Abstract
During studies examining the rate of human immunodeficiency virus type 1 (HIV-1) mutation in a single cycle of replication, the 5′ long terminal repeat of one progeny provirus was found to contain an insertion of 147 bp including an entire tRNA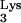 sequence as well as an additional 66 bp insertion of nonviral origin. Database searches revealed that 65 of 66 bp aligned with the human CpG island sequence found on chromosomes 6, 14, and 17. Therefore it seems probable that it is of human cellular sequence origin and was transduced by HIV-1. This is the first demonstration that HIV-1 can capture a cellular sequence. The site of integration of the parental provirus was mapped to chromosome 1p32.1. Sequence with homology to the transduced CpG island was not found on chromosome 1, suggesting that the transduced cellular sequence was not linked to the site of viral integration.
sequence as well as an additional 66 bp insertion of nonviral origin. Database searches revealed that 65 of 66 bp aligned with the human CpG island sequence found on chromosomes 6, 14, and 17. Therefore it seems probable that it is of human cellular sequence origin and was transduced by HIV-1. This is the first demonstration that HIV-1 can capture a cellular sequence. The site of integration of the parental provirus was mapped to chromosome 1p32.1. Sequence with homology to the transduced CpG island was not found on chromosome 1, suggesting that the transduced cellular sequence was not linked to the site of viral integration.
Retroviruses were originally studied because of the ability of some strains to efficiently induce tumors in animals, and as such they have served as important tools for studying oncogenesis (27). It was later found that the acutely transforming retroviruses harbored oncogenes (v-onc genes), that were acquired via transduction of cellular counterparts termed proto-oncogenes (c-onc genes). The most often described mechanism of transduction by retroviruses proposes that the parental provirus integrates next to the sequence that is transduced (3, 24). Readthrough transcription then occurs, followed by either direct packaging of the chimeric virus/cellular mRNA into a virion or aberrant splicing yielding a chimeric mRNA which is then encapsidated into a virion (12, 19). In both cases it is then thought that recombination occurs during reverse transcription, resulting in incorporation of the cellular sequence into the progeny provirus, which is typically, although not always, defective owing to the loss of some viral sequence (7). It has also been suggested that transduction of cellular sequence can occur after cellular RNA is randomly copackaged into a virion followed by recombination again during reverse transcription (8, 22). However, this mechanism does not represent the prevalent theory concerning transduction.
Although transduction of cellular oncogenes by oncoretroviruses is well established, it has not been reported that lentiviruses such as human immunodeficiency virus type 1 (HIV-1) are also capable of transducing cellular sequences. In this study, we report the identification of an HIV-1 vector provirus which harbors sequences of cellular origin. The transduced sequence includes a full-length tRNA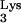 and cellular sequence which is 66 bp long and is an identical match in 65 of 66 bp to sequences on human chromosomes 6, 14, and 17. This clone was obtained after restricting a HIV-1 vector virus to a single cycle of replication at a low multiplicity of infection. Thus, it would appear that the nonviral sequence was captured from human genomic DNA by a lentivirus, indicating that this class of retrovirus can also acquire cellular sequences. Furthermore, it was found that the transduced cellular sequence was not linked to the integration site of the parental provirus.
and cellular sequence which is 66 bp long and is an identical match in 65 of 66 bp to sequences on human chromosomes 6, 14, and 17. This clone was obtained after restricting a HIV-1 vector virus to a single cycle of replication at a low multiplicity of infection. Thus, it would appear that the nonviral sequence was captured from human genomic DNA by a lentivirus, indicating that this class of retrovirus can also acquire cellular sequences. Furthermore, it was found that the transduced cellular sequence was not linked to the integration site of the parental provirus.
Cellular sequence embedded within an HIV-1 vector provirus.
We have established a single-cycle system to study HIV-1 mutational events. The system is based on the use of a defective viral vector in conjunction with a packaging cell line. This approach allows one to confine retroviral replication to a single cycle and identify mutations in authentic viral sequences, so that after one round of replication the newly formed provirus essentially represents a “fossil record,” reflecting events the virus experienced while it underwent replication (9, 15, 18). For this project, an HIV-1 vector based on the HIV-1HXB2 strain (20), HIV-gpt, was employed (14). It has a large deletion of env that is replaced by the Escherichia coli gpt gene under transcriptional control of the simian virus 40 (SV40) early-gene promoter. HIV-gpt was used in conjunction with the HIV-1 env-inducible cell line, #69TIRevEnv (30), to produce vector virus. This HIV-gpt vector virus was used to infect HeLaT4 cells, and its replication was restricted to one cycle as previously described (9, 15).
