Abstract
We used the simian immunodeficiency virus (SIV) molecular clone SIVmac239 to generate a deletion construct, termed SD2, in which we eliminated 22 nucleotides at positions +398 to +418 within the putative dimerization initiation site (DIS) stem. This SD2 deletion severely impaired viral replication, due to adverse effects on the packaging of viral genomic RNA, the processing of Gag proteins, and viral protein patterns. However, long-term culture of SD2 in either C8166 or CEMx174 cells resulted in restoration of replication capacity, due to two different sets of three compensatory point mutations, located within both the DIS and Gag regions. In the case of C8166 cells, both a K197R and a E49K mutation were identified within the capsid (CA) protein and the p6 protein of Gag, respectively, while the other point mutation (A423G) was found within the putative DIS loop. In the case of CEMx174 cells, two compensatory mutations were present within the viral nucleocapsid (NC) protein, E18G and Q31K, in addition to the same A423G substitution as observed with C8166 cells. A set of all three mutations was required in each case for restoration of replication capacity, and either set of mutations could be substituted for the other in both the C8166 and CEMx174 cell lines.
The 5′ untranslated leader sequences of both simian immunodeficiency virus (SIV) and human immunodeficiency virus type 1 (HIV-1) possess distinct functional domains that include elements required for transactivation of transcription, initiation of reverse transcription, packaging of viral RNA, and integration of the proviral genome (7). Leader sequences downstream of the U5 region include the major splice donor site and are involved in encapsidation of full-length viral genomic RNA as well as viral gene expression (11, 14–19). The secondary structure of the HIV-1 leader sequence, located downstream of U5, includes four stem-loop RNA motifs, termed SL1, SL2, SL3, and SL4 (5, 13). Each of the SL1, SL3, and SL4 elements has been shown to be involved in packaging of viral genomic RNA (7). In addition, SL1 contains a palindromic loop sequence that is thought to serve as a dimerization initiation site (DIS) (6, 25). Mutations within SL1 have been shown to severely diminish viral infectivity (23).
The 5′ leader sequence of SIV has little sequence similarity to that of HIV-1; however, similar secondary structures are predicted for both (11, 24). A stem-loop structure that serves as a putative DIS has been identified in the leader sequences of both SIV and HIV-1. Our group has previously shown that deletions within the DIS stem-loop of each of the viruses HIV-1 and SIV severely impaired viral replication and that this led to impairment of both Gag protein processing and packaging of viral genomic RNA (11, 12, 19, 20). Reversion to wild-type viral replication has been observed following deletions within the DIS of HIV-1, and this has been attributed to a series of four point mutations within distinct Gag proteins, i.e., matrix (MA), capsid (CA), p2, and nucleocapsid (NC) (21). In the case of SIV, deletion of presumed DIS sequences at positions +398 to +418 yielded a construct termed SD2. This virus was initially impaired in replication ability in C8166 cells; nonetheless, long-term passage led to a restoration of viral replication that was shown to be due to three point mutations, located in both the putative DIS (an A423G substitution) and Gag regions (K197R and G49K) (11). However, the mechanisms whereby these mutations might contribute to the observed rebound of viral replication capacity were not characterized.
We have now extended our studies of the reversion of deleted SD2 viruses in both C8166 cells and CEMx174 cells and identified a different set of compensatory mutations, i.e., Glu18Gln (E18G) and Gln31Lys (Q31K), both located in the nucleocapsid protein of Gag, and the same A423G substitution in the putative DIS, responsible for restored viral growth.
All three point mutations, i.e., A423G, K197R, and E49K, are required for full replication of the SD2 variant.
