. The list of antibodies used in the study is included in the Supplementary Table 2. Tissue slides were visualized under a microscope at a magnification of × 200, and the relative positive cell ratio was determined by NIH ImageJ software.
RNA extraction and real-time RT-PCR analysis
Total RNA was extracted from 8505C, 8505C-WTp53 and 8505C-MTp53 cells or tumors induced by 8505C, 8505C-WTp53 or 8505C-MTp53 by using TRIzol (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Real-time RT-PCR analysis was carried as described [23] and the primers used were listed in Supplementary Table 2.
Single-cell dissociation from xenografts and single-cell transcriptomic analysis
Xenograft tumors were dissociated into single cells using a tumor dissociation kit. Briefly, the tumors induced by 8505C, 8505C-WTp53 or 8505C-MTp53 were dissected and cut into small pieces of 2-4 mm, and these tissues were transferred into the gentleMACS C Tube containing an enzyme mix (2.35 mL of RPMI-1640, 100 µL of Enzyme D, 50 µL of Enzyme R, and 12.5 µL of Enzyme A), followed by the tumor dissociation using a gentleMACS Dissociator. The dissociated cells were shortly centrifuged, resuspended, and applied to a cell strainer (70 µm) placed on a 15 mL tube, followed by washing the cell strainer with 10 mL of RPMI-1640. The cells were then centrifuged at 300 × g for seven minutes, resuspended with the AKC lysing buffer to remove erythrocytes, washed phosphate buffer saline, and subjected to the scRNA-seq. Further detailed methods and bioinformatics analyses are included in Supplementary Materials.
Statistical analysis
Continuous variables were compared using a two-tailed Student’s t-test, and categorical data were compared using a two-tailed χ2 test or Fisher’s exact test. Pearson correlation coefficients were used for the multiple correlation analyses between the THRB gene and stemness-related genes. To illustrate the results from these analyses, RStudio 1.2 program was used. Two-way analysis of variance (ANOVA) with Bonferroni’s post-hoc test was used to compare tumor growth in xenograft model according to the time and groups. Disease-free or overall survival was compared between high and low THRB groups using a Kaplan-Meier estimator. Statistical analysis was performed using SPSS version 23.0 or GraphPad Prism. Data are presented as the mean ± SD. All P values reported throughout the manuscript are two-sided, and those less than 0.05, were considered statistically significant.
Further detailed methods are included in Supplementary Materials.
Results
WTp53 abrogates the tumor phenotypes of anaplastic thyroid cancer cells
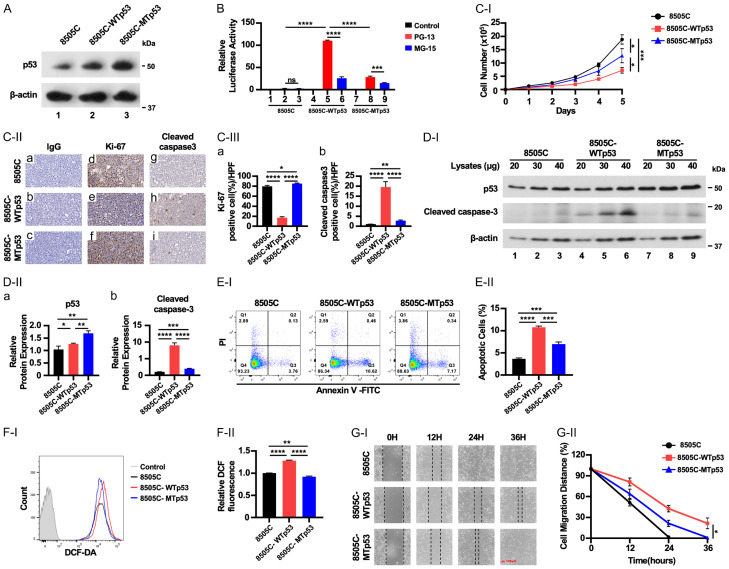
To determine if WTp53 could counter act the oncogenic action of mutant p53 expressed in the 8505C cells, 8505C cells stably expressing either WTp53 (8505C-WTp53) or mutant p53C742G (8505C-MTp53) were generated. To ensure comparable results between 8505C-WTp53 or 8505C-MTp53 cells, clones expressing similar levels of WTp53 and MTp53 were used in these studies (Figure 1A). For 8505C-WTp53, WTp53 was confirmed to be functional, transcriptionally, by inducing expression of a luciferase reporter driven by a promoter containing p53 binding elements (Figure 1B, bar 5), but not in a reporter which had mutated p53 binding element (Figure 1B, bar 6). Conversely, only a low basal reporter activity was detected in the presence of MTp53 (bars 8 and 9), validating that both the exogenously expressed WTp53 and MTp53 were functional.
Figure 1.
Exogenously expressed WTp53 inhibits cell growth of ATC cells. (A) Representative western blots showing p53 protein levels in 8505C, 8505C-WTp53, 8505C-MTp53 cells. (B) WTp53, but not MTp53, activated the luciferase activity mediated by WTp53 cis elements (PG13), but not mutated cis-elements (MG-13). Significant differences are indicated by asterisks (****P<0.0001). Data represent the mean ± SD (n=3). (C-I) Cell proliferation curves for 8505C, 8505C-WTp53, 8505C-MTp53 cells, respectively. Cells were seeded in six-well plates at a density of 50,000 cells per well and counted daily for 5 days. (C-II) Cells were expanded for 3-7 days before pelleting for IHC analysis. Representative IHC images and quantitative analysis of IHC results for Ki-67 (d-f) and cleaved caspase-3 (g-i). (C-III) Cell stained positively with Ki-67 (a) or cleaved caspase 3 were counted and graphed (n=3). Significant differences are indicated by asterisks ****P<0.0001. Data represent the mean ± SD (n=3). (D) Representative western blot analysis showing high expression of WTp53 and cleaved caspase-3 proteins in ATC cells (n=3). (E) Elevated WTp53 proteins in ATC cells increases apoptotic cell death. (E-II) Quantitative analysis of apoptotic cells (n=3) in 8505C, 8505C-WTp53, and 8505C-MTp53 cells. (F-I) Elevated WTp53 protein increases reactive oxygen species (ROS) in ATC cells analyzed by flow cytometry analysis. (F-II) Quantitative analysis of ROS (n=3) in 8505C, 8505C-WTp53, and 8505C-MTp53 cells. (G-I) WTp53, inhibits cell migration of ATC cells. (G-II) Quantitative analysis of migration distances from the wound scratch sites. Cell migration capacity was analyzed for 8505C, 8505C-WTp53, and 8505C-MTp53 cells, respectively. Scale bar (red bar) =100 µm. Significant differences are indicated by asterisks (*P<0.05, **P<0.01, ***P<0.001, and ****P<0.0001). Data represent the mean ± SD (n=3).