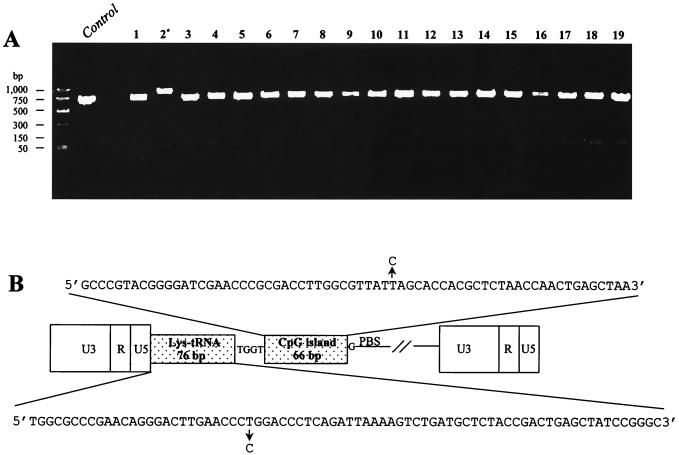
Proviruses that had undergone a single cycle of replication were analyzed for long terminal repeat (LTR) mutations to assess mechanisms of HIV-1 mutagenesis at different stages of reverse transcription. To analyze the proviral LTRs, genomic DNA was isolated (2) from 215 independent cell clones and then the corresponding LTRs were subjected to PCR amplification. The 5′ LTRs of proviral DNA were amplified by using primers B8+ (5′-GCTAATTCACTCCCAAAGAAGC-3′), located close to the 5′ end of U3, and B732 (5′-CCCTCGCCTCTTGCCGTGC-3′), located at the 3′ end of the primer-binding site (PBS). The amplification was anticipated to yield a fragment of 713 bp. The reaction conditions were as follows: 1 cycle at 94°C for 1 min, 30 cycles at 94°C for 30 s, 55°C for 40 s, and 72°C for 50 s, and a final cycle at 72°C for 5 min. The PCR products were then subjected to electrophoresis in a 1.2% agarose gel and visualized by staining with ethidium bromide. The PCR products which displayed a shift in mobility were expected to have mutations. It was noted that the PCR product from the 5′ LTR of one of the proviruses was larger than anticipated (Fig. 1A). Further analysis of the other LTRs are described elsewhere (P. K. O'Neil, G. Sun, J. P. Dougherty, and B. D. Preston, unpublished data).
FIG. 1.
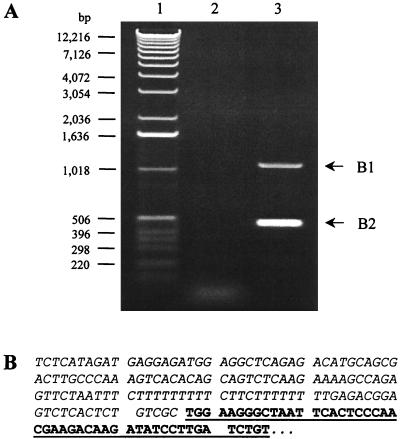
(A) Examples of mutation screening by PCR of the 5′ LTR of progeny proviruses. The band in lane 2 is shifted up (∗) compared to the control and other progeny proviruses (lanes 1 and 3 to 19), indicating an insertion. (B) The insertion causing the change in electrophoretic mobility of the sample from lane 2 in panel A is depicted. The whole insertion is composed of an entire tRNA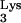 with its sequence denoted below the figure, including one T-to-C substitution, followed by 4 bp (TGGT) of unknown origin, 66 bp of human CpG sequence, with its sequence depicted above the diagram, including one T-to-C substitution, and another G of unknown origin. Note that the figure is not drawn to scale, as indicated by //.
with its sequence denoted below the figure, including one T-to-C substitution, followed by 4 bp (TGGT) of unknown origin, 66 bp of human CpG sequence, with its sequence depicted above the diagram, including one T-to-C substitution, and another G of unknown origin. Note that the figure is not drawn to scale, as indicated by //.