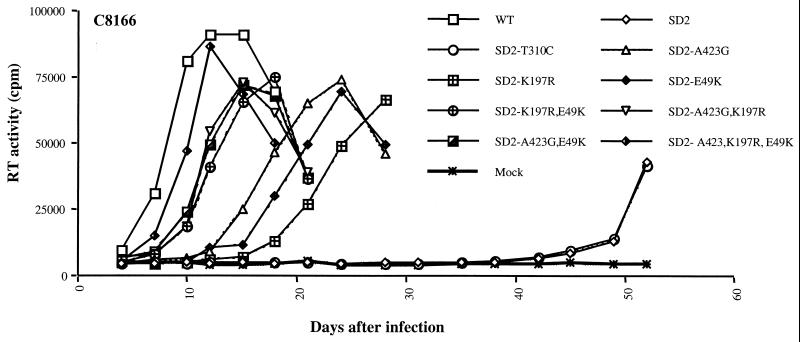
Research involving cell lines, viral stocks, viral replication, viral protein analysis, and packaging of viral genomic RNA was carried out as described elsewhere (11). To shed further light on the role of these mutations in the restoration of replication, a series of seven SD2 derivatives were generated that represent all of the combinations of the three point mutations previously identified. These viruses contained either one, two, or all three of the above-mentioned mutations, as follows: SD2-A423G; SD2-K197R; SD2-E49K; SD2-A423G,K197R; SD2-A423G,E49K; SD2-K197R,E49K; and SD2-A423G,K197R,E49K. As shown in Fig. 1, each of the above-mentioned point mutations was able to partially compensate for the SD2 deletion in C8166 cells; however, all three mutations in tandem were required for full replication capacity, and a previously observed polymorphism, i.e., T310C (12), located upstream of the putative DIS, was unable to restore even minimal viral replication.
FIG. 1.
Growth curves of reverted viruses in C8166 cells. Compensatory mutations located within both the DIS, i.e., A423G, and the Gag region, i.e., K197R and E49K, were introduced into the SD2 construct. The resultant constructs were then transfected into COS-7 cells. Equivalent amounts of recovered viruses, based on levels of p27 antigen (10 ng/106 cells), were used to infect C8166 cells, Viral replication was monitored by reverse transcriptase (RT) assay of culture fluids. Mock infection denotes exposure of cells to heat-inactivated wild-type (WT) virus as a negative control.
Reversion of SD2 virus in CEMx174 cells.
We also passaged the SD2 virus over multiple generations in the CEMx174 cell line; these studies led to recovery of a virus with wild-type replication properties. The viral DNA fragment from the U5 region to the end of the gag gene of this reverted virus was amplified and cloned as described previously, and six of these clones (from two independent series of passages) were sequenced (11) The results show that the original SD2 deletion had been retained in each case but that three additional point mutations were also present. One of these was the same as that previously identified in the aftermath of SD2 reversion in C8166 cells, i.e., A423G, but two distinct mutations located in the NC region were also identified, i.e., E18Q and Q31K. The E18Q mutation (Glu→Gln) was located within the first zinc finger domain of the NC protein, while the Q31K substitution (Gln-Lys) was found further downstream within NC.
Site-directed mutagenesis was then performed to introduce each of these NC mutations, alone, together, and in combination with the A423G substitution, into the SD2 genome. This was performed using the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) (11, 21, 22) and the following primer pairs, nc1-1 (5′-GTGTTGGAATTGTGGGAAACAGGGACACTCTGCAAGGC-3′) and nc1-2 (5′-GCCTTGCAGAG TG TCCC TG T T TCCCACAAT TCCAACAC - 3′) for E18Q and nc2-1 (5′-GCAGAGCCCCAAGAAGAAAGGGATGCTGGAAATGTGG-3′) and nc2-2 (5′-CCACATTTCCAGCATCCCTTTCTTCTTGGGGCTCTGC-3′) for Q31K. Different combinations of these point mutations were also generated using the same methods. The resultant constructs, termed SD2-E18Q; SD2-Q31K; SD2-E18Q,Q31K; SD2-A423G,E18Q; SD2-A423G,Q31K; and SD2-A423G,E18Q,Q31K, were then transfected into COS-7 cells, and the virus particles thereby recovered were assayed for viral replication capacity in CEMx174 cells. The results in Fig. 2 show that various of these mutations could individually contribute to recovered viral growth but that full restoration of replication capacity required the combination of all three mutations, i.e., SD2-A423G,E18Q,Q31K.
FIG. 2.
Growth curves of reverted viruses in CEMx174 cells. The second set of compensatory mutations, located in the DIS, i.e., A423G, and the NC region, i.e., E18Q and Q31K, were introduced, either alone (A) or combined (B), into the SD2 construct. The resultant constructs were transfected into COS-7 cells. Equivalent amounts of virus thereby recovered were used to infect C8166 cells based on levels of p27 antigen (10 ng/106 cells). Viral replication was monitored by reverse transcriptase (RT) assay of culture fluids. Mock infection denotes exposure of cells to heat-inactivated wild-type (WT) virus as a negative control.