We next demonstrated that compared to 8505C-MTp53 cells, 8505C-WTp53 grew slower, indicating that WTp53 acted to suppress cell proliferation (Figure 1C-I). IHC analysis demonstrated that parental 8505C and 8505C-MTp53 cells were expressing high Ki-67 proliferation marker, but lower Ki-67-stained cells were detected in 8505C-WTp53 cells (Figure 1C-II, Figure 1C-II-a-c as IgG controls, compare Figure 1C-II-e versus Figure 1C-II-d and 1C-II-f; also Figure 1C-III-a). WTp53 is known to induce apoptosis, which was evident by IHC analysis, in which cells markedly stained with cleaved caspase 3 was found in 8505C-WTp53 cells (Figure 1C-II-h) as compared with 8505C and 8505C-MTp53 (Figure 1C-II-g, 1C-II-i, 1C-III-b). The increased expression of cleaved caspase 3 was further demonstrated by western blot analysis in which a dose-dependent increased cleaved caspase 3 protein levels were correlated with increased WTp53 protein levels (Figure 1D-I, 1D-II, 1D-II-a and 1D-II-b). Using flowcytometry analysis (FACS), we further showed increased apoptotic cells in 8505C cells expressing WTp53 (Figure 1E-I and 1E-II). Further, consistent with the reports showing that ROS could act as an upstream signal to trigger WTp53 activation to induce apoptosis [8], we found an increase in intracellular levels of ROS (Figure 1F-I and 1F-II). We further demonstrated that expressed WTp53 diminished the capacity of 8505C tumor cells migration by using wound healing assays. WTp53 clearly lessened the capacity of 8505C cells shown by delayed wound healing of 8505C-WTp53 cells as compared with 8505C cells (Figure 1G-I and 1G-II). Taken together, these data indicate that elevated WTp53 could act to counter act the deleterious mutant p53 actions by decreasing tumor cell proliferation, inducing apoptosis, and mitigating cell migration.
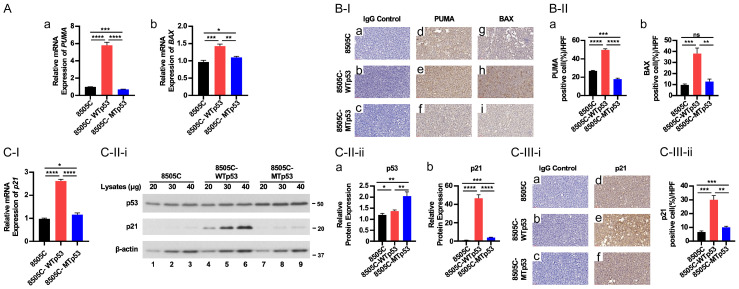
We next explored WTp53 downstream target the genes involved in apoptosis and cell cycle regulation. Among the pro-apoptotic regulators in the mitochondria pathways, the expression of PUMA is induced by p53 [9-11]. Once PUMA is expressed, it activates the pro-apoptotic effectors BAX and BAK [12]. We found that WTp53 activated the mRNA expression of PUMA (Figure 2A-a) and BAX (Figure 2A-b), but MTp53 did not. Consistently, IHC showed that these pro-apoptotic regulators were elevated at the protein levels in 8505C-WTp53 cells (Figure 2B-I, panel Figure 2B-I-e and panel Figure 2B-I-h for PUMA and BAX protein, respectively; also see Figure 2B-II-a and 2B-II-b).
Figure 2.
WTp53 increases the expression of PUMA, BAX, and p21 in ATC cells. (A) Comparison of the expression of the PUMA and BAX genes at the mRNA by RT-qPCR analysis in 8505C, 8505C-WTp53, and 8505C-MTp53 cells respectively (n=3). (B) Comparison of protein levels of PUMA and BAX in 8505C, 8505C-WTp53, and 8505C-MTp53 cells by IHC analysis [B-I, images; B-II, quantitation of cell positively immuno-stained with PUMA (d-f of B-I) or BAX (g-i of B-I)]. (C-I) In ATC cells, p53 expression increases the expression of p21 at the mRNA and protein levels. (C-II) Representative western blot analysis showing high expression of p53 and p21 proteins in ATC cells (II-ii, n=3). (C-III) Representative IHC images (i) and quantitative analysis of IHC results (ii, n=3) for p21 (d-f). Significant differences are indicated by asterisks (*P<0.05, **P<0.01, ***P<0.001, and ****P<0.0001). Data represent the mean ± SD (n=3).
We further examined cell cycle regulators that are regulated by WTp53. p21, which acts as a negative regulator in the G1 phase of cell cycle progression, is a direct target of WTp53 [13]. We found that WTp53 activated the expression of p21 at the mRNA level (Figure 2C-I). The protein levels of p21 were elevated in concert with the increased levels of WTp53 as shown by western blot analysis (Figure 2C-II-i; quantitation data shown in Figure 2C-II-ii) and by IHC (Figure 2C-III-i and 2C-III-ii). Notably, MTp53 did not activate the expression p21 [(expression was not altered in 8505C-MTp53 cells (Figure 2C-I, 2C-II and 2C-III)]. These results show that the exogenously introduced WTp53 activates its downstream direct target genes to induce apoptosis and inhibit cell cycle progression in 8505C-WTp53 cells.