Automated DNA sequencing demonstrated that 147 bp of nonviral sequence was inserted between the 5′ U5 and the PBS of the progeny vector provirus, which yielded the larger-than-anticipated PCR product (Fig. 1B). Through database searches and sequence analyses, it was found that the first 76 bp of the inserted sequence was a complete reverse read (antisense orientation) of the tRNA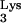 primer. The transduced tRNA
primer. The transduced tRNA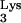 sequence differs in only 1 nucleotide from the reported wild-type sequence, containing a T-to-C change at position 50 (Fig. 1B). Another fragment in the inserted sequences, 66 bp in length, was found to have 95 to 98% identity to CpG island sequences located on human chromosomes 6p22.1 (accession no. AL121934 and AL021808), 6p21.32 (accession no. AL121936), 14q32.33 (accession no. AL352978), and 17 (accession no. AC015734). Database searches were performed by using standard nucleotide-nucleotide BLAST [blastn] (version 2.0; National Center for Biotechnology Information, National Institutes of Health, Bethesda, Md.; http://www.ncbi.nlm.nih.gov/BLAST) against databases nr, htgs, and est_human (31). The chromosomal locations of interesting sequences were found by submitting a query against the whole genome through the National Center for Biotechnology Information's Human Genome Map Viewer (for details, go to http://www.ncbi.nlm.nih.gov/genome/guide/human).
sequence differs in only 1 nucleotide from the reported wild-type sequence, containing a T-to-C change at position 50 (Fig. 1B). Another fragment in the inserted sequences, 66 bp in length, was found to have 95 to 98% identity to CpG island sequences located on human chromosomes 6p22.1 (accession no. AL121934 and AL021808), 6p21.32 (accession no. AL121936), 14q32.33 (accession no. AL352978), and 17 (accession no. AC015734). Database searches were performed by using standard nucleotide-nucleotide BLAST [blastn] (version 2.0; National Center for Biotechnology Information, National Institutes of Health, Bethesda, Md.; http://www.ncbi.nlm.nih.gov/BLAST) against databases nr, htgs, and est_human (31). The chromosomal locations of interesting sequences were found by submitting a query against the whole genome through the National Center for Biotechnology Information's Human Genome Map Viewer (for details, go to http://www.ncbi.nlm.nih.gov/genome/guide/human).
CpG islands contain a high percentage of CG base pairs and are most often found around promoters (10). Almost all housekeeping genes and many tissue-specific genes have a CpG island at their 5′ end (5, 11), and there are instances when they are transcribed (21, 29). Our database search against the est_human database also revealed that the transduced sequence was expressed in human mRNA (accession no. AI470805). This expressed mRNA is a 100% match with a sequence on chromosome 17p13.2. There was also a 4-bp stretch of unknown origin, TGGT, located between the tRNA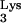 and CpG island sequences (Fig. 1B). Another base pair of unknown origin was also inserted between the CpG island sequence and the PBS (Fig. 1B).
and CpG island sequences (Fig. 1B). Another base pair of unknown origin was also inserted between the CpG island sequence and the PBS (Fig. 1B).
Although the captured sequence is 66 bp in length, which is relatively short, its origin is almost certainly human genomic DNA. The probability of randomly obtaining a particular stretch of 66 nucleotides is 1 in 466 or 1 in 5.4 × 1039 nucleotides. Thus, given the size of the human genome (3 × 109 nucleotides) and the match obtained with the human cellular sequence, the chance that the captured sequence is not of cellular origin is approximately 0.5 × 10−30 (3 × 109 /5.4 × 1039). We conclude that this is human sequence that was transduced into the HIV-1 genome during a single cycle of replication.
Mapping of the HIV-1 integration site by inverse PCR.
Knowing the transduced cellular sequence and having the parental producer cell clone allows one to glean additional information about the mechanism of this particular transduction event. More specifically, it provides the opportunity to examine whether the parental provirus is integrated next to the transduced cellular sequence, as has been proposed in most models of retroviral capture of cellular sequence. The availability of human genome databases can assist with this approach by potentially allowing one to identify the site of integration of the parental provirus within the cellular genome.