Our findings on the involvement of mutations in NC are consistent with studies of HIV-1 that showed that the NC protein is the major domain within Gag that recognizes the encapsidation signals present within leader sequences (2–4). The NC protein contains two zinc finger motifs that contribute to its specific interactions with viral RNA (8–10), and our data point to a role for NC in interactions between Gag and RNA leader sequences. Of course, the p6 protein is also important in the incorporation of viral proteins into virions and is involved in encapsidation of viral RNA (1, 9, 22). Mechanistic studies are in progress to assess the role of each of the mutations identified above in restoration of viral replication.
The two sets of compensatory mutations can restore viral replication equally in each of the cell lines C8166 and CEMx174.
We next tested the infectiousness of viruses containing each of the above-described sets of mutations in both cell types. Figure 3 demonstrates that the mutations that had emerged in CEMx174 cells, i.e., A423G, E18Q, and Q31K, were also able to rescue the SD2 deletion in C8166 cells and that the A423G, K197R, and E49K mutations rescued viral replication in CEMx174 cells.
FIG. 3.
Replication capacity of wild-type (WT) and mutated viruses in C8166 cells or CEMx174 cells. Equivalent amounts of virus were used to infect cells based on levels of p27 antigen (10 ng/106 cells). Viral replication was monitored by reverse transcriptase (RT) assay of culture fluids. Mock infection denotes exposure of cells to heat-inactivated wild-type virus as a negative control. (A) Growth curves of the second set of SD2 reverted viruses in C8166 cells. (B) Growth curves of the first set of SD2 reverted viruses in CEMx174 cells.
Moreover, the infectiousness of SD2 deleted viruses, containing either of these sets of mutations, but not individual substitutions, was similar to that of wild-type virus in CEMx174 cells, as determined by assay of 50% tissue culture infective dose per nanogram of p27 antigen (results not shown).
Thus, we have shown that both sets of compensatory mutations selected in these studies were able to act interchangeably to restore viral replication, regardless of the cell type in which the virus was grown. Of course, it is conceivable that either the same mutational patterns or different ones may have been observed in either of the cell lines studied had either additional replication studies been performed or different conditions for cell growth been employed. Nonetheless, it is not trivial that the escape mechanisms for lentiviruses, grown under conditions of stress as demonstrated here, are apparently not restricted to single pathways, and this is the first demonstration of such plasticity on the part of a lentiviral genome.
Acknowledgments
This research was supported by grant RO1 AI43878-01 to M.A.W. from the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
The CEMx174 cell line was obtained from Peter Cresswell through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. We thank Maureen Oliveira for technical assistance.
REFERENCES
- 1.Accola M A, Bukovsky A A, Jones M S, Gottlinger H G. A conserved dileucine-containing motif in p6gag governs the particle association of Vpx and Vpr of simian immunodeficiency viruses SIVmac and SIVagm. J Virol. 1999;73:9992–9999. doi: 10.1128/jvi.73.12.9992-9999.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldovini A, Young R A. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J Virol. 1990;64:1920–1926. doi: 10.1128/jvi.64.5.1920-1926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berkowitz R D, Goff S P. Analysis of binding elements in the human immunodeficiency virus type 1 genomic RNA and nucleocapsid protein. Virology. 1994;202:233–246. doi: 10.1006/viro.1994.1339. [DOI] [PubMed] [Google Scholar]
- 4.Berkowitz R D, Ohagen A, Hoglund S, Goff S P. Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag polyproteins during RNA packaging in vivo. J Virol. 1995;69:6445–6456. doi: 10.1128/jvi.69.10.6445-6456.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clever J, Sassetti C, Parslow T G. RNA secondary structure and binding sites for gag gene products in the 5′ packaging signal of human immunodeficiency virus type 1. J Virol. 1995;69:2101–2109. doi: 10.1128/jvi.69.4.2101-2109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clever J L, Wong M L, Parslow T G. Requirements for kissing-loop-mediated dimerization of human immunodeficiency virus RNA. J Virol. 1996;70:5902–5908. doi: 10.1128/jvi.70.9.5902-5908.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coffin J M, Hughes S H, Varmus H E. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 8.Dannull J, Surovoy A, Jung G, Moelling K. Specific binding of HIV-1 nucleocapsid protein to PSIRNA in vitro requires N-terminal zinc finger and flanking basic amino acid residues. EMBO J. 1994;13:1525–1533. doi: 10.1002/j.1460-2075.1994.tb06414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dettenhofer M, Yu X-F. Proline residues in human immunodeficiency virus type 1 p6Gag exert a cell type-dependent effect on viral replication and virion incorporation of Pol proteins. J Virol. 1999;73:4696–4704. doi: 10.1128/jvi.73.6.4696-4704.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorfman T, Luban J, Goff S P, Haseltine W A, Gottlinger H G. Mapping of functionally important residues of a cysteine-histidine box in the human immunodeficiency virus type 1 nucleocapsid protein. J Virol. 1993;67:6159–6169. doi: 10.1128/jvi.67.10.6159-6169.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guan Y, Whitney J B, Diallo K, Wainberg M A. Leader sequences downstream of the primer binding site are important for efficient replication of simian immunodeficiency virus. J Virol. 2000;74:8854–8860. doi: 10.1128/jvi.74.19.8854-8860.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guan Y, Whitney J B, Liang C, Wainberg M A. Novel live attenuated simian immunodeficiency virus constructs containing major deletions in leader RNA sequences. J Virol. 2001;75:2776–2785. doi: 10.1128/JVI.75.6.2776-2785.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrison G P, Lever A M L. The human immunodeficiency virus type 1 packaging signal and major splice donor region have a conserved stable secondary structure. J Virol. 1992;66:4144–4153. doi: 10.1128/jvi.66.7.4144-4153.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrison G P, Miele G, Hunter E, Lever A M L. Functional analysis of the core human immunodeficiency virus type 1 packaging signal in a permissive cell line. J Virol. 1998;72:5886–5896. doi: 10.1128/jvi.72.7.5886-5896.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaye J F, Lever A M L. Human immunodeficiency virus types 1 and 2 differ in the predominant mechanism used for selection of genomic RNA for encapsidation. J Virol. 1999;73:3023–3031. doi: 10.1128/jvi.73.4.3023-3031.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim H J, Lee K, O'Rear J J. A short sequence upstream of the 5′ major splice site is important for encapsidation of HIV-1 genomic RNA. Virology. 1994;198:336–340. doi: 10.1006/viro.1994.1037. [DOI] [PubMed] [Google Scholar]
- 17.Lenz C, Scheid A, Schaal H. Exon 1 leader sequences downstream of U5 are important for efficient human immunodeficiency virus type 1 gene expression. J Virol. 1997;71:2757–2764. doi: 10.1128/jvi.71.4.2757-2764.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lever A, Gottlinger H, Haseltine W, Sodroski J. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J Virol. 1989;63:4085–4087. doi: 10.1128/jvi.63.9.4085-4087.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang C, Li X, Quan Y, Langhrea M, Kleiman L, Hiscott J, Wainberg M A. Sequence elements downstream of human immunodeficiency virus type 1 long terminal repeat are required for efficient viral gene transcription. J Mol Biol. 1997;272:167–177. doi: 10.1006/jmbi.1997.1239. [DOI] [PubMed] [Google Scholar]
- 20.Liang C, Rong L, Cherry E, Kleiman L, Laughrea M, Wainberg M A. Deletion mutagenesis within the dimerization initiation site of human immunodeficiency virus type 1 results in delayed processing of the p2 peptide from precursor proteins. J Virol. 1999;73:6147–6151. doi: 10.1128/jvi.73.7.6147-6151.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang C, Rong L, Quan Y, Laughrea M, Kleiman L, Wainberg M A. Mutations within four distinct Gag proteins are required to restore replication of human immunodeficiency virus type 1 after deletion mutagenesis within the dimerization initiation site. J Virol. 1999;73:7014–7020. doi: 10.1128/jvi.73.8.7014-7020.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ott D E, Chertova E N, Busch L K, Coren L V, Gagliardi T D, Johnson D G. Mutational analysis of the hydrophobic tail of the human immunodeficiency virus type 1 p6Gag protein produces a mutant that fails to package its envelope protein. J Virol. 1999;73:19–28. doi: 10.1128/jvi.73.1.19-28.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poon D T K, Wu J, Aldovini A. Changed amino acid residues of human immunodeficiency virus type 1 nucleocapsid p7 protein involved in RNA packaging and infectivity. J Virol. 1996;70:6607–6616. doi: 10.1128/jvi.70.10.6607-6616.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rizvi T A, Panganiban A T. Simian immunodeficiency virus RNA is efficiently encapsidated by human immunodeficiency virus type 1 particles. J Virol. 1993;67:2681–2688. doi: 10.1128/jvi.67.5.2681-2688.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skripkin E, Pailart J C, Marquet R, Ehresmann B, Ehresmann C. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc Natl Acad Sci USA. 1994;91:4945–4949. doi: 10.1073/pnas.91.11.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]