WTp53 suppresses in vivo tumor growth induced by 8505C cells in xenograft mouse models
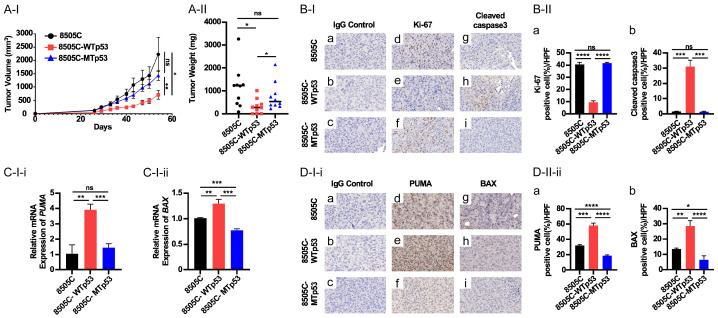
We next carried out in vivo studies to ascertain the effects of WTp53 on tumor growth. Nude mice, injected with 8505C cells, developed large tumors which were not significantly different in the growth rate from those induced by 8505C-MTp53 cells (Figure 3A-I and 3A-II). In contrast, the tumors induced by 8505C-WTp53 cells grew significantly slower (Figure 3A-I) and with 69% reduced weight than those induced by parental 8505C cells (Figure 3A-II). IHC analysis also showed that proportion of cells that were positively stained with Ki-67 was the lowest in 8505C-WTp53 cells (Figure 3B-I-d-f and 3B-II-a, 3B-I-a-c as IgG controls), indicating inhibition of tumor cell proliferation by WTp53. The suppression of tumor growth could also be due to the induction of apoptosis as evidenced by highly elevated apoptosis marker; cleaved caspase 3 (Figure 3B-I-g-i and 3B-II-b) as well as increased the mRNA expression of pro-apoptotic markers, PUMA and BAX in 8505C-WTp53 cells (Figure 3C-I-i and 3C-I-ii, respectively). Consistent with mRNA expression, the protein levels of these two pro-apoptotic regulators were also elevated as evidenced by IHC analysis (Figure 3D-I-i-d-f and 3D-I-i-g-i, for PUMA and BAX, respectively. 3D-I-i-a-c as IgG controls). Quantitative analysis showed the protein levels were increased 1.5- and 1.6-fold as compared with parental 8505C cells (Figure 3D-II-ii-a and 3D-II-ii-b, respectively). It is worthy to point out that the exogenously expressed MTp53 in 8505C cells did not further exacerbate the tumor phenotype due to the exogenous mutated p53 as no significant changes were observed in the cells immuno-stained with Ki-67 and cleaved caspase 3 in 8505C-MTp53 cells as compared with 8505C cells (Figure 3B-I and 3B-II). Similarly, no marked changes in the expression of pro-apoptotic markers, PUMA and BAX, either at the mRNA levels or protein levels, were observed when compared with those in parental 8505C cells. These results indicate that exogenously expressed WTp53 could counter act the oncogenic promoting actions of mutated p53 to reverse the tumor progression, but exogenously expressed MTp53 does not further exacerbate the tumor phenotypes in 8505C cells.
Figure 3.
WTp53 suppresses xenograft tumor growth induced by 8505C cells. (A) Comparison of xenograft tumors induced by 8505C, 8505C-WTp53, and 8505C-MTp53 cells. Growth curve (I), and weight (II) of induced xenograft tumors. (B-I) Immunostaining of the proliferation marker, Ki-67 (d-f) and apostolic marker, cleaved caspase 3 (g-i) in tumors induced by 8505C, 8505C-WTp53, and 8505C-MTp53, respectively. (B-II) Quantitation of cell positively immune-stained cells with Ki-67 (a) and cleaved caspase 3 (b). (C-I) Comparison of the expression of PUMA (i) and BAX (ii) by q/PCR in tumors induced by 8505C, 8505C-WTp53, and 8505C-MTp53 (n=3). (D-I) Representative IHC images (i) and quantitative analysis of IHC results (ii, n=3) for PUMA (d-f) and BAX (g-i). Significant differences are indicated by asterisks (*P<0.05, **P<0.01, ***P<0.001, and ****P<0.0001). Data represent the mean ± SD (n=3).
Multiple altered signaling pathways in 8505C induced WTp53 unveiled by single cell RNA-sequencing
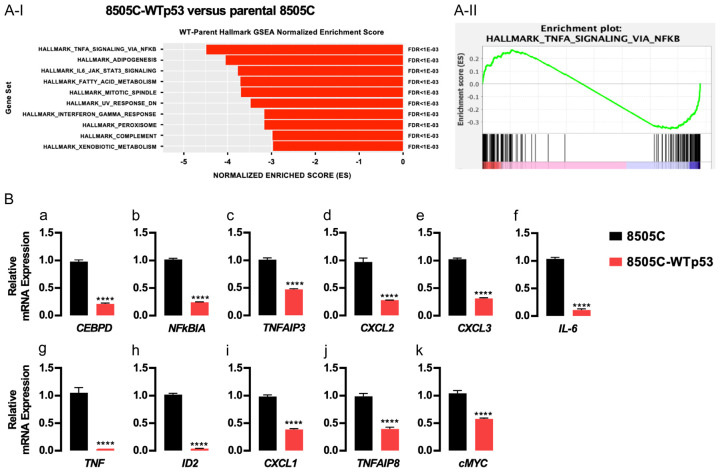
These results state that WTp53 functioned to induce changes in tumor phenotypes as shown above in vitro and in vivo. To probe the global genomic changes induced by the exogenously expressed WTp53 in 8505C cells, we carried out scRNA-seq. Single cells were prepared from tumors induced by either 8505C parental cells, 8505C-WTp53 or 8505C-MTp53 cells. Sufficient numbers of cells were obtained for sequencing (Supplementary Table 3) and consistent read counts were obtained from tumor cells for analysis (Supplementary Figure 1A-I and 1A-II). To verify that the majority of the data were obtained from living normal cells, it was found that <3% of all reads mapped to mitochondria transcripts (Supplementary Figure 1A-III). UMAP profiling to compare the cell distribution of tumors induced by 8505C-WTp53 and 8505C parental cells (Supplementary Figure 1B) or induced by 8505C-MTp53 and 8505C parental cells showed sub-regional distinction (Supplementary Figure 1C). Differential gene expression was then performed between 8505C-WTp53 and 8505C parental cells (Supplementary Figure 1B) as well as between 8505C-MTp53 and 8505C parental cells (Supplementary Figure 1C). Differential gene expression results were then used to produce a gene enrichment score that accounted for significance, fold change, and overall expression of the gene across samples. These scores were then used to rank the genes in the context of overall difference and processed through a pre-ranked Gene Set Enrichment Analysis (GSEA).