The flanking region of the HIV-gpt provirus was identified by using two rounds of inverse PCR (25). A 1-μg portion of producer cell genomic DNA was digested with 20 U of HindIII (New England Biolabs) in a final volume of 20 μl for 6 h. HindIII was then inactivated by heating at 65°C for 20 min. The digested genomic DNA was diluted to 10 ng/μl in a final volume of 100 μl and circularized by adding 10 U of T4 DNA ligase (Boehringer Mannheim Inc.) followed by incubation at 16°C overnight. The self-ligation product was subjected to two rounds of inverse PCR. The first round of inverse PCR was performed with 40 ng of self-ligation product, using primers p1 (5′-GAAGTAGCCTTGTGTGTG-3′) and p1-1 (5′-AGCCGCCTAGCATTTCATCAC-3′). Amplification conditions for the first round were as follows: 1 cycle at 95°C for 2 min, 30 cycles at 94°C for 45 s, at 51°C for 1 min, and 72°C for 1 min 30 s, and a final cycle of extension at 72°C for 10 min. To reduce the amount of nonspecific PCR product, a 2-μl aliquot from the first round of inverse PCR was amplified with nested primers p2 (5′-GTCTTCGTTGGGAGTGAATTAG-3′) and p2-1 (5′-AGCCCTCAGATCCTGCATATAAG-3′). The second round of inverse PCR was carried out under the same conditions as the first, except that the annealing temperature was increased from 51 to 58°C. All of these primer pairs are complementary to the opposite strands and are located in both HIV-1 LTRs, facing away from each other.
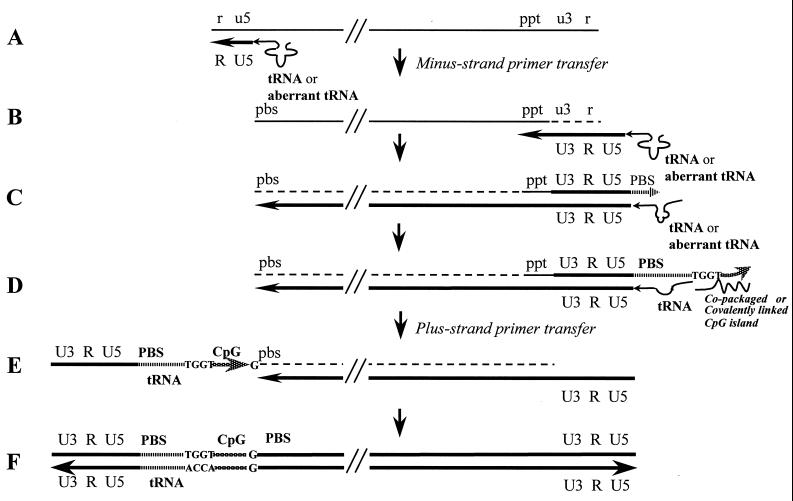
The PCR products were then subjected to gel electrophoresis in a 1% agarose gel. At least two bands were expected from this amplification since the primers would be able to anneal to two different sequences: the 5′ LTR linked to cellular flanking sequence, yielding a PCR product of unknown size, and the 3′ LTR linked to internal viral vector sequence, anticipated to yield a 1.1-kbp band. Two bands were observed (Fig. 2A). One band, B1, was 1.1 kbp, and the second prominent band, B2, was approximately 450 bp (Fig. 2A). The DNA from the two bands was isolated and cloned into the pGEM-T Easy vector (Promega) for subsequent DNA sequencing. Automated DNA sequencing confirmed that the 1.1-kbp B1 DNA was self-ligated HIV-1 virus sequence as expected. The B2 DNA was composed of HIV-1 LTR sequence linked to 315 bp of nonviral DNA sequence (Fig. 2B), which yielded a 100% match with a sequence on human chromosome 1p32.1 (accession no. AC025928).
FIG. 2.
Products and DNA sequence obtained from inverse PCR of the producer cell provirus. (A) Lanes: 1, DNA markers; 2, template-negative PCR control; 3, second-round inverse PCR products with HindIII-digested and self-ligated genomic DNA as templates. Band B1 was amplified from the HIV-1 self-ligated product. Band B2 was anticipated to contain the HIV-1 integration site and flanking producer cell genome sequence because of its strong intensity. Both bands were cloned and sequenced. (B) Flanking human genomic sequence at the 5′ end of the HIV-1 integration site. Bold underlined letters represent the first 48 bp of the HIV-1 provirus 5′ sequence. Italic letters represent the human chromosome 1 sequence adjacent to the integration site.
The most widely accepted retrovirus transduction model proposes that the captured cellular sequence is adjacent to the retrovirus integration site (23). However, sequence matches with the transduced CpG island were found on chromosomes 6, 14, and 17, but none was found on chromosome 1. It is of course possible that a sequence match might be found in a yet unsequenced gap on this chromosome, but the nearest gap is located approximately 200 kbp away from the 3′ end of the integrated provirus based on the contig map and associated information (accession no. NT_004873). This indicates that the parental provirus integrated into chromosome 1 is not linked to the transduced sequences found in the progeny provirus. Therefore, in this particular case it appears that the mechanism of transduction differs somewhat from that most typically proposed.
Putative transduction mechanism.
How did the CpG island sequence become available as a template for reverse transcription? There are two possible scenarios. The first is that RNA containing a CpG island was copackaged into the budding virus. It was previously reported that cellular RNAs can be randomly encapsidated into virions, although not as efficiently as full-length viral RNA, which contains packaging signals (1, 6). The second possibility is that the CpG island sequence was covalently linked to the tRNA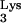 primer and was encapsidated because of this linkage. tRNA
primer and was encapsidated because of this linkage. tRNA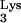 sequences are located in a number of positions throughout the genome, and two of them are on the same chromosomes as the CpG island sequences. The closest linkage occurs on chromosome 6p22.1, where a tRNA
sequences are located in a number of positions throughout the genome, and two of them are on the same chromosomes as the CpG island sequences. The closest linkage occurs on chromosome 6p22.1, where a tRNA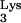 (accession no. AL021918) is about 22 kb from a CpG island (accession no. AL031229). Reverse transcriptase PCR (RT-PCR) using primer pairs with specificity for the CpG island and tRNA
(accession no. AL021918) is about 22 kb from a CpG island (accession no. AL031229). Reverse transcriptase PCR (RT-PCR) using primer pairs with specificity for the CpG island and tRNA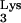 sequences failed to detect such a chimeric RNA in the parental cell line (data not shown). However, such a chimeric RNA might be rare and below the level of detection.
sequences failed to detect such a chimeric RNA in the parental cell line (data not shown). However, such a chimeric RNA might be rare and below the level of detection.
In light of these findings, Fig. 3 outlines what would seem to be the most likely mechanism for production of this progeny provirus. Given the position of the insertion within the virus genome, it seems that it was incorporated during the early phase of plus-strand synthesis (Fig. 3). Instead of copying only the PBS sequence from the tRNA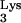 primer, RT was able to read through the entire tRNA
primer, RT was able to read through the entire tRNA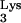 (Fig. 3D). It is possible that the base change observed in the transduced tRNA
(Fig. 3D). It is possible that the base change observed in the transduced tRNA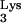 sequence may affect readthrough past the viral PBS sequence (4, 13). After this, a 4-nucleotide sequence was added via an unknown mechanism. Next, either continued readthrough into the CpG island occurred or an aberrant transfer to the CpG island-containing RNA transpired (Fig. 3D). After 66 bp of CpG island sequence was copied, a single G was inserted (Fig. 3E). This extra base might have been incorporated with the CpG island-containing RNA during transcription and RNA processing. Another possibility is that it arose from a nontemplated addition on either the plus-strand or minus-strand growing points (16, 28). In either event, strand transfer back to the PBS of the newly synthesized minus-strand DNA then occurred, resulting in duplication of the PBS (Fig. 3E). Finally, after the plus-strand primer transfer, reverse transcription was completed via the usual mechanism (Fig. 3F).
sequence may affect readthrough past the viral PBS sequence (4, 13). After this, a 4-nucleotide sequence was added via an unknown mechanism. Next, either continued readthrough into the CpG island occurred or an aberrant transfer to the CpG island-containing RNA transpired (Fig. 3D). After 66 bp of CpG island sequence was copied, a single G was inserted (Fig. 3E). This extra base might have been incorporated with the CpG island-containing RNA during transcription and RNA processing. Another possibility is that it arose from a nontemplated addition on either the plus-strand or minus-strand growing points (16, 28). In either event, strand transfer back to the PBS of the newly synthesized minus-strand DNA then occurred, resulting in duplication of the PBS (Fig. 3E). Finally, after the plus-strand primer transfer, reverse transcription was completed via the usual mechanism (Fig. 3F).