When comparing the 8505C-WTp53 and 8505C parental cells, GSEA identified signaling pathways which were suppressed by the exogenously expressed WTp53 in 8505C cells. Figure 4A-I lists the top 10 pathways, indicating the changes in the signaling of necrosis factor-α (TNF-α)/NFκB, adipogenesis, IL-6_Jak_STAT3, fatty acid metabolism, mitotic spindle, UV response, peroxisome, complement, and xenobiotic metabolism. Further analysis showed that 86 genes were significantly involved in WTp53-mediated attenuating TNFα/NFκB signaling in parental 8505C cells, with an overall normalized enrichment score of -4.48 and FDR value of <1E-03 (Figure 4A-II; Table 1 and Supplementary Table 4). We then validated the suppression of the top 11 genes that have reported functions by q/PCR. Indeed, we found that WTp53 acted to suppress the expression of key regulatory genes of inflammation, such as CEBPD, NFκB1A, TNFAIP3, CXCL2, CXCL3, IL-6, TNF, ID2, CXCL1, TNFAIP8, and cMYC (Figure 4B-a-k). While it is known that WTp53 can suppress inflammation in many human tissues, our studies have uncovered the novel genes that could be activated by WTp53 to mitigate thyroid carcinogenesis.
Figure 4.
Expression of WTp53 attenuates oncogenic actions of mutant p53 in 8505C cells. A-I. Gene Set Enrichment Analysis of scRNA-seq identified top pathways contributing to counteracting oncogenic actions of mutant p53. A-II. Gene enrichment plot of TNFα signaling via NFkB analyzed by differential expression genes between WTp53 and 8505C cells. B. Validation of the top 11 genes in mediating signaling TNFα/NFkB by q/PCR by comparing the extent of mRNA expression of the 11 genes as shown. The gene names are as listed. Significant differences are indicated by asterisks (*P<0.05, **P<0.01, ***P<0.001, and ****P<0.0001). Data represent the mean ± SD (n=3).
Table 1.
Differential expression statistics for core genes in TNFα-NFκb signaling between 8505c-WTp53 and 8505c parental cells
| Gene | Raw P-value | Avg Log2FC | pct.WT | pct.Parent | Adj. P-value |
|---|---|---|---|---|---|
| CEBPD | 0 | -2.5915813 | 0.946 | 0.99 | 0 |
| NAMPT | 0 | -1.4692919 | 0.921 | 0.979 | 0 |
| NFKBIA | 0 | -2.0753695 | 0.88 | 0.963 | 0 |
| PTX3 | 0 | -3.0933264 | 0.515 | 0.898 | 0 |
| TNFAIP3 | 0 | -1.5424508 | 0.747 | 0.884 | 0 |
| CXCL2 | 0 | -2.0364498 | 0.585 | 0.838 | 0 |
| CXCL3 | 0 | -1.8039134 | 0.487 | 0.805 | 0 |
| PLPP3 | 0 | -1.3903519 | 0.328 | 0.782 | 0 |
| IL6 | 0 | -2.2673918 | 0.205 | 0.706 | 0 |
| TNF | 0 | -2.5374013 | 0.044 | 0.61 | 0 |
| G0S2 | 9.52E-317 | -2.0542562 | 0.576 | 0.865 | 3.71E-312 |
| ID2 | 0 | -0.9355528 | 0.022 | 0.497 | 0 |
| SOCS3 | 1.22E-286 | -1.6612173 | 0.445 | 0.77 | 4.76E-282 |
| CXCL1 | 5.77E-224 | -1.2845055 | 0.603 | 0.83 | 2.25E-219 |
| RIPK2 | 6.10E-197 | -0.8597438 | 0.696 | 0.892 | 2.38E-192 |
| IRF1 | 1.61E-198 | -1.4097428 | 0.572 | 0.767 | 6.29E-194 |
| IER3 | 4.99E-161 | -1.1702329 | 0.899 | 0.943 | 1.95E-156 |
| SOD2 | 1.98E-156 | -1.2099059 | 0.923 | 0.965 | 7.71E-152 |
| BCL6 | 4.04E-227 | -0.8547331 | 0.25 | 0.623 | 1.58E-222 |
| JUN | 4.40E-138 | -0.8528791 | 0.953 | 0.983 | 1.71E-133 |
| TNC | 3.65E-137 | -0.7571754 | 0.893 | 0.964 | 1.42E-132 |
| ZFP36 | 5.27E-136 | -1.3147865 | 0.903 | 0.931 | 2.05E-131 |
| TNFAIP8 | 2.09E-170 | -0.6956557 | 0.376 | 0.71 | 8.13E-166 |
| MYC | 8.39E-129 | -0.780564 | 0.805 | 0.917 | 3.27E-124 |
| MARCKS | 3.79E-109 | -0.5002077 | 0.947 | 0.981 | 1.48E-104 |
| SMAD3 | 6.60E-126 | -0.5205715 | 0.496 | 0.774 | 2.57E-121 |
| CCNL1 | 8.76E-105 | -0.6105337 | 0.802 | 0.918 | 3.41E-100 |
| NINJ1 | 1.24E-109 | -0.7184984 | 0.611 | 0.795 | 4.83E-105 |
| BCL2A1 | 5.60E-108 | -0.6656719 | 0.441 | 0.71 | 2.18E-103 |
| VEGFA | 5.24E-97 | -0.6462843 | 0.498 | 0.746 | 2.04E-92 |
| IL1B | 1.41E-145 | -0.3528941 | 0.193 | 0.488 | 5.50E-141 |
| NFKB1 | 1.38E-90 | -0.6491981 | 0.575 | 0.768 | 5.37E-86 |