FIG. 3.
Proposed model for generating the mutant progeny. Thin lines represent viral RNA. Thick lines represent viral DNA. Dashed lines represent RNase H-digested RNA. Striped lines denote DNA copied from tRNA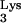 . Stippled lines indicate the captured cellular CpG island sequence. (A) The minus-strand synthesis was primed by either normal tRNA
. Stippled lines indicate the captured cellular CpG island sequence. (A) The minus-strand synthesis was primed by either normal tRNA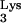 or an aberrant tRNA
or an aberrant tRNA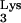 which has the CpG island sequence linked to it. (B) Minus-strand synthesis continued normally. (C) The plus-strand strong-stop DNA product did not terminate at the end of PBS as is typical. If there was a conventional tRNA
which has the CpG island sequence linked to it. (B) Minus-strand synthesis continued normally. (C) The plus-strand strong-stop DNA product did not terminate at the end of PBS as is typical. If there was a conventional tRNA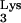 used to prime reverse transcription, it continued to read through this entire tRNA
used to prime reverse transcription, it continued to read through this entire tRNA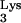 . (D) An extra 4-bp stretch (TGGT) of unknown origin was added by RT. After that, the plus-strand strong-stop DNA growing point either continued to utilize the chimeric RNA as template or annealed to a copackaged human RNA (CpG) and copied 66 bases. (E) Then an additional base was added (see the text), and this was followed by strand transfer to the minus-strand PBS. (F) Reverse transcription was subsequently completed in typical fashion. The figure is not drawn to scale, as indicated by //. ppt, polypurine tract.
. (D) An extra 4-bp stretch (TGGT) of unknown origin was added by RT. After that, the plus-strand strong-stop DNA growing point either continued to utilize the chimeric RNA as template or annealed to a copackaged human RNA (CpG) and copied 66 bases. (E) Then an additional base was added (see the text), and this was followed by strand transfer to the minus-strand PBS. (F) Reverse transcription was subsequently completed in typical fashion. The figure is not drawn to scale, as indicated by //. ppt, polypurine tract.
This scenerio specifies that at least two and possibly five aberrant events occurred over a short stretch of sequence. It has previously been observed both for Moloney murine leukemia virus and for spleen necrosis virus that a number of mutations can occur over a short stretch of sequence, so the current findings are consistent with such occurrences in other types of retroviruses (17, 26). It should also be noted that in approximately 40% of acutely transforming retroviruses, relatively short insertions of unknown origin are found at the junction between the viral and oncogene sequences, which is similar to what is observed in this case with the 4-bp insertion of unknown origin between the tRNA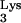 and CpG island sequences (32).
and CpG island sequences (32).
Although it seems that the mechanism for transduction in this particular instance differs from what has been proposed for oncoretroviruses at the initial stage of acquiring the template, the additional steps resulting in capture of the cellular sequence into the proviral genome are likely to be similar. Obviously, only a single example is reported here, so the mechanism of transduction in this case may represent an exception to the rule. However, these data show that at some frequency HIV-1 can acquire sequence from a different chromosomal location than the site of parental provirus integration.
Summary.
The findings described above demonstrate that HIV-1 can transduce cellular sequence, as had been previously found for oncoretroviruses. Furthermore, the transduced cellular sequence does not seem to be linked to the site of parental provirus integration, indicating that transduction can occur in the absence of transcriptional readthrough at the site of parental provirus.
Acknowledgments
G.S. and P.K.O. contributed equally to this work.
We thank Martin Adelson, Chiann-Chyi Chen, Malvika Kaul, Annmarie Pacchia, and Amariliz Rivera for constructive comments on the manuscript and helpful discussions.
This work was supported by National Institutes of Health grants CA50777, NS38272, AI43886, and AI34834.
REFERENCES
- 1.Anderson D J, Stone J, Lum R, Linial M L. The packaging phenotype of the SE21Q1b provirus is related to high proviral expression and not trans-acting factors. J Virol. 1995;69:7319–7323. doi: 10.1128/jvi.69.11.7319-7323.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 3.Coffin J M. Genetic diversity and evolution of retroviruses. Curr Top Microbiol Immunol. 1992;176:143–164. doi: 10.1007/978-3-642-77011-1_10. [DOI] [PubMed] [Google Scholar]
- 4.Colicelli J, Goff S P. Structure of a cloned circular retroviral DNA containing a tRNA sequence between the terminal repeats. J Virol. 1986;57:674–677. doi: 10.1128/jvi.57.2.674-677.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cross S H, Bird A P. CpG islands and genes. Curr Opin Genet Dev. 1995;5:309–314. doi: 10.1016/0959-437x(95)80044-1. [DOI] [PubMed] [Google Scholar]
- 6.Dornburg R, Temin H M. Presence of a retroviral encapsidation sequence in nonretroviral RNA increases the efficiency of formation of cDNA genes. J Virol. 1990;64:886–889. doi: 10.1128/jvi.64.2.886-889.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Felder M P, Eychene A, Barnier J V, Calogeraki I, Calothy G, Marx M. Common mechanism of retrovirus activation and transduction of c-mil and c-Rmil in chicken neuroretina cells infected with Rous-associated virus type 1. J Virol. 1991;65:3633–3640. doi: 10.1128/jvi.65.7.3633-3640.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hajjar A M, Linial M L. A model system for nonhomologous recombination between retroviral and cellular RNA. J Virol. 1993;67:3845–3853. doi: 10.1128/jvi.67.7.3845-3853.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jetzt A E, Yu H, Klarmann G J, Ron Y, Preston B D, Dougherty J P. High rate of recombination throughout the human immunodeficiency virus type 1 genome. J Virol. 2000;74:1234–1240. doi: 10.1128/jvi.74.3.1234-1240.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klenova E M, Fagerlie S, Filippova G N, Kretzner L, Goodwin G H, Loring G, Neiman P E, Lobanenkov V V. Characterization of the chicken CTCF genomic locus, and initial study of the cell cycle-regulated promoter of the gene. J Biol Chem. 1998;273:26571–26579. doi: 10.1074/jbc.273.41.26571. [DOI] [PubMed] [Google Scholar]
- 11.Larsen F, Gundersen G, Lopez R, Prydz H. CpG islands as gene markers in the human genome. Genomics. 1992;13:1095–1107. doi: 10.1016/0888-7543(92)90024-m. [DOI] [PubMed] [Google Scholar]
- 12.Nilsen T W, Maroney P A, Goodwin R G, Rottman F M, Crittenden L B, Raines M A, Kung H J. c-erbB activation in ALV-induced erythroblastosis: novel RNA processing and promoter insertion result in expression of an amino-truncated EGF receptor. Cell. 1985;41:719–726. doi: 10.1016/s0092-8674(85)80052-0. [DOI] [PubMed] [Google Scholar]
- 13.Olsen J C, Bova-Hill C, Grandgenett D P, Quinn T P, Manfredi J P, Swanstrom R. Rearrangements in unintegrated retroviral DNA are complex and are the result of multiple genetic determinants. J Virol. 1990;64:5475–5484. doi: 10.1128/jvi.64.11.5475-5484.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Page K A, Landau N R, Littman D R. Construction and use of a human immunodeficiency virus vector for analysis of virus infectivity. J Virol. 1990;64:5270–5276. doi: 10.1128/jvi.64.11.5270-5276.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parthasarathi S, Varela-Echavarria A, Ron Y, Preston B D, Dougherty J P. Genetic rearrangements occurring during a single cycle of murine leukemia virus vector replication: characterization and implications. J Virol. 1995;69:7991–8000. doi: 10.1128/jvi.69.12.7991-8000.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patel P H, Preston B D. Marked infidelity of human immunodeficiency virus type 1 reverse transcriptase at RNA and DNA template ends. Proc Natl Acad Sci USA. 1994;91:549–553. doi: 10.1073/pnas.91.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pathak V K, Temin H M. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: substitutions, frameshifts, and hypermutations. Proc Natl Acad Sci USA. 1990;87:6019–6023. doi: 10.1073/pnas.87.16.6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Preston B D, Dougherty J P. Mechanisms of retroviral mutation. Trends Microbiol. 1996;4:16–21. doi: 10.1016/0966-842x(96)81500-9. [DOI] [PubMed] [Google Scholar]