| DUSP1 | 1.65E-70 | -0.6861371 | 0.991 | 0.991 | 6.45E-66 |
| ATP2B1 | 3.46E-77 | -0.6608909 | 0.779 | 0.886 | 1.35E-72 |
| CEBPB | 6.94E-66 | -0.4675741 | 0.881 | 0.951 | 2.70E-61 |
| BCL3 | 5.79E-71 | -0.6245673 | 0.674 | 0.797 | 2.26E-66 |
| CCL20 | 2.49E-141 | -1.4003929 | 0.081 | 0.33 | 9.72E-137 |
| B4GALT5 | 7.86E-64 | -0.312425 | 0.564 | 0.768 | 3.06E-59 |
| IFNGR2 | 1.96E-53 | -0.3199083 | 0.826 | 0.926 | 7.63E-49 |
| NFIL3 | 2.40E-64 | -0.5343927 | 0.551 | 0.731 | 9.36E-60 |
| IL1A | 2.52E-116 | -0.750101 | 0.126 | 0.373 | 9.80E-112 |
| TANK | 5.18E-57 | -0.3006104 | 0.604 | 0.788 | 2.02E-52 |
| ZC3H12A | 3.71E-70 | -0.4791044 | 0.354 | 0.579 | 1.45E-65 |
| B4GALT1 | 4.02E-46 | -0.3784296 | 0.716 | 0.83 | 1.57E-41 |
| KLF9 | 3.65E-100 | -0.395157 | 0.112 | 0.328 | 1.42E-95 |
| BIRC2 | 6.62E-48 | -0.1877034 | 0.516 | 0.696 | 2.58E-43 |
| TNIP1 | 3.08E-37 | -0.4036281 | 0.859 | 0.915 | 1.20E-32 |
| PNRC1 | 6.75E-35 | -0.5489594 | 0.916 | 0.905 | 2.63E-30 |
| FOSL1 | 7.54E-35 | -0.1783856 | 0.928 | 0.837 | 2.94E-30 |
| NFKB2 | 5.61E-43 | -0.3218467 | 0.454 | 0.624 | 2.19E-38 |
| BIRC3 | 6.07E-77 | -0.4626289 | 0.131 | 0.328 | 2.37E-72 |
| FOSL2 | 8.09E-29 | -0.2154694 | 0.754 | 0.86 | 3.15E-24 |
| ZBTB10 | 6.54E-61 | -0.3388142 | 0.172 | 0.339 | 2.55E-56 |
| EHD1 | 1.68E-25 | -0.2147203 | 0.748 | 0.851 | 6.54E-21 |
| RELB | 8.06E-26 | -0.2419431 | 0.65 | 0.769 | 3.14E-21 |
| SERPINE1 | 2.53E-22 | -0.2691889 | 0.866 | 0.785 | 9.85E-18 |
| PDE4B | 3.15E-24 | -0.2330746 | 0.661 | 0.775 | 1.23E-19 |
| PTGER4 | 1.91E-65 | -0.2494643 | 0.077 | 0.229 | 7.42E-61 |
| IL6ST | 5.12E-20 | -0.1931439 | 0.793 | 0.874 | 2.00E-15 |
| LIF | 1.49E-64 | -0.3495302 | 0.071 | 0.218 | 5.82E-60 |
| EFNA1 | 6.95E-27 | -0.1795794 | 0.458 | 0.596 | 2.71E-22 |
| TRIP10 | 1.02E-19 | -0.0830324 | 0.771 | 0.862 | 3.98E-15 |
| DENND5A | 3.23E-22 | -0.1084268 | 0.466 | 0.585 | 1.26E-17 |
| CSF1 | 7.86E-33 | -0.3920333 | 0.243 | 0.354 | 3.06E-28 |
| TGIF1 | 7.59E-16 | -0.2247837 | 0.842 | 0.877 | 2.96E-11 |
| TIPARP | 2.83E-17 | -0.2214694 | 0.667 | 0.764 | 1.10E-12 |
| REL | 8.33E-28 | -0.2483838 | 0.253 | 0.364 | 3.25E-23 |
| NFKBIE | 6.67E-32 | -0.2463917 | 0.225 | 0.306 | 2.60E-27 |
| STAT5A | 7.54E-33 | -0.1834649 | 0.147 | 0.27 | 2.94E-28 |
| TNFAIP2 | 8.51E-68 | -0.2869743 | 0.014 | 0.111 | 3.32E-63 |
| BTG3 | 3.25E-13 | -0.1251713 | 0.644 | 0.731 | 1.27E-08 |
| GEM | 8.59E-11 | -0.170898 | 0.869 | 0.912 | 3.35E-06 |
| CLCF1 | 1.95E-21 | -0.0866145 | 0.216 | 0.313 | 7.61E-17 |
| GCH1 | 3.32E-40 | -0.2598598 | 0.055 | 0.139 | 1.29E-35 |
| NFE2L2 | 3.98E-10 | -0.1231324 | 0.869 | 0.915 | 1.55E-05 |
| ICAM1 | 5.88E-12 | -0.3088566 | 0.499 | 0.572 | 2.29E-07 |
| ABCA1 | 1.13E-38 | -0.1694072 | 0.023 | 0.096 | 4.42E-34 |
| GADD45B | 4.27E-09 | -0.1885803 | 0.756 | 0.693 | 0.00016645 |
| TRAF1 | 2.06E-24 | -0.1549647 | 0.044 | 0.113 | 8.03E-20 |
| PPP1R15A | 2.24E-07 | -0.1055692 | 0.939 | 0.922 | 0.00874136 |
| SQSTM1 | 1.24E-06 | -0.1282598 | 0.999 | 0.998 | 0.04813949 |
| TNFAIP6 | 1.71E-26 | -0.5904362 | 0.016 | 0.061 | 6.68E-22 |
| JUNB | 1.81E-06 | -0.1341125 | 0.955 | 0.947 | 0.07061945 |
| SLC2A6 | 9.68E-11 | -0.1229614 | 0.156 | 0.214 | 3.77E-06 |
| MAP3K8 | 1.28E-09 | -0.0898817 | 0.224 | 0.219 | 4.98E-05 |
| IFIT2 | 3.47E-12 | -0.1058913 | 0.06 | 0.111 | 1.35E-07 |
| TNFRSF9 | 4.91E-18 | -0.0693328 | 0.016 | 0.056 | 1.91E-13 |
Raw p-value represents the p-value calculated using the MAST algorithm in Seurat. Avg Log2FC represents the average log2 fold change calculated between 8505c-WTp53 (positive fold change) and the 8505C parent. Pct.WT indicates the fraction of cells within the 8505C-WTp53 sample that expressed the gene. Pct.Parent indicates the fraction of cells within the 8505C parental sample that expressed the gene in question. Adjusted p-value is calculated from the raw p-value using False Discovery Rate.