- 19.Raines M A, Maihle N J, Moscovici C, Crittenden L, Kung H J. Mechanism of c-erbB transduction: newly released transducing viruses retain poly(A) tracts of erbB transcripts and encode C-terminally intact erbB proteins. J Virol. 1988;62:2437–2443. doi: 10.1128/jvi.62.7.2437-2443.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ratner L, Fisher A, Jagodzinski L L, Mitsuya H, Liou R S, Gallo R C, Wong-Staal F. Complete nucleotide sequences of functional clones of the AIDS virus. AIDS Res Hum Retroviruses. 1987;3:57–69. doi: 10.1089/aid.1987.3.57. [DOI] [PubMed] [Google Scholar]
- 21.Shigemoto K, Kubo S, Maruyama N, Yamada S, Obata K, Kikuchi K, Kondo I. Identification and characterization of 5′ extension of mammalian agrin cDNA, the exons and the promoter sequences. Biochim Biophys Acta. 2000;1494:170–174. doi: 10.1016/s0167-4781(00)00214-1. [DOI] [PubMed] [Google Scholar]
- 22.Stuhlmann H, Dieckmann M, Berg P. Transduction of cellular neo mRNA by retrovirus-mediated recombination. J Virol. 1990;64:5783–5796. doi: 10.1128/jvi.64.12.5783-5796.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swain A, Coffin J M. Mechanism of transduction by retroviruses. Science. 1992;255:841–845. doi: 10.1126/science.1371365. [DOI] [PubMed] [Google Scholar]
- 24.Temin H M. Retrovirus variation and reverse transcription: abnormal strand transfers result in retrovirus genetic variation. Proc Natl Acad Sci USA. 1993;90:6900–6903. doi: 10.1073/pnas.90.15.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tonjes R R, Czauderna F, Kurth R. Genome-wide screening, cloning, chromosomal assignment, and expression of full-length human endogenous retrovirus type K. J Virol. 1999;73:9187–9195. doi: 10.1128/jvi.73.11.9187-9195.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Varela-Echavarria A, Prorock C M, Ron Y, Dougherty J P. High rate of genetic rearrangement during replication of a Moloney murine leukemia virus-based vector. J Virol. 1993;67:6357–6364. doi: 10.1128/jvi.67.11.6357-6364.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Varmus H E. Oncogenes and transcriptional control. Science. 1987;238:1337–1339. doi: 10.1126/science.2825348. [DOI] [PubMed] [Google Scholar]
- 28.Wu W, Blumberg B M, Fay P J, Bambara R A. Strand transfer mediated by human immunodeficiency virus reverse transcriptase in vitro is promoted by pausing and results in misincorporation. J Biol Chem. 1995;270:325–332. doi: 10.1074/jbc.270.1.325. [DOI] [PubMed] [Google Scholar]
- 29.Young J, Biden K G, Simms L A, Huggard P, Karamatic R, Eyre H J, Sutherland G R, Herath N, Barker M, Anderson G J, Fitzpatrick D R, Ramm G A, Jass J R, Leggett B A. HPP1: a transmembrane protein-encoding gene commonly methylated in colorectal polyps and cancers. Proc Natl Acad Sci USA. 2001;98:265–270. doi: 10.1073/pnas.98.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu H, Rabson A B, Kaul M, Ron Y, Dougherty J P. Inducible human immunodeficiency virus type 1 packaging cell lines. J Virol. 1996;70:4530–4537. doi: 10.1128/jvi.70.7.4530-4537.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang C, Rasmussen C, Chang L J. Cell cycle inhibitory effects of HIV and SIV Vpr and Vpx in the yeast Schizosaccharomyces pombe. Virology. 1997;230:103–112. doi: 10.1006/viro.1997.8459. [DOI] [PubMed] [Google Scholar]
- 32.Zhang J, Temin H M. 3′ junctions of oncogene-virus sequences and the mechanisms for formation of highly oncogenic retroviruses. J Virol. 1993;67:1747–1751. doi: 10.1128/jvi.67.4.1747-1751.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]