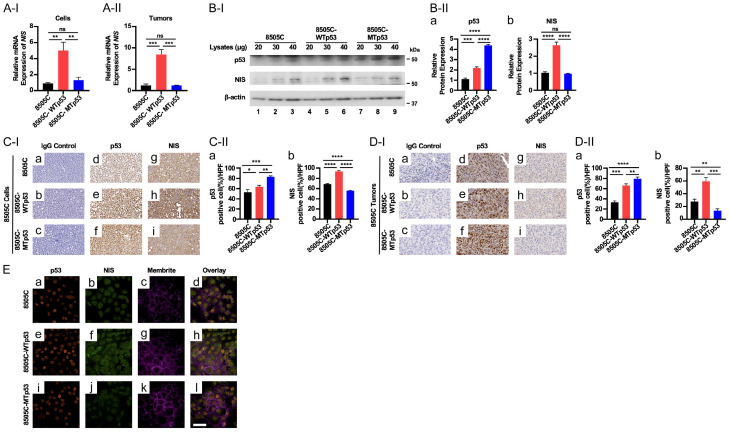
WTp53 induces NIS expression in 8505C cells
In anaplastic thyroid cancer, loss of the differentiation marker, sodium iodide symporter (NIS; encoded by the SLC5A5 gene), has made radio-iodine treatment ineffective in the treatment of ATC patients. Thus, we evaluated whether the WTp53 introduced into 8505C cells could induce the expression of the SLC5A5 gene. Excitingly, we found that WTp53 reactivated the expression of the SLC5A5 at the mRNA level in 8505C-WTp53 cells (Figure 5A-I) and tumors induced by 8505C-WTp53 cells (Figure 5A-II). Western blot analysis showed that NIS protein levels were concordantly elevated when the WTp53 was increased (Figure 5B-I, 5B-II-a and 5B-II-b). These findings were further confirmed by IHC analysis in which NIS protein levels were higher in WTp53 as compared with 8505C parental and MTp53 cells (Figure 5C-I and 5C-II). Importantly, IHC analysis indicated that a higher NIS protein level was associated with an elevated WTp53 protein amount in 8505C-WTp53-induced tumors (Figure 5C-I-e, 5C-I-g, 5C-II-a and 5C-II-b). We further demonstrated that the NIS protein levels were elevated in tumors induced by 8505C-WTp53 cells (Figure 5D-I, compare Figure 5D-I-e with Figure 5D-I-d and quantitation in Figure 5D-II). To ascertain the cellular localization of the elevated NIS, we carried out confocal imaging. We found that p53 was localized in the nuclei in 8505C, 8505C-WTp53, and 8505C-MTp53 cells (Figure 5E-a, 5E-e and 5E-i, respectively, red images), while NIS proteins were detected on the membrane and cytoplasm (Figure 5E-b, 5E-f and 5E-j, green images). Previously, NIS has been reported to be localized in the cytoplasm [14,15]. We used MemBrite 640 to mark the cell membrane (Figure 5E-c, 5E-g and 5E-k, magenta images). The overlay merged images showed that some NIS proteins was localized to the membrane (Figure 5E-d, 5E-h and 5E-l). It is of importance to point out that consistent with western blot and IHC analyses, elevated NIS protein levels were detected in 8505C-WTp53 cells (compare Figure 5E-f with Figure 5E-b and 5E-j). Taken together, these data indicated that WTp53 induced the re-expression of NIS.
Figure 5.
WTp53 increases the expression of the SLC5A5 gene at the mRNA and protein levels. (A) Comparison of the expression of the SLC5A5 gene at the mRNA levels by q/PCR analysis in 8505C, 8505C-WTp53, and 8505C-MTp53 cells (A-I) and tumors induced by 8505C, 8505C-WTp53, 8505C-MTp53 cells (A-II). (B) Representative western blots showing increased p53 (top row) and NIS proteins (middle row) in 8505C, 8505C-WTp53, and 8505C-MTp53 cells. B-actin was used as loading controls (bottom row) (B-I). Quantitation of the band intensities in (B-II) showing the protein levels of p53 (a) and NIS protein (b). (C and D) Analysis of protein levels of p53 and NIS by IHC analysis [(C-I and D-I), representative images of IHC from cultured cell pellets and tumors, respectively; (C-II and D-II), quantitation of the cells immuno-stained for p53 (a) and NIS proteins (b)]. Significant differences are indicated by asterisks (*P<0.05, **P<0.01, ***P<0.001, and ****P<0.0001). Data represent the mean ± SD (n=3). (E) Confocal images of 8505C cells (top row), 8505C cells expressing WTp53 (middle row) and 8505C cells expressing mutant p53 (bottom row) immunolabeled against p53 (red; a, e and i) or NIS (green; b, f and j) and counterstained with MemBrite 640 plasma membrane stain (magenta). The merged of p53, NIS and MemBrite 640 (d, e and l). Scale bar =50 µm.
Discussion
High throughput sequencing of ATC tumors has shown extensive cellular disruption including previously unidentified genetic changes such as SWI/SNF, and histone methyltransferases [7]. In another study, sequencing of additional ATC tumors uncovered other genetic changes such as copy losses and mutated CDKN2A and CDKN2B, and amplification of several receptor tyrosine kinases genes and immune evasion genes [16]. However, a high frequency of TP53 mutations has been the major occurrence in late stage of ATC development. At present, how the mutations of TP53 occur at the late stage of ATC is not clear, but it is reasonable to postulate that TP53 mutations could contribute to the lethal outcome at the advanced stage of ATC patients. The question arose whether the oncogenic actions of mutant p53 could be reversed by elevated WTp53, when exogenously introduced into cells. In the present study, we tested this possibility. We found that indeed, stably expressing WTp53 in 8505C cells led to decreased proliferation and migration of tumor cells, induction of apoptosis in vitro, and suppression of xenograft tumor growth in vivo.
To gain a deeper understanding of how WTp53 functioned to reverse cancer phenotypes, we carried out scRNA-seq. Analysis of data unveiled that the reversal of the cancer phenotypes, was not limited to the inhibition of tumor cell growth. It also indicated the participation of multiple pathways to counter act the tumorigenic effects of mutant p53 in 8505C cells. On the top of the identified pathways was the TNFα-NFκb signaling, involving the alterations of 87 genes (see Table 1). We performed q/PCR analysis for the top 11 genes involved in TNFα-NFκb signaling individually to validate altered gene expression by scRNA-seq analysis (Figure 4). These genes were reported to have functions related to inflammatory and immune responses in thyroid cancer [17,18]. Recent studies have shown that WTp53 emerges as one major regular of cancer-immune system interactions [19]. Loss of canonical functions of WTp53 (i.e., mutations of TP53) favors an inflammatory microenvironment, leading to limited immune surveillances and responses to benefit cancer cells. The present studies showed that the exogenously expressed WTp53 in 8505C cells resulted in the dampening the inflammatory responses as evidenced by the suppressed expression of critical genes in the TNFα-NFκb signaling. In addition to causing immune evasion, mutations of p53 also broadly affect metabolic reprogramming, lipogenesis, and other cellular functions [18,20]. Indeed, analysis of scRNA-seq data also identified other pathways impacted by the exogenously expressed WTp53 including adipogenesis, Il6_Jak_STAT3 signaling, fatty acid metabolism, mitotic spindle, UV responses, interferon gamma response, peroxisome signaling, and xenobiotic_metabolism. Other significant pathways that could also contribute to mitigate the mutated p53’s cancer promoting effects are listed in Supplementary Table 4. These results demonstrated that exogenously expressed WTp53 could counteract the oncogenic actions of MTp53 via multiple pathways.
To validate the potential of p53 protein expression in inducing the re-differentiation of ATC cells, we conducted experiments demonstrating the re-expression of NIS in p53WT-expressing cells. NIS plays a crucial role in iodine absorption for the biosynthesis of thyroid hormones. We found that WTp53 reactivated the expression of NIS at the mRNA and protein levels. The western blot analysis revealed a notable 1.7-fold increase in NIS protein levels in 8505c-p53WT cells compared to control cells (Figure 5). Confocal microscopy further confirmed that the NIS protein levels were higher in 8505C-WTp53 cells. Extending our observations to tumors induced by 8505c, 8505c-p53WT, and 8505c-p53MT cells, we found an elevated NIS protein presence in tumors induced by 8505c-p53WT cells (Figure 5D-I and 5D-II). These findings collectively indicate that WTp53-induced re-differentiation in ATC cells leads to the re-expression of the SLC5A5 gene, suggesting a potential avenue for enhancing treatment strategies in ATC patients. This re-differentiation process holds promise for restoring critical functional pathways in thyroid cells and may bear significant implications for the therapeutic management of ATC.
As shown above, our extensive molecular and genetic characterization of 8505C-WTp53 exploring the functional consequences of exogenously expressed WTp53 has expanded our understanding of oncogenic actions of mutant p53 in ATC cells. Importantly, the findings that exogenously expressed WTp53 could reverse some of the oncogenic phenotypes of mutant p53 has opened a possibility in considering gene editing as a novel treatment options. The advancement in using CRISPR/Cas9-based genomic editing could be considered for editing the mutant TP53 allele or knocking in extra WT TP53 allele into the genome. Such approach could correct the potentially lethal outcome of mutant p53, which could allow time for using other therapies such as more effective radio-iodide treatment as we have shown that exogenously expressed WTp53 could reactivate the expression of the NIS gene (Figure 5). In summary, our demonstration that the exogenously expressed WTp53 could partially rescue the cancer phenotype incurring by mutant p53 has opened new opportunity for treatment of ATC patients.
Acknowledgements
The bioinformatic analysis of scRNA-seq has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, Department of Health and Human Services, under Contract No. 75N91019D00024. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. We thank Drs. Zachary Rae, Michael Kelly, and Kimia Dadkhah for carrying out single-cell RNA sequencing at the Single Cell Analysis Facility, Cancer Research Technology Program, Frederick National Laboratory (Leidos Biomed), NCI. We also thank Ms Swati Priya for manuscript editing assistance.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Boucai L, Zafereo M, Cabanillas ME. A review of thyroid cancer-reply. JAMA. 2024;331:1863–1864. doi: 10.1001/jama.2024.6001. [DOI] [PubMed] [Google Scholar]
- 2.Subbiah V, Kreitman RJ, Wainberg ZA, Cho JY, Schellens JHM, Soria JC, Wen PY, Zielinski C, Cabanillas ME, Urbanowitz G, Mookerjee B, Wang D, Rangwala F, Keam B. Dabrafenib and trametinib treatment in patients with locally advanced or metastatic BRAF V600-mutant anaplastic thyroid cancer. J. Clin. Oncol. 2018;36:7–13. doi: 10.1200/JCO.2017.73.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cleere EF, Prunty S, O’Neill JP. Anaplastic thyroid cancer: improved understanding of what remains a deadly disease. Surgeon. 2024;22:e48–e53. doi: 10.1016/j.surge.2023.10.002. [DOI] [PubMed] [Google Scholar]
- 4.Abdullah MI, Junit SM, Ng KL, Jayapalan JJ, Karikalan B, Hashim OH. Papillary thyroid cancer: genetic alterations and molecular biomarker investigations. Int J Med Sci. 2019;16:450–460. doi: 10.7150/ijms.29935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prete A, Borges de Souza P, Censi S, Muzza M, Nucci N, Sponziello M. Update on fundamental mechanisms of thyroid cancer. Front Endocrinol (Lausanne) 2020;11:102. doi: 10.3389/fendo.2020.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Armstrong MJ, Yang H, Yip L, Ohori NP, McCoy KL, Stang MT, Hodak SP, Nikiforova MN, Carty SE, Nikiforov YE. PAX8/PPARgamma rearrangement in thyroid nodules predicts follicular-pattern carcinomas, in particular the encapsulated follicular variant of papillary carcinoma. Thyroid. 2014;24:1369–1374. doi: 10.1089/thy.2014.0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Landa I, Ibrahimpasic T, Boucai L, Sinha R, Knauf JA, Shah RH, Dogan S, Ricarte-Filho JC, Krishnamoorthy GP, Xu B, Schultz N, Berger MF, Sander C, Taylor BS, Ghossein R, Ganly I, Fagin JA. Genomic and transcriptomic hallmarks of poorly differentiated and anaplastic thyroid cancers. J Clin Invest. 2016;126:1052–1066. doi: 10.1172/JCI85271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu B, Chen Y, St Clair DK. ROS and p53: a versatile partnership. Free Radic Biol Med. 2008;44:1529–1535. doi: 10.1016/j.freeradbiomed.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hikisz P, Kilianska ZM. PUMA, a critical mediator of cell death--one decade on from its discovery. Cell Mol Biol Lett. 2012;17:646–669. doi: 10.2478/s11658-012-0032-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu J, Zhang L. PUMA, a potent killer with or without p53. Oncogene. 2008;27(Suppl 1):S71–83. doi: 10.1038/onc.2009.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li M. The role of P53 up-regulated modulator of apoptosis (PUMA) in ovarian development, cardiovascular and neurodegenerative diseases. Apoptosis. 2021;26:235–247. doi: 10.1007/s10495-021-01667-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green DR. The coming decade of cell death research: five riddles. Cell. 2019;177:1094–1107. doi: 10.1016/j.cell.2019.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat Rev Cancer. 2009;9:749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hingorani M, Spitzweg C, Vassaux G, Newbold K, Melcher A, Pandha H, Vile R, Harrington K. The biology of the sodium iodide symporter and its potential for targeted gene delivery. Curr Cancer Drug Targets. 2010;10:242–267. doi: 10.2174/156800910791054194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faria M, Domingues R, Bugalho MJ, Silva AL, Matos P. Analysis of NIS plasma membrane interactors discloses key regulation by a SRC/RAC1/PAK1/PIP5K/EZRIN pathway with potential implications for radioiodine re-sensitization therapy in thyroid cancer. Cancers (Basel) 2021;13:5460. doi: 10.3390/cancers13215460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pozdeyev N, Gay LM, Sokol ES, Hartmaier R, Deaver KE, Davis S, French JD, Borre PV, LaBarbera DV, Tan AC, Schweppe RE, Fishbein L, Ross JS, Haugen BR, Bowles DW. Genetic analysis of 779 advanced differentiated and anaplastic thyroid cancers. Clin Cancer Res. 2018;24:3059–3068. doi: 10.1158/1078-0432.CCR-18-0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guarino V, Castellone MD, Avilla E, Melillo RM. Thyroid cancer and inflammation. Mol Cell Endocrinol. 2010;321:94–102. doi: 10.1016/j.mce.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 18.Alvarado-Ortiz E, de la Cruz-Lopez KG, Becerril-Rico J, Sarabia-Sanchez MA, Ortiz-Sanchez E, Garcia-Carranca A. Mutant p53 gain-of-function: role in cancer development, progression, and therapeutic approaches. Front Cell Dev Biol. 2020;8:607670. doi: 10.3389/fcell.2020.607670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blagih J, Buck MD, Vousden KH. p53, cancer and the immune response. J Cell Sci. 2020;133:jcs237453. doi: 10.1242/jcs.237453. [DOI] [PubMed] [Google Scholar]
- 20.Zhu G, Pan C, Bei JX, Li B, Liang C, Xu Y, Fu X. Mutant p53 in cancer progression and targeted therapies. Front Oncol. 2020;10:595187. doi: 10.3389/fonc.2020.595187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fozzatti L, Alamino VA, Park S, Giusiano L, Volpini X, Zhao L, Stempin CC, Donadio AC, Cheng SY, Pellizas CG. Interplay of fibroblasts with anaplastic tumor cells promotes follicular thyroid cancer progression. Sci Rep. 2019;9:8028. doi: 10.1038/s41598-019-44361-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee WK, Kim WG, Fozzatti L, Park S, Zhao L, Willingham MC, Lonard D, O’Malley BW, Cheng SY. Steroid receptor coactivator-3 as a target for anaplastic thyroid cancer. Endocr Relat Cancer. 2020;27:209–220. doi: 10.1530/ERC-19-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimura T, Doolittle WKL, Kruhlak M, Zhao L, Hwang E, Zhu X, Tang B, Wolcott KM, Cheng SY. Inhibition of MEK signaling attenuates cancer stem cell activity in anaplastic thyroid cancer. Thyroid. 2024;34:484–495. doi: 10.1089/thy.2023.0